Abstract
Background
Peroxisome proliferator-activated receptors (PPARs) are transcriptional factors involved in several biological processes such as inflammation, cancer growth, progression and apoptosis that are important in lung and upper aero-digestive tract (UADT) cancer outcomes. Nonetheless, there are no published studies of the relationship between PPARs gene polymorphisms and survival of patients with lung cancer or UADT cancers.
Methods
1,212 cancer patients (611 lung, 303 oral, 100 pharyngeal, 90 laryngeal, and 108 esophageal) were followed for a median duration of 11 years. We genotyped three potentially functional single nucleotide polymorphisms (SNPs) using Taqman--rs3734254 of the gene PPARD and rs10865710 and rs1801282 of the gene PPARG--and investigated their associations with lung and UADT cancer survival using Cox regression. A semi-Bayesian shrinkage approach was used to reduce the potential for false positive findings when examining multiple associations.
Results
The variant homozygote CC (vs. TT) of PPARD rs3734254 was inversely associated with mortality of both lung cancer (adjusted hazard ratio [aHR] = 0.63, 95% confidence interval [CI] = 0.42, 0.96) and UADT cancers (aHR = 0.51, 95% CI = 0.27, 0.99). Use of the semi-Bayesian shrinkage approach yielded a posterior aHR for lung cancer of 0.66 (95% posterior limits = 0.44, 0.98) and a posterior aHR for UADT cancers of 0.58 (95% posterior limits = 0.33, 1.03).
Conclusion
Our findings suggest that lung-cancer patients with the CC variant of PPARD rs3734254 may have a survival advantage over lung-cancer patients with other gene variants.
Keywords: Single nucleotide polymorphism, peroxisome proliferator-activated receptors, survival, lung cancer, upper aero-digestive tract cancers
Introduction
Lung cancer and upper aero-digestive tract (UADT) cancers are common cancers that are responsible for serious morbidity and mortality. According to Globocan 2012, lung cancer ranks third in incidence and first in mortality of cancers in the world; and third in incidence and first in mortality of cancers in the United States (U.S.) [1]. The overall 5-year survival of lung cancer is 16.6% in the U.S. in 2013 [2, 3]. UADT cancers comprise the cancers of the airway and upper digestive tract, specifically, oral cavity, pharynx, larynx and esophagus, which are contiguous and commonly exposed to inhaled and sometimes swallowed substances. Collectively, UADT cancers rank fifth in incidence and fourth in mortality of cancers in the world and seventh in incidence and sixth in mortality of cancers in the U.S. in 2012 [1]. The overall 5-year survival is 61% for head and neck cancer (HNC) and 17.3% for esophagus cancer in the U.S. in 2013 [2]. Despite advances in understanding the pathogenesis of lung and UADT cancers, improvements in surgical procedures and the introduction of newer treatment regimens, survival, especially for lung and esophageal cancers has not changed much [2, 4]. At the present time, stage, histology and treatment are thought to be related to cancer prognosis [5]; however there is only a limited amount of information available on genetic factors and cancer prognosis for both lung cancer and UADT cancers.
Peroxisome proliferator-activated receptors (PPARs) are ligand-activated transcription factors belonging to the nuclear hormone receptor superfamily [6–8]. Initially, PPARs are found to regulate genes that control glucose and lipid metabolism and are linked to type 2 diabetes mellitus risk [9, 10]. Later it is discovered that they are involved in many other important biological functions, including early development, inflammation, cell differentiation, proliferation, and apoptosis [6, 11].
Several studies on lung cancer illustrate that the activation of PPAR-γ could constrain cancer cell growth and oral administration of synthesized PPAR-γ ligand could inhibit lung cancer progression and metastases [12–14]. Compared to extensive research on PPAR-γ, studies on PPAR-δ are fewer and focus mostly on the digestive system, with inconsistent results [15–17]. Limited studies of non-small cell lung cancer (NSCLC) show that the function of PPAR-δ is different from that of PPAR-γ and that the former increases cancer proliferation [18]. Considering the cancer-related links with PPARs and the lack of epidemiologic studies examining their relation with lung- and UADT- cancer survival, we conducted this study to explore the association between PPARs gene polymorphisms and survival among patients with lung and UADT cancers.
Methods
Study Population
The study population consisted of newly diagnosed cancer cases identified for the University of California at Los Angeles (UCLA) and University of Southern California (USC) population-based case-control study between 1999 and 2004 in Los Angeles County (LAC) [19]. These subjects included pathologically confirmed new cases of lung cancer or UADT cancers (oral cancer, pharyngeal cancer, laryngeal cancer and esophageal cancer) identified by the rapid ascertainment system of the LA County Cancer Registry under the Cancer Surveillance Program at USC [19]. Recurrent cancer cases were excluded from our study.
Recruitment occurred from 1999 to 2004. All participants were residents of LAC, aged 18 to 65 years old at the time of diagnosis and able to speak either English or Spanish or had a translator on site. Vital status was determined through linking the cases with the Social Security Death Index. The survival time was calculated as the interval between the date of diagnosis and the date of death, or the date of the last follow-up which was July 13th, 2012. The median follow-up time was 11.1 years in all cases, and 11.5 years in lung cancer cases and 10.8 years in UADT cancer cases, respectively.
Recruitment rates among eligible cancer patients were 39%, 54%, 45%, 42% and 35% for lung, oral, pharyngeal, laryngeal and esophageal cancer, respectively. A total of 1,212 patients were included in our study. There were 611 patients with lung cancer, including 95 squamous carcinomas (SQC), 297 adenocarcinomas (ADC), 115 large cell carcinomas (LCC), 75 small cell lung cancers (SCLC) and 29 others. There were a total of 601 patients with UADT cancers, including 303 patients with oral cancer, 100 with pharyngeal cancer and 90 with laryngeal cancer; and 108 esophageal cancer patients. 497 UADT cancer patients were squamous carcinomas, 74 were esophageal adenocarcinomas, and 30 were other cell types.
Data Collection
Trained interviewers used study specific standardized questionnaires to collect subject-reported data including age, gender, ethnicity, education level, smoking and alcohol drinking. Tobacco smokers were defined as those who smoked more than 100 cigarettes in their lifetimes. Alcohol drinkers were defined as those who drank at least one alcoholic drink per month for a period of at least six months. Cumulative level of smoking was measured by pack-years, which were calculated by summing packs per day times the number of years that a subject smoked that amount prior to the diagnosis of cancer. One pack-year is equivalent to smoking one pack per day for one year. Alcohol drinking was measured by the average number of drinks (including wine, beer or liquor) consumed per day. Interviews occurred within six months of diagnoses for 89% of cases.
Buccal cells were collected for DNA analysis by asking subjects to brush their buccal mucosa and rinse with mouthwash. Response rates for interviewed participants providing buccal cells were 89%, 68%, 88%, and 90% for lung, oral and pharyngeal, laryngeal and esophageal cancer cases, respectively.
