SUMMARY
The oocyte-to-embryo transition marks the onset of development. The initial phase of this profound change from the differentiated oocyte to the totipotent embryo occurs in the absence of both transcription and mRNA degradation. Here we combine global polysome profiling, ribosome-footprint profiling, and quantitative mass spectrometry in a comprehensive approach to delineate the translational and proteomic changes at this important transition in Drosophila. Our results show that PNG kinase is a critical regulator of the extensive changes in the translatome, acting uniquely at this developmental window. Analysis of the proteome in png mutants provided insights into the contributions of translation to changes in protein levels, revealing a compensatory dynamic between translation and protein turnover during proteome remodeling at the return to totipotency. The proteome changes additionally suggested new regulators of meiosis and early embryogenesis, including the conserved H3K4 demethylase LID, which we demonstrated is required during this period despite transcriptional inactivity.
INTRODUCTION
The oocyte-to-embryo transition marks the onset of development for any multicellular organism. This dramatic change involves the completion of meiosis in the oocyte, sperm entry, fusion of the male and female pronucleus, and the start of mitotic divisions (Horner and Wolfner, 2008b). These events accompany the profound developmental change from the differentiated oocyte into a totipotent embryo.
Studies on the restoration of cell potency have focused on the regulation of transcription (Young, 2011), but the oocyte-to-embryo transition necessitates a fundamentally different control mechanism. Following the primary arrest in prophase I, oocytes are transcriptionally silent, and in all animals at least the first embryonic division occurs prior to the initiation of zygotic transcription (Tadros and Lipshitz, 2009). Organisms such as insects, fish, and amphibians rely on stockpiled maternal mRNAs. These organisms proceed through several hours of embryonic development, i.e 12–13 division cycles in Xenopus and Drosophila, prior to the onset of zygotic transcription, which also triggers turnover of maternal mRNAs (Anderson and Lengyel, 1979; Zalokar, 1976). In Drosophila, a pathway to degrade maternal mRNAs is not active until two hours after egg laying (Tadros et al., 2007; Tadros et al., 2003). Thus, the oocyte-to-embryo transition and early embryogenesis occur with constant mRNA levels (Tadros and Lipshitz, 2009).
Although many aspects of translational regulation remain to be elucidated in the oocyte-to-embryo transition, the effect of translation on meiotic progression has been more extensively analyzed in two other developmental contexts. First, in yeast meiosis, which proceeds without developmentally programmed arrests and in the presence of transcriptional control, extensive translational regulation nevertheless occurs (Brar et al., 2012;Carlile and Amon, 2008; Chu and Herskowitz, 1998). Second, in metazoans, translational control at oocyte maturation has been intensively investigated. At maturation, the oocyte exits the primary arrest in prophase I and progresses into the meiotic divisions. Experiments in amphibians, mice, and marine invertebrates demonstrated a role for cytoplasmic polyadenylation in activating translation at oocyte maturation and showed precisely timed translation of several mRNAs to be required for progression through the meiotic divisions (Charlesworth et al., 2013; Chen et al., 2011; Gebauer et al., 1994; Tay et al., 2000).
Here we define the regulatory steps of gene expression at the oocyte-to-embryo transition of Drosophila. In Drosophila, as in most animals, the mature oocyte is arrested at a secondary point in meiosis (Sagata, 1996). In insects this arrest point is metaphase of meiosis I. The trigger for the oocyte-to-embryo transition is egg activation. Egg activation and exit from meiosis in Drosophila take place as the oocyte passes into the uterus, regardless of whether it is fertilized. Instead of sperm entry, mechanical pressure as well as osmotic and Ca2+ changes are thought to initiate egg activation in Drosophila (Horner and Wolfner, 2008a).
Egg activation is presumably accompanied by changes in translation, as poly(A) tails on mRNAs are lengthened, and proteins involved in developmental patterning and cell cycle control are synthesized (Horner and Wolfner, 2008b). In addition, proteins are actively subject to degradation during egg activation. Release of the metaphase I arrest and completion of meiosis requires the Anaphase Promoting Complex/Cyclosome (APC/C) to target Cyclin B for degradation (Pesin and Orr-Weaver, 2007; Swan and Schupbach, 2007). We recently showed that a meiosis-specific form of the APC/C also contributes to the change from meiosis in the oocyte to mitosis in the embryo by mediating degradation of the meiotic protein Matrimony (Mtrm) (Whitfield et al., 2013).
The Drosophila PNG kinase is required for the onset of mitotic divisions in the embryo (Fenger et al., 2000; Shamanski and Orr-Weaver, 1991). This Ser/Thr kinase is a complex of a catalytic subunit, encoded by the png gene, and two activating subunits, the proteins GNU and PLU (Lee et al., 2003). PNG kinase complex is present and acts solely at the oocyte-to-embryo transition. By promoting the translation of cyclin B after egg activation, PNG kinase complex leads to Cyclin B/Cdk1 reactivation and entry into the first embryonic mitosis (Fenger et al., 2000; Vardy and Orr-Weaver, 2007). PNG has an indirect role later in embryogenesis in promoting degradation of maternal mRNAs. This degradation requires Smaug (SMG), whose translation also is dependent on PNG at egg activation (Tadros et al., 2007).
Here, we report the first quantitation of the changes in translation and the proteome during the oocyte-to-embryo transition in any organism, leading to identification of new regulators of this key developmental change. Our comprehensive approach allowed us to determine the extent to which translational changes are reflected in the proteome. These studies reveal that both extensive translational and posttranslational regulatory mechanisms sculpt the proteome at the developmental change from oocyte to embryo, and they emphasize the value of examining both the translatome and the proteome.
RESULTS
Translational changes at egg activation
To assess globally the translational changes at egg activation, we performed polysome profiling and ribosome footprinting of mature oocytes and activated eggs (Figure 1A). We used unfertilized Drosophila eggs instead of embryos because they undergo all the events of activation, including the completion of meiosis and the onset of expression of proteins controlling embryonic patterning (Horner and Wolfner, 2008b). Unfertilized eggs offered an advantage over embryos: egg activation could be monitored without the potentially confounding changes in protein synthesis and degradation that occur during the subsequent embryonic divisions of fertilized embryos.
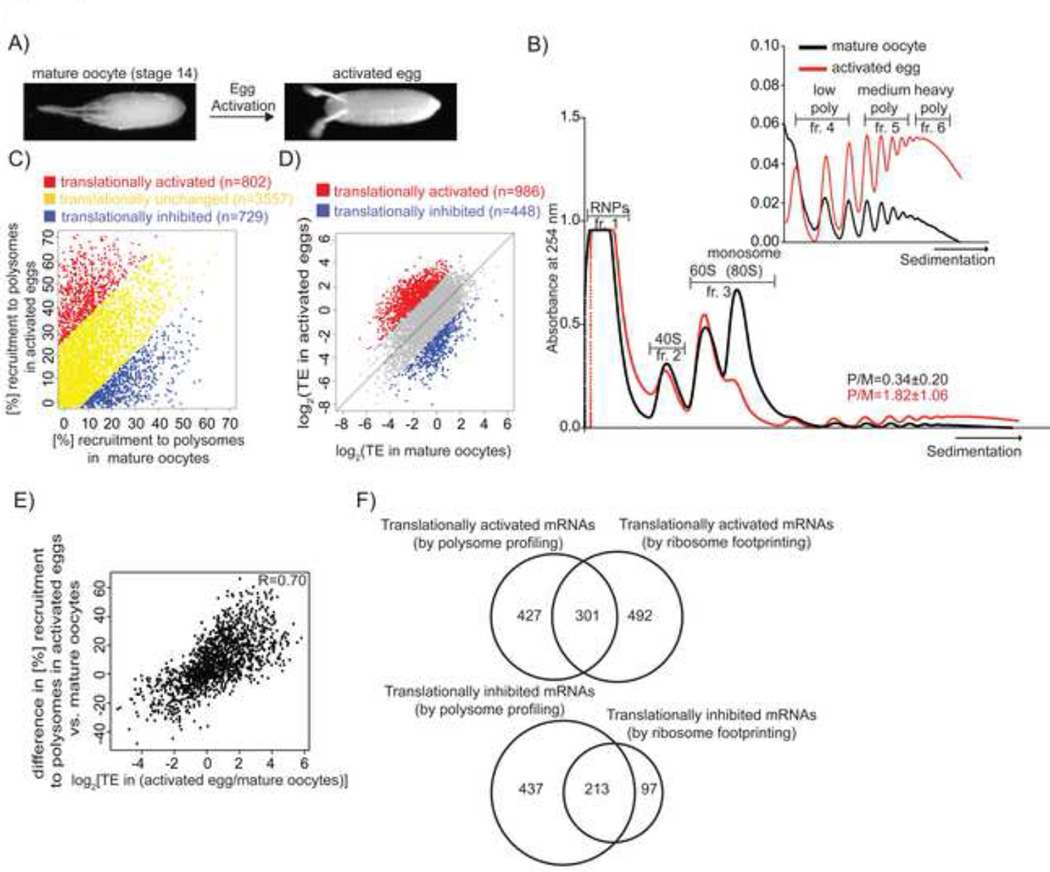
Figure 1. Translational changes at egg activation.
A) A micrograph of a mature, stage 14 oocyte (pre-activation state) and an activated egg (post-activation state). Anterior is on the left and dorsal is up.
