Abstract
The genetic mechanisms that regulate CFTR, the gene responsible for cystic fibrosis, have been widely investigated in cultured cells. However, mechanisms responsible for tissue-specific and time-specific expression are not completely elucidated in vivo. Through the survey of public databases, we found that the promoter of CFTR was associated with bivalent chromatin in human embryonic stem (ES) cells. In this work, we analyzed fetal (at different stages of pregnancy) and adult tissues and showed that, in digestive and lung tissues, which expressed CFTR, H3K4me3 was maintained in the promoter. Histone acetylation was high in the promoter and in two intronic enhancers, especially in fetal tissues. In contrast, in blood cells, which did not express CFTR, the bivalent chromatin was resolved (the promoter was labeled by the silencing mark H3K27me3). Cis-regulatory sequences were associated with lowly acetylated histones. We also provide evidence that the tissue-specific expression of CFTR is not regulated by dynamic changes of DNA methylation in the promoter. Overall, this work shows that a balance between activating and repressive histone modifications in the promoter and intronic enhancers results in the fine regulation of CFTR expression during development, thereby ensuring tissue specificity.
Keywords: bivalent chromatin, enhancers, promoter, DNA methylation, histone modifications, cystic fibrosis, fetal tissues
Introduction
Cystic fibrosis (OMIM 602421) is a life-threatening disease characterized by recurrent pulmonary infections, permanent inflammation, pancreatic insufficiency, and male infertility. This inherited autosomal recessive condition results from mutations in the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. CFTR encodes a chloride channel that is responsible for anion transport across the apical membrane of epithelial cells. The defective protein results in thick, sticky, and obstructive mucus. CFTR expression is generally low and confined primarily to specific epithelial cell types in tissues and organs that are affected in cystic fibrosis. The gene is highly expressed in the pancreatic duct and in the crypts of the small intestine but weakly expressed in the epididymis. In the lung, its expression is high during fetal life and low postnatally.1
The molecular mechanisms responsible for CFTR transcription are complex: the promoter mainly accounts for basal and the AMPc-induced transcription, whereas multiple long-distance cis-regulatory sequences contribute to temporal and spatial regulation.2,3 Critical sequences have been identified in the promoter of CFTR, such as CArG-like and CCAAT-like elements and a variant AMPc response element (CRE).4-7 Promoter regulatory sequences bind ubiquitously-expressed factors that either activate or repress transcription.2 Long-distance cis-regulatory sequences interact with the CFTR promoter via a looping mechanism and bind trans-activating factors that are expressed in a restricted number of tissues. Among the best characterized, two intestinal enhancers are present in introns 1 and 11.2,3,8-11 Another CFTR cis-regulatory sequence, which enhances gene transcription in airway cell lines, maps 35 kb upstream of the transcription start site (TSS).12 Finally, the CFTR gene is flanked by insulators that bind the CCCTC-factor (CTCF) protein and promote chromatin loop formation.13,14 Great advances have been made in the comprehension of CFTR regulation, but the mechanisms responsible for tissue- and time-specific expression are not yet completely elucidated. Current knowledge is based largely on the study of cultured cells, and regulation is poorly characterized in vivo. In this work, we address how epigenetic modifications contribute to the complex regulation of CFTR transcription in human tissues. Compelling evidence suggests that epigenetic mechanisms are relevant. The promoter of CFTR is highly methylated in lung, liver, and bladder cancers, showing that the gene may acquire DNA methylation during cell transformation.15-17 Moreover, most of the factors that either activate or repress CFTR transcription are associated with HATs (Histone acetyltransferases) and HDACs (Histone deacetylases), respectively.7,18
Since epigenetic modifications are profoundly altered by cell culture, we addressed the regulation of CFTR in a physiological context by analyzing human tissues from non-cystic fibrosis donors. To detect developmental changes, we compared fetal and adult tissues.
Results
CFTR expression in human adult and fetal tissues
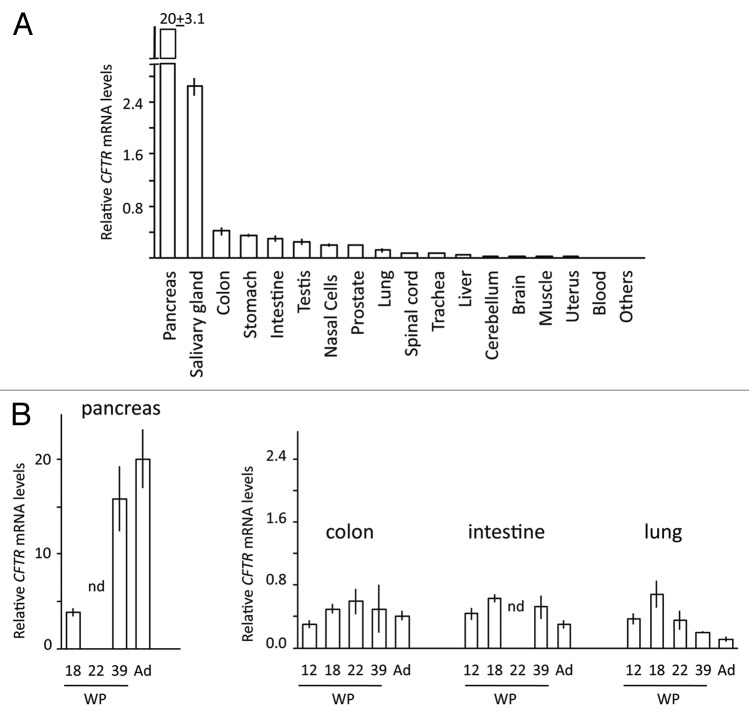
We quantified the relative expression of CFTR in 22 human adult tissues (mRNAs pooled from 1 to 64 individuals) and in 33 fetal samples corresponding to nine tissues from women at different stages in their pregnancies. Adult donors were not affected by cystic fibrosis and fetuses were genotyped to exclude the presence of any known pathogenic mutation in the CFTR gene.
