Abstract
The arthropod cuticle is a composite, bipartite system, made of chitin filaments embedded in a proteinaceous matrix. The physical properties of cuticle are determined by the structure and the interactions of its two major components, cuticular proteins (CPs) and chitin. The proteinaceous matrix consists mainly of structural cuticular proteins. The majority of the structural proteins that have been described to date belong to the CPR family, and they are identified by the conserved R&R region (Rebers and Riddiford Consensus). Two major subfamilies of the CPR family RR-1 and RR-2, have also been identified from conservation at sequence level and some correlation with the cuticle type. Recently, several novel families, also containing characteristic conserved regions, have been described. The package HMMER v3.0 [http://hmmer.janelia.org/] was used to build characteristic profile Hidden Markov Models based on the characteristic regions for 8 of these families, (CPF, CPAP3, CPAP1, CPCFC, CPLCA, CPLCG, CPLCW, Tweedle). In brief, these families can be described as having: CPF (a conserved region with 44 amino acids); CPAP1 and CPAP-3 (analogous to peritrophins, with 1 and 3 chitin-binding domains, respectively); CPCFC (2 or 3 C-x(5)-C repeats); and four of five low complexity (LC) families, each with characteristic domains. Using these models, as well as the models previously created for the two major subfamilies of the CPR family, RR-1 and RR-2 (Karouzou et al., 2007), we developed CutProtFam-Pred, an on-line tool (http://bioinformatics.biol.uoa.gr/CutProtFam-Pred) that allows one to query sequences from proteomes or translated transcriptomes, for the accurate detection and classification of putative structural cuticular proteins. The tool has been applied successfully to diverse arthropod proteomes including a crustacean (Daphnia pulex) and a chelicerate (Tetranychus urticae), but at this taxonomic distance only CPRs and CPAPs were recovered.
Keywords: arthropod cuticle, cuticular proteins, profile Hidden Markov Models (pHMMs), structural cuticular protein families
1. Introduction
The arthropod cuticle is a composite, bipartite system, made of chitin filaments embedded in a proteinaceous matrix, and acts as protection and as structural and mechanical support in arthropods (Neville, 1975). The physical properties of cuticle are determined by the structure and the interactions of its two major components, cuticular proteins (CPs) and chitin (Neville, 1993).
The proteinaceous matrix consists mainly of structural cuticular proteins (Willis, 2010; Willis et al., 2012). The majority of the structural cuticular proteins that have been discovered to date belong to the CPR family, and they are identified by the conserved Rebers and Riddiford (R&R) Consensus (Rebers and Riddiford, 1988). The original consensus was G-x(8)-G-x(6)-Y-x(2)-A-x-E-x-G-Y-x(7)-P-x-P and the modified PROSITE pattern (Sigrist et al., 2013) (PS00233) is G-x(7)-[DEN]-G-x(6)-[FY]-x-A-[DNG]-x(2,3)-G-[FY]-x-[APV]. Two subfamilies of the CPR family RR-1 and RR-2, have also been identified with further conservation at sequence level and some correlation with the cuticle type(Andersen, 1998; Andersen et al., 1997). A third, far smaller subfamily, RR-3, is less well defined, and no discriminating features could be identified (Andersen, 2000). Some proteins containing the RR-1 motif were found in soft (flexible) cuticles, while the proteins containing the RR-2 motif were found in hard (rigid) cuticles, but this distinction is not firmly established (Andersen, 2000). The “chitin_bind_4” profile (PF00379), included in Pfam database (Punta et al., 2012), identifies proteins that belong to the CPR family, but since it was based on both RR-1 and RR-2 sequences, it matches none of them particularly well. The cuticleDB webpage <http://bioinformatics.biol.uoa.gr/cuticleDB/hmmfind_form.jsp> (Karouzou et al., 2007) offers two distinct pHMMs, one for each subfamily (RR-1 and RR-2) of the CPR family proteins. These pHMMs are more accurate than the Pfam profile and are able to discriminate between RR-1 and RR-2 proteins, therefore, making the annotation of structural cuticular proteins more specific (Karouzou et al., 2007).
Several additional families of structural cuticular proteins have been described. Some of these families contain characteristic conserved regions. Willis (2010) and Willis et al. (2012) offer insights for all thirteen families and describe extensively each family’s features including the arthropod groups where family members have been identified. Members of all of these families have been identified either in proteins extracted from manually cleaned cuticles or from MS analyses of cuticles left behind after molting, thus confirming that family members are indeed authentic cuticular proteins (See Willis (2010) and Willis et al. (2012) for references).
The CPF consensus was first recognized as having 51 aa by Andersen et al. (1997) but examination of the sequence in additional species resulted in its reduction to 42–44 aa (Togawa et al., 2007). It now is: (A-[LIV]-x-[SA]-[QS]-x-[SQ]-x-[IV]- [LV]-R-S-x-G-[NG]-x(3)-V-S-x-Y-[ST]-K-[TA]-[VI]-D-[TS]-[PA]-[YF]-S-S-V-x-K-x-D-x-R-[VI]-[TS]-N-x-[GA]). Another feature of these CPF proteins is the similarity of their C-terminals (Andersen et al., 1997; Togawa et al., 2007). The CPFL family (CPF-like) members share a conserved C-terminal region similar to the one present in the CPF family, but lack the 44 amino acid residue defining region (Togawa et al., 2007).
The CPG family (Cuticular Proteins rich in Glycines) members have many repeats of GGGG and GGxGG motifs along their sequence (Futahashi et al., 2008).
Five low complexity families of structural cuticular proteins have been recognized: the Tweedle family, named after a mutant phenotype in Drosophila melanogaster that reminded the authors of Tweedledee from “Alice through the Looking-glass”, has a conserved region consisting of four conserved blocks in a continuous stretch of about 100 amino acid residues (Guan et al., 2006); the CPLCA family (Cuticular Proteins of Low-Complexity with Alanine residues) contains about 13–26 % alanine residues and has a conserved region that looks like the retinin domain (Cornman and Willis, 2009); the CPLCG family (Cuticular Proteins of Low-Complexity with conserved Glycine residues) has the conserved signature motif G-x(2)-H-x-A-P-x(2)-G-H that extends in a longer stretch of 35 amino acids (Cornman and Willis, 2009); the CPLCW family (Cuticular Proteins of Low-Complexity with invariant W residue) has an invariant tryptophan in a longer stretch of 29 amino acids and seems to be restricted to mosquitoes (Cornman and Willis, 2009). A final low complexity family, CPLCP, (Cuticular Proteins of Low-Complexity with Proline residues) contains a high density of PV and PY repeats (Cornman and Willis, 2009). While only a few of the 27 annotated have been detected in MS/MS analyses of Anopoheles cuticle (Cornman and Willis, 2009), several have been identified in the cuticle of Tribolium castaneum (Dittmer et al., 2012) and some, not yet named as such, in Bombyx mori (Fu et al., 2011).
The CPAP3 and the CPAP1 families (Cuticular Proteins Analogous to Peritrophins) contain three and one chitin-binding domains, respectively. Each chitin-binding domain contains 6 cysteine residues, assumed to form three disulfide bridges, and, in its general form, can be described by the Pfam Chitin-binding Peritrophin A domain (CBM_14 – PF01607, previously known as ChtBD2). The chitin-binding domains of these two families, which have been shown to be cuticular and not peritrophic membrane components, have distinct spacing of the cysteines (C-x(11–24)-C-x(5)-C-x(9–14)-C-x(12–16)-C-x(6–8)-C) within each chitin binding domain. For the CPAP3 members, the spacing between the three repeats of the domain is also specific (Jasrapuria et al., 2010).
The CPCFC family (Cuticular Proteins with 2 or 3 C-x(5)-C repeats) is the third family with conserved cysteines along the sequence and was first recognized in a protein from cuticle from Blaberus craniifer BCNCP1 (Jensen et al., 1997). Members contain three repeats of the C-x(5)-C motif, except for the moths and beetles in which the middle repeat is missing (Willis, 2010; Willis et al., 2012).
