Abstract
Aberrant DNA methylation is a feature of human cancer affecting gene expression and tumor phenotype. Here, we quantified promoter methylation of candidate genes and global methylation in 44 small intestinal-neuroendocrine tumors (SI-NETs) from 33 patients by pyrosequencing. Findings were compared with gene expression, patient outcome and known tumor copy number alterations. Promoter methylation was observed for WIF1, RASSF1A, CTNNB1, CXCL14, NKX2–3, P16, LAMA1, and CDH1. By contrast APC, CDH3, HIC1, P14, SMAD2, and SMAD4 only had low levels of methylation. WIF1 methylation was significantly increased (P = 0.001) and WIF1 expression was reduced in SI-NETs vs. normal references (P = 0.003). WIF1, NKX2–3, and CXCL14 expression was reduced in metastases vs. primary tumors (P < 0.02). Low expression of RASSF1A and P16 were associated with poor overall survival (P = 0.045 and P = 0.011, respectively). Global methylation determined by pyrosequencing of LINE1 repeats was reduced in tumors vs. normal references, and was associated with loss in chromosome 18. The tumors fell into three clusters with enrichment of WIF1 methylation and LINE1 hypomethylation in Cluster I and RASSF1A and CTNNB1 methylation and loss in 16q in Cluster II. In Cluster III, these alterations were low-abundant and NKX2-3 methylation was low. Similar analyses in the SI-NET cell lines HC45 and CNDT2 showed methylation for CDH1 and WIF1 and/or P16, CXCL14, NKX2-3, LAMA1, and CTNNB1. Treatment with the demethylating agent 5-azacytidine reduced DNA methylation and increased expression of these genes in vitro. In conclusion, promoter methylation of tumor suppressor genes is associated with suppressed gene expression and DNA copy number alterations in SI-NETs, and may be restored in vitro.
Keywords: neuroendocrine, carcinoid, SI-NET, DNA methylation, pyrosequencing
Introduction
Small intestinal neuroendocrine tumors (SI-NET) originate from enterochromaffin cells in the ileal mucosa. The incidence of the disease has been increasing and is presently more than 1.1 per 100 000.1,2 Based on genome-wide studies, a few recurrent regions of copy number alterations have been identified.3 Most commonly, this involves loss in chromosome 18;4 however, no target gene has so far been precisely identified. Genes located within recurrently altered chromosomal regions, along with deregulated genes reported in expression profiling studies,5 are candidates for being involved in the tumor development.
Studies of epigenetic modifications, predominantly DNA methylation, may further reveal aberrations contributing to tumor development. Aberrant methylation of promoter CpG islands with effects on gene expression has been linked to many cancer types. Moreover, global genomic hypomethylation may contribute to genomic instability, a characteristic of human cancer.6 Global methylation may be assessed by different approaches, for example, by analysis of long interspersed element 1 (LINE1), which constitutes 17% of the human genome7 and has also been linked to genomic stability.8 In SI-NETs, several genes have been shown to be methylated using qualitative methodologies, e.g., RASSF1A and CTNNB1.9,10 Global hypomethylation of LINE1 and Alu repeats has been demonstrated by quantitative pyrosequencing in a limited number of SI-NETs.11 Recently, a genome-wide profiling study reported several methylated genes in SI-NETs with a general tendency to relative hypomethylation in lymph node metastases compared with primary tumors.12 However, little is known about tumor-specific alterations, the relationship between methylation density and gene expression in tumors, and whether the alterations may be reversed in vitro.
In this study, promoter methylation densities were quantified in a panel of SI-NETs and normal references for candidate tumor suppressor genes located in chromosomal regions with recurrent copy number losses or showing downregulation in expression profiling studies. Global methylation was assessed for LINE1 repeats and by ELISA-based detection of 5-methyl cytosines. The expression of methylated genes was further investigated in vitro after treatment with a demethylating agent. Associations to global DNA methylation, DNA copy number status, tumor characteristics and patient outcomes were also investigated.
Results
Gene-specific promoter methylation in SI-NETs
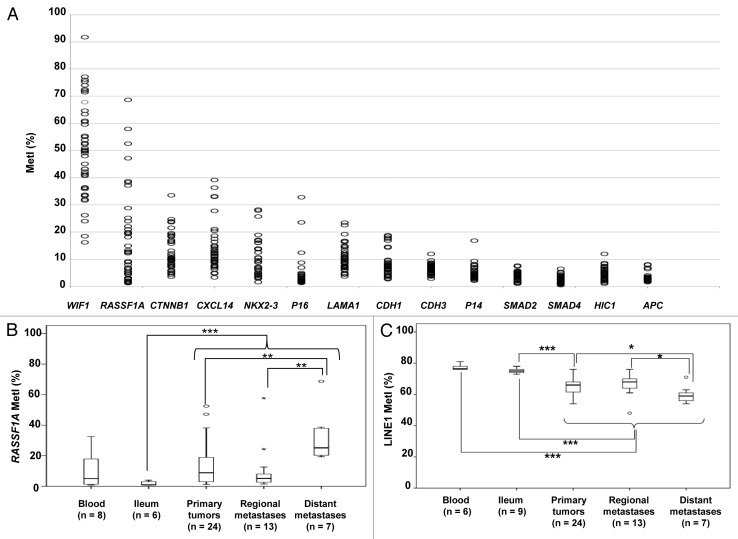
We quantified promoter methylation of 14 candidate genes in a panel of SI-NETs, together with blood and normal ileum used as references (Table 1). The genes were selected based on known tumor suppressor function and reported aberrant methylation in neuroendocrine tumors (RASSF1A, CTNNB1, P16, P14, HIC1, and APC),9,10,13 other tumors (WIF1),14 location within frequently deleted regions in SI-NETs (CDH1, LAMA1, CDH3, SMAD2, SMAD4),15 or downregulation in SI-NETs (CXCL14 and NKX2–3).5 Eight genes were found methylated in SI-NETs with frequent individual methylation indices (MetI) > 10%, including WIF1, RASSF1A, CTNNB1, CXCL14, NKX2–3, P16, LAMA1, and CDH1 (Fig. 1A; Table S1). The highest methylation in SI-NETs was detected for WIF1, with a mean MetI of 50% (range 16–92%). By contrast, the mean MetI was <10% (with individual MetI > 10% rarely observed) for APC, CDH3, HIC1, P14, SMAD2, and SMAD4, and they were therefore excluded from further analysis (Fig. 1A).