All specimens were transported and stored at −70°C in the Molecular Epidemiology Laboratory at UCLA, Fielding School of Public Health. DNA samples were isolated by using a modified phenol-chloroform assay [20]. We selected single nucleotide polymorphisms (SNPs) of which the minor allele frequencies (MAFs) in Caucasians were ≥ 5%; when the pairwise linkage disequilibrium (LD) r2 was ≥ 0.8, we picked non-synonymous SNPs or SNPs located in regions regulating gene transcriptions, such as promoter areas from the National Center for Biotechnology Information SNP database. A total of one SNP in the gene PPARD (rs3734254) and four SNPs in the gene PPARG (rs10865710, rs1801282, rs3856806, and rs13306747) were selected. SNP genotyping was done using TaqMan (Applied Biosystems (ABI, Foster City, CA) 7900HT). Samples were first held at 92°C for 10 minutes; then underwent 60 thermocycles of denaturing at 92°C for 15 seconds and annealing at 62°C for 80 seconds. After PCR amplification, end-point fluorescence was read using the ABI 7900HT Sequence Detection System, and genotypes were coded using the SDS 2.3 Allelic Discrimination Software. SNPs that did not meet the criteria of Hardy-Weinberg equilibrium (HWE) p-value ≥ Bonferroni-adjusted p-value of 0.01 and a genotyping call rate ≥ 95% were excluded, leaving rs3734254, rs10865710 and rs1801282 in the analysis.
Statistical Analysis
We first analyzed SNP genotypes (TT, TC, CC or CC, CG, GG) as dummy variables. These results were used to decide the appropriateness of the dominant or recessive model. Cox regression was used to estimate crude and adjusted hazard ratios (cHRs and aHRs) and their corresponding 95% confidence intervals (CIs). SNP genotypes were also treated as ordinal variables to be tested for p-trends. For lung cancer, we adjusted for age, gender, ethnicity, education level, and smoking; we also adjusted for cell differentiation and morphology that included lung SQC, lung ADC, LCC and SCLC. For UADT cancers, we adjusted for age, gender, ethnicity, education level, and smoking and alcohol drinking; we also adjusted for morphology including UADT SQC and ADC, and cell differentiation. Proportionality assumptions were checked in each model, and no noteworthy violations were detected. We also performed semi-Bayesian shrinkage to adjust for potential false discoveries caused by multiple comparisons [21, 22]. Due to lack of previous relevant epidemiology studies and based on existing biology evidence, we applied a Normal coefficient prior with mean 0 and variance 0.5 (corresponding to HR = 1.0, 95% prior limits = 0.25, 4.00, after exponentiation) for the adjustment. All statistical analyses were conducted using SAS v9.3 software (SAS Institute Inc., Cary, NC). We used R 2.15.1 (The R Foundation for Statistical Computing) to construct Kaplan-Meier curves for each SNP.
The study was approved by the institutional review boards of UCLA and USC. Informed consents were obtained from all participants.
Results
Study Population
Table 1 shows the demographic characteristics of the deaths and censored survivors of lung cancer and UADT cancers separately. For lung cancer, the median survival time was 2.5 years; while the median survival time of UADT cancers was 9.4 years.
Table 1.
Demographic characteristics in lung and UADT cancer cases, respectively.
| Lung cancer (N = 611) |
UADT (N = 601) |
|||||||
|---|---|---|---|---|---|---|---|---|
| All, n | Death, n (%) | Censored, n (%) | Median survival years | All, n | Death, n (%) | Censored, n (%) | Median survival years | |
| Survival | 611 | 406 (66) | 205 (34) | 2.5 | 601 | 248 (41) | 353 (59) | 9.4 |
| Age of Diagnosis, mean (SD) | 52.6±5.3 | 51.5±5.7 | 51.2±7.3 | 49.8±7.7 | ||||
| <45 | 61 | 38 (62) | 23 (33) | 3.0 | 109 | 38 (35) | 71 (65) | 10.1 |
| 45–54 | 301 | 188 (62) | 113 (38) | 2.9 | 267 | 105 (39) | 162 (61) | 9.6 |
| 55+ | 249 | 180 (72) | 69 (28) | 2.2 | 225 | 105 (47) | 120 (53) | 9.0 |
| Sex | ||||||||
| Male | 303 | 215 (71) | 88 (29) | 2.0 | 454 | 191 (42) | 263 (53) | 9.3 |
| Female | 308 | 191 (62) | 117 (38) | 3.7 | 147 | 57 (39) | 90 (61) | 9.7 |
| Ethnicity | ||||||||
| Caucasian | 359 | 245 (68) | 114 (32) | 2.4 | 341 | 135 (40) | 206 (60) | 9.4 |
| Hispanic | 70 | 44 (63) | 26 (37) | 2.2 | 109 | 46 (42) | 63 (58) | 9.4 |
| Aincan-Amerian | 96 | 60 (62) | 36 (38) | 3.0 | 69 | 39 (56) | 30 (43) | 5.6 |
| Asian-American | 70 | 46 (66) | 24 (34) | 2.8 | 64 | 21 (33) | 43 (67) | 99 |
| Other | 15 | 10 (67) | 5 (33) | 4.1 | 16 | 6 (33) | 10 (62) | 9.0 |
| Morphology | ||||||||
| Squamous cell | 95 | 53 (56) | 42 (44) | 5.8 | 497 | 195 (39) | 302 (61) | 9.6 |
| Adenocarcinoma | 297 | 186 (63) | 111 (34) | 3.4 | 74 | 42 (57) | 32 (43) | 3.6 |
| Large cell | 115 | 85 (74) | 30 (26) | 2.1 | - | - | ||
| Small cell | 75 | 60 (80) | 15 (20) | 1.4 | - | - | ||
| Other | 29 | 22 (76) | 7 (24) | 1.5 | 30 | 11 (37) | 19 (63) | 9.9 |
| Education (years of schooling) | 13.2±3.3 | 13.4±3.5 | 12.9±3.6 | 13.3±3.7 | ||||
| 0–12 | 265 | 181 (68) | 84 (32) | 2.6 | 273 | 117 (43) | 156 (57) | 9.3 |
| 13–16 | 275 | 181 (66) | 94 (34) | 2.4 | 259 | 110 (42) | 149 (58) | 9.4 |
| >16 | 71 | 44 (62) | 27 (38) | 2.8 | 69 | 21 (30) | 48 (70) | 10.1 |
| Tumor cell differentiation | ||||||||
| Well to moderate | 168 | 90 (22) | 78 (38) | 7.1 | 397 | 171 (43) | 226 (57) | 9.3 |
| Poor to very poor | 222 | 154 (38) | 68 (33) | 2.0 | 121 | 42 (35) | 79 (65) | 9.8 |
| Undeteimined | 219 | 161 (40) | 58 (28) | 2.0 | 81 | 34 (42) | 47 (58) | 9.6 |
| Smoking (pack-years) | 33.11±24.78 | 27.89±25.35 | 28.3±25.8 | 18.4±22.5 | ||||
| Never | 110 | 61 (55) | 49 (44) | 5.5 | 182 | 53 (29) | 129 (71) | 10.1 |
| Ever | 501 | 345 (69) | 156 (31) | 2.2 | 419 | 195 (46) | 224 (53) | 9.0 |
| less than 20 | 98 | 63 (64) | 35 (36) | 2.6 | 145 | 51 (35) | 94 (65) | 9.7 |
| 20 – 40 | 201 | 139 (69) | 62 (31) | 2.5 | 146 | 71 (49) | 75 (51) | 8.8 |
| 40 or more | 202 | 143 (71) | 59 (29) | 1.9 | 128 | 73 (57) | 55 (43) | 6.8 |
| Alcohol drinking status (drinks-day) | 1.62±3.08 | 1.63±5.06 | 3.1±4.6 | 2.1±4.6 | ||||
| Never | 170 | 111 (65) | 59 (34) | 2.8 | 117 | 45 (38) | 72 (62) | 9.8 |
| Ever | 440 | 294 (67) | 146 (33) | 2.5 | 482 | 202 (42) | 280 (58) | 9.4 |
| < 2 drinks/day >=2 | 302 | 200 (66) | 102 (34) | 2.4 | 279 | 97 (35) | 182 (65) | 9.8 |
| drinks/day | 138 | 94 (68) | 44 (32) | 2.8 | 203 | 105 (52) | 93 (48) | 8.2 |
For lung cancer, the proportion of deaths was higher in the age group 55 or older (72%), comparing to those younger than 55 years of age (62%). It was higher in the male (71%) than in the female (62%). Among different morphological types, SCLC was associated with the highest mortality (80%), and lung SQC, the lowest (56%). Mortality increased with cell differentiation, varying from 54% to 74%. More deaths occurred in smokers than in nonsmokers (69% vs. 55%), and the proportion increased slightly with increasing number of pack-years.
