Abstract
Background
The molecular mechanisms involved in plant tolerance to either drought or cold have been extensively studied in many plant species. However, few studies have focused on their comparisons especially using non-model plants with strong tolerance to both stresses. Ammopiptanthus mongolicus (Maxim. ex Kom.) Cheng f. is the only evergreen broadleaf shrub grown in the central Asian desert and it has very strong cold and drought tolerance. To provide further insights into plant tolerance, the transcriptome profiles of drought- and cold-treated A. mongolicus seedlings were analyzed using Illumina technology and differentially expressed genes (DEGs) were compared.
Results
A comprehensive transcriptome of A. mongolicus was sequenced using pooled mRNA extracted from drought-, cold-stressed and unstressed seedlings as well as leaves from naturally grown shrub. These sequences were assembled into 86058 unigenes, of which 51014 unigenes had an annotated function and 2440 encoded transcription factors (TFs). Transcriptome profiles were analyzed in A. mongolicus seedlings after drought and cold treatments at three time points (2, 8 and 24 h). Between 3917 and 6102 unigenes were identified as DEGs at a single time point in both stresses. Among these DEGs 2028 and 2026 DEGs were common across the three time points of drought and cold treatments respectively, and 971 DEGs were co-regulated by both stresses. Functional enrichment analyses identified many common or specific biological processes and gene sets in response to drought and cold stresses. The most pronounced findings are that flavonoid biosynthesis genes were enriched in the DEGs co-up-regulated by both stresses; while membrane protein genes and genes related to chloroplast were abundant in the DEGs specifically up-regulated by drought or cold, respectively. Furthermore, the DREB, ERF, NAC and WRKY TFs were predominantly co-up-regulated by both stresses.
Conclusions
The present study provides the most comprehensive transcriptome resource and the first dynamic transcriptome profiles of A. mongolicus under drought and cold stresses. This information will deepen our understanding of plant tolerance to drought and cold. The up-regulated DEGs will be valuable for further investigations of key genes and molecular mechanisms involved in the adaptation of A. mongolicus to harsh environments.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2164-15-671) contains supplementary material, which is available to authorized users.
Keywords: Ammopiptanthus mongolicus, Drought, Cold, Transcriptome profiling, Illumina sequencing
Background
Drought and low temperature are two major environmental stresses that greatly affect plant growth and crop production worldwide. This is particularly the case in arid and cold areas. To improve the tolerance of crops and other economic plants to these stresses, it is essential to know how plants respond to these stresses and which genes and biological pathways are involved in the stress tolerance. Over the past decades, thousands of genes and dozens of metabolic and signaling pathways have been identified in plants during drought and cold stresses [1–4]. Some of these genes have been confirmed to significantly improve plant tolerance to drought, cold and/or salt stresses [5, 6]. In addition, many genes have been found to be commonly or specifically regulated under drought and cold in Arabidopsis [7], rice [8] and Brachypodium [9] at transcriptional level. However, similar studies are very limited in other plants, and the molecular mechanisms controlling plant tolerance to both drought and cold remain largely unknown.
Ammopiptanthus is an endangered survivor from the Tethys in the Tertiary Period, and is the only evergreen broadleaf shrub grown in the central Asian desert. The genus Ammopiptanthus (Leguminosae) comprises of two species: A. mongolicus (Maxim. ex Kom.) Cheng f. and A. nanus (M. Pop.) Cheng f. Both species are diploid with 18 chromosomes and have high stress tolerance [10]. The habitats of A. mongolicus are marked by arid climate with an annual precipitation less than 200 mm but a mean annual evaporation up to 3000 mm. Its natural distribution areas are also characterized as sandy or stony soil with high salinity, intense ultraviolet irradiation, and seasonally extreme temperature from about –30°C in winter to more than 40°C in summer. The extreme tolerance of this species to harsh environments makes it invaluable for exploring key stress-tolerant genes and mechanisms, especially those involved in cold and drought tolerance.
The transcriptomes of A. mongolicus roots (treated by 20% PEG-6000 for 72 hours) and seedlings (cultured at normal condition or 4°C for 14 days) were recently sequenced by 454 pyrosequencing [11] and Illumina technology [12], respectively. These studies generated 29056 and 82795 unigenes and identified 32728 cold-regulated genes. However, transcriptome profiling of this species under drought stress and its comparison with cold stress have not been reported in the literatures.
In the present study, we firstly performed a comprehensive transcriptome sequencing of A. mongolicus using pooled mRNA extracted from drought-treated, cold-treated and control seedlings as well as young leaves of plants grown in desert in both summer and winter. We then applied RNA-Seq (RNA sequencing) to investigate seven cDNA libraries derived from the seedling samples exposed to drought and cold stresses for 2 h, 8 h and 24 h, and the non-stressed control seedlings respectively. Finally, we analyzed the DEGs and identified common and specific DEGs during drought and cold treatments. Our study provided the first transcriptome profile of A. mongolicus under drought stress and the comparison of it with the transcriptome profile under cold stress.
Results
Sequencing, de novoassembly and functional annotation of the A. mongolicustranscriptome
Illumina sequencing of the pooled mRNA generated 47.29 million (M) clean paired-end reads with Q20 over 98%. De novo assembly of the clean reads resulted in 86058 unigenes with an average length of 756 bp and N50 length of 1279 bp (Table 1). Approximately 41.4% and 22.6% of these unigenes had a length ≥ 500 bp or ≥ 1000 bp, respectively (Figure 1).
Table 1.
Overview of the transcriptome sequencing and assembly
| Item | Reads | Contigs | Unigenes |
|---|---|---|---|
| Total number of raw reads | 50477558 | - | - |
| Total number of clean reads | 47291846 | - | - |
| Total clean nucleotides (bp) | 4256266140 | - | - |
| Q20 percentage (%) | 98.36 | - | - |
| Percentage (%) of assembled reads | 99.84 | - | - |
| Number of contigs or unigenes | - | 190185 | 86058 |
| Average length (bp) | - | 325 | 756 |
| N50 (bp) | - | 525 | 1279 |
Figure 1.
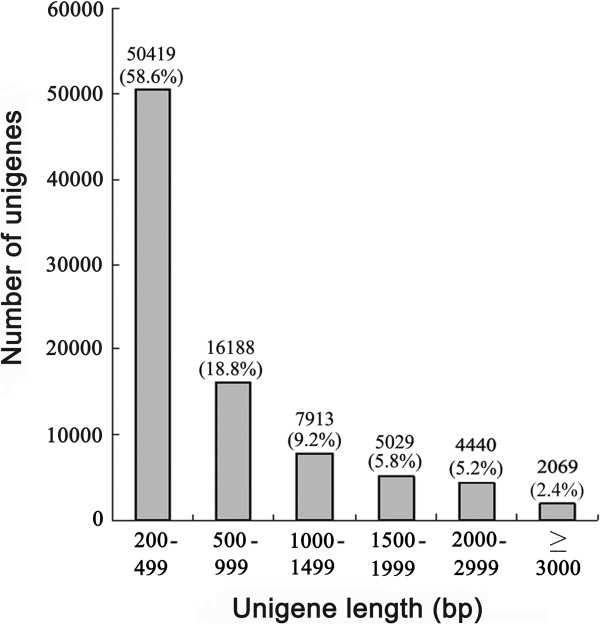
Size distribution of the unigene sequences. The numbers of six groups of unigenes with different lengths (200 to 499 bp, 500 to 999 bp, 1000 to 1499 bp, 1500 to 1999 bp, 2000 to 2999 bp and ≥ 3000 bp) were shown, respectively. The percentages of the unigenes in each group out of the total unigenes (86058) were also indicated.
Of the 86058 sequences, 51014 (59.3%) unigenes had at least one significant match in the NCBI protein databases (E-value ≤ 1e-5). Distribution of the annotated unigenes in each database was shown in the Additional file 1. By searching against the PlantTFDB (http://planttfdb.cbi.edu.cn) (E-value ≤ 1e-5) in combination with annotations in the NR (non-redundant) and Swiss-Prot databases, 2440 unigenes were annotated as transcription factors (TFs), representing 4.8% of the annotated unigenes and covering 57 TF families (Table 2). Furthermore, 47486 unigenes had significant hits in the TIGR Plant Transcript Assemblies database (E-value ≤ 1e-5). Most of these hits were from Glycine max, one of its closest relatives, followed by Medicago truncatula, Lotus japonicus, Malus x, Glycine soja, and others. Surprisingly, only 919 (1.9%) and 787 (1.7%) unigenes had significant similarity to the sequences of Arabidopsis thaliana and Oryza sativa, respectively (Additional file 2).
Table 2.
Transcription factor (TF) gene families identified in the A. mongolicus transcriptome*
| TF family | Number of unigenes | TF family | Number of unigenes | TF family | Number of unigenes |
|---|---|---|---|---|---|
| bHLH | 243 | GTF | 39 | Nin-like | 9 |
| MYB | 196 | HSF | 38 | HB-PHD | 8 |
| AP2/EREBP | 172 | Dof | 35 | WOX | 7 |
| C2H2 | 156 | JUMONJI | 33 | YABBY | 7 |
| MYB-related | 155 | TCP | 32 | BES1 | 6 |
| WRKY | 149 | LBD | 27 | Whirly | 6 |
| bZIP | 127 | NF-YC | 25 | GeBP | 4 |
| C3H | 82 | SBP | 24 | SRS | 4 |
| GRAS | 74 | NF-YA | 18 | PLATZ | 4 |
| G2-like | 68 | ZF-HD | 18 | BBR/BPC | 3 |
| NAC | 60 | GRF | 17 | EIL | 3 |
| CAMTA | 59 | CO-like | 16 | S1Fa-like | 3 |
| FARI | 57 | NF-YB | 16 | VOZ | 3 |
| TALE | 57 | DBB | 15 | LIM | 2 |
| HD-ZIP | 56 | E2F-DP | 15 | NF-X1 | 1 |
| ARF | 54 | M-type | 14 | STAT | 1 |
| Trihelix | 52 | MIKC | 12 | KAN | 1 |
| B3/ABI3 | 43 | ARR-B | 11 | ||
| GATA | 42 | TUB | 11 | ||
| HB-other | 40 | CPP | 10 |
*A total of 2440 TF unigenes were identified, covering 57 TF families.
