Abstract
Superfamily 2 helicase proteins are ubiquitous in RNA biology and have an extraordinarily broad set of functional roles. Central among these roles are to promote rearrangements of structured RNAs and to remodel RNA-protein complexes (RNPs), allowing formation of native RNA structure or progression through a functional cycle of structures. While all superfamily 2 helicases share a conserved helicase core, they are divided evolutionarily into several families, and it is principally proteins from three families, the DEAD-box, DEAH/RHA and Ski2-like families, that function to manipulate structured RNAs and RNPs. Strikingly, there are emerging differences in the mechanisms of these proteins, both between families and within the largest family (DEAD-box), and these differences appear to be tuned to their RNA or RNP substrates and their specific roles. This review outlines basic mechanistic features of the three families and surveys individual proteins and the current understanding of their biological substrates and mechanisms.
Keywords: ATPase, RNA folding, RNA structure, RNA unwinding, Self-splicing intron
INTRODUCTION
Structured RNAs function throughout biology in processes such as translation, pre-mRNA splicing, RNA processing, chromosome end maintenance, and the regulation of gene expression. To function, these RNAs must fold to specific three-dimensional structures that include secondary structure (RNA helices) and tertiary structure, and they commonly include proteins to form ribonucleoprotein complexes (RNPs). The recent discovery of pervasive transcription of eukaryotic genomes has revealed further the large number of non-coding, and potentially structured, RNAs produced in cells (1–3).
The underlying physical properties of RNA result in a universal need for proteins to accelerate RNA structural transitions. It has been known for decades that RNAs face substantial challenges in folding efficiently to specific structures and in transitioning between structured conformations (4–6). Local secondary structure can be independently stable and long-lived, such that transitions requiring changes in secondary structure can require assistance in vivo (7). Further, because of the limited diversity of RNA side chains, non-native base pairs are likely to form during folding, resulting in kinetically trapped structures in vitro and in vivo. Indeed, even mRNAs, which must be unstructured over much of their length to be translated into protein, are likely to form local structures that require assistance from proteins.
While many diverse proteins possess RNA chaperone activity and have been shown to function as chaperones in some instances (8), superfamily 2 (SF2) helicase proteins stand out for their ubiquitous roles in manipulating structured RNAs in vivo (9–11). This expansive collection of proteins comprises multiple families, including many proteins that function on DNA instead of RNA. It also includes proteins that lack conventional helicase activity, despite the ‘helicase’ designation, which was coined decades ago from sequence similarity with the limited number of known helicases at the time (12). As described below, some SF2 proteins can unwind RNA locally but do not translocate significantly along RNA, while others, not covered here, translocate along RNA but do not unwind it. For simplicity, we refer to all of these proteins as helicases, despite recognizing that not all of them possess helicase activity.
A sizeable subset of these proteins, representing three families, function to manipulate structured RNAs. In parallel with the diversity inherent to structured RNAs and their transitions, SF2 helicases use diverse mechanisms to promote RNA rearrangements. Strikingly, emerging evidence suggests that the mechanisms track to a large extent with the family delineations. In this review, we summarize the mechanistic capabilities of these three protein families, as deduced principally from structures and biochemical studies using simple, defined RNA substrates. We then survey biological processes that require these proteins, with emphasis on the specific RNA and RNP substrates, the conformational transitions and, if known, the mechanisms used to promote these transitions.
HELICASE FAMILIES INVOLVED IN RNA CHAPERONING AND RNP REMODELING
SF2 helicases have been divided based on comparisons of their sequences and limited structural information into nine families and one smaller ‘group’ (13), refining and extending earlier classification schemes (12, 14). All SF2 proteins share a conserved core that binds a nucleoside triphosphate and a nucleic acid and consists of two domains (D1 and D2), each of which resembles RecA (Figure 1A). The core includes at least 12 identified sequence motifs, which are more highly conserved within families than between families. These motifs contact both ligands and play important roles in the processes mediated by these proteins, as has been reviewed elsewhere (13, 15, 16). In addition to the helicase core, many SF2 proteins include ancillary domains. For some families, these additional domains are conserved, while for others they are highly variable (13).
Figure 1. SF2 families involved in RNA chaperoning and RNP remodeling.
(a) Arrangement of conserved structural domains. Conserved motifs of the helicase core are shown in dark gray. The domain arrangements of DEAH/RHA and Ski2-like families shown are based on Mtr4 and Prp43 structures, respectively (domains not to scale). The winged helix and ratchet domains are conserved in DEAH/RHA and Ski2-like helicases and the OB fold domain is conserved in DEAH/RHA helicases, while the arch domain is present in only a subset of Ski2-like helicases. Individual proteins from all three families may include other, non-conserved domains that are not shown in the figures.
(b) Crystal structures of Ski2-like (Mtr4)(28), DEAH/RHA (Prp43)(30) and DEAD-box (Mss116)(54) helicases. Domains are colored as in (a). The nucleotide (ADP for Mtr4 and Prp43, and ADPNP for Mss116) is shown in red and co-crystallized ssRNA bound to Mtr4 and Mss116 is shown in black. The β hairpin within D2 in Ski2-like and DEAH/RHA helicases that is thought to function as a pin during RNA unwinding is highlighted in pink.
Most of the proteins known to function as RNA chaperones or RNP remodelers are found within two families, the DEAD-box and the DEAH/RHA families, and a few are found within the Ski2-like family. We therefore focus below on these three families, and representative schematics and structures are shown in Figure 1A and 1B, respectively. Although the DEAD-box and DEAH/RHA families have at times been described collectively as DExD/H-box proteins, recent analysis showed that these two families are no more closely related to each other than to other SF2 families, and they are correspondingly indicated here as two distinct families (13). This is an important change in viewpoint because the molecular properties and mechanisms of these two families appear to differ substantially from each other.
ATPase ACTIVITY AND RNA UNWINDING
As a step toward defining their mechanisms in RNA chaperoning and remodeling, the capabilities of individual SF2 helicases have been probed using simple, defined RNA substrates. Although these studies have revealed differences between families, there are also central properties that are shared by proteins in these families as well as in other SF2 families and even in the more distantly related SF1 helicases. A hallmark of these proteins is DNA- and/or RNA-dependent nucleotide hydrolysis activity, which is derived from energetic coupling between these ligands. As illustrated most dramatically by structures of DEAD-box proteins, the two core domains have substantial flexibility relative to each other (17–19). Upon binding of the nucleotide, principally to D1, the domains have increased affinity for each other and are found in more intimate contact, at least in the presence of bound nucleic acid. Nucleotide hydrolysis and product release reset the protein to the open complex. The consequences of the cycle vary, as outlined below.
Directional RNA translocation and unwinding by DEAH/RHA and Ski2-like proteins
Conventional helicase mechanisms include translocation, or directional movement of the protein along one of the nucleic acid strands. This movement is achieved for SF1 and SF2 helicases at the level of a protein monomer and is directly linked to unwinding, because movement along one strand results in displacement of the complementary strand.
The mechanistic steps involved in translocation have been described in detail for the SF1A DNA helicases PcrA and Rep (14). Crystallographic and biochemical approaches have provided high-resolution views of intermediates and revealed dynamics in the translocation process. When bound to a DNA that includes a 3’ extension, these proteins form extensive contacts to single-stranded and double-stranded regions of the DNA, essentially encircling the ssDNA region. This encapsulation is made possible by domain insertions in D1 and D2 that provide additional DNA binding surfaces. Translocation occurs as ATP binding induces closure of the two core domains (D1 and D2), resulting in movement of D1 while D2 remains stationary on the DNA. ATP hydrolysis and product release allow a transient loosening of D2 from contacts with both ssDNA and D1, resulting in domain opening and movement of D2 by one nucleotide in the direction of translocation, 3’ to 5’ for these proteins. Reformation of contacts by D2 then resets the core for ATP binding and another turn of the cycle.
A similar reaction cycle appears to operate in some SF2 helicases. The viral helicases NS3 and NPH-II (from hepatitis C and vaccinia virus respectively), which form their own group within superfamily 2, unwind RNA with moderate processivity by translocating from 3’ to 5’ (20–22). Although structural and biochemical data suggest some differences in the detailed mechanism of these proteins relative to the well-studied SF1 proteins, such as differences in the identities of amino acids that contact the nucleobases of unwound RNA and possible differences in chemical or kinetic step sizes, the basic features of translocation and unwinding are maintained. Thus, SF2 RNA helicases are capable of functioning much like SF1 DNA helicases. However, it should be noted that these viral helicases evolved to function in the context of viral genomes, where processivity and directionality are probably required for function. In contrast, helicase proteins that manipulate cellular RNAs are much less likely to encounter long, continuous helices, and one could envision that their mechanisms would be correspondingly different.
Compared to the SF1 and SF2 families above, there is less structural and mechanistic information for DEAH/RHA and Ski2-like proteins. However, such information is now accumulating at a rapid rate. The first crystal structure of a Ski2-like protein was Hel308, an archaeal DNA helicase (23, 24), and since then structures have been obtained for the S. cerevisiae RNA helicases Brr2, Mtr4, and Ski2 (25–29) (Figure 1B). The first crystal structure of a DEAH/RHA helicase was the S. cerevisiae protein Prp43 (30, 31), which functions to remodel the spliceosome and the pre-ribosome. Although it was not recognized by sequence analysis, the structures revealed that these protein families share two domains outside the core, referred to as a winged-helix domain and a ratchet domain. These domains make key contacts with substrate RNA and are proposed to play key roles in RNA unwinding (32).
DEAH/RHA and Ski2-like proteins are thought to unwind RNA helices by a mechanism that is fundamentally similar to SF1 and viral SF2 helicases. Unwinding by Mtr4 of a 20-bp duplex requires a 3’-ssRNA extension, indicating that Mtr4 binds to the extension and unwinds RNA by translocating 3’ to 5’ (33), and Mtr4 can unwind a helix as long as 36 bp when in complex with partner proteins of TRAMP (34). 3’ overhangs were also shown to enhance unwinding by Brr2 (25). Similarly, directional unwinding is indicated for Prp22, a DEAH/RHA protein that functions in pre-mRNA splicing (35). The unwinding efficiencies appear to decline with increasing helix length, suggesting that the level of processivity is much lower than for SF1 helicases and lower than for the viral SF2 proteins noted above. However, the lower processivity probably does not reflect a difference in mechanism, but rather simply a change in the rate constant for further translocation relative to that for dissociation from the helix, which defines processivity (36). The mechanistic features of DEAH/RHA and Ski2-like helicases suggest a general requirement for a 3’ single-stranded extension to allow for helicase loading, which most likely generates important constraints on their physiological substrates and specific functions.
Non-processive RNA unwinding by DEAD-box proteins
DEAD-box proteins make up the largest family of SF2 helicases and they function throughout biology to promote RNA rearrangements and RNP remodeling. Although DEAD-box proteins display RNA unwinding activity, they use a non-conventional mechanism. Early work using mammalian eIF4A protein showed that DEAD-box proteins can unwind RNA helices up to 15 base pairs but with efficiencies that decrease with increases in helix length or stability (37), suggesting very low processivity. In addition, early studies showed that DEAD-box proteins do not require either 5’ or 3’ extensions, implying that they can interact directly with a duplex region and at least initiate unwinding without translocating (38–41). This point was underscored by the finding that although DEAD-box proteins do not perform ATP-dependent unwinding of DNA helices, the S. cerevisiae protein Ded1 can unwind a DNA helix that is substituted in one strand with a small number of internal RNA nucleotides, implying that the unwinding is initiated and centered upon the RNA segment (42). The lack of translocation was highlighted by findings that complete unwinding of RNA helices can occur in a single cycle of ATP binding and hydrolysis (43–45). Further, complete unwinding is possible with the ATP analog ADP-BeF3 (46), indicating that unwinding can precede ATP hydrolysis and the release of the hydrolysis products. These steps then promote release of the tightly bound RNA strand after unwinding, and they may also contribute to unwinding of longer or more stable helices (47, 48).