For UADT cancers, the mortality was higher in the patients aged 55 years or older (47%) than in the younger patients (35% – 39%); and higher in esophageal adenocarcinomas (57%) than other morphological types (37% – 39%). Higher proportion of deaths was observed among smokers than among nonsmokers (46% vs. 29%); also more in alcohol drinkers than in non-alcohol drinkers (42% vs. 38%).
SNP Analysis
Table 2 presents the crude and adjusted hazard ratios for lung cancer mortality and corresponding adjusted posterior estimates after semi-Bayesian shrinkage of selected SNPs, stratified by morphological types.
Table 2.
Selected crude and adjusted hazard ratio of PPAR SNPs and lung cancer survival, stratified by morphological types*.
|
PPARD rs3734254 T>C |
PPARG rs10865710 C>G |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Death/All | cHR (95% CI) | aHR** (95% CI) | Posterior aHR** (95% posterior limits) | Genotype | Death/All | cHR (95% CI) | aHR** (95% CI) | Posterior aHR** (95% posterior limits) |
| Lung Cancer (N=611) | |||||||||
| TT | 193/289 | 1.00 | 1.00 | 1.00 | CC | 186/286 | 1.00 | 1.00 | 1.00 |
| TC | 112/168 | 1.02(0.81, 1.29) | 1.07(0.83, 1.37) | 1.08(0.84, 1.37) | CG | 130/192 | 1.11(0.88, 1.38) | 1.06(0.84, 1.34) | 1.06(0.85, 1.33) |
| CC | 29/55 | 0.65(0.44, 0.96) | 0.63(0.42, 0.96) | 0.66(0.44, 0.98) | GG | 24/39 | 0.99(0.65, 1.51) | 1.11(0.71, 1.73) | 1.10(0.72, 1.68) |
| Ptrend | 0.10 | 0.15 | 0.16 | Ptrend | 0.62 | 0.54 | 0.54 | ||
| Recessive | 29/55 | 0.64(0.44, 0.94) | 0.61(0.41, 0.92) | 0.64(0.43, 0.94) | Recessive | 24/39 | 0.95(0.63, 1.44) | 1.07(0.70, 1.66) | 1.07(0.71, 1.62) |
| Non-SCLC (N=536) | |||||||||
| TT | 162/251 | 1.00 | 1.00 | 1.00 | CC | 162/254 | 1.00 | 1.00 | 1.00 |
| TC | 97/150 | 1.04(0.81, 1.34) | 1.04(0.80, 1.36) | 1.05(0.80, 1.36) | CG | 105/162 | 1.05(0.82, 1.34) | 1.03(0.80, 1.33) | 1.03(0.80, 1.32) |
| CC | 24/46 | 0.69(0.45, 1.06) | 0.69(0.44, 1.09) | 0.72(0.47, 1.10) | GG | 22/36 | 1.00(0.64, 1.55) | 1.14(0.71, 1.33) | 1.12(0.72, 1.76) |
| Ptrend | 0.24 | 0.28 | 0.28 | Ptrend | 0.83 | 0.63 | 0.63 | ||
| Recessive | 24/46 | 0.68(0.45, 1.03) | 0.68(0.44, 1.05) | 0.70(0.47, 1.06) | Recessive | 22/36 | 0.98(0.63, 1.51) | 1.12(0.71, 1.78) | 1.11(0.72, 1.72) |
| SQC (N=95) | |||||||||
| TT | 23/43 | 1.00 | 1.00 | 1.00 | CC | 18/35 | 1.00 | 1.00 | 1.00 |
| TC | 16/27 | 1.22(0.65, 2.32) | 1.51(0.74, 3.05) | 1.40(0.75, 2.60) | CG | 22/37 | 1.27(0.68, 2.37) | 1.35(0.68, 2.67) | 1.26(0.69, 2.29) |
| CC | 5/10 | 0.94(0.36, 2.46) | 0.91(0.30, 2.71) | 0.92(0.39, 2.13) | GG | 4/8 | 0.95(0.32, 2.80) | 1.21(0.35, 4.19) | 1.09(0.43, 2.75) |
| Ptrend | 0.87 | 0.75 | 0.77 | Ptrend | 0.76 | 0.50 | 0.53 | ||
| Recessive | 5/10 | 0.86(0.34, 2.20) | 0.74(0.26, 2.09) | 0.82(0.36, 1.86) | Recessive | 4/8 | 0.84(0.30, 2.34) | 1.01(0.31, 3.27) | 1.01(0.41, 2.47) |
| ABC (N=297) | |||||||||
| TT | 82/137 | 1.00 | 1.00 | 1.00 | CC | 83/141 | 1.00 | 1.00 | 1.00 |
| TC | 54/87 | 1.13(0.80, 1.60) | 1.19(0.82, 1.73) | 1.18(0.83, 1.68) | CG | 57.89 | 1.22(0.87, 1.71) | 1.11(0.77, 1.58) | 1.09(0.78, 1.54) |
| CC | 13/23 | 0.88(0.49, 1.58) | 0.99(0.51, 1.92) | 0.99(0.55, 1.79) | GG | 13/20 | 1.37(0.76, 2.46) | 1.24(0.65, 2.33) | 1.19(0.66, 2.12) |
| Ptrend | 0.97 | 0.64 | 0.64 | Ptrend | 0.16 | 0.45 | 0.46 | ||
| Recessive | 13/23 | 0.84(0.48, 1.49) | 0.90(0.48, 1.69) | 0.92(0.52, 1.63) | Recessive | 13/20 | 1.27(0.72, 2.24) | 1.18(0.64, 2.17) | 1.15(0.65, 2.02) |
| LCC (N=115) | |||||||||
| TT | 45/56 | 1.00 | 1.00 | 1.00 | CC | 50/64 | 1.00 | 1.00 | 1.00 |
