Abstract
Previous studies have investigated the associations between FEN1 -69G>A (rs174538) and 4150G>T (rs4246215) polymorphisms and cancer risk in Chinese population. However, the results were controversial. We therefore carried out a meta-analysis to derive a more precise estimation of the associations. PubMed Database was systematically searched to identify potentially eligible literatures. Crude odds ratios (ORs) and their 95% confidence intervals (CIs) were used to assess the strength of associations between FEN1 -69G>A and 4150G>T polymorphisms and cancer risk in Chinese population. A total of 4 articles, including 5,108 cases and 6,382 controls, were used to evaluate the effect of the two polymorphisms on cancer risk. The pooled ORs indicated that FEN1 -69G>A and 4150G>T polymorphisms were significantly associated with cancer risk in Chinese population. In stratified analyses by cancer type, significant associations were also observed in digestive system cancer. In addition, haplotypes consisting of -69G>A and 4150G>T polymorphisms were closely associated with cancer risk. Interestingly, significantly correlation between FEN1 -69G>A polymorphism and mRNA expression was observed. In conclusion, this meta-analysis suggests that FEN1 -69G>A and 4150G>T polymorphisms may be associated with cancer susceptibility in Chinese population. However, further investigation on large population and different ethnicities are warranted.
Cancer is one of serious diseases threatening public health. About 12.7 million cases were diagnosed and 7.6 million patients died from cancer in 20081. The pathogenesis of cancer remains unknown, but evidence from epidemiological and genetic studies has indicated significant association between inherited and environmental factors and cancer development2,3,4.
DNA repair systems play critical roles in protecting against mutations and are essential for maintaining the integrity of the genome. Reduced DNA repair capacity (DRC) is reportedly related to an increased risk of cancer5, and single nucleotide polymorphisms (SNPs) located in DNA repair genes may affect gene function and thereby debilitate DRC6.
Flap endonuclease 1 (FEN1) is an evolutionarily conserved component of DNA replication in humans. Based on in vitro results, it processes Okazaki fragments during replication and is involved in base excision repair. FEN1 removes the last primer ribonucleotide on the lagging strand and it cleaves a 5-prime flap that may result from strand displacement during replication or during base excision repair7,8,9. In addition, the conserved FEN1 carboxyl terminus binds proliferating cell nuclear antigen and positions FEN1 to act primarily as an exonuclease in DNA replication, in contrast to its endonuclease activity in DNA repair. Therefore, FEN1 play a vital role in maintaining genomic stability and protecting against carcinogenesis.
FEN1 gene is located on the chromosome 11q12.2 region and has some common genetic polymorphisms identified. Among them, -69G>A (rs174538) and 4150G>T (rs4246215) polymorphisms have been gaining great attention. Previous studies have suggested that the two polymorphisms in FEN1 may function as biomarkers for cancer risk in Chinese population, including breast cancer, glioma, hepatocellular carcinoma, esophageal cancer, gastric cancer, colorectal cancer, etc10,11,12,13. However, these studies had relatively small sample sizes, and may lack enough power to assess the associations between FEN1 -69G>A and 4150G>T polymorphisms and cancer risk in Chinese population. Hence, we performed this meta-analysis from all eligible case-control studies to provide a more precise estimation of the associations.
Results
Characteristics of Studies
Figure 1 and Table 1 show the study selection process and main characteristics of included studies, respectively. A total of 9 articles were retrieved based on the search criteria10,11,12,13,14,15,16,17,18. Among them, 3 articles were excluded through screening title and abstract14,17,18. Then, 2 articles without FEN1 -69G>A and 4150G>T polymorphisms and cancer risk were excluded15,16. Thus, a total of 4 articles with 5,108 cancer cases and 6,382 controls were included in the meta-analysis10,11,12,13. For the -69G>A polymorphism, 7 studies were available, including a total of 5,108 cases and 6,381 controls. For the 4150G>T polymorphism, 7 studies involved a total of 5,071 cases and 6,381 controls. Among them, 4 studies focused on digestive system cancer including hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer, 3 involved with other system cancer, such as breast cancer, glioma and lung cancer. Frequency distribution of the FEN1 -69G>A and 4150G>T haplotypes is shown in Supplement Table S1. A total of 4 studies with 10,669 cases and 12,761 controls were combined to test the difference in the frequencies of haplotypes. In addition, there are 2 articles involved FEN1 -69G>A polymorphism and FEN1 mRNA expression in normal tissues (Supplement Table S2). In the two articles including five studies, the same assay method (SYBR-Green real-time quantity PCR) and internal reference (β-actin) were used to examine FEN1 mRNA levels. Thus, they were pooled to assess the genotype-mRNA expression correlation.