Of the 86058 unigenes, 23834 unigenes were assigned to 44 GO (Gene Ontology) terms. The terms “Cell”, “Cell part”, “Organelle”, “Binding”, “Catalytic activity”, “Metabolic process”, “Cellular process” and “Response to stimulus” were mostly dominant (Figure 2). Moreover, 17100 unigenes were classified into 25 COG (Clusters of Orthologous Groups) classes. The class “General function prediction only” represented the largest group, followed by the classes “Transcription”, “Signal transduction mechanisms” and “Amino acid transport and metabolism”, etc. Notably, nearly 1600 of the annotated unigenes were assigned to the class of “Function unknown” (Additional file 3).
Figure 2.
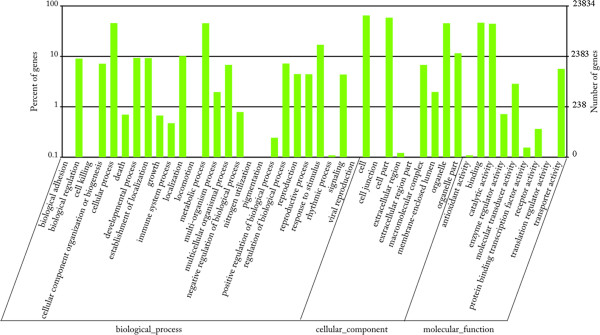
Histogram of GO classification of the annotated unigenes. Unigenes with best BLAST hits were aligned to GO database. In total, 23834 unigenes were assigned to at least one GO term and were grouped into three main GO categories and 44 GO terms. Left Y-axis represented the percentages of unigenes in each main category. Right Y-axis indicated the numbers of unigenes in each GO term.
To gain insights into function interactions among the unigenes, 26999 of the 86058 unigenes were assigned to 125 KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways. Of the top 10 pathways (Table 3), “Metabolic pathways” were mostly represented, followed by “Biosynthesis of secondary metabolites”, “Plant-pathogen interaction” and “Plant hormone signal transduction”, etc. Unigenes encoding core components in the plant hormone signal transduction pathways were present, including ABA (abscisic acid), BR (brassinosteroid) and ethylene signaling pathways. These pathways were well-documented to participate in the response and adaptation of plants to stressful environments [13, 14].
Table 3.
The top 10 enriched pathways in the A. mongolicus transcriptome*
| Pathway | Number of unigenes | Percentage (%) | Pathway ID |
|---|---|---|---|
| Metabolic pathways | 5623 | 20.8 | ko01100 |
| Biosynthesis of secondary metabolites | 2723 | 10.1 | ko1110 |
| Plant-pathogen interaction | 1753 | 6.5 | ko04626 |
| Plant hormone signal transduction | 1740 | 6.4 | ko04075 |
| Purine metabolism | 1483 | 5.5 | ko00230 |
| RNA transport | 1124 | 4.2 | ko03013 |
| Endocytosis | 1004 | 3.7 | ko04144 |
| Splicesome | 945 | 3.5 | ko03040 |
| Ribosome | 908 | 3.4 | ko03010 |
| Glycerophospholipid metabolism | 904 | 3.4 | ko00564 |
*Percentage (%): number of the unigenes assigned to certain pathway/total 26999 unigenes assigned to 125 pathways.
Identification and validation of differentially expressed genes (DEGs)
Transcriptome changes of the A. mongolicus seedlings under drought and cold stresses (2 h, 8 h and 24 h) and unstressed control were analyzed using Illumina technology. Between 8.29 and 8.87 M clean reads (50 bp in length) were obtained from the seven libraries. Approximately 7.18 to 7.78 M of the clean reads in each library were mapped to the 86058 unigenes assembled earlier in this study (mismatch ≤ 2 bp), accounting for 85.0% to 88.4% of the total clean reads (Table 4). In the D1, D2 and D3 libraries (2, 8 and 24 h after drought treatment), 2565, 2703 and 2589 unigenes were up-regulated and 2193, 3160 and 3513 unigenes were down-regulated, respectively. Similar results were observed in the C1, C2 and C3 libraries (2, 8 and 24 h after cold treatment), including 2027, 1894 and 1924 unigenes with up-regulation and 2011, 2023 and 2489 unigenes with down-regulation, respectively (Figure 3). Taken together, 9487 and 6524 unigenes were identified as DEGs with an absolute value of log2 Ratio ≥ 1 and FDR (false discovery rate) ≤ 0.001 at one or more time points during drought and cold stresses, respectively.To validate the RNA-Seq results, 11 DEGs with different expression patterns were selected for RT-qPCR (real-time quantitative PCR) analysis. These genes encode NAC (NAM, ATAF and CUC) and DREB (dehydration responsive element binding) TFs, HSP (heat shock protein), GLP (germin-like protein), TPP (trehalose-6-phosphate phosphatase) and CCD (carotenoid cleavage dioxygenase). Nine (81.8%) of these genes showed similar expression patterns to those detected by the RNA-Seq data (Figure 4), while other two genes had similar expression trends at one to two time points. Thus, the RNA-Seq results were considerably reliable for the identification of DEGs during drought and cold stresses in this study.
Table 4.
Statistics of seven libraries and their reads mapping*
| Library | CK | D1 | D2 | D3 | C1 | C2 | C3 |
|---|---|---|---|---|---|---|---|
| Raw reads (M) | 8.91 | 8.34 | 8.66 | 8.92 | 8.59 | 8.38 | 8.37 |
| Clean reads (M) | 8.86 | 8.29 | 8.62 | 8.87 | 8.52 | 8.32 | 8.32 |
| Mapped reads (M) | 7.53 | 7.31 | 7.62 | 7.78 | 7.42 | 7.18 | 7.23 |
| % of mapped reads | 85.0 | 88.2 | 88.4 | 87.7 | 87.1 | 86.3 | 86.9 |
| Perfectly matched reads (M) | 6.25 | 6.12 | 6.37 | 6.53 | 6.22 | 5.90 | 5.98 |
| % of perfectly matched reads | 71.6 | 73.8 | 73.9 | 74.2 | 72.9 | 70.6 | 71.9 |
| Mismatched reads (M) | 1.28 | 1.19 | 1.25 | 1.25 | 1.20 | 1.29 | 1.24 |
| % of mismatched reads | 14.7 | 14.4 | 14.5 | 14.2 | 14.1 | 15.5 | 15.0 |
| Uniquely matched reads (M) | 5.60 | 5.27 | 5.54 | 5.75 | 5.46 | 5.27 | 5.31 |
| % of uniquely matched reads | 63.2 | 63.6 | 64.3 | 64.8 | 64.1 | 63.3 | 63.8 |
| Unmapped reads (M) | 1.2 | 0.98 | 1.00 | 1.02 | 1.10 | 1.14 | 1.09 |
| % of unmapped reads | 13.5 | 11.8 | 11.6 | 11.5 | 12.9 | 13.7 | 13.1 |
| Total unigenes | 60279 | 61029 | 60149 | 59789 | 61231 | 62693 | 61568 |
| Coverage (%) | 70.0 | 70.9 | 69.9 | 69.5 | 71.2 | 72.9 | 71.5 |
*CK: untreated control; D1, D2 and D3: 2, 8 and 24 h after drought treatment; C1, C2 and C3: 2, 8 and 24 h after cold treatment; M: million; Mismatch: ≤ 2 bp; Coverage (%): total unigenes in each library/86058 assembled unigenes.
Figure 3.
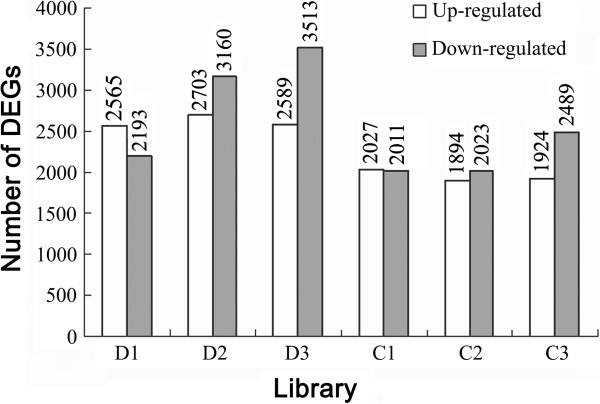
The numbers of DEGs up- and down-regulated in each library. The DEGs (differentially expressed genes) were identified using a threshold of FDR (false discovery rate) ≤ 0.001 and absolute value of log2 Ratio ≥ 1. D1, D2 and D3 represented three drought-treated libraries, corresponding to 2, 8 and 24 h after drought treatment; C1, C2 and C3 represented three cold-treated libraries, corresponding to 2, 8 and 24 h after cold treatment. The numbers of the DEGs in each library were shown in terms of up-regulation and down-regulation, respectively.
Figure 4.