| TC | 23/31 | 0.82(0.50, 1.35) | 0.68(0.37, 1.25) | 0.75(0.44, 1.30) | CG | 19/27 | 0.74(0.44, 1.26) | 0.85(0.48, 1.48) | 0.87(0.52, 1.45) |
| CC | 4/9 | 0.34(0.12, 0.95) | 0.32(0.09, 1.06) | 0.50(0.22, 1.15) | GG | 4/7 | 0.55(0.20, 1.52) | 0.87(0.26, 2.92) | 0.93(0.38, 2.29) |
| Ptrend | 0.04 | 0.03 | 0.04 | Ptrend | 0.14 | 0.59 | 0.61 | ||
| Recessive | 4/9 | 0.37(0.13, 1.01) | 0.36(0.11, 1.20) | 0.54(0.24, 1.23) | Recessive | 4/7 | 0.60(0.22, 1.65) | 0.94(0.28, 3.07) | 0.96(0.39, 2.36) |
| SCLC (N=75) | |||||||||
| TT | 31/38 | 1.00 | 1.00 | 1.00 | CC | 24/32 | 1.00 | 1.00 | 1.00 |
| TC | 15/18 | 1.04(0.56, 1.92) | 1.22(0.61, 2.45) | 1.19(0.64, 2.22) | CG | 25/30 | 1.28(0.73, 2.25) | 1.03(0.54, 1.95) | 1.03(0.58, 1.84) |
| CC | 5/9 | 0.43(0.17, 1.10) | 0.67 (0.20, 2.18) | 0.78(0.32, 1.91) | GG | 2/3 | 1.17(0.28, 4.93) | 0.80(0.17, 3.69) | 0.90(0.33, 2.46) |
| Ptrend | 0.13 | 0.84 | 0.85 | Ptrend | 0.45 | 0.91 | 0.92 | ||
| Recessive | 5/9 | 0.42(0.17, 1.07) | 0.63(0.20, 2.03) | 0.76(0.32, 1.84) | Recessive | 2/3 | 1.04(0.25, 4.27) | 0.79(0.18, 3.43) | 0.89(0.33, 2.39) |
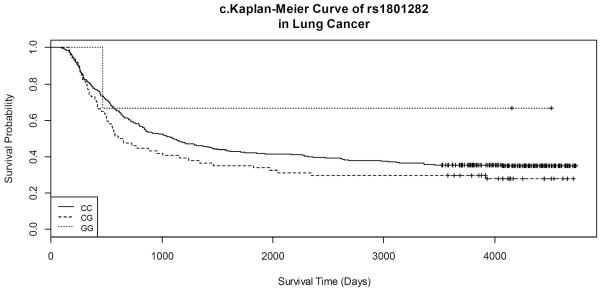
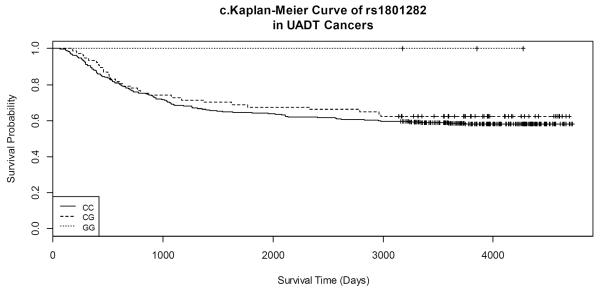
Due to many 0 counts of PPARG rs1801282, the corresponding results were presented in supplementary tables.
Adjusted by age, gender, ethnicity, education level, smoking as packyears and tumor cell differentiation and morphology including squamous carcinoma, adenocarcinoma and large cell carcinoma and small cell carcinoma, if applicable.
For lung cancer overall, only the SNP of the gene PPARD was associated with mortality. The CC (vs. TT) variant of rs3734254 was inversely associated with mortality (cHR = 0.65; 95% CI = 0.44, 0.96). After adjusting for age, gender, ethnicity, education levels, pack-years of smoking, tumor cell differentiation and morphology, the association changed little (aHR = 0.63; 95% CI = 0.42, 0.96). Moreover, the association persisted after semi-Bayesian shrinkage (posterior aHR = 0.66; 95% posterior limits = 0.44, 0.98). We observed similar associations in the recessive model (CC vs. TT+TC), both from the actual data (cHR = 0.64; 95% CI = 0.44, 0.94 and aHR = 0.61; 95% CI = 0.41, 0.92) and the posterior estimations (posterior aHR = 0.64; 95% posterior limits = 0.43, 0.94). When stratified by morphological types, we observed the CC (vs. TT) genotype of rs3734254 was associated with decreased risk of death in LCC in the crude model (cHR = 0.34; 95% CI = 0.12, 0.95) but not in the adjusted model (aHR = 0.32; 95% CI = 0.09, 1.06) or in the recessive models (CC vs. TT+TC). The crude association was weakened after semi-Bayesian shrinkage (posterior cHR = 0.49; 95% posterior limits = 0.23, 1.03) (presented in the supplementary table 1).
Similar to the lung cancer findings, the CC (vs. TT) variant of rs3734254 was inversely associated with UADT cancer mortality (aHR = 0.51; 95% CI = 0.27, 0.99). However, semi-Bayesian shrinkage weakened the association and increased the width of the posterior interval (posterior aHR = 0.58; 95% posterior limits = 0.33, 1.03).
No associations were observed between SNP variants of the PPARG gene and lung cancer or UADT cancer mortality.
We further combined genotypes of all three SNPs (rs3734254, rs10865710 and rs1801282) based on the results from recessive models to evaluate joint effects on survival. However, the combined PPAR genotypes did not provide additional information on survival of lung or UADT cancer patients (data not shown).