B) A representative profile of 254nm absorbance for wild-type mature oocytes (black) and wild-type activated eggs (red). The inset is an enlargement of the polysomal section of the profile (starting from disomes). The six gradient fractions that were sequenced are labeled. Low polysomes correspond to 2–4 ribosomes, medium 5–9 and heavy 10 and higher. Polysome/monosome ratios (P/M) averaged from three biological replicate experiments are represented as mean ± SD.
C) Comparison of mRNAs associated with polysomes (n≥5 ribosomes) in mature oocytes and activated eggs. Data were corrected for the presence of mRNAs in the same regions of the gradient after fractionation of puromycin-treated samples. mRNAs were categorized as translationally inhibited (blue) if they had at least 9.1% higher polysomal recruitment in mature oocytes than activated eggs. Translationally activated (red) mRNAshad at least 26.4% higher polysomal recruitment inactivated eggs than mature oocytes. The cutoffs werechosen because they are 1SD from the mean differencefor all the identified mRNAs. The remaining, translationally unchanged, mRNAs are shown in yellow. 5088 mRNAs are represented as the mean of two biological replicates.
D) Translational efficiencies (TE, where TE = rpkm ofribosome protected fragments/rpkm for mRNAabundance) in mature oocytes and activated eggs for5842 mRNAs. 986 translationally activated mRNAs (red)have ~4.3 fold higher TE (1SD above the median ratio forall identified mRNAs in both replicates) in activated eggsthan mature oocytes, whereas for 448 translationallyinhibited mRNAs (blue) the TE ratio is ~4.3 fold lower inboth replicates. The mean of two biological replicates isshown.
E) Correspondence between the two complimentarymethods to measure the translational status of mRNAs inactivated eggs versus mature oocytes. 4580 mRNAs,detected by both approaches, are shown as the average of two biological replicates for both experiments. The Spearman R value is indicated.
F) Upper Venn diagram compares the number of mRNAs identified by polysome profiling (left) or ribosome footprinting (right) as translationally upregulated at egg activation, applying the criteria described in Figures 1C and 1D, respectively. Lower Venn diagram presents translationally inhibited mRNAs. Here, and for all the other Venn diagrams, only factors identified by both approaches (or in all compared samples) are represented.
In polysome profiling, mRNA-protein complexes are separated by fractionation through linear sucrose gradients (Beilharz and Preiss, 2004). The position of an mRNA within the sucrose gradient reflects its translational status: co-sedimentation of mRNAs with RNPs or ribosomal subunits suggests their lack of translation, whereas co-sedimentation with polysomes indicates their active translation. An abundant monosome peak present in polysome profiles of mature oocytes was lost at egg activation, accompanied by a more than 5-fold gain in the relative abundance of polysomes (Figure 1B). To define the translational changes for each mRNA, we isolated mRNAs from six regions within the gradient (Figure 1B) and quantified their abundance by mRNA sequencing (RNA-seq).
5088 mRNAs (~35% of the annotated genes) were identified in sucrose gradient fractions in both independent polysome-profiling experiments (Table S1). As expected, mRNA abundance in unfractionated lysates of mature oocytes and activated eggs, which are devoid of mRNA transcription and degradation, was highly correlated (R2=0.89) (Figure S1A). This result also indicated that although cytoplasmic polyadenylation is reported to accompany egg activation, poly(A) selection is a reliable method for mRNA isolation in mature oocytes and activated eggs (Horner and Wolfner, 2008b; Salles et al., 1994). Importantly, the mRNA abundance within the same fraction from two independent polysome-profiling experiments was highly correlated (R2 = 0.72–0.99, Figures S1B–C), allowing us to calculate for every mRNA its average percentage distribution across the six fractions.
To confirm that the mRNAs co-sedimenting with the polysomal peaks (defined here as ≥5 ribosomes) were actively engaged in translation, lysates of mature oocytes and activated eggs were treated with puromycin ex-vivo, which causes premature termination of elongating ribosomes (Blobel and Sabatini, 1971). Puromycin treatment of mature oocytes and activated eggs was efficient, as it led to complete disassembly of polysomes as measured by OD254 (Figures S2A and S2B). Fractionation of puromycin-treated lysates of mature oocytes, but not of activated eggs, revealed that many mRNAs migrated within sucrose gradients in a translation-independent manner (Figures S2C and S2D). Following puromycin treatment of mature oocyte lysates, the median association of an mRNA with fractions corresponding to polysomes equaled a surprisingly high 25% of the total abundance (Figure S2C). In contrast, in activated eggs after puromycin treatment, on average only 9% of the total mRNA co-sedimented with the gradient regions corresponding to polysomes (Figure S2D). The large fraction of mRNA from mature oocytes that was not actually associated with actively translating polysomes, but co-sedimented, highlights the necessity of the puromycin control in genome-wide polysome profiling experiments. After correcting for translation-independent migration in the region corresponding to polysomes (see Supplemental Experimental Procedures), we classified 802 mRNAs as recruited onto polysomes, 729 mRNAs as released off the polysomes, and 3557 mRNAs as unchanged during egg activation (Figure 1C and S2E, Table S4).
The discovery of a large number of translationally regulated mRNAs prompted us to employ an alternative genome-wide method to analyze the change in translational status of mRNAs at egg activation. Ribosome footprint profiling measures the number of ribosome-protected fragments (RPFs) derived from the mRNAs of each gene, which when normalized to mRNA abundance results in a single value for the translational efficiency of each gene (Ingolia et al., 2009). Moreover, ribosome footprinting reports the position of ribosomes on mRNAs. We observed an unusual accumulation of ribosomes on start codons specifically in mature oocytes, despite no cycloheximide being used (Figure S2F). This feature, however, seems unrelated to translational regulation for two reasons: 1) even in mature oocytes, RPFs at the start codon comprised only a small fraction of the RPFs for each gene (Figure S2F); and 2) initiation codon occupancy was not predictive of translational status in the activated egg (Figures S2G and S2H).
Two independent ribosome footprinting experiments yielded translational efficiency measurements for 5842 mRNAs in mature oocytes and activated eggs (Figure S2I) and revealed drastic translational changes at egg activation (R = 0.37) (Figure 1D and S2J). Mtrm, the regulator of timing of oocyte maturation, and 447 other mRNAs were translationally repressed at egg activation with ~4.3-fold (one standard deviation from the mean) lower translational efficiency in activated eggs than mature oocytes. 986 mRNAs, including positive controls such as embryonic patterning mRNAs bcd, hb, cad, tor, Tl, and smg as well as mRNAs encoding cell-cycle regulators (cycA, cycB and stg), had ~4.3-fold higher translational efficiency in activated eggs than mature oocytes and are translationally activated at egg activation (Tables S2 and S4). 968 of the 986 mRNAs categorized as translationally activated at egg activation and 390 of the 448 translationally repressed mRNAs are significantly translationally regulated with a p-value <0.05 between the two replicate experiments. If only a p-value cut-off of <0.05 is applied, 2435 mRNAs would be considered translationally activated and 989 translationally repressed. Because the mRNAs identified as translationally regulated at egg activation using a ~4.3-fold-change cut-off were largely contained in the set selected by p-value analysis, we chose the former approach for subsequent analyses, as it is more stringent.
The translational changes, observed by ribosome footprinting and polysome profiling, were in broad agreement (Spearman R = 0.70, Figure 1E), especially considering that these methods measure slightly different translational properties. Polysome profiling reveals the translationally active fraction of an mRNA. Ribosome footprinting provides information on the average number of ribosomes on each mRNA, with no insights as to whether only a small fraction of that mRNA is very efficiently translated and the bulk repressed.
301 and 213 mRNAs were identified by both methods as translationally upregulated or downregulated at egg activation, respectively (Figure 1F). Our dataset of 301 mRNAs translationally upregulated at egg activation was significantly enriched for GO-term categories (FDR <0.05) such as ‘RNA processing’, ‘mRNA binding’ and ‘chromatin organization’ (Figure S2K). Thus translational activation might help prepare for the maternal-to-zygotic transition or these factors might serve transcription-independent roles during the first two hours of embryogenesis. Although hundreds of mRNAs were reported as translationally inhibited at egg activation in mice (Potireddy et al., 2006), none had been described in Drosophila. The 213 mRNAs we identified as translationally inhibited at egg activation showed enrichment in several GO-term categories, including ‘cell cycle process’, consistent with egg activation involving completion of meiosis. Other enriched GO-categories such as ‘pyruvate metabolic process’, ‘tRNA aminoacylation’ and ‘nucleotide (ATP) binding’, suggest that changes in metabolism or translation could be occurring at the exit of meiosis and entry into embryonic cycles (Figure S2L).
The PNG kinase is a major translational regulator at egg activation
Having identified hundreds of mRNAs as translationally regulated at egg activation, our next goal was to discover key regulatory factors. One candidate was PNG, a Ser/Thr kinase previously demonstrated to promote translation of cycB and smg at egg activation (Tadros et al., 2007; Vardy and Orr-Weaver, 2007).
A slightly lower polysome-monosome ratio (P/M) in the png than wild-type activated eggs suggested a more systemic translational activation failure in png eggs (Figure 2A). These differences between wild-type and png activated eggs could not be attributed to mRNA expression levels, as mRNA abundance was highly correlated between these two samples (Pearson R = 0.99, Figure S3A). To identify translational targets of PNG, we performed ribosome footprinting of png mutant mature oocytes and activated eggs. Translational efficiencies of the positive controls, cycB, cycA and smg were lower in png activated eggs than in wild-type (Figure 2B and Table S2). In png eggs the translational efficiencies of all 986 mRNAs normally translationally upregulated at egg activation were closer to those of either wild-type or png mature oocytes than wild-type activated eggs (Figure 2C).