Additional work has led to a physical model for this non-processive RNA unwinding. Crystal structures of various DEAD-box proteins showed that the two RecA-like domains have significant flexibility relative to each other in the absence of ligands (17–19, 49–51). However, with a bound ATP analog and ssRNA, the two domains are brought close together and position the ssRNA in the interface in a manner that would be incompatible with the presence of a partner strand (18, 19, 52–55). Adenosine nucleotide binding is generally coupled energetically with RNA binding by DEAD-box proteins (45, 56–61), and this cooperative binding is proposed to lead to domain closure and separation of the two RNA strands. This domain closure also activates the ATPase activity, which allows release of the tightly bound RNA strand after unwinding and resetting of the DEAD-box protein. Unlike the families described above, DEAD-box proteins lack conserved domains that surround the ssRNA and they lack a ‘pin’ element that may function as a wedge in unwinding (Figure 1B)(13). Either or both of these differences likely leads to their lack of processivity.
A recent study provided additional insight into the steps that lead to RNA unwinding in DEAD-box proteins (62). Fluorescence studies and crystallography of the S. cerevisiae protein Mss116 showed that D2 alone can bind dsRNA without unwinding it, and this study supported and extended previous results indicating that D1 can bind ATP independently (63–66). The simultaneous presence of ATP and dsRNA is proposed to result in closing of the domains, resulting in the exclusion of one strand, bending of the other strand, and consequent local unwinding of the helix.
In the context of this model, it remains possible that there are intermediates in which the domains are in contact but unwinding is not achieved, and that these intermediates accumulate under conditions that do not promote unwinding. For example, binding of a 154-mer RNA and AMP-PNP to the B. subtilis protein YxiN leads to favorable domain closure, as monitored by smFRET, but AMP-PNP does not give detectable unwinding of a model RNA duplex (61). Analogously, binding of an ATPase-deficient mutant of YxiN to a closely related RNA and AMP-PNP or ATP gives domain closure but not RNA unwinding (67). Alternatively, this domain closure may reflect binding of YxiN to a single-stranded segment within the large RNA, whereas domain closure may not be achieved upon binding dsRNA.
In addition to accelerating local unwinding of RNA, some DEAD-box proteins can accelerate the reverse reaction, the annealing of two strands to form a dsRNA helix (41, 68–70). Strand annealing activity does not depend on ATP (41, 69). A recent analysis suggested that DEAD-box proteins can be divided into two broad groups based on ATPase activity and corresponding RNA unwinding activity (71). Interestingly, Rok1, one of the proteins in the ‘low’ ATPase/unwinding group, is suggested to function by promoting an RNA annealing reaction in ribosome biogenesis (see below).
Unstructured, basic ‘tails’ of DEAD-box proteins as tethers
Despite their ability to interact directly with a duplex to initiate unwinding, several DEAD-box proteins are activated for unwinding by ssRNA extensions, which can be either 5’ or 3’ extensions (40, 41, 71–74). The puzzling ability of DEAD-box proteins to use extensions of either polarity led to them being described as bi-directional, but it is now clear that the extensions do not increase unwinding efficiency by providing starting points for translocation but instead provide binding sites for additional monomers or ancillary regions of the same monomer, which can tether the helicase core in proximity to a substrate helix and remain bound during the local RNA unwinding reaction (16, 75).
Several lines of evidence support this conclusion. The extension is not required to be contiguous with the duplex to be unwound; for Ded1, the extension can even be connected by a bridging streptavidin protein as long as it allows proximal binding of an additional protein molecule (40). For other DEAD-box proteins, the extension does not have to be single-stranded, as a dsRNA or DNA extension is at least as effective as ssRNA for the Neurospora crassa protein CYT-19 (76). Deletion of an unstructured C-terminal region of CYT-19 leaves the unwinding activity of the helicase core intact but abrogates the stimulation by DNA or RNA extensions, indicating that this ‘C-tail’ is responsible for binding the extension and tethering the core for unwinding of an adjacent helix (77, 78).
There is significant variation in the properties of these tethering interactions. Some DEAD-box proteins function as general RNA chaperones, and interactions of the C-tail are correspondingly versatile (75). There is no evidence for RNA sequence or structure specificity, and the C-tail is very flexible relative to the core, accounting for the enhanced RNA unwinding from extensions regardless of their polarity (78). On the other hand, some yeast DEAD-box proteins have C-terminal basic regions that may function similarly but with less flexibility between the tail and the core, such that a preference for polarity is observed (71). The Thermus thermophilus protein Hera includes a bipartite extension with an RRM domain that binds ssRNA with a preference for guanosine and a short unstructured region that binds dsRNA (79, 80). Other DEAD-box proteins are targeted to specific substrates by interactions with ancillary domains (81, 82) or surfaces within the helicase core (83, 84).
Remodeling of RNP complexes
While much is known about the abilities of SF2 RNA helicases to unwind duplexes, the physiological substrates for many of these proteins include assembled proteins, and a few studies have probed the abilities of certain SF2 helicases to remodel simple, defined RNP complexes. The viral helicase NPH-II can remove several protein complexes from ssRNA or dsRNA in an ATP-dependent manner (85, 86), most likely by translocating 3’ to 5’ along one RNA strand and displacing the protein(s) as well as the complementary RNA strand, if present (87). Ded1 is also able to remove proteins from RNA (86, 88), despite unwinding RNA non-processively as described above. Interestingly, Ded1 displays selectivity, accelerating dissociation of some protein complexes but relying on spontaneous dissociation of the tight-binding U1A protein before unwinding the two RNA strands that form the U1A binding site (88). The physical origins of this selectivity remain to be elucidated (87).
RIBOSOME BIOGENESIS
Here and in the sections that follow, we outline the biological functions of SF2 helicase proteins in rearranging RNAs and remodeling RNPs. Ribosome biogenesis is an elaborate process that involves regulated cleavage, modification and folding of several large ribosomal RNAs, as well as extensive assembly with proteins and changes in RNP constitution throughout the process (89, 90). Large fractions of the DEAD-box proteins of E. coli and S. cerevisiae function in ribosome biogenesis (4 of 5 and 15 of 26, respectively), and in S. cerevisiae several DEAH/RHA helicases and a Ski2-like helicase also participate. In bacteria, the RNA helicases seem to be required only at low temperatures, presumably due to increased lifetimes of non-native RNA structures (91, 92). However, in eukaryotes most of the helicases involved in ribosome biogenesis are essential, likely reflecting the increased complexity of the eukaryotic ribosome and a greater need for regulation.
Ribosome biogenesis in E. coli
Ribosome biogenesis in E .coli involves four DEAD-box helicases: DbpA, SrmB, CsdA and RhlE. DbpA (YxiN in B. subtilis) was originally implicated based on specific stimulation of its ATPase activity by 23S rRNA, which later was narrowed down to hairpin 92 (hp92) of the peptidyltransferase center (93, 94). The recognition of hp92 is mediated by a C-terminal RRM domain, which is flexibly linked to the helicase core and enhances unwinding of helices positioned either 3’ or 5’ from hp92 (81, 82, 95, 96). Functional and structural evidence supports a model in which DbpA rearranges RNA structure around hp92 in the 50S precursor while remaining tethered via the RRM domain. Despite this specific hp92 recognition, ΔdbpA mutants show no obvious defects, and defective ribosome assembly is only observed with a dominant negative DbpA mutant that lacks ATP-dependent unwinding activity but retains RNA binding (97, 98). Thus, in vivo, DbpA-mediated rearrangements appear to be redundant with other pathways but can be blocked by bound DbpA (98).
Recent evidence also suggests specific sites for SrmB. The absence of functional SrmB leads to accumulation of 40S intermediates that contain immature 23S rRNA and reduced amounts of some ribosomal proteins (99). Tandem affinity purification revealed SrmB interactions with two ribosomal proteins, L4 and L24, which bind a 5’ region of 23S rRNA (100). However, rRNA mutations that suppress the requirement of SrmB map to domain II of 23S rRNA, which is distant in the mature ribosome. Some suppressors map to a section of 5S rRNA with base complementary to domain II of 23S rRNA, suggesting that SrmB may prevent 5S rRNA annealing and promote native 23S rRNA folding and protein assembly within domain II (101).
CsdA, aka DeaD, is induced at low growth temperatures and several lines of evidence indicate that it functions as a general RNA chaperone in ribosome biogenesis and other RNA-related processes, including mRNA decay and translation (102, 103). CsdA functions at multiple stages of ribosome biogenesis and can suppress defects in ΔsrmB strains, suggesting functional overlap (104). Interestingly, mutations in CsdA can be suppressed by overexpression of another cold shock protein and general chaperone, CspA, – a small (70 aa), unrelated protein that can destabilize RNA helices and functions in transcription antitermination (105–107). CsdA contains a predicted RRM in its C-terminus that may contribute to general RNA recognition, possibly by recognizing a commonly occurring structural element (39).
RhlE appears to be largely but not completely redundant with CsdA. While growth defects are not observed in ΔrhlE cells, removal of RhlE and CsdA enhances the cold-sensitive phenotype of ΔcsdA cells and increases the accumulation of 50S precursors (108). However, unlike CsdA, RhlE cannot substitute for SrmB, and its overexpression even increases the accumulation of 50S precursors observed in ΔsrmB cells (108). Together the results suggest a complex network of functions for SrmB, CsdA, and RhlE, with extensive but incomplete overlap between them.
Ribosome biogenesis in S. cerevisiae
Nineteen yeast helicases have been implicated in ribosome biogenesis. Seventeen of them are essential, and several have been linked to distinct steps in small or large subunit biogenesis based on the sizes and compositions of ribosome intermediates that accumulate upon mutation or depletion of the helicase protein (Figure 2A)(109). While it can be difficult to separate direct from indirect effects, as some of the blocked steps may be downstream from those mediated directly by the helicase (89), these studies have provided important insights and constraints on the helicase functions in ribosome biogenesis. A distribution of the proposed roles of S. cerevisiae helicases in ribosome biogenesis is shown in Figure 2B. Below we discuss examples of proteins implicated in each of these roles, illustrating the mechanistic diversity of SF2 helicases, particularly within the DEAD-box family.
Figure 2. Helicases that function in ribosome biogenesis.
(a) S. cerevisiae helicases that function in biogenesis of the large and small ribosomal subunits. SSU: small subunit, LSU: large subunit. Essential helicases are shown in bold. DEAD-box, DEAH/RHA and Ski2-like helicases are shown in green, blue and orange, respectively.
(b) Known and proposed roles of RNA helicases in ribosome biogenesis. SnoRNA-related roles include promoting binding and dissociation of snoRNAs, as well as rearrangements associated with snoRNA-guided cleavage events. Helicases indicated as promoting non-snoRNA-mediated rearrangements facilitate pre-rRNA cleavage at specific, snoRNA-independent processing sites (see text for details).
Several helicases have been suggested to regulate nucleolytic processing of pre-rRNA into mature rRNA by remodeling structures adjacent to cleavage sites. The DEAD-box protein Dbp3 promotes site A3 cleavage by RNase MRP during 25S rRNA maturation and was suggested to disrupt an adjacent hairpin to increase access of the RNase (110). The DEAH/RHA protein Prp43 (see below) is recruited near the site of Nob1-mediated 3’ cleavage of 18S pre-rRNA by the cofactor Pfa1, which binds the OB-fold domain of Prp43 and stimulates its helicase activity (31, 111–113). 3’ end processing of 5.8 S rRNA requires the Ski2-like helicase Mtr4 (114), which functions as part of the TRAMP complex (see below) and is also involved in general quality control during ribosome biogenesis (89).