The Apidermin family is the last known family of structural cuticular proteins. Members of this family were first found in Apis mellifera (Kucharski et al., 2007). No sequence conservation was identified in this family and its members are recognized only by chromosomal linkage (Willis, 2010; Willis et al., 2012).
Some of the families are restricted to specific orders or even smaller groups, others like the CPRs appear in all arthropods, and as more genomes are sequenced, it will be of interest to learn more about their distribution. There are, of course, other non-enzymatic proteins that have been verified to be in arthropod cuticle that do not belong to any of these families. But the vast majority can be assigned to these families and identifying them in proteomes will facilitate annotation. Hence, a new, more complete, tool for their detection would be valuable. This paper describes the development of CutProtFam-Pred <http://bioinformatics.biol.uoa.gr/CutProtFam-Pred/>, a tool which allows the accurate detection and classification of putative structural cuticular proteins from sequence alone.
2. Methods
2.1 Data collection
In order to collect sequences belonging to one of the new families, an extensive literature search was conducted. In the case of the CPCFC family, unpublished data were also used. The protein sequences for all CPR family members were retrieved from CuticleDB <http://bioinformatics.biol.uoa.gr/cuticleDB/> (update: 20 Oct 2009) (Magkrioti et al., 2004). The full dataset of structural cuticular proteins consists of 1796 protein sequences; the distribution of sequences in their respective family and the source of data are listed in Table 1.
Table 1.
Dataset summary
| Family Type | Sequences | Source | |
|---|---|---|---|
| CPR | 1099 | 1099 | cuticleDB - Magkrioti et al., 2004 |
| Apidermin | 8 | 8 | Kucharski et al., 2007 |
| CPAP1 | 10 | 10 | Jasrapuria et al., 2010 |
| CPAP3 | 58 | 58 | Jasrapuria et al., 2010 |
| CPCFC | 33 | 11 | Willis, 2010 |
| 22 | unpublished data | ||
| CPF | 48 | 29 | Togawa et al., 2007 |
| 18 | Cornman, 2009 | ||
| 1 | Futahashi et al., 2008 | ||
| CPFL | 39 | 35 | Togawa et al., 2007 |
| 4 | Futahashi et al., 2008 | ||
| CPG | 18 | 18 | Futahashi et al., 2008 |
| CPLCA | 21 | 20 | Cornman and Willis, 2009 |
| 1 | Willis et al., 2012 | ||
| CPLCG | 104 | 86 | Cornman and Willis, 2009 |
| 18 | Cornman, 2009 | ||
| CPLCP | 93 | 93 | Cornman and Willis, 2009 |
| CPLCW | 27 | 27 | Cornman and Willis, 2009 |
| Tweedle | 234 | 68 | Cornman and Willis, 2009 |
| 162 | Cornman, 2009 | ||
| 4 | Willis, 2010 |
2.2 Selection and Preparation of Training Sets
The preferable sequences of each training set came from one species in order to avoid redundancy. When more than one species had more than three sequences available, all the possible training sets were tested. If this was either not possible, or the model did not perform well, sequences from various species were used, always, based on a published characteristic alignment of the family.
For CPAP1, CPAP3, CPCFC, CPF, CPFL, CPLCA, CPLCG, CPLCW and Tweedle only the conserved part of the protein that characterizes the family was used in the training set, and not the whole sequence. For CPAP3, CPCFC and Tweedle, a continuous stretch that contained all the repeats that corresponded to each family, was used in each training set. The conserved regions were chosen based on manually curated published alignments (Table 2). For Apidermin, CPG and CPLCP, there were no characteristic conserved domains, since these families are mostly characterized by repeats, so the whole sequence was used as a training set.
Table 2.
References for conserved regions, for each family
| Family Type | Source of Conserved Region | |
|---|---|---|
| CPAP1 | Jasrapuria et al., 2010 | Fig. 2C |
| CPAP3 | Jasrapuria et al., 2010 | Fig. 2B |
| CPCFC | Willis et al., 2012 | Fig. 3B |
| CPF | Togawa et al., 2007 | Fig. 1A |
| CPFL | Togawa et al., 2007 | Fig. 3 |
| CPLCA | Willis, 2010 | Fig. 4A |
| CPLCG | Willis, 2010 | Fig. 3B |
| CPLCW | Willis, 2010 | Fig. 3C |
| Tweedle | Willis, 2010 | Fig. 3A |
Profile Hidden Markov Models (pHMMs) are statistical models, describing a multiple sequence alignment by capturing position-specific information (Eddy, 1998; Krogh et al., 1994). Compared to the classic tool BLAST (Altschul et al., 1990; Altschul et al., 1997), profile HMMs can be more accurate and more able to detect remote homologs. Instead of a single sequence, they include more information, using a statistical representation of a multiple sequence alignment.
For each family of structural cuticular proteins, a multiple sequence alignment of the training set was created using ClustalW2 (Larkin et al., 2007) with default settings. Each alignment was then used as an input to the hmmbuild program of HMMER v3.0 <http://hmmer.janelia.org/> (Eddy, 1998) and, using default settings, a profile Hidden Markov Model was created for each family. The conversion of the alignment file from Clustal to Stockholm format, which is the default input format for HMMER v3.0, was performed with a homemade Perl script.
2.3 Selection of Test Sets
In order to evaluate the constructed pHMMs, test sets were created. For each family, all its members, as described in Table 1, were used as the positive training set, while the members of the remaining twelve families were used as negative test set. In the cases of CPAP1 and CPAP3, an extra negative test set was used. Since these families are analogous to peritrophins and contain common chitin binding domains, the additional negative set consisted of peritrophic matrix proteins and chitin metabolic enzymes, in order to check whether the pHMMs erroneously identify as positive results such non-cuticular proteins, due to their similarity. All the peritrophic matrix proteins and chitin metabolic enzymes used, came from Tribolium castaneum, where a precise identification and discrimination between those protein families has been done (Jasrapuria et al., 2010).
2.4 Evaluation Method for Models
The probability parameters in a profile HMM are converted to additive log–odds scores before aligning and scoring a query sequence (Barrett et al., 1997). The scores for aligning a residue to a profile match state are, therefore, comparable to the derivation of BLAST or FASTA scores (Eddy, 1998).
For each family, the respective pHMM was applied against both the positive and the negative test sets, using the hmmsearch program of HMMER v3.0 <http://hmmer.janelia.org/> (Eddy, 1998). Subsequently, the standard statistical measures for the performance of binary (a protein either belongs to a family or not) classification tests, specificity and sensitivity, were calculated for a range of 5 units of score cutoff for each model: Specificity = TN / (TN + FP) and Sensitivity = TP / (TP + FN), where TP is the number of True Positive predictive values, TN the number of True Negatives, FP the number of False Positives and FN the number of False Negatives. To estimate the cutoff for each family, a plot of specificity and sensitivity against the different scores was designed for each pHMM. As cutoff, we selected the score where sensitivity and specificity were at a balance. As an extra step for specificity control, all models, with the cutoff calculated above, were applied against UniProt/SwissProt (UniProt, 2013) to check if there are any non-specific hits.
2.5 Modification of Evaluation Method for CPAP3 and CPAP1 Families
The two families that are analogous to peritrophins, CPAP3 and CPAP1, contain three and one chitin-binding domains, respectively. This specific domain CBM_14 (Chitin binding Peritrophin-A domain – PF01607 in Pfam (Punta et al., 2012)) is also found in other families that bind chitin, such as peritrophic matrix proteins of insects and animal chitinases. We found single instances of three repeats in proteins that clearly were not cuticular. In order to discriminate CPAP3 and CPAP1, the distinct spacing between their conserved cysteines was valuable. As mentioned in section 2.2, in the case of CPAP3, all 3 repeats were treated as one continuous domain instead of 3 repeats of the same domain, to take advantage of the distinct spacing between the domain repeats.