Table 1. Summary of results from gene specific promoter and global methylation quantification.
| Gene symbol |
Chromosomal location |
Tumors MetI | Normal Ileum MetI | Blood MetI | |||
|---|---|---|---|---|---|---|---|
| n = | mean (range) % | n = | mean (range) % | n = | mean (range) % | ||
| Gene specific promoter methylation | |||||||
| WIF1 | 12q14.3 | 44 | 50 (16 - 92) | 9 | 49 (27 - 75) | 8 | 25 (20 - 30) |
| RASSF1A | 3p21.31 | 44 | 16 (1 - 69) | 6 | 2 (1 - 4) | 8 | 11 (1 - 33) |
| CTNNB1 | 3p22.1 | 44 | 13 (4 - 34) | 9 | 14 (4 - 30) | 8 | 8 (5 - 11) |
| CXCL14 | 5q31.1 | 44 | 14 (3 - 39) | 9 | 14 (10 - 20) | 7 | 14 (11 - 16) |
| NKX2–3 | 10q24.2 | 44 | 10 (2 - 28) | 9 | 10 (6 - 16) | 7 | 5 (1 - 8) |
| P16 (CDKN2A) | 9p21.3 | 41 | 4 (1 - 33) | 5 | 2 (1 - 4) | 4 | 2 (1 - 3) |
| LAMA1 | 18p11.31 | 44 | 10 (4 - 24) | 9 | 15 (8 - 22) | 8 | 14 (9 - 21) |
| CDH1 | 16q22.1 | 44 | 8 (3 - 22) | 9 | 11 (5 - 20) | 8 | 22 (13 - 29) |
| CDH3 | 16q22.1 | 44 | 6 (3 - 12) | 9 | 9 (4 - 29) | 8 | 9 (7 - 12) |
| P14 (CDKN2A) | 9p21.3 | 42 | 5 (2 - 17) | 5 | 5 (4 - 6) | 4 | 3 (2 - 3) |
| SMAD2 | 18q21.1 | 44 | 4 (1 - 8) | 9 | 5 (2 - 15) | 7 | 6 (4 - 8) |
| SMAD4 | 18q21.2 | 44 | 3 (1 - 6) | 9 | 3 (1 - 8) | 7 | 4 (2 - 5) |
| HIC1 | 17p13.3 | 44 | 5 (1 - 12) | 9 | 9 (2 - 19) | 7 | 12 (5 - 19) |
| APC | 5q22.2 | 41 | 3 (2 - 8) | 5 | 5 (4 - 8) | 4 | 4 (3 - 6) |
| Global methylation | |||||||
| LINE1 | 44 | 65 (48 - 76) | 9 | 75 (73- 78) | 6 | 77 (76 - 81) | |
| ELISA | 27 | 1.1 (0.32 - 1.83) | 8 | 1.2 (0.69 - 1.93) | - | - | |
Met I, Methylation Index.
Figure 1. Results from quantifications of gene specific promoter methylation global methylation. (A) Scatter plot showing MetI values (mean methylation of the CpGs assessed in %) for each gene promoter in individual samples of SI-NETs. (B) Comparison of RASSF1A methylation in normal references, primary tumors and regional and distant metastases. (C) LINE1 global methylation in the different sample groups. Outliers are indicated by open circles and extremes by asterisks. P values for statistical comparisons between groups are indicated for suggestive P values ≤ 0.050 (*) or ≤ 0.01 (**), and for significant P-values ≤ 0.001 (***).
Comparison of MetIs between tumor groups revealed increased methylation of RASSF1A in distant metastasis vs. primary tumors (U = 325, P = 0.005) and vs. regional metastasis (U = 9, P = 0.004) (Fig. 1B). For WIF1, CTNNB1, CXCL14, NKX2-3, P16, LAMA1, and CDH1 no differences in MetI values were observed between tumor groups (P > 0.05). Some of these eight genes were also found to be methylated in normal ileum and/or blood samples (Table 1). As compared with reference samples, statistically significant higher methylation was found for WIF1 in SI-NETs vs. blood (U = 22, P < 0.001), and for RASSF1A in tumors vs. normal ileum (U = 21, P = 0.001). For comparison, in the two tumor samples from the colorectal carcinoid case, none of the 14 analyzed genes showed increased methylation as compared with blood and ileum (data not shown).
For the eight methylated gene promoters, we associated MetI with patient gender, age at diagnosis, size of the primary tumor, and presence and type of metastasis. This revealed higher RASSF1A methylation in SI-NETs from female (mean MetI 21%; range 3–69%) vs. male cases (mean MetI 8%; range 1–29%) (U = 113, P = 0.004). To further investigate this difference, pooled normal blood DNA from 10 unaffected males and 10 unaffected females were compared and showed that RASSF1A MetIs were similar in female and male blood samples (18% and 15%, respectively). No correlations were detected between MetIs and clinical characteristics for the other seven genes.
Global hypomethylation in SI-NETs
Overall, SI-NETs showed significantly lower LINE1 MetI (54–76%) than normal ileum (U = 20, P < 0.001) and reference blood (U = 0, P < 0.001). Lower LINE1 methylation was observed in distant metastasis compared with primary tumors (U = 41, P = 0.041) or regional metastasis (U = 18, P = 0.029) (Fig. 1C). The finding of global hypomethylation in SI-NETs was also verified using an ELISA-based methodology with 5-methyl cytosine antibodies (Table 1). Statistically significant associations to patient characteristics were not revealed.
Correlations and clustering of gene-specific and global methylation
We compared the results from promoter methylation analyses with global methylation measurements using Spearman rank order analysis. While LINE1 methylation was significantly correlated with CDH1 and LAMA1 methylation (r = 0.574, P < 0.001 and r = 0.510, P < 0.001, respectively), LINE1 methylation was significantly inversely correlated with methylation of WIF1 (r = –0.470, P < 0.001) and inversely correlated with RASSF1A methylation (r = –0.292, P = 0.026) (Table S2). Global methylation examined by ELISA also showed a positive correlation with LAMA1 methylation (r = 0.498, P = 0.002) and an inverse correlation with methylation of WIF1 (r = –0.491, P = 0.003).
