Abstract
Toxins from the bacterium Bacillus thuringiensis (Bt) are used widely for insect control in sprays and transgenic plants, but their efficacy is reduced when pests evolve resistance. Previous work showed that mutations in a gene encoding the transporter protein ABCC2 are linked with resistance to Bt toxins Cry1Ab, Cry1Ac or both in four species of Lepidoptera. Here we compared the ABCC2 gene of Helicoverpa armigera (HaABCC2) between susceptible strains and a laboratory-selected strain with >1,000-fold resistance to Cry1Ac relative its susceptible parent strain. We discovered a 73-base pair (bp) insertion in the cDNA of the resistant strain that generates a premature stop codon expected to yield a truncated ABCC2 protein. Sequencing of genomic DNA revealed that this insertion is an intron that is not spliced out because of a 6-bp deletion at its splicing site. Analysis of progeny from crosses revealed tight genetic linkage between HaABCC2 and resistance to Cry1Ac. These results provide the first evidence that mis-splicing of a gene encoding an ABCC2 protein confers resistance to a Bt toxin.
Insecticidal proteins from the soil bacterium Bacillus thuringiensis (Bt) have been used extensively for insect control in sprays and genetically engineered crops1,2. These Bt proteins kill some devastating insect pests, but are not toxic to most other organisms, including people1,2,3. The area planted to Bt crops worldwide increased to 76 million hectares (ha) in 2013, with a cumulative total of >560 million ha since 19964. Although Bt crops have provided substantial environmental and economic benefits5,6,7,8,9,10 evolution of resistance to Bt proteins by pests can cut short these benefits11,12,13,14.
Understanding the mechanisms that confer resistance to Bt toxins can facilitate efforts to sustain the advantages of these proteins2,15,16,17,18. The most common and most potent mechanisms of resistance to Bt toxins in Lepidoptera entail reduced binding of the toxins to midgut proteins including cadherin and aminopeptidase N2,15. Resistance to Bt toxins Cry1Ab, Cry1Ac, or both is also linked with mutations affecting the ATP-binding cassette transporter protein ABCC2 in four strains of Lepidoptera, each from a different species (Heliothis virescens, Plutella xylostella Trichoplusia ni, and Bombyx mori19,20,21).
Here we evaluated the role of the ABCC2 gene in resistance to Cry1Ac of a laboratory-selected strain (LF60) of Helicoverpa armigera from China. Although Bt cotton producing Cry1Ac remains effective against this major pest in China, many strains of this species have been selected for high levels of resistance to Cry1Ac in the laboratory and “early warning” of increases in the frequency of resistance to Cry1Ac have been reported from field populations in northern China exposed intensively to Bt cotton22,23,24. We found a 6-bp deletion in the genomic DNA (gDNA) of the ABCC2 gene of the resistant strain that interferes with splicing, introduces a premature stop codon in transcripts, and is genetically linked with resistance to Cry1Ac.
Results
Resistance to Cry1Ac in the LF60 strain
The concentration of Cry1Ac killing 50% of larvae (LC50) was >1000 times higher for the laboratory-selected resistant strain LF60 relative to its unselected susceptible parent strain (LF) in diet bioassays with either protoxin or activated toxin (Table 1). The LC50 of Cry1Ac did not differ significantly between the two susceptible strains (LF and 96S) for either protoxin or activated toxin (Table 1).
Table 1. Responses to Cry1Ac of two susceptible strains (LF and 96S) and a resistant strain (LF60) of H. armigera.
| Strain | Form of Cry1Ac | LC50 (95% fiducial limits) (µg Cry1Ac per ml diet) | Resistance ratio* |
|---|---|---|---|
| LF | protoxin | 0.009 (0.002–0.020) | 1.0 |
| 96S | protoxin | 0.021 (0.008–0.036) | 2.3 |
| LF60 | protoxin | 12.4 (8.0–17) | 1400 |
| LF | activated toxin | 0.008 (0.002–0.014) | 1.0 |
| 96S | activated toxin | 0.013 (0.007–0.021) | 1.6 |
| LF60 | activated toxin | 9.15 (5.8–13) | 1100 |
*LC50 of each strain divided by the LC50 of the susceptible LF strain.