Instead of using the “full sequence” score, which takes into account the whole query sequence, for those two families, the “best 1 domain” score was used. The “best 1 domain” score takes into account only the part of the sequence that matches better with the model <http://hmmer.janelia.org/> (Eddy, 1998). A non-cuticular protein with many repeats of the chitin-binding domain could match the model multiple times and therefore have a very high “full sequence” score.
The “best 1 domain” score, however, is expected to be high only for cuticular proteins because of the conservation of spacing.
2.6 Application of the pHMMs on arthropod proteomes
In order to test the performance of successful models in real-world data, the models were applied to twelve insect proteomes and two from non-insect arthropods.
The following proteomes were used:
the fruit fly Drosophila melanogaster (FlyBase, http://flybase.org/, Drosophila melanogaster, Dmel_r5.56) (St Pierre et al., 2014), which had 30307 peptides,
the tsetse fly Glossina morsitans (VectorBase, http://www.vectorbase.org/, Glossina morsitans Yale annotation, GmorY1.3) (Megy et al., 2012), which had 12449 peptides,
the southern house mosquito Culex quinquefasciatus (VectorBase, http://www.vectorbase.org/, Culex quinquefasciatus Johannesburg JHB annotation, CpipJ1.4) (Arensburger et al., 2010; Megy et al., 2012), which had 19019 peptides,
the yellow fever mosquito Aedes aegypti (VectorBase, http://www.vectorbase.org/, Aedes aegypti Liverpool LVP annotation, AaegL2.2) (Megy et al., 2012; Nene et al., 2007), which had 17143 peptides,
the African malaria mosquito Anopheles gambiae (VectorBase, http://www.vectorbase.org/, Anopheles gambiae PEST annotation, AgamP3.8) (Megy et al., 2012), which had 14667 peptides,
the domesticated silkmoth Bombyx mori (SilkDB, http://silkworm.genomics.org.cn/, Bombyx mori) (Wang et al., 2005; Xia et al., 2004), which had 14623 peptides,
the monarch butterfly Danaus plexippus (MonarchBase, http://monarchbase.umassmed.edu/, Dp_OGS2.0) (Zhan and Reppert, 2013), which had 15130 peptides,
the red flour beetle Tribolium castaneum (BeetleBase, http://beetlebase.org/, Tribolium castaneum, Tcas3.0 OGS) (Kim et al., 2010; Richards et al., 2008; Wang et al., 2007), which had 16645 peptides,
the honey bee Apis mellifera (BeeBase, http://hymenopteragenome.org/beebase/, Apis mellifera, Amel_4.5 OGSv3.2) (Elsik et al., 2014; The Honeybee Genome Sequencing Consortium, 2006), which had 15314 peptides,
the jewel wasp Nasonia vitripennis (NasoniaBase, http://www.hymenopteragenome.org/nasonia/, Nasonia vitripennis, Nvit_OGSv1.2) (Werren et al., 2010), which had 18822 peptides,
the pea aphid Acyrthosiphon pisum (AphidBase, http://www.aphidbase.com/, Acyrthosiphon pisum, ACYPI v2.1b)2010, which had 36195 peptides,
the human body louse Pediculus humanus (VectorBase, http://www.vectorbase.org/, Pediculus humanus USDA annotation, PhumU1.3) (Megy et al., 2012), which had 10775 peptides,
the water flea Daphnia pulex (wFleaBase, http://wfleabase.org/, Daphnia pulex, Gene Set 2.0 beta3) (These sequence data were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/ in collaboration with the Daphnia Genomics Consortium http://daphnia.cgb.indiana.edu/.), which had 47712 peptides, and
the two-spotted spider mite Tetranychus urticae (UniProtKB Complete Proteome, http://www.uniprot.org/) (Grbic et al., 2011; UniProt, 2013), which had 18082 peptides.
2.7 Website implementation
The web page was implemented using the following technologies: the HTML markup language and the CSS style sheet language for the page layout and design, the PHP scripting language for server side functions and the pre-processing of the results, the JavaScript programming language (using jQuery and AJAX) for dynamic effects to the web page, and finally the HMMER v3.0 suite <http://hmmer.janelia.org/> (Eddy, 1998) which runs the searches on the server.
3. Results and Discussion
3.1 Selection of Models
Out of all the models built, one – the most characteristic – was selected for each family. The design of models that perform well was possible for only eight (CPAP1, CPAP3, CPCFC, CPF, CPLCA, CPLCG, CPLCW and Tweedle) of the twelve CP families, since the other four families (Apidermin, CPFL, CPG, CPLCP) did not have enough conservation in sequence level and none of the models that were tested passed the evaluation steps. The number of sequences used in each set for the successful models, are listed in Table 3.
Table 3.
Number of sequences in the training and test set for each family
| Family | Training Set | Test Set | Positive Test Set | Negative Test Set |
|---|---|---|---|---|
| CPAP1 | 10 | 1816 | 10 | 1806 |
| CPAP3 | 6a | 1816 | 58 | 1758 |
| CPCFC | 12a | 1792 | 33 | 1759 |
| CPF | 4 | 1792 | 48 | 1744 |
| CPLCA | 11 | 1792 | 21 | 1771 |
| CPLCG | 86 | 1792 | 104 | 1688 |
| CPLCW | 9 | 1792 | 27 | 1765 |
| Tweedle | 27a | 1792 | 234 | 1558 |
A continuous stretch that contained all repeats was used in the training set.
The training sets that were finally selected for each family came from all the available sequences from only one species, except for CPLCG and CPCFC, where a mixed training set with sequences from various organisms was used. For Tweedle, CPLCA and CPAP3 the training sets were created with sequences from Drosophila melanogaster, for CPF and CPLCW from Anopheles gambiae, and for CPAP1 from Tribolium castaneum. For CPLCG the training set was created using sequences from various species, and, specifically the sequences used to create the characteristic alignment and logo by Willis (2010). This was necessary since the construction of a characteristic model from sequences that came from one species only was not possible, indicating a possible higher degree of variation in sequence between proteins of different species than in other families. For CPCFC, the training set was also created using sequences from various species, specifically, those used to create the characteristic alignment and logo by Willis et al. (2012), since there were not enough sequences available from one species only (1 or 2 sequences available from each species). For the alignments used as training sets see Supplementary Data File 1.
3.2 Estimation of Cutoffs
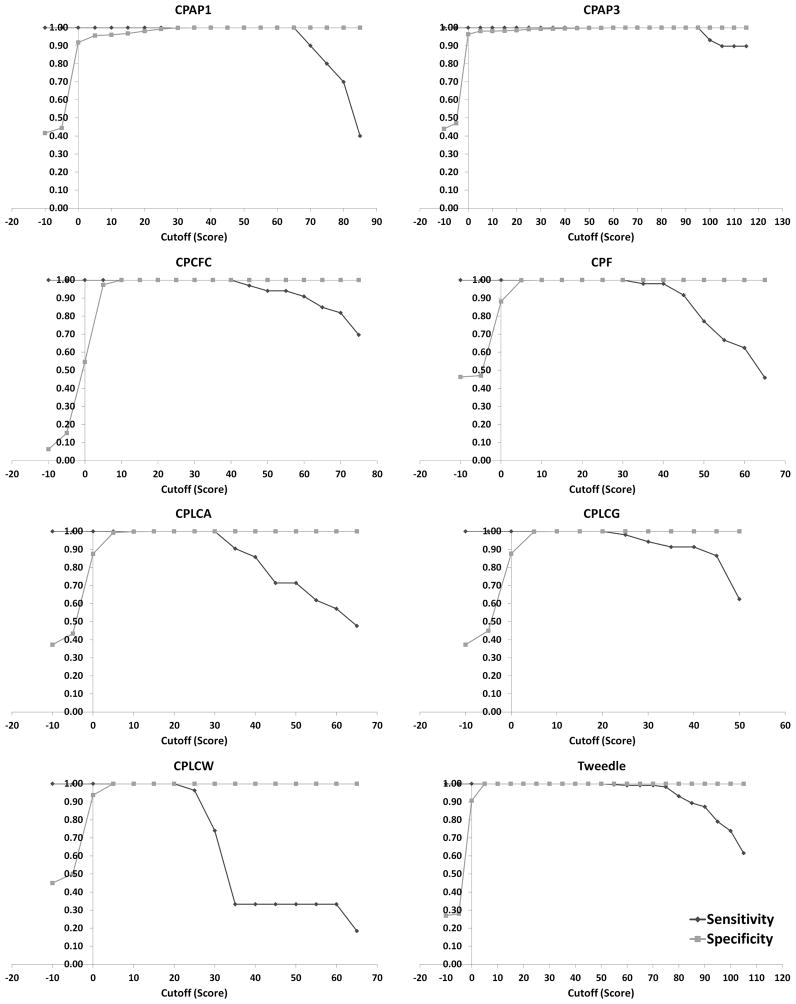
The cutoff score for each model was estimated as the middle value of the range where specificity meets sensitivity. This was preferred based on the hypothesis that with larger separation between protein sequences that belong to the family type described by the model and protein sequences that don’t, we are more likely to avoid misclassifications. We noticed that the scores of true positives and false negatives of each pHMM did not overlap. Thus, in all cases both specificity and sensitivity were equal to 1, in the score that was assigned as a cutoff, as shown in Fig. 1. All cutoffs, in addition to the lowest score for the proteins of the same type as the one described by the model, and the highest score for the proteins of different type are listed in Table 4. The models with their corresponding cutoffs were applied in UniProt/SwissProt (UniProt, 2013) and none of them found unspecific hits, either in arthropods or in other species.