In adult tissues, CFTR expression was high in digestive samples, the highest levels being found in pancreas and salivary gland samples; CFTR expression was lower in respiratory samples and very low or absent in other tissues (i.e., blood, thymus, and heart) (Fig. 1A). In fetuses, CFTR was expressed in the same tissues, but the relative expression levels differed (Fig. 1B). In fetal pancreatic samples, expression increased steadily from the second to the third trimester of pregnancy and to adulthood. No first trimester samples were collected because the pancreas is very small at this stage. A less striking, but still significant change was observed in the lung, where CFTR expression was higher in first trimester (end) and second trimester samples, lower in a third trimester sample, and very low in adults. No expression was detected in fetal thymus, heart, muscle, skin, and liver samples.
Figure 1. Relative expression of CFTR gene: the obtained values were normalized to the expression (considered equal to 1) of CFTR in the colon cancer cell line T84. (A) Adult tissues (mRNA pools comprising 1 to 64 different individuals); “Others” are tissues (heart, thymus, spleen, and bone marrow) whose expression was < 0.01. (B) Fetal tissues (mRNA from single individuals): WP, weeks of pregnancy; Ad, adult tissues (same data as in top figure); nd, not determined.
Low DNA methylation in the CFTR promoter irrespective of transcription
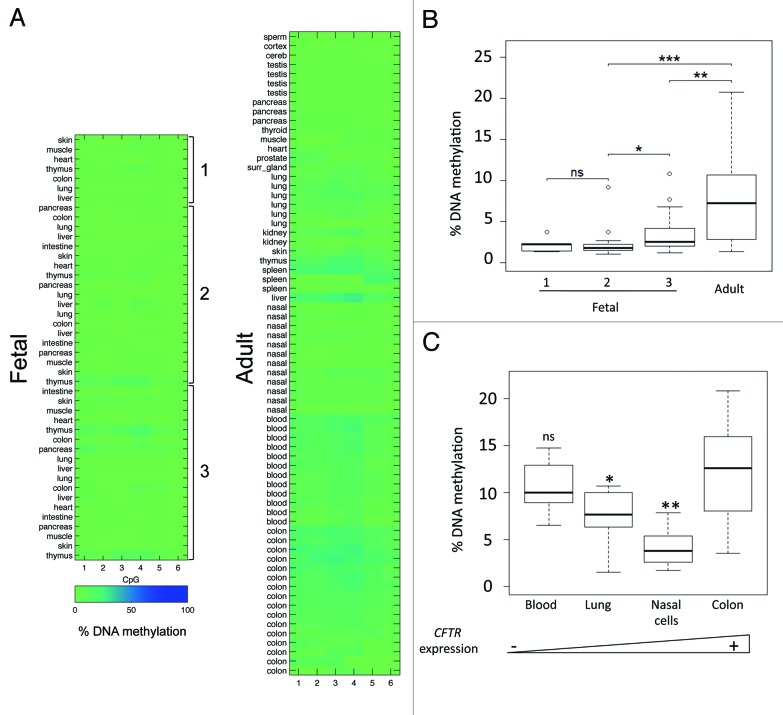
The human CFTR gene is associated with a 1-kb CpG island (GC = 55%, Observed/Expected = 0.65) that overlaps the minimal promoter (Fig. 2). Differences in the level of expression across tissues and during fetal development may result from dynamic changes of DNA methylation in the promoter. Using pyrosequencing, we measured DNA methylation in 44 fetal samples collected from 9 organs, and 68 adult samples collected from 19 organs (Fig. 3A and B). In fetal tissues, DNA methylation was very low; little variation was observed across individuals or across tissues. However, it was slightly higher in fetal tissues from third than first or second trimester pregnancies. DNA methylation was higher in adult tissues than in fetal tissues. Inter-tissue and inter-individual variations were evaluated in four tissues for which at least six adult samples were available: DNA methylation was higher in colon and blood samples than in lung and nasal epithelial samples (Fig. 3C). Variations did not correlate with the donor’s age (the Spearman test; Table S1). Also, they did not correlate with the levels of expression: DNA methylation was not significantly different in colon and blood samples, although the former expressed CFTR and the latter did not. We concluded that tissue-specific expression of CFTR is not regulated by dynamic changes of DNA methylation in the promoter.
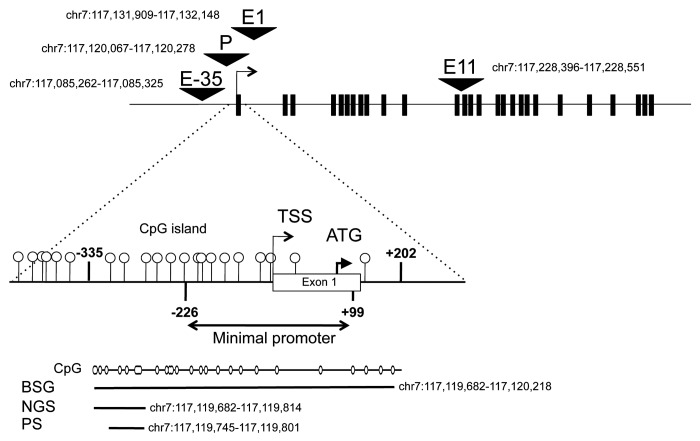
Figure 2. Schematic representation of the CFTR gene (exon-intron structure) showing the promoter (P) and three enhancers located 35-kb 5′ (upstream) from the gene (E-35), in intron 1 (E1) and intron 11 (E11). Zoom on the 5′ region where DNA methylation was analyzed by either bisulfite and genomic sequencing (BGS), or next-generation sequencing (NGS), or pyrosequencing (PS). The three assays measured methylation in 29, 10, and 6 overlapping CpG dinucleotides, respectively. TSS is the transcription start site, and ATG is the first methionine codon. Shown are the hg19 coordinates of the analyzed regions.