Figure 1. The flow chart of literature search and study selection.
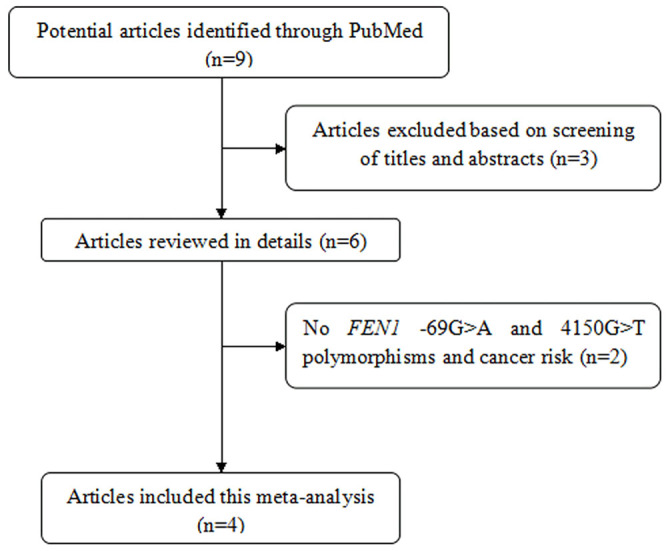
Table 1. Main Characteristics of Included Studies in the Meta-Analysis.
| First author | Year | Country | Cancer type | Genotyping | Source of control | -69G>A rs174538 | 4150G>T rs4246215 | ||
|---|---|---|---|---|---|---|---|---|---|
| Case AA/GA/GG | Control AA/GA/GG | Case TT/GT/GG | Control TT/GT/GG | ||||||
| Lv, Z. | 2014 | China | Breast cancer | PCR-RFLP | Community cancer-screening program for early detection of cancer | 116/437/547 | 246/639/515 | 134/449/517 | 246/651/503 |
| Chen, Y. D. | 2013 | China | Glioma | PCR-RFLP | Hospital | 35/122/160 | 130/356/316 | 34/120/160 | 130/363/309 |
| Liu, L. | 2012 | China | Hepatocellular carcinoma | PCR-RFLP | Community cancer-screening program for early detection of cancer | 68/290/290 | 134/334/270 | 73/295/280 | 129/335/274 |
| Esophageal cancer | 62/249/244 | 128/331/263 | 58/255/225 | 125/336/262 | |||||
| Gastric cancer | 41/182/189 | 76/209/169 | 42/177/183 | 76/212/166 | |||||
| Colorectal cancer | 28/104/104 | 54/144/109 | 27/102/100 | 52/145/109 | |||||
| Yang, M. | 2009 | China | Lung cancer | PCR-RFLP,TaqMan | Community nutritional survey, Community screening program for early detection of cancer and chronic noninfectious disease | 253/796/791 | 354/880/724 | 271/815/754 | 358/883/717 |
Quantitative data synthesis
Results of FEN1 -69G>A and 4150G>T polymorphisms and cancer risk are presented in Table 2 and Figure 2. For the -69G>A polymorphism, significant association was observed in all cancer type combined study, including breast cancer, glioma, hepatocellular carcinoma, esophageal cancer, gastric cancer, colorectal cancer and lung cancer (A vs. G: OR = 0.73, 95%CI: 0.69–0.77, PH = 0.20; AA vs. GG: OR = 0.54, 95%CI: 0.48–0.61, PH = 0.31; AG vs. GG: OR = 0.76, 95%CI: 0.70–0.82, PH = 0.39; (AA+AG) vs. GG: OR = 0.70, 95%CI: 0.65–0.75, PH = 0.26; AA vs. (AG+GG): OR = 0.62, 95%CI: 0.56–0.69, PH = 0.51). Subgroup analysis by cancer type showed that statistically significant associations were also found among digestive system cancer (A vs. G: OR = 0.72, 95%CI: 0.66–0.79, PH = 0.99; AA vs. GG: OR = 0.50, 95%CI: 0.41–0.61, PH = 0.96; AG vs. GG: OR = 0.80, 95%CI: 0.70–0.91, PH = 0.99; (AA+AG) vs. GG: OR = 0.71, 95%CI: 0.63–0.81, PH = 1.00; AA vs. (AG+GG): OR = 0.56, 95%CI: 0.47–0.68, PH = 0.94).
