ABSTRACT
Serum amyloid A (SAA) proteins are acute-phase proteins and are classified into multiple isoforms; however, the biological functions of each SAA isoform are not fully understood. In this study, to clarify the roles of SAA3 in the intestine, we characterized mRNA expression in mouse colonic epithelial CMT-93 cells treated with rotavirus, Toxoplasma, Staphylococcus aureus, and Escherichia coli, as well as lipopolysaccharide (LPS) and recombinant murine SAAs (rSAAs). E. coli together with LPS, but not the other pathogens, enhanced SAA3 mRNA expression. The mRNA expression of SAA3 by dead E. coli was higher than that by living E. coli, and the mRNA expression by E. coli and LPS increased in a dose-dependent manner. In contrast, mRNA expressions of SAA1 and/or SAA2 were not stimulated by any of the treatments. In comparisons of cell treatments with rSAA1 or rSAA3, rSAA3 significantly up-regulated the mRNA expression of mucin 2 (MUC2), a major component of the mucus layer of the intestines that acts as an epithelial cell barrier against pathogens, while MUC2 mRNA expression was not significantly increased by E. coli and LPS. Furthermore, treatment with rSAAs intensively induced tumor necrosis factor-α mRNA expression. These results suggest that SAA3 plays a role in host innate immunity in the colon by up-regulating MUC2 mucin production, which builds a physiological barrier of colonic epithelia against bacterial invasion.
Keywords: colon, MUC2, mucin, SAA3, serum amyloid A
Serum amyloid A (SAA) proteins are acute-phase proteins [21, 25] and are precursors of amyloid A (AA) fibrils in AA amyloidosis [14]. The concentration of SAAs in plasma dramatically increases up to 1,000-fold during inflammation [25], and the increase of SAA in plasma is used as a clinical biomarker of inflammation and infection. Although SAA is usually degraded rapidly, continuously elevated SAA levels in plasma are considered a high risk factor for AA amyloidosis, which often results from a severe complication of chronic diseases, such as rheumatoid arthritis and autoinflammatory diseases [14].
In a variety of mammalian species, multiple SAA isoforms have been described, such as SAA1, 2, 3 and 4 in humans and mice. SAA1 and SAA2 are the major acute-phase isoforms and are mainly expressed in hepatocytes [23, 25], and SAA1 is considered the main precursor of AA fibrils [4, 9, 13]. Murine SAA1 was formerly called SAA2 until a revision of the nomenclature in 1999 [19]. In mice, SAA1 and SAA2 genes (GenBank accession nos. BC087933 and M11130) share 95.1% and 92.6% sequence identities in 369 nucleotides and 122 amino acids, respectively. In contrast to the similarity between SAA1 and SAA2, nucleotide and amino acid sequence identities between SAA1 and SAA3 (NM011315) are 74.3% and 64.7%, respectively (Supplemental Fig. S1). SAA3 in mice is expressed from extrahepatic cells, such as macrophages and adipocytes [12, 25], and SAA3 does not contribute to circulating SAA levels in plasma [1]. Interestingly, human SAA3 is a pseudogene and is not expressed [7, 18]. These divergent biological and genetic characteristics between SAA1/2 and SAA3 indicate that the functions of SAA3 in hosts are not the same as those of SAA1/2. However, the roles of SAA3 are not fully understood.
Recently, Reigstad et al. [17] reported that SAA3 expression is increased in the mouse colon surface in the presence of microbiota. Moreover, SAA3 expression is strongly induced by Escherichia coli lipopolysaccharide (LPS) in murine colonic epithelial cell lines, whereas LPS does not up-regulate SAA1/2 expression [2]. These studies on the SAA3 response against bacteria suggest the involvement of SAA3 in intestinal immunity. Furthermore, previous studies have shown that SAA is produced in response to inflammatory cytokines, particularly tumor necrosis factor (TNF)- α, interleukin (IL)-1β and IL-6 [24], and also reported that SAA proteins have cytokine-like activities, such as the ability to induce IL-1β, IL-6 and TNF-α [15, 20]. However, the exact role of SAA3 has not been determined. In the present study, we investigated mRNA expression in mouse colonic epithelial cells, focusing especially on mucin 2 (MUC2), after treatment with several pathogens and recombinant SAAs (rSAAs). MUC2 is a major mucin of mucus in the colon [16, 28], and MUC2 mucin builds a mucus barrier that separates bacteria from the colon epithelia [6].