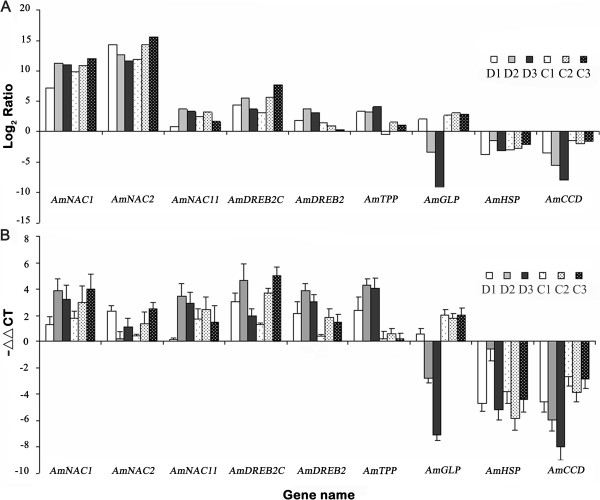
Validation of expression patterns of nine DEGs by RT-qPCR assay. Nine DEGs (differentially expressed genes) showed similar expression patterns between RNA-Seq data (A) and RT-qPCR assay (B). These genes included six genes up-regulated under drought and cold stresses: AmNAC1 (NAM, ATAF and CUC), AmNAC2, AmNAC11, AmDREB2C (dehydration responsive element binding 2C), AmDREB2 and AmTPP (trehalose-6-phosphate phosphatase, not significantly down-regulated in C1 library); two genes down-regulated under both stresses: AmHSP (heat shock protein) and AmCCD (carotenoid cleavage dioxygenase); and one gene with different expression trends: AmGLP (germin-like protein). D1, D2 and D3 represented 2, 8 and 24 h after drought treatment; C1, C2 and C3 represented 2, 8 and 24 h after cold treatment. The total RNAs used for the transcriptome profile analyses using RNA-Seq were used in the RT-qPCR assay, including three biological replicates. The relative expression levels of the selected genes were calculated using the 2–ΔΔCT method. Error bars represented the standard deviation of the mean expression values.
Differentially expressed genes in response to drought stress
Of 4758 to 6102 DEGs identified at the three time points of drought treatment, 2028 DEGs (21.4% of total DEGs) were common across all three time points, including 779 DEGs with up-regulation, 1185 DEGs with down-regulation (Figure 5) and 64 DEGs with complex regulation. The common DEGs represented reproducible changes at each time point. Between 1218 and 1874 DEGs (12.8% to 19.8% of total DEGs) were specifically regulated at a single time point, including 600 to 1178 DEGs with up-regulation and 591 to 1038 DEGs with down-regulation. Besides, many DEGs were common at two of the three time points (Figure 5). The top 100 DEGs commonly regulated by drought stress were listed in the Additional file 4.
Figure 5.
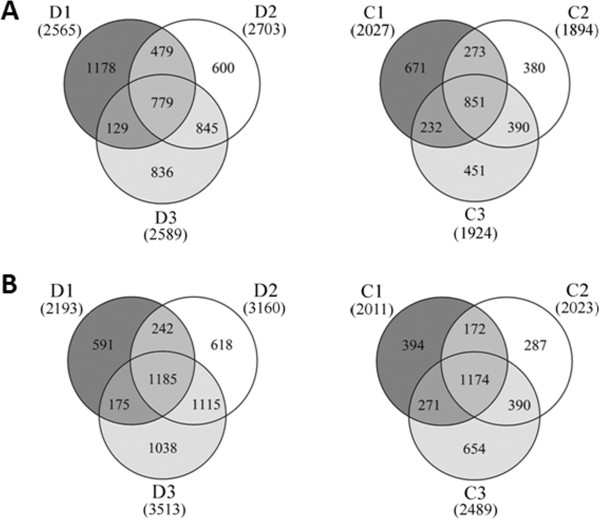
Venn diagrams showing the numbers of common and specific DEGs at different time points of drought and cold treatments. The up-regulated (A) and down-regulated (B) DEGs (differentially expressed genes) identified at 2, 8 and 24 h after drought (D1, D2 and D3) and cold (C1, C2 and C3) treatments were analyzed. The numbers of common and specific DEGs at different time points were shown in the overlapping and non-overlapping regions, respectively. The total numbers of up- and down-regulated DEGs at each time point were indicated in parentheses.
Among the common DEGs up-regulated at the three time points, genes known to respond to drought or other stresses were largely represented, such as those encoding ripening-related proteins, LEA (late embryogenesis abundant) proteins, peroxidases, transporters, enzymes in the flavonoid biosynthetic pathways, protein kinases, and ethylene receptors. Many TFs in multiple families were also up-regulated, including the AP2/EREBP (APETALA2/ethylene-responsive element binding protein), NAC, WRKY, MYB (myeloblastosis oncogene), bHLH (basic helix-loop-helix), C2H2 (Cys2/His2-type zinc finger protein 2), bZIP (basic-domain leucine-zipper protein), C3H (Cys3/His-type zinc finger protein), GRAS (GAT, RGA and SCR), TCP (TB1, CYC and PCF), COL (constans-like), Dof (DNA binding with one finger), and GRF (general regulatory factor) families, etc. Approximately 17.2% (134) of the common DEGs were functional unknowns. Similarly, the common DEGs down-regulated at the three time points of drought stress had a wide range of functions. For example, genes encoding aquaporin NIP, polyol transporter, gibberellin, small heat shock proteins, calcium-binding proteins, and some bHLH TFs appeared in this group. Interestingly, the unknowns in the down-regulated DEGs (397, 33.5%) were almost twice as those in the commonly up-regulated DEGs.
Based on GO enrichment analysis, a large number of the DEGs commonly up-regulated in drought were mostly enriched in two similar biological processes: “Response to stimulus” and “Response to stress”, followed by “Organic substance transport”, “Cellular aromatic compound metabolic process” and “Protein phosphorylation”. In contrast, the commonly down-regulated DEGs were significantly enriched in the biological processes “Monocarboxylic acid metabolism”, “Small molecule biosynthesis” and “Nucleobase transport”. In the cellular component and molecular function categories, the commonly up- and down-regulated DEGs were significantly enriched in similar terms, such as “Membrane” and “Oxidoreductase activity” (Additional file 5). KEGG pathway enrichment analysis revealed that genes involved in microbial metabolism in diverse environments and some secondary metabolite degradation were significantly enriched in both the up- and down-regulated DEGs. However, genes in the pathways for nitrogen and phenylalanine metabolisms were specifically enriched in the up-regulated DEGs; while genes involved in metabolic pathways and the pathways for starch and sucrose metabolism as well as carotenoid biosynthesis were only enriched in the down-regulated DEGs (Additional file 6).
As expected, many regulatory genes were specifically up-regulated at the first time point (2 h), such as genes encoding some types of protein kinases, calcium-binding proteins, TFs, as well as enzymes or components in plant hormone (especially ABA) biosyntheses or signaling pathways. After the treatment was prolonged (8 h to 24 h), many of these regulatory genes were no longer differentially expressed. Instead, a large number of functional genes were up-regulated, such as genes encoding osmoprotectant synthetases, dehydration-protective proteins, PR (pathogenesis-related) proteins, hydrolases, and detoxification enzymes.
Differentially expressed genes in response to cold stress
Between 3917 and 4413 DEGs were identified at a single time point during cold treatment, and 2026 DEGs were common across the three time points, including 851 with up-regulation and 1174 with down-regulation (Figure 5). The top 100 DEGs commonly regulated by cold stress were listed in the Additional file 7. Similar to drought stress, many commonly up-regulated DEGs were annotated to well-known stress- or cold-induced genes. Some of these genes are identical to the drought-induced genes; while others encode different types of proteins, such as fatty acid desaturases, chitinases and proteins or enzymes located in chloroplast. The common DEGs down-regulated at three time points of cold stress also encode various functional and regulatory proteins. Notably, 16.8% (143) and 40.3% (473) of the commonly up- or down-regulated DEGs were unknowns, respectively.
A large number of the DEGs up-regulated under cold stress were significantly enriched in the biological processes “Response to stimulus”, “Response to stress”, “Defense response” and “Cellular aromatic compound metabolic process”. In the down-regulated DEGs, the mostly represented GO term of biological process was also “Response to stimulus”, followed by “Lipid biosynthetic process”, “Organic acid biosynthetic process” and “Lipid localization”. Interestingly, in the cellular component category, the up-regulated DEGs were mostly enriched in “Plastid” and “Chloroplast”; while the down-regulated DEGs were not significantly enriched in any terms of this category (Additional file 8). KEGG pathway analysis revealed that the metabolic pathways and the pathways for secondary metabolite biosynthesis or degradation were significantly enriched in both the up- and down-regulated DEGs. However, the pathways involved in phenylalanine metabolism and photosynthesis were exclusively enriched in the up-regulated DEGs. In contrast, the pathways related to fatty acid metabolism, carotenoid biosynthesis and several amino acid degradations were only enriched in the down-regulated DEGs (Additional file 9).
Differentially expressed genes co-regulated during drought and cold stresses
Among the 9487 drought-regulated and 6524 cold-regulated DEGs with significant expression changes (absolute value of log2 Ratio ≥ 1 and FDR ≤ 0.001) at one or more time points, 4515 DEGs were co-regulated by both stresses, accounting for 47.6% and 69.2% of the total DEGs, respectively. Of the co-regulated DEGs, 971 DEGs had significant expression changes at all drought- and cold-treated time points, including 334 with up-regulation, 618 with down-regulation and 19 with complex regulation patterns. The results clearly showed an overlap between the DEGs in response to drought and cold. The top 100 co-regulated DEGs were listed in the Additional file 10.
Of the 971 co-regulated DEGs, 69 DEGs (33 up- and 36 down-regulated) were annotated as TFs, including the AP2/EREBP, NAC, WRKY, MYB, bHLH, C2H2, bZIP, GRAS, C3H, Dof, TCP, COL and GRF families. In addition, 46 co-regulated DEGs were annotated as protein kinases and phosphatases, and 17 co-regulated DEGs as synthetases or components in plant hormone biosynthesis or signaling. Besides, many stress-related functional genes appeared in the co-up-regulated DEGs, such as those encoding the enzymes in flavonoid biosynthetic pathways, ripening related proteins, thaumatin-like or osmotin-like proteins, transporters, cell wall-associated proteins, and some types of PR proteins.
The co-up-regulated DEGs were significantly aggregated into two GO biological processes: response to stress and secondary metabolite (mainly aromatic compounds or phenylpropanoids) biosynthesis or metabolism. However, the co-down-regulated DEGs were significantly enriched in carboxylic acid metabolic process and fatty acid or lipid biosynthetic process. In the molecular function category, the co-up- or co-down-regulated DEGs were mainly related to catalytic activity or oxidoreductase activity. Unexpectedly, both the up- and down-regulated DEGs were not significantly enriched in any cellular component terms (Table 5).
Table 5.