Table 2. Results of meta-analysis for FEN1 -69G>A and 4150G>T polymorphisms and cancer risk.
| Polymorphism | Type | Allele model | Homozygous model | Heterozygous model | Dominant model | Recessive model | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A vs. G | AA vs. GG | AG vs. GG | (AA+AG) vs. GG | AA vs. (AG+GG) | |||||||
| OR (95%CI) | PH | OR (95%CI) | PH | OR (95%CI) | PH | OR (95%CI) | PH | OR (95%CI) | PH | ||
| -69G>A | Total | 0.73 (0.69–0.77) | 0.20 | 0.54 (0.48–0.61) | 0.31 | 0.76 (0.70–0.82) | 0.39 | 0.70 (0.65–0.75) | 0.26 | 0.62 (0.56–0.69) | 0.51 |
| DSC | 0.72 (0.66–0.79) | 0.99 | 0.50 (0.41–0.61) | 0.96 | 0.80 (0.70–0.91) | 0.99 | 0.71 (0.63–0.81) | 1.00 | 0.56 (0.47–0.68) | 0.94 | |
| Other | 0.72 (0.61–0.84) | 0.02 | 0.56 (0.49–0.65) | 0.05 | 0.74 (0.67–0.82) | 0.07 | 0.67 (0.55–0.81) | 0.03 | 0.65 (0.57–0.75) | 0.21 | |
| T vs. G | TT vs. GG | TG vs. GG | (TT+TG) vs. GG | TT vs. (TG+GG) | |||||||
| OR (95%CI) | PH | OR (95%CI) | PH | OR (95%CI) | PH | OR (95%CI) | PH | OR (95%CI) | PH | ||
| 4150G>T | Total | 0.77 (0.73–0.81) | 0.19 | 0.59 (0.53–0.67) | 0.37 | 0.79 (0.73–0.86) | 0.16 | 0.74 (0.68–0.79) | 0.13 | 0.67 (0.60–0.75) | 0.61 |
| DSC | 0.76 (0.69–0.83) | 0.95 | 0.54 (0.44–0.66) | 0.98 | 0.83 (0.73–0.95) | 0.82 | 0.75 (0.66–0.86) | 0.87 | 0.60 (0.50–0.72) | 0.98 | |
| Other | 0.75 (0.64–0.87) | 0.02 | 0.62 (0.54–0.72) | 0.09 | 0.74 (0.60–0.91) | 0.02 | 0.69 (0.56–0.86) | 0.01 | 0.71 (0.63–0.81) | 0.40 | |
Allele model (A vs. G): (AA*2+GA) vs. (GG*2+GA); DSC: Digestive system cancer; PH: P value for the heterogeneity test.
Figure 2. Forest plots for FEN1 -69G>A and 4150G>T polymorphisms and cancer risk in the heterozygous model.
(A: FEN1 -69G>A polymorphism; B: FEN1 4150G>T polymorphism; Liu, L. 1-4 indicate Liu, L.'s study of hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer, respectively).
For the 4150G>T polymorphism, significant associations with cancer risk were observed in all cancer type combined study (T vs. G: OR = 0.77, 95%CI: 0.73–0.81, PH = 0.19; TT vs. GG: OR = 0.59, 95%CI: 0.53–0.67, PH = 0.37; TG vs. GG: OR = 0.79, 95%CI: 0.73–0.86, PH = 0.16; (TT+TG) vs. GG: OR = 0.74, 95%CI: 0.68–0.79, PH = 0.13; TT vs. (TG+GG): OR = 0.67, 95%CI: 0.60–0.75, PH = 0.61). In the subgroup analysis by cancer type, statistically significant associations were found in digestive system cancer (T vs. G: OR = 0.76, 95%CI: 0.69–0.83, PH = 0.95; TT vs. GG: OR = 0.54, 95%CI: 0.44–0.66, PH = 0.98; TG vs. GG: OR = 0.83, 95%CI: 0.73–0.95, PH = 0.82; (TT+TG) vs. GG: OR = 0.75, 95%CI: 0.66–0.86, PH = 0.87; TT vs. (TG+GG): OR = 0.60, 95%CI: 0.50–0.72, PH = 0.98).