MATERIALS AND METHODS
Recombinant murine SAA1 and SAA3: The coding region of murine SAA (amino acids 20 to 122, Supplemental Fig. S1) was amplified from hepatic mRNA by reverse transcription-polymerase chain reaction (RT-PCR) as follows. Total RNA was extracted from the liver of a normal mouse by an RNeasy Mini kit (Qiagen, Hilden, Germany). Genes of SAA1 and SAA3 were amplified by RT-PCR using a Titan One Tube RT-PCR Kit (Roche Diagnostics, Mannheim, Germany). Primers specific for SAA1 and SAA3 were designed based on the sequences (BC087933 and NM011315), and primer sequences that included Sac I and Kpn I restriction endonuclease sites were as follows:
SAA1 F: 5′-CCC CCC GAG CTC GGG TTT TTT TCA TTT GTT-3′
SAA1 R: 5′-A AAC GGT ACC TCA GTA TTT GTC AGG CAG-3′
SAA3 F: 5′-CCC CCC GAG CTC AGA TGG GTC CAG TTC ATG-3′
SAA3 R: 5′-A AAC GGT ACC TCA GTA TCT TTT AGG CAG-3′
The amplified PCR fragments of SAA1 and SAA3 were digested with Sac I and Kpn I (Toyobo, Osaka, Japan) and cloned between the Sac I and Kpn I sites of the pRSET A expression vector (Invitrogen, Carlsbad, CA, U.S.A.). Sequences of the cloned plasmids were confirmed by sequencing. Plasmid DNA was transformed into E. coli BL21 (DE3) pLysS (Invitrogen), and expressed proteins were purified by Ni2+ affinity chromatography using Chelating Sepharose Fast Flow (GE Healthcare, Buckinghamshire, U.K.) according to the manufacturer’s instructions. Proteins were eluted from the Ni2+ column with guanidine buffer at pH 7.8, pH 4.9 and pH 4.3, and their purity was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and coomassie brilliant blue (CBB) staining and western blot (WB) analysis. Then, purified fractions were dialyzed against phosphate buffered saline (PBS) with 0.5 M NaCl. Dialyzed solutions were concentrated by Amicon Ultra Centrifugal Filters Ultracel-10K (Merck Millipore, Cork, Ireland) and used as recombinant SAAs (rSAAs).
CBB staining and WB analysis: After SDS-PAGE, gels were stained with CBB staining solution (0.25% CBB, 5% methanol, and 7.5% acetic acid) for 30 min. Then, gels were destained by destaining solution (25% methanol and 7.5% acetic acid) several times. For WB analysis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P, Merck Millipore, Billerica, MA, U.S.A.) blocked with 5% nonfat milk in PBS supplemented with 0.1% Tween 20 (PBST) and incubated for 30 min at room temperature. Subsequently, the membrane was incubated with Anti-Xpress antibody (1:5,000, R910-25, Invitrogen) in 1% nonfat milk in PBST for 1 hr at room temperature. After washing three times with PBST, the membrane was incubated with horseradish peroxidase-conjugated anti-mouse IgG antibody (GE Healthcare) in 1% nonfat milk in PBST for 1 hr at room temperature. After washing three times, the peroxidase activity was detected using ECL Western Blotting Detection Reagents (GE Healthcare) and visualized by X-ray film (Fujifilm, Tokyo, Japan).
Cells: Murine colonic epithelial cells, CMT-93, were purchased from ECACC (Salisbury, U.K.) and used in this study. Murine fibroblast cells, NIH3T3, were also used. Both were maintained in Dulbecco’s modified Eagle’s minimal essential medium (DMEM, Wako, Osaka, Japan) containing 100 U/ml penicillin, 100 µg/ml streptomycin and 10% fetal bovine serum (PAA Laboratories GmbH, Pasching, Austria). Cells were seeded at 1 × 106 cells in 60-mm dishes and incubated in a humidified atmosphere of 5% CO2 at 37°C for 15 ± 3 hr before experiments.
Preparation of pathogens: Rotavirus strain Wa [22] was kindly provided by Dr. M. Sugiyama (Gifu University, Gifu, Japan). Viruses were treated with 10 µg/ml trypsin in DMEM at 37°C for 30 min. Then, viruses were inoculated to CMT-93 cells. Toxoplasma gondii strain PLK was kindly provided by Dr. Y. Takashima (Gifu University). Toxoplasma was collected by centrifugation at 600 × g for 10 min and washed with PBS. This procedure was repeated twice, and then, Toxoplasma was inoculated to CMT-93 cells. E. coli strain O157 lacking the production of Vero-toxin and Staphylococcus aureus strain 209P were used as Gram-positive and Gram-negative bacteria, respectively, and were grown in Luria-Bertani (LB) medium at 37°C. After overnight incubations, bacteria were collected by centrifugation at 6,300 × g for 5 min, and the pellets were washed with PBS. This procedure was repeated twice, and then, bacteria were aliquoted. One suspension was boiled for 15 min, and the other was placed on ice to retard the bacterial growth until use. After the boiling or cooling step, samples were adjusted to 37°C and inoculated to cells. To estimate cell counts of E. coli and S. aureus, bacteria were serially diluted with PBS and plated on LB agar at 37°C for 24 hr.