Significantly enriched GO terms in the DEGs co-regulated by drought and cold*
| GO term | DEGs a | Unigenes b | Corrected P-value |
|---|---|---|---|
| Co-up-regulated | |||
| Biological process | |||
| Response to stress | 36 (21.4%) | 2565 (11.8%) | 0.05017 |
| Cellular aromatic compound metabolic process | 15 (8.9%) | 418 (1.9%) | 0.00018 |
| Defense response | 15 (8.9%) | 580 (2.7%) | 0.00935 |
| Secondary metabolic process | 11 (6.5%) | 232 (1.1%) | 0.00040 |
| Aromatic compound biosynthetic process | 10 (6.0%) | 154 (0.7%) | 6.91E-05 |
| Phenylpropanoid biosynthetic process | 9 (5.4%) | 147 (0.7%) | 0.00044 |
| Phenylpropanoid metabolic process | 9 (5.4%) | 200 (0.9%) | 0.00535 |
| Cellular component | |||
| External encapsulating structure | 9 (8.1%) | 857 (4.1%) | 1 |
| Chloroplast | 7 (6.3%) | 614 (2.9%) | 1 |
| Cell wall | 6 (5.4%) | 329 (1.6%) | 0.38977 |
| Plant-type vacuole | 3 (2.7%) | 109 (0.5%) | 0.97678 |
| Chloroplast stroma | 2 (1.8%) | 56 (0.3%) | 1 |
| Molecular function | |||
| Catalytic activity | 135 (76.7%) | 15838 (64.3%) | 0.03081 |
| Polysaccharide binding | 3 (1.7%) | 14 (0.1%) | 0.01477 |
| Pattern binding | 3 (1.7%) | 18 (0.1%) | 0.03243 |
| Carbon-nitrogen lyase activity | 3 (1.7%) | 27 (0.1%) | 0.11089 |
| Alkene binding | 2 (1.1%) | 3 (0.0%) | 0.01818 |
| Co-down-regulated | |||
| Biological process | |||
| Monocarboxylic acid metabolic process | 27 (6.5%) | 520 (2.4%) | 0.00096 |
| Fatty acid metabolic process | 24 (5.8%) | 364 (1.7%) | 5.22E-05 |
| Small molecule biosynthetic process | 23 (5.6%) | 324 (1.5%) | 2.53E-05 |
| Organic acid biosynthetic process | 22 (5.3%) | 284 (1.3%) | 1.04E-05 |
| Lipid biosynthetic process | 19 (4.6%) | 311 (1.4%) | 0.00316 |
| Fatty acid biosynthetic process | 17 (4.1%) | 138 (0.6%) | 4.13E-07 |
| Nucleobase transport | 9 (2.2%) | 41 (0.2%) | 2.18E-05 |
| Purine nucleoside transport | 5 (1.2%) | 24 (0.1%) | 0.02775 |
| Cellular component | |||
| Intrinsic to membrane | 27 (16.3%) | 2357 (11.2%) | 1 |
| Cytoplasmic vesicle | 17 (10.2%) | 1240 (5.9%) | 0.99779 |
| Vesicle | 17 (10.2%) | 1256 (6.0%) | 1 |
| Microbody part | 2 (1.2%) | 33 (0.2%) | 1 |
| Peroxisomal part | 2 (1.2%) | 33 (0.2%) | 1 |
| Molecular function | |||
| Oxidoreductase activity | 44 (18.6%) | 2608 (10.6%) | 0.01967 |
| Nucleobase transmembrane transporter activity | 6 (2.5%) | 43 (0.2%) | 0.00043 |
| Arsenate reductase activity | 3 (1.3%) | 4 (0.0%) | 0.00044 |
*The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term.
According to KEGG pathway analysis, the co-up-regulated DEGs were significantly enriched in eight pathways. Most of these pathways are completely or highly overlapped and actually belong to two pathways: microbial metabolism in diverse environments and biosyntheses of secondary metabolites. In contrast, the co-down-regulated DEGs were mainly assigned to metabolic pathways and starch/sucrose metabolic pathway. The DEGs involved in carotenoid biosynthesis and bisphenol degradation were also significantly enriched (Table 6). Furthermore, about 30% of the co-regulated DEGs were unknowns or unannotated genes. They may play unique roles in response and adaptation to drought and cold environments of A. mongolicus.
Table 6.
Significantly enriched KEGG pathways in the DEGs co-regulated by drought and cold*
| Pathway | DEGs a | Unigenes b | Corrected P-value | Pathway ID |
|---|---|---|---|---|
| Co-up-regulated | ||||
| Microbial metabolism in diverse environments | 29 (15.3%) | 2079 (8.8%) | 0.03272 | ko01120 |
| Biosynthesis of secondary metabolites | 28 (14.7%) | 1995 (8.4%) | 0.03272 | ko01110 |
| Limonene and pinene degradation | 17 (9.0%) | 1009 (4.3%) | 0.03274 | ko00903 |
| Aminobenzoate degradation | 17 (9.0%) | 1030 (4.3%) | 0.03274 | ko00627 |
| Bisphenol degradation | 9 (4.7%) | 221 (0.9%) | 0.00497 | ko00363 |
| Polycyclic aromatic hydrocarbon degradation | 9 (4.7%) | 248 (1.0%) | 0.00594 | ko00624 |
| Chloroalkane and chloroalkene degradation | 6 (3.2%) | 187 (0.8%) | 0.03274 | ko00625 |
| Phenylalanine metabolism | 5 (2.6%) | 108 (0.5%) | 0.03272 | ko00360 |
| Co-down-regulated | ||||
| Metabolic pathways | 55 (26.8%) | 4071 (17.2%) | 0.00961 | ko01100 |
| Starch and sucrose metabolism | 13 (6.3%) | 594 (2.5%) | 0.04050 | ko00500 |
| Carotenoid biosynthesis | 7 (3.4%) | 84 (0.4%) | 0.00049 | ko00906 |
| Bisphenol degradation | 7 (3.4%) | 221 (0.9%) | 0.04684 | ko00363 |
*The significantly enriched KEGG pathways were determined using a corrected P-value ≤ 0.05. anumber of DEGs assigned to certain KEGG pathway; bnumber of all reference unigenes assigned to certain KEGG pathway.
Differentially expressed genes specifically responding to drought or cold stress
In total, 4972 and 2009 DEGs were specifically regulated in at least one time point during drought or cold stress, respectively. Of these DEGs, 1057 (445 up-regulated, 566 down-regulated and 46 complicatedly regulated) or 1055 (514 up-regulated, 541 down-regulated) had significant expression changes across all three time points under drought or cold stress, respectively. The top 100 drought- or cold-specific DEGs were listed in the Additional files 11 and 12.
Among the 1057 drought-specific DEGs, 91 DEGs encode TFs, 66 encode protein kinases and phosphatases, and 24 are involved in hormone or cell signaling. Compared to the co-regulated DEGs, more TF gene families were found in the drought-specific DEGs. In the 1055 cold-specific DEGs, however, fewer genes encode regulatory proteins. Some distinct differences appeared between the drought- and cold-specific DEGs. For instance, genes encoding trehalose-6-phosphate synthase, trehalose-6-phosphate phosphatase and early responsive to dehydration protein were only found in the DEGs specifically up-regulated in drought; while genes encoding acidic endochitinase, protochlorophyllide reductase, protein chloroplast import apparatus 2 and chloroplast omega-3 fatty acid desaturase appeared only in the DEGs specifically up-regulated in cold. In addition, about 23.7% of the drought-specific DEGs and 32.3% of the cold-specific DEGs were functional unknowns.
The DEGs specifically up-regulated in drought were mostly enriched in the GO terms “Response to stress”, “Response to stimulus”, “Membrane” and “Antioxidant activity” (Additional file 13). However, the DEGs specifically up-regulated in cold were mostly enriched in the terms “Aminoglycan catabolic process”, “Plastid”, “Chloroplast” and “pattern binding” (Additional file 14). On the other hand, the DEGs specifically down-regulated in drought were more enriched in the GO terms “Small molecule biosynthetic process”, “Organic acid biosynthetic process” and “Iron ion binding” (Additional file 13); while the DEGs specifically down-regulated in cold were highly enriched in the GO terms “Lipid localization” and “Antioxidant activity” (Additional file 14). These results indicated that distinct molecular mechanisms were involved in A. mongolicus seedlings under drought compared to those in cold stress.
Discussion
A. mongolicus is an emerging model species for studying the response and tolerance of plants to extreme cold and drought environments. By using Illumina technology, 9309 up-regulated and 23419 down-regulated DEGs were previously identified in the cold-acclimated A. mongolicus seedlings [12]. Using an EST approach, 120 abiotic stress-responsive genes were identified in this species, and 82 genes were confirmed to be cold- and/or drought-inducible by qRT-PCR [15]. In the present study, we performed comparative transcriptome profiling of A. mongolicus in response to drought and cold and identified thousands of the DEGs regulated by each stress. In particular, 971 co-regulated DEGs by both stresses and about 1050 DEGs specifically regulated by a single stress were identified, suggesting common and distinct molecular mechanisms underlying the response to drought and cold environments. These results were similar to those found in Arabidopsis [7], rice [8] and Brachypodium [9] despite of some new information also observed in the present study. The co-regulated DEGs could represent a subset of basal drought and cold responsive genes in A. mongolicus and thus may play essential roles in its adaptation to adverse environments; while the specifically regulated DEGs might underline the distinct mechanisms of this species in response to different stresses. Functional enrichment analyses of these DEGs provided clues to the molecular bases of A. mongolicus combating drought and cold environments.