Figure 3. (A) Heat maps represent DNA methylation of the CFTR promoter in normal fetal and adult tissues (1, first; 2, second; 3, third trimester of pregnancy). Rows represent individual samples and columns the percentage of DNA methylation measured by pyrosequencing in 6 CpG within the promoter of CFTR. (B) Boxplots show DNA methylation distribution in fetal and adult tissues. Asterisks indicate statistical significance in two-by-two comparisons (*P = 9 × 10−3; **P = 7.4 × 10−7, ***P = 1.2 × 10−12, “ns” is not significant (P = 0.75). (C) Boxplots represent inter-individual DNA methylation variations in blood (n = 12), lung (n = 6), nasal cell (n = 12) and colon (n = 16) samples. In two-by-two comparisons with colon samples, ns P = 0.547; *P = 0.011; **P = 2.04 10−7. The triangle at the bottom represents (from left to right) increasing levels of CFTR expression in the tissues.
DNA methylation of CFTR increases in a subset of cells during lifetime
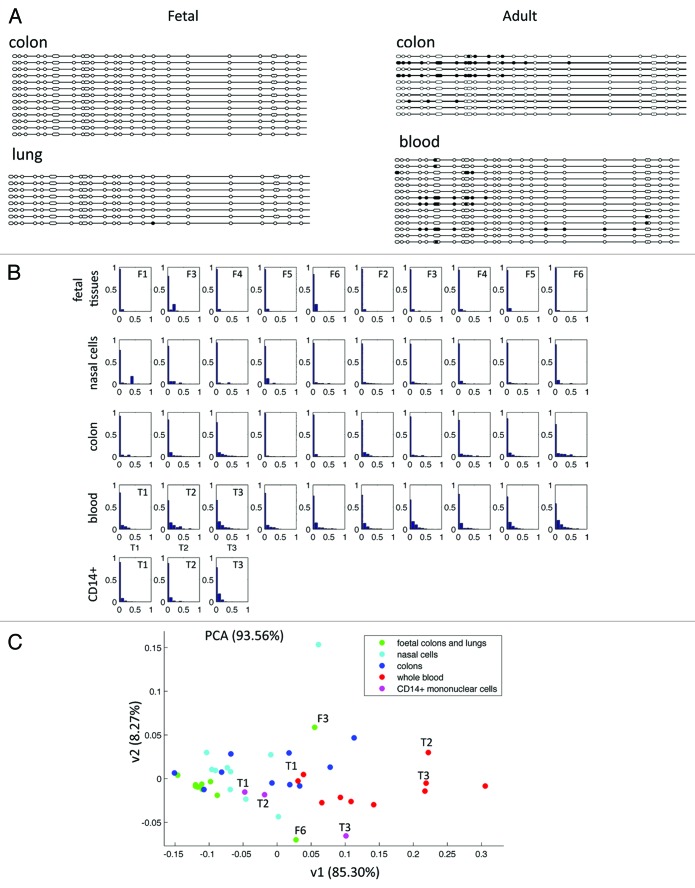
Next, using bisulfite and genomic sequencing (BGS), we measured DNA methylation of a 537-bp region in the promoter of CFTR (Fig. 2). The analysis of eight adult and four fetal samples confirmed that DNA methylation was higher in adult samples and, of importance, showed that methylated CpG dinucleotides were clustered in a few clones (Fig. 4A). Having excluded incomplete bisulfite conversion (no methylation was found in CpA, CpT, and CpC sites—data not shown), we concluded that adult tissues comprised a majority of unmethylated cells mixed with a few heavily methylated cells. To corroborate this finding in a larger number of samples, we analyzed DNA methylation by bisulfite and next-generation sequencing (NGS), an approach that allowed us to multiplex several samples (Fig. 4B). In principal component analysis, the DNA methylation profiles of 30 adult (10 colon, 10 nasal epithelial, and 10 blood) samples were scattered, revealing the presence of a variable percentage of heavily methylated cells that contributed to inter-individual variations (Fig. 4C). In contrast, eight fetal samples clustered: their methylation was low and homogeneous. Very interestingly, two fetal colon samples from third trimester pregnancies were slightly more methylated and more heterogeneous (Fig. 4C). CFTR hypermethylation in a subset of cells may correspond to specific cell types within the tissue sample. To address this issue, we compared CD14+-sorted mononuclear cells and whole blood of three adult donors (Fig. 4C). In mononuclear cells, average DNA methylation and cellular heterogeneity were lower than in whole blood from the same donors, showing that increased cell complexity contributed to methylation heterogeneity in adult tissues.
Figure 4. (A) DNA methylation of a 537-bp region in the promoter of CFTR was analyzed by bisulfite and genomic sequencing. Full and open circles represent methylated and unmethylated CpG, respectively. Fetal tissues have no methylation (left side); in adult tissues some sequences are heavily methylated (right side). (B) Next-generation sequencing of an amplicon containing 10 CpG dinucleotides from the promoter of CFTR: DNA methylation distribution (eleven 0–100% bins) was calculated in >500 sequences/sample. Fetal tissues (first row) are five fetal colon samples on the left side and five fetal lung samples on the right side; CD14+ (last row) are sorted mononuclear cells. (C) Principal component analysis on the same data as 4B. The x-axis is a good estimate of the average methylation (higher values for adult samples than for fetal samples). The y-axis, which has a large weight on the fifth bin (corresponding to sequences characterized by 40–50% of DNA methylation), quantifies the presence of highly methylated sequences. F3 and F6 are two third trimester-colon samples characterized by high cellular DNA methylation heterogeneity. T1, T2, and T3 are either whole blood (red dots) or CD14+ mononuclear cells (pink dots) from the same individual.