Fig. 1.
Plots of sensitivity and specificity against different cutoff scores in order to find the threshold for each profile HMM. The scores for each sequence were generated by hmmsearch <http://hmmer.janelia.org/> (Eddy, 1998). The analysis was performed repeatedly, by setting a different cutoff score, in a range of 5 units of score, each time, and calculating the specificity and sensitivity values. As mentioned in section 2.3, the cutoff score for each model was estimated as the middle value of the range where specificity meets sensitivity, which in all cases was where both were equal to 1. Intuitively, this was chosen so that the cutoff score will represent the largest separation between protein sequences that belong to the family described by the model, and protein sequences that do not. The larger the margin, the lower the errors of the classification will be. In order to capture how specificity and sensitivity change, the range of cutoff scores between the families differs and, consequently, the number of repeats of the procedure, also, differs.
Table 4.
Cutoff estimation and overlap check
| Family | Cutoff | Lowest True Positive | Highest False Negative |
|---|---|---|---|
| CPR_RR-1a | 35 | 45.9 | 21.2 |
| CPR_RR-2a | 37.5 | 51.0 | 22.8 |
| CPAP1 | 50 | 66.7 | 31 |
| CPAP3 | 77.5 | 97.2 | 59.3 |
| CPCFC | 27.5 | 42.7 | 13.3 |
| CPF | 20 | 34.3 | 7.3 |
| CPLCA | 22.5 | 31.9 | 14.6 |
| CPLCG | 15 | 21 | 5.8 |
| CPLCW | 12.5 | 23.7 | 3.5 |
| Tweedle | 30 | 52.1 | 7 |
Cutoffs and scores calculated by Karouzou et al. (2007). We have set scores to 0 to allow all CPRs to be identified. It is necessary to use the cutoff scores, shown above, for the correct assignment of the CPR subfamily.
For RR-1 and RR-2 subfamilies, the cutoffs were calculated by Karouzou et al. (2007). But if one wants to retrieve all of the proteins belonging to the CPR family, we recommend setting the cutoff for both RR-2 and RR-1 at 0, as was also suggested in the cuticleDB’s RR-Find tool (Karouzou et al., 2007). This way, all proteins that contain the R&R Consensus, including RR-3, should be retrieved. Since discrimination between the two classes of CPRs is lost when 0 is used as a score cutoff, we characterized all proteins below the two assigned cutoffs as “unclassified”. Also, because the RR-1 and RR-2 domain are very similar, it will be necessary to identify duplicates and save the one with the best score and assign it to the proper class using the highest score. Selection of the best score occurs automatically if one searches for “all profiles.”
3.3 Application in Proteomes
The eight new profiles, plus the two old ones for the CPRs RR-1 and RR-2 families, were applied to fourteen available arthropod proteomes: ten from holometabolous species, two from hemimetabolous, plus two from non-insect arthropods. Protein sequences, which either had not been annotated at all (proteins with unknown function) or annotated simply as cuticle proteins without indication of their family type, were found by the models. The results are summarized in Table 5, and the accession codes for each CP family member in each tested species are provided in Supplementary Data File 2. A comparison of Table 5 versus Table 1 shows some inconsistency in some numbers. This is easily explained by the fact that the initial dataset, as described in section 2.1, was collected mainly from papers published by experts in this field, since they were considered the most accurate source, but not all of these protein sequences have been incorporated in the proteomes we used even though they were the most up-to-date versions.
Table 5.
Proteinsa identified from 14 arthropod proteomes using CutProtFam-Pred (see section 2.6)
| Family |
Drosophila melanogaster |
Glossina morsitans |
Culex quinquefasciatus |
Aedes aegypti |
Anopheles gambiae |
Bombyx mori |
Danaus plexippus |
Tribolium castaneum |
Apis mellifera |
Nasonia vitripennis |
Acyrthosiphon pisum |
Pediculus humanus corporis |
Daphnia pulex |
Tetranychus urticae |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPR_RR-1b | 61 | 33c | 49 | 66 | 43 | 47 | 47 | 34 | 13 | 19 | 9 | 9 | 101c | 0 |
| CPR_RR-2b | 42 | 27c | 97 | 150 | 103 | 78 | 57 | 55 | 15 | 32 | 84 | 15 | 36 | 7 |
| CPR_Uncl.b | 34a | 17 | 30 | 28c | 21c | 19 | 18c | 21c | 10 | 18c | 20c | 17 | 152c | 31c |
| CPAP1 | 29 | 11 | 10 | 14 | 13 | 13 | 16 | 13 | 15 | 16 | 10 | 12 | 20 | 14 |
| CPAP3 | 10 | 6 | 8 | 9 | 10 | 6 | 10 | 7 | 7 | 6 | 8 | 6 | 12 | 5 |
| CPCFC | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| CPF | 5 | 1 | 5 | 3 | 4 | 1 | 1 | 5 | 4 | 5 | 2 | 1 | 0 | 0 |
| CPLCA | 13 | 9 | 3 | 3 | 3 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CPLCG | 4 | 4 | 41 | 18 | 25 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| CPLCW | 0 | 0 | 0 | 7 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tweedle | 29 | 9 | 9 | 6 | 12 | 4 | 5 | 3 | 2 | 2 | 3 | 2 | 0 | 0 |
|
| ||||||||||||||
| Total | 228 | 118 | 253 | 305 | 245 | 171 | 157 | 142 | 66 | 98 | 137 | 63 | 321 | 57 |
Numbers include all splice variants. See Supplementary File 2 and Supplementary File 3 for values based on gene numbers.
Sequences that score equal or above the assigned score cutoffs for the CPR_RR-1 and CPR_RR-2 models are classified in the corresponding family, while sequences that score below the assigned score cutoffs and above 0 for one of the models are characterized as “unclassified” (for more details, see Supplementary File 2).
Contain a few sequences lacking PF00379 (marked with red background in Supplementary File 2).
We also attempted to compare our results to those from manual annotations of the genomes of the same species. Most of the published annotations were carried out on searches of genomic data and some identified sequences are not present in the proteomes we used and new proteins have been added. Nonetheless, we thought it useful to provide what comparative data exist, to indicate that our tool appears to be effective in quickly identifying cuticular proteins. We have thus compared data from Table 3 (Willis et al., 2012) to the data in Table 5, and present the results in Supplementary Data File 3. For this purpose, we counted all splice variants as a single gene. The results confirm our conclusion that the tool is an efficient first step in manual annotation of proteomes, across a broad spectrum of insects.