Flavonoids may contribute greatly to the adaptation of A. mongolicusto drought and cold environments
Flavonoids constitute a large group of polyphenolic secondary metabolites in plants, which have antioxidant activity and are of prime importance for plant defense against pathogens and UV stress [16]. Recent studies have showed that flavonoids accumulated to high levels in high alpine and polar plants [17] and in A. mongolicus [18] during cold acclimation, suggesting their important roles in cold adaptation of these plants. The biosyntheses of flavonoids require the enzymes involved in the general phenylpropanoid pathway and its flavonoid branch pathways, including phenylalanine ammonia-lyase (PAL), cinnamate-4-hydroxylase (C4H), 4-coumarate CoA ligase (4CL), chalcone synthase (CHS), chalcone isomerase (CHI), chalcone reductase (CHR), flavanone 3-hydroxylase (F3H), dihydroflavonol-4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), anthocyanidin synthase (ANS), isoflavone synthase (IFS), 2-hydroxyisoflavanone dehydratase (HID), isoflavone reductase (IFR), isoflavone-3’-hydroxylase (IF3H), flavonol synthase (FLS), O-methyltransferase (OMT) and UDP-glycosyltransferase (UGT), etc [19, 20]. In the present study, genes encoding almost all of these enzymes were not only moderately or highly expressed in A. mongolicus seedlings in normal condition (average RPKM is 25.3), but also were significantly up-regulated by drought and cold stresses. GO terms and pathways relevant to flavonoids were highly enriched in the co-up-regulated DEGs identified in both drought and cold stresses (Tables 5 and 6). The phenylpropanoid or flavonoid biosynthetic pathways are usually regulated by MYB and/or bHLH TFs [21, 22]. Genes encoding these TFs were also included in the co-up-regulated DEGs identified in the present study. Although their exact functions need to be characterized, our results suggest that genes involved in flavonoid biosyntheses may be crucial in the adaptation of A. mongolicus to drought and cold environments. An increased production of reactive oxygen species is a common consequence in plants under abiotic stresses, which usually damage cellular membranes and other cellular components resulting in oxidative stress and eventually cell death [23]. It is likely that a high level of flavonoids may protect A. mongolicus cells from oxidative stress originated from drought and cold, and hence they contribute to stress tolerance.
Membrane proteins may play important roles in drought adaptation of A. mongolicus
In plants, membrane transport and perception systems have essential roles in maintaining cellular homeostasis under adverse environmental stresses through cell-to-cell and/or organ-to-organ communication [24, 25]. An increased expression of multiple transporter and channel protein genes in response to drought has been found in some plant species [7, 24, 26]. Over-expressing some of these genes, such as aquaporin OsPIP2-2 [27] and ABC-transporter AtABCG36 [28], enhanced plant tolerance to drought and/or salt stress. In the present study, the GO term “Membrane” was highly enriched in the DEGs up-regulated in drought (Additional files 5 and 13), and approximately 70 membrane protein genes were found with up-regulation under drought stress. Most of these genes were annotated to encode transporters and channel proteins, such as those for ions, sugars, organic acids, water, nitrate and phosphate, suggesting their involvement in drought adaptation of A. mongolicus.
Receptor-like protein kinases (RLKs), another group of membrane proteins, are important signaling components that mediate plant responses to developmental and environmental stimuli [25]. Recent studies have shown that some RLKs, such as Arabidopsis RPK1, GHR1 and AtCRK45, responded to drought, ABA, salt and/or cold stresses and they were implicated in ABA and BR signaling pathways [29, 30]. Genes encoding cysteine-rich receptor-like kinases and LRR-RLKs (leucine-rich receptor-like protein kinases) were highly induced in chilling-treated Chorispora bungeana [31]. In the present study, several RLK genes, such as those encoding LRR-RLKs, lectin-like receptor kinase, leucine-rich repeat receptor-like tyrosine-protein kinase and G-type lectin S-receptor-like serine/threonine-protein kinase, were up-regulated in drought-stressed A. mongolicus seedlings. Although their functions and detailed molecular mechanisms are not clear, an increased transcription of RLK genes suggests they may play important roles in drought adaptation of A. mongolicus.
Chloroplast plays a central role in cold adaptation of A. mongolicus
Physiological studies have confirmed that chloroplast plays a central role in plant cold acclimation and freezing tolerance, which acts as sensor of plants responding to cold and light stress as well as the target of many cold acclimation processes [32]. In the present study, genes related to chloroplast were specifically enriched in the DEGs up-regulated in cold (Additional files 8, 9 and 14), covering almost all aspects of chloroplast. The largest group of these genes was involved in photosynthesis, encoding chlorophyll a-b binding proteins, photosystem II and photosystem I components, cytochrome b6/f complex subunit VIII, thioredoxins, ribulose-1, 5-bisphosphate carboxylase/oxygenase large and small subunits, as well as protochlorophyllide reductase and Mg-chelatase. This result reinforced previous findings in woody or shrub plants with strong cold tolerance, including A. mongolicus [15], rhododendron [33] and Populus euphratica [34], in which several photosynthesis-related genes were up-regulated in cold acclimation. However, it was different from those observed in herbaceous plants like Arabidopsis [35], Brachypodium [9], barley [36] and tomato [37] . In these plants with moderate cold tolerance or cold sensitivity, significant suppression of photosynthesis-related genes was observed under cold stress. An up-regulated transcription of the photosynthesis-related genes might be an important mechanism underlying the maintenance of normal or higher photosynthetic efficiency of A. mongolicus during extreme cold winter.
The second group of the up-regulated genes was annotated to encode early light-induced proteins (ELIPs), 20 kDa chaperonin, chaperone protein DnaJ 11, protease Do-like 8 (DEGP8) and lipoxygenase 4 (LOX 4). ELIPs might protect chloroplasts from light-induced damage or photooxidative damage [38]. Chaperonins or chaperone proteins participate in proper folding of protein substrates, protein disaggregation and degradation [39]. DEGP8 located in the thylakoid lumen [40] is probably involved in the cleavage of photodamaged proteins. LOX 4 is involved in the production of jasmonic acid and functions in biotic stress response [41]. To our knowledge, these genes (except for chaperone protein DnaJ 11) were firstly reported with up-regulation in A.mongolicus under cold stress.
Two other chloroplast-related genes up-regulated in cold stress encode chloroplast omega-3 desaturase and glycerol-3-phosphate acyltransferase. Increasing evidence has indicated that both genes participate in the desaturation of fatty acids in chloroplast lipids and play crucial roles in cold response and tolerance [42, 43]. An increased expression of these genes might help to maintain suitable fluidity and stability of chloroplast membranes of A. mongolicus in cold environment. The remaining chloroplast-related genes up-regulated under cold stress may have roles in protein import, ion transport, ATP synthesis, starch degradation, protein synthesis and processing. All these genes could provide structural and physiological mechanisms for the protection of photosynthetic apparatus, and thus for extreme tolerance of A. mongolicus in very cold winter.
Transcriptional regulatory network is involved in the adaptation of A. mongolicusto drought and cold environments
TFs are crucial components in stress signal transduction pathways, which directly control the expression of specific sets of stress-responsive genes [44, 45]. Of the A. mongolicus transcriptome, at least 160 TFs in 13 TF families were identified as DEGs during drought- and/or cold treatments, such as AP2/EREBP, NAC, WRKY and bHLH families.
DREBs and ERFs (ethylene responsive factors) are two subfamilies in the AP2/EREBP superfamily [46]. It is well-known that DREBs play central roles in the regulation of cold- and drought-responsive genes and are important to the related stress tolerance in many plant species [47]. ERFs have been recently found to participate in the response of plants to abiotic stresses, and some ERFs can enhance drought, salt and/or freezing tolerance of their transgenic plants [48, 49]. In the present study, 9 DERBs and 8 ERFs were up-regulated in the drought- and cold-stressed A. mongolicus seedlings respectively, highlighting the importance of these TFs in the adaptation of this species to drought and cold environments.
The NAC family is another group of TFs involved in the tolerance of plants to abiotic stresses, especially to drought and high salinity [50, 51]. Our study identified 8 putative NAC genes up-regulated under drought and cold. Similar results were obtained in the WRKY family, a well-known family involved in biotic-stress tolerance in plants. Recent evidence showed that the WRKY family also functions in response to abiotic stresses [52]. Our findings provide supports to these results, suggesting an important role of the NAC and WRKY families in drought and cold tolerance.
The MYB, bHLH and C2H2 families are also involved in plant abiotic stress tolerance [5, 45, 53]. In the present study, different members of these families were either up- or down-regulated under drought and cold treatments. This differed obviously from the three TF families discussed earlier which were almost up-regulated during the stress treatments. In summary, sophisticated transcriptional regulation could participate in the adaptation of A. mongolicus to adverse environments, and several families, especially the AP2/EREBP (DREBs and ERFs), NAC and WRKY families might be important in the processes.
Conclusions
A comprehensive transcriptome resource is generated for A. mongolicus and transcriptome profiles under drought and cold stresses are characterized. Comparative transcriptome analysis identified many genes and pathways commonly or specifically regulated in the two stresses, implicating molecular basis of drought and cold tolerance in A. mongolicus. A number of drought- and cold-regulated genes were firstly identified in this study, including some unknown genes. These newly found genes could be important to A. mongolicus in combating extremely cold and drought environments in desert. Our data will facilitate further molecular studies on stress tolerance of A. mongolicus and provide new insights into plant adaptation to harsh environments.
Methods
Plant stress treatment and sampling
A. mongolicus seeds were collected from a single shrub grown in the desert area in Dengkou County, Inner mongolia, China. These seeds were surface-sterilized with Clorox solution and soaked in water for three days at 25°C and then sown in pots filled with sand. The pots were placed in a greenhouse at 25°C under long-day condition (16 h light/8 h dark cycle). The seedlings were watered every three days with 1/5 strength of Hoagland’s solution. Three months after germination, the seedlings were treated as follows.