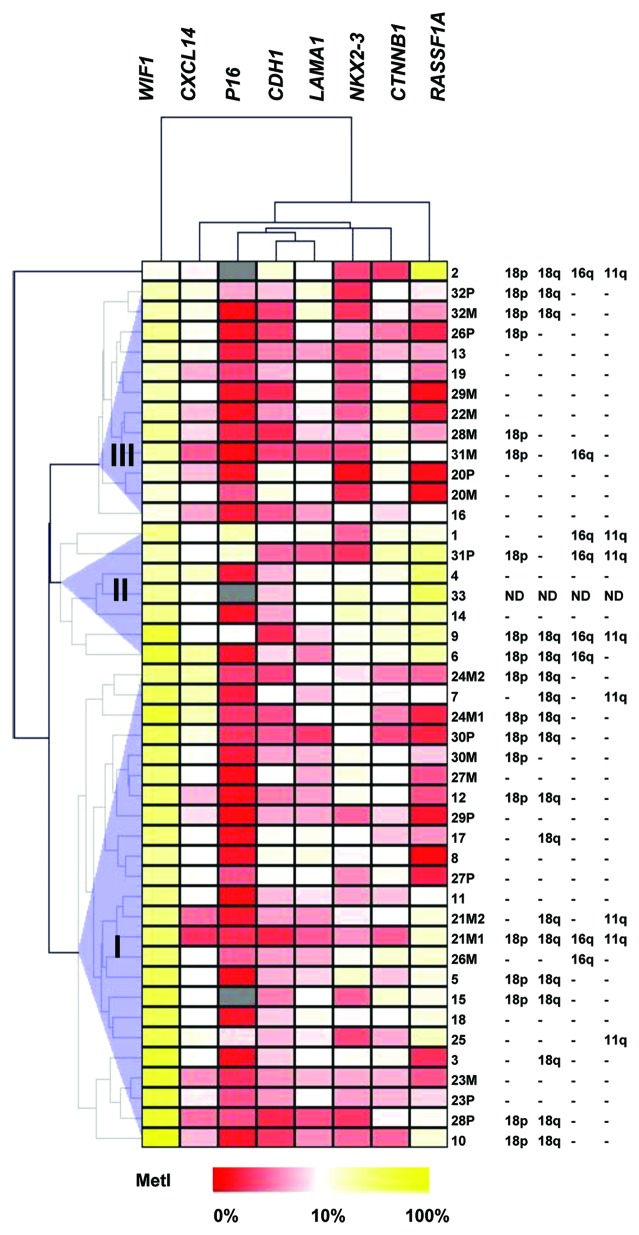
Three clusters were identified from unsupervised hierarchical clustering of the eight methylated genes (Fig. 2). The clusters were characterized by the following differences (P < 0.05): WIF1 had higher MetIs in Cluster I than Cluster II and III; RASSF1A and CTNNB1 showed higher MetIs in Cluster II than in I or III; NKX2–3 methylation was lower in Cluster III compared with I and II and; Cluster I had lower global methylation by ELISA than Clusters II and III, and lower LINE1 methylation compared with Cluster III.
Figure 2. Unsupervised hierarchical clustering of Pyrosequencing data for genes that were found methylated in SI-NETs. MetI values of 0–100% were converted to a range of 0 to 1 and subjected to Euclidean hierarchical clustering. The three tumor clusters are indicated as I, II and III. Gray indicates lack of data. Previously published15 data for copy number loss determined by qPCR for loci representing 11q (SDHD), 16q (CDH1), 18p (EMILIN2) and 18q (CDH19) in case 1–32 are indicated to the right for comparison.
Association of MetI with DNA copy number aberrations
Results from methylation analysis were compared with DNA copy numbers based on TaqMan copy number analysis of loci representing chromosomal regions 11q, 16q, 18p, and 18q15 published for case 1–32 in the same tumor series. We found that low levels of LAMA1 methylation were associated with loss in 18p (U = 146, P = 0.045). Furthermore, low global LINE1 methylation was associated with loss in 18p and 18q (U = 134, P = 0.022 and U = 92, P = 0.003, respectively).
With respect to the three identified clusters, loss in 18q was more frequent in Cluster I than in Cluster III (P = 0.030), 11q loss occurred more often in Cluster II compared with Cluster III (P = 0.025), and Cluster II included more samples with loss on 16q compared with Clusters I and III (P = 0.007 and P = 0.022, respectively).
mRNA expression levels of methylated genes in SI-NETs
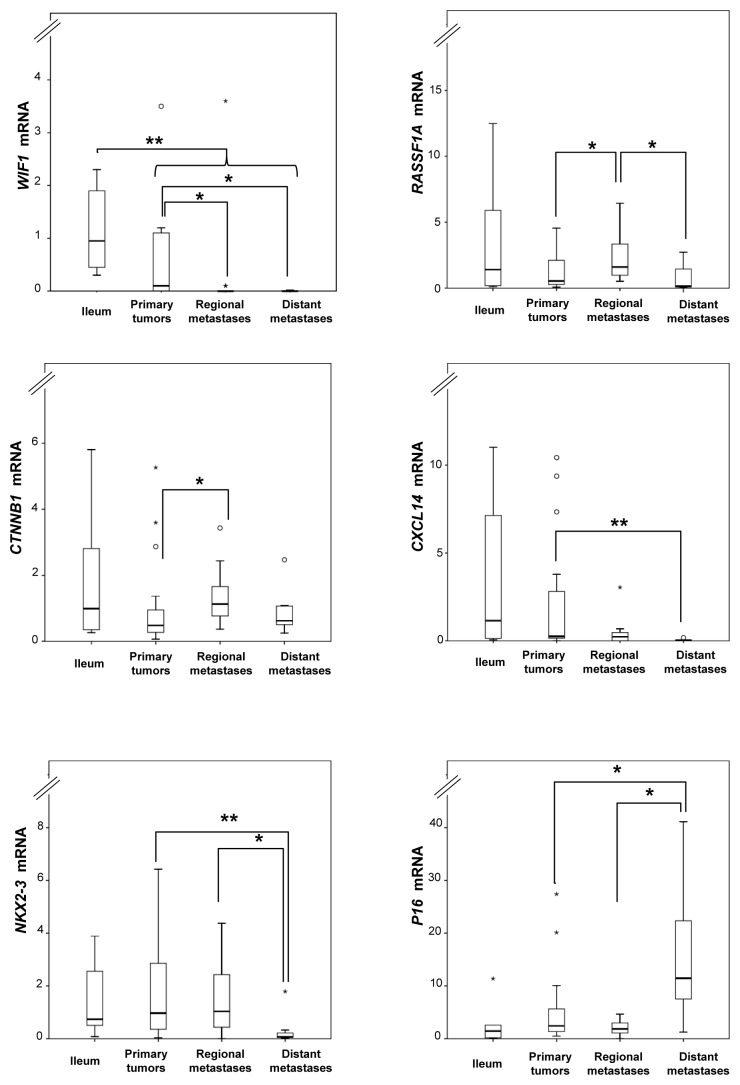
Relative mRNA expression levels were determined for the methylated genes WIF1, RASSF1A, CTNNB1, CXCL14, NKX2–3, P16, LAMA1, and CDH1 using TaqMan-based qRT-PCR and evaluated against MetI values and between sample groups. Overall, statistically significant correlations were not observed between mRNA levels and MetI values of individual genes in SI-NETs. As compared with the normal references, WIF1 expression was lower in tumors compared with normal ileum (U = 60, P = 0.003). In addition, several genes showed differential expression levels between tumor sample groups. Lower expression was observed for WIF1 in metastases compared with primary tumors (U = 118, P = 0.004); for CXCL14, in metastasis compared with primary tumors (U = 138, P = 0.016); for CXCL14, in distant metastasis compared with all other tumor groups (U = 51, P = 0.013); and for NKX2-3, in distant metastasis compared with the other tumor groups (U = 45, P = 0.008) (Fig. 3). In contrast, increased expression was revealed for CTNNB1 in metastases compared with primary tumors (U = 146, P = 0.041); for P16, in distant metastasis in comparison to other tumors (U = 52, P = 0.015); and for RASSF1A, in regional metastasis compared with the other tumors (U = 94, P = 0.008) (Fig. 3).