The possibility of an interesting divergence of DEAD-box protein functions is introduced by recent studies of Rok1 in small subunit biogenesis. Rok1 promotes a structural change to facilitate cleavage at site A2 by the Nob1 nuclease. Interestingly, in vitro experiments suggest that rather than disrupting structure, Rok1 specifically promotes the temporary formation of a helix that allows correct processing of pre-18S rRNA (115, 116). The specificity for this “pre-A2” helix is conferred by cofactor Rrp5, which stimulates Rok1-mediated annealing of pre-A2 by ~10-fold relative to other helices. Studies using model helices, noted above, showed that Rok1 has low ATPase and RNA unwinding activities relative to other DEAD-box proteins (71), suggesting an intriguing link between the capabilities and functions of Rok1. Nevertheless, Rok1 does retain a basal level of nucleotide-dependent RNA unwinding activity (71), and it is also suggested to function in snoRNP dissociation (see below).
Several helicases have been shown to function in rearrangements involving snoRNAs. snoRNAs are small structured RNAs that base pair with pre-rRNAs to guide site-specific cleavage, methylation and pseudouridylation, and can direct pre-rRNA folding (117). There are 75 snoRNAs in yeast, from structurally distinct box-C/D and H/ACA classes, with three essential ones (U3, U14, snR30) that guide cleavage events and 72 non-essential ones that specify nucleotide modifications (117). Three DEAD-box proteins are implicated in dissociation of certain snoRNAs. Has1 is required primarily for dissociation of the essential U3 and U14 snoRNAs, although several other snoRNAs are also affected upon Has1 depletion (118), while snR30 dissociation requires Rok1 (119). Interestingly, dissociation of U14 requires Dbp4 in addition to Has1 (120), indicating that removal of a given snoRNA can require direct or indirect activities of more than one helicase. Several helicases have also been suggested to function in U3 snoRNP-guided cleavage and rearrangements of pre-rRNA. The DEAH/RHA helicases Dhr1 and Dhr2 and DEAD-box proteins Dbp8, Rrp3 and Rok1 coprecipitate U3 snoRNA and are required for U3-guided cleavages (121). One or more of these helicases may also function in U3-assisted formation of the central pseudoknot in the 18S rRNA (122).
Of the helicases in eukaryotic ribosome biogenesis, the DEAH/RHA protein Prp43p is particularly versatile, playing multiple roles while also functioning in pre-mRNA splicing (see below). In addition to promoting Nob1-mediated cleavage of 18S rRNA, as noted above, Prp43 co-precipitates with snoRNAs (primarily box-C/D) and rRNA sequences near snoRNA binding sites, and is required for binding or release of snoRNAs from the pre-ribosome (112, 123–125). The broad spectrum of RNA substrates indicates that Prp43 recognizes general features of structured RNAs, either intrinsically or through cofactors such as Pfa1 (113). Surprisingly, in vitro studies showed that Prp43 does not require a 3’ extension for RNA unwinding, which would seem to be at odds with the 3’-5’ directionality observed for other DEAH/RHA helicases (126, 127). Together, these results have led to the suggestion that Prp43 could function similarly to DEAD-box proteins by forming tethering interactions with structured RNA via a cofactor protein or an ancillary domain and disrupting RNA structure locally (128), which would be an important exception from the general differences in functional mechanisms between DEAH/RHA and DEAD-box proteins.
SPLICEOSOME ASSEMBLY AND FUNCTION
After ribosome biogenesis, eukaryotic pre-mRNA splicing is the process that requires the most RNA helicases [reviewed in (129, 130)]. Eight helicases function in splicing in S. cerevisiae, including three DEAD-box, one Ski2-like and four DEAH/RHA proteins. These helicases have been implicated in RNA secondary structure rearrangements and protein displacement, allowing the spliceosome to assemble, rearrange and dissociate in a controlled manner (Figure 3). Splicing helicases have also been shown to function in proofreading, antagonizing the forward progression of slowly-reacting pre-mRNAs and intermediates and promoting their dissociation (131). Below, we summarize the current knowledge of helicase-mediated rearrangements of the S. cerevisiae spliceosome and discuss how the properties of each helicase family correspond to their roles in pre-mRNA splicing.
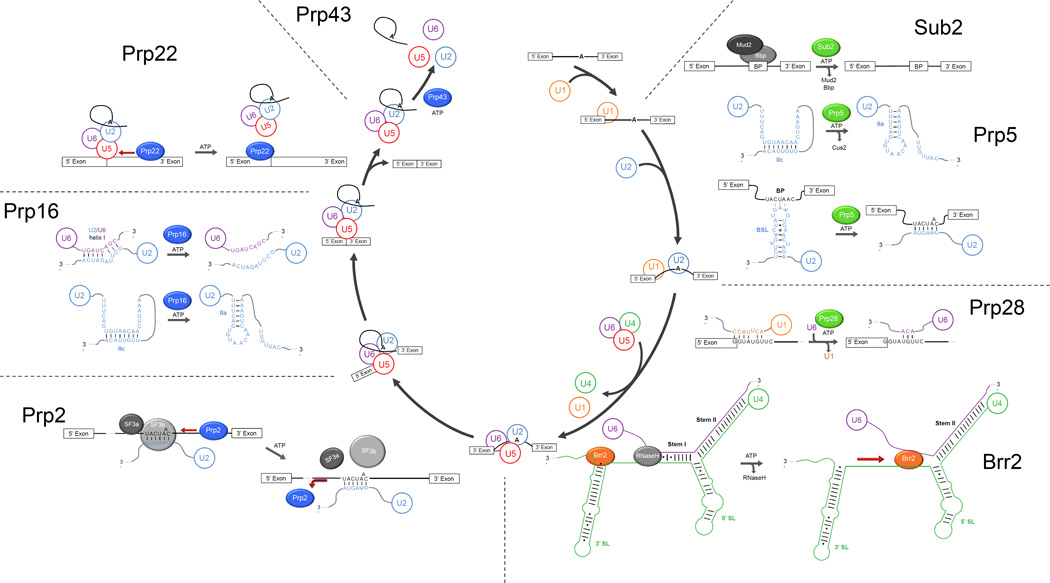
Figure 3. Helicase-mediated rearrangements in splicing.
DEAD-box proteins are green, DEAH/RHA proteins are blue, and Ski2-like proteins are orange. Abbreviations in the figure are BP (branchpoint, shown as A in the cycle), Bbp (BP binding protein, aka Msl5), BSL (branchpoint stemloop), and SL (stemloop). For Prp2, Prp22, and Brr2, experimental evidence indicates interaction with a specific RNA strand and a direction of translocation (see text), which are shown by red arrows.
DEAD-box helicases: chaperoning spliceosome assembly
Two DEAD-box helicases, Sub2 and Prp5, are required for branchpoint (BP) recognition by U2 snRNP and promote rearrangements that expose and position complementary sequences of BP and U2 snRNA for base pairing (132, 133). Genetic studies have implicated Sub2 in dissociation of the Mud2/Bbp protein heterodimer from the BP sequence, and consistent with a role in protein displacement, studies of the human homolog UAP56 suggest that ATPase activity, but not RNA unwinding, is required for BP exposure (133, 134). Prp5 promotes a transition between alternative conformations of the U2 snRNP, which involves direct or indirect disruption of the 8-bp helix IIc, formation of alternative helix IIa, and ejection of the Cus2 protein (135–137) (Figure 3). This transition increases the accessibility of the BP-recognition sequence of U2 snRNA, located in the loop of a conserved branchpoint interaction stemloop (BSL)(135, 138, 139). Upon initial recognition of the BP sequence by the BSL, Prp5 has also been implicated in unwinding the BSL stem to allow stable association of U2 snRNP with the BP (139) (Figure 3).
Following stable U2 snRNP binding, the Prp28 helicase is required for unwinding of the 6-bp helix between U1 snRNP and the 5’ splice site (5’-ss), facilitating release of U1 snRNP and exposing the 5’-ss for base pairing with the U6 snRNP (140) (Figure 3). Although Prp28 and Prp5 have been described as lacking unwinding activity (135, 141), the reported experiments have used helices longer than the 10–15 bp that are efficiently unwound by other DEAD-box proteins. Overall, the localized rearrangements by all three DEAD-box helicases are consistent with the general capabilities of DEAD-box proteins and in contrast with the generally more extensive remodeling events mediated by the helicases described below.
DEAH/RHA helicases: activation for catalysis, product release and snRNP recycling
The four spliceosomal DEAH/RHA-family helicases catalyze rearrangements that accompany or follow the chemical steps of splicing, beginning with activation of the spliceosome for the first step of splicing. Strikingly, these rearrangements generally involve more extensive base-pairing interactions and/or displacement of proteins than the changes promoted by DEAD-box proteins, and the current data suggest a strong correspondence of the functional roles of these proteins with the capabilities of the protein family as a whole, as established from crystallographic and biochemical studies described above.
Prp2 promotes dissociation of proteins from the BP region, exposing the BP adenosine for attack on the 5’ss (142, 143). Prp2 requires 23–33 nt of pre-mRNA in the 3’ direction from the BP, suggesting 3’ to 5’ translocation (144). Along the way, Prp2 is suggested to dislodge the BP-binding SF3a and SF3b proteins and a number of others, contributing to catalytic activation of the spliceosome (142, 143, 145) (Figure 3). Following lariat formation, Prp16 promotes rearrangement of the spliceosome for the second catalytic step. The rearrangement involves refolding of U2 snRNP to the IIa state, destabilization of U2/U6 helix 1 (Figure 3), and displacement of several proteins (137, 146–149). Following splicing, Prp22 releases the mRNA by disrupting base pairs between the U5 snRNP and the mRNA (150, 151) (Figure 3). Prp22 initiates translocation ~23 nts downstream from the splice junction, suggesting 3’-5’ translocation (35, 152). Finally, Prp43 mediates the extensive rearrangements that allow release of the intron lariat and recycling of the U2, U5 and U6 snRNPs (153–155). The activity of Prp43 in spliceosome disassembly is stimulated by cofactor Ntr1, which also enhances in vitro unwinding activity of the helicase (127, 155).
Ski2-like helicase Brr2: remodeling U4/6 snRNP
Like the DEAH/RHA proteins, the Ski2-like helicase Brr2 promotes a large-scale rearrangement, acting early in the splicing cycle to disrupt an extensive base-pairing network between the U4 and U6 snRNAs (156, 157) (Figure 3). The disruption releases the 5’ss-recognition sequence within U6 snRNA and allows it to form catalytically important intra-U6 and U2/U6 interactions. The mechanism of U4/U6 snRNP dissociation is not fully understood, but recent data suggest that Brr2 binds the 3’ end of the U4 snRNA and translocates 3’ to 5’ to unwind U4/U6 stem I and presumably stem II (158–160). While Brr2 readily unwinds protein-free U4/U6 snRNA in vitro, it is unknown how proteins that coat the RNA in the U4/U6 snRNP affect unwinding (156, 158).
Proofreading by spliceosomal helicases
In analogy with GTPases in translation, several splicing helicases use ATP to enhance fidelity by proofreading the splicing process (131, 161). Productive steps are in kinetic competition with a non-productive branch in which a helicase uses ATP to reject the pre-mRNA substrate (162, 163). For a substrate with a non-consensus sequence, the reduced rate of the forward step increases flux through the non-productive branch, leading to rejection and eventual discard of the unspliced substrate or a splicing intermediate. In addition, the helicases may act preferentially on stalled splicing complexes because of their decreased stability and enhanced dynamics, analogous to DEAD-box proteins that act as chaperones by resolving misfolded RNA structures [(131, 164); see Self-splicing introns below].