For de novo transcriptome sequencing and assembly: the seedlings were placed into a low temperature-programmable incubator (BD-PRX-1000A; -10°C to 50°C) for cold treatment at 4°C to -8°C under a 16 h dim light/8 h dark cycle in a gradual cooling way: 4°C for 6 h, 2°C for 14 h, 0°C for 4 h, -2°C for 12 h, -5°C for 8 h, and -8°C for 4 h, in total for 48 h; the cooling rate was 0.5°C per minute. This cold treatment followed by a 3-day recovery period can lead to a slight lethal damage to some leaves of the A. mongolicus seedlings according to our preliminary experiments (unpublished data). The seedlings were carefully removed from sand at 1 h (4°C), 3 h (4°C), 8 h (4°C 6 h + 2°C 2 h), 16 h (4°C 6 h + 2°C 10 h), 24 h (4°C 6 h + 2°C 14 h + 0°C 4 h) and 48 h (cold treatment finished) after the initiation of the cold treatment at 4°C respectively, and washed with pre-cooled water (in a refrigerator at 3°C) and collected. Drought treatment was performed as follows: the seedlings were carefully removed from sand, washed with tap water, placed on filter paper and transferred in the same incubator for dehydration at 25°C under a 16 h dim light/8 h dark cycle. The seedlings were collected at the same time points as cold stress (after 1, 3, 8, 16, 24 and 48 h of dehydration). The untreated seedlings (0 h) were used as control (CK).
For transcriptome profile analysis: the seedlings were placed into the same low temperature-programmable incubator for cold treatment in a gradual cooling way: 4°C for 4 h, 2°C for 12 h, 0°C for 2 h, -2°C for 2 h, -4°C for 2 h, and -6°C for 2 h, in total for 24 h. The drought treatment was performed using dehydration on a filter paper as described earlier. The seedlings were sampled at 2, 8 and 24 h after the initiation of the cold or drought treatment respectively. The untreated seedlings (0 h) were used as control (CK). For each time point, four seedlings were sampled. To minimize the bias of random changes from a single sample, each time point was sampled from three independent stress treatments and the total RNA was pooled equally for further experiment.
Young leaves were sampled from a single A. mongolicus plant grown in the desert area mentioned above in summer (late July) and winter (late December). All samples from stress treatments, controls and the young leaves were immediately frozen in liquid nitrogen and then stored at –76°C for RNA extraction.
RNA extraction and Illumina sequencing
Total RNA was extracted using a modified Trizol method developed by our laboratory (Trizol agent plus 2% β-ME and 3 M KAc with pH 4.8). The quantity of RNAs was verified by an ultraviolet spectrometer (OD260/OD280 ratios with 1.84 to 2.04). RNAs were dissolved in DEPC-treated H2O. For de novo transcriptome sequencing and assembly, total RNAs extracted from each sample were pooled equally, and 20 μg of the pooled RNA was used for the isolation of poly (A) + mRNA. For transcriptome profile analysis, poly (A) + mRNA was purified from 10 μg total RNA from each cold- or drought-stressed seedlings and control, respectively.
The mRNAs were isolated by using oligo (dT) magnetic beads and was interrupted into short fragments by adding fragmentation buffer. Taking these short fragments as templates, the first-strand cDNA was synthesized using random hexamer-primer and reverse transcriptase. The second-strand cDNA was synthesized using buffer, dNTPs, RNase H and DNA polymerase I. After the end reparation and ligation of adaptors, the products were amplified by PCR and purified using the QiaQuick PCR extraction kit (Qiagen, Valencia, CA) to create cDNA libraries. The cDNA libraries were sequenced by Beijing Genomics Institute (BGI)-Shenzhen (Shenzhen, China) on an Illumina HiSeq™ 2000 according to the manufacturer’s protocols. For de novo transcriptome assembly, the 90 PE (paired-end) strategy was adopted; while for transcriptome profile analysis, the 50 SE (single-end) strategy was used.
Transcriptome assembly
Raw reads from the library were filtered to remove the dirty raw reads if they contain adaptors, over 5% of unknown nucleotides or over 50% of low quality bases (quality value ≤ 5). Clean reads were used for de novo assembly with Trinity, a short read assembling program released recently [54]. Briefly, Trinity firstly combined reads with certain length of overlap to form contigs. These reads were then mapped back to contigs. Finally, Trinity connected the contigs and generated unigenes, i.e. the sequences that cannot be extended on either end.
Functional annotation and classification of the unigenes
The unigenes were aligned to protein databases in the NCBI in the priority order of NR, Swiss-Prot, KEGG and COG by BLASTX algorithm [55], with a cut-off E-value ≤ 1e-5. Proteins with the highest sequence similarity were retrieved as functional annotations for a given unigene. For unigenes that did not align to any of the above databases, ESTScan software was used to predict their coding regions and corresponding amino acid sequences [56]. To identify putative TFs, all unigenes were blasted against the plantTFDB (http://planttfdb.cbi.edu.cn) with a cut-off E-value ≤ 1e-5, and the top hit for each query was applied. Unigenes with no significant hits in the plantTFDB but with annotations as TFs in the NR and Swiss-Prot databases (E-value ≤ 1e-10) were manually selected and classified into the corresponding TF families. To ascertain the sequence similarity of A. mongolicus to other plant species, total unigenes were searched by BLASTN against the PlantGDB-generated Unique Transcripts (http://www.plantgdb.org/prj/ESTCluster/) (similarity ≥ 80% and E-value ≤ 1e-5).
The GO annotations of the unigenes were obtained by using Blast2GO program [57]. The WEGO software was applied to perform GO functional classification for all annotated unigenes and to show the distribution of gene functions of the species at macro level [58]. After aligning the unigenes to the COG database, the COG functional annotations were acquired. The KEGG pathway annotations were obtained by aligning the unigenes to the KEGG database [59].
Transcriptome profile analyses
The clean reads from each library were mapped to the assembled transcriptome sequences (used as reference genes) using SOAPaligner/SOAP2 [60]. Mismatches with no more than two bases were allowed in the alignment, and ambiguous reads with multi-position match were excluded. Gene expression levels were measured by the RPKM (the numbers of reads per kilobase of exon region in a given gene per million mapped reads) values [61]. If there were two or more transcripts for a gene, the longest one was used to calculate its RPKM. To identify genes responding to drought and cold, the number of reads and RPKM for each coding region in the control and each stress-treated library were determined, and the ratios of RPKM value for each gene in six stress-treated libraries to that in control library were calculated. Statistical significance of the differential expression value for each gene was determined according to Audic and Claverie [62].
Genes were selected as DEGs according to the FDR values ≤ 0.001 and at least a two-fold change (the absolute value of log2 Ratio ≥ 1) in expression level between two libraries. GO enrichment analysis was performed by mapping all the DEGs to GO terms in the GO database (http://www.geneontology.org/) and the number of unigenes included in each term was calculated. Hypergeometric test [63] was used to identify significantly enriched GO terms in DEGs compared with the assembled transcriptome of A. mongolicus using p-value ≤ 0.05. Pathway enrichment was analyzed by mapping DEGs to the KEGG database (http://www.genome.jp/kegg). The number of unigenes involved in each pathway was calculated. By comparing with the assembled transcriptome of A. mogolicus, the significantly enriched pathways were identified using p-value ≤ 0.05.
Validation of DEGs by RT-qPCR
Eleven genes with different expression patterns were chosen for the validation. The template cDNAs were synthesized from 1.5 μg of total RNAs (pre-treated with DNase I) from the same samples as those for the transcriptome profile analysis using M-MLV Reverse Transcriptase (Promega, USA). The 2 × SYBR-Green I RT-PCR Master Mix (Takara, Japan) was used as labeling agent, and α-tubulin of A. mongolicus was served as an internal reference gene. The reaction mixture (20 μL) contained 10 μL 2× Master Mix, 10 μM forward and reverse primers (Additional file 15) (0.4 μL each) and 1 μL template cDNA (diluted 10 folds with deionized water). The PCR program was as follows: 95°C for 30 sec; the first step: 40 cycles of 95°C for 5 sec, 55 to 60° for 20 sec and 72°C for 20 sec; the second step: 1 cycle of 95°C for 15 sec, 55 to 60° for 1 min, and 72°C for 15 sec. The reaction was performed on a Statagene Mx3000 PCR system. Three independent biological replicates were performed for each sample. The relative expression levels of the selected unigenes were calculated using the relative 2–ΔΔCT method [64]. Results represent mean standard deviations of the three experimental replicates.
Availability of supporting data
The data set supporting the results of this article is available in the NCBI SRA (short read archive) repository under the accession number of SRS526744 (NCBI SRA, http://www.ncbi.nlm.nih.gov/sra?term= SRS526744).
Electronic supplementary material
Additional file 1: Distribution of the annotated unigenes in each database. aannotated or unannotated unigenes/total unigenes × 100%; bannotated unigenes in each database/total annotated unigenes × 100%. (XLS 19 KB)
Additional file 2: Species distribution of A. mongolicus sequences in the TIGR Plant Transcript Assemblies database. All unigenes were searched by BLASTN against the TIGR Plant Transcript Assemblies database. In total, 47486 unigenes had significant hits (similarity ≥ 80% and E-value ≤ 1e-5), and the sequence similarities to 12 species were shown in the figure. (TIFF 3 MB)
Additional file 3: Histogram of COG classification of the annotated unigenes. In total, 17100 annotated unigenes were assigned to 25 COG classes. (TIFF 3 MB)
Additional file 4: The top 100 DEGs commonly regulated by drought stress. The unigenes with absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 were identified as DEGs. (XLS 40 KB)
Additional file 5: The significantly enriched GO terms in the DEGs commonly regulated by drought stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 32 KB)
Additional file 6: The significantly enriched KEGG pathways in the DEGs commonly regulated by drought stress. The significantly enriched KEGG pathways were determined using a corrected P-value ≤ 0.05. anumber of DEGs assigned to certain KEGG pathway; bnumber of all reference unigenes assigned to certain KEGG pathway. (XLS 20 KB)
Additional file 7: The top 100 DEGs commonly regulated by cold stress. The unigenes with absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 were identified as DEGs. (XLS 38 KB)
Additional file 8: The significantly enriched GO terms in the DEGs commonly regulated by cold stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 37 KB)
Additional file 9: The significantly enriched KEGG pathways in the DEGs commonly regulated by cold stress. The significantly enriched KEGG pathways were determined using a corrected P-value ≤ 0.05. anumber of DEGs assigned to certain KEGG pathway; bnumber of all reference unigenes assigned to certain KEGG pathway. (XLS 24 KB)
Additional file 10: The top 100 DEGs co-regulated by drought and cold. These DEGs had absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 at all drought- and cold-treated time points. (XLS 48 KB)
Additional file 11: The top 100 DEGs specifically regulated by drought stress. These DEGs had absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 at three drought-treated time points. (XLS 40 KB)
Additional file 12: The top 100 DEGs specifically regulated by cold stress. These DEGs had absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 at three cold-treated time points. (XLS 36 KB)
Additional file 13: The significantly enriched GO terms in the DEGs specifically regulated by drought stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 24 KB)
Additional file 14: The significantly enriched GO terms in the DEGs specifically regulated by cold stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 29 KB)
Additional file 15: Primer sequences used in the RT-qPCR assay. The primer pairs for 9 selected unigenes were shown. (XLS 20 KB)
Acknowledgments
This work was funded by the Major Project of Natural Science Fund of Inner Mongolia Autonomous Region (2012ZD02), the National Natural Science Foundation of China (30960158), the Major Project of Cultivating New Varieties of Transgenic Organisms (2009ZX08009-019B), and the Innovative Research Group Fund of Inner Mongolia Agriculture University (NDPYTD2010-37). We would like to thank the staff in Beijing Genomics Institute-Shenzhen for their supports in the transcriptome sequencing and data analysis.