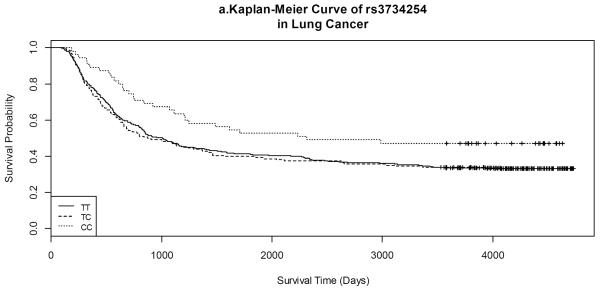
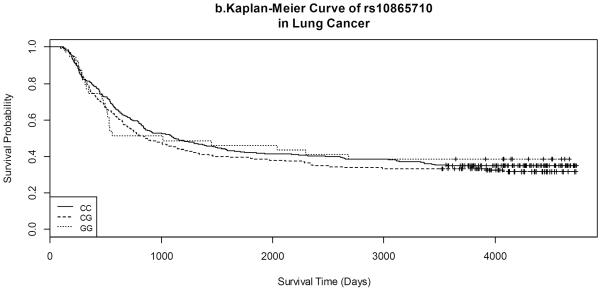
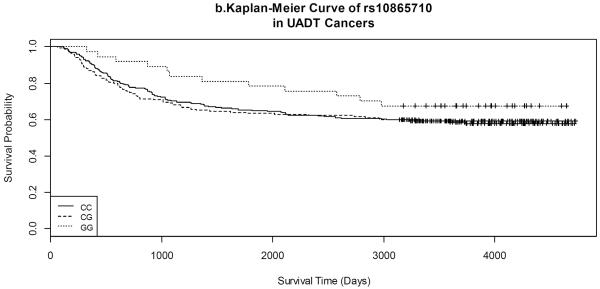
Kaplan-Meier survival curves by gene variants of each SNP for both lung and UADT cancers are shown in Figures 1 and 2. Lung cancer patients with the CC variant of rs3734254 showed a clear survival advantage over the lung cancer patients of the other two genotypes (Log-rank test: p = 0.02, Figure 1a). In UADT cancers, better survival was observed for the rs10865710 GG variant carriers, in comparison with that of the CC/CG carriers; however, the log rank p-value was 0.20 (Figure 2b).
Figure 1.
Kaplan-Meier Curves of selected SNPs in lung cancer.
Figure 2.
Kaplan-Meier Curves of selected SNPs in UADT cancers.
Gene-environment interactions
Due to the importance of smoking in lung cancer and UADT cancer development, its potential interactions with gene polymorphisms were also explored. We noted that the inverse association of the CC variant of rs3734254 with cancer mortality was observed primarily in lung-cancer smokers (in the recessive model CC vs. TT+TC: aHR = 0.55; 95% CI = 0.35, 0.87). Among lung-cancer non-smokers, the associations were much weaker (in the recessive model CC vs. TT+TC: aHR = 0.84; 95% CI = 0.31, 2.28); and p-value for the interaction between ever vs. never smokers and the CC vs. TT+TC variants was 0.21. We also explored the potential discrepancies of SNP-survival associations between ever and never drinkers in UADT cancer patients and did not observe any apparent variations.
Discussion
We found that the CC variant of rs3734254 of the gene PPARD was inversely associated with mortality in lung cancer and UADT cancers, and the association persisted in lung cancer with semi-Bayesian shrinkage which reduced the issue of false positive findings resulting from multiple comparisons. To date, most published studies have explored the relationship between the gene polymorphisms of PPARG with the incidence of cancer. Among those studies that examined multiple types of cancer, one study on lung cancer in the Chinese population shows that rs2972162 of PPARG is inversely associated with lung cancer, and this association differs little between smokers and non-smokers [23]. Our study was the first to examine the association of polymorphism of PPARs on survival of lung and UADT cancers and we did not find any associations between selected SNPs of PPARG and survival from either lung cancer or UADT cancers. Although rs1801282 coding non-synonymous protein resulting Pro-to-Ala exchange of PPAR-γ2--one isoform of PPAR-γ--causes reduced transcriptional activity, the lack of expression of PPAR-γ2 in the lung/UADT tissues may have resulted in the null findings [24–27]. Also, several studies have suggested that PPAR-γ may play a role in the early stage of lung cancer development rather than its progression and metastasis [12, 28].
PPARD rs3734254 is located on chromosome 6 and at a 3 prime untranslated region(3'-UTR) [29]. Two other SNPs of the gene PPARD are in LD with rs3734254. One is rs2076167 that results in a synonymous change to an asparagine residue and the other is rs1053049 within the PPARD 3'-UTR. [30, 31]. The synonymous SNP (rs2076167) may alter the mRNA structure that might influence protein translation and folding [32]. SNPs at 3'-UTR (rs3734254 included in our study and rs1053049 in LD) might impact the stability of the corresponding mRNAs, all of which may affect the activity of PPAR-δ and influence the fate of cancer cells.
PPAR-δ is ubiquitously expressed in multiple tissues [33, 34]. Studies have shown that PPAR-δ has a compound effect on cancer progression of which the exact mechanism is still under investigations [35–37]. For example, in a lung cancer model, down-regulation of phosphatase and tensin homolog (PTEN) and the activation of the PI3 kinase/AKT pathway have been postulated as underlying mechanisms connecting PPAR-δ and cancer progression[37–40]. In addition, studies have found that PPAR-δ impacts angiogenesis via vascular endothelial growth factor receptors (VEGFRs) depending on the concentration of its agonist [41–43]. PPAR-δ could also directly interact with DNA response elements of target genes and repress transcription activities, in turn to suppress the cell cycle, initiate cell differentiation or apoptosis [44–47]. Inflammation may provide another link between PPAR-δ and cancer. The role of inflammation in lung and UADT cancer progression, metastasis and prognosis has been extensively investigated and acknowledged [48–52]. The NF-κB pathway and the COX2 pathway have been identified as main connections[53, 54]. PPAR-δ is inversely associated with inflammation-related genetic factors, including NF-κB and COX2; also PPAR-δ can inhibit NF-κB-dependent signaling by combining with the p65 subunit of NF-κB, suggesting its potential anti-inflammatory function [47, 55, 56].
Since PPAR-δ plays a pivotal role from energy and blood supply and inflammation reactions to cell differentiation, growth and apoptosis [9, 10, 57–60], it is reasonable to assume that the CC genotype of rs37342574 at 3'-UTR of PPARD identified by our study could affect the transcription of PPARD and change the expression of PPAR-δ, eventually increase survival through the interactive network PPAR-δ entangled. However, future studies employing functional assays and deep sequencing of the PPARD gene would help elucidate the precise polymorphism of functional consequence and the characteristics of a mutated gene product. The inverse association between the CC (vs. TT) genotype of rs37342574 and morphology specific cancer survival was strongest for large cell carcinomas (LCC). However, the association between the CC (vs TT) genotype of rs37342574 and LCC mortality was weakened when adjusted for multiple testing and the 95% posterior limits contained the null after introducing a moderate prior to the actual data, which might indicate that the observation was simply due to chance.