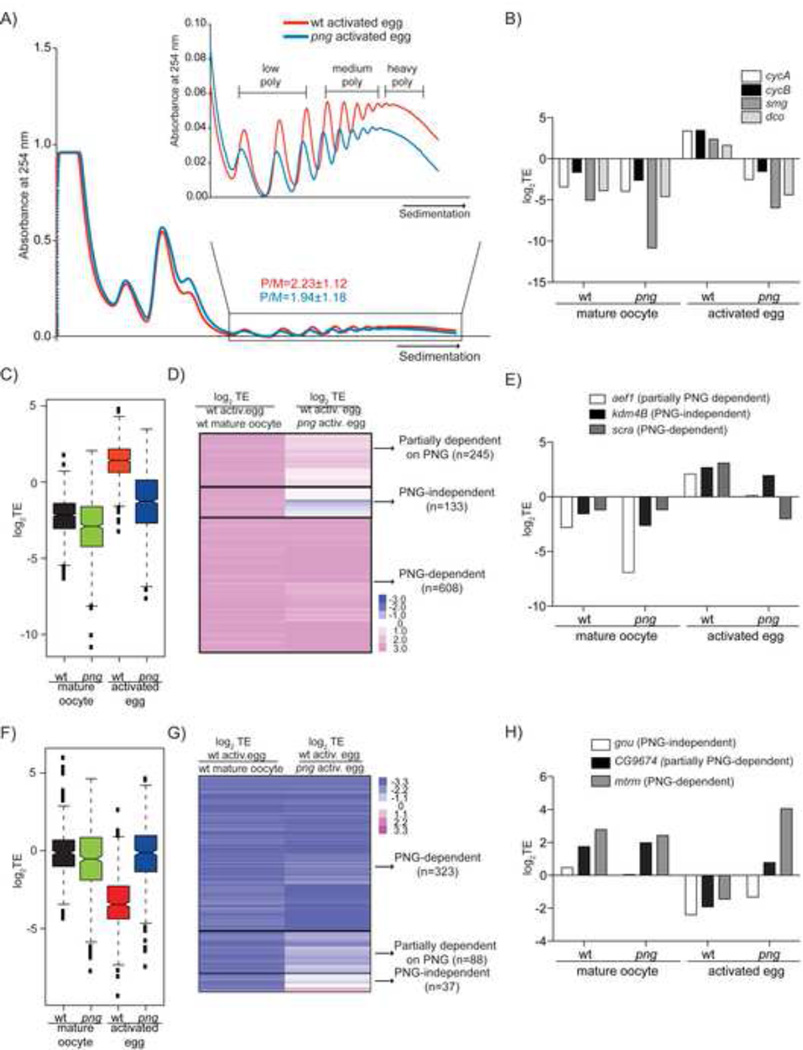
Figure 2. PNG kinase regulates the translational status of the majority of mRNAs at egg activation.
A) A representative profile of 254nm absorbance for wild-type (red) and png activated eggs (blue) as in Fig. 1B, except the polysome/monosome ratios are from two independent experiments.
B) Graph showing the TE of four mRNAs translationally upregulated at egg activation in wild-type (wt) but not in png mutants (png).
C) Box plot showing TE in wild-type mature oocytes (black), png mature oocytes (green), wild-type (red) and png activated eggs (blue) for 986 mRNAs translationally upregulated at wild-type egg activation. The black lines within each box indicate the median, the edges of the boxes show the first and third quartiles of the values, and whiskers extend to the minimum and maximum values. The average of two replicates is shown.
D) Heatmap for 986 translationally activated mRNAscompares the TE ratios of wild-type activated eggsversus mature oocytes with the ratios of wild-type versus png activated eggs. Three classes defining dependenceon PNG for translational activation at egg activationemerged (n=number of mRNAs in each category).TE=mean of two biological replicates.
E) TE, in wild-type and png mature oocytes as well asactivated eggs, of three mRNAs representative of thethree groups described in D).
F) and G) Same as in C) and D), except 448 translationally inhibited mRNAs are shown.
H) Same as in E) except three mRNAs representative of the three groups classifying dependence on PNG for translational repression at egg activation are shown.
A heat map depicting the differences in translational efficiency in png activated eggs compared to wild-type for each of the 986 translationally upregulated mRNAs shows that many,but not all, mRNAs require PNG function for activation (Figure 2D). 608 mRNAs were dependent on PNG for their translational activation (e.g. scra) (Figure 2E, Tables S2 and S4). In contrast,133 mRNAs were identified as PNG-independent (e.g. kdm4B)and 245 as partially PNG-dependent (e.g. aef-1) (Figures 2D and 2E). 38% of translationally activated mRNAs were at least partially independent of PNG suggesting that there are one or more pathways acting in parallel to PNG to govern translation at egg activation, possibly serving divergent goals (Table S4). The PNG-independent pathway(s) may control translational activation of mRNAs encoding proteins involved in chromatin modifications (Figure S3B), whereas the PNG-dependent pathway seems to regulate mRNAs encoding protein kinases and factors involved in cell or tissue morphogenesis as well as embryonic development (Figure S3C).
We also found that many mRNAs translationally repressed at egg activation were dependent on PNG (Figure 2F). Based on differences in translational efficiencies in png versus wild-type activated eggs, 323 mRNAs translationally inhibited at egg activation were classified as dependent on PNG (e.g. mtrm), 37 as PNG-independent (e.g. gnu), and 88 mRNAs as partially PNG-dependent (e.g. CG9674) (Figures 2G and 2H, Tables S2 and S4). PNG-independent mRNAs were not enriched in any particular GO-term category (FDR >0.05).
To examine the developmental timing when PNG is active, we tested whether PNG impacts the translational status of its target mRNAs prior to egg activation. The difference in translational efficiency for both translationally activated and repressed mRNAs was more pronounced between wild-type and png activated eggs than between mutant and wild-type mature oocytes (p-value<0.0001, Figure 2C and 2F). Combining polysome profiling and qRT-PCR we found a similar distribution of four different candidate mRNAs within the sucrose gradients of wild-type and png mature oocytes (Figure S3D). These results indicate that mRNAs are translationally regulated by PNG at but not prior to egg activation.
Translational upregulation can strongly contribute to increased protein levels at egg activation
Translational activity is well correlated with protein levels in S. cerevisiae growing in nutrient-rich media (Ingolia et al., 2009). However, the extent of coupling between the translatome and proteome has not been investigated in a developmental context. To assess proteome changes accompanying Drosophila egg activation we compared unfertilized eggs to mature oocytes with a quantitative proteomics approach. Between 3648 and 4187 proteins were quantified in three biological replicate experiments (Figure S4A). At egg activation, comparing the change in protein levels to either a change in polysomal recruitment (Figures S4B and S4C) or a change in translational efficiency (Figures 3A and S4D) of the encoding mRNAs, revealed a relatively poor correlation (R = 0.17 or 0.2, respectively). There were many examples of robust changes in translational efficiency corresponding to undetectable changes in protein. The correlation was not improved by considering changes in RPFs instead of changes in translational efficiency (Figure S4E). This is a consequence of egg activation occurring in the presence of constant mRNA levels (Figure S1A), thus resulting in a high correlation between changes in translational efficiency and changes in RPF (Spearman R=0.99, Figure S4F).
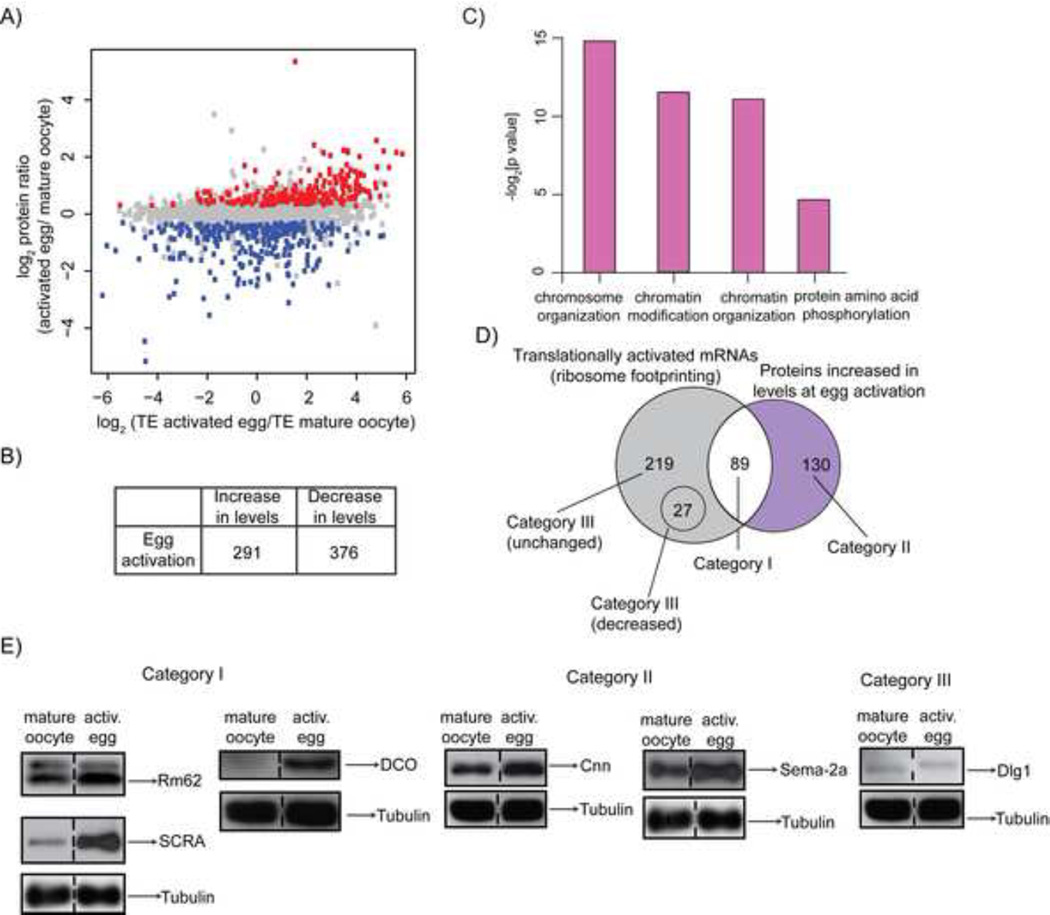
Figure 3. Protein remodeling during the oocyte-to-embryo transition.