Inoculation of pathogens, LPS and rSAAs: CMT-93 cells were washed twice with PBS, inoculated with pathogens rotavirus (8.0 × 104 tissue culture infective dose for 50% (TCID50)/ml), Toxoplasma (2.7 × 106 cells/ml), S. aureus (5.2 × 107 colony forming units (CFU)/ml) and E. coli(~7.3 × 108 CFU/ml) suspended in DMEM and incubated at 37°C for 2 hr. Cells were also incubated with 0.1–100 µg/ml LPS from E. coli O111:B4 (L2630, Sigma Aldrich, St. Louis, MO, U.S.A.), rSAA1 (34 µg/ml) and rSAA3 (100 µg/ml) at 37°C for 2 hr. Control cells were not inoculated with any pathogens or rSAAs. After incubation, cells were washed twice with PBS, and total RNA was extracted immediately. Isolated RNA was stored at −80°C until use.
Real-time PCR: RNA was quantified using a Gene Quant 100 (GE Healthcare) and depleted of contaminating DNA with DNase I (Invitrogen). cDNA was synthesized using a SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) according to the manufacturer’s instructions. Quantitative real-time PCR was carried out in 48-well plates using the final concentrations of 250 nmol each of forward and reverse primes, 1 × Fast SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, U.S.A.) and 5 ng of cDNA. Thermal cycling conditions were 20 sec at 95°C followed by 40 cycles at 95°C for 3 sec and 60°C for 30 sec. Data were collected using a StepOne analytical thermal cycler (Applied Biosystems). mRNA expressions of SAA1/2, SAA3, IL-1β, IL-6, TNF-α, MUC2 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were investigated by using specific primers for real-time PCR. Primer sequences are shown in Table 1. Data were normalized to the expression of GAPDH mRNA as an endogenous gene, and fold-changes relative to control levels were determined by the ΔΔCt method [32]. A melting-curve analysis of amplification products was performed at the end of each PCR reaction. All experiments were replicated at least three times.
Table 1. Primers used for real-time PCR.
| Target * | Sequence (5’-3’) | Reference |
|---|---|---|
| SAA1/2 F | CTGCCTGCCAAATACTGAGAGTC | Eckhardt et al., 2010 [2] |
| SAA1/2 R | CCACTTCCAAGTTCCTGTTTATTAC | |
| SAA3 F | GCTGGCCTGCCTAAAAGATACTG | Eckhardt et al., 2010 [2] |
| SAA3 R | GCATTTCACAAGTATTTATTCAGC | |
| IL-1β F | CCCAAGCAATACCCAAAGAA | Li et al., 2009 [8] |
| IL-1β R | CATCAGAGGCAAGGAGGAAA | |
| IL-6 F | TTCCATCCAGTTGCCTTCTT | Yang et al., 2006 [31] |
| IL-6 R | ATTTCCACGATTTCCCAGAG | |
| TNF-α F | GCCTCTTCTCATTCCTGCTT | Yang et al., 2006 [31] |
| TNF-α R | CACTTGGTGGTTTGCTAGGA | |
| MUC2 F | GCTGACGAGTGGTTGGTGAATG | Wlodarska et al., 2011 [30] |
| MUC2 R | GATGAGGTGGCAGACAGGAGAC | |
| GAPDH F | TGCACCACCACCAACTGCTTAG | This study |
| GAPDH R | GGATGCAGGGATGATGTTC | Accession No. GU214026 |
*SAA, serum amyloid A; IL, interleukin; TNF-α, tumor necrosis factor-α; MUC2, mucin 2; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Statistical analysis: Data were expressed as means ± standard errors and analyzed for statistical significance by unpaired two-tailed t-tests.
RESULTS
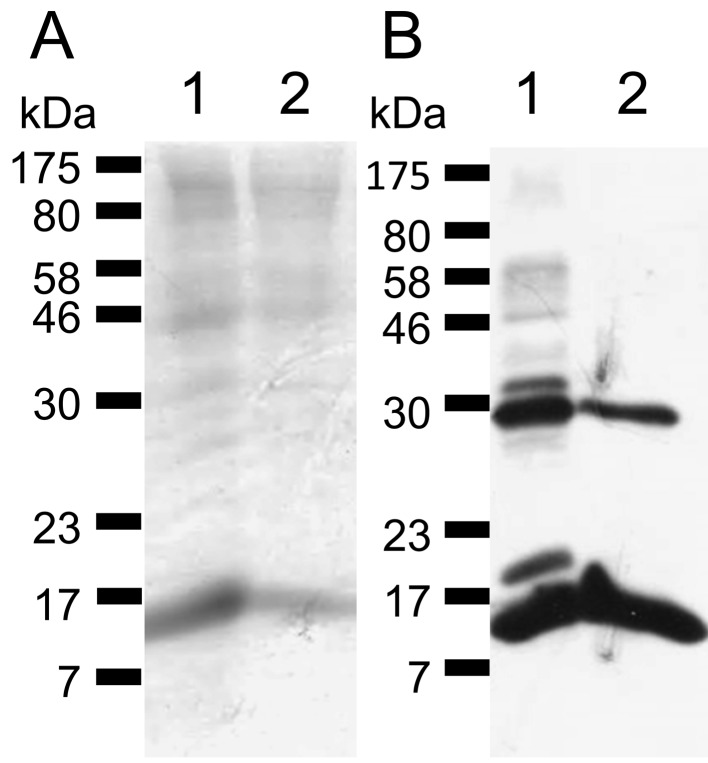
Expression of recombinant SAA1 and SAA3: The molecular weights of expressed rSAA1 and rSAA3 were approximately 16.1 kDa, and both CBB staining and WB analysis detected the expected sized bands (Fig.1). The approximately 30 kDa-sized proteins seemed to be dimers of rSAAs.