In addition to the twelve insect species, we obtained data for both the crustacean, Daphnia pulex, and the chelicerate Tetranychus urticae. Analyses of cuticular proteins had been carried out for each of these two species (Colbourne et al., 2007; Grbic et al., 2011) (<http://server7.wfleabase.org/prerelease4/gene-predictions/>). We only identified CPRs and CPAP1 and CPAP3 family members. While Daphnia had good representation of both RR-1 and RR-2 proteins, Tetranychus had only RR-2 or non-classified CPRs.
As a final check, since we used a score of zero to distinguish between RR-1 and RR-2, we checked whether the proteins identified by these two models actually had the PF00379 (chitin_bind_4) domain. Very few hits, even with 0 as the cutoff score, were for proteins lacking PF00379. They are highlighted in red in Supplementary Data File 2.
It is worth noting that sequences belonging to the CPR family constitute, in each case, the larger proportion of the structural cuticular proteins. This could indicate a more general role for this family, in comparison to the rest, which may have more specific functions. Also, as was expected, since chitin is a basic component of the arthropod cuticle, families that have been demonstrated experimentally to bind chitin [CPR (Rebers and Willis, 2001); CPAP1, CPAP3 (Arakane et al., 2003; Nisole et al., 2010) and Tweedle (Tang et al., 2010)] seem to have more members and a wider taxonomic distribution in insects than the rest.
A quantification analysis, which was performed in order to estimate the percentage of structural cuticular proteins in proteomes, is presented in Table 6. It was found that structural cuticular protein sequences can comprise about 0.32% – 1.94% (with a mean value of 0.94% and a standard deviation of 0.55%) of an insect’s proteome. Of course the results present only a minimum estimate of the number of cuticular proteins in a species. Few proteomes have received thorough manual curation. They were rather created using automatic prediction programs. So it is certain, that not all genes/peptides have been found or they are not correctly predicted, and there are certain to be errors in the predictions. Also, these data represent a lower limit as sequences belonging to 4 known families (CPFL, CPG, CPLCP and Apidermins) are not included in our analysis, nor are the cuticular proteins that have not yet been assigned to families. Some of the proteomes we used report more than one protein from a gene, i.e. splice variants, PA, PB. Many splice variants only differ in their 5’UTRs, so they may not be indicating protein diversity. In Supplementary Data File 2, we have provided a tab that gives the total number of proteins assigned to each class (Total number including splice variants) and the percent of CPs in the proteome (% Total Proteome). We have also calculated the number of CP genes in each class obtained by removing all the splice variants (Total minus Splice Variants) and their fraction of the proteome reduced to one entry per gene (% Reduced Proteome).
Table 6.
Percentage of cuticular proteins found in arthropod proteomes combining results from all families
| Species | Cuticular Proteinsa | Peptidesa | % |
|---|---|---|---|
| Drosophila melanogaster | 228 | 30307 | 0.75 |
| Glossina morsitans | 118 | 12449 | 0.95 |
| Culex quinquefasciatus | 253 | 19019 | 1.33 |
| Aedes aegypti | 305 | 17143 | 1.78 |
| Anopheles gambiae | 245 | 14667 | 1.67 |
| Bombyx mori | 171 | 14623 | 1.17 |
| Danaus plexippus | 157 | 15130 | 1.04 |
| Tribolium castaneum | 142 | 16645 | 0.85 |
| Apis mellifera | 66 | 15314 | 0.43 |
| Nasonia vitripennis | 98 | 18822 | 0.52 |
| Acyrthosiphon pisum | 137 | 36195 | 0.38 |
| Pediculus humanus corporis | 63 | 10775 | 0.58 |
| Daphnia pulex | 321 | 47712 | 0.67 |
| Tetranychus urticae | 57 | 18082 | 0.32 |
Numbers include all splice variants. See Supplementary File 2 and Supplementary File 3 for values based on gene numbers.
It is also worth noting that the models seem to perform well in the well-studied flies, mosquitoes, moths and beetles. As more proteomes that belong to other species become available, it will be necessary to re-test the tool’s performance and make possible modifications. This is especially true for the two non-insect species where the fraction of unclassified CPRs was quite high. It is likely that other CP sequences and families exist in various groups, but this can only be established with protein sequences obtained from manually cleaned cuticles, cast skins, or other methods, such as immunolocalization that can verify that a particular protein actually reside within the cuticle. Recognition of cuticular proteins began with this way and has been reviewed by Andersen et al. (1995). More recent studies are by He et al. (2007); Fu et al. (2011); Dittmer et al. (2012). Work of this type on non-insect arthropods has been even more limited and produced few sequences (Andersen and Roepstorff, 2005; Ditzel et al., 2003; Norup et al., 1996; Otte et al., 2014).
3.4 CutProtFam-Pred Website Development
The eight new profiles created in this work, in addition to the two old ones from cuticleDB (Karouzou et al., 2007) were used in the development of CutProtFam-Pred <http://bioinformatics.biol.uoa.gr/CutProtFam-Pred/>, an on-line freely available tool for the detection and classification of structural cuticular proteins from sequence alone.
The CutProtFam-Pred website has the following menu options in tab form: Home, Search, Manual and Contact.
The “Search” tab contains the form in which the user can either paste the query sequence or sequences in the textbox area or upload a file that contains them, in both cases the sequences must be in fasta format. Next, the user can either search against the library of the available models and get a prediction for each sequence based on the assigned default cutoffs, that were calculated as described in section 2.5, or search against a specific profile, where either the score or the e-value cutoff can be changed (the assigned cutoff appears when the user selects this option as a default value). In the results page, the user can see the predictions for each sequence, along with the corresponding e-value and score, and can also download one file in fasta format for each searched family, containing all the protein sequences that were predicted to belong to it. If the user searches against all the families, the fasta files are combined in one file. Also available are the raw HMMER output and/or the results in tab-delimited format.
The tool aims to be user-friendly, making the whole prediction process easy, even for someone without extensive knowledge in the field. Apart from the predictions given using the assigned cutoff, it also provides the option of changing it, for more experienced users.
3.5 Limitations of the Tool
The tool queries proteomes that have been produced by automated annotation. These programs sometimes combine closely linked genes into a single protein and many cuticular proteins are tightly clustered on chromosomes. Hence, the data produced using the tool must be considered preliminary. Indeed, we regard this tool as an aid to annotation, and not a substitute for manual annotation. HMMER produces scores based on the number of hits as well as their quality <http://hmmer.janelia.org/> (Eddy, 1998). Since only rarely does a CPR protein have more than a single R&R Consensus region, high ranking proteins are apt to have been incorrectly annotated due to combining adjacent genes. We found that by setting the score to 0 for RR-1 and RR-2 searches, more CPRs were identified, including some that had been classified as RR-3, but of course, there will be considerable overlap between RR-1 and RR-2 because the discriminating score was not used. So, in case the RR-1 or RR-2 profiles are run separately, we recommend using the score of 0, and then sorting out the duplicate hits to identify those which score best as RR-1 or RR-2. When a search against all families is selected, if a protein matches with both profiles, it will be, by default, indicate that the protein belongs in the family against which it has a higher score. Less than 1% of the sequences we recovered with this low setting appear on inspection to be something other than a CPR (Supplementary Data File 2).
4. Conclusions
In this paper we introduce CutProtFam-Pred, an on-line tool for the identification of putative structural cuticular proteins and their classification into the respective families, from sequence alone. We hope that implementation of these pHMMs via CutProtFam-Pred, for nine of the thirteen families of structural cuticular proteins identified to date, will be useful in the functional annotation of arthropod proteomes.
Supplementary Material
All the alignments that were used to build the profile Hidden Markov Models given in Clustal format. The numbers at the end of each row in the alignments, correspond to the last amino acid as numbered in the characteristic region, not in the original sequence.
Accession numbers of sequences identified by our tool. Each tab contains a family and each column a species. Red background: no PF00379 domain found in that CPR (the hmmsearch was performed with all filters off). Dark pink font: unclassified CPR scored highest as a CPR_RR-1, dark blue font: unclassified CPR scored highest as a CPR_RR-2. Yellow background: peptides than came from the same gene not counted in “Total minus Splice Variants” and “% Reduced Proteome” tabs, at the end of this file.