A) Changes in protein levels and translational efficiency at egg activation. Scatterplot of ratio of protein levels in activated eggs versus mature oocytes (mean of three biological replicates) and TE (mean of two biological replicates). The proteins scored as upregulated are shown in red and those downregulated are in blue. 2934 data points are presented (identified both by quantitative mass spectrometry and ribosome fooprinting).
B) Table summarizing number of proteins that increase or decrease in levels during egg activation.
C) Gene Ontology (GO) term categories (FDR p-value < 0.05) for proteins more abundant in activated eggs than mature oocytes.
D) Venn diagram comparing the number of translationally activated mRNAs (Figure 1D) that encode proteins upregulated at egg activation (Category I). Proteins whose levels increase at egg activation according to quantitative mass spectrometry are show in Category II (violet). mRNAs identified by ribosome footprinting as translationally upregulated at egg activation but encoding proteins with unchanged or decreased levels at egg activation are Category III (grey). Only factors identified by both approaches are represented.
E) Western blot validation of candidates belonging to the categories described in D). Rm62, SCRA (same membrane reprobed) and DCO are validation examples for Category I; Cnn and Sema-2a (reprobed on Rm62/SCRA membrane) for Category II; and Dlg1 (reprobed on Cnn membrane) is representative of Category III. In this and all subsequent Western blots tubulin was used as a loading control. Dashed line marks that one lane from the original blot is not shown.
We found that 291 proteins significantly increased after egg activation (FDR<0.05, Figures 3A and 3B, Table S4). Among them were well-characterized regulators of early embryogenesis that served as our positive controls, such as the Cdc25 phosphatase String (Stg), as well as embryonic patterning factors Toll and SMG (Edgar et al., 1994; Salles et al., 1994; Tadros et al., 2007) (Tables S3 and S4). The list of proteins that increased at egg activation was enriched in some of the same GO-term categories as mRNAs translationally activated at egg activation, i.e. chromatin organization and modification as well as protein amino acid phosphorylation (Figure 3C). Additionally, the proteins upregulated at egg activation participated in a variety of other processes, supporting the complexity of this developmental transition (Table S3).
Approximately 40% of proteins whose levels increased at egg activation were encoded by mRNAs with a significant increase (>4.3-fold) in translation efficiency (Category I, 89 proteins, Figure 3D, Table S4). Increased levels of several Category I proteins was confirmed by immunoblots (Figures 3E and S4G). Overall, translational upregulation strongly contributed to increases in protein levels at egg activation, as the log2 median fold-change in translation efficiency for mRNAs encoding these proteins was 1.32 as compared to 0.14 for the entire quantified proteome. However, mechanisms other than translational regulation also play a role. For example, Category II contains mRNAs that are not significantly translationally upregulated yet encode proteins whose levels are increased (Figures 3D and S4H, Table S4). In these cases, protein accumulation might be caused by post-translational or other changes that stabilize these proteins and increase their half-life even in the absence of new synthesis. Alternatively, these proteins might be inherently more stable.
Surprisingly, only 25% of mRNAs identified as translationally activated by ribosome footprinting (or 20% in the case of polysome profiling) encoded proteins whose levels increased at egg activation (Figures 3A, 3D, S4B–S4D and S4H, Table S4). The rest of the translationally activated mRNAs, Category III, encoded proteins that were either unchanged (e.g. Dlg1, Figure 3E) or actually decreased (e.g. Dup, Figure S4G). For example, the level of Dlg1 remained unchanged at egg activation, despite a 23.6-fold increase in translational efficiency of its mRNA (Figure 3E). The level of Dup decreased 1.4-fold at egg activation, although the translational efficiency of its mRNA increased 2.6-fold (Figure S4G and Tables S2 and S3).
One possible mechanism for the existence of Category III (unchanged) proteins is that increased protein degradation offsets higher synthesis rates in activated eggs. An alternative explanation is that pre-existing stores of these proteins greatly exceed the newly synthesized protein, such that increases from translation make up an undetectably small fraction of the total protein pool. We note that if pre-existing protein stores generally obscured our ability to observe newly synthesized protein, this effect was independent of the starting amounts of protein, as we observed no significant difference in the abundance of Category III as compared to Category I proteins in mature oocytes (Two-sample Wilcoxon test p = 0.09, Fig. S4I–S4K). A significant difference in mRNA abundance also was not detected between these two categories (Two sample Wilcoxon test p = 0.17, Fig. S4L).
Regardless of the mechanism, the imperfect relationship between changes in protein levels and changes in translational efficiency highlight the value of our integrated approach. In considering translational changes most relevant for egg activation, acquiring proteomics data enables a focus on those that have greatest impact on protein levels (Category I). Moreover, proteomics measurements revealed some changes that were not detectable when examining translation changes (Category II), whereas translation measurements uncovered translationally activated mRNAs not detectable when examining only protein changes (Category III).
Lid activity is required in early embryogenesis
The limited number of proteins that increase at activation may represent factors that are critical for early embryonic development. It was interesting that the levels of three histone demethylases significantly increased at egg activation, given that the early embryonic development in Drosophila is transcriptionally silent. The levels of Little Imaginal Discs (Lid) were increased 2.7-fold at egg activation, whereas those of Kdm4A and Kdm4B increased 3.6- and 2.3-fold, respectively (Table S3). Lid is a conserved and essential histone H4-me3-specific demethylase that has been shown previously to regulate gene expression, particularly of Hox genes (Gildea et al., 2000; Lloret-Llinares et al., 2008), but it also may be involved in growth control through its association with dMyc (Secombe et al., 2007).
To study the role of Lid in the oocyte-to-embryo transition, we depleted Lid in the female germline by expressing RNAi against lid using the UAS-GAL4 system. qRT-PCR analysis of lid levels in mature oocytes of two different lid RNAi expressing lines revealed that they were about 99% lower than in the control, demonstrating efficient knockdown (Figure S5A). Importantly, meiotic progression was not perturbed upon lid knockdown, as evidenced by metaphase I plate morphology that was comparable to that of the control (Figures S5B and S5C).
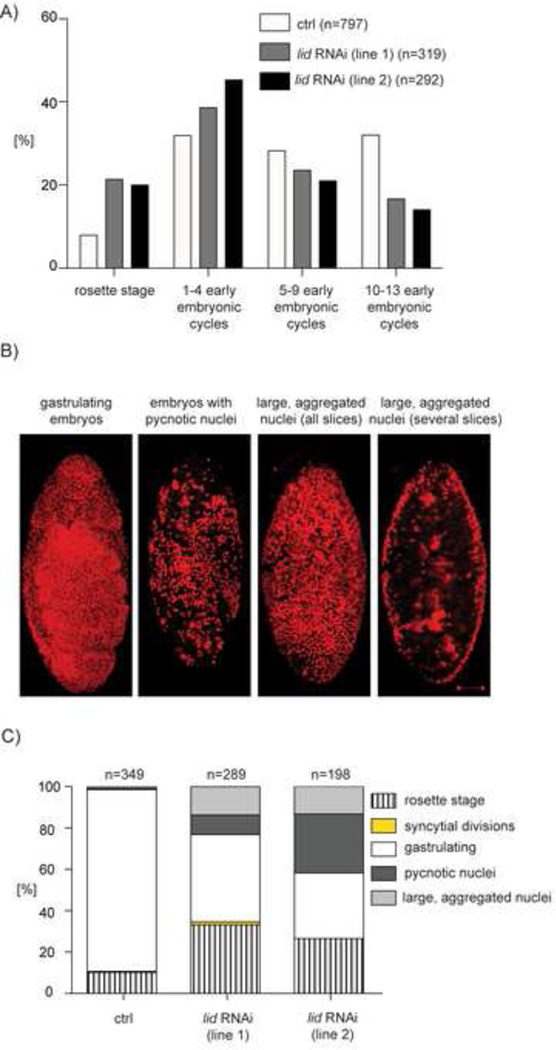
To examine the consequence of lid ablation on the early embryonic mitotic divisions, we measured the progression through early embryonic cycles by counting the percentage of laid embryos that: i) completed meiosis, and thus contained polar bodies arranged into rosettes, but did not initiate embryonic divisions; ii) completed one to four embryonic cycles; iii) completed five to nine embryonic cycles; or iv) reached the syncytial blastoderm, corresponding to division cycles 10–13 (Figures 4A and S5D). The embryos from lid RNAi-ablated mothers showed slower progression through embryogenesis than the control. In particular, increased numbers of embryos were in the rosette stage or going through the first-to-fourth embryonic cycles. Consequently, fewer embryos reached later cycles (Figure 4A). This developmental delay led to subsequent embryonic defects including a reduction in the number of gastrulating embryos and the appearance of embryos with pycnotic or aberrantly aggregated nuclei (Figures 4B and 4C). These data indicate that Lid has a role unrelated to transcription that is required during early embryogenesis for proper further embryonic development.