Abbreviations
- TF
Transcription factor
- DEG
Differentially expressed gene
- NR
Non-redundant
- GO
Gene Ontology
- COG
Clusters of Orthologous Groups
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- ABA
Abscisic acid
- BR
Brassinosteroid
- FDR
False discovery rate
- RT-qPCR
Real-time quantitative PCR
- NAC
NAM, ATAF and CUC
- DREB
Dehydration responsive element binding
- HSP
Heat shock protein
- GLP
Germin-like protein
- TPP
Trehalose-6-phosphate phosphatase
- CCD
Carotenoid cleavage dioxygenase
- LEA
Late embryogenesis abundant
- AP2/EREBP
APETALA2/ethylene-responsive element binding protein
- MYB
Myeloblastosis oncogene
- bHLH
Basic helix-loop-helix
- C2H2
Cys2/His2-type zinc finger protein 2
- bZIP
Basic-domain leucine-zipper protein
- C3H
Cys3/His-zinc finger protein
- GRAS
GAT, RGA and SCR
- TCP
TB1, CYC and PCF
- COL
Constans-like
- Dof
DNA binding with one finger
- GRF
General regulatory factor
- PR
Pathogenesis-related
- PAL
Phenylalanine ammonia-lyase
- C4H
Cinnamate-4-hydroxylase
- 4CL
4-coumarate CoA ligase
- CHS
Chalcone synthase
- CHI
Chalcone isomerase
- CHR
Chalcone reductase
- F3H
Flavanone 3-hydroxylase
- DFR
Dihydroflavonol-4-reductase
- LDOX
Leucoanthocyanidin dioxygenase
- ANS
Anthocyanidin synthase
- IFS
Isoflavone synthase
- HID
2-Hydroxyisoflavanone dehydratase
- IFR
Isoflavone reductase
- IF3H
Isoflavone-3’-hydroxylase
- FLS
Flavonol synthase
- OMT
O-methyltransferase
- UGT
UDP-glycosyltransferase
- RLK
Receptor-like protein kinase
- LRR-RLK
Leucine-rich receptor-like protein kinase
- ELIP
Early light-induced protein
- DEGP8
Protease Do-like 8
- LOX 4
Lipoxygenase
- ERF
Ethylene responsive factor
- RPKM
Reads per kilobase per million reads
- SRA
Short read archive.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YW and WW participated in the design of the study, sampled the shrub leaves in the wild, analyzed and submitted data, and drafted the manuscript. XP and XW cultivated, treated and sampled seedlings, and carried out RNA extraction. HZ and BD participated in RNA extraction and performed RT-qPCR assay. YX participated in data analysis. XL participated in analysis and interpretation of data, and critically revised the manuscript. MW conceived of the study, participated in its design and data interpretation, and revised the manuscript critically. All authors read and approved the final manuscript.
Contributor Information
Yaqi Wu, Email: wuyaqi88@hotmail.com.
Wei Wei, Email: hjwei85@hotmail.com.
Xinyue Pang, Email: pangxy@hotmail.com.
Xuefeng Wang, Email: 1184213789@qq.com.
Huiling Zhang, Email: 226377182@qq.com.
Bo Dong, Email: 5958490@163.com.
Yanping Xing, Email: xyping8315@163.com.
Xinguo Li, Email: xinguo.li@csiro.au.
Maoyan Wang, Email: wangmaoyan@163.com.
References
- 1.Flower S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 2002;14(8):1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei H, Dhanaraj AL, Arora R, Rowland LJ, Fu Y, Sun L. Identification of cold acclimation-responsive Rhododendron genes for lipid metabolism, membrane transport and lignin biosynthesis: importance of moderately abundant ESTs in genomic studies. Plant Cell Environ. 2006;29(4):558–570. doi: 10.1111/j.1365-3040.2005.01432.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Wang X, Jiao Y, Qin Y, Liu X, He K, Chen C, Ma L, Wang J, Xiong L, Zhang Q, Fan L, Deng XW. Global genome expression analysis of rice in response to drought and high-salinity stresses in shoot, flag leaf, and panicle. Plant Mol Biol. 2007;63(5):591–608. doi: 10.1007/s11103-006-9111-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molina C, Rotter B, Horres R, Udupa SM, Besser B, Bellarmino L, Baum M, Matsumura H, Terauchi R, Kahl G, Winter P. SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics. 2008;9:553. doi: 10.1186/1471-2164-9-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hussain SS, Kayani MA, Amjad M. Transcription factors as tools to engineer enhanced drought stress tolerance in plants. Biotechnol Prog. 2011;27(2):297–306. doi: 10.1002/btpr.514. [DOI] [PubMed] [Google Scholar]
- 6.Sanghera GS, Wani SH, Hussain W, Singh NB. Engineering cold stress tolerance in crop plants. Curr Genomics. 2011;12(1):30–43. doi: 10.2174/138920211794520178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, Kamiya A, Nakajima M, Enju A, Sakurai T, Satou M, Akiyama K, Taji T, Yamaguchi-Shinozaki K, Carninci P, Kawai J, Hayashizaki Y, Shinozaki K. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold, and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002;31(3):279–292. doi: 10.1046/j.1365-313X.2002.01359.x. [DOI] [PubMed] [Google Scholar]
- 8.Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003;133(4):1755–1767. doi: 10.1104/pp.103.025742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Priest HD, Fox SE, Rowley ER, Murray JR, Michael TP, Mockler TC. Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS One. 2014;9(1):e87499. doi: 10.1371/journal.pone.0087499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan BR, Huang SP. Cytological study of the genus Ammopiptanthus. Acta Botanica Sinica. 1993;35:314–317. [Google Scholar]
- 11.Zhou Y, Gao F, Liu R, Feng J, Li H. De novo sequencing and analysis of root transcriptome using 454 pyrosequencing to discover putative genes associated with drought tolerance in Ammopiptanthus mongolicus. BMC Genomics. 2012;13:266. doi: 10.1186/1471-2164-13-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pang T, Ye CY, Xia X, Yin W. De novo sequencing and transcriptome analysis of the desert shrub, Ammopiptanthus mongolicus, during cold acclimation using Illumina/Solexa. BMC Genomics. 2013;14:488. doi: 10.1186/1471-2164-14-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuteja N. Abscisic acid and abiotic stress signaling. Plant Signal Behav. 2007;2(3):135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gruszka D. The brassinosteroid signaling pathway-new key players and interconnections with other signaling networks crucial for plant development and stress tolerance. Int J Mol Sci. 2013;14(5):8740–8774. doi: 10.3390/ijms14058740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M, Shi J, Lu C. Identification of stress-responsive genes in Ammopiptanthus mongolicus using ESTs generated from cold- and drought-stressed seedlings. BMC Plant Biol. 2013;13(4):88. doi: 10.1186/1471-2229-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Chen S, Yu O. Metabolic engineering of flavonoids in plants and microorganisms. Appl Microbiol Biotechnol. 2011;91:949–956. doi: 10.1007/s00253-011-3449-2. [DOI] [PubMed] [Google Scholar]
- 17.Lütz C. Cell physiology of plants growing in cold environments. Protoplasma. 2010;244:53–73. doi: 10.1007/s00709-010-0161-5. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Chen Y, Lu C, Zhang H, Yin W. Cold acclimation induced accumulation of phenolic compounds and freezing tolerance in Ammopiptanthus mongolicus. For Stud China. 2007;9(3):203–207. doi: 10.1007/s11632-007-0032-0. [DOI] [Google Scholar]
- 19.Du H, Huang YB, Tang YX. Genetic and metabolic engineering of isoflavonoid biosynthesis. Appl Microbiol Biotechnol. 2010;86(5):1293–1312. doi: 10.1007/s00253-010-2512-8. [DOI] [PubMed] [Google Scholar]
- 20.Vogt T. Phenylpropanoid biosynthesis. Mol Plant. 2010;3(1):2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 21.Czemmel S, Heppel SC, Bogs J. R2R3 MYB transcription factors: key regulators of the flavonoid biosynthetic pathway in grapevine. Protoplasma. 2012;249(Suppl 2):S109–S118. doi: 10.1007/s00709-012-0380-z. [DOI] [PubMed] [Google Scholar]
- 22.Li ST, Zachgo S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013;76(6):901–913. doi: 10.1111/tpj.12348. [DOI] [PubMed] [Google Scholar]
- 23.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7(9):405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 24.Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 2014;202(1):35–49. doi: 10.1111/nph.12613. [DOI] [PubMed] [Google Scholar]
- 25.Osakabe Y, Yamaguchi-Shinozaki K, Shinozaki K, Tran LS. Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J Exp Bot. 2013;64(2):445–458. doi: 10.1093/jxb/ers354. [DOI] [PubMed] [Google Scholar]
- 26.Hadiarto T, Tran LS. Progress studies of drought-responsive genes in rice. Plant Cell Rep. 2011;30(3):297–310. doi: 10.1007/s00299-010-0956-z. [DOI] [PubMed] [Google Scholar]
- 27.Guo L, Wang ZY, Lin H, Cui WE, Chen J, Liu M. Expression and functional analysis of the rice plasma membrane intrinsic protein gene family. Cell Res. 2006;16(3):277–286. doi: 10.1038/sj.cr.7310035. [DOI] [PubMed] [Google Scholar]
- 28.Kim DY, Jin JY, Alejandro S, Martinoia E, Lee Y. Overexpression of AtABCG36 improves drought and salt stress resistance in Arabidopsis. Physiol Plant. 2010;139(2):170–180. doi: 10.1111/j.1399-3054.2010.01353.x. [DOI] [PubMed] [Google Scholar]
- 29.Li J, Tax FE. Receptor-like kinases: key regulators of plant development and defense. J Integr Plant Biol. 2013;55(12):1184–1187. doi: 10.1111/jipb.12129. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Yang G, Shi R, Han X, Qi L, Wang R, Xiong L, Li G. Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses. Plant Physiol Biochem. 2013;67:189–198. doi: 10.1016/j.plaphy.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Z, Tan L, Dang C, Zhang H, Wu Q, An L. Deep-sequencing transcriptome analysis of chilling tolerance mechanisms of a subnival alpine plant, Chorispora bungeana. BMC Plant Biol. 2012;12:222. doi: 10.1186/1471-2229-12-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosatti C, Rizza F, Badeck FW, Mazzucotelli E, Cattivelli L. Harden the chloroplast to protect the plant. Physiol Plant. 2013;147(1):55–63. doi: 10.1111/j.1399-3054.2012.01689.x. [DOI] [PubMed] [Google Scholar]