Figure 3. Relative mRNA expression levels determined by qRT-PCR for selected loci in normal ileum references and tumor subgroups. Fold changes are represented as compared with the mean of ileum (assigned the arbitrary value of 1). P values for statistical comparisons between groups are indicated for suggestive P values ≤ 0.050 (*) or ≤ 0.01 (**).
RASSF1A expression was associated with global methylation by ELISA (r = 0.423, P = 0.011), while P16 expression was negatively associated with LINE1 methylation (r = -0.382, P = 0.006).
Comparison to clinical findings revealed that CTNNB1 expression was correlated with tumor size (r = 0.339, P = 0.035) and tumor stage and did not have a confounding effect on this correlation. Associations between gene expression levels and gender, age at diagnosis, or metastasis were not observed. The association that was found between gender and RASSF1A methylation was not observed for the corresponding mRNA expression.
Survival analysis
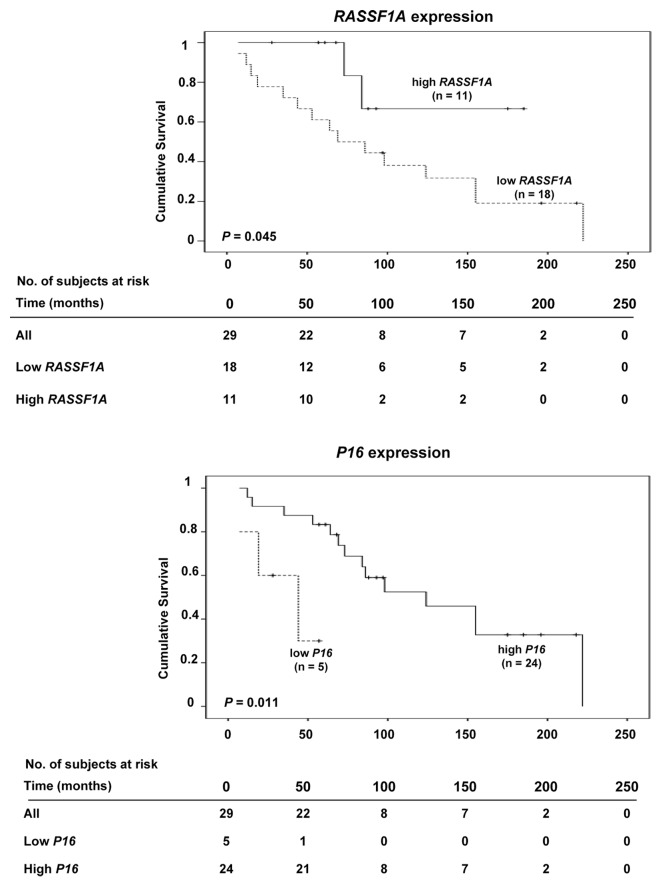
Gene-specific promoter methylation or global methylation levels were not found to influence patient outcome in terms of survival. However, low mRNA expression levels of RASSF1A and P16 were each associated with short survival (P = 0.045 and P = 0.011, respectively) (Fig. 4). In multivariate analysis, the clinical variables gender, tumor size and age at diagnosis did not affect the survival function of RASSF1A expression. However, age at diagnosis had an impact on the association between P16 expression and survival (β = 0.071, P = 0.034).
Figure 4. Kaplan-Meier survival curves comparing overall survival in SI-NET cases with high or low mRNA expression of RASSF1A (top) and P16 (bottom).
Restoration of mRNA expression after demethylation in vitro
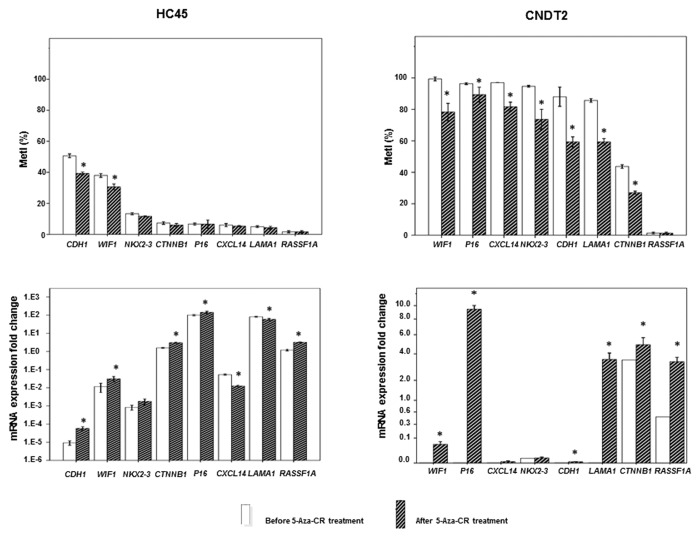
To investigate the effect of DNA methylation on gene expression in vitro, the SI-NET cell lines HC45 and CNDT2 were treated with the demethylating agent 5-azacytidine (5-aza-CR) for 4 days. Before treatment MetIs > 20% were revealed for WIF1, P16, CXCL14, NKX2–3, CDH1, LAMA1, and CTNNB1 in CNDT2 cells and for CDH1 and WIF1 in HC45 cells (Table S1); these methylation levels were reduced (P ≤ 0.050) after 5-aza-CR treatment in both cell lines (Fig. 5). In CNDT2 cells, 5-aza-CR treatment was associated with increased gene expression of WIF1, P16, CDH1, LAMA1, and CTNNB1. RASSF1A expression was also increased; however, the methylation level was constitutionally low and could therefore not be affected by 5-aza-CR. In HC45 cells, 5-aza-CR treatment led to lower promoter methylation density for CDH1 and WIF1, as well as corresponding increase in gene expression levels (P ≤ 0.050). We also observed increased expression of CTNNB1, P16, and RASSF1A and reduced expression of CXCL14 and LAMA1 after 5-aza-CR treatment; however, the corresponding methylation levels were low both before and after treatment (Fig. 5). Global LINE1 methylation was reduced after treatment with 5-aza-CR in both cell lines (data not shown).