Comparison between Updated Willis et al. (2012), part of Table 3, and CutProtFam-Pred results, in gene numbers.
Highlights.
pHMMs created for 8 of the 12 newly characterized cuticular protein families
Detection of CPR, CPAP1, CPAP3, CPCFC, CPF, CPLCA, CPLCG, CPLCW, Tweedle proteins
4 other families did not have enough conservation for sequence-based models
Development of CutProtFam-Pred, a publicly available on-line web tool
CutProtFam-Pred will be useful in the functional annotation of arthropod proteomes
Acknowledgments
We thank Prof. Don Gilbert for advice on non-insect proteomes. We should like to thank the handling editor and the reviewers of this manuscript for their very useful and constructive criticism. Work by JHW was supported by a grant from the U.S. National Institutes of Health R01AI055624.
Appendix. Supplementary data
Three files of supplementary data associated with this article can be found in the online version at doi:().
Footnotes
Availability and Requirements
The CutProtFam-Pred is freely available at <http://bioinformatics.biol.uoa.gr/CutProtFam-Pred/>. The website has been tested with Internet Explorer, Firefox, Chrome, Opera and Safari browsers.
Competing interests
The authors report no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen SO. Amino acid sequence studies on endocuticular proteins from the desert locust, Schistocerca gregaria. Insect Biochem Mol Biol. 1998;28:421–434. doi: 10.1016/s0965-1748(98)00028-9. [DOI] [PubMed] [Google Scholar]
- Andersen SO. Studies on proteins in post-ecdysial nymphal cuticle of locust, Locusta migratoria, and cockroach, Blaberus craniifer. Insect Biochem Mol Biol. 2000;30:569–577. doi: 10.1016/s0965-1748(00)00029-1. [DOI] [PubMed] [Google Scholar]
- Andersen SO, Hojrup P, Roepstorff P. Insect cuticular proteins. Insect biochemistry and molecular biology. 1995;25:153–176. doi: 10.1016/0965-1748(94)00052-j. [DOI] [PubMed] [Google Scholar]
- Andersen SO, Rafn K, Roepstorff P. Sequence studies of proteins from larval and pupal cuticle of the yellow meal worm, Tenebrio molitor. Insect Biochem Mol Biol. 1997;27:121–131. doi: 10.1016/s0965-1748(96)00076-8. [DOI] [PubMed] [Google Scholar]
- Andersen SO, Roepstorff P. The extensible alloscutal cuticle of the tick, Ixodes ricinus. Insect biochemistry and molecular biology. 2005;35:1181–1188. doi: 10.1016/j.ibmb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Arakane Y, Zhu Q, Matsumiya M, Muthukrishnan S, Kramer KJ. Properties of catalytic, linker and chitin- binding domains of insect chitinase. Insect Biochem Mol Biol. 2003;33:631–648. doi: 10.1016/s0965-1748(03)00049-3. [DOI] [PubMed] [Google Scholar]
- Arensburger P, Megy K, Waterhouse RM, Abrudan J, Amedeo P, Antelo B, Bartholomay L, Bidwell S, Caler E, Camara F, Campbell CL, Campbell KS, Casola C, Castro MT, Chandramouliswaran I, Chapman SB, Christley S, Costas J, Eisenstadt E, Feschotte C, Fraser-Liggett C, Guigo R, Haas B, Hammond M, Hansson BS, Hemingway J, Hill SR, Howarth C, Ignell R, Kennedy RC, Kodira CD, Lobo NF, Mao C, Mayhew G, Michel K, Mori A, Liu N, Naveira H, Nene V, Nguyen N, Pearson MD, Pritham EJ, Puiu D, Qi Y, Ranson H, Ribeiro JM, Roberston HM, Severson DW, Shumway M, Stanke M, Strausberg RL, Sun C, Sutton G, Tu ZJ, Tubio JM, Unger MF, Vanlandingham DL, Vilella AJ, White O, White JR, Wondji CS, Wortman J, Zdobnov EM, Birren B, Christensen BM, Collins FH, Cornel A, Dimopoulos G, Hannick LI, Higgs S, Lanzaro GC, Lawson D, Lee NH, Muskavitch MA, Raikhel AS, Atkinson PW. Sequencing of Culex quinquefasciatus establishes a platform for mosquito comparative genomics. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C, Hughey R, Karplus K. Scoring hidden Markov models. Computer applications in the biosciences : CABIOS. 1997;13:191–199. doi: 10.1093/bioinformatics/13.2.191. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Eads BD, Shaw J, Bohuski E, Bauer DJ, Andrews J. Sampling Daphnia’s expressed genes: preservation, expansion and invention of crustacean genes with reference to insect genomes. BMC Genomics. 2007;8:217. doi: 10.1186/1471-2164-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornman RS, Willis JH. Annotation and analysis of low-complexity protein families of Anopheles gambiae that are associated with cuticle. Insect Mol Biol. 2009;18:607–622. doi: 10.1111/j.1365-2583.2009.00902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer NT, Hiromasa Y, Tomich JM, Lu N, Beeman RW, Kramer KJ, Kanost MR. Proteomic and transcriptomic analyses of rigid and membranous cuticles and epidermis from the elytra and hindwings of the red flour beetle, Tribolium castaneum. J Proteome Res. 2012;11:269–278. doi: 10.1021/pr2009803. [DOI] [PubMed] [Google Scholar]
- Ditzel N, Andersen SO, Hojrup P. Cuticular proteins from the horseshoe crab, Limulus polyphemus. Comparative biochemistry and physiology Part B, Biochemistry & molecular biology. 2003;134:489–497. doi: 10.1016/s1096-4959(02)00291-9. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics (Oxf) 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, Childers CP, de Graaf DC, Debyser G, Deng J, Devreese B, Elhaik E, Evans JD, Foster LJ, Graur D, Guigo R, Hoff KJ, Holder ME, Hudson ME, Hunt GJ, Jiang H, Joshi V, Khetani RS, Kosarev P, Kovar CL, Ma J, Maleszka R, Moritz RF, Munoz-Torres MC, Murphy TD, Muzny DM, Newsham IF, Reese JT, Robertson HM, Robinson GE, Rueppell O, Solovyev V, Stanke M, Stolle E, Tsuruda JM, Vaerenbergh MV, Waterhouse RM, Weaver DB, Whitfield CW, Wu Y, Zdobnov EM, Zhang L, Zhu D, Gibbs RA. Finding the missing honey bee genes: lessons learned from a genome upgrade. BMC Genomics. 2014;15:86. doi: 10.1186/1471-2164-15-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li P, Xu Y, Zhang S, Jia L, Zha X, Xiang Z, He N. Proteomic analysis of larval integument, trachea and adult scale from the silkworm, Bombyx mori. Proteomics. 2011;11:3761–3767. doi: 10.1002/pmic.201000506. [DOI] [PubMed] [Google Scholar]