ABCC2 cDNA in susceptible and resistant strains
We give the name HaABCC2 (GeneBank accession no. KF479231) to the gene encoding the ABCC2 protein of H. armigera. The cDNA of HaABCC2 from the susceptible LF strain had 4,017 bp encoding a predicted ABCC2 protein of 1,338 amino acids (Fig. 1). Compared with predicted HaABCC2 proteins from other Lepidoptera, the predicted HaABCC2 protein shares 93% amino acid identity with Heliothis virescens (GenBank accession nos. ADH16740.1) and Heliothis subflexa (ADH16744.1), 72% from Bombyx mori (BAK82126.1) and 66% from Plutella xylostella (AEI27592.1). Similar to the structures of other lepidopteran ABCC2 proteins (Fig. 1), the proposed structure of HaABCC2 protein includes twelve transmembrane segments and two ATP-binding domains (Fig. 1).
Figure 1. Predicted amino acid sequences of the ABCC2 proteins from a susceptible strain (LF) and a resistant strain (LF60) of H. armigera compared with susceptible strains from B. mori (BmABCC2, GenBank BAK82126.1) and H. virescens (HvABCC2, GenBank ADH16740.1).
Walker A and B sequences, C motifs, and transmembrane (TM) domains were predicted by TMHMM ver.2.0 (http://www.cbs.dtu.dk/services/TMHMM/). Arrows show seven amino acid differences between LF and LF60.
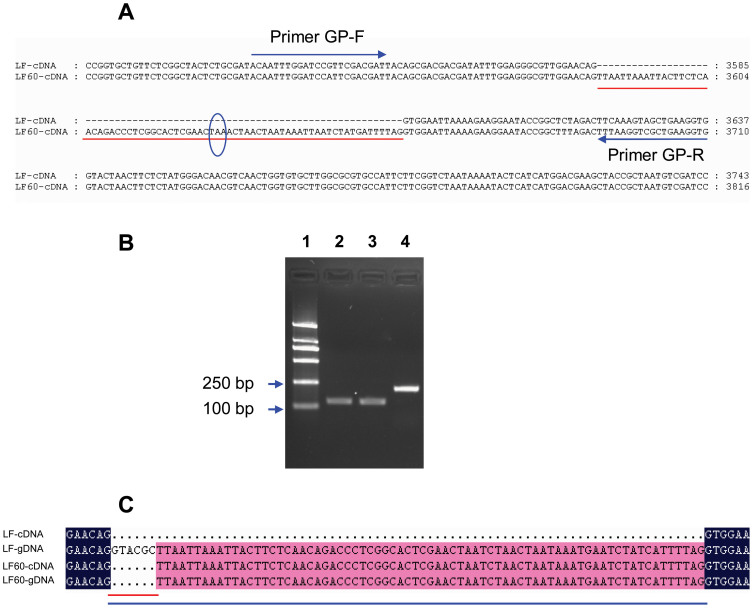
The HaABCC2 cDNA from the resistant strain LF60 has a 73-bp insertion between bp 3582 and 3583 that introduces a premature stop codon (Figs. 1 and 2A). As a result, the amino acid sequence encoded by the resistant allele not only lacks the C-terminal 143 amino acids that occur in the susceptible strain (from 1196 amino acid to the end 1338 amino acid), but also has 15 amino acids that are not encoded by the susceptible allele (after amino acid 1196) (Fig. 1). We also found 124 single nucleotide polymorphisms (SNPs) indicating differences between the cDNA from the resistant and susceptible strains (Fig. S1) of which seven cause amino acid changes (Fig. 1). The relative quantity of transcripts of HaABCC2 did not differ significantly between LF and LF60 (Fig. S2).
Figure 2. Comparison of HaABCC2 cDNA and genomic DNA between susceptible and resistant strains.