Figure 5. Promoter methylation and gene expression in SI-NET cell lines treated by 5µM 5-aza-CR. At the top is shown MetI in HC45 and CNDT2 cell lines treated with 5-aza-CR. Corresponding mRNA expression fold changes are shown below. The values are in relation to SI-NET 30P in which all genes were expressed (assigned the arbitrary value 1.0). Results before and after 5-aza-CR treatments are indicated by white and striped bars, respectively. P values for statistical comparisons between groups are indicated for suggestive P values ≤ 0.050 (*).
Discussion
Here we demonstrate global hypomethylation and gene-specific promoter methylation in SI-NETs using a quantitative approach by pyrosequencing.16 For some genes, differential methylation as well as corresponding mRNA expression was observed between tumor groups and in association with survival. Methylation could be reverted and expression restored upon treatment with a demethylating agent in vitro. Applying a cut-off of 10% for biologically significant promoter methylation we found promoter methylation for WIF1, RASSF1A, CTNNB1, CXCL14, NKX2-3, P16, LAMA1, and CDH1. These results confirm previous findings for RASSF1A, CTNNB1, and P16.10,17
The most heavily methylated gene, WIF1 (wnt inhibitory factor), was downregulated in metastases compared with primary tumors and ileum. Furthermore, tumors with WIF1 hypermethylation preferentially fell into Cluster I, which was also characterized by global hypomethylation and loss in 18q (the most common genomic alteration in SI-NETs). WIF1 was highly methylated in both CNDT2 (MetI 99%) and HC45 (MetI 38%) cell lines, and treatment with 5-aza-CR reduced methylation density and restored gene expression. WIF1 is a Wnt antagonist that inhibits the interaction of Wnt with its receptor, named Frizzled. This in turn prevents cytoplasmic accumulation and nuclear translocation of CTNNB1 (β-catenin), which would otherwise activate transcription of oncogenic factors such as c-Myc and TERT, the catalytic subunit of telomerase.18 Epigenetic silencing of WIF1 has been reported in squamous cell carcinoma of the cervix,19 breast cancer,20 bladder cancer,21 colorectal cancer,22 nasopharyngeal and esophageal carcinoma,23 and non-small-cell lung cancer.14 In pancreatic and lung carcinoid cell lines absent or reduced WIF1 expression was observed without accompanying promoter methylation.24
We found increased methylation of RASSF1A in distant metastases together with reduced gene expression, in agreement with previous studies.10,17 Moreover, low RASSF1A expression was associated with shorter survival. RASSF1A hypermethylation has been shown in different cancer types including SI-NETs.10 It contributes to cancer development basically through modulation of cyclin D1 accumulation, inhibition of the JNK pathway25 and pro-apoptotic activities achieved by binding to MST1 (Mammalian Sterile Twenty 1) and other apoptotic agents.26 RASSF1A has a role in the modulation of tubulin dynamics and its localization to centrosomes and mitotic spindles.27 It also inhibits cell motility and promotes cell-cell adhesion,28 which is in line with our observation of its epigenetic inactivation in distant metastasis. We also identified higher MetI in SI-NETs from female as compared with male cases. However, we did not observe a similar gender-specific difference between normal blood samples arguing against a constitutional difference. Analysis of larger numbers of matched blood and SI-NETs samples could establish whether the alteration is a tumor-specific event. Furthermore, it may be noted that RASSF1A methylation was one of the alterations distinguishing Cluster I, which could be partly attributed to differences in gender distribution between clusters.
High methylation and downregulation was observed for CTNNB1 in a subset of tumors, in agreement with a previous report.10 However, this was only observed in a small subset of cases, and never in metastases, suggesting that CTNNB1 does not have a major role as tumor suppressor in SI-NETs.
Our study confirms a previous report of downregulated mRNA expression for CXCL14 and NKX2–3 in progressive SI-NETs,5 and also proposes an epigenetic background for this observation. The chemokine CXCL14 is downregulated in prostate cancer cells and restoration of its expression by 5-Aza-2-deoxycytidine increases chemo-attraction for dendritic immune cells.29 In addition, forced expression of CXCL14 in lung cancer cells, where it is constitutively silenced by DNA methylation, rendered up to 90% necrosis and tumor shrinkage in xenograft models.30 Promoter hypermethylation of NKX2–3, encoding a homeodomain-containing transcription factor, has been reported in melanoma cell lines.31 Silencing of NKX2–3 by siRNA have suggested a role for Nkx2–3 in colorectal cancer development by regulation of the Wnt signaling pathway.32
The possibility of epigenetic inactivation of P16 in SI-NETs is controversial.9,13,17 We detected increased P16 promoter methylation in a few tumors only, suggesting a limited role of inactivation by promoter methylation. P16 overexpression has been suggested to keep tumors in “oncogene-induced senescence” and low-proliferative states, and P16 inhibition may contribute to malignant transformation.33,34 The current study supports this hypothesis in that a worse patient outcome was observed for cases with P16 downregulation. Age at diagnosis was identified as a confounding factor in agreement with observations of altered P16 in relation to age.35
The CDH1 gene is located in 16q and LAMA1 in 18p, chromosomal regions that are frequently affected by copy number losses as shown in our previous study of the same samples.15 Epigenetic inactivation by methylation and a reduction in expression would be consistent with bi-allelic inactivation according to Knudson’s two-hit hypothesis.36 However, the methylation density was not found to be higher in tumors than in normal ileum and blood controls for these two genes. Hence, DNA methylation is not expected to be the only mechanism involved. CDH1 and LAMA1 MetIs were positively correlated, and low MetIs as well as increased CDH1 mRNA expression were associated with loss in chromosome 18p. Furthermore, hypomethylation of CDH3, located adjacent to its homolog CDH1, has previously been observed in cancer.37
We found global hypomethylation in SI-NETs and, particularly, in distant metastases, a phenomenon attributed to overriding transposon activity control and genome integrity. Both ELISA and LINE1 pyrosequencing results were valuable indicators of global methylation, acting in a firm pattern of correlations with promoter methylation alterations. While LINE1 analysis directly represents retrotransposon methylation and silent status, ELISA gives an overall ratio of 5-methyl cytosine over other nucleotides in the genome. Hydroxymethyl cytosines, which were recently implicated in cancer,38 are not included in the estimations by ELISA but are taken into account by pyrosequencing. Using either method, hypomethylation was detected in a large proportion of samples. Hypomethylation of LINE1 and Alu in a cohort of pancreatic and ileal and non-ileal NETs has previously been reported by Choi et al.11 In that study, hypomethylation was more common in ileal NETs than in other NETs. Our study confirms the association of SI-NETs with global hypomethylation, RASSF1A hypermethylation and chromosome 18 loss. Furthermore, LINE1 methylation was correlated with CDH1 and LAMA1 and inversely correlated with WIF1 MetI. Hypomethylation by ELISA was also correlated with WIF1 and inversely to LAMA1 MetI. In addition, distant metastases were hypomethylated compared with the primary tumors.