In haplotype analysis (Table 3), the FEN1 haplotype (G-69G4150) comprising the major alleles was taken as reference. The pooled results indicated that G-69T4150, A-69G4150 and A-69T4150 haplotypes were significantly associated with cancer risk (G-69T4150 vs. G-69G4150: OR = 0.68, 95%CI: 0.55–0.83, PH = 0.00; A-69G4150 vs. G-69G4150: OR = 0.41, 95%CI: 0.32–0.51, PH = 0.00; A-69T4150 vs. G-69G4150: OR = 0.76, 95%CI: 0.70–0.82, PH = 0.00).
Table 3. Results of meta-analysis for FEN1 haplotypes and cancer risk.
| vs. G-69G4150 | ||
|---|---|---|
| Haplotypes | OR (95%CI) | PH |
| G-69T4150 | 0.68 (0.55–0.83) | 0.00 |
| A-69G4150 | 0.41 (0.32–0.51) | 0.00 |
| A-69T4150 | 0.76 (0.70–0.82) | 0.00 |
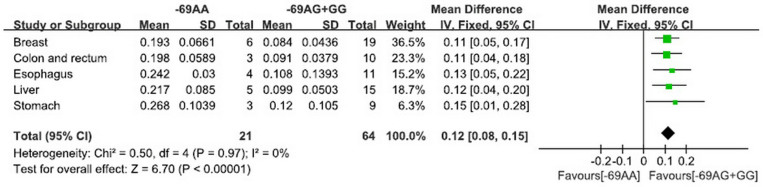
For FEN1 -69G>A polymorphism and FEN1 mRNA expression (Figure 3), the pooled analysis revealed that FEN1 mRNA levels were different between subjects with the -69AA genotype and those with the -69GG and GA genotypes in all included normal tissues (P < 0.001).
Figure 3. The pooled results of FEN1 -69G>A polymorphism and FEN1 mRNA expression in recessive model.
(-69AA: FEN1 mRNA levels in subjects with the -69AA genotype; -69AG+GG: FEN1 mRNA levels in subjects with the -69AG and -69GG genotype).
Sensitivity analysis
The sensitivity analysis indicated that no single study influenced the pooled OR value, either for FEN1 -69G>A polymorphism or for FEN1 4150G>T polymorphism, suggesting that the results of this meta-analysis are stable (data not shown).
Publication bias
Publication bias of the included articles was assessed using Begg's funnel plot. As shown in Figure 4, the shape of the funnel plot was symmetrical and did not show an obvious publication bias for FEN1 -69G>A or 4150G>T polymorphism.
Figure 4. Funnel plots of FEN1 -69G>A and 4150G>T polymorphisms in the heterozygous model.
(A: FEN1 -69G>A polymorphism; B: FEN1 4150G>T polymorphism).
Discussion
To the best of our knowledge, this is the first meta-analysis providing comprehensive insights into the effects of FEN1 -69G>A and 4150G>T polymorphisms on the risk of cancer. For the -69G>A polymorphism in the FEN1 promoter region, the overall results suggested that the subjects with A allele caused an decreased susceptibility to cancer in Chinese populations. For the 4150G>T polymorphism in the FEN1 3′-untranslated region, results showed that subjects with T allele were associated with the decreased risk of cancer in Chinese populations. However, the previous results showed no associations between the two polymorphisms and the risk of colorectal cancer11. Considering that the previous single-institution study for colorectal cancer has a small sample size and may not justify the significance of current work, further studies are needed to clarify the effect of the two polymorphisms on the risk of colorectal cancer. In addition, the current results were consistent with the biological function studies. Transient transfection and luciferase assays showed that FEN1, a tumor suppressor gene, -69A or 4150T variant has heightened expression in vitro13. According to cancer type, significantly decreased risks were also observed in digestive system cancer. In addition, similar results were observed in FEN1 haplotype analysis. Namely, G-69T4150, A-69G4150 and A-69T4150 haplotypes were significantly associated with the decreased risk of cancer in Chinese populations. Interestingly, -69G>A polymorphism is significantly associated with not only cancer risk but also FEN1 mRNA levels in normal tissues. Subjects with the -69AA genotype had significantly higher FEN1 mRNA levels than those with the -69GG and GA genotypes.