(A) Alignment of cDNA sequences from LF and LF60. The red line shows the 73 bp insertion in the resistant strain. Blue arrows show specific primers that distinguish between cDNA from the LF and LF60 strains. The blue oval shows the premature stop codon in LF60. (B) HaABCC2 cDNA bands in the LF, 96S and LF60 strains amplified by primers GP-F and GP-R. Lane 1: DNA2000 markers, Lane 2: LF, Lane 3: 96S, and Lane 4: LF60. As expected, lanes 2 and 3 from the susceptible strains show a band of about 130 bp, while lane 4 from the resistant strain shows a band of about 200 bp, which reflects the 73-bp insertion. (C) Alignment of gDNA and cDNA sequences. The blue line indicates the intron sequence and the red line indicates the deletion in the splicing site in LF60.
We developed an allele-specific RT-PCR gel analysis using specific primers (GF and GR) flanking the 73-bp insertion to distinguish between the transcripts in the susceptible and resistant strains (Fig. 2). As expected, the band amplified was about 70 bp longer in the resistant strain than in the susceptible strain (Fig. 2B).
ABCC2 gDNA in susceptible and resistant strains
Alignment of the gDNA and cDNA sequences of HaABCC2 from the two strains revealed that the 73-bp insertion in the cDNA sequence of the resistant allele is an intron, which was spliced out in the transcript from the susceptible strain but not from the resistant strain (Fig. 2C). Comparison of the gDNA sequences indicates that LF60 lacks the first 6 bp (“GTACGC”) of this intron, including the 5′ “GT” dinucleotide intron signature (Fig. 2C), which leads to failure to splice out this intron. In 100 sequenced gDNA samples from each strain, all samples in LF60 were homozygous for the presence of this 6-bp deletion and all samples from LF lacked this deletion. All F1 progeny from a cross between LF60 and LF were heterozygous for the deletion.
Linkage between HaABCC2 mis-splicing and Cry1Ac resistance
Sequencing of the gDNA fragment flanking the 6-bp deletion of larvae from 10 backcross families tested in bioassays shows tight genetic linkage between HaABCC2 and resistance to Cry1Ac (Table 2). For the 1200 larvae genotyped by sequencing gDNA (120 per backcross family), all 600 survivors on treated diet were homozygous for the 6-bp deletion, but of the 600 larvae on untreated diet, 48% were homozygous for the 6-bp deletion and 52% were heterozygous (Table 2, Fisher's exact test, P < 0.0001 for each family).
Table 2. Genetic linkage between HaABCC2 and resistance to Cry1Ac.
| Larvae with insertion (%) | ||
|---|---|---|
| Backcross family | Control diet | Treated diet |
| A1 | 47 | 100 |
| A2 | 45 | 100 |
| A3 | 53 | 100 |
| A4 | 50 | 100 |
| A5 | 52 | 100 |
| B6 | 52 | 100 |
| B7 | 43 | 100 |
| B8 | 52 | 100 |
| B9 | 40 | 100 |
| B10 | 50 | 100 |
| Mean | 48 | 100 |
Backcross families A1–A5 were generated by crossing a resistant male with an F1 female and families B6–B10 by crossing an F1 male with a resistant female. For each of the 10 backcross families, genotype was determined for 120 larvae (60 fed control diet and 60 fed diet treated with Cry1Ac). Genomic DNA sequencing method was used to determine genotype. The proportion of larvae with the insertion was significantly higher on treated than control diet (Fisher's exact test, P < 0.0001 for each family). On control diet, the proportion of larvae with the insertion did not differ significantly from the expected proportion of 0.5 (P > 0.35 for each family).