As expected from this mechanism, the proofreading checkpoints largely overlap with the step(s) mediated by a given helicase in the productive splicing cycle. Thus, Prp5 rejects pre-mRNAs with mutations in the branch site (165), Prp28 and Prp16 suppress splicing of 5’-ss mutants (166–169), and Prp22 primarily proofreads the 3’-ss (170). Some of the helicases may also act with some promiscuity on structures that become exposed due to their lower stability or to a lack of bound proteins. Thus, Prp43 can reject or discard stalled intermediates in at least two steps of the spliceosome cycle, in addition to disassembling the spliceosome to complete the native cycle (171, 172), while Prp22 can reject BP and 5’-ss mutants, although less efficiently than 3’-ss mutants (170).
SELF-SPLICING INTRONS: DEAD-BOX PROTEINS AS GENERAL RNA CHAPERONES
Group I and group II introns are autocatalytic RNAs that are broadly distributed in nature and function by splicing themselves out of precursor RNAs, with some retaining ancestral roles as mobile genetic elements (173). Several mitochondrial group I introns in N. crassa require the DEAD-box protein CYT-19 for efficient folding (174), and all of the S. cerevisiae mitochondrial group I and group II introns depend on Mss116, a CYT-19 homolog (175, 176). Group II introns in plant mitochondria and chloroplasts also require DEAD-box proteins, suggesting a widespread role (177, 178). The versatility of these DEAD-box proteins indicates that they are general RNA chaperones (174, 176), and biochemical studies of these proteins and introns have been instrumental in elucidating general features of the mechanisms by which DEAD-box proteins manipulate structured RNAs.
Model group I introns have been used to probe how DEAD-box proteins promote transitions away from misfolded structure. A ribozyme derivative of the Tetrahymena thermophila group I intron (from the large subunit rRNA) misfolds to a single, dominant conformation that is compact and includes extensive native structure but is thought to have a non-native topology (179, 180). This misfolded conformation undergoes a slow ‘refolding’ rearrangement to the native state that is rate limited by extensive unfolding (180, 181). CYT-19 accelerates refolding in an ATP-dependent manner, indicating that it can use ATP to accelerate unfolding of the misfolded ribozyme (76). The function of CYT-19 as a general chaperone, along with the non-cognate nature of the Tetrahymena ribozyme, would suggest that CYT-19 accelerates this refolding reaction without recognizing specific features of the misfolded ribozyme, and indeed CYT-19 can also unfold the native ribozyme (164). CYT-19 also accelerates refolding of a misfolded group I intron from the bacterium Azoarcus (182), and it promotes splicing of its cognate introns in vitro, most likely by accelerating refolding transitions (174). These refolding processes are known or suggested to require unfolding of tertiary structure, but DEAD-box proteins have not been shown to disrupt RNA tertiary contacts. Recent work indicates that CYT-19 disrupts tertiary structure in the Tetrahymena ribozyme using a ‘helix capture’ mechanism by trapping helical elements after they lose tertiary contacts spontaneously and then using RNA unwinding activity to separate the strands of the trapped helix (C. Pan, I.J., and R.R., in preparation).
Group I introns have also been valuable for probing how directionality in folding can be conferred upon the inherently non-specific activity of these DEAD-box proteins. For the Tetrahymena ribozyme, there is a correlation between global RNA stability and the efficiency of unfolding by CYT-19, with less stable conformations being unfolded more efficiently. The effect is that net folding proceeds from the less stable misfolded conformation to the more stable native state despite the absence of specific recognition by CYT-19 (164). If the native structure is destabilized by mutations or by lowering Mg2+ concentration, it is unfolded efficiently by CYT-19 and instead of the native ribozyme accumulating, the misfolded conformation or unfolded conformations accumulate (164). Mss116, which has greater RNA unwinding activity than CYT-19, can readily unfold the native structure under a broader range of conditions (183, 184). Mss116 can also function as a chaperone to promote conformational transitions between or following the two steps of splicing for a model intron from yeast (185), most likely by disrupting structure non-specifically, with directionality conferred by the functionally irreversible dissociation of the intron from the ligated exons.
Group II introns also undergo slow folding transitions to the native state, and these transitions can be accelerated in vitro by DEAD-box proteins (186). Two yeast mitochondrial introns have been used most extensively in these studies. The bI1 intron requires Mss116 and ATP for native folding (187). Mss116 is dispensable after the native state is reached, establishing that it chaperones folding of the RNA (187). A second intron, aI5γ and its ribozyme derivative (D135), fold slowly under near-physiological conditions (183, 188), with D135 folding through multiple pathways on time scales from minutes to hours (189, 190). Mss116 accelerates native folding of both the intron and D135 ribozyme (183, 188, 190, 191), and further work with D135 has shown that Mss116 accelerates two transitions, only one of which requires ATP (190, 192). The ATP-dependent step is suggested to be the disruption of kinetically trapped structure, analogous to the roles of DEAD-box proteins in folding of other group I and II introns (192, 193), but it may also or instead reflect nucleotide-dependent dissociation of Mss116 from a folding intermediate (192). The ATP-independent step is earlier in folding and thought to include compaction of the largest domain of the intron, a step that can limit folding for a subpopulation of the ribozyme (189, 194).
GENERAL RNA CHAPERONES IN TRANSLATION
Eukaryotic translation requires multiple helicases, largely for rearrangements of the 5’ untranslated region (5’-UTR) in cap-dependent initiation. In this process, the pre-initiation complex (PIC) is recruited to the 5’ cap through interactions with the eIF4F complex, followed by 5’ to 3’ scanning along the 5’ UTR in search of the start codon (195). Both PIC loading and scanning, the latter process involving up to thousands of nucleotides, require disruption of RNA structures within the 5’UTR. Three helicases, eIF4A, Ded1 and DHX29, are thought to clear the way for the PIC by resolving these structures.
The DEAD-box protein eIF4A is a component of the eIF4F complex, which also includes eIF4G and the cap-binding protein eIF4E. Due to its localization to the cap, eIF4A is thought to promote PIC loading by disrupting RNA secondary structures in the vicinity of the 5’ terminus (195, 196). A poor helicase on its own, eIF4A is enhanced for RNA unwinding by interactions with other initiation factors (197). eIF4G binds and modulates the orientation of the eIF4A core domains, and yeast eIF4G can bias eIF4A to unwind helices with 5’ overhangs, potentially generating an overall 5’ to 3’ directionality for eIF4A-mediated rearrangements of the 5’UTR (198–201). eIF4B binds to D2 of eIF4A and can interact with ssRNA via its RRM, potentially trapping ssRNA produced by eIF4A (195, 199, 202). eIF4B may also promote RNA unwinding by tethering eIF4A to RNA, analogous to the ancillary RNA-binding domains and extensions found in other helicases (202).
Following PIC loading, eIF4A is also thought to disrupt downstream 5’UTR structures to facilitate scanning. Consistent with this model, eIF4A and eIF4B crosslink to the mRNA up to 52 nt downstream from the cap (203). The much higher intracellular concentration of eIF4A than the other initiation factors and ribosomes suggests that eIF4A may carry out some functions independently, and that multiple eIF4A monomers can act per mRNA (195, 204). However, Ded1p and DHX29 are also involved in translation and are more potent helicases, suggesting that eIF4A may not be the primary helicase during scanning (195). In addition to cap-dependent translation initiation, eIF4G and 4A are required for initiation at Type 1 and Type 2 IRES elements in viral mRNAs and were shown to induce conformational rearrangements necessary for PIC loading and initiation (205, 206).
Both the helicase and RNP remodeling activities of Ded1 are likely to play roles in translation initiation. Ded1 interacts with eIF4G, and this interaction can both stimulate and repress translation initiation (207). While the role of Ded1 in repression is ATP-independent, ATP is required for Ded1-mediated release of mRNAs from stress granules, which reverses repression (207). As a helicase, Ded1 is thought to disrupt 5’UTR helices to promote PIC loading and scanning of the 5’UTR, although these functions remain to be established experimentally (195).
The DEAH/RHA helicase DHX29 is required for efficient translation of mRNAs with highly structured 5’UTRs and is also important for overall translation (208, 209). DHX29 interacts with the 40S ribosome in the PIC near the mRNA entry channel and promotes disruption of double-stranded segments, allowing these sequences to enter the ribosome (209). DHX29 may unwind these mRNA structures directly, or it may act indirectly by remodeling the entry channel of the ribosome (209).
Another helicase that promotes translation of specific groups of mRNAs is the DEAH/RHA helicase RNA helicase A (RHA), which interacts with mRNAs containing the 150 nt post-transcriptional control element (PCE) (210, 211). RHA also interacts with other structures in 5’UTRs, including G-quadruplexes (see G4 box), and it presumably destabilizes these structures to give the observed increases in polysome loading and translation.
Finally, Vasa (DDX4) promotes translation of mRNAs encoding proteins that are involved in embryogenesis and germline development in animals (212). Vasa interacts with the initiation factor eIF5B, which mediates ribosomal subunit joining (213, 214), and the role of the helicase appears to be to promote eIF5B loading to the PIC, perhaps by removing repressors from the eIF5B binding site (213, 214).
Although helicase functions have been best characterized in translation initiation, a handful of RNA helicases function at later steps. The Ski2-like protein Slh1 associates with translating ribosomes and is important for translation (215). The DEAD-box protein Dbp5 and its cofactors Gle1 and IP6 function in translation termination (216, 217). Based on genetic data, Dbp5 is suggested to promote binding of release factors eRF1 and 3, likely by promoting rearrangements of the ribosome (218). The translating ribosome may also disrupt mRNA structures directly during elongation, analogous to the prokaryotic ribosome (219, 220), limiting the requirement for helicases at this stage.
RNA DECAY
Stable RNA structures and bound proteins can inhibit enzymatic RNA degradation, and unsurprisingly RNases are often found to recruit RNA helicases (221). The DEAD-box helicase RhlB is a component of the E. coli degradosome and promotes degradation of RNAs with stable secondary structures by the exonuclease PNPase (222, 223). Interactions with the degradosome have also been reported for other DEAD-box poteins in E.coli, and DEAD-box components are found in the degradosomes of other bacteria (103, 221).
The eukaryotic exosome primarily utilizes helicases from two regulatory complexes: Ski2 helicase of the cytoplasmic Ski complex and the closely related Ski2-like helicase Mtr4 of the nuclear complex TRAMP (224). Similarly to RhlB, both helicases appear to unwind structured RNAs and channel ssRNA into the exosome for degradation (34, 225). Directional 3’-5’ unwinding by these Ski2-like helicases is thought to promote coupling of unwinding and degradation, which also proceeds 3’ to 5’. An interesting interplay has been observed between Mtr4 and the poly(A) polymerase component of TRAMP. The polymerase promotes Mtr4 loading on RNAs by adding oligo(A) tails, and once bound, Mtr4 inhibits the polymerase, thus controlling the length of the tail (34, 226). The RHAU helicase can also transiently interact with the exosome, promoting degradation of a specific group of mRNAs, likely by disrupting RNA structures or protein binding (227).
OTHER RNA CHAPERONE AND REMODELING ACTIVITIES
Transcription
It has long been known that the DEAD-box proteins p68 and p72 function in transcription by recruiting transcription regulators to RNA polymerase II, including the tumor suppressor p53 (228). This function appears to be carried out at the level of DNA, but recent results suggest a general RNA chaperone function for the S. cerevisiae homolog Dbp2. Dbp2 was shown to promote clearance of cryptic RNA transcripts from chromatin, apparently by remodeling RNA co-transcriptionally and promoting binding of decay factors (229). Dbp2 probably remodels the RNA indiscriminately, as it also promotes loading of export factors to native transcripts (230). It is currently unknown whether corresponding roles are played by p68 and/or p72, but there is evidence for their localization to regions of elongation in transcription (228).