Fig. 1.
Expressed recombinant SAA1 and SAA3. (A) CBB staining. (B) WB analysis using an anti-Xpress antibody. Expected sized rSAA1 and rSAA3 (approximately 16.1 kDa) were detected. Approximately 30 kDa proteins seemed to be dimers of SAAs. Lane 1, rSAA1; Lane 2, rSAA3.
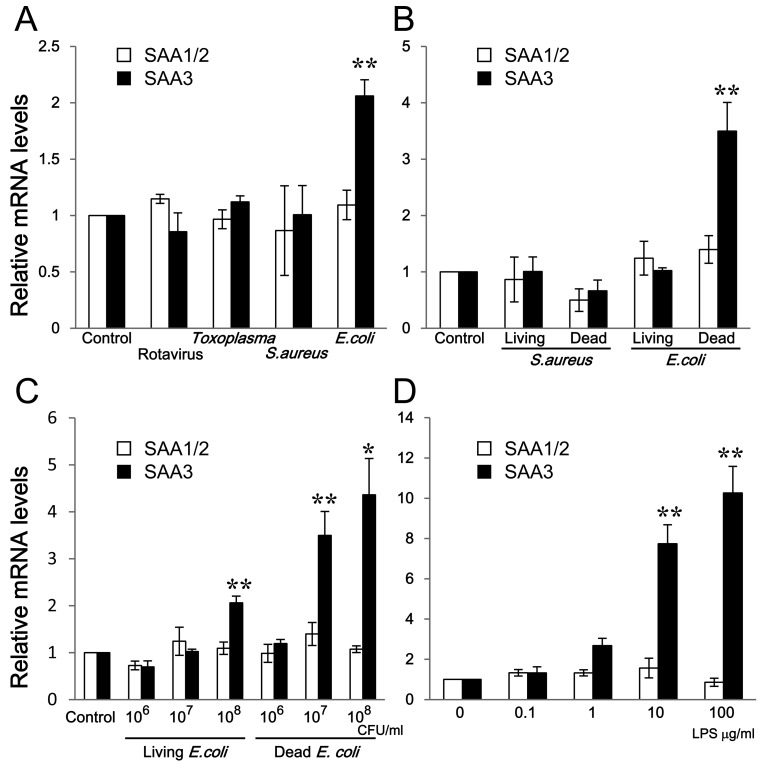
Comparison of mRNA expression of SAAs: To assess whether representative intestinal pathogens affected the mRNA expression of SAAs, rotavirus, Toxoplasma, S. aureus and E. coli were inoculated to CMT-93 cells. As mentioned above, since the sequences of SAA1 and SAA2 genes are highly conserved, we used primers for the detection both of SAA1 and SAA2 mRNAs as an indistinguishable SAA1/2 mRNA as described previously [2]. SAA1/2 mRNA expression in CMT-93 cells was not changed significantly by inoculation with the four different pathogens compared with the control (Fig. 2A). However, SAA3 mRNA expression was induced by inoculation with E. coli (Fig. 2A). SAA1/2 mRNA expression was not changed by inoculation with either Gram-positive S. aureus or Gram-negative E. coli, whether living or dead, at the same bacterial concentrations (107 CFU/ml) (Fig. 2B). In contrast, SAA3 mRNA expression was apparently increased by dead E. coli (Fig. 2B). To analyze the influence of living or dead bacteria in detail, living and dead E. coli at different amounts were inoculated to CMT-93 cells. Both living and dead E. coli induced SAA3 mRNA expression in a dose-dependent manner, whereas SAA1/2 mRNA expression was not enhanced (Fig. 2C).
Fig. 2.