CFTR cis-regulatory sequences association with permissive chromatin in digestive and lung tissues
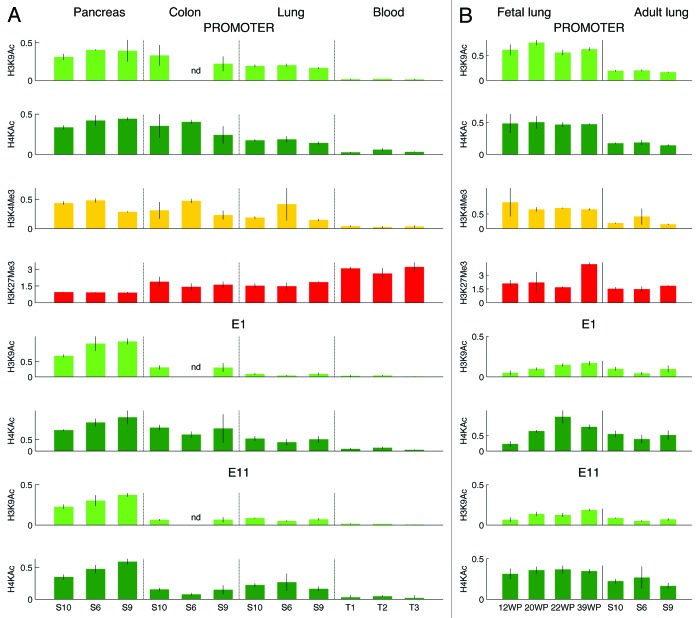
Next, we asked whether chromatin organization in cis-regulatory sequences accounted for different levels of CFTR expression in human tissues. Using ChIP, we quantified histone posttranslational modifications in the promoter, in two intestinal enhancers (one in intron 1 and the other in intron 11—hereafter named E1 and E11, respectively), and in an airway enhancer that maps 35 kb 5′ to the gene (hereafter named E-35) (Fig. 2).7-11 We analyzed whole blood cells and tissue samples from pancreas, colon, and lung. Samples were collected in three healthy adult donors.
A permissive chromatin was observed in the promoter of digestive and lung tissues: histones were acetylated and the TSS was associated with high levels of H3K4me3; the promoter was also enriched in H3K27me3 (Fig. 5A). Because CFTR was highly expressed, we reasoned that the co-presence of activating and repressive histone modifications in pancreas and colon tissues resulted from cellular heterogeneity. By contrast, in lung samples, where CFTR was very weakly expressed, H3K4me3 and H3K27me3 could have been in the same cells, thus representing bivalent chromatin.19 To assess whether bivalent chromatin was associated with the promoter of CFTR in adult lung samples, we performed sequential chromatin precipitation using antibodies directed against H3K4me3 and H3K27me3. No bivalent chromatin was detected in adult lung samples (Fig. S1).
Figure 5. Histone modifications in adult tissues (A) and in fetal and adult lung samples (B). The promoter (top lines) and two enhancers of CFTR, E1 (middle lines) and E11 (bottom lines), were analyzed. Bars represent relative DNA enrichment (means ± sd calculated on ≥3 independent replicates). The amount of immunoprecipitated DNA was expressed relative to the amount of histone modifications either in GAPDH (H3K9Ac, H4KAc, H3K4me3) or in the STS JB10 (H3K27me3) as previously described.30,32 On the x-axis, adult tissues are indicated by the donor code, fetal tissues by weeks of pregnancy (WP).
In digestive and lung tissues, high levels of histone acetylation were found in E1 and E11 enhancers. In E1, histone acetylation was significantly higher in digestive than in lung samples. In contrast, in E11, acetylation was maximal in pancreatic samples and slightly higher in lung than in colon samples. Histone acetylation in the E-35 enhancer was very low in all tissues, including the lung (Fig. S2).
Silent chromatin was found in the promoter and in the three enhancers in blood samples: all sequences were hypoacetylated, no H3K4me3 was found in the TSS, and H3K27me3 in the promoter was significantly higher than in digestive and lung samples.
In summary, digestive and lung tissues were characterized by the presence of permissive chromatin in the promoter and in two intragenic enhancers of CFTR, whereas blood samples showed silent chromatin in all regulatory regions. The levels of expression were positively correlated with the level of H3K4me3 in the TSS and with acetylation in the promoter and in the E1 enhancer; in addition, they were negatively correlated with levels of H3K27me3 in the promoter (Fig. S3).
Histone modifications in CFTR regulatory regions change during development
Since CFTR expression was higher in fetal than in adult lung tissues, we asked whether different levels of expression were associated with histone changes during development. We analyzed four fetuses from third trimester pregnancies. In fetal lung tissues, the promoter was more acetylated and associated with higher levels of H3K4me3 in the TSS and E1 and E11 enhancers were more acetylated than in adult tissues (Fig. 5B). The histone profile was preserved throughout fetal development, except for acetylation in the E1 enhancer, which was maximal in a 22-wk fetal lung and lower before and after this stage.
Histone modifications were analyzed in three fetal colon samples from second trimester pregnancies (Fig. S4). In fetal tissues, CFTR expression was slightly higher than in adult colon tissues and, consistently, acetylation was much higher in the promoter and in the E1 and E11 enhancers, whereas H3K4me3 and H3K27me3 was similar in fetal and adult samples. Histone acetylation was variable among fetal samples: the highest levels were found in the promoter and in the E1 enhancer of a 22-wk colon sample.
Histone acetylation of the E-35 enhancer was as low in fetal samples (lung and colon) as in adult tissues (Fig. S2).