Nuclear export
Several DEAD-box proteins have been implicated in the RNP remodeling events that occur as mRNA is exported through the nuclear pore complex (NPC). Dbp5 is localized to the cytoplasmic face of the NPC and is required for dissociation of several export factors, contributing to the directionality of mRNA export (231, 232). The ATPase activity of Dbp5, which is required for protein displacement, is regulated by the activator Gle1 and co-activator IP6, while the nuclear pore component Nup159 promotes recycling of nucleotide-free Dbp5 after ATP hydrolysis (83). Inside the nucleus, UAP56 (Sub2) plays an incompletely understood, ATP-dependent role in mRNA export as a component of the TREX complex (233, 234). Furthermore, multiple human helicases are used by viruses during export of viral RNAs, including DEAD-box proteins DDX3 (the human homolog of Ded1) and DDX1, as well as the DEAH/RHA helicase RHA (235).
Editing
Illustrating the diversity of SF2 helicase-dependent processes, RNA helicases have been implicated in small RNA-guided pre-mRNA editing that occurs in the mitochondria of some protozoa (236). During editing, the 5’ region of a short guide RNA (gRNA) pairs with pre-mRNA, while the partially complementary 3’ region serves as template for uridine insertions and deletions by the editosome. Editing creates new complementarity between the pre-mRNA and the 5’ region of another gRNA, resulting in a 3’-5’ cascade of editing events. In Trypanosoma brucei, the sequential binding and dissociation of gRNAs is promoted by the DEAD-box helicase REH1, which appears to dissociate gRNAs after editing (237). A second helicase, REH2 (related to DEAH/RHA and Ski2-like families), associates with the editosome and may promote RNA rearrangements during editing (238).
CONCLUSIONS
It is now clear that SF2 helicase proteins function throughout RNA biology, playing a diverse set of roles and using a diverse set of mechanisms. The list of helicases that function to rearrange or remodel RNAs or RNPs is large and growing. Still, despite remarkable progress in recent years, we have very limited knowledge of the interaction sites for helicases on their biological RNA and RNP substrates, and we know little about the structural rearrangements that they promote. Mechanistically, the available data suggest a general model in which DEAD-box proteins act locally, disrupting or in some cases promoting RNA structure without significant translocation, while DEAH/RHA and Ski2-like proteins load onto ssRNA and translocate 3’-5’, disrupting RNA structures and displacing proteins as they translocate. In the coming years, it will be fascinating to continue to learn about the structures and dynamics of the RNA and RNP substrates and the mechanisms used by helicases to manipulate them.
SUMMARY POINTS.
Helicase proteins from three families within superfamily 2 – the DEAD-box, DEAH/RHA, and Ski2-like families – function as RNA chaperones and RNP remodelers and act upon a wide variety of physiological substrates.
The available data indicate that DEAH/RHA and Ski2-like helicases unwind model RNA substrates with a mechanism that includes 3’-to-5’ translocation and a modest amount of processivity. DEAD-box proteins initiate RNA unwinding internally and do not translocate significantly.
Intriguingly, the differences in mechanistic properties between families show a good correspondence with differences in their in vivo roles. Thus, DEAH/RHA and Ski2-like proteins require 3’ overhangs for helicase loading and then unwind RNA and/or displace proteins directionally, while DEAD-box proteins initiate limited RNA unwinding internally.
Large, structured RNPs such as the ribosome and spliceosome rely heavily on the activities of RNA helicase proteins for assembly and function. In prokaryotes, DEAD-box proteins play extensively overlapping roles in ribosome assembly, while in eukaryotes, larger numbers of helicase proteins play more specific roles, both in the contexts of the ribosome and the spliceosome.
The biological roles of SF2 helicases in RNA and RNP rearrangements represent a remarkably diverse set of activities, including unwinding of dsRNA, annealing of ssRNA, displacement of proteins, and disruption of highly stable non-canonical RNA structures.
FUTURE ISSUES.
It is highly likely that there will turn out to be exceptions to summary point 3 above. That is, there will be proteins that do not interact with and unwind RNA using the mechanism expected for their SF2 family. A current candidate is Prp43, which is a DEAH/RHA family protein but displays some behaviors that resemble those of DEAD-box proteins.
It is not at all clear how DEAD-box helicases displace proteins from RNPs. DEAH/RHA and Ski2-like proteins, as well as proteins from other SF2 families, most likely displace proteins by translocating along the RNA substrate. However, DEAD-box proteins function extensively as RNP remodelers, removing proteins while promoting RNA rearrangements, yet there is no evidence for significant translocation by any DEAD-box proteins.
For most SF2 RNA helicases, the exact RNA and RNP rearrangements they promote in vivo remain unclear. A combination of in vivo and in vitro approaches will be required to enhance the understanding of the biological roles of these proteins.
It remains possible or even likely that there are physiological roles and physical properties for RNA helicase proteins that remain to be discovered, especially with the continuing discoveries of vast new functional classes of noncoding RNAs.
SIDEBAR (to be included next to GENERAL RNA CHAPERONES IN TRANSLATION).
Disruption of G-quadruplex structures
G-quadruplexes (G4s) are four-stranded structures comprised of stacked G-quartets, in which four guanosines interact by Hoogsteen interactions. G-rich sequences that adopt G4s in vitro are abundant in cellular RNAs, and G4s have been suggested to play regulatory roles throughout RNA metabolism (239). G4 structures are extremely stable due to the extensive network of H-bonds and π-π interactions, as well as cation coordination in the G4 interior. Thus, it is not surprising that helicases have been implicated in G4 remodeling.
The human DEAH/RHA helicases RHAU and RHA efficiently disrupt RNA G4s in vitro and both are likely to unwind G4s in the cell. RHAU has been proposed to disrupt a G4 within the telomerase RNA in order to promote its native folding (240, 241). In addition, RHAU interacts with many mRNAs that contain G4 motifs (241). G4 recognition is linked to an N-terminal accessory domain of RHAU, but the mechanism of disruption is unknown (240–242). The second helicase, RHA, is proposed to unwind DNA and RNA G4s during transcription and can resolve other non-canonical structures (243). It remains to be explored whether G4-unwinding activity is common among RNA helicases and how many of them disrupt G4s physiologically.
ACKNOWLEDGMENTS
Even with the generous length and reference limits, it is not possible to cite or discuss all of the excellent work in this field, and we would like to apologize to all of our colleagues whose work was omitted. Research on RNA folding and chaperones in the Russell lab is supported by grants from NIGMS (GM070456) and the Welch Foundation (F-1563).
DEFINITIONS
- RNA chaperone
a protein that interacts with RNA to promote RNA folding and/or conformational rearrangements without remaining bound to the folded or rearranged RNA.
- Pre-initiation complex (PIC)
a complex consisting of the 40S ribosome, eIF2-GTP with initiator tRNA and eIFs 1, 1A, 3 and 5.
- Exosome
a barrel-shaped multiprotein complex that functions in RNA processing and decay in eukaryotes and Archaea.
Footnotes
RELATED RESOURCES
RNA Helicase Database: http://www.rnahelicase.org/.
Contributor Information
Inga Jarmoskaite, Email: i_jarmoskaite@utexas.edu.
Rick Russell, Email: rick_russell@cm.utexas.edu.
LITERATURE CITED
- 1.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sigler PB. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 5.Karpel RL, Swistel DG, Miller NS, Geroch ME, Lu C, Fresco JR. Acceleration of RNA renaturation by nucleic acid unwinding proteins. Brookhaven Symp Biol. 1975:165–174. [PubMed] [Google Scholar]
- 6.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 7.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch M, Konrat R, Bläsi U, Schroeder R. RNA chaperones, RNA annealers and RNA helicases. RNA Biol. 2007;4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 9.Pan C, Russell R. Roles of DEAD-box proteins in RNA and RNP Folding. RNA Biol. 2010;7:667–676. doi: 10.4161/rna.7.6.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jankowsky E. RNA helicases at work: binding and rearranging. Trends Biochem Sci. 2011;36:19–29. doi: 10.1016/j.tibs.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12:505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 12.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Current Opinion in Structural Biology. 1993;3:419–429. [Google Scholar]
- 13.Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 15.Cordin O, Banroques J, Tanner N, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 16.Hilbert M, Karow AR, Klostermeier D. The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem. 2009;390:1237–1250. doi: 10.1515/BC.2009.135. [DOI] [PubMed] [Google Scholar]
- 17.Caruthers JM, Johnson ER, McKay DB. Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci U S A. 2000;97:13080–13085. doi: 10.1073/pnas.97.24.13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Andersen C, Ballut L, Johansen J, Chamieh H, Nielsen K, Oliveira C, Pedersen J, Séraphin B, Le Hir H, Andersen G. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 20.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]
- 21.Pang PS, Jankowsky E, Planet PJ, Pyle AM. The hepatitis C viral NS3 protein is a processive DNA helicase with cofactor enhanced RNA unwinding. EMBO J. 2002;21:1168–1176. doi: 10.1093/emboj/21.5.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serebrov V, Pyle AM. Periodic cycles of RNA unwinding and pausing by hepatitis C virus NS3 helicase. Nature. 2004;430:476–480. doi: 10.1038/nature02704. [DOI] [PubMed] [Google Scholar]
- 23.Büttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007;14:647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 24.Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, Carter L, Naismith JH, White MF. Structure of the DNA repair helicase Hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008;283:5118–5126. doi: 10.1074/jbc.M707548200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Lührmann R, Wahl MC. Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell. 2009;35:454–466. doi: 10.1016/j.molcel.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009;16:731–739. doi: 10.1038/nsmb.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson RN, Klauer AA, Hintze BJ, Robinson H, van Hoof A, Johnson SJ. The crystal structure of Mtr4 reveals a novel arch domain required for rRNA processing. EMBO J. 2010;29:2205–2216. doi: 10.1038/emboj.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weir JR, Bonneau F, Hentschel J, Conti E. Structural analysis reveals the characteristic features of Mtr4, a DExH helicase involved in nuclear RNA processing and surveillance. Proc Natl Acad Sci U S A. 2010;107:12139–12144. doi: 10.1073/pnas.1004953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halbach F, Rode M, Conti E. The crystal structure of S. cerevisiae Ski2, a DExH helicase associated with the cytoplasmic functions of the exosome. RNA. 2012;18:124–134. doi: 10.1261/rna.029553.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He Y, Andersen GR, Nielsen KH. Structural basis for the function of DEAH helicases. EMBO Rep. 2010;11:180–186. doi: 10.1038/embor.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J. 2010;29:2194–2204. doi: 10.1038/emboj.2010.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson SJ, Jackson RN. Ski2-like RNA helicase structures: common themes and complex assemblies. RNA Biol. 2013;10:33–43. doi: 10.4161/rna.22101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernstein J, Patterson DN, Wilson GM, Toth EA. Characterization of the essential activities of Saccharomyces cerevisiae Mtr4p, a 3'->5' helicase partner of the nuclear exosome. J Biol Chem. 2008;283:4930–4942. doi: 10.1074/jbc.M706677200. [DOI] [PubMed] [Google Scholar]