Figure 4. The histone H3K4 demethylase lid is required for proper early embryonic development.
A) Quantification of the percentage of 0–2h embryos present in different cycles of embryogenesis after the completion of meiosis (as shown in Figure S5D). Embryos were laid either by mothers with only the maternal-tubulin-Gal4 driver (control) or by mothers in which a lid RNAi line was expressed using the mata-tubulin-Gal4 driver. lid RNAi line 1 is BL35706; line 2 is BL36652. One representative experiment is shown. n=the number of embryos scored.
B) Representative images of 0–2h embryos that were aged for an additional three hours and stained with a DNA stain (propidium iodide, red). Gastrulating embryos (control) and embryos with pycnotic or aggregated nuclei laid by mothers expressing lid RNAi. Shown are maximal intensity projections of Z-stacks. For embryos with large, aggregated nuclei, shown are the maximal intensity projections of the entire embryo as well as the optical sections in which aggregated nuclei are particularly visible. In all panels the dorsal side of the embryo is shown with anterior at the top. Scale bar is 50 µm.
C) Percentage of properly developed (gastrulating) and aberrantly developed 0–2h embryos after three hours of aging. Aberrantly developed embryos were classified as: still at the rosette stage, in syncytial divisions, gastrulating, or displaying pycnotic or aggregated nuclei (as shown in B). The same genotypes as in A) were examined. n=the number of embryos scored.
PNG-mediated translational upregulation provides insights into the balance of translation and protein degradation for a subset of proteins
It was striking that at egg activation the majority of translationally activated mRNAs encoded proteins whose levels remained equal or even decreased (Figures 3D, and S4B–S4D). To test the importance of translational upregulation for those mRNAs, we compared the proteomes of wild-type activated eggs with eggs laid by mothers mutant for the translational regulator, PNG (Figure S6A, Table S4). The majority of proteins that increased at egg activation and are encoded by translationally upregulated mRNAs had lower levels in png eggs than wild-type eggs (46 out of 75, Table S4) (Figure 5A). Consistently, ~80% of these 46 mRNAs completely depended on PNG for translational upregulation at egg activation (Figure 5B). The other 29 mRNAs encode proteins with unchanged levels in png activated eggs as compared to wild-type (Figures 5A and S6B, Table S4), most likely because a large fraction of them, ~59%, was at least partially independent of PNG for their translational activation at egg activation (Figure 5B).
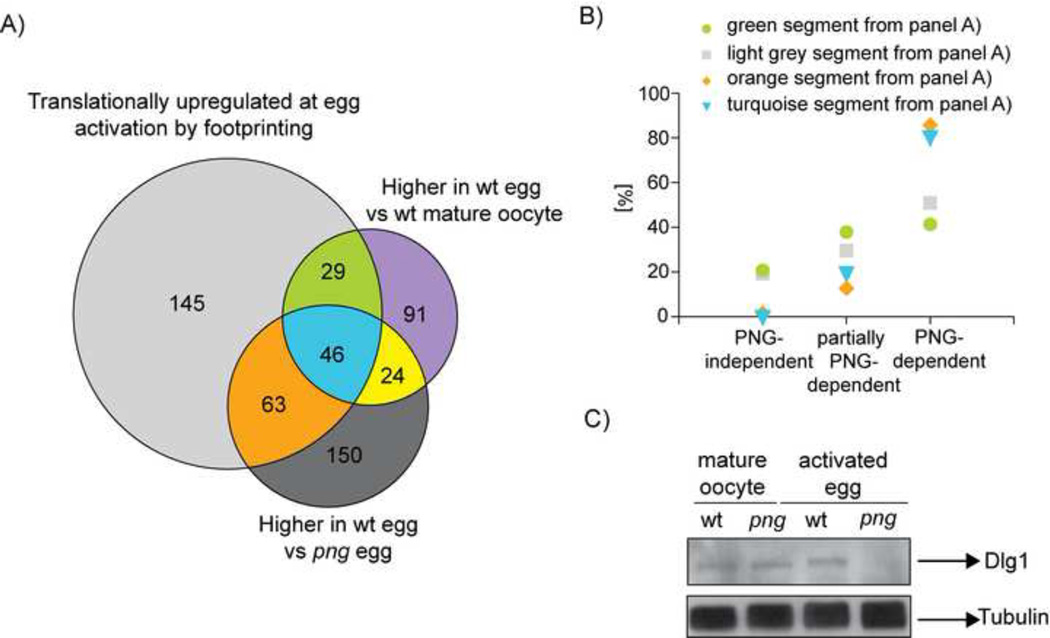
Figure 5. The translational regulator PNG reveals the importance of translational regulation for homeostasis of protein levels for a subset of proteins at egg activation.
A) Same as Figure 3D, except that the comparison included the proteins whose levels are higher in wild-type than png activated eggs according to mass spectrometry. Only factors identified in all compared samples are represented. The translationally upregulated mRNAs are from wild type.
B) Percentage of mRNAs encoding proteins belonging to the light grey, orange, green or turquoise segments in panel A that are independent, partially dependent, or dependent on PNG for translational upregulation at egg activation.
C) Western blot validation of Dlg1, a candidate from the orange segment of Venn diagram in panel A. Dlg1 levels do not change at egg activation in wild type, although the protein shows altered mobility. In contrast, protein levels are decreased following activation of png mutants (reprobed on the same membrane as in Figure 6F).
Interestingly, there were 63 translationally upregulated mRNAs that encode proteins with stable levels at egg activation but decreased abundance in png as compared to wild-type activated eggs (orange segment in Figures 5A and S6B, Table S4). 86% of these 63 mRNAs are dependent on PNG for their translational upregulation at egg activation (Figure 5B). This group of proteins, represented by Dlg1, suggested that translational upregulation at egg activation might even be influencing proteins whose abundance does not change (Figure 5C). Another possibility was that inefficient maternal protein deposition in png mature oocytes leads to the lower levels of proteins in the orange segment of Figure 5A in png activated eggs versus wild-type, although this is not the case for Dlg1 (Figure 5C). In addition, the majority of these 63 proteins have lower levels in png activated eggs as compared to png mature oocytes (Figures S6B and S6C, Table S3). Collectively, these results are consistent with the possibility that, for a subset of proteins, translational upregulation compensates for protein turnover occurring at egg activation to maintain rather than increase their levels at this developmental transition.
Downregulation of protein levels at egg activation is largely post-translationally driven
Comparison of the proteomes of mature oocytes and activated eggs revealed that 376 proteins became significantly less abundant at egg activation (p <0.05) (Figures 3A and 3B, Table S4). This group was not enriched in any particular GO category (FDR >0.05). Downregulation of proteins can be at least partially attributed to the activation of the APC/C at the completion of meiosis resulting in the proteasomal degradation of its substrates, such as cell-cycle regulators CycB3 and Mtrm (Pesin and Orr-Weaver, 2007; Whitfield et al., 2013). In addition, other degradation pathways are likely to be active.
For 265 of 376 significantly downregulated proteins, we obtained translational efficiency information of their mRNAs and found that 43 were translationally downregulated (Category IV, Figure 6A, Table S4). In agreement with the proteomics data, immunoblot showed that the levels of Giant Nuclei (GNU, Category IV) decrease after egg activation (Figure 6B). This overlap between downregulated proteins and translationally inhibited mRNAs is significant, as only 6 of translationally repressed mRNAs encode proteins whose levels increased during egg activation (p <10−7, Figure 6A). The levels of 226 proteins encoded by translationally inhibited mRNAs were not decreased at egg activation, presumably because the maternal stockpiles of these proteins are stable. We obtained comparable results when the translation status at egg activation was measured as a change in polysomal recruitment (Figure S6D).