Discussion
In this work, we address how epigenetic modifications contribute to the complex regulation of CFTR transcription in human tissues. Using real-time PCR, we measured expression in various human tissues. CFTR was more highly expressed in digestive than airway tissues, which is consistent with previous semiquantitative analyses.20-22 In human fetuses, CFTR was expressed in the same tissues, but transcript levels were generally higher than in adult tissues, especially in the lung, but also in the colon and the intestine. In the lung, CFTR expression decreased gradually during the third trimester to reach low postnatal levels. A similar gradual reduction of CFTR expression was observed in the ovine lung.23
We showed that promoter DNA methylation did not regulate the tissue-specific expression of CFTR, or variations of transcript levels during fetal development. Similarly, it was reported that dynamic changes of DNA methylation in intronic enhancers were not correlated with levels of CFTR expression.24 However, we observed a slight and progressive increase in fetal promoter DNA methylation from third trimester pregnancies into adulthood. Interestingly, the methylation of genes that are important for immune response and metabolism was also higher during the third trimester of pregnancy.25 In adult tissues and some fetal samples from third trimester pregnancies, DNA methylation of CFTR was higher due to the presence of few heavily methylated cells mixed with a majority of unmethylated cells. Heavily methylated cells may correspond to specific cell types, or may result from random accumulation of epimutations due to environmental cues and/or stochastic changes during lifetime.
To further investigate the regulation of CFTR expression in vivo, we analyzed histone posttranslational modifications. In digestive and lung tissues, which expressed CFTR, the promoter showed permissive chromatin and was characterized by the co-presence of active (H3K4me3) and repressive (H3K27me3) marks, which were suggestive of bivalent chromatin. A bivalent chromatin was first described in embryonic stem cells in the promoter of genes that are poised at this stage and become either active or repressed after lineage commitment.19 Through the survey of public databases, we found that human CFTR and mouse Cftr have bivalent chromatin in ES cells.26,27 Here, we show that in endoderm-derived tissues (lung, colon, and pancreas), the H3K4me3 mark was maintained. Very likely, the co-presence of H3K27me3 was due to cellular heterogeneity. In contrast, in blood cells, which are derived from the mesodermal layer, the bivalent chromatin resolved in the silencing mark H3K27me3 and all regulatory sequences were associated with low histone acetylation. Because bivalent domains were also found in adult muscle stem cells,28 it would be of interest to determine whether bivalent chromatin marks the CFTR promoter in adult stem cells of endoderm-derived tissues.
Most genes that are hypermethylated in human cancers have a bivalent chromatin in ES cells.29 Interestingly, the CFTR promoter was hypermethylated in cancer tissues of endodermic origin.15-17 Also of note, genes associated with bivalent chromatin in ES cells are prone to acquiring DNA methylation during aging.30 Based upon these findings, we suggest that the progressive decline of the pulmonary function in cystic fibrosis patients is, at least partly, a consequence of CFTR methylation in bronchial epithelial cells.
Two intestinal-specific enhancers were previously described in introns 1 and 11 of CFTR.8-11 Here, we show that (1) these intronic enhancers were acetylated in human tissues that express CFTR, including the lung; (2) the levels of acetylation of the enhancers correlated positively with the expression levels; (3) in fetal lung, acetylation of E11 was slightly higher than in fetal colon and much higher than in adult lung samples. Collectively, these data suggest that E1 and E11 enhancers are active not only in digestive, but also in human pulmonary tissues and play a role during fetal development. Lack of enhancer activity in the mouse lung suggests that E1 is not conserved in this species.8 In human pulmonary cell lines, E1 and E11 may have been inactivated by genetic and/or epigenetic changes that occurred during cell culture.8 E-35, a cis-regulatory sequence that maps 5′ to CFTR, enhanced transcription in airway cell lines.11 Low histone acetylation in various adult and fetal tissue samples showed that E-35 was associated with condensed chromatin, suggesting that this regulatory sequence was not active in vivo. Together, these data highlight the importance of using human tissues to validate cis-regulatory sequences in vivo.
It was previously shown that ectopic transcription of CFTR was stimulated by trans-activating factors that bring HATs favoring histone acetylation of the promoter and/or other cis-regulatory sequences in human cell lines.3,7,18 Conversely, CFTR transcription was suppressed by repressors that were associated with HDAC activities.18 The present work shows that similar dynamic changes of histone acetylation occur in regulatory sequences of CFTR in human tissues. Overall, we provide evidence that a balance between activating and repressive histone modifications in the promoter and in intronic enhancers confines CFTR expression to specific tissues and modulates transcript levels during development.
Methods
Human fetal and adult tissues
The collection and analysis of fetal tissues was approved by the “Agence de la Biomédecine.” Fetal tissues (from 12 to 39 wk of pregnancy) were obtained from spontaneous abortions and fetal death of fetuses with no detectable malformations (Table S2). In one case (fetus F5), pregnancy was medically interrupted at the parents’ request and in accordance with French law, after the detection of a severe cardiac malformation; no other malformations were detected in this fetus. Fetuses were genotyped using SCAP (single condition amplification primers) sequencing, to exclude the presence of known pathogenic mutations in the CFTR gene.31 Adult digestive and respiratory tissue samples were provided by Amsbio. Peripheral blood and nasal epithelial cells were collected on adult healthy volunteers who provided their informed written consent (approved by the Nîmes Hospital Ethics Committee #2013.02.01bis). Prior to DNA and RNA extraction, 10–20 mg of adult and fetal tissue samples were ground using 1.4 mm ceramic beads (Precellys R Lysing kit CK14; Bertin Technologies) and Mixer Mill MM30 Retsch. For chromatin preparation, about 200 mg of tissue were used.
Cell purification and sorting
Blood Mononuclear Peripheral Cells (BMPCs) were collected using Histopaque-1077 (#10771) from Sigma Aldrich according to the manufacturer's instructions. BMPCs were then stained using a mouse anti-human anti-CD14 antibody (Ab) conjugated to phycoerythrin (A07764 Clone #RMO52) from Beckman Coulter. Isotype-matched mouse Ab recognizing no human antigen were used as controls. The monocyte fraction (CD14+) was sorted with a multicolor fluorescence-activated cell sorting (FACS) Aria device from BD with a purity ≥ 95%.