- 34.Jia H, Wang X, Anderson JT, Jankowsky E. RNA unwinding by the Trf4/Air2/Mtr4 polyadenylation (TRAMP) complex. Proc Natl Acad Sci U S A. 2012;109:7292–7297. doi: 10.1073/pnas.1201085109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwer B. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell. 2008;30:743–754. doi: 10.1016/j.molcel.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 37.Rogers GW, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 38.Rogers GJ, Lima W, Merrick W. Further characterization of the helicase activity of eIF4A. Substrate specificity. J Biol Chem. 2001;276:12598–12608. doi: 10.1074/jbc.M007560200. [DOI] [PubMed] [Google Scholar]
- 39.Bizebard T, Ferlenghi I, Iost I, Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- 40.Yang Q, Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]
- 41.Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz A. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol. 2007;365:835–855. doi: 10.1016/j.jmb.2006.09.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Q, Del Campo M, Lambowitz AM, Jankowsky E. DEAD-box proteins unwind duplexes by local strand separation. Mol Cell. 2007;28:253–263. doi: 10.1016/j.molcel.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Chen Y, Potratz J, Tijerina P, Del Campo M, Lambowitz A, Russell R. DEAD-box proteins can completely separate an RNA duplex using a single ATP. Proc Natl Acad Sci U S A. 2008;105:20203–20208. doi: 10.1073/pnas.0811075106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henn A, Cao W, Licciardello N, Heitkamp SE, Hackney DD, De La Cruz EM. Pathway of ATP utilization and duplex rRNA unwinding by the DEAD-box helicase, DbpA. Proc Natl Acad Sci U S A. 2010;107:4046–4050. doi: 10.1073/pnas.0913081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao W, Coman MM, Ding S, Henn A, Middleton ER, Bradley MJ, Rhoades E, Hackney DD, Pyle AM, De La Cruz EM. Mechanism of Mss116 ATPase reveals functional diversity of DEAD-Box proteins. J Mol Biol. 2011;409:399–414. doi: 10.1016/j.jmb.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci U S A. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Henn A, Bradley MJ, De La Cruz EM. ATP utilization and RNA conformational rearrangement by DEAD-Box proteins. Annu Rev Biophys. 2012;41:247–267. doi: 10.1146/annurev-biophys-050511-102243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell R, Jarmoskaite I, Lambowitz AM. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol. 2012;10:44–55. doi: 10.4161/rna.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Story RM, Li H, Abelson JN. Crystal structure of a DEAD box protein from the hyperthermophile Methanococcus jannaschii. Proc Natl Acad Sci U S A. 2001;98:1465–1470. doi: 10.1073/pnas.98.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci U S A. 2004;101:17628–17633. doi: 10.1073/pnas.0408172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng Z, Coller J, Parker R, Song H. Crystal structure and functional analysis of DEAD-box protein Dhh1p. RNA. 2005;11:1258–1270. doi: 10.1261/rna.2920905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 53.Collins R, Karlberg T, Lehtiö L, Schütz P, van den Berg S, Dahlgren LG, Hammarström M, Weigelt J, Schüler H. The DEXD/H-box RNA helicase DDX19 is regulated by an α-helical switch. J Biol Chem. 2009;284:10296–10300. doi: 10.1074/jbc.C900018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Del Campo M, Lambowitz A. Structure of the Yeast DEAD box protein Mss116p reveals two wedges that crimp RNA. Mol Cell. 2009;35:598–609. doi: 10.1016/j.molcel.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat Struct Mol Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 56.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1. A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 57.Iost I, Dreyfus M, Linder P. Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem. 1999;274:17677–17683. doi: 10.1074/jbc.274.25.17677. [DOI] [PubMed] [Google Scholar]
- 58.Polach KJ, Uhlenbeck OC. Cooperative binding of ATP and RNA substrates to the DEAD/H protein DbpA. Biochemistry. 2002;41:3693–3702. doi: 10.1021/bi012062n. [DOI] [PubMed] [Google Scholar]
- 59.Cordin O, Tanner NK, Doère M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Banroques J, Cordin O, Doère M, Linder P, Tanner NK. A conserved phenylalanine of motif IV in superfamily 2 helicases is required for cooperative, ATP-dependent binding of RNA substrates in DEAD-box proteins. Mol Cell Biol. 2008;28:3359–3371. doi: 10.1128/MCB.01555-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Theissen B, Karow A, Köhler J, Gubaev A, Klostermeier D. Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc Natl Acad Sci U S A. 2008;105:548–553. doi: 10.1073/pnas.0705488105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mallam AL, Del Campo M, Gilman B, Sidote DJ, Lambowitz AM. Structural basis for RNA-duplex recognition and unwinding by the DEAD-box helicase Mss116p. Nature. 2012;490:121–125. doi: 10.1038/nature11402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benz J, Trachsel H, Baumann U. Crystal structure of the ATPase domain of translation initiation factor 4A from Saccharomyces cerevisiae--the prototype of the DEAD box protein family. Structure. 1999;7:671–679. doi: 10.1016/s0969-2126(99)80088-4. [DOI] [PubMed] [Google Scholar]
- 64.Rudolph MG, Heissmann R, Wittmann JG, Klostermeier D. Crystal structure and nucleotide binding of the Thermus thermophilus RNA helicase Hera N-terminal domain. J Mol Biol. 2006;361:731–743. doi: 10.1016/j.jmb.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 65.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc Natl Acad Sci U S A. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schütz P, Karlberg T, van den Berg S, Collins R, Lehtiö L, Högbom M, Holmberg-Schiavone L, Tempel W, Park HW, Hammarström M, Moche M, Thorsell AG, Schüler H. Comparative structural analysis of human DEAD-box RNA helicases. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Karow AR, Klostermeier D. A conformational change in the helicase core is necessary but not sufficient for RNA unwinding by the DEAD box helicase YxiN. Nucleic Acids Res. 2009;37:4464–4471. doi: 10.1093/nar/gkp397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rössler OG, Straka A, Stahl H. Rearrangement of structured RNA via branch migration structures catalysed by the highly related DEAD-box proteins p68 and p72. Nucleic Acids Res. 2001;29:2088–2096. doi: 10.1093/nar/29.10.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 70.Uhlmann-Schiffler H, Jalal C, Stahl H. Ddx42p--a human DEAD box protein with RNA chaperone activities. Nucleic Acids Res. 2006;34:10–22. doi: 10.1093/nar/gkj403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia I, Albring MJ, Uhlenbeck OC. Duplex destabilization by four ribosomal DEAD-box proteins. Biochemistry. 2012;51:10109–10118. doi: 10.1021/bi301172s. [DOI] [PubMed] [Google Scholar]
- 72.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang Y, Liu ZR. The ATPase, RNA unwinding, and RNA binding activities of recombinant p68 RNA helicase. J Biol Chem. 2002;277:12810–12815. doi: 10.1074/jbc.M200182200. [DOI] [PubMed] [Google Scholar]
- 74.Rocak S, Emery B, Tanner NK, Linder P. Characterization of the ATPase and unwinding activities of the yeast DEAD-box protein Has1p and the analysis of the roles of the conserved motifs. Nucleic Acids Res. 2005;33:999–1009. doi: 10.1093/nar/gki244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jarmoskaite I, Russell R. DEAD-box proteins as RNA helicases and chaperones. WIREs RNA. 2011;2:135–152. doi: 10.1002/wrna.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tijerina P, Bhaskaran H, Russell R. Nonspecific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD-box RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:16698–16703. doi: 10.1073/pnas.0603127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grohman J, Del Campo M, Bhaskaran H, Tijerina P, Lambowitz A, Russell R. Probing the mechanisms of DEAD-box proteins as general RNA chaperones: the C-terminal domain of CYT-19 mediates general recognition of RNA. Biochemistry. 2007;46:3013–3022. doi: 10.1021/bi0619472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mallam AL, Jarmoskaite I, Tijerina P, Del Campo M, Seifert S, Guo L, Russell R, Lambowitz AM. Solution structures of DEAD-box RNA chaperones reveal conformational changes and nucleic acid tethering by a basic tail. Proc Natl Acad Sci U S A. 2011;108:12254–12259. doi: 10.1073/pnas.1109566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steimer L, Wurm JP, Linden MH, Rudolph MG, Wöhnert J, Klostermeier D. Recognition of two distinct elements in the RNA substrate by the RNA-binding domain of the T. thermophilus DEAD box helicase Hera. Nucleic Acids Res. 2013;41:6259–6272. doi: 10.1093/nar/gkt323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Klostermeier D. Rearranging RNA structures at 75 °C? Toward the molecular mechanism and physiological function of the Thermus thermophilus DEAD-box helicase Hera. Biopolymers. 2013;99:1137–1146. doi: 10.1002/bip.22316. [DOI] [PubMed] [Google Scholar]
- 81.Kossen K, Karginov FV, Uhlenbeck OC. The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA. J Mol Biol. 2002;324:625–636. doi: 10.1016/s0022-2836(02)01140-3. [DOI] [PubMed] [Google Scholar]
- 82.Hardin JW, Hu YX, McKay DB. Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif. J Mol Biol. 2010;402:412–427. doi: 10.1016/j.jmb.2010.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–548. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Andreou AZ, Klostermeier D. The DEAD-box helicase eIF4A: paradigm or the odd one out? RNA Biol. 2013;10:19–32. doi: 10.4161/rna.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 86.Fairman M, Maroney P, Wang W, Bowers H, Gollnick P, Nilsen T, Jankowsky E. Protein displacement by DExH/D "RNA helicases" without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 87.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006;34:4181–4188. doi: 10.1093/nar/gkl410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bowers HA, Maroney PA, Fairman ME, Kastner B, Lührmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strunk BS, Karbstein K. Powering through ribosome assembly. RNA. 2009;15:2083–2104. doi: 10.1261/rna.1792109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shajani Z, Sykes MT, Williamson JR. Assembly of bacterial ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 91.Jagessar KL, Jain C. Functional and molecular analysis of Escherichia coli strains lacking multiple DEAD-box helicases. RNA. 2010;16:1386–1392. doi: 10.1261/rna.2015610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lehnik-Habrink M, Rempeters L, Kovács Á, Wrede C, Baierlein C, Krebber H, Kuipers OP, Stülke J. DEAD-Box RNA helicases in Bacillus subtilis have multiple functions and act independently from each other. J Bacteriol. 2013;195:534–544. doi: 10.1128/JB.01475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fuller-Pace FV, Nicol SM, Reid AD, Lane DP. DbpA: a DEAD box protein specifically activated by 23s rRNA. EMBO J. 1993;12:3619–3626. doi: 10.1002/j.1460-2075.1993.tb06035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tsu C, Kossen K, Uhlenbeck O. The Escherichia coli DEAD protein DbpA recognizes a small RNA hairpin in 23S rRNA. RNA. 2001;7:702–709. doi: 10.1017/s1355838201010135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Diges C, Uhlenbeck O. Escherichia coli DbpA is an RNA helicase that requires hairpin 92 of 23S rRNA. EMBO J. 2001;20:5503–5512. doi: 10.1093/emboj/20.19.5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang S, Hu Y, Overgaard MT, Karginov FV, Uhlenbeck OC, McKay DB. The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold. RNA. 2006;12:959–967. doi: 10.1261/rna.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Elles LM, Uhlenbeck OC. Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype. Nucleic Acids Res. 2008;36:41–50. doi: 10.1093/nar/gkm926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sharpe Elles L, Sykes M, Williamson J, Uhlenbeck O. A dominant negative mutant of the E. coli RNA helicase DbpA blocks assembly of the 50S ribosomal subunit. Nucleic Acids Res. 2009;37:6503–6514. doi: 10.1093/nar/gkp711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol Microbiol. 2003;48:1253–1265. doi: 10.1046/j.1365-2958.2003.03513.x. [DOI] [PubMed] [Google Scholar]
- 100.Trubetskoy D, Proux F, Allemand F, Dreyfus M, Iost I. SrmB, a DEAD-box helicase involved in Escherichia coli ribosome assembly, is specifically targeted to 23S rRNA in vivo. Nucleic Acids Res. 2009;37:6540–6549. doi: 10.1093/nar/gkp685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Proux F, Dreyfus M, Iost I. Identification of the sites of action of SrmB, a DEAD-box RNA helicase involved in Escherichia coli ribosome assembly. Mol Microbiol. 2011;82:300–311. doi: 10.1111/j.1365-2958.2011.07779.x. [DOI] [PubMed] [Google Scholar]