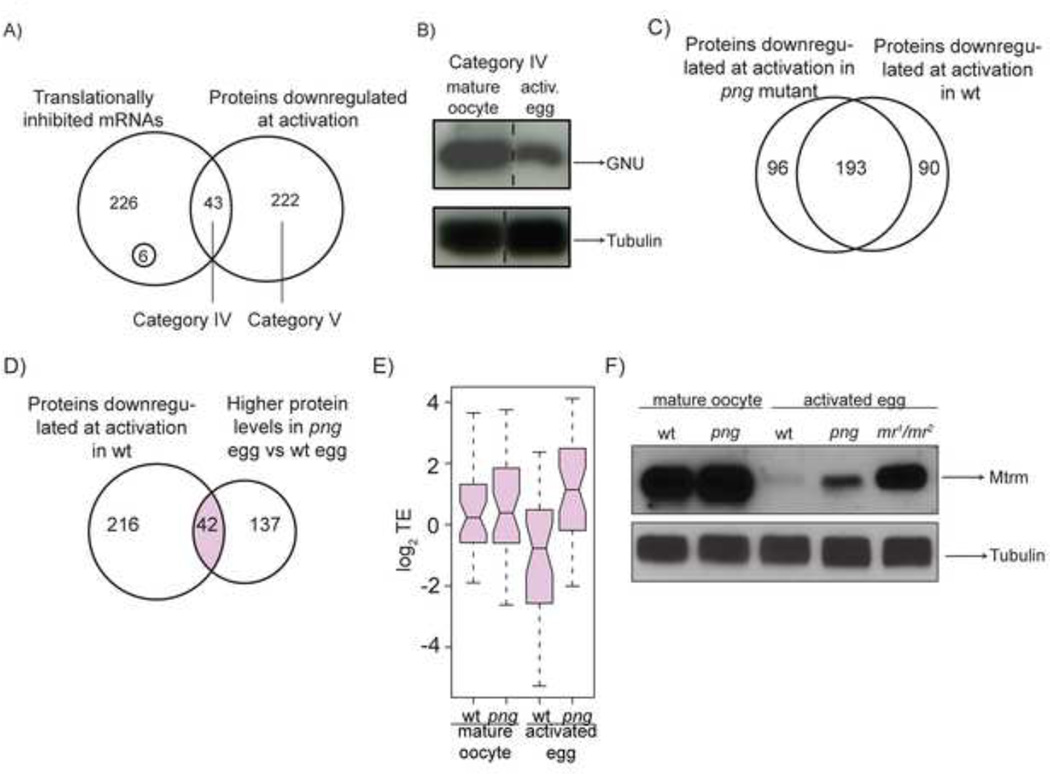
Figure 6. Absence of translational shut down at egg activation may interfere with efficient downregulation of a subset of proteins.
A) Venn diagram showing the number of translationally inhibited mRNAs (Figure 1D) that encode proteins downregulated at egg activation (Category IV). Otherproteins whose levels decrease at egg activationaccording to quantitative mass spectrometry are inCategory V. Among translationally inhibited mRNAs there are two groups: the majority of mRNAs encode proteins with unchanged levels at egg activation, whereas only six encode proteins whose levels increase. Only factors identified in both samples are represented.
B) Western blot validation of GNU, a protein from Category IV.
C) And D) Venn diagrams comparing the number of proteins that according to mass spectrometry are downregulated at egg activation in wild-type or png activated eggs (C) or downregulated at activation in wild-type but more abundant in png activated eggs than wild-type (D). Only factors identified in both compared samples are represented.
E) Box plot showing translational efficiencies of mRNAsencoding for 36 proteins in the overlap zone (shown inlight pink) of Venn diagram in panel D. 36 out of 42mRNAs are presented because the other 6 were notidentified by ribosome footprinting.
F) Western blot comparing Mtrm levels at egg activation in wild-type and png mutant background as well as in the activated eggs laid by mothers transheterozygous for female-sterile alleles of the APC2 subunit of APC/C, morula (mr1/mr2). In png mutants in which translational inhibition of mtrm does not occur, Mtrm protein levels are elevated, although to a lesser extent than when the APC/C is mutated.
Category V proteins decrease at egg activation despite retaining their translational status (Table S4). For example,CycB3 protein was reduced 5.6-fold although the translational efficiency of its encoding mRNA increased 7.6-fold (Tables S2 and S3). Most proteins downregulated at egg activation have not been studied for their role in meiosis, and many are completely uncharacterized. In addition to revealing candidate regulators of the oocyte-to-embryo transition, these result sindicate that for most proteins downregulated at egg activation,post-translational rather than translational control governs the decrease in protein levels at egg activation. Most of these downregulated proteins decreased without translational inhibition of their mRNAs.
Failure in translational inhibition for a subset of mRNAs interferes with successful removal of the proteins they encode at the oocyte-to-embryo transition
The modest contribution of translational inhibition to decreased protein levels at egg activation suggests that many proteins reduced at egg activation in wild-type would also be reduced in the png mutant background. Indeed, 289 proteins were significantly reduced in png activated eggs as compared to png mature oocytes, and 193 of them were among the 283 proteins that were significantly reduced in wild-type eggs at activation (Figure 6C). Nonetheless, 42 proteins reduced at egg activation in wild-type had higher levels in png versus wild-type activated eggs (Figure 6D). Approximately 31% of these proteins, including Mtrm, were encoded by mRNAs that were dependent on PNG for their translational shutoff. Consequently, their translational efficiencies were overall significantly higher in png than in wild-type eggs (Mann Whitney test p <0.0001, Figure 6E and Table S4).
The absence of translational shut-off in png eggs allowed us to investigate the role of translational inhibition to decreased protein levels at egg activation, using Mtrm as an example. In parallel, we employed the mr mutant, which carries a mutation in the APC2 subunit, to weigh the contributions of APC/C-mediated proteasomal degradation versus translational inhibition to Mtrm protein levels (Kashevsky et al., 2002). Western blotting confirmed the proteome data showing that although there was a substantial decrease in Mtrm levels both in wild-type and png activated eggs as compared to mature oocytes, there was persistent Mtrm protein in png eggs (Figure 6F). Because the Mtrm levels remaining in png activated eggs were lower than in mr activated eggs, we conclude that Mtrm levels are primarily regulated by targeting via the APC/C for proteasomal degradation. Thus, optimal protein decrease at egg activation in addition to protein degradation may require PNG-dependent shut off of translation.
DISCUSSION
Relationship between the translatome and proteome
This study provides a first demarcation of the contribution of translational and post-translational regulation to proteome remodeling during a key developmental transition from oocyte to embryo. This transition requires a change from meiosis to mitosis as well as resetting of the oocyte to restore totipotency in the embryo. The absence of transcription and mRNA degradation in the oocyte and early embryo leads to exclusively translational and post-translational control of gene expression. In this study, the oocyte-to-embryo transition was assessed through quantitative proteomics analysis combined with complementary translatome measurements. We found that translational upregulation strongly contributes to increased protein levels at egg activation. The effect of translational shutdown on decreased protein levels is more modest, with decreased levels appearing to be largely driven by protein degradation.
The striking translational changes at egg activation, with as many as 802–986 translationally upregulated and 448–729 inhibited mRNAs, are comparable to those observed in other developmental studies. Polysome profiling at oocyte maturation and egg activation in mouse or in different stages of Drosophila embryogenesis also showed that hundreds of mRNAs were both released from and recruited to the polysomes (Chen et al., 2011; Chen et al., 2014; Potireddy et al., 2006; Qin et al., 2007). Thus, this window of development is highly dynamic across species, and it relies on translational control.
The conclusions from our analysis of developmental control of the proteome at Drosophila egg activation contrast with two previous studies where protein levels were quantified. In mouse fibroblasts, the contribution of protein degradation to protein abundance is minor (Schwanhausser et al., 2011). Similarly, in S. cerevisiae RPFs and protein abundance are well correlated, suggesting that also in this system translational control largely shapes the proteome (Ingolia et al., 2009). Unlike Drosophila activated eggs, mouse fibroblast and S. cerevisiae are transcriptionally active, steady-state systems that lack maternally loaded proteins, potentially resulting in different requirements for translational regulation in controlling protein levels.
Mechanism of translational control at egg activation
We observed both activation and inhibition of mRNA translation at the oocyte-to-embryo transition. Translational inhibition at egg activation may not be an actively regulated process. Rather, the onset of increased translational activity observed at egg activation may limit the available ribosomes resulting in a reduced access to ribosomes and consequent translational repression for the subset of mRNAs.
The Ser/Thr kinase PNG had been previously demonstrated to activate the translation of cycA, cycB and smg, but given the specificity of the mutant phenotypes and rescue experiments with overexpression of cycB was assumed to influence a circumscribed set of targets (Lee et al., 2001). In contrast, we find that PNG is a global regulator of translational status of mRNAs exclusively at egg activation. In addition, although PNG has been thought to activate translation, our analysis reveals that it acts both positively and negatively, controlling translational activation of at least 60% of mRNAs and inhibition of 70%. Despite the critical role of PNG in controlling the translational status of the majority of mRNAs at egg activation, it is unlikely that the primary function of PNG kinase is in an activation event upstream in the egg activation pathway. Many of the activation events such as eggshell hardening (Tadros et al., 2003) and downregulation of protein levels occur in png mutants.
We postulate that many of the PNG-dependent translational effects reflect a direct role of the kinase. It will be interesting to investigate PNG substrates for candidates whose phosphorylation affects translational efficiencies of mRNAs. It is likely, however, that translation of some mRNAs may be affected as a secondary or indirect effect of PNG-activity.
One possible direct target of PNG that affects translation is the translational repressor, Pumilio (PUM). The png embryonic cell cycle defect is suppressed by pum mutations, which restore Cyclin B protein levels (Vardy and Orr-Weaver, 2007). PNG-PUM antagonism does not appear to be an exclusive mechanism through which PNG mediates translational activation, as there is no enrichment of PUM binding sites in the 3’UTRs of PNG-dependent translationally activated mRNAs as compared to PNG-independent mRNAs (see Supplemental Discussion for additional 5’ and 3’ UTR analysis). This suggests that PNG controls translation via multiple targets. Some of its targets may also be translational regulators, as CG17514, a putative activator of translation, is translationally downregulated, whereas the translational repressor Cup is translationally upregulated, in png activated eggs.