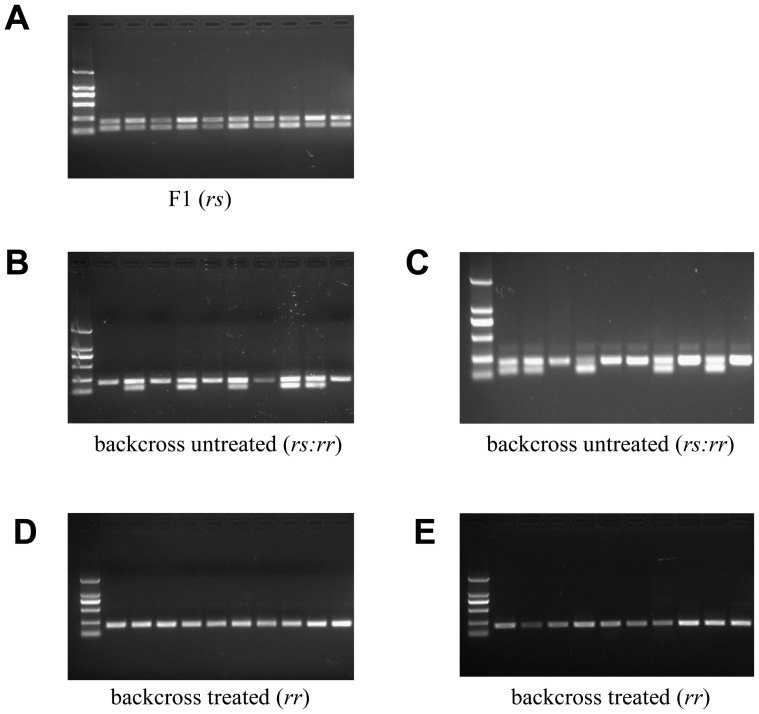
Results from larvae genotyped by RT-PCR with cDNA (Fig. 3) confirm the results of the linkage analysis based on sequencing of gDNA (Table 2). The RT-PCR results show that all of the F1 larvae tested were heterozygous for the 73-bp insertion (Fig. 3A). In backcross families A1 and B6 analyzed by RT-PCR, 20 larvae that survived on treated diet were homozygous for the 73-bp insertion (Fig. 3D and 3E), whereas 20 larvae from untreated treated diet were either heterozygous or homozygous for the insertion in equal proportions (Fig. 3B and 3C).
Figure 3. PCR analysis of HaABCC2 cDNA amplified by allele-specifc primers GP-F and GP-R.
(A) F1 (rs) progeny from a cross between the resistant LF60 strain and the susceptible LF strain. (B and C) Progeny from backcross larvae reared on untreated diet; genotypes are either rs (two bands) or rr (one band). (D and E) Progeny from backcross larvae reared on diet treated with Cry1Ac; all are rr (one band). Backcrosses were done with single-pair crosses either with a resistant male and an F1 female (B and D) or an F1 male and a resistant female (C and E).
Discussion
The results here showing tight genetic linkage between a mutation in the ABCC2 gene of Helicoverpa armigera (HaABCC2) and resistance to Cry1Ac confirm the importance of the ABCC2 protein in toxicity of Cry1Ac against lepidopteran larvae. Previous work revealed linkage between the ABCC2 gene and resistance to Cry1Ab, Cry1Ac, or both in strains of four lepidopteran species19,20,21. In Bombyx mori, a mutation in the gene encoding ABCC2 was associated with reduced binding of Cry1Ab in transformed Sf9 cells25, but not in brush border membrane vesicles isolated from midguts of resistant and susceptible larvae21. It has been hypothesized that for some Bt toxins, binding to ABCC2 proteins is essential for toxicity; thus disruption of this binding causes resistance19,26. However, as far as we know, direct evidence of binding of Bt toxins to ABCC2 proteins has not been reported.
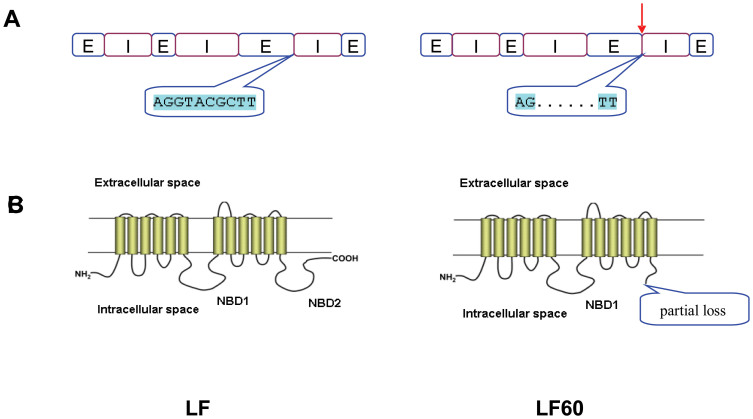
In H. armigera, the association between toxin binding and the ABCC2 mutation remains to be determined. In resistant strain LF60 of H. armigera, the ABCC2 gene is missing six base pairs in one of its intron-exon boundaries, including the conserved 5′ splicing dinucleotide “GT” (Fig. 4A). Because of this mutation, an intron is not spliced out of the ABCC2 transcript, which introduces a stop codon. This premature codon is predicted to cause the loss of 143 amino acids, including a C motif and Walker B sequence, which could affect the protein's conformation (Fig. 4B), potentially interfering with binding of ATP as well as with Bt toxins.