Comparison of mRNA expressions of SAA1/2 and SAA3 in CMT-93 cells. (A) CMT-93 cells were inoculated with rotavirus, Toxoplasma, S. aureus and E. coli for 2 hr at 37°C. (B) CMT-93 cells were inoculated with living or dead S. aureus, and living or dead E. coli at the same bacterial concentrations (107 CFU/ml) for 2 hr at 37°C. (C) CMT-93 cells were inoculated with different amounts of living or dead E. coli for 2 hr at 37°C. (D) CMT-93 cells were inoculated with different amounts of LPS for 2 hr at 37°C. The relative mRNA expression levels of SAA1/2 and SAA3 were corrected with GAPDH and then compared with the control. Data are the means of 8 independent observations with standard errors represented by vertical bars. Asterisks indicate significant differences compared with the control. *, P<0.05; **, P<0.01.
Gram-negative E. coli, but not Gram-positive S. aureus, induced SAA3 mRNA expression, and dead E. coli up-regulated SAA3 mRNA expression higher than did living E. coli (Fig. 2C). LPS induced SAA3 mRNA expression in a dose-dependent manner, while SAA1/2 mRNA expression did not change by exposure to LPS (Fig. 2D). SAA3 mRNA expression was strongly induced by LPS even more so than by dead E. coli (Fig. 2C and 2D).
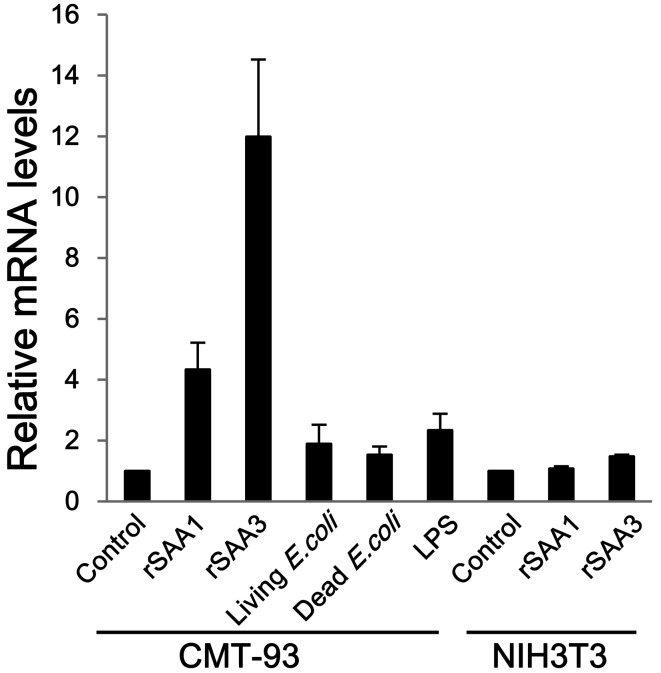
Induction of MUC2 mRNA expression: CMT-93 and NIH3T3 cells were incubated with rSAA1, rSAA3, E. coli and LPS. rSAA1 and rSAA3 induced MUC2 mRNA expression in CMT-93 cells about 4.3-fold and 12.0-fold, respectively, whereas inducible expression was not observed in NIH3T3 cells (Fig. 3). Living and dead E. coli and LPS did not significantly stimulate MUC2 mRNA expression (Fig. 3). These results suggest that SAA proteins have the ability to up-regulate MUC2 mRNA expression.
Fig. 3.
Comparison of MUC2 mRNA expression in CMT-93 and NIH3T3 cells. CMT-93 cells were inoculated with rSAA1, rSAA3, living or dead E. coli, or LPS. Murine fibroblast NIH3T3 cells were used as control cells. The relative mRNA expression levels of MUC2 were corrected with GAPDH and then compared with controls. Data are the means of at least 3 observations with standard errors represented by vertical bars.
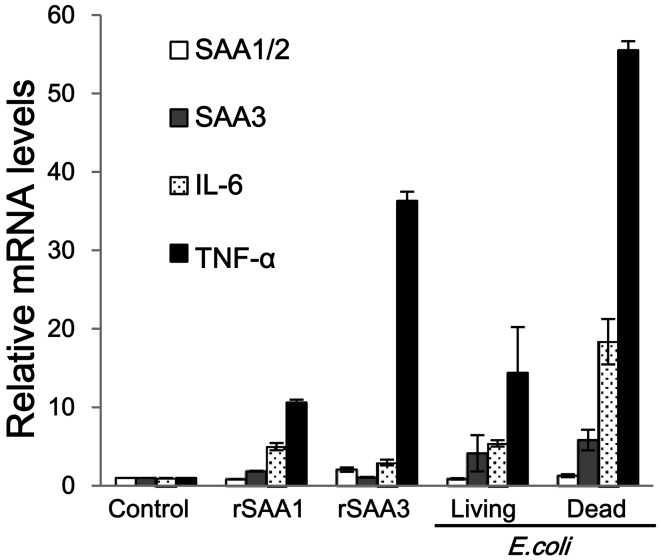
Induction of mRNA expression of cytokines: The mRNA expression of inflammatory cytokines was also measured in CMT-93 cells treated with rSAAs and E. coli. Both rSAA1 and rSAA3 enhanced TNF-α mRNA expression, as consistent with previous reports [15, 20] (Fig. 4). On the other hand, IL-1β mRNA expression was not detected (data not shown). In comparisons between rSAA1 and rSAA3 treatment, rSAA3 induced TNF-α mRNA expression intensively. mRNA expressions of SAA1/2 and SAA3 did not change significantly by treatment with rSAAs (Fig. 4). Both living and dead E. coli stimulated mRNA expressions of IL-6 and TNF-α, and the expression level was higher than that by rSAAs (Fig. 4). IL-1β mRNA expression was not stimulated by E. coli (data not shown).