- 102.Jones PG, Mitta M, Kim Y, Jiang W, Inouye M. Cold shock induces a major ribosomal-associated protein that unwinds double-stranded RNA in Escherichia coli. Proc Natl Acad Sci U S A. 1996;93:76–80. doi: 10.1073/pnas.93.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iost I, Bizebard T, Dreyfus M. Functions of DEAD-box proteins in bacteria: current knowledge and pending questions. Biochim Biophys Acta. 2013;1829:866–877. doi: 10.1016/j.bbagrm.2013.01.012. [DOI] [PubMed] [Google Scholar]
- 104.Charollais J, Dreyfus M, Iost I. CsdA, a cold-shock RNA helicase from Escherichia coli is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 2004;32:2751–2759. doi: 10.1093/nar/gkh603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 106.Awano N, Xu C, Ke H, Inoue K, Inouye M, Phadtare S. Complementation analysis of the cold-sensitive phenotype of the Escherichia coli csdA deletion strain. J Bacteriol. 2007;189:5808–5815. doi: 10.1128/JB.00655-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Phadtare S. Unwinding activity of cold shock proteins and RNA metabolism. RNA Biol. 2011;8:394–397. doi: 10.4161/rna.8.3.14823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jain C. The E. coli RhlE RNA helicase regulates the function of related RNA helicases during ribosome assembly. RNA. 2008;14:381–389. doi: 10.1261/rna.800308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Martin R, Straub AU, Doebele C, Bohnsack MT. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol. 2013;10:4–18. doi: 10.4161/rna.21879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Weaver PL, Sun C, Chang TH. Dbp3p, a putative RNA helicase in Saccharomyces cerevisiae, is required for efficient pre-rRNA processing predominantly at site A3. Mol Cell Biol. 1997;17:1354–1365. doi: 10.1128/mcb.17.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem. 2009;284:35079–35091. doi: 10.1074/jbc.M109.040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, Caizergues-Ferrer M, Grigoriev M, Henry Y. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J. 2009;28:3808–3819. doi: 10.1038/emboj.2009.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3' end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lamanna AC, Karbstein K. An RNA conformational switch regulates pre-18S rRNA cleavage. J Mol Biol. 2011;405:3–17. doi: 10.1016/j.jmb.2010.09.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Young CL, Khoshnevis S, Karbstein K. Cofactor-dependent specificity of a DEAD-box protein. Proc Natl Acad Sci U S A. 2013;110:E2668–E2676. doi: 10.1073/pnas.1302577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA. 2012;3:397–414. doi: 10.1002/wrna.117. [DOI] [PubMed] [Google Scholar]
- 118.Liang XH, Fournier MJ. The helicase Has1p is required for snoRNA release from pre-rRNA. Mol Cell Biol. 2006;26:7437–7450. doi: 10.1128/MCB.00664-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bohnsack MT, Kos M, Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep. 2008;9:1230–1236. doi: 10.1038/embor.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kos M, Tollervey D. The Putative RNA Helicase Dbp4p Is Required for Release of the U14 snoRNA from Preribosomes in Saccharomyces cerevisiae. Mol Cell. 2005;20:53–64. doi: 10.1016/j.molcel.2005.08.022. [DOI] [PubMed] [Google Scholar]
- 121.Granneman S, Bernstein KA, Bleichert F, Baserga SJ. Comprehensive mutational analysis of yeast DEXD/H box RNA helicases required for small ribosomal subunit synthesis. Mol Cell Biol. 2006;26:1183–1194. doi: 10.1128/MCB.26.4.1183-1194.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Colley A, Beggs JD, Tollervey D, Lafontaine DL. Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol Cell Biol. 2000;20:7238–7246. doi: 10.1128/mcb.20.19.7238-7246.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase Prp43p is a component of multiple preribosomal particles. Mol Cell Biol. 2005;25:9269–9282. doi: 10.1128/MCB.25.21.9269-9282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Combs DJ, Nagel RJ, Ares M, Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol. 2006;26:523–534. doi: 10.1128/MCB.26.2.523-534.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol. 2006;26:513–522. doi: 10.1128/MCB.26.2.513-522.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tanaka N, Schwer B. Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry. 2006;45:6510–6521. doi: 10.1021/bi052656g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev. 2007;21:2312–2325. doi: 10.1101/gad.1580507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Guenther UP, Jankowsky E. Helicase multitasking in ribosome assembly. Mol Cell. 2009;36:537–538. doi: 10.1016/j.molcel.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 129.Cordin O, Beggs JD. RNA helicases in splicing. RNA Biol. 2013;10:83–95. doi: 10.4161/rna.22547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang TH, Tung L, Yeh FL, Chen JH, Chang SL. Functions of the DExD/H-box proteins in nuclear pre-mRNA splicing. Biochim Biophys Acta. 2013;1829:764–774. doi: 10.1016/j.bbagrm.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 131.Semlow DR, Staley JP. Staying on message: ensuring fidelity in pre-mRNA splicing. Trends Biochem Sci. 2012;37:263–273. doi: 10.1016/j.tibs.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 133.Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15:42–49. doi: 10.1101/gad.851301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Shen H, Zheng X, Shen J, Zhang L, Zhao R, Green MR. Distinct activities of the DExD/H-box splicing factor hUAP56 facilitate stepwise assembly of the spliceosome. Genes Dev. 2008;22:1796–1803. doi: 10.1101/gad.1657308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.O'Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- 136.Perriman R, Barta I, Voeltz GK, Abelson J, Ares M. ATP requirement for Prp5p function is determined by Cus2p and the structure of U2 small nuclear RNA. Proc Natl Acad Sci U S A. 2003;100:13857–13862. doi: 10.1073/pnas.2036312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Perriman RJ, Ares M. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev. 2007;21:811–820. doi: 10.1101/gad.1524307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Abu Dayyeh BK, Quan TK, Castro M, Ruby SW. Probing interactions between the U2 small nuclear ribonucleoprotein and the DEAD-box protein, Prp5. J Biol Chem. 2002;277:20221–20233. doi: 10.1074/jbc.M109553200. [DOI] [PubMed] [Google Scholar]
- 139.Perriman R, Ares M. Invariant U2 snRNA nucleotides form a stem loop to recognize the intron early in splicing. Mol Cell. 2010;38:416–427. doi: 10.1016/j.molcel.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Staley JP, Guthrie C. An RNA switch at the 5' splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- 141.Strauss EJ, Guthrie C. Prp28, a 'DEAD-box' protein, is required for the first step of mRNA splicing in vitro. Nucleic Acids Res. 1994;22:3187–3193. doi: 10.1093/nar/22.15.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol. 2009;16:1237–1243. doi: 10.1038/nsmb.1729. [DOI] [PubMed] [Google Scholar]
- 143.Lardelli RM, Thompson JX, Yates JR, Stevens SW. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA. 2010;16:516–528. doi: 10.1261/rna.2030510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Liu HL, Cheng SC. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol. 2012;32:5056–5066. doi: 10.1128/MCB.01109-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ohrt T, Prior M, Dannenberg J, Odenwälder P, Dybkov O, Rasche N, Schmitzová J, Gregor I, Fabrizio P, Enderlein J, Lührmann R. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA. 2012;18:1244–1256. doi: 10.1261/rna.033316.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ohrt T, Odenwälder P, Dannenberg J, Prior M, Warkocki Z, Schmitzová J, Karaduman R, Gregor I, Enderlein J, Fabrizio P, Lührmann R. Molecular dissection of step 2 catalysis of yeast pre-mRNA splicing investigated in a purified system. RNA. 2013;19:902–915. doi: 10.1261/rna.039024.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Mefford MA, Staley JP. Evidence that U2/U6 helix I promotes both catalytic steps of pre-mRNA splicing and rearranges in between these steps. RNA. 2009;15:1386–1397. doi: 10.1261/rna.1582609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hilliker AK, Mefford MA, Staley JP. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev. 2007;21:821–834. doi: 10.1101/gad.1536107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Tseng CK, Liu HL, Cheng SC. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA. 2011;17:145–154. doi: 10.1261/rna.2459611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Schwer B, Gross CH. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 1998;17:2086–2094. doi: 10.1093/emboj/17.7.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tanaka N, Schwer B. Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry. 2005;44:9795–9803. doi: 10.1021/bi050407m. [DOI] [PubMed] [Google Scholar]
- 153.Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci U S A. 1997;94:11798–11802. doi: 10.1073/pnas.94.22.11798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem. 2002;277:17743–17750. doi: 10.1074/jbc.M200762200. [DOI] [PubMed] [Google Scholar]
- 155.Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 2005;19:2991–3003. doi: 10.1101/gad.1377405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Laggerbauer B, Achsel T, Lührmann R. The human U5-200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci U S A. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- 158.Mozaffari-Jovin S, Santos KF, Hsiao HH, Will CL, Urlaub H, Wahl MC, Lührmann R. The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26:2422–2434. doi: 10.1101/gad.200949.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hahn D, Kudla G, Tollervey D, Beggs JD. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26:2408–2421. doi: 10.1101/gad.199307.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Nielsen KH, Staley JP. Spliceosome activation: U4 is the path, stem I is the goal, and Prp8 is the keeper. Let's cheer for the ATPase Brr2! Genes Dev. 2012;26:2461–2467. doi: 10.1101/gad.207514.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zaher HS, Green R. Fidelity at the molecular level: lessons from protein synthesis. Cell. 2009;136:746–762. doi: 10.1016/j.cell.2009.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Hopfield JJ. Kinetic proofreading: a new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc Natl Acad Sci U S A. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ninio J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- 164.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xu YZ, Query CC. Competition between the ATPase Prp5 and branch region-U2 snRNA pairing modulates the fidelity of spliceosome assembly. Mol Cell. 2007;28:838–849. doi: 10.1016/j.molcel.2007.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang F, Wang XY, Zhang ZM, Pu J, Fan YJ, Zhou J, Query CC, Xu YZ. Splicing proofreading at 5' splice sites by ATPase Prp28p. Nucleic Acids Res. 2013;41:4660–4670. doi: 10.1093/nar/gkt149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Burgess S, Couto JR, Guthrie C. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell. 1990;60:705–717. doi: 10.1016/0092-8674(90)90086-t. [DOI] [PubMed] [Google Scholar]
- 168.Konarska MM, Query CC. Insights into the mechanisms of splicing: more lessons from the ribosome. Genes Dev. 2005;19:2255–2260. doi: 10.1101/gad.1363105. [DOI] [PubMed] [Google Scholar]
- 169.Villa T, Guthrie C. The Isy1p component of the NineTeen complex interacts with the ATPase Prp16p to regulate the fidelity of pre-mRNA splicing. Genes Dev. 2005;19:1894–1904. doi: 10.1101/gad.1336305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Mayas RM, Maita H, Staley JP. Exon ligation is proofread by the DExD/H-box ATPase Prp22p. Nat Struct Mol Biol. 2006;13:482–490. doi: 10.1038/nsmb1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc Natl Acad Sci U S A. 2010;107:10020–10025. doi: 10.1073/pnas.0906022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Koodathingal P, Novak T, Piccirilli JA, Staley JP. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5' splice site cleavage during pre-mRNA splicing. Mol Cell. 2010;39:385–395. doi: 10.1016/j.molcel.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lambowitz AM, Zimmerly S. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol. 2011;3:a003616. doi: 10.1101/cshperspect.a003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mohr S, Stryker J, Lambowitz A. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 175.Séraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 176.Huang H, Rowe C, Mohr S, Jiang Y, Lambowitz A, Perlman P. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci U S A. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Köhler D, Schmidt-Gattung S, Binder S. The DEAD-box protein PMH2 is required for efficient group II intron splicing in mitochondria of Arabidopsis thaliana. Plant Mol Biol. 2010;72:459–467. doi: 10.1007/s11103-009-9584-9. [DOI] [PubMed] [Google Scholar]