A demethylation study on two SI-NET cell line models was conducted. Both cell lines were highly methylated for some of the genes found methylated in tumors. 5-aza-CR treatment reduced methylation density of global LINE1 repeats and of individual promoter CpG islands. The inhibition of promoter methylation was accompanied by restoration of gene expression, suggesting a causative effect of promoter methylation on gene expression levels.
In conclusion, global hypomethylation and promoter methylation was demonstrated in SI-NETs with related effects on gene expression observed in vitro. Continued studies to explore SI-NET development may focus on WIF1 inactivation by methylation and on RASSF1A and P16 expression as potential prognostic markers for refined clinical handling of SI-NETs.
Materials and Methods
SI-NET and reference samples
A total number of 44 fresh frozen sporadic SI-NETs from 33 patients were obtained from Karolinska University Hospital biobank (Table S3, case 1–33). Twenty-four tumors were primary and 20 were metastasis including 7 distant (5 liver and 2 ovarian) and 13 regional metastases. From 9 patients paired primary tumor and metastasis were available. A detailed description of the clinical and histopathological characteristics of case 1–32 has been published, together with data on copy number alterations.15 In addition, a regional metastasis from a patient with family history of SI-NET (case 33) was included. Two metastases from a patient with colorectal carcinoid were also included for comparison. All samples were obtained with informed oral consent as documented in the patient’s medical record and the collection and study of patient material was approved by the local Ethics Committee. Nine DNA samples from anonymized normal ileum, peripheral blood from 6 of the SI-NET cases, and pooled samples of peripheral blood from 10 female or 10 male healthy individuals (Promega, G1521 and G1471) were used as references.
Cell lines
The cell lines HC45 and CNDT2 are both of human ileal origin derived from liver metastases. HC45 was a kind gift from Professor RV Lloyd, Mayo Clinic,39 and CNDT2 was kindly provided by Professor LM Elis, MD Anderson Cancer Center.40 The authenticity was demonstrated by genotyping of short tandem repeats (STRs) at Biosynthesis Inc. (Table S4).
Extraction of DNA and RNA
Genomic DNA and RNA were extracted simultaneously from fresh frozen tissues and cell pellets following the manufacturer’s instructions (Allprep DNA/RNA/Protein, Qiagen) and measured by Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies).
Pyrosequencing
500 ng genomic DNA from each sample was bisulfite converted following the protocol of the manufacturer (Epitect Bisulfite, Qiagen, or EZ DNA methylation kit, Zymo Research) and quantified with Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies) to apply 17 ng in the PCR amplification (Pyromark PCR, Qiagen). PCR conditions were 95 °C activation step for 15 min, followed by 40–45 cycles of amplification (denaturation at 94 °C, annealing at 50 °C, 55 °C, or 56 °C, and extension at 72 °C, each for 30 s) and a final extension of 72 °C for 10 min. Annealing temperature, concentration of MgCl2 and quality of PCR product were optimized and assessed for each assay by electrophoresis in 1.5% agarose gels. Commercially available assays (Table S5) were used together with a PyroMark Q24 instrument and PyroMark Gold Q24 Reagents according to the manufacturer’s instructions (Qiagen). Controls were run for each assay including: 0, 50 and 100% methylated DNA (Qiagen) to confirm the fidelity of Pyrogram peaks; a PCR without template DNA to confirm lack of interaction between primers and contamination; a PCR with template DNA without sequencing primer to confirm lack of template looping background; and reaction with sequencing primer without template to confirm lack of sequencing primer hairpin background. Data were analyzed using the PyroMark Q24 software. For each gene and sample a mean methylation density was calculated based on the values for the individual CpGs assayed. Samples with MetI ≤ 10% were regarded as having a low methylation of experimental and biological low significance. Samples with MetI > 10% were classified as methylated. Samples were assigned as hypermethylated if the MetI was above the mean for the entire group of normal ileal samples. Thus the cut-off for methylation was 10% or higher. The following genes were studied: WIF1, RASSF1A, CTNNB1, CXCL14, NKX2–3, P16, LAMA1, CDH1, CDH3, P14, SMAD2, SMAD4, HIC1, and APC (Table S5).
LINE1 was similarly analyzed by Pyrosequencing as an indicator of global DNA methylation. SI-NETs with a MetI below the mean of normal ileum were regarded as hypomethylated.
ELISA-based quantification of global DNA methylation
Antibody against 5-methyl cytosine was applied in an ELISA-based global DNA methylation quantification method following the manufacturer’s instructions (MethylFlashTM Methylated DNA Quantification, Colorimetric, Epigentek). Briefly, DNA was bound to the strip wells with high DNA affinity and was incubated with capture and detection antibodies. The absorbance was read at 450 nm using a microplate reader (VERSAmax; Molecular Devices). The absolute quantity of 5-methyl cytosine in the global DNA was determined by subtracting the sample OD from the negative control divided by the slope of a standard curve, which was obtained from positive control DNA with known 5-methyl cytosine content, running on the same plate in duplicate.
Real Time Quantitative PCR (qRT-PCR)
Reverse transcription was performed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). 100 ng RNA was used as described before.41 The quality and concentration of cDNA was assessed by spectrophotometry. 100–1500 ng of cDNA per reaction was applied to the TaqMan expression assays (Table S5) following the manufacturer’s instructions (Applied Biosystems). ACTB (Beta-actin) was run as endogenous control in the same plate for each assay. Each sample was run in triplicate and non-template control was included in each plate. Plates were run using Applied Biosystems 7900 instrument and analyzed with the SDS 2.4 software. Mean CT values were normalized against ACTB to calculate ΔCT for each sample and fold changes were calculated by the 2−ΔΔCT method, as ΔΔCT = ΔCT (sample) - ΔCT (mean of normal ileum).42
In vitro demethylation
Each cell line was cultured in DMEM, with L-glutamine and high glucose supplemented with 10% FBS (Invitrogen, 41965039 and 10500056) in T25 flasks and was maintained at 37 °C, humidified incubator with 5% CO2 in triplicate. After 24 h the cells were at 40% confluency and 5-aza-CR (Sigma Aldrich, A2385) treatment was started for 4 consecutive days. The medium was replaced every day by freshly prepared 0 or 5 µM 5-aza-CR. The concentration of drug was optimized beforehand, by ELISA in 0, 10 and 20 nM 5-aza-CR and refined using LINE1 assay in a range of 0 to 10 µM. Accordingly, a concentration of 5 µM 5-aza-CR was used in the further experiments. Doubling time was already measured as 2 days and accordingly in 4 days 75% of DNA molecules were expected to be unmethylated. The cell pellet harvested for extraction by Trypsin/PBS (1%, Invitrogen, 15400054) after washing with PBS. DNA and RNA extracted from treated and untreated cells were analyzed by Pyrosequencing and qRT-PCR.