Fig. 4.
Comparison of mRNA expressions of SAAs and cytokines in CMT-93 cells. CMT-93 cells were incubated with rSAA1, rSAA3, or living or dead E. coli for 2 hr at 37°C. mRNA expression levels of SAA1/2, SAA3, IL-1β, IL-6 and TNF-α were measured. IL-1β mRNA expression was not detected. The relative mRNA expression levels of SAA1/2, SAA3, IL-6 and TNF-α were corrected with GAPDH and then compared with controls. Data are the means of at least 3 observations with standard errors represented by vertical bars.
DISCUSSION
In this study, we demonstrated that the Gram-negative bacterium E. coli, but not the other pathogens, induced SAA3 mRNA expression significantly in mouse colonic epithelial CMT-93 cells, whereas SAA1/2 mRNA expression was not enhanced by treatment with these pathogens. SAA3 mRNA expression by dead E. coli was higher than that by living E. coli. This might have been caused by the influence of LPS, because LPS is a major component of Gram-negative bacterial membranes. The bursting of E. coli membranes by boiling results in increased amounts of LPS. We observed increased expression of SAA3 mRNA by LPS in a dose-dependent manner as previously described [2, 17]. Furthermore, MUC2 mRNA expression was significantly up-regulated by SAAs, especially by SAA3, but not by E. coli and LPS. It might be because of a delayed effect of SAA3 protein production after SAA3 mRNA expression. These results suggest that SAA3 plays a role in innate immunity in the colon by up-regulating MUC2 mucin production.
Intestinal epithelial cells together with the mucus layer act as a physiological barrier against pathogenic bacteria, viruses and protozoa, as well as harmless commensal bacteria [3, 10]. In the human colon, 1013-1014 microbes cohabitate peacefully with host immunity and establish intestinal homeostasis [3, 29]. MUC2 mucin-deficient mice do not build a mucus layer above the epithelial cells in the colon, and direct contact of bacteria with epithelial cells, invasion into the crypts [6] and spontaneous inflammation [26, 27] are observed. Consistent with a previous report [2], we observed that SAA1/2 mRNA expression was not inducible by any treatment, but rather that SAA1/2 mRNA was constantly expressed. Although the stimulation levels of rSAA1 for MUC2 mRNA expression were lower than those of rSAA3, both rSAA1 and rSAA3 apparently function to augment MUC2 mRNA expression. Additionally, rSAA3 and E. coli strongly enhanced the mRNA expression of the inflammatory cytokine TNF-α. It has been reported that SAA3 mRNA expression is induced by TNF-α in CMT-93 cells [17] and also that TNF-α induces MUC2 mRNA expression in human colonic epithelial HT-29 cells [5]. These results suggest that SAA1/2 may continuously stimulate MUC2 mucin production and build a mucosal layer for “peaceful cohabitation” with harmless commensal bacteria under intestinal homeostasis. When bacteria attack colonic epithelial cells, mRNA expressions of SAA3 and inflammatory cytokines, such as TNF-α, are induced, TNF-α also induces SAA3 mRNA expression, and then, produced SAA3 protein up-regulates MUC2 mucin production in cooperation with TNF-α, which leading to the protection of colonic epithelial cells from bacterial invasion.
Up-regulation of MUC2 mRNA expression by rSAAs was demonstrated in this study. Contrary to our results, Mack et al. [11] demonstrated no differences in MUC2 mRNA expression by various kinds of peptides from bovine mammary-associated SAA3 (M-SAA3) in human colonic epithelial HT-29 cells. There are at least two possibilities for this discrepancy. First, it may be due to the length of the peptides. Here, we used rSAA3 as a mature-length protein. On the other hand, Mack et al. used 4- to 10-mer peptides from bovine M-SAA3, which may not have full biological activity. Second, it may be due to species differences. This study showed the effects of murine SAA3 on murine colonic intestinal CMT-93 cells, while Mack et al. applied bovine M-SAA3 on human colonic epithelial HT-29 cells. To clarify this difference, further investigations are needed.