The large number of puromycin-insensitive mRNAs in mature oocytes was surprising and suggests that many mRNAs are in cytoplasmic granules or heavy RNPs at this developmental stage. The inefficient translation of these mRNAs in mature oocytes was corroborated by ribosome footprinting analysis, as they were found to associate with significantly fewer ribosomes compared to puromycin-sensitive mRNAs (data not shown). As puromycin only displaces actively translocating ribosomes, a fraction of puromycin-insensitive mRNAs may localize to cytosolic compartments where they are associated with stalled and non-elongating polysomes.
There is precedent for specific mRNAs to be localized to cytoplasmic granules in oocytes. In Drosophila, the maternally-deposited mRNAs for three patterning genes, osk, grk and bcd, localize to large RNP complexes (Chekulaeva et al., 2006; Weil et al., 2012). It has been suggested that these cytoplasmic RNPs control both activation and repression of translation: grk mRNA is on the periphery of the complex and translated, whereas the internally localized bcd mRNA is repressed (Weil et al., 2012). Cytosolic granule association also may serve as a timer for translational activation. In zebrafish and mouse oocytes, association of cycB mRNA with cytosolic granules is not necessary for translational repression, but release out of the granules leads to premature CycB synthesis (Kotani et al., 2013).
Insights into key regulators of the oocyte-to-embryo transition
By identifying translationally regulated mRNAs and proteins whose levels change at egg activation this work also highlights potential key processes in the oocyte-to-embryo transition and reveals previously unrecognized regulators. As a proof of principle, we demonstrated that one of the 291 proteins upregulated at egg activation, histone H3-K4 demethylase, LID, is required for timely progression through early embryonic cycles and proper further embryonic development. Other upregulated proteins may be required immediately at the onset of embryogenesis.
Downregulation of specific proteins at the oocyte-to-embryo transition also is important. This has been demonstrated for the Polo kinase inhibitor Mtrm whose degradation is required for proper embryogenesis (Whitfield et al., 2013). The microtubule severing enzyme katanin subunit, Kat80, is controlled similarly (Tables S1 and S2). In C. elegans katanin (MEI-1) is required for assembly of the meiotic spindle but failure to decrease its levels before embryonic mitosis leads to spindle defects (Quintin et al., 2003; Stitzel et al., 2006). The translational inhibition applied to these mRNAs may combine with proteasomal degradation for faster and more complete removal of meiotic proteins.
“Resetting” the proteome at the oocyte-to-embryo transition
Our translatome and proteome survey of png eggs provided an unexpected insight, the possible coupling between translational regulation and protein degradation at egg activation. There is a class of 63 mRNAs that are translationally upregulated without a perceptible increase in their protein levels in wild-type eggs. These proteins seem to be properly maternally deposited in png mutant oocytes, but their mRNAs require PNG for translational upregulation at egg activation. Lower levels of these proteins in png versus wild-type activated eggs showed that failure of translational activation resulted in easily detected decreases in protein levels. Hence, the apparently constant levels of these proteins in the wild-type background were not merely a consequence of high maternal stores overwhelming the translational activation. Moreover, the proteome changes in png mutants are unlikely to result from an additional independent effect of PNG on protein stability, as quantitative mass spectrometry data show that downregulation of protein levels occurs comparably in wild-type and png mutants eggs.
Thus, the simplest explanation for these results is that translational activation compensates for protein degradation, leading to no net change in protein abundance in the wild-type background. Although the png mutant allowed us to demonstrate this for 63 proteins, it is likely that additional proteins are reset. An additional possibility is that for some of these proteins there is a spatial control in the embryo, with synthesis at one site and degradation at another. The increased protein degradation normally occurring at egg activation probably is supported by mechanisms in addition to the APC/C, as we found at least 11 E2 Ubiquitin-conjugating enzymes, E3 Ubiquitin ligases or their putative regulators among the proteins upregulated at egg activation.
Why would a subset of proteins in activated eggs undergo an energetically costly process of increased synthesis and degradation only to maintain the same levels as in mature oocytes? We suggest that the developmental coordination between translation and protein stability restores the proteome, exchanging those proteins from their “oogenesis” to their “embryogenesis” form. As some post-translational modifications might interfere with embryonic functions of a given protein, an expedient mechanism to remove these modifications might be to degrade and then resynthesize the proteins. Such a “resetting” of this subset of the proteome at egg activation resembles a phenomenon occurring later in embryogenesis at the mid-blastula stage, in which the transcriptome is reset to allow zygotic control of development (Tadros and Lipshitz, 2009).
MATERIALS AND METHODS
Quantitative mass spectrometry
Mature oocytes were hand-dissected in Grace’s Unsupplemented Insect Media (Gibco) from four-day old flies that had been fattened for three days with wet yeast at 22°C. 0–2h collections of activated eggs (laid by wild-type, Oregon R, females mated with spermless twine HB5 males) were dechorionated, lysed, sonicated, and supernatants frozen. Digestion of the proteins and stable isotope labeling of the peptides (peptide dimethylation) were performed as previously described (Wisniewski et al., 2009) (Boersema et al., 2009). The labeled peptides were fractionated, desalted and separated using the nanoAcquity UPLC system (Waters) from which they were directed to an LTQ Orbitrap Velos (Thermo Fisher Scientific) using a Proxeon nanospray source. The mass spectrometric raw data were processed using MaxQuant (version 1.1.1.25) (Cox and Mann, 2008) and MS/MS spectra were searched using the Andromeda search engine (Cox et al., 2011) against a Uniprot Drosophila melanogaster database. Statistical analysis of mass spectrometric data was performed using the Limma package in R/Bioconductor (Gentleman et al., 2004). Further details are explained in the Supplemental Experimental Procedures.
Western Blots
Samples were lysed and Western blots performed as previously described (Whitfield et al., 2013). Antibodies used are described in Supplemental Experimental Procedures.
Polysome Analysis, Ribosome Footprint Profiling, RNA Isolation and mRNA Sequencing
Samples were lysed as described and flash frozen (Mermod and Crippa, 1978). For puromycin treatment, samples were prepared as described previously (Clark et al., 2000). Samples were run on 10–50% linear sucrose gradients with 0.5 mg/ml cycloheximide. Cycloheximide was excluded for the gradients in the puromycin-treatment experiments. Following centrifugation and fractionation, prior to RNA extraction, in vitro transcribed 5ng of Firefly luciferase (Promega) and 5ng of S. cerevisae mRNA were added to each pooled fraction to allow for normalization between the fractions.
RNA was isolated from whole lysates of mature oocytes or activated eggs by homogenizing them in TRIzol (Invitrogen) according to manufacturer’s instructions. To extract RNA from sucrose gradient fractions, 0.5% SDS and 200 µg/ml of Proteinase K (Sigma) were added for 30 minutes at 50°C, followed by RNA isolation using the hot acid phenol method.
For ribosome footprint profiling, samples were thawed on ice and then triturated four times with a 26-gauge needle. After centrifuging to clear the lysate, ribosome profiling and RNA-seq were performed as described (Subtelny et al., 2014), with a detailed protocol available at http://bartellab.wi.mit.edu/protocols.html. A replicate of each sample was prepared with cycloheximide excluded from all solutions.
To sequence mRNAs, 1 µg of total RNA was poly(A)-selected using Sera-Mag magnetic oligo(dT) magnetic particles (Thermo Scientific). Barcoded mRNA seq libraries were made according to the manufacturer’s instructions (Illumina), with the exception that mRNA was fragmented using a RNA fragmentation kit (Ambion). mRNA sequencing analysis is described in Supplemental Experimental Procedures.
The average translational efficiency (TE) from two independent ribosome footprinting experiments obtained by our method of isolating and preparing lysates from 0–2h unfertilized eggs correlated well (Spearmen R=0.89) with TEs recently published for fertilized 0–2h embryos cryolysed in the absence of dechorionation (Dunn et al., 2013).
Immunofluorescence and Imaging
Embryos were collected at 25°C for 2 hours (or aged for additional 3 hours at 25°C), dechorionated, fixed and stained as described (Pesin and Orr-Weaver, 2007). Images were acquired on LSM 700 microscope (Carl Zeiss) and processed using ImageJ software.
Supplementary Material
The translatome is extensively altered at egg activation
PNG kinase is a major regulator of translational status of mRNAs at egg activation
Subset of proteins is “reset” at Drosophila oocyte-to--embryo transition
Catalog of candidate new regulators of early embryogenesis, i.e. H3K4 demethylase Lid
ACKNOWLEDGEMENTS
We thank A. Amon, W. Gilbert, J. Richter, M. Hara, B. Petrova, O. Rissland and I. Ulitsky for helpful comments on the manuscript. We are grateful to J. Kirkpatrick from the EMBL Proteomics Core Facility for the help in processing the samples for quantitative mass spectrometry analysis. We thank G. Bell for the help with bioinformatics analysis and T. DiCesare for help with the graphical abstract. We acknowledge A. Imhof, J. Brill, J. Price, T. Kaufman, E. Wahle and M. Asano for the kind gifts of antibodies. The anti-Sema-2a and Dlg1 antibodies developed by C. Goodman were obtained from the Developmental Studies Hybridoma Bank. We thank the TRiP at Harvard Medical School (NIH/NIGMS R01-GM084947) and the Bloomington Stock Center for providing transgenic RNAi fly stocks used in this study. IK was supported by the Feodor Lynen Postdoctoral Fellowship by the Alexander von Humboldt Foundation. This research was funded by NIH grant GM39341 to TO-W. DB is an HHMI investigator and TO-W is an American Cancer Society Research Professor.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Polysome profiling and ribosome footprinting sequencing data were deposited in the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) under accession number GSE52799.