Figure 4. Summary of differences between ABCC2 in a susceptible strain (LF) and a resistant strain (LF60) of H. armigera.
(A) gDNA. E: exon, I: intron, red arrow shows 6-bp deletion in LF60. (B) Schematic structure of HaABCC2 protein21 showing location of predicted loss of amino acids in the ATP-binding domain 2 of LF60.
If the mutation in HaABCC2 does disrupt ATP binding, this might reduce the fitness of resistant insects relative to susceptible insects. However, the lack of a significant deficit of resistant homozygotes in the backcross larvae fed untreated diet (Table 2) indicates that a major fitness cost affecting survival was not detected in our experiments. Major fitness costs affecting survival also were not detected in similar backcross experiments on diet with H. virescens, P. xylostella and T. ni, but this does not exclude the possibility of fitness costs affecting survival or other traits when larvae develop on plants in the field19,20.
Mis-splicing of cadherin genes is associated with resistance to Cry1Ac in Pectinophora gossypiella and H. armigera27,28,29. As far as we know, the results reported here are the first showing that mis-splicing of an ABCC2 gene is associated with resistance to a Bt toxin. Based on the results here and previous reports, the mechanisms of resistance to Cry1Ac in laboratory- and field-selected strains of H. armigera are diverse, including the recessive mutation disrupting ABCC2 found here, recessive and dominant mutations disrupting the extracellular and intracellular domains of cadherin, reduced transcription of a protease that converts protoxin to toxin, reduced transcription of membrane-bound alkaline phosphatase, reduced activity of aminopeptidase N, elevated immune response, and non-recessive mutations in unidentified genes17,23,24,30,31,32,33,34,35,36. This diversity will continue to provide challenges for understanding, monitoring, and managing resistance of H. armigera to Bt cotton.
Methods
Insects
We used three laboratory strains of H. armigera: susceptible strains 96S and LF, and resistant strain LF60. The 96S strain was started with 20 pairs of adults collected from conventional cotton in Xinxiang, Henan Province, China in 199637,38. The LF strain was started with about 100 third to sixth instars collected from Bt cotton in Langfang, Hebei Province, China in 199839,40. Both susceptible strains were reared in the laboratory on artificial diet without exposure to Bt toxins or other insecticides. We generated the resistant strain LF60 by selecting insects from the susceptible LF strain with MVPII (Dow AgroSciences), a commercial formulation of CryAc protoxin incorporated in diet41,42. Selection was conducted for more than a decade with progressively increasing concentrations: 1, 5, 10, 30 and 60 µg Cry1Ac protoxin per g diet. Insects were reared at 27 ± 2°C and 75 ± 10% relative humidity with a photoperiod of 14L:10D.
Bioassays
We used diet incorporation bioassays to evaluate susceptibility to Cry1Ac protoxin and Cry1Ac activated toxin. Cry1Ac protoxin was extracted and purified from the HD73 strain of B. thuringiensis subsp. kurstaki by the Biotechnology Group in Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China. To obtain activated Cry1Ac toxin, Cry1Ac protoxin was incubated 2 h at 37°C with a 25:1 ratio of trypsin (Sigma) to protoxin, and the soluble trypsinized toxin was purified by a Superdex 200 HR 10/30 column (Amersham Biosciences) on a fast protein liquid chromatography (FPLC) system.
Various concentrations of Cry1Ac protoxin and activated toxin were added and thoroughly mixed with diet to obtain the desired concentrations. After mixing, the diet solidified and we put pieces of solid diet (1 g) into each well of a 24-well plate. We put one first instar in each well of a 24-well plate for each replicate, with three replicates per treatment (total n = 72 per treatment). Larvae were considered dead if they died or did not reach third instar after 7 days.