We showed that SAA3 mRNA expression was induced by E. coli and LPS, and that SAA3 protein was capable of up-regulating MUC2 mRNA expression in mouse colonic epithelial cells. Since we did not detect MUC2 mucin production in this study, further investigations both in vitro and in vivo are needed to elucidate the biological functions of SAA3 in the colon.
Supplementary
Acknowledgments
We are grateful to Dr. M. Sugiyama and Dr. Y. Takashima (Gifu University, Gifu, Japan) for providing rotavirus and Toxoplasma, respectively. This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- 1.Chiba T., Han C. Y., Vaisar T., Shimokado K., Kargi A., Chen M.H., Wang S., McDonald T. O., O’Brien K. D., Heinecke J. W., Chait A.2009. Serum amyloid A3 does not contribute to circulating SAA levels. J. Lipid Res. 50: 1353–1362. doi: 10.1194/jlr.M900089-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eckhardt E. R. M., Witta J., Zhong J., Arsenescu R., Arsenescu V., Wang Y., Ghoshal S., de Beer M. C., de Beer F. C., de Villiers W. J. S.2010. Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol. 10: 133–141. doi: 10.1186/1471-230X-10-133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goto Y., Kiyono H.2012. Epithelial barrier: an interface for the cross-communication between gut flora and immune system. Immunol. Rev. 245: 147–163. doi: 10.1111/j.1600-065X.2011.01078.x [DOI] [PubMed] [Google Scholar]
- 4.Hoffman J. S., Ericsson L. H., Ericsen N., Walsh K. A., Benditt E. P.1984. Murine tissue amyloid protein AA NH2-terminal sequence identity with only one of two serum amyloid protein (ApoSAA) gene products. J. Exp. Med. 159: 641–646. doi: 10.1084/jem.159.2.641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iwashita J., Sato Y., Sugaya H., Takahashi N., Sasaki H., Abe T.2003. mRNA of MUC2 is stimulated by IL-4, IL-13 or TNF-α through a mitogen-activated protein kinase pathway in human colon cancer cells. Immunol. Cell Biol. 81: 275–282. doi: 10.1046/j.1440-1711.2003.t01-1-01163.x [DOI] [PubMed] [Google Scholar]
- 6.Johansson M. E., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C.2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105: 15064–15069. doi: 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kluve-Beckerman B., Drumm M. L., Benson M. D.1991. Nonexpression of the human serum amyloid A three (SAA3) gene. DNA Cell Biol. 10: 651–661. doi: 10.1089/dna.1991.10.651 [DOI] [PubMed] [Google Scholar]
- 8.Li Y., Limmon G. V., Imani F., Teng C.2009. Induction of lactoferrin gene expression by innate immune stimuli in mouse mammary epithelial HC-11 cells. Biochimie 91: 58–67. doi: 10.1016/j.biochi.2008.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liepnieks J. J., Kluve-Beckerman B., Benson M. D.1995. Characterization of amyloid A protein in human secondary amyloidosis: the predominant deposition of serum amyloid A1. Biochim. Biophys. Acta 1270: 81–86. doi: 10.1016/0925-4439(94)00076-3 [DOI] [PubMed] [Google Scholar]
- 10.Linden S. K., Sutton P., Karlsson N. G., Korolik V., McGuckin M. A.2008. Mucins in the mucosal barrier to infection. Mucosal Immunol. 1: 183–197. doi: 10.1038/mi.2008.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mack D. R., McDonald T. L., Larson M. A., Wei S., Weber A.2003. The Conserved TFLK Motif of mammary-associated serum amyloid A3 is responsible for up-regulation of intestinal MUC3 mucin expression in vitro. Pediatr. Res. 53: 137–142. doi: 10.1203/00006450-200301000-00023 [DOI] [PubMed] [Google Scholar]
- 12.Meek R. L., Benditt E. P.1986. Amyloid A gene family expression in different mouse tissues. J. Exp. Med. 164: 2006–2017. doi: 10.1084/jem.164.6.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meek R. L., Hoffman J. S., Benditt E. P.1986. Amyloidogenesis. One serum amyloid A isotype is selectively removed from the circulation. J. Exp. Med. 163: 499–510. doi: 10.1084/jem.163.3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obici L., Merlini G.2012. AA amyloidosis: basic knowledge, unmet needs and future treatments. Swiss Med. Wkly. 142: 13580–13588 [DOI] [PubMed] [Google Scholar]
- 15.Patel H., Fellowes R., Coade S., Woo P.1998. Human serum amyloid A has cytokine-like properties. Scand. J. Immunol. 48: 410–418. doi: 10.1046/j.1365-3083.1998.00394.x [DOI] [PubMed] [Google Scholar]