Expression analyses
Total mRNA from normal adult tissues was provided by Clontech, or by Amsbio or extracted from tissue samples. Total RNA was extracted using the following kits according to the manufacturer’s protocol: RNAeasy Plus (Qiagen) for solid tissues and PAXgene Blood DNA (Qiagen) for blood samples. One μg of total RNA from each sample was added to RNase free water up to 8 μl and treated with DNase I for 15 min at room temperature. Each sample was then added to a reverse transcription mix containing 4 μl of first strand 5X buffer, 2 μl of 10× dithiothreitol, 1 μl of 10 mM dNTP mix, 300 ng/μl of hexaprimer (random primers), 20–40 U/μl of RNasin® enzyme, and 200 U/μl of MMLV-RT enzyme. Reverse transcription reaction program consisted of three steps: 10 min at 25 °C, 50 min at 37 °C, and 15 min at 70 °C. Two independent reverse transcriptions were done for each RNA extraction. mRNA expression was evaluated by RT-quantitative PCR, using a LightCycler 480 real-time PCR system and SYBR Green I Master mix® (Roche Diagnostics) (primers are listed in Table S3). Standard curves for CFTR and β-actin, the latter used as reference gene, were generated by serial dilution of T84 cell cDNAs of the corresponding reverse transcription, and included in all qPCR runs. After normalization of threshold cycle values with the amount of β-actin, gene expression levels were expressed as ratios relative to that of T84 cells. Each sample was run in triplicate.
DNA extraction and Bisulfite treatment
Genomic DNA was extracted using the following kits according to the manufacturer’s protocol: QIAamp DNA Mini for solid tissues, QIAamp DNA Micro for nasal epithelial cells, and Flexigen for blood samples (Qiagen). Then, 300 ng of genomic DNA were treated with sodium bisulfite as previously described.30
Pyrosequencing
A 133-bp PCR product was amplified using 1 μl of bisulfite-treated genomic DNA and the PyroMark PCR kit (#978703, Qiagen), 10 μM forward and reverse primers (Table S3), and a final volume of 25 μl. The PCR program was 94 °C for 15 min, followed by 94 °C 30 s, 57 °C for 30 s, 72 °C for 30 s repeated 45 cycles, and 72 °C for 10 min. PCR products were purified using 2μl of Streptavidin Sepharose HP (GE Healthcare) and the WorkStation (Qiagen). Then, 10 μl of single strand DNA were pyrosequenced with the PyroMark Gold Q24 reagents (#970802), 7.5 μM sequencing primer (Table S3), and using a PyroMarkQ24 (Qiagen) following the manufacturer’s protocol. We quantified DNA methylation with the Pyromark Q24 software 2.0.
Bisulfite and genomic sequencing
Bisulfite and genomic sequencing was done as previously described.32 Primers are listed in Table S3.
Next-generation sequencing
A 133-bp product was amplified using the PyroMark PCR kit (#978703, Qiagen) kit, and 10μM forward and reverse primers (Table S3) in a 25-μl final volume. Fusion primers for 454 sequencing of Amplicons were designed with MIDs (Multiplex Identifiers, Roche) to computationally screen each sample after analysis. PCR conditions were 95 °C for 15 min, followed by 94 °C for 30 s, 56 °C for 30 s, 72 °C for 30 s repeated 45 cycles, and 72 °C for 10 min. CFTR amplicons obtained on different tissue samples were purified with QIAquick PCR Purification Kit (Qiagen) and quantified on NanoDrop 2000 Spectrophotometer (Thermo Scientific) and QuBit 2.0 fluorometer (Life Technologies). Purified amplicons were pooled in equimolar amounts. Emulsion PCR and subsequent sequencing were done according to the GS Junior emPCR Amplification Method Manual-Lib-A and GS Junior Sequencing Method Manual, (#05996520001 and #05996554001 Roche, respectively). Raw sequencing data were analyzed with the Galaxy Tools (http://main.g2.bx.psu.edu/) and DNA methylation was quantified using BiQ Analyzer HT.33
Native chromatin immunoprecipitation
ChIP experiments were done in 2–3 biological replicates as previously described.32,34 The following antibodies were purchased from Upstate (Millipore): anti-H3K4me3 (#07-473), anti-H3K27me3 (#07-449; #ABE44), anti-hyperacetylated histone H4 (penta) (#06-946), and anti-acetylated H3K9 (#07-352). Real-Time PCR was done using a LightCycler 480 real-time PCR system and SYBR Green I Master mix® (Roche Diagnostics). Primers are listed in Table S3. The euchromatic GAPDH locus and a heterochromatic STS (JB 10) served as references to normalize the amount of precipitated DNA. For a detailed description of normalization see Grunau et al.33 In brief, ratios of precipitated DNA in each gene to that in the reference sequence (either GAPDH or STS JB10) were calculated as follows: enrichment factor = [ng GENE(B)/ng GAPDH(B)]/[ng GENE(I)/ng GAPDH(I)] with (B) for antibody bound and (I) for input. The unbound fraction of mock-treated chromatin (i.e., precipitation in parallel with the other ChIP but without antibody) was considered as input. DNA immunoprecipitated with anti-trimethylated H3K27 was normalized with STS (JB 10); DNA immunoprecipitated with anti-hyperacetylated histone H4 (penta), anti-acetylated H3K9, and anti-trimethylated H3K4 was normalized with GAPDH.