Cloning and sequence analysis of HaABCC2
We analyzed 20 larvae (5th instars, 12 days old), 10 from LF and 10 from LF60. Midguts were dissected under a dissecting microscope, washed with cold 0.7% physiological saline, briefly dried on filter paper, pooled for each strain in a centrifuge tube (1.5 ml), frozen in liquid nitrogen, and stored at −80°C for subsequent RNA extraction.
Total RNA was extracted from the sample homogenates according to the standard TRIzol reagent protocols (Invitrogen). RNA purity was evaluated by 260/280 and 260/230 ratios measured in a NanoDrop 3300 (Thermo). Genomic DNA was eliminated from the samples by treatment with DNaseI (Fermentas). Total RNA (4 μg) was reverse transcribed into cDNA in a 20-μl reaction using SuperScript™ III First-Strand Synthesis kit (Invitrogen) according to the manufacturer's instructions. The resultant cDNA was stored at −20°C for subsequent homology-based cloning of ABCC2 cDNA sequences from the two H. armigera strains, and then quantitative polymerase chain reaction (qPCR) analysis of ABCC2 expression in the two strains.
Cloning of H. armigera ABCC2 (named HaABCC2) cDNA sequence was initiated with PCR amplification of a partial fragment using the cDNA sample from LF as the template and the primers VP -F and VP -R (Table S1) designed based on the nucleotide sequence got from data of RNA sequence experiment. The PCR conditions were 4 min denaturation at 95°C, followed by 35 circles of 30 sec denaturation at 94°C, 30 sec annealing at 60°C and 90 sec extension at 72°C, and a final 10 min extension at 72°C. The amplified partial fragment was eluted from the agarose gel, cloned into the pMD20-T Vector (TaKaRa), and sequenced by Sangon Biotech Company.
Based on the sequence of the partial fragment, four gene-specific primers (5′RACE-R1, 5′RACE-R2, 3′RACE-F1 and 3′RACE-F2 in Table S1) were designed to nest PCR-amplify the 5′ (5′RACR-R1 and 5′RACE-R2) and 3′-ends (3′RACE-F1 and 3′RACE-F2) of HaABCC2 cDNA with four general primers (5′RACE outer primer, 5′RACE inner primer, 3′RACE outer primer and 3′RACE inner primer TaKaRa), using the cDNA sample prepared from LF RNA as the template and the 5′- and 3′-Full RACE Kit (TaKaRa) following manufacturer's instructions. The resultant 5′ and 3′ ends were cloned, sequenced, and aligned with the partial fragment to yield the full-length cDNA of HaABCC2 from LF. We then designed a pair of primers HaABCC2 F and HaABCC2 R based on the above full-length cDNA of HaABCC2 to PCR-amplify the ORF (open reading frame) of HaABCC2 from LF and LF60 respectively. The ORF PCR conditions were 4 min denaturation at 95°C, followed by 35 circles of 30 sec denaturation at 94°C, 30 sec annealing at 55°C and 3 min extension at 72°C, and a final 10 min extension at 72°C.
The programs ClustalX and DNAMAN were used to align the sequences. TMHMM ver.2.0 (http://www.cbs.dtu.dk/services/TMHMM/) was used to predict the HaABCC2 domains.
qPCR analysis of HaABCC2
Three cDNA samples per strain prepared as above from 3 pools of 10 larval midguts each were use as the templates to compare the expression level of HaABCC2 between LF and LF60 by qPCR. Two H. armigera housekeeping genes, beta actin (β-actin) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), were used as the dual reference genes to normalize the expression of HaABCC2. qPCR (TaqMan) of the three genes were individually performed in a 20-μl reaction containing 2X Maxima probe/ROX qPCR Master Mix (10 μl), 10 μmol forward primer and reverse primer (0.6 μl each), 10 μmol probe (0.4 μl), 2 μl LF or LF60 template cDNA, and 6.4 μl nuclease-free water. All qPCR reactions were performed in 96-well optical plates using the Applied Biosystems 7500 FAST qPCR system (ABI7500 Fast) under the following conditions: 50°C, 2 min; 95°C, 10 min; 40 cycles: 95°C, 15 second, 60°C, 1 min. qPCR of each of the three genes were repeated three times per cDNA sample and no-template nuclease-free water control. Data were processed using the 2-ΔΔct method43. All the primers and FAM probes used for the three genes are shown in Table S1.