Statistical and clustering analysis
The Mann Whitney U test was used to compare mean values between all double groups of continuous variables and Chi-square to compare mean values of every two or several categorical groups. Log-rank test was used to evaluate the effect of each variable on the survival and illustrated by Kaplan-Meier plots. Multivariate Cox regression was used to evaluate cofounders involvement in survival analysis and multiple regressions to evaluate the hypothesis of tumor stage being cofounding factor in CTBBN1 overexpression correlation with tumor size. For paired samples only laboratory data of the metastasis were considered for survival analysis and comparison to clinical characteristics. All statistical analyses were performed using the statistical software SPSS v 16.0. Observed differences with P values ≤ 0.050 were reported. Given the large number of comparisons performed, a conservative approach was taken in the interpretation of statistically significant differences. P values ≤ 0.001 were reported as statistically significant and P values ≤ 0.050 were regarded as being of suggestive significance.
Methylation densities of the methylated genes in tumor samples were standardized in a range of 0 to 1 and subjected to unsupervised hierarchical clustering, Euclidian method using MeV 43.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the Swedish Cancer Society, the Swedish Research Council, the Gustav V Jubilee Foundation, the Cancer Society in Stockholm, Karolinska Institutet, and Stockholm County Council.
Acknowledgments
The authors wish to thank Ms. Lisa Ånfalk, Karolinska University Hospital for excellent assistance in tissue collection and the medical genetics group for valuable discussions.
References
- 1.Landerholm K, Falkmer S, Järhult J. Epidemiology of small bowel carcinoids in a defined population. World J Surg. 2010;34:1500–5. doi: 10.1007/s00268-010-0519-z. [DOI] [PubMed] [Google Scholar]
- 2.Pape UF, Perren A, Niederle B, Gross D, Gress T, Costa F, Arnold R, Denecke T, Plöckinger U, Salazar R, et al. Barcelona Consensus Conference participants ENETS Consensus Guidelines for the management of patients with neuroendocrine neoplasms from the jejuno-ileum and the appendix including goblet cell carcinomas. Neuroendocrinology. 2012;95:135–56. doi: 10.1159/000335629. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham JL, Díaz de Ståhl T, Sjöblom T, Westin G, Dumanski JP, Janson ET. Common pathogenetic mechanism involving human chromosome 18 in familial and sporadic ileal carcinoid tumors. Genes Chromosomes Cancer. 2011;50:82–94. doi: 10.1002/gcc.20834. [DOI] [PubMed] [Google Scholar]
- 4.Kytölä S, Höög A, Nord B, Cedermark B, Frisk T, Larsson C, Kjellman M. Comparative genomic hybridization identifies loss of 18q22-qter as an early and specific event in tumorigenesis of midgut carcinoids. Am J Pathol. 2001;158:1803–8. doi: 10.1016/S0002-9440(10)64136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leja J, Essaghir A, Essand M, Wester K, Öberg K, Tötterman TH, Lloyd R, Vasmatzis G, Demoulin JB, Giandomenico V. Novel markers for enterochromaffin cells and gastrointestinal neuroendocrine carcinomas. Mod Pathol. 2009;22:261–72. doi: 10.1038/modpathol.2008.174. [DOI] [PubMed] [Google Scholar]
- 6.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, Kotsinas A, Gorgoulis V, Field JK, Liloglou T. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124:81–7. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 9.Chan AO, Kim SG, Bedeir A, Issa JP, Hamilton SR, Rashid A. CpG island methylation in carcinoid and pancreatic endocrine tumors. Oncogene. 2003;22:924–34. doi: 10.1038/sj.onc.1206123. [DOI] [PubMed] [Google Scholar]
- 10.Zhang HY, Rumilla KM, Jin L, Nakamura N, Stilling GA, Ruebel KH, Hobday TJ, Erlichman C, Erickson LA, Lloyd RV. Association of DNA methylation and epigenetic inactivation of RASSF1A and beta-catenin with metastasis in small bowel carcinoid tumors. Endocrine. 2006;30:299–306. doi: 10.1007/s12020-006-0008-1. [DOI] [PubMed] [Google Scholar]
- 11.Choi IS, Estecio MR, Nagano Y, Kim H, White JA, Yao JC, Issa JP, Rashid A. Hypomethylation of LINE-1 and Alu in well-differentiated neuroendocrine tumors (pancreatic endocrine tumors and carcinoid tumors) Mod Pathol. 2007;20:802–10. doi: 10.1038/modpathol.3800825. [DOI] [PubMed] [Google Scholar]
- 12.Delgado Verdugo A, Crona J, Starker LF, Stålberg P, Åkerström G, Westin G, Hellman P, Björklund P. Global DNA methylation patterns in small intestinal neuroendocrine tumors (SI-NETs) Endocr Relat Cancer. 2013 doi: 10.1530/ERC-13-0481. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 13.Arnold CN, Sosnowski A, Schmitt-Gräff A, Arnold R, Blum HE. Analysis of molecular pathways in sporadic neuroendocrine tumors of the gastro-entero-pancreatic system. Int J Cancer. 2007;120:2157–64. doi: 10.1002/ijc.22569. [DOI] [PubMed] [Google Scholar]
- 14.Mazieres J, He B, You L, Xu Z, Lee AY, Mikami I, Reguart N, Rosell R, McCormick F, Jablons DM. Wnt inhibitory factor-1 is silenced by promoter hypermethylation in human lung cancer. Cancer Res. 2004;64:4717–20. doi: 10.1158/0008-5472.CAN-04-1389. [DOI] [PubMed] [Google Scholar]
- 15.Hashemi J, Fotouhi O, Sulaiman L, Kjellman M, Höög A, Zedenius J, Larsson C. Copy number alterations in small intestinal neuroendocrine tumors determined by array comparative genomic hybridization. BMC Cancer. 2013;13:505. doi: 10.1186/1471-2407-13-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kiss NB, Geli J, Lundberg F, Avci C, Velazquez-Fernandez D, Hashemi J, Weber G, Höög A, Ekström TJ, Bäckdahl M, et al. Methylation of the p16INK4A promoter is associated with malignant behavior in abdominal extra-adrenal paragangliomas but not pheochromocytomas. Endocr Relat Cancer. 2008;15:609–21. doi: 10.1677/ERC-07-0285. [DOI] [PubMed] [Google Scholar]