Sequential ChIP
About 200 mg of adult lung were ground and treated with 1.5% formaldehyde (20 min at room temperature) to cross-link chromatin. Then sequential ChIP was performed as previously described.35 After three 1X PBS and PIC washes, tissues were lysed in SDS lysis buffer (#20-163 Millipore) and then sonicated to obtain 0.5- to 1-kb DNA fragments. Chromatin was pre-cleared for 2 h with protein A agarose beads (#16-157 Millipore), then incubated in dilution buffer (0.01% SDS, 1.1% Triton, 1.2 mM EDTA, 167 mM Tris pH 8.0, 167 mM NaCl) overnight at 4 °C with the first antibody directed against either H3K4Me3 or H3K27Me3. The antibodies (5 μg) used in the first precipitation had been cross-linked to 50 μl of protein A agarose beads (Millipore) using 2.5 mM BS3 (Bis-sulfosuccinimidyl suberate) (ProteoChem #c1103). Beads were washed with 1 × RIPA, high salt buffer (0.5M NaCl; 0.5M Tris pH 8, 0.1% SDS, 1%NP40), LiCl buffer (0.5M LiCl, 0.5M Tris pH 8.0, 0.5% Na deoxycholate, 1% NP40) and 1× TE (10 mM Tris pH = 8 and 1mM EDTA). Chromatin was then eluted with 500 μl of 0.1 M NaHCO3 and 1% SDS, diluted (1:10) and incubated overnight at 4 °C with the second antibody. The second antibody was not immobilized to agarose beads. Washes and elution were performed as in the first precipitation. Chromatin crosslink was reversed with 20 mM NaCl at 65 °C overnight. After RNase and proteinase K treatment, immunoprecipitated DNA was extracted with phenol/chloroform and precipitated with isopropanol. Primers specific to the promoter of CFTR were used to detect DNA in input chromatin, chromatin precipitated by one antibody, chromatin precipitated by two consecutive antibodies, and chromatin precipitated by one antibody followed by a mock antibody.
Statistical analysis
Prior to ANOVAs, in general, the Barlett test was used to verify variance homogeneity.
Relative expression of CFTR was analyzed using ANOVA to determine the effect of the developmental stage, and Student t tests for subsequent two-by-two comparisons.
To homogenize the variance of pyrosequencing DNA methylation data, we analyzed the logarithm of these values. The effect of developmental stage was analyzed using a two-way ANOVA, with a fixed effect factor, the stage, and a random effect factor, the tissue. In adults, the effect of the tissue (lung, nasal cells, blood, and colon only) was analyzed with a fixed effect one way ANOVA. Subsequent two-by two comparisons were performed with Student t tests.
NGS DNA methylation data were represented using 11 bins (0%; 0–10%; 10–20%; …; 90–100%) as descriptors for a Principal Component Analysis (PCA).
ChIP data were analyzed with two different ANOVAs. Among adults, for each histone modification and each locus, the effect of the tissue was established with a two-way ANOVA without interaction, with the tissue as fixed effect factor and the individual as random effect factor, the two factors being crossed. Subsequent two-by-two comparisons were performed with Student t tests. For each histone modification and each locus, in lung and in colon, the effect of the developmental stage (adult or fetus) was investigated with a two-way ANOVA, with the stage as fixed effect factor and the individual as random effect factor (the individual were nested in the developmental stage).
P < 0.05 was regarded as statistically significant.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
M.M. was supported by Ciência Sem Fronteiras Program, A.S.H. by an Averroès Fellowship, and J.V. by Agence de la Biomédecine. We thank Jean-Pierre Altieri (Montpellier, France) for SCAP analysis, Marco De Gobbi (Torino, Italy), Francis Poulat (Montpellier, France), and Matthieu Schoenhals (Montpellier, France) for helpful discussion.
Glossary
Abbreviations:
- ANOVA
Analyses Of Variance
- BGS
Bisulfite and genomic sequencing
- CD14+
CD14-positive mononuclear cells
- CFTR
Cystic Fibrosis Transmembrane conductance Regulator
- ChIP
chromatin immunoprecipitation
- ES cells
Embryonic Stem cells
- E1
enhancer localized in intron 1
- E11
enhancer localized in intron 11
- E-35
enhancer localized 35 kb 5′ the TSS
- NGS
next-generation sequencing
- TSS
transcription start site
References
- 1.McCarthy VA, Harris A. The CFTR gene and regulation of its expression. Pediatr Pulmonol. 2005;40:1–8. doi: 10.1002/ppul.20199. [DOI] [PubMed] [Google Scholar]
- 2.Ott CJ, Blackledge NP, Leir SH, Harris A. Novel regulatory mechanisms for the CFTR gene. Biochem Soc Trans. 2009;37:843–8. doi: 10.1042/BST0370843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerschner JL, Harris A. Transcriptional networks driving enhancer function in the CFTR gene. Biochem J. 2012;446:203–12. doi: 10.1042/BJ20120693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McDonald RA, Matthews RP, Idzerda RL, McKnight GS. Basal expression of the cystic fibrosis transmembrane conductance regulator gene is dependent on protein kinase A activity. Proc Natl Acad Sci U S A. 1995;92:7560–4. doi: 10.1073/pnas.92.16.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews RP, McKnight GS. Characterization of the cAMP response element of the cystic fibrosis transmembrane conductance regulator gene promoter. J Biol Chem. 1996;271:31869–77. doi: 10.1074/jbc.271.50.31869. [DOI] [PubMed] [Google Scholar]
- 6.René C, Taulan M, Iral F, Doudement J, L’Honoré A, Gerbon C, Demaille J, Claustres M, Romey MC. Binding of serum response factor to cystic fibrosis transmembrane conductance regulator CArG-like elements, as a new potential CFTR transcriptional regulation pathway. Nucleic Acids Res. 2005;33:5271–90. doi: 10.1093/nar/gki837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paul T, Li S, Khurana S, Leleiko NS, Walsh MJ. The epigenetic signature of CFTR expression is co-ordinated via chromatin acetylation through a complex intronic element. Biochem J. 2007;408:317–26. doi: 10.1042/BJ20070282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowntree RK, Vassaux G, McDowell TL, Howe S, McGuigan A, Phylactides M, Huxley C, Harris A. An element in intron 1 of the CFTR gene augments intestinal expression in vivo. Hum Mol Genet. 2001;10:1455–64. doi: 10.1093/hmg/10.14.1455. [DOI] [PubMed] [Google Scholar]