Allele-specific RT-PCR
Based on a 73-bp insertion in the ORF of HaABCC2 LF60 allele relative to HaABCC2 LF allele revealed by sequence alignment (Fig. 2A), we developed an allele-specific PCR to genotypy individuals of the susceptible (LF, 96S) and resistant (LF60) strains as well as of genetic crossings (see the linkage analysis section below). A pair of primers named GF and GR (Genotyping F and Genotyping R) (Table S1) was designed to PCR-amplify the cDNA flanking this 73-bp insertion from individual larvae. The allele-specific PCR conditions were 4 min denaturation at 95°C, followed by 35 circles of 30 sec denaturation at 94°C, 30 sec annealing at 60°C and 50 sec extension at 72°C, and a final 10 min extension at 72°C. The resultant PCR band from the susceptible individuals of 96S and LF strains was about 130 bp, but the PCR band from the resistant individuals of LF60 was about 200 bp (Fig. 2B).
Cloning and sequencing of a HaABCC2 gDNA fragment
Genomic DNA (gDNA) was extracted from individual larvae from LF, LF60 and their F1 offspring using the DNAzol extraction kit of TIANGEN Company. HaABCC2 gDNA fragments flanking the indel were PCR-amplified from the three gDNA samples, respectively, using the primers Genotyping F (GF)and Genotyping (GR) R (Table S1). The PCR conditions were the same as the allele-specific PCR described above except the cDNA templates were replaced with the gDNA samples. The resultant PCR products from the gDNA samples were sequenced directly. The gDNA sequences obtained were aligned to locate the gDNA mutations responsible for the 73-bp insertion in cDNA.
Genetic linkage between HaABCC2 and resistance to Cry1Ac
To test for genetic linkage between HaABCC2 and Cry1Ac resistance, we generated F1 progeny from a single-pair cross between a male from the susceptible LF strain and a female from the resistant strain. We generated 10 backcross families of two different types: backcross families A1-A5 were each produced by a single-pair cross of a female F1 with a male from the resistant strain, and families B6-B10 were each produced by a single-pair cross of a male F1 with a female from the resistant strain. From each of the 10 backcross families, 120 larvae were tested using the bioassay method described above; 60 from each family on diet that had no toxin (control) and 60 from each family on diet treated with 2 µg Cry1Ac per ml activated toxin to kill susceptible larvae.
We used the allele-specific PCR described above to determine the genotype of larvae from F1 and backcross families. We also sequenced the HaABCC2 gDNA fragment flanking the 73-bp insertion.
A total of 40 larvae from families A1 and B6 were analyzed with PCR; 10 from each family fed diet without toxin (control) and 10 from each family fed diet treated with 2 µg Cry1Ac activated toxin per ml diet to kill susceptible larvae. Ten F1 larvae were also analyzed with PCR.
Statistical analysis
Data processing system (DPS:a software package on analysis of statistical data. Zhejiang University. China) was used to get LC50s44. For each of ten backcross families, we used a separate Fisher's exact test (http://graphpad.com/quickcalcs/contingency1.cfm) to determine if the proportion of genotypes (rs and rr) differed significantly between larvae reared on treated versus untreated diet and to determine if the proportion of the genotypes (rs and rr) for larvae fed untreated diet differed significantly from the expected proportion of 0.5 for each genotype.
Author Contributions
K.W. and Y.X. designed the study. Y.X., T.Z. and C.L. performed the experiments. K.W., Y.X. and B.E.T. analyzed the data. K.W., Y.X., B.E.T., D.G.H. and X.L. wrote the manuscript. All authors have read and approved the manuscript for publication.