Some limitations should be considered when interpreting our results. Firstly, our results were based on unadjusted estimates, since more accurate outcomes should be adjusted by other confounders including gender, age, body mass index, lifestyle, etc. Secondly, all eligible studies included in this analysis were insufficient, especially in subgroup analysis. Thus, potential publication bias was very likely to exist, in spite of no significant publication bias in our meta-analysis. Thirdly, language of all included studies was limited to English, which may result in potential language bias. Last but not least, the interaction of gene-environment had not been evaluated owing to the absence of original data. However, some advantages should be highlighted in this meta-analysis. The current research could shed lights on the relationship of FEN1 -69G>A and 4150G>T polymorphisms and the decreased susceptibility to cancer in Chinese population, comprehensively and systematically. Additionally, the present research, which had a larger sample size, could enhance the power and persuasion of our conclusion when compared with previous study.
In summary, our meta-analysis suggests that FEN1 -69G>A and 4150G>T polymorphisms and their haplotypes are associated with cancer risk, and FEN1 -69G>A polymorphism are associated with FEN1 mRNA expression in normal tissues. However, larger sample sizes of different ethnic populations are required to confirm our findings.
Methods
Search strategy and inclusion criteria
We systematically searched literature from PubMed for all relevant studies using the following terms: “flap endonuclease or FNE1” “polymorphism” and “cancer” (up to June 14, 2014). In addition, the references of all retrieved articles were also manually searched for additionally relevant publications. Criteria for eligible studies were as follows: (a) case-control studies, (b) evaluating the association between FEN1 -69G>A and/or 4150G>T polymorphisms and cancer risk in Chinese population, (c) and sufficient information for calculating the pooled odds ratios (ORs) with 95% confidence intervals (CIs).
Data extraction
The re-sampling method was performed as described previously19. Two authors independently extracted data from eligible studies10,11,12,13. The following information was collected from each study: first author's name, year of publication, country of origin, cancer type, source of control, genotyping methods, the frequency distribution of -69G>A and 4150G>T genotypes/alleles/haplotypes. In addition, data of correlations between FEN1 -69G>A polymorphism and FEN1 mRNA expression in normal tissues were also extracted from eligible studies11,12. Any disagreements were resolved by discussion with a third investigator.
Statistical Methods
All statistical analyses were carried out with the Stata software version 12.0 (Stata Corporation, College Station, TX) and RevMan 5.0 software. Heterogeneity among studies was tested using the Chi square-based Q-test, and P value for the heterogeneity test (PH) <0.05 was regarded as statistically significant. Based on the heterogeneity test, the pooled OR was calculated using the fixed (PH ≥ 0.05) or random (PH < 0.05) effects model. The strength of the associations between FEN1 -69G>A and 4150G>T polymorphisms and their haplotypes and cancer risk was measured by ORs with 95% CIs. The pooled ORs were also estimated for -69G>A by homozygous (AA vs. GG), heterozygous (AG vs. GG), dominant [(AA+AG) vs. GG] and recessive models [AA vs. (AG+GG)] as well as allele model (A vs. G) and so was 4150G>T by homozygous (TT vs. GG), heterozygous (TG vs. GG), dominant [(TT+TG) vs. GG] and recessive models [TT vs. (TG+GG)] as well as allele model (T vs. G). For FEN1 -69G>A and 4150G>T haplotypes, the pooled ORs were estimated by comparison with G-69G4150 haplotype (G-69T4150 vs. G-69G4150, A-69G4150 vs. G-69G4150, A-69T4150 vs. G-69G4150). In addition, ORs and 95% CIs were used to assess the associations between FEN1 -69G>A polymorphism and FEN1 mRNA expression in normal tissues. Subsequently, in order to ensure that no individual study was entirely responsible for the combined results, sensitivity analysis was performed by omitting each study in turn to assess the quality and consistency of the results. Begg's funnel plot was used to evaluate possible publication bias of literatures. All the P values < 0.05 were considered significant.