- 9.Ott CJ, Suszko M, Blackledge NP, Wright JE, Crawford GE, Harris A. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med. 2009;13:680–92. doi: 10.1111/j.1582-4934.2008.00621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ott CJ, Blackledge NP, Kerschner JL, Leir SH, Crawford GE, Cotton CU, Harris A. Intronic enhancers coordinate epithelial-specific looping of the active CFTR locus. Proc Natl Acad Sci U S A. 2009;106:19934–9. doi: 10.1073/pnas.0900946106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gheldof N, Smith EM, Tabuchi TM, Koch CM, Dunham I, Stamatoyannopoulos JA, Dekker J. Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic Acids Res. 2010;38:4325–36. doi: 10.1093/nar/gkq175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Z, Leir SH, Harris A. Immune mediators regulate CFTR expression through a bifunctional airway-selective enhancer. Mol Cell Biol. 2013;33:2843–53. doi: 10.1128/MCB.00003-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackledge NP, Carter EJ, Evans JR, Lawson V, Rowntree RK, Harris A. CTCF mediates insulator function at the CFTR locus. Biochem J. 2007;408:267–75. doi: 10.1042/BJ20070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blackledge NP, Ott CJ, Gillen AE, Harris A. An insulator element 3′ to the CFTR gene binds CTCF and reveals an active chromatin hub in primary cells. Nucleic Acids Res. 2009;37:1086–94. doi: 10.1093/nar/gkn1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding S, Gong BD, Yu J, Gu J, Zhang HY, Shang ZB, Fei Q, Wang P, Zhu JD. Methylation profile of the promoter CpG islands of 14 “drug-resistance” genes in hepatocellular carcinoma. World J Gastroenterol. 2004;10:3433–40. doi: 10.3748/wjg.v10.i23.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu J, Zhu T, Wang Z, Zhang H, Qian Z, Xu H, Gao B, Wang W, Gu L, Meng J, et al. A novel set of DNA methylation markers in urine sediments for sensitive/specific detection of bladder cancer. Clin Cancer Res. 2007;13:7296–304. doi: 10.1158/1078-0432.CCR-07-0861. [DOI] [PubMed] [Google Scholar]
- 17.Son JW, Kim YJ, Cho HM, Lee SY, Lee SM, Kang JK, Lee JU, Lee YM, Kwon SJ, Choi E, et al. Promoter hypermethylation of the CFTR gene and clinical/pathological features associated with non-small cell lung cancer. Respirology. 2011;16:1203–9. doi: 10.1111/j.1440-1843.2011.01994.x. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld EJ, LeLeiko NS, Walsh MJ. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803–15. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–26. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 20.Harris A, Chalkley G, Goodman S, Coleman L. Expression of the cystic fibrosis gene in human development. Development. 1991;113:305–10. doi: 10.1242/dev.113.1.305. [DOI] [PubMed] [Google Scholar]
- 21.Trezise AE, Chambers JA, Wardle CJ, Gould S, Harris A. Expression of the cystic fibrosis gene in human foetal tissues. Hum Mol Genet. 1993;2:213–8. doi: 10.1093/hmg/2.3.213. [DOI] [PubMed] [Google Scholar]
- 22.Engelhardt JF, Zepeda M, Cohn JA, Yankaskas JR, Wilson JM. Expression of the cystic fibrosis gene in adult human lung. J Clin Invest. 1994;93:737–49. doi: 10.1172/JCI117028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broackes-Carter FC, Mouchel N, Gill D, Hyde S, Bassett J, Harris A. Temporal regulation of CFTR expression during ovine lung development: implications for CF gene therapy. Hum Mol Genet. 2002;11:125–31. doi: 10.1093/hmg/11.2.125. [DOI] [PubMed] [Google Scholar]
- 24.Kerschner JL, Gosalia N, Leir SH, Harris A. Chromatin remodeling mediated by the FOXA1/A2 transcription factors activates CFTR expression in intestinal epithelial cells. Epigenetics. 2014;9:557–65. doi: 10.4161/epi.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics. 2011;12:529. doi: 10.1186/1471-2164-12-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Cheung TH, Charville GW, Hurgo BM, Leavitt T, Shih J, Brunet A, Rando TA. Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 2013;4:189–204. doi: 10.1016/j.celrep.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez J, Muñoz M, Vives L, Frangou CG, Groudine M, Peinado MA. Bivalent domains enforce transcriptional memory of DNA methylated genes in cancer cells. Proc Natl Acad Sci U S A. 2008;105:19809–14. doi: 10.1073/pnas.0810133105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010;20:434–9. doi: 10.1101/gr.103101.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bareil C, Guittard C, Altieri JP, Templin C, Claustres M, des Georges M. Comprehensive and rapid genotyping of mutations and haplotypes in congenital bilateral absence of the vas deferens and other cystic fibrosis transmembrane conductance regulator-related disorders. J Mol Diagn. 2007;9:582–8. doi: 10.2353/jmoldx.2007.070040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brun ME, Lana E, Rivals I, Lefranc G, Sarda P, Claustres M, Mégarbané A, De Sario A. Heterochromatic genes undergo epigenetic changes and escape silencing in immunodeficiency, centromeric instability, facial anomalies (ICF) syndrome. PLoS One. 2011;6:e19464. doi: 10.1371/journal.pone.0019464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lutsik P, Feuerbach L, Arand J, Lengauer T, Walter J, Bock C. BiQ Analyzer HT: locus-specific analysis of DNA methylation by high-throughput bisulfite sequencing. Nucleic Acids Res. 2011;39:W551-6. doi: 10.1093/nar/gkr312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grunau C, Buard J, Brun ME, De Sario A. Mapping of the juxtacentromeric heterochromatin-euchromatin frontier of human chromosome 21. Genome Res. 2006;16:1198–207. doi: 10.1101/gr.5440306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Gobbi M, Garrick D, Lynch M, Vernimmen D, Hughes JR, Goardon N, Luc S, Lower KM, Sloane-Stanley JA, Pina C, et al. Generation of bivalent chromatin domains during cell fate decisions. Epigenetics Chromatin. 2011;4:9. doi: 10.1186/1756-8935-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.