- 33.Wei H, Dhanaraj AL, Rowland LJ, Fu Y, Krebs SL, Arora R. Comparative analysis of expressed sequence tags from cold-acclimated and non-acclimated leaves of Rhododendron catawbiense Michx. Planta. 2005;221(3):406–416. doi: 10.1007/s00425-004-1440-1. [DOI] [PubMed] [Google Scholar]
- 34.Chen J, Tian Q, Pang T, Jiang L, Wu R, Xia X, Yin W. Deep-sequencing transcriptome analysis of low temperature perception in a desert tree, Populus euphratica. BMC Genomics. 2014;15:326. doi: 10.1186/1471-2164-15-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hannah MA, Heyer AG, Hincha DK. A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 2005;1(2):e26. doi: 10.1371/journal.pgen.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensson JT, Crosatti C, Campoli C, Bassi R, Stanca AM, Close TJ, Cattivelli L. Transcriptome analysis of cold acclimation in barley albina and xantha mutants. Plant Physiol. 2006;141(1):257–270. doi: 10.1104/pp.105.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Ouyang B, Zhang J, Wang T, Li H, Zhang Y, Yu C, Ye Z. Differential modulation of photosynthesis, signaling, and transcriptional regulation between tolerant and sensitive tomato genotypes under cold stress. PLoS One. 2012;7(11):e50785. doi: 10.1371/journal.pone.0050785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welling A, Palva ET. Molecular control of cold acclimation in trees. Physiol Plant. 2006;127(2):167–181. doi: 10.1111/j.1399-3054.2006.00672.x. [DOI] [Google Scholar]
- 39.Wang W, Vinocur B, Shoseyov O, Altman A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004;9(5):244–252. doi: 10.1016/j.tplants.2004.03.006. [DOI] [PubMed] [Google Scholar]
- 40.Adam Z, Adamska I, Nakabayashi K, Ostersetzer O, Haussuhl K, Manuell A, Zheng B, Vallon O, Rodermel SR, Shinozaki K, Clarke AK. Chloroplast and mitochondrial proteases in Arabidopsis. A proposed nomenclature. Plant Physiol. 2001;125(4):1912–1918. doi: 10.1104/pp.125.4.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nalam VJ, Keeretaweep J, Sarowar S, Shah J. Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell. 2012;24(4):1643–1653. doi: 10.1105/tpc.111.094110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Domínguez T, Hernández ML, Pennycooke JC, Jiménez P, Martínez-Rivas JM, Sanz C, Stockinger EJ, Sánchez-Serrano JJ, Sanmartín M. Increasing omega-3-desaturase expression in tomato results in altered aroma profile and enhanced resistance to cold stress. Plant Physiol. 2010;153(2):655–665. doi: 10.1104/pp.110.154815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sui N, Li M, Zhao SJ, Li F, Liang H, Meng QW. Overexpression of glycerol-3-phosphate acyltransferase gene improves chilling tolerance in tomato. Planta. 2007;226(5):1097–1108. doi: 10.1007/s00425-007-0554-7. [DOI] [PubMed] [Google Scholar]
- 44.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakashima K, Ito Y, Yamaguchi-Shinozaki K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009;149(1):88–95. doi: 10.1104/pp.108.129791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold- inducible gene expression. Biochem Biophys Res Commun. 2002;290(3):998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- 47.Lata C, Prasad M. Role of DREBs in regulation of abiotic stress responses in plants. J Exp Bot. 2011;62(14):4731–4748. doi: 10.1093/jxb/err210. [DOI] [PubMed] [Google Scholar]
- 48.Zhang JY, Broeckling CD, Sumner LW, Wang ZY. Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol Biol. 2007;64(3):265–278. doi: 10.1007/s11103-007-9150-2. [DOI] [PubMed] [Google Scholar]
- 49.Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerance to salt, drought and diseases in transgenic tobacco. J Exp Bot. 2009;60(13):3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc Natl Acad Sci U S A. 2006;103(35):12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Redillas MC, Jeong JS, Kim YS, Jung H, Bang SW, Choi YD, Ha SH, Reuzeau C, Kim JK. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant Biotechnol J. 2012;10(7):792–805. doi: 10.1111/j.1467-7652.2012.00697.x. [DOI] [PubMed] [Google Scholar]
- 52.Chen L, Song Y, Li S, Zhang L, Zou C, Yu D. The role of WRKY transcription factors in plant abiotic stresses. Biochim Biophys Acta. 2012;1819(2):120–128. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Umezawa T, Fujita M, Fujita Y, Yamaguchi-Shinozaki K, Shinozaki K. Engineering drought tolerance in plants: discovering and tailoring genes to unlock the future. Curr Opin Biotechnol. 2006;17(2):113–122. doi: 10.1016/j.copbio.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 54.Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng QD, Chen ZH, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. Proc Int Conf Intell Syst Mol Biol. 1999;99:138–148. [PubMed] [Google Scholar]
- 57.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 58.Ye J, Fang L, Zheng H, Zhang Y, Chen J, Zhang Z, Wang J, Li S, Li R, Bolund L, Wang J. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 2006;34:W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6:60. doi: 10.1186/1471-2164-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 61.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 62.Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 63.Hao DC, Ge GB, Xiao PG, Zhang YY, Yang L. The first insight into the tissue specific Taxus transcriptome via Illumina second generation sequencing. PLoS One. 2011;6(6):e21220. doi: 10.1371/journal.pone.0021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Distribution of the annotated unigenes in each database. aannotated or unannotated unigenes/total unigenes × 100%; bannotated unigenes in each database/total annotated unigenes × 100%. (XLS 19 KB)
Additional file 2: Species distribution of A. mongolicus sequences in the TIGR Plant Transcript Assemblies database. All unigenes were searched by BLASTN against the TIGR Plant Transcript Assemblies database. In total, 47486 unigenes had significant hits (similarity ≥ 80% and E-value ≤ 1e-5), and the sequence similarities to 12 species were shown in the figure. (TIFF 3 MB)
Additional file 3: Histogram of COG classification of the annotated unigenes. In total, 17100 annotated unigenes were assigned to 25 COG classes. (TIFF 3 MB)
Additional file 4: The top 100 DEGs commonly regulated by drought stress. The unigenes with absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 were identified as DEGs. (XLS 40 KB)
Additional file 5: The significantly enriched GO terms in the DEGs commonly regulated by drought stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 32 KB)
Additional file 6: The significantly enriched KEGG pathways in the DEGs commonly regulated by drought stress. The significantly enriched KEGG pathways were determined using a corrected P-value ≤ 0.05. anumber of DEGs assigned to certain KEGG pathway; bnumber of all reference unigenes assigned to certain KEGG pathway. (XLS 20 KB)
Additional file 7: The top 100 DEGs commonly regulated by cold stress. The unigenes with absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 were identified as DEGs. (XLS 38 KB)
Additional file 8: The significantly enriched GO terms in the DEGs commonly regulated by cold stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 37 KB)
Additional file 9: The significantly enriched KEGG pathways in the DEGs commonly regulated by cold stress. The significantly enriched KEGG pathways were determined using a corrected P-value ≤ 0.05. anumber of DEGs assigned to certain KEGG pathway; bnumber of all reference unigenes assigned to certain KEGG pathway. (XLS 24 KB)
Additional file 10: The top 100 DEGs co-regulated by drought and cold. These DEGs had absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 at all drought- and cold-treated time points. (XLS 48 KB)
Additional file 11: The top 100 DEGs specifically regulated by drought stress. These DEGs had absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 at three drought-treated time points. (XLS 40 KB)
Additional file 12: The top 100 DEGs specifically regulated by cold stress. These DEGs had absolute values of log2 Ratio ≥ 1 and FDR ≤ 0.001 at three cold-treated time points. (XLS 36 KB)
Additional file 13: The significantly enriched GO terms in the DEGs specifically regulated by drought stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 24 KB)
Additional file 14: The significantly enriched GO terms in the DEGs specifically regulated by cold stress. The significantly enriched GO terms were determined using a corrected P-value ≤ 0.05. If no significantly enriched GO terms were identified, the top five terms were listed. anumber of DEGs assigned to certain GO term; bnumber of all reference unigenes assigned to certain GO term. (XLS 29 KB)
Additional file 15: Primer sequences used in the RT-qPCR assay. The primer pairs for 9 selected unigenes were shown. (XLS 20 KB)