Author Contributions
Conceived and designed the experiments: H.G., G.X. Performed the experiments: H.G., G.X., Z.S. Analyzed the data: G.X., Z.S. Contributed reagents/material/analysis tools: H.G., G.X., Z.S. Wrote the main manuscript text: H.G., G.X. Reference collection and data management: H.G., G.X., Z.S., Y.Y. Statistical analyses and paper writing: H.G., G.X., Z.S. Study design: H.G., G.X., Z.S., Y.Y. Prepared figures 1–4: Y.Y. All authors reviewed the manuscript.
Supplementary Material
Table S1,Table S2
References
- Jemal A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011). [DOI] [PubMed] [Google Scholar]
- Gao X. et al. Insertion/deletion polymorphisms in the promoter region of BRM contribute to risk of hepatocellular carcinoma in Chinese populations. PloS one 8, e55169 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Huang S., Chen W., Song Z. & Cai S. NFKB1 -94 insertion/deletion polymorphism and cancer risk: a meta-analysis. Tumour Biol 35, 5181–5187 (2014). [DOI] [PubMed] [Google Scholar]
- Ying H., Wang J. & Gao X. CCL5-403, CCR5-59029, and Delta32 polymorphisms and cancer risk: a meta-analysis based on 20,625 subjects. Tumour Biol 35, 5895–5904 (2014). [DOI] [PubMed] [Google Scholar]
- Shen H. et al. Smoking, DNA repair capacity and risk of nonsmall cell lung cancer. Int J Cancer 107, 84–88 (2003). [DOI] [PubMed] [Google Scholar]
- Xi T., Jones I. M. & Mohrenweiser H. W. Many amino acid substitution variants identified in DNA repair genes during human population screenings are predicted to impact protein function. Genomics 83, 970–979 (2004). [DOI] [PubMed] [Google Scholar]
- Harrington J. J. & Lieber M. R. The characterization of a mammalian DNA structure-specific endonuclease. EMBO J 13, 1235–1246 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M. R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays 19, 233–240 (1997). [DOI] [PubMed] [Google Scholar]
- Shen B. et al. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays 27, 717–729 (2005). [DOI] [PubMed] [Google Scholar]
- Chen Y. D. et al. Functional FEN1 genetic variants and haplotypes are associated with glioma risk. J Neurooncol 111, 145–151 (2013). [DOI] [PubMed] [Google Scholar]
- Liu L. et al. Functional FEN1 genetic variants contribute to risk of hepatocellular carcinoma, esophageal cancer, gastric cancer and colorectal cancer. Carcinogenesis 33, 119–123 (2012). [DOI] [PubMed] [Google Scholar]
- Lv Z. et al. Association of functional FEN1 genetic variants and haplotypes and breast cancer risk. Gene 538, 42–45 (2014). [DOI] [PubMed] [Google Scholar]
- Yang M. et al. Functional FEN1 polymorphisms are associated with DNA damage levels and lung cancer risk. Hum Mutat 30, 1320–1328 (2009). [DOI] [PubMed] [Google Scholar]
- Chen Y. et al. A NEIL1 single nucleotide polymorphism (rs4462560) predicts the risk of radiation-induced toxicities in esophageal cancer patients treated with definitive radiotherapy. Cancer 119, 4205–4211 (2013). [DOI] [PubMed] [Google Scholar]
- Corral R. et al. Genetic variation in the base excision repair pathway, environmental risk factors, and colorectal adenoma risk. PloS one 8, e71211 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. Q. et al. Genetic variants in DNA repair pathway genes and risk of esophageal squamous cell carcinoma and gastric adenocarcinoma in a Chinese population. Carcinogenesis 34, 1536–1542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Jin Q., Forsti A., Hemminki K. & Kumar R. Single nucleotide polymorphism analyses of the human proliferating cell nuclear antigen (pCNA) and flap endonuclease (FEN1) genes. Int J Cancer 88, 938–942 (2000). [DOI] [PubMed] [Google Scholar]
- Mitra A. K. et al. Association of polymorphisms in base excision repair genes with the risk of breast cancer: a case-control study in North Indian women. Oncol Res 17, 127–135 (2008). [DOI] [PubMed] [Google Scholar]
- Li J. et al. Identification of high-quality cancer prognostic markers and metastasis network modules. Nat Commun 1:34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1,Table S2