We also explored the potential interactions between gene polymorphisms and environmental risk factors including alcohol drinking and cigarette smoking. The inverse association between the CC (vs. TT) genotype of rs37342574 and cancer death was much stronger in lung-cancer smokers than in non-smokers, but the power for testing this interaction in our data was low. This is the first study on PPARD gene polymorphisms and lung/UADT cancer survival. A major limitation of our study is the partial gene coverage. We analyzed only three SNPs and might have failed to identify functionally important markers. However, our SNP selection strategy of focusing on potential functional SNPs of >=5% MAF increased the chance for identifying relevant markers at the population-level. Another drawback is that there was no tumor-node-metastasis (TNM) cancer staging information in our database. Although we treated the cell differentiation as a proxy, it could not fully substitute for TNM staging and might introduce measurement errors. Measurement errors could also arise from genotyping and disease diagnoses, which would bias the results; unfortunately, we cannot predict the direction or magnitude of such bias. The recruitment rates were relatively low in our study. Nonparticipation of eligible cancer cases due to early death or sickness, especially for lung and esophageal cancers with poor prognoses, might favor less severe patients included in the study. The potential selection bias limits our ability to generalize our findings to all lung cancer patients and might distort our observed associations. It is difficult to predict the direction of the selection bias or to correct it in the analysis, since the imputation of the SNP distribution in the nonparticipants is problematic. However, by employing the semi-Bayesian shrinkage approach with a null prior, the estimated associations were conservative and the validity of our results would be reliable. In fact, for the CC variant of PPARD rs3734254 that was still inversely related with lung cancer death even after semi-Bayesian shrinkage with a null prior, the missing of severe lung cancer patients with shorter survival would bias the association towards the null. The small numbers of certain cancers or morphological subtypes also limit the power and precision with which we could estimate the effects of low-penetrance SNPs. Finally, the biological mechanisms involving PPAR-δ are still not fully understood and controversial [61]. Various mechanisms mentioned above on PPAR-δ have been proposed by different researchers, but they have not been replicated by independent laboratories [57], which prevent an definite explanation about our findings.
Conclusion
We found that the CC variant of PPARD rs3734254 was inversely associated with lung-cancer mortality among diagnosed cases. If replicated in larger populations with higher recruitment rates, these findings may be valuable in lung cancer treatment and prognostic predictions.
Supplementary Material
Highlights.
We explore single nucleotide polymorphisms and cancer survival.
Peroxisome Proliferative Activated Receptor Beta and Delta are major interests.
We apply semi-Bayesian shrinkage to adjust for potential false discoveries.
Lung-cancer cases with the CC variant of rs3734254 may have a survival advantage.
Table 3.
Selected results of crude and adjusted hazard ratio of PPAR SNPs and UADT cancer survival, stratified by morphological types*.
|
PPARD rs3734254 T>C |
PPARG rs10865710 C>G |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Genotype | Death/All | cHR (95% CI) | aHR** (95% CI) | Posterior aHR** (95% posterior limits) | Genotype | Death/All | cHR (95% CI) | aHR** (95% CI) | Posterior aHR** (95% posterior limits) |
| UADT (N=601) | |||||||||
| TT | 123/286 | 1.00 | 1.00 | 1.00 | CC | 96/235 | 1.00 | 1.00 | 1.00 |
| TC | 51/134 | 0.88 (0.63, 1.21) | 0.90 (0.64, 1.27) | 0.92 (0.66, 1.27) | CG | 73/175 | 1.06 (0.78, 1.43) | 1.07 (0.78, 1.46) | 1.07 (0.79, 1.45) |
| CC | 11/54 | 0.71(0.33, 1.31) | 0.51 (0.27, 0.99) | 0.58 (0.33, 1.03) | GG | 12/37 | 0.70 (0.38, 1.28) | 0.79 (0.42, 1.46) | 0.82 (0.47, 1.43) |
| Ptrend | 0.20 | 0.07 | 0.07 | Ptrend | 0.52 | 0.77 | 0.78 | ||
| Recessive | 11/34 | 0.74(0.40, 1.35) | 0.53 (0.28, 1.01) | 0.59 (0.34, 1.04) | Recessive | 12/57 | 0.69 (0.38, 1.23) | 0.76 (0.42, 1.40) | 0.80 (0.46, 1.38) |
| SQC (N=497) | |||||||||
| TT | 91/227 | 1.00 | 1.00 | 1.00 | CC | 71/187 | 1.00 | 1.00 | 1.00 |
| TC | 34/106 | 0.78 (0.53, 1.16) | 0.84 (0.56, 1.26) | 0.86 (0.59, 1.27) | CG | 52/143 | 0.99 (0.69, 1.42) | 0.98 (0.68, 1.41) | 0.98 (0.69, 1.39) |
| CC | 11/31 | 0.87 (0.47, 1.63) | 0.60 (0.30, 1.18) | 0.66 (0.37, 1.20) | GG | 11/30 | 0.91 (0.48, 1.71) | 0.92 (0.48, 1.77) | 0.94 (0.52, 1.68) |
| Ptrend | 0.32 | 0.12 | 0.12 | Ptrend | 0.80 | 0.81 | 0.81 | ||
| Recessive | 11/31 | 0.94 (0.51, 1.73) | 0.63 (0.33, 1.23) | 0.69 (0.38, 1.24) | Recessive | 11/30 | 0.91 (0.49, 1.69) | 0.93 (0.50, 1.76) | 0.95 (0.53, 1.67) |
| Oral and oropharyngeal (N=335) | |||||||||
| TT | 53/145 | 1.00 | 1.00 | 1.00 | CC | 42/116 | 1.00 | 1.00 | 1.00 |
| TC | 20/67 | 0.80 (0.48, 1.35) | 1.05 (0.62, 1.79) | 1.05 (0.64, 1.72) | CG | 28.91 | 0.85 (0.52, 1.37) | 0.86 (0.52, 1.44) | 0.89 (0.55, 1.43) |
| CC | 6/15 | 1.18 (0.51, 2.74) | 0.91 (0.37, 2.27) | 0.94 (0.44, 2.00) | GG | 7/19 | 0.98 (0.44, 2.19) | 0.75 (0.32, 1.74) | 0.81 (0.40, 1.65) |
| Ptrend | 0.80 | 0.96 | 057 | Ptrend | 0.69 | 0.43 | 0.44 | ||
| Recessive | 6/15 | 1.26 (0.55, 2.90) | 0.90 (0.36, 2.21) | 0.93 (0.44, 1.96) | Recessive | 7/19 | 1.05 (0.48, 2.29) | 0.79 (0.35, 1.81) | 0.84 (0.42, 1.69) |
| Laryngeal (N=90) | |||||||||
| TT | 16/39 | 1.00 | 1.00 | 1.00 | CC | 16/41 | 1.00 | 1.00 | 1.00 |
| TC | 6/24 | 0.58 (0.23, 1.48) | 0.62 (0.20, 1.94) | 0.69 (0.30, 1.59) | CG | 11/29 | 1.04 (0.48, 2.24) | 0.98 (0.42, 2.31) | 1.01 (0.49, 2.09) |
| CC | 5/12 | 1.07 (0.39, 2.91) | 2.11 (0.57, 7.83) | 1.57 (0.60, 4.07) | GG | 1/5 | 0.43 (0.06, 3.24) | 0.50 (0.05, 4.87) | 0.81 (0.26, 2.53) |
| Ptrend | 0.76 | 0.50 | 0.55 | Ptrend | 0.60 | 0.69 | 0.72 | ||
| Recessive | 5/12 | 1.28 (0.48, 3.38) | 2.69 (0.81, 8.91) | 1.74 (0.68, 4.44) | Recessive | 1/5 | 0.42 (0.06, 3.11) | 0.50 (0.05, 4.76) | 0.81 (0.26, 2.52) |
Due to the very small sample size of esophageal cancer and many 0 counts of PPARG rs1801282, the corresponding results were presented in supplementary tables.