- 178.Asakura Y, Galarneau E, Watkins KP, Barkan A, van Wijk KJ. Chloroplast RH3 DEAD box RNA helicases in maize and Arabidopsis function in splicing of specific group II introns and affect chloroplast ribosome biogenesis. Plant Physiol. 2012;159:961–974. doi: 10.1104/pp.112.197525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Russell R, Millett I, Doniach S, Herschlag D. Small angle X-ray scattering reveals a compact intermediate in RNA folding. Nat Struct Biol. 2000;7:367–370. doi: 10.1038/75132. [DOI] [PubMed] [Google Scholar]
- 180.Russell R, Das R, Suh H, Travers K, Laederach A, Engelhardt M, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 181.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 182.Sinan S, Yuan X, Russell R. The Azoarcus group I intron ribozyme misfolds and is accelerated for refolding by ATP-dependent RNA chaperone proteins. J Biol Chem. 2011 doi: 10.1074/jbc.M111.287706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Del Campo M, Tijerina P, Bhaskaran H, Mohr S, Yang Q, Jankowsky E, Russell R, Lambowitz A. Do DEAD-box proteins promote group II intron splicing without unwinding RNA? Mol Cell. 2007;28:159–166. doi: 10.1016/j.molcel.2007.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Del Campo M, Mohr S, Jiang Y, Jia H, Jankowsky E, Lambowitz A. Unwinding by local strand separation is critical for the function of DEAD-box proteins as RNA chaperones. J Mol Biol. 2009;389:674–693. doi: 10.1016/j.jmb.2009.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bifano A, Caprara M. A DExH/D-box protein coordinates the two steps of splicing in a group I intron. J Mol Biol. 2008;383:667–682. doi: 10.1016/j.jmb.2008.08.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Russell R, Jarmoskaite I, Lambowitz AM. Toward a molecular understanding of RNA remodeling by DEAD-box proteins. RNA Biol. 2013;10:44–55. doi: 10.4161/rna.22210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mohr S, Matsuura M, Perlman P, Lambowitz A. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci U S A. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Solem A, Zingler N, Pyle A. A DEAD protein that activates intron self-splicing without unwinding RNA. Mol Cell. 2006;24:611–617. doi: 10.1016/j.molcel.2006.10.032. [DOI] [PubMed] [Google Scholar]
- 189.Fedorova O, Solem A, Pyle AM. Protein-facilitated folding of group II intron ribozymes. J Mol Biol. 2010;397:799–813. doi: 10.1016/j.jmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Potratz JP, Del Campo M, Wolf RZ, Lambowitz AM, Russell R. ATP-dependent roles of the DEAD-box protein Mss116p in group II intron splicing in vitro and in vivo. J Mol Biol. 2011;411:661–679. doi: 10.1016/j.jmb.2011.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Zingler N, Solem A, Pyle AM. Dual roles for the Mss116 cofactor during splicing of the ai5γ group II intron. Nucleic Acids Res. 2010;38:6602–6609. doi: 10.1093/nar/gkq530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Karunatilaka KS, Solem A, Pyle AM, Rueda D. Single-molecule analysis of Mss116-mediated group II intron folding. Nature. 2010;467:935–939. doi: 10.1038/nature09422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Potratz JP, Russell R. RNA catalysis as a probe for chaperone activity of DEAD-box helicases. Methods Enzymol. 2012;511:111–130. doi: 10.1016/B978-0-12-396546-2.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Waldsich C, Pyle AM. A kinetic intermediate that regulates proper folding of a group II intron RNA. J Mol Biol. 2008;375:572–580. doi: 10.1016/j.jmb.2007.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol. 2012;19:568–576. doi: 10.1038/nsmb.2303. [DOI] [PubMed] [Google Scholar]
- 196.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5' secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Rogers GW, Richter NJ, Lima WF, Merrick WC. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem. 2001;276:30914–30922. doi: 10.1074/jbc.M100157200. [DOI] [PubMed] [Google Scholar]
- 198.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 2011;39:2260–2270. doi: 10.1093/nar/gkq1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5'-overhangs. J Biol Chem. 2012;287:20301–20312. doi: 10.1074/jbc.M112.347278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Sun Y, Atas E, Lindqvist L, Sonenberg N, Pelletier J, Meller A. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucleic Acids Res. 2012;40:6199–6207. doi: 10.1093/nar/gks278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lindqvist L, Imataka H, Pelletier J. Cap-dependent eukaryotic initiation factor-mRNA interactions probed by cross-linking. RNA. 2008;14:960–969. doi: 10.1261/rna.971208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.von der Haar T, McCarthy JE. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 205.Kolupaeva VG, Lomakin IB, Pestova TV, Hellen CU. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosomal entry site. Mol Cell Biol. 2003;23:687–698. doi: 10.1128/MCB.23.2.687-698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.de Breyne S, Yu Y, Unbehaun A, Pestova TV, Hellen CU. Direct functional interaction of initiation factor eIF4G with type 1 internal ribosomal entry sites. Proc Natl Acad Sci U S A. 2009;106:9197–9202. doi: 10.1073/pnas.0900153106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hilliker A, Gao Z, Jankowsky E, Parker R. The DEAD-box protein Ded1 modulates translation by the formation and resolution of an eIF4F-mRNA complex. Mol Cell. 2011;43:962–972. doi: 10.1016/j.molcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Abaeva IS, Marintchev A, Pisareva VP, Hellen CU, Pestova TV. Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning. EMBO J. 2011;30:115–129. doi: 10.1038/emboj.2010.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hartman TR, Qian S, Bolinger C, Fernandez S, Schoenberg DR, Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nat Struct Mol Biol. 2006;13:509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 211.Ranji A, Shkriabai N, Kvaratskhelia M, Musier-Forsyth K, Boris-Lawrie K. Features of double-stranded RNA-binding domains of RNA helicase A are necessary for selective recognition and translation of complex mRNAs. J Biol Chem. 2011;286:5328–5337. doi: 10.1074/jbc.M110.176339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lasko P. The DEAD-box helicase Vasa: evidence for a multiplicity of functions in RNA processes and developmental biology. Biochim Biophys Acta. 2013;1829:810–816. doi: 10.1016/j.bbagrm.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 213.Carrera P, Johnstone O, Nakamura A, Casanova J, Jäckle H, Lasko P. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell. 2000;5:181–187. doi: 10.1016/s1097-2765(00)80414-1. [DOI] [PubMed] [Google Scholar]
- 214.Johnstone O, Lasko P. Interaction with eIF5B is essential for Vasa function during development. Development. 2004;131:4167–4178. doi: 10.1242/dev.01286. [DOI] [PubMed] [Google Scholar]
- 215.Daugeron MC, Prouteau M, Lacroute F, Séraphin B. The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1. Nucleic Acids Res. 2011;39:2221–2233. doi: 10.1093/nar/gkq898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli J, Seedorf M, Cole C, Krebber H. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 217.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tieg B, Krebber H. Dbp5 - from nuclear export to translation. Biochim Biophys Acta. 2013;1829:791–798. doi: 10.1016/j.bbagrm.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 219.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 220.Qu X, Wen JD, Lancaster L, Noller HF, Bustamante C, Tinoco I. The ribosome uses two active mechanisms to unwind messenger RNA during translation. Nature. 2011;475:118–121. doi: 10.1038/nature10126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Hardwick SW, Luisi BF. Rarely at rest: RNA helicases and their busy contributions to RNA degradation, regulation and quality control. RNA Biol. 2013;10:56–70. doi: 10.4161/rna.22270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Py B, Higgins CF, Krisch HM, Carpousis AJ. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 223.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3' exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Schneider C, Tollervey D. Threading the barrel of the RNA exosome. Trends Biochem Sci. 2013;38:485–493. doi: 10.1016/j.tibs.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Halbach F, Reichelt P, Rode M, Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- 226.Jia H, Wang X, Liu F, Guenther UP, Srinivasan S, Anderson JT, Jankowsky E. The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell. 2011;145:890–901. doi: 10.1016/j.cell.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 228.Fuller-Pace FV. The DEAD box proteins DDX5 (p68) and DDX17 (p72): multi-tasking transcriptional regulators. Biochim Biophys Acta. 2013;1829:756–763. doi: 10.1016/j.bbagrm.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 229.Cloutier SC, Ma WK, Nguyen LT, Tran EJ. The DEAD-box RNA helicase Dbp2 connects RNA quality control with repression of aberrant transcription. J Biol Chem. 2012;287:26155–26166. doi: 10.1074/jbc.M112.383075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ma WK, Cloutier SC, Tran EJ. The DEAD-box Protein Dbp2 Functions with the RNA-Binding Protein Yra1 to Promote mRNP Assembly. J Mol Biol. 2013 doi: 10.1016/j.jmb.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lund MK, Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 232.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 233.Taniguchi I, Ohno M. ATP-dependent recruitment of export factor Aly/REF onto intronless mRNAs by RNA helicase UAP56. Mol Cell Biol. 2008;28:601–608. doi: 10.1128/MCB.01341-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Strässer K, Masuda S, Mason P, Pfannstiel J, Oppizzi M, Rodriguez-Navarro S, RondÛn AG, Aguilera A, Struhl K, Reed R, Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 235.Fullam A, Schröder M. DExD/H-box RNA helicases as mediators of anti-viral innate immunity and essential host factors for viral replication. Biochim Biophys Acta. 2013;1829:854–865. doi: 10.1016/j.bbagrm.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Kruse E, Voigt C, Leeder WM, Göringer HU. RNA helicases involved in U-insertion/deletion-type RNA editing. Biochim Biophys Acta. 2013;1829:835–841. doi: 10.1016/j.bbagrm.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 237.Li F, Herrera J, Zhou S, Maslov DA, Simpson L. Trypanosome REH1 is an RNA helicase involved with the 3'–5' polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc Natl Acad Sci U S A. 2011;108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Hernandez A, Madina BR, Ro K, Wohlschlegel JA, Willard B, Kinter MT, Cruz-Reyes J. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J Biol Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Millevoi S, Moine H, Vagner S. G-quadruplexes in RNA biology. Wiley Interdiscip Rev RNA. 2012;3:495–507. doi: 10.1002/wrna.1113. [DOI] [PubMed] [Google Scholar]
- 240.Sexton AN, Collins K. The 5' guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol Cell Biol. 2011;31:736–743. doi: 10.1128/MCB.01033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Lattmann S, Stadler MB, Vaughn JP, Akman SA, Nagamine Y. The DEAH-box RNA helicase RHAU binds an intramolecular RNA G-quadruplex in TERC and associates with telomerase holoenzyme. Nucleic Acids Res. 2011;39:9390–9404. doi: 10.1093/nar/gkr630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RHAU-specific motif in the recognition and resolution of guanine quadruplex-RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Chakraborty P, Grosse F. Human DHX9 helicase preferentially unwinds RNA-containing displacement loops (R-loops) and G-quadruplexes. DNA Repair (Amst) 2011;10:654–665. doi: 10.1016/j.dnarep.2011.04.013. [DOI] [PubMed] [Google Scholar]