- 16.Pearson J. P., Brownlee I. A.2010. The interaction of large bowel microflora with the colonic mucus barrier. Int. J. Inflam. 2010: 321426. doi: 10.4061/2010/321426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reigstad C. S., Lundén G. Ö., Felin J., Bäckhed F.2009. Regulation of serum amyloid A3 (SAA3) in mouse colonic epithelium and adipose tissue by the intestinal microbiota. PLoS ONE 4: e5842. doi: 10.1371/journal.pone.0005842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sellar G. C., Whitehead A. S.1993. Localization of four human serum amyloid A (SAA) protein superfamily genes to chromosome 11p: characterization of a fifth SAA-related gene sequence. Genomics 16: 774–776. doi: 10.1006/geno.1993.1265 [DOI] [PubMed] [Google Scholar]
- 19.Sipe J.1999. Revised nomenclature for serum amyloid A (SAA). Amyloid 6: 67–70. doi: 10.3109/13506129908993291 [DOI] [PubMed] [Google Scholar]
- 20.Song C., Hsu K., Yamen E., Yan W., Fock J., Witting P. K., Geczy C. L., Freedman S. B.2009. Serum amyloid A induction of cytokines in monocytes/macrophages and lymphocytes. Atherosclerosis 207: 374–383. doi: 10.1016/j.atherosclerosis.2009.05.007 [DOI] [PubMed] [Google Scholar]
- 21.Suffredini A. F., Fantuzzi G., Badolato R., Oppenheim J. J., O’Grady N. P.1999. New insights into the biology of the acute phase response. J. Clin. Immunol. 19: 203–214. doi: 10.1023/A:1020563913045 [DOI] [PubMed] [Google Scholar]
- 22.Suzuki H., Kutsuzawa T., Konno T., Ebina T., Ishida N.1981. Morphogenesis of human rotavirus type 2 Wa strain in MA 104 cells. Arch. Virol. 70: 33–41. doi: 10.1007/BF01320791 [DOI] [PubMed] [Google Scholar]
- 23.Uhlar C. M., Burgess C. J., Sharp P. M., Whitehead A. S.1994. Evolution of the serum amyloid A (SAA) protein superfamily. Genomics 19: 228–235. doi: 10.1006/geno.1994.1052 [DOI] [PubMed] [Google Scholar]
- 24.Uhlar C. M., Grehan S., Steel D. M., Steinkasserer A., Whitehead A. S.1997. Use of the acute phase serum amyloid A2 (SAA2) gene promoter in the analysis of pro- and anti-inflammatory mediators: differential kinetics of SAA2 promoter induction by IL-1β and TNF-α compared to IL-6. J. Immunol. Methods 203: 123–130. doi: 10.1016/S0022-1759(96)00220-7 [DOI] [PubMed] [Google Scholar]
- 25.Uhlar C. M., Whitehead A. S.1999. Serum amyloid A, the major vertebrate acute-phase reactant. Eur. J. Biochem. 265: 501–523. doi: 10.1046/j.1432-1327.1999.00657.x [DOI] [PubMed] [Google Scholar]
- 26.Van der Sluis M., De Koning B. A. E., De Bruijn A. C. J. M., Velcich A., Meijerink J. P. P., Van Goudoever J. B., Büller H. A., Dekker J., Van Seuningen I., Renes I. B., Einerhand A. W. C.2006. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131: 117–129. doi: 10.1053/j.gastro.2006.04.020 [DOI] [PubMed] [Google Scholar]
- 27.Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L.2002. Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295: 1726–1729. doi: 10.1126/science.1069094 [DOI] [PubMed] [Google Scholar]
- 28.Weiss A. A., Babyatsky M. W., Ogata S., Chen A., Itzkowitz S. H.1996. Expression of MUC2 and MUC3 mRNA in human normal, malignant, and inflammatory intestinal tissues. J. Histochem. Cytochem. 44: 1161–1166. doi: 10.1177/44.10.8813081 [DOI] [PubMed] [Google Scholar]
- 29.Whitman W. B., Coleman D. C., Wiebe W. J.1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95: 6578–6583. doi: 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wlodarska M., Willing B., Keeney K. M., Menendez A., Bergstrom K. S., Gill N., Russell S. L., Vallance B. A., Finlay B. B.2011. Antibiotic treatment alters the colonic mucus layer and predisposes the host to exacerbated Citrobacter rodentium-induced colitis. Infect. Immun. 79: 1536–1545. doi: 10.1128/IAI.01104-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y. H., Morand E. F., Getting S. J., Paul-Clark M., Liu D. L., Yona S., Hannon R., Buckingham J. C., Perretti M., Flower R. J.2004. Modulation of inflammation and response to dexamethasone by Annexin 1 in antigen-induced arthritis. Arthritis Rheum. 50: 976–984. doi: 10.1002/art.20201 [DOI] [PubMed] [Google Scholar]
- 32.Yuan J. S., Reed A., Chen F., Stewart C. N., Jr2006. Statistical analysis of real-time PCR data. BMC Bioinformatics 7: 85–97. doi: 10.1186/1471-2105-7-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.