Adjusted by age, gender, ethnicity, education, smoking as packyears, alcohol drinking as alcoholic drinks per day and tumor cell differentiation and morphology including squamous carcinoma and adenocarcinoma, if applicable.
Acknowledgments
The authors appreciate all participants of the Los Angeles Study for their valuable time, supports and contributions to this study.
This research was partially supported by the National Institutes of Health [Grant Numbers ES011667, CA90833, CA09142, DA11386] and the Alper Research Center for Environmental Genomics of the UCLA Jonsson Comprehensive Cancer Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Statement None declared.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [Internet] International Agency for Research on Cancer; Lyon, France: [accessed on 29/01/2014]. 2013. Available from: http://globocan.iarc.fr. [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.SEER Cancer Statistics Factsheets: Lung and Bronchus Cancer . National Cancer Institute. Bethesda, MD: http://seer.cancer.gov/statfacts/html/lungb.html. [Google Scholar]
- 4.Reka AK, et al. Molecular cross-regulation between PPAR-gamma and other signaling pathways: implications for lung cancer therapy. Lung Cancer. 2011;72(2):154–9. doi: 10.1016/j.lungcan.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol. 2010;17(6):1471–4. doi: 10.1245/s10434-010-0985-4. [DOI] [PubMed] [Google Scholar]
- 6.Kota BP, Huang TH, Roufogalis BD. An overview on biological mechanisms of PPARs. Pharmacol Res. 2005;51(2):85–94. doi: 10.1016/j.phrs.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 7.Willson TM, et al. The PPARs: from orphan receptors to drug discovery. ChemInform. 2000;31(22) doi: 10.1021/jm990554g. no-no. [DOI] [PubMed] [Google Scholar]
- 8.Mangelsdorf DJ, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luquet S, et al. Roles of PPAR delta in lipid absorption and metabolism: a new target for the treatment of type 2 diabetes. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2005;1740(2):313–317. doi: 10.1016/j.bbadis.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann JM, et al. An antidiabetic thiazolidinedione is a high affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ) Journal of Biological Chemistry. 1995;270(22):12953–12956. doi: 10.1074/jbc.270.22.12953. [DOI] [PubMed] [Google Scholar]
- 11.Misra P, et al. Phosphorylation of Transcriptional Coactivator Peroxisome Proliferator-activated Receptor (PPAR)-binding Protein (PBP) Journal of Biological Chemistry. 2002;277(50):48745–48754. doi: 10.1074/jbc.M208829200. [DOI] [PubMed] [Google Scholar]
- 12.Chang TH, Szabo E. Induction of differentiation and apoptosis by ligands of peroxisome proliferator-activated receptor γ in non-small cell lung cancer. Cancer research. 2000;60(4):1129–1138. [PubMed] [Google Scholar]
- 13.Sasaki H, et al. Decreased perioxisome proliferator-activated receptor gamma gene expression was correlated with poor prognosis in patients with lung cancer. Lung Cancer. 2002;36(1):71–76. doi: 10.1016/s0169-5002(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 14.Spagnolo A, et al. Prolonged survival of mice with established intracerebral glioma receiving combined treatment with peroxisome proliferator-activated receptor-γ thiazolidinedione agonists and interleukin-2-secreting syngeneic/allogeneic fibroblasts. Journal of neurosurgery. 2007;106(2):299–305. doi: 10.3171/jns.2007.106.2.299. [DOI] [PubMed] [Google Scholar]
- 15.He T-C, et al. PPARδ is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99(3):335–345. doi: 10.1016/s0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barak Y, et al. Effects of peroxisome proliferator-activated receptor δ on placentation, adiposity, and colorectal cancer. Proceedings of the National Academy of Sciences. 2002;99(1):303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harman FS, et al. Peroxisome proliferator–activated receptor-δ attenuates colon carcinogenesis. Nature medicine. 2004;10(5):481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 18.Keshamouni VG, Han S, Roman J. Peroxisome proliferator-activated receptors in lung cancer. PPAR Res. 2007;2007:90289. doi: 10.1155/2007/90289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashibe M, et al. Marijuana use and the risk of lung and upper aerodigestive tract cancers: results of a population-based case-control study. Cancer Epidemiology Biomarkers & Prevention. 2006;15(10):1829–1834. doi: 10.1158/1055-9965.EPI-06-0330. [DOI] [PubMed] [Google Scholar]
- 20.Cao W, et al. Comparison of methods for DNA extraction from paraffin-embedded tissues and buccal cells. Cancer detection and prevention. 2003;27(5):397–404. doi: 10.1016/s0361-090x(03)00103-x. [DOI] [PubMed] [Google Scholar]
- 21.Greenland S. Bayesian perspectives for epidemiological research. II. Regression analysis. Int J Epidemiol. 2007;36(1):195–202. doi: 10.1093/ije/dyl289. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan SG, Greenland S. Bayesian regression in SAS software. Int J Epidemiol. 2013;42(1):308–17. doi: 10.1093/ije/dys213. [DOI] [PubMed] [Google Scholar]
- 23.Chen D, et al. Genetic variants in peroxisome proliferator-activated receptor-gamma gene are associated with risk of lung cancer in a Chinese population. Carcinogenesis. 2008;29(2):342–50. doi: 10.1093/carcin/bgm285. [DOI] [PubMed] [Google Scholar]
- 24.Sundvold H, Brzozowska A, Lien S. Characterisation of Bovine Peroxisome Proliferator-Activated Receptors γ1 and γ2: Genetic Mapping and Differential Expression of the Two Isoforms. Biochemical and biophysical research communications. 1997;239(3):857–861. doi: 10.1006/bbrc.1997.7564. [DOI] [PubMed] [Google Scholar]
- 25.Deeb SS, et al. A Pro12Ala substitution in PPARγ2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nature genetics. 1998;20(3):284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 26.Fajas L, et al. The organization, promoter analysis, and expression of the human PPARγ gene. Journal of Biological Chemistry. 1997;272(30):18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 27.Database of Single Nucleotide Polymorphisms (dbSNP) National Center for Biotechnology Information, National Library of Medicine; Bethesda (MD): dbSNP accession:{rs1801282}, (dbSNP Build ID: {89/138}). Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 28.Theocharis S, et al. Expression of peroxisome proliferator activated receptor-gamma in non-small cell lung carcinoma: correlation with histological type and grade. Lung Cancer. 2002;36(3):249–255. doi: 10.1016/s0169-5002(02)00013-2. [DOI] [PubMed] [Google Scholar]
- 29.Database of Single Nucleotide Polymorphisms (dbSNP) National Center for Biotechnology Information, National Library of Medicine; Bethesda (MD): dbSNP accession:{rs3734254}, (dbSNP Build ID: {107/138}). Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 30.Database of Single Nucleotide Polymorphisms (dbSNP) National Center for Biotechnology Information, National Library of Medicine; Bethesda (MD): dbSNP accession:{rs2076167}, (dbSNP Build ID: {96/138}). Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 31.Database of Single Nucleotide Polymorphisms (dbSNP) National Center for Biotechnology Information, National Library of Medicine; Bethesda (MD): dbSNP accession:{rs1053049}, (dbSNP Build ID: {86/138}). Available from: http://www.ncbi.nlm.nih.gov/SNP/ [Google Scholar]