- Futahashi R, Okamoto S, Kawasaki H, Zhong YS, Iwanaga M, Mita K, Fujiwara H. Genome-wide identification of cuticular protein genes in the silkworm, Bombyx mori. Insect Biochem Mol Biol. 2008;38:1138–1146. doi: 10.1016/j.ibmb.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Grbic M, Van Leeuwen T, Clark RM, Rombauts S, Rouze P, Grbic V, Osborne EJ, Dermauw W, Ngoc PC, Ortego F, Hernandez-Crespo P, Diaz I, Martinez M, Navajas M, Sucena E, Magalhaes S, Nagy L, Pace RM, Djuranovic S, Smagghe G, Iga M, Christiaens O, Veenstra JA, Ewer J, Villalobos RM, Hutter JL, Hudson SD, Velez M, Yi SV, Zeng J, Pires-daSilva A, Roch F, Cazaux M, Navarro M, Zhurov V, Acevedo G, Bjelica A, Fawcett JA, Bonnet E, Martens C, Baele G, Wissler L, Sanchez-Rodriguez A, Tirry L, Blais C, Demeestere K, Henz SR, Gregory TR, Mathieu J, Verdon L, Farinelli L, Schmutz J, Lindquist E, Feyereisen R, Van de Peer Y. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Middlebrooks BW, Alexander S, Wasserman SA. Mutation of TweedleD, a member of an unconventional cuticle protein family, alters body shape in Drosophila. Proc Natl Acad Sci U S A. 2006;103:16794–16799. doi: 10.1073/pnas.0607616103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Botelho JM, McNall RJ, Belozerov V, Dunn WA, Mize T, Orlando R, Willis JH. Proteomic analysis of cast cuticles from Anopheles gambiae by tandem mass spectrometry. Insect biochemistry and molecular biology. 2007;37:135–146. doi: 10.1016/j.ibmb.2006.10.011. [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasrapuria S, Arakane Y, Osman G, Kramer KJ, Beeman RW, Muthukrishnan S. Genes encoding proteins with peritrophin A-type chitin-binding domains in Tribolium castaneum are grouped into three distinct families based on phylogeny, expression and function. Insect Biochem Mol Biol. 2010;40:214–227. doi: 10.1016/j.ibmb.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Jensen UG, Rothmann A, Skou L, Andersen SO, Roepstorff P, Hojrup P. Cuticular proteins from the giant cockroach, Blaberus craniifer. Insect Biochem Mol Biol. 1997;27:109–120. doi: 10.1016/s0965-1748(96)00074-4. [DOI] [PubMed] [Google Scholar]
- Karouzou MV, Spyropoulos Y, Iconomidou VA, Cornman RS, Hamodrakas SJ, Willis JH. Drosophila cuticular proteins with the R&R Consensus: annotation and classification with a new tool for discriminating RR-1 and RR-2 sequences. Insect Biochem Mol Biol. 2007;37:754–760. doi: 10.1016/j.ibmb.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Kim HS, Murphy T, Xia J, Caragea D, Park Y, Beeman RW, Lorenzen MD, Butcher S, Manak JR, Brown SJ. BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic acids research. 2010;38:D437–442. doi: 10.1093/nar/gkp807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Brown M, Mian IS, Sjolander K, Haussler D. Hidden Markov models in computational biology. Applications to protein modeling. J Mol Biol. 1994;235:1501–1531. doi: 10.1006/jmbi.1994.1104. [DOI] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Maleszka R. Novel cuticular proteins revealed by the honey bee genome. Insect Biochem Mol Biol. 2007;37:128–134. doi: 10.1016/j.ibmb.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics (Oxf) 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Magkrioti CK, Spyropoulos IC, Iconomidou VA, Willis JH, Hamodrakas SJ. cuticleDB: a relational database of Arthropod cuticular proteins. BMC bioinformatics. 2004;5:138. doi: 10.1186/1471-2105-5-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megy K, Emrich SJ, Lawson D, Campbell D, Dialynas E, Hughes DS, Koscielny G, Louis C, Maccallum RM, Redmond SN, Sheehan A, Topalis P, Wilson D. VectorBase: improvements to a bioinformatics resource for invertebrate vector genomics. Nucleic acids research. 2012;40:D729–734. doi: 10.1093/nar/gkr1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nene V, Wortman JR, Lawson D, Haas B, Kodira C, Tu ZJ, Loftus B, Xi Z, Megy K, Grabherr M, Ren Q, Zdobnov EM, Lobo NF, Campbell KS, Brown SE, Bonaldo MF, Zhu J, Sinkins SP, Hogenkamp DG, Amedeo P, Arensburger P, Atkinson PW, Bidwell S, Biedler J, Birney E, Bruggner RV, Costas J, Coy MR, Crabtree J, Crawford M, Debruyn B, Decaprio D, Eiglmeier K, Eisenstadt E, El-Dorry H, Gelbart WM, Gomes SL, Hammond M, Hannick LI, Hogan JR, Holmes MH, Jaffe D, Johnston JS, Kennedy RC, Koo H, Kravitz S, Kriventseva EV, Kulp D, Labutti K, Lee E, Li S, Lovin DD, Mao C, Mauceli E, Menck CF, Miller JR, Montgomery P, Mori A, Nascimento AL, Naveira HF, Nusbaum C, O’Leary S, Orvis J, Pertea M, Quesneville H, Reidenbach KR, Rogers YH, Roth CW, Schneider JR, Schatz M, Shumway M, Stanke M, Stinson EO, Tubio JM, Vanzee JP, Verjovski-Almeida S, Werner D, White O, Wyder S, Zeng Q, Zhao Q, Zhao Y, Hill CA, Raikhel AS, Soares MB, Knudson DL, Lee NH, Galagan J, Salzberg SL, Paulsen IT, Dimopoulos G, Collins FH, Birren B, Fraser-Liggett CM, Severson DW. Genome sequence of Aedes aegypti, a major arbovirus vector. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville AC. Biology of the arthropod cuticle. Springer-Verlag; Berlin ; New York: 1975. [Google Scholar]
- Neville AC. Biology of fibrous composites : development beyond the cell membrane. Cambridge University Press; New York, NY, USA: 1993. [Google Scholar]
- Nisole A, Stewart D, Bowman S, Zhang D, Krell PJ, Doucet D, Cusson M. Cloning and characterization of a Gasp homolog from the spruce budworm, Choristoneura fumiferana, and its putative role in cuticle formation. J Insect Physiol. 2010;56:1427–1435. doi: 10.1016/j.jinsphys.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Norup T, Berg T, Stenholm H, Andersen SO, Hojrup P. Purification and characterization of five cuticular proteins from the spider Araneus diadematus. Insect biochemistry and molecular biology. 1996;26:907–915. doi: 10.1016/s0965-1748(96)00052-5. [DOI] [PubMed] [Google Scholar]
- Otte KA, Frohlich T, Arnold GJ, Laforsch C. Proteomic analysis of Daphnia magna hints at molecular pathways involved in defensive plastic responses. BMC Genomics. 2014;15:306. doi: 10.1186/1471-2164-15-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol. 1988;203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- Rebers JE, Willis JH. A conserved domain in arthropod cuticular proteins binds chitin. Insect Biochem Mol Biol. 2001;31:1083–1093. doi: 10.1016/s0965-1748(01)00056-x. [DOI] [PubMed] [Google Scholar]
- Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen CJ, Klingler M, Lorenzen M, Roth S, Schroder R, Tautz D, Zdobnov EM, Muzny D, Attaway T, Bell S, Buhay CJ, Chandrabose MN, Chavez D, Clerk-Blankenburg KP, Cree A, Dao M, Davis C, Chacko J, Dinh H, Dugan-Rocha S, Fowler G, Garner TT, Garnes J, Gnirke A, Hawes A, Hernandez J, Hines S, Holder M, Hume J, Jhangiani SN, Joshi V, Khan ZM, Jackson L, Kovar C, Kowis A, Lee S, Lewis LR, Margolis J, Morgan M, Nazareth LV, Nguyen N, Okwuonu G, Parker D, Ruiz SJ, Santibanez J, Savard J, Scherer SE, Schneider B, Sodergren E, Vattahil S, Villasana D, White CS, Wright R, Park Y, Lord J, Oppert B, Brown S, Wang L, Weinstock G, Liu Y, Worley K, Elsik CG, Reese JT, Elhaik E, Landan G, Graur D, Arensburger P, Atkinson P, Beidler J, Demuth JP, Drury DW, Du YZ, Fujiwara H, Maselli V, Osanai M, Robertson HM, Tu Z, Wang JJ, Wang S, Song H, Zhang L, Werner D, Stanke M, Morgenstern B, Solovyev V, Kosarev P, Brown G, Chen HC, Ermolaeva O, Hlavina W, Kapustin Y, Kiryutin B, Kitts P, Maglott D, Pruitt K, Sapojnikov V, Souvorov A, Mackey AJ, Waterhouse RM, Wyder S, Kriventseva EV, Kadowaki T, Bork P, Aranda M, Bao R, Beermann A, Berns N, Bolognesi R, Bonneton F, Bopp D, Butts T, Chaumot A, Denell RE, Ferrier DE, Gordon CM, Jindra M, Lan Q, Lattorff HM, Laudet V, von Levetsow C, Liu Z, Lutz R, Lynch JA, da Fonseca RN, Posnien N, Reuter R, Schinko JB, Schmitt C, Schoppmeier M, Shippy TD, Simonnet F, Marques-Souza H, Tomoyasu Y, Trauner J, Van der Zee M, Vervoort M, Wittkopp N, Wimmer EA, Yang X, Jones AK, Sattelle DB, Ebert PR, Nelson D, Scott JG, Muthukrishnan S, Kramer KJ, Arakane Y, Zhu Q, Hogenkamp D, Dixit R, Jiang H, Zou Z, Marshall J, Elpidina E, Vinokurov K, Oppert C, Evans J, Lu Z, Zhao P, Sumathipala N, Altincicek B, Vilcinskas A, Williams M, Hultmark D, Hetru C, Hauser F, Cazzamali G, Williamson M, Li B, Tanaka Y, Predel R, Neupert S, Schachtner J, Verleyen P, Raible F, Walden KK, Angeli S, Foret S, Schuetz S, Maleszka R, Miller SC, Grossmann D. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Sigrist CJ, de Castro E, Cerutti L, Cuche BA, Hulo N, Bridge A, Bougueleret L, Xenarios I. New and continuing developments at PROSITE. Nucleic Acids Res. 2013;41:D344–347. doi: 10.1093/nar/gks1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Pierre SE, Ponting L, Stefancsik R, McQuilton P. FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic acids research. 2014;42:D780–788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L, Liang J, Zhan Z, Xiang Z, He N. Identification of the chitin-binding proteins from the larval proteins of silkworm, Bombyx mori. Insect Biochem Mol Biol. 2010;40:228–234. doi: 10.1016/j.ibmb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T, Augustine Dunn W, Emmons AC, Willis JH. CPF and CPFL, two related gene families encoding cuticular proteins of Anopheles gambiae and other insects. Insect Biochem Mol Biol. 2007;37:675–688. doi: 10.1016/j.ibmb.2007.03.011. [DOI] [PubMed] [Google Scholar]
- UniProt C. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 2013;41:D43–47. doi: 10.1093/nar/gks1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xia Q, He X, Dai M, Ruan J, Chen J, Yu G, Yuan H, Hu Y, Li R, Feng T, Ye C, Lu C, Li S, Wong GK, Yang H, Xiang Z, Zhou Z, Yu J. SilkDB: a knowledgebase for silkworm biology and genomics. Nucleic acids research. 2005;33:D399–402. doi: 10.1093/nar/gki116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li Y, Paradesi MS, Brown SJ. BeetleBase: the model organism database for Tribolium castaneum. Nucleic acids research. 2007;35:D476–479. doi: 10.1093/nar/gkl776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK, Beukeboom LW, Desplan C, Elsik CG, Grimmelikhuijzen CJ, Kitts P, Lynch JA, Murphy T, Oliveira DC, Smith CD, van de Zande L, Worley KC, Zdobnov EM, Aerts M, Albert S, Anaya VH, Anzola JM, Barchuk AR, Behura SK, Bera AN, Berenbaum MR, Bertossa RC, Bitondi MM, Bordenstein SR, Bork P, Bornberg-Bauer E, Brunain M, Cazzamali G, Chaboub L, Chacko J, Chavez D, Childers CP, Choi JH, Clark ME, Claudianos C, Clinton RA, Cree AG, Cristino AS, Dang PM, Darby AC, de Graaf DC, Devreese B, Dinh HH, Edwards R, Elango N, Elhaik E, Ermolaeva O, Evans JD, Foret S, Fowler GR, Gerlach D, Gibson JD, Gilbert DG, Graur D, Grunder S, Hagen DE, Han Y, Hauser F, Hultmark D, Hunter HCt, Hurst GD, Jhangian SN, Jiang H, Johnson RM, Jones AK, Junier T, Kadowaki T, Kamping A, Kapustin Y, Kechavarzi B, Kim J, Kiryutin B, Koevoets T, Kovar CL, Kriventseva EV, Kucharski R, Lee H, Lee SL, Lees K, Lewis LR, Loehlin DW, Logsdon JM, Jr, Lopez JA, Lozado RJ, Maglott D, Maleszka R, Mayampurath A, Mazur DJ, McClure MA, Moore AD, Morgan MB, Muller J, Munoz-Torres MC, Muzny DM, Nazareth LV, Neupert S, Nguyen NB, Nunes FM, Oakeshott JG, Okwuonu GO, Pannebakker BA, Pejaver VR, Peng Z, Pratt SC, Predel R, Pu LL, Ranson H, Raychoudhury R, Rechtsteiner A, Reese JT, Reid JG, Riddle M, Robertson HM, Romero-Severson J, Rosenberg M, Sackton TB, Sattelle DB, Schluns H, Schmitt T, Schneider M, Schuler A, Schurko AM, Shuker DM, Simoes ZL, Sinha S, Smith Z, Solovyev V, Souvorov A, Springauf A, Stafflinger E, Stage DE, Stanke M, Tanaka Y, Telschow A, Trent C, Vattathil S, Verhulst EC, Viljakainen L, Wanner KW, Waterhouse RM, Whitfield JB, Wilkes TE, Williamson M, Willis JH, Wolschin F, Wyder S, Yamada T, Yi SV, Zecher CN, Zhang L, Gibbs RA. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH. Structural cuticular proteins from arthropods: annotation, nomenclature, and sequence characteristics in the genomics era. Insect Biochem Mol Biol. 2010;40:189–204. doi: 10.1016/j.ibmb.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis JH, Papandreou NC, Iconomidou VA, Hamodrakas SJ. 5 - Cuticular Proteins. In: Gilbert LI, editor. Insect Molecular Biology and Biochemistry. Academic Press; San Diego: 2012. pp. 134–166. [Google Scholar]
- Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qian J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, Yu M, Shen W, Wu D, Xiang Z, Yu J, Wang J, Li R, Shi J, Li H, Su J, Wang X, Zhang Z, Wu Q, Li J, Zhang Q, Wei N, Sun H, Dong L, Liu D, Zhao S, Zhao X, Meng Q, Lan F, Huang X, Li Y, Fang L, Li D, Sun Y, Yang Z, Huang Y, Xi Y, Qi Q, He D, Huang H, Zhang X, Wang Z, Li W, Cao Y, Yu Y, Yu H, Ye J, Chen H, Zhou Y, Liu B, Ji H, Li S, Ni P, Zhang J, Zhang Y, Zheng H, Mao B, Wang W, Ye C, Wong GK, Yang H. A draft sequence for the genome of the domesticated silkworm (Bombyx mori) Science. 2004;306:1937–1940. doi: 10.1126/science.1102210. [DOI] [PubMed] [Google Scholar]
- Zhan S, Reppert SM. MonarchBase: the monarch butterfly genome database. Nucleic acids research. 2013;41:D758–763. doi: 10.1093/nar/gks1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
All the alignments that were used to build the profile Hidden Markov Models given in Clustal format. The numbers at the end of each row in the alignments, correspond to the last amino acid as numbered in the characteristic region, not in the original sequence.
Accession numbers of sequences identified by our tool. Each tab contains a family and each column a species. Red background: no PF00379 domain found in that CPR (the hmmsearch was performed with all filters off). Dark pink font: unclassified CPR scored highest as a CPR_RR-1, dark blue font: unclassified CPR scored highest as a CPR_RR-2. Yellow background: peptides than came from the same gene not counted in “Total minus Splice Variants” and “% Reduced Proteome” tabs, at the end of this file.
Comparison between Updated Willis et al. (2012), part of Table 3, and CutProtFam-Pred results, in gene numbers.