REFERENCES
- Anderson KV, Lengyel JA. Rates of synthesis of major classes of RNA in Drosophila embryos. Dev Biol. 1979;70:217–231. doi: 10.1016/0012-1606(79)90018-6. [DOI] [PubMed] [Google Scholar]
- Beilharz TH, Preiss T. Translational profiling: the genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic. 2004;3:103–111. doi: 10.1093/bfgp/3.2.103. [DOI] [PubMed] [Google Scholar]
- Blobel G, Sabatini D. Dissociation of mammalian polyribosomes into subunits by puromycin. Proc Natl Acad Sci USA. 1971;68:390–394. doi: 10.1073/pnas.68.2.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, Weissman JS. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Meiosis I is established through division-specific translational control of a cyclin. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth A, Meijer HA, de Moor CH. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA. 2013;4:437–461. doi: 10.1002/wrna.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes & Dev. 2011;25:755–766. doi: 10.1101/gad.2028911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Dumelie JG, Li X, Cheng MH, Yang Z, Laver JD, Siddiqui NU, Westwood JT, Morris Q, Lipshitz HD, et al. Global regulation of mRNA translation and stability in the early Drosophila embryo by the Smaug RNA-binding protein. Genome Biol. 2014;15:R4. doi: 10.1186/gb-2014-15-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Herskowitz I. Gametogenesis in yeast is regulated by a transcriptional cascade dependent on Ndt80. Mol Cell. 1998;1:685–696. doi: 10.1016/s1097-2765(00)80068-4. [DOI] [PubMed] [Google Scholar]
- Clark IE, Wyckoff D, Gavis ER. Synthesis of the posterior determinant Nanos is spatially restricted by a novel cotranslational regulatory mechanism. Curr Biol. 2000;10:1311–1314. doi: 10.1016/s0960-9822(00)00754-5. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. Elife. 2013;2:e01179. doi: 10.7554/eLife.01179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Sprenger F, Duronio RJ, Leopold P, O’Farrell PH. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis. Genes & Dev. 1994;8:440–452. doi: 10.1101/gad.8.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenger DD, Carminati JL, Burney-Sigman DL, Kashevsky H, Dines JL, Elfring LK, Orr-Weaver TL. PAN GU: a protein kinase that inhibits S phase and promotes mitosis in early Drosophila development. Development. 2000;127:4763–4774. doi: 10.1242/dev.127.22.4763. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Xu W, Cooper GM, Richter JD. Translational control by cytoplasmic polyadenylation of c-mos mRNA is necessary for oocyte maturation in the mouse. EMBO Journal. 1994;13:5712–5720. doi: 10.1002/j.1460-2075.1994.tb06909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gildea JJ, Lopez R, Shearn A. A screen for new trithorax group genes identified little imaginal discs, the Drosophila melanogaster homologue of human retinoblastoma binding protein 2. Genetics. 2000;156:645–663. doi: 10.1093/genetics/156.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner VL, Wolfner MF. Mechanical stimulation by osmotic and hydrostatic pressure activates Drosophila oocytes in vitro in a calcium-dependent manner. Dev Biol. 2008a;316:100–109. doi: 10.1016/j.ydbio.2008.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner VL, Wolfner MF. Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev Dyn. 2008b;237:527–544. doi: 10.1002/dvdy.21454. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashevsky H, Wallace JA, Reed BH, Lai C, Hayashi-Hagihara A, Orr-Weaver TL. The anaphase promoting complex/cyclosome is required during development for modified cell cycles. Proc. Natl. Acad. Sci. USA. 2002;99:11217–11222. doi: 10.1073/pnas.172391099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani T, Yasuda K, Ota R, Yamashita M. Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol. 2013;202:1041–1055. doi: 10.1083/jcb.201302139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LA, Elfring LK, Bosco G, Orr-Weaver TL. A genetic screen for suppressors and enhancers of the Drosophila PAN GU cell cycle kinase identifies cyclin B as a target. Genetics. 2001;158:1545–1556. doi: 10.1093/genetics/158.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LA, Van Hoewyk D, Orr-Weaver TL. The Drosophila cell cycle kinase PAN GU forms an active complex with PLUTONIUM and GNU to regulate embryonic divisions. Genes & Dev. 2003;17:2979–2991. doi: 10.1101/gad.1132603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret-Llinares M, Carre C, Vaquero A, de Olano N, Azorin F. Characterization of Drosophila melanogaster JmjC+N histone demethylases. Nucl Acids Res. 2008;36:2852–2863. doi: 10.1093/nar/gkn098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod JJ, Crippa M. Variations in the amount of polysomes in mature oocytes of Drosophila melanogaster. Dev Biol. 1978;66:586–592. doi: 10.1016/0012-1606(78)90264-6. [DOI] [PubMed] [Google Scholar]
- Pesin JA, Orr-Weaver TL. Developmental role and regulation of cortex, a meiosis-specific anaphase-promoting complex/cyclosome activator. PLoS Genet. 2007;3:e202. doi: 10.1371/journal.pgen.0030202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol. 2006;298:155–166. doi: 10.1016/j.ydbio.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Qin X, Ahn S, Speed TP, Rubin GM. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol. 2007;8:R63. doi: 10.1186/gb-2007-8-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintin S, Mains PE, Zinke A, Hyman AA. The mbk-2 kinase is required for inactivation of MEI-1/katanin in the one-cell Caenorhabditis elegans embryo. EMBO reports. 2003;4:1175–1181. doi: 10.1038/sj.embor.7400029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagata N. Meiotic metaphase arrest in animal oocytes: its mechanisms and biological significance. Trends Cell Biol. 1996;6:22–28. doi: 10.1016/0962-8924(96)81034-8. [DOI] [PubMed] [Google Scholar]
- Salles FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S. Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science. 1994;266:1996–1999. doi: 10.1126/science.7801127. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Secombe J, Li L, Carlos L, Eisenman RN. The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth. Genes & Dev. 2007;21:537–551. doi: 10.1101/gad.1523007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamanski FL, Orr-Weaver TL. The Drosophila plutonium and pan gu genes regulate entry into S phase at fertilization. Cell. 1991;66:1289–1300. doi: 10.1016/0092-8674(91)90050-9. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Pellettieri J, Seydoux G. The C. elegans DYRK Kinase MBK-2 Marks Oocyte Proteins for Degradation in Response to Meiotic Maturation. Curr Biol. 2006;16:56–62. doi: 10.1016/j.cub.2005.11.063. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan A, Schupbach T. The Cdc20 (Fzy)/Cdh1-related protein, Cort, cooperates with Fzy in cyclin destruction and anaphase progression in meiosis I and II in Drosophila. Development. 2007;134:891–899. doi: 10.1242/dev.02784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Goldman AL, Babak T, Menzies F, Vardy L, Orr-Weaver T, Hughes TR, Westwood JT, Smibert CA, Lipshitz HD. SMAUG is a major regulator of maternal mRNA destabilization in Drosophila and its translation is activated by the PAN GU kinase. Dev Cell. 2007;12:143–155. doi: 10.1016/j.devcel.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Tadros W, Houston SA, Bashirullah A, Cooperstock RL, Semotok JL, Reed BH, Lipshitz HD. Regulation of maternal transcript destabilization during egg activation in Drosophila. Genetics. 2003;164:989–1001. doi: 10.1093/genetics/164.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. Setting the stage for development: mRNA translation and stability during oocyte maturation and egg activation in Drosophila. Dev Dyn. 2005;232:593–608. doi: 10.1002/dvdy.20297. [DOI] [PubMed] [Google Scholar]
- Tadros W, Lipshitz HD. The maternal-to-zygotic transition: a play in two acts. Development. 2009;136:3033–3042. doi: 10.1242/dev.033183. [DOI] [PubMed] [Google Scholar]
- Tay J, Hodgman R, Richter JD. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev Biol. 2000;221:1–9. doi: 10.1006/dbio.2000.9669. [DOI] [PubMed] [Google Scholar]
- Vardy L, Orr-Weaver TL. The Drosophila PNG kinase complex regulates the translation of cyclin B. Dev Cell. 2007;12:157–166. doi: 10.1016/j.devcel.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C, et al. Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol. 2012;14:1305–1313. doi: 10.1038/ncb2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ZJ, Chisholm J, Hawley RS, Orr-Weaver TL. A Meiosis-Specific Form of the APC/C Promotes the Oocyte-to-Embryo Transition by Decreasing Levels of the Polo Kinase Inhibitor Matrimony. PLoS Biol. 2013;11:e1001648. doi: 10.1371/journal.pbio.1001648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- Young RA. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalokar M. Autoradiographic study of protein and RNA formation during early development of Drosophila eggs. Dev Biol. 1976;49:425–437. doi: 10.1016/0012-1606(76)90185-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.