- 17.Liu L, Broaddus RR, Yao JC, Xie S, White JA, Wu TT, Hamilton SR, Rashid A. Epigenetic alterations in neuroendocrine tumors: methylation of RAS-association domain family 1, isoform A and p16 genes are associated with metastasis. Mod Pathol. 2005;18:1632–40. doi: 10.1038/modpathol.3800490. [DOI] [PubMed] [Google Scholar]
- 18.Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–4. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- 19.Delmas AL, Riggs BM, Pardo CE, Dyer LM, Darst RP, Izumchenko EG, Monroe M, Hakam A, Kladde MP, Siegel EM, et al. WIF1 is a frequent target for epigenetic silencing in squamous cell carcinoma of the cervix. Carcinogenesis. 2011;32:1625–33. doi: 10.1093/carcin/bgr193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ai L, Tao Q, Zhong S, Fields CR, Kim WJ, Lee MW, Cui Y, Brown KD, Robertson KD. Inactivation of Wnt inhibitory factor-1 (WIF1) expression by epigenetic silencing is a common event in breast cancer. Carcinogenesis. 2006;27:1341–8. doi: 10.1093/carcin/bgi379. [DOI] [PubMed] [Google Scholar]
- 21.Urakami S, Shiina H, Enokida H, Kawakami T, Tokizane T, Ogishima T, Tanaka Y, Li LC, Ribeiro-Filho LA, Terashima M, et al. Epigenetic inactivation of Wnt inhibitory factor-1 plays an important role in bladder cancer through aberrant canonical Wnt/beta-catenin signaling pathway. Clin Cancer Res. 2006;12:383–91. doi: 10.1158/1078-0432.CCR-05-1344. [DOI] [PubMed] [Google Scholar]
- 22.Roperch JP1, Incitti R, Forbin S, Bard F, Mansour H, Mesli F, Baumgaertner I, Brunetti F, Sobhani I. Aberrant methylation of NPY, PENK, and WIF1 as a promising marker for blood-based diagnosis of colorectal cancer. BMC cancer. 2013;13:566. doi: 10.1182/blood-2006-09-047043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan SL, Cui Y, van Hasselt A, Li H, Srivastava G, Jin H, Ng KM, Wang Y, Lee KY, Tsao GS, et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab Invest. 2007;87:644–50. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 24.Kim JT, Li J, Jang ER, Gulhati P, Rychahou PG, Napier DL, Wang C, Weiss HL, Lee EY, Anthony L, et al. Deregulation of Wnt/β-catenin signaling through genetic or epigenetic alterations in human neuroendocrine tumors. Carcinogenesis. 2013;34:953–61. doi: 10.1093/carcin/bgt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whang YM, Kim YH, Kim JS, Yoo YD. RASSF1A suppresses the c-Jun-NH2-kinase pathway and inhibits cell cycle progression. Cancer Res. 2005;65:3682–90. doi: 10.1158/0008-5472.CAN-04-2792. [DOI] [PubMed] [Google Scholar]
- 26.Cheung WL, Ajiro K, Samejima K, Kloc M, Cheung P, Mizzen CA, Beeser A, Etkin LD, Chernoff J, Earnshaw WC, et al. Apoptotic phosphorylation of histone H2B is mediated by mammalian sterile twenty kinase. Cell. 2003;113:507–17. doi: 10.1016/S0092-8674(03)00355-6. [DOI] [PubMed] [Google Scholar]
- 27.Song MS, Chang JS, Song SJ, Yang TH, Lee H, Lim DS. The centrosomal protein RAS association domain family protein 1A (RASSF1A)-binding protein 1 regulates mitotic progression by recruiting RASSF1A to spindle poles. J Biol Chem. 2005;280:3920–7. doi: 10.1074/jbc.M409115200. [DOI] [PubMed] [Google Scholar]
- 28.Dallol A, Agathanggelou A, Tommasi S, Pfeifer GP, Maher ER, Latif F. Involvement of the RASSF1A tumor suppressor gene in controlling cell migration. Cancer Res. 2005;65:7653–9. doi: 10.1158/0008-5472.CAN-05-0247. [DOI] [PubMed] [Google Scholar]
- 29.Song EY, Shurin MR, Tourkova IL, Gutkin DW, Shurin GV. Epigenetic mechanisms of promigratory chemokine CXCL14 regulation in human prostate cancer cells. Cancer Res. 2010;70:4394–401. doi: 10.1158/0008-5472.CAN-10-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tessema M, Klinge DM, Yingling CM, Do K, Van Neste L, Belinsky SA. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159–70. doi: 10.1038/onc.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tellez CS, Shen L, Estécio MR, Jelinek J, Gershenwald JE, Issa JP. CpG island methylation profiling in human melanoma cell lines. Melanoma Res. 2009;19:146–55. doi: 10.1097/CMR.0b013e32832b274e. [DOI] [PubMed] [Google Scholar]
- 32.Yu W, Lin Z, Pastor DM, Hegarty JP, Chen X, Kelly AA, Wang Y, Poritz LS, Koltun WA. Genes regulated by Nkx2-3 in sporadic and inflammatory bowel disease-associated colorectal cancer cell lines. Dig Dis Sci. 2010;55:3171–80. doi: 10.1007/s10620-010-1138-0. [DOI] [PubMed] [Google Scholar]
- 33.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/S0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 34.Romagosa C, Simonetti S, López-Vicente L, Mazo A, Lleonart ME, Castellvi J, Ramon y Cajal S. p16(Ink4a) overexpression in cancer: a tumor suppressor gene associated with senescence and high-grade tumors. Oncogene. 2011;30:2087–97. doi: 10.1038/onc.2010.614. [DOI] [PubMed] [Google Scholar]
- 35.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–11. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 36.Knudson AG., Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Milicic A, Harrison LA, Goodlad RA, Hardy RG, Nicholson AM, Presz M, Sieber O, Santander S, Pringle JH, Mandir N, et al. Ectopic expression of P-cadherin correlates with promoter hypomethylation early in colorectal carcinogenesis and enhanced intestinal crypt fission in vivo. Cancer Res. 2008;68:7760–8. doi: 10.1158/0008-5472.CAN-08-0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–30. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stilling GA, Zhang H, Ruebel KH, Leontovich AA, Jin L, Tanizaki Y, Zhang S, Erickson LA, Hobday T, Lloyd RV. Characterization of the functional and growth properties of cell lines established from ileal and rectal carcinoid tumors. Endocr Pathol. 2007;18:223–32. doi: 10.1007/s12022-007-9001-3. [DOI] [PubMed] [Google Scholar]
- 40.Van Buren G, II, Rashid A, Yang AD, Abdalla EK, Gray MJ, Liu W, Somcio R, Fan F, Camp ER, Yao JC, et al. The development and characterization of a human midgut carcinoid cell line. Clin Cancer Res. 2007;13:4704–12. doi: 10.1158/1078-0432.CCR-06-2723. [DOI] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–8. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.