- 32.Hunt R, et al. Single Nucleotide Polymorphisms. Springer; 2009. Silent (synonymous) SNPs: should we care about them? pp. 23–39. [DOI] [PubMed] [Google Scholar]
- 33.Wagner KD, Wagner N. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) acts as regulator of metabolism linked to multiple cellular functions. Pharmacol Ther. 2010;125(3):423–35. doi: 10.1016/j.pharmthera.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Braissant O, et al. Differential expression of peroxisome proliferator-activated receptors (PPARs): tissue distribution of PPAR-alpha,-beta, and-gamma in the adult rat. Endocrinology. 1996;137(1):354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 35.Müller Brüsselbach S, et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARβ-deficient mice. The EMBO journal. 2007;26(15):3686–3698. doi: 10.1038/sj.emboj.7601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muller-Brusselbach S, et al. Growth of transgenic RAF-induced lung adenomas is increased in mice with a disrupted PPARss/delta gene. International journal of oncology. 2007;31(3):607. [PubMed] [Google Scholar]
- 37.Pedchenko TV, et al. Peroxisome Proliferator–Activated Receptor β/δ Expression and Activation in Lung Cancer. Am J Respir Cell Mol Biol. 2008;39(6):689. doi: 10.1165/rcmb.2007-0426OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han S, et al. Activation of Peroxisome Proliferator-activated Receptor β/δ (PPARβ/δ) Increases the Expression of Prostaglandin E2 Receptor Subtype EP4 THE ROLES OF PHOSPHATIDYLINOSITOL 3-KINASE AND CCAAT/ENHANCER-BINDING PROTEIN β. Journal of Biological Chemistry. 2005;280(39):33240–33249. doi: 10.1074/jbc.M507617200. [DOI] [PubMed] [Google Scholar]
- 39.Han S, et al. PPARβ/δ agonist stimulates human lung carcinoma cell growth through inhibition of PTEN expression: the involvement of PI3K and NF-κB signals. American Journal of Physiology-Lung Cellular and Molecular Physiology. 2008;294(6):L1238–L1249. doi: 10.1152/ajplung.00017.2008. [DOI] [PubMed] [Google Scholar]
- 40.Tan NS, et al. The nuclear hormone receptor peroxisome proliferator-activated receptor β/δ potentiates cell chemotactism, polarization, and migration. Molecular and cellular biology. 2007;27(20):7161–7175. doi: 10.1128/MCB.00436-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meissner M, et al. PPARdelta agonists suppress angiogenesis in a VEGFR2-dependent manner. Arch Dermatol Res. 2011;303(1):41–7. doi: 10.1007/s00403-010-1091-y. [DOI] [PubMed] [Google Scholar]
- 42.Piqueras L, et al. Activation of PPARβ/δ induces endothelial cell proliferation and angiogenesis. Arteriosclerosis, Thrombosis, and Vascular Biology. 2007;27(1):63–69. doi: 10.1161/01.ATV.0000250972.83623.61. [DOI] [PubMed] [Google Scholar]
- 43.Shirotani M, et al. U-61, 431F, a stable prostacyclin analogue, inhibits the proliferation of bovine vascular smooth muscle cells with little antiproliferative effect on endothelial cells. Prostaglandins. 1991;41(2):97–110. doi: 10.1016/0090-6980(91)90023-9. [DOI] [PubMed] [Google Scholar]
- 44.Michalik L, Desvergne B, Wahli W. Peroxisome-proliferator-activated receptors and cancers: complex stories. Nature Reviews Cancer. 2004;4(1):61–70. doi: 10.1038/nrc1254. [DOI] [PubMed] [Google Scholar]
- 45.Tachibana K, Yamasaki D, Ishimoto K. The role of PPARs in cancer. PPAR Res. 2008;2008 doi: 10.1155/2008/102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Hon M, Evans RM. The peroxisome proliferator-activated receptor δ, an integrator of transcriptional repression and nuclear receptor signaling. Proceedings of the National Academy of Sciences. 2002;99(5):2613–2618. doi: 10.1073/pnas.052707099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters JM, Hollingshead HE, Gonzalez FJ. Role of peroxisome-proliferator-activated receptor β/δ (PPARβ/δ) in gastrointestinal tract function and disease. Clinical science (London, England: 1979) 2008;115(4):107. doi: 10.1042/CS20080022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? The Lancet. 2001;357(9255):539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 49.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5(10):749–59. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 50.Scott H, et al. The systemic inflammatory response, weight loss, performance status and survival in patients with inoperable non-small cell lung cancer. Br J Cancer. 2002;87(3):264–267. doi: 10.1038/sj.bjc.6600466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Forrest LM, et al. A prospective longitudinal study of performance status, an inflammation-based score (GPS) and survival in patients with inoperable non-small-cell lung cancer. Br J Cancer. 2005;92(10):1834–6. doi: 10.1038/sj.bjc.6602591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JM, et al. Inflammation in lung carcinogenesis: new targets for lung cancer chemoprevention and treatment. Critical reviews in oncology/hematology. 2008;66(3):208. doi: 10.1016/j.critrevonc.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mann JR, Backlund MG, DuBois RN. Mechanisms of disease: Inflammatory mediators and cancer prevention. Nature Clinical Practice Oncology. 2005;2(4):202–210. doi: 10.1038/ncponc0140. [DOI] [PubMed] [Google Scholar]
- 54.Ulrich CM, Bigler J, Potter JD. Non-steroidal anti-inflammatory drugs for cancer prevention: promise, perils and pharmacogenetics. Nature Reviews Cancer. 2006;6(2):130–140. doi: 10.1038/nrc1801. [DOI] [PubMed] [Google Scholar]
- 55.Wu CW, Yu J, Sung J. Peroxisome proliferator-activated receptor delta and gastric cancer (Review) Oncology reports. 2009;22(3):451. [PubMed] [Google Scholar]
- 56.Fucci A, et al. The role of peroxisome proliferator-activated receptors in the esophageal, gastric, and colorectal cancer. PPAR Res. 2012;2012:242498. doi: 10.1155/2012/242498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peters JM, Gonzalez FJ. Sorting out the functional role (s) of peroxisome proliferator-activated receptor-β/δ (PPARβ/δ) in cell proliferation and cancer. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2009;1796(2):230–241. doi: 10.1016/j.bbcan.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bishop-Bailey D, Swales KE. The role of PPARs in the endothelium: implications for cancer therapy. PPAR Res. 2008;2008 doi: 10.1155/2008/904251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Müller R, Rieck M, Müller-Brüsselbach S. Regulation of Cell Proliferation and Differentiation by PPAR. PPAR Res. 2008;2008 doi: 10.1155/2008/614852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Barish GD, Narkar VA, Evans RM. PPARdelta: a dagger in the heart of the metabolic syndrome. Journal of Clinical Investigation. 2006;116(3):590. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peters JM, Shah YM, Gonzalez FJ. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat Rev Cancer. 2012;12(3):181–95. doi: 10.1038/nrc3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.