Supplementary Material
Supporting Information
Acknowledgments
We thank Juan Luis Jurat-Fuentes,Wanna Zhang and Gemei Liang for their invaluable assistance with the experiments, Jie Zhang for kindly providing Cry1Ac protoxin and Cuirong Zhang for rearing insects. This work was supported by the National Natural Science Funds (No. 31321004), the Key Project for Breeding Genetic Modified Organisms (2014ZX0812-004), the Max-Planck-Gesellschaft and U. S. Department of Agriculture Biotechnology Risk Assessment Grant 2011-33522-30729.
References
- Sanahuja G., Banakar R., Twyman R. M., Capell T. & Christou P. Bacillus thuringiensis: a century of research, development and commercial applications. Plant Biotechnol J 9, 283–300 (2011). [DOI] [PubMed] [Google Scholar]
- Pardo-Lopez L., Soberon M. & Bravo A. Bacillus thuringiensis insecticidal three-domain Cry toxins: mode of action, insect resistance and consequences for crop protection. FEMS Microbiol Rev 37, 3–22 (2013). [DOI] [PubMed] [Google Scholar]
- Comas C., Lumbierres B., Pons X. & Albajes R. No effects of Bacillus thuringiensis maize on nontarget organisms in the field in southern Europe: a meta-analysis of 26 arthropod taxa. Transgenic Res 23, 135–143 (2014). [DOI] [PubMed] [Google Scholar]
- James C. Global status of commercialized biotech/GM Crops. ISAAA Briefs 46, (ISAAA, Ithaca, NY, 2013) (2013). [Google Scholar]
- Carpenter J. E. Peer-reviewed surveys indicate positive impact of commercialized GM crops. Nat Biotechnol 28, 319–321 (2010). [DOI] [PubMed] [Google Scholar]
- Hutchison W. D. et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science 330, 222–225 (2010). [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E. et al. Suppressing resistance to Bt cotton with sterile insect releases. Nat Biotechnol 28, 1304–1307 (2010). [DOI] [PubMed] [Google Scholar]
- Edgerton M. D. et al. Transgenic insect resistance traits increase corn yield and yield stability. Nat Biotechnol 30, 493–496 (2010). [DOI] [PubMed] [Google Scholar]
- Kathage J. & Qaim M. Economic impacts and impact dynamics of Bt (Bacillus thuringiensis) cotton in India. Proc Natl Acad Sci U S A 109, 11652–11656 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Wu K., Jiang Y., Guo Y. & Desneux N. Widespread adoption of Bt cotton and insecticide decrease promotes biocontrol services. Nature 487, 362–365 (2012). [DOI] [PubMed] [Google Scholar]
- Bagla P. India. Hardy cotton-munching pests are latest blow to GM crops. Science 327, 1439 (2010). [DOI] [PubMed] [Google Scholar]
- Storer N. P., Kubiszak M. E., Ed King J., Thompson G. D. & Santos A. C. Status of resistance to Bt maize in Spodoptera frugiperda: lessons from Puerto Rico. J Invertebr Pathol 110, 294–300 (2012). [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E., Brevault T. & Carriere Y. Insect resistance to Bt crops: lessons from the first billion acres. Nat Biotechnol 31, 510–521 (2013). [DOI] [PubMed] [Google Scholar]
- Gassmann A. J. et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. Proc Natl Acad Sci U S A 111, 5141–5146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre J. & Van Rie J. Biochemistry and genetics of insect resistance to Bacillus thuringiensis. Annu Rev Entomol 47, 501–533 (2002). [DOI] [PubMed] [Google Scholar]
- Heckel D. G. et al. The diversity of Bt resistance genes in species of Lepidoptera. J Invertebr Pathol 95, 192–197 (2007). [DOI] [PubMed] [Google Scholar]
- Jurat-Fuentes J. L. et al. Reduced levels of membrane-bound alkaline phosphatase are common to lepidopteran strains resistant to Cry toxins from Bacillus thuringiensis. PLoS One 6, e17606 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabashnik B. E. et al. Efficacy of genetically modified Bt toxins against insects with different genetic mechanisms of resistance. Nat Biotechnol 29, 1128–U1198 (2011). [DOI] [PubMed] [Google Scholar]
- Gahan L. J., Pauchet Y., Vogel H. & Heckel D. G. An ABC Transporter Mutation Is Correlated with Insect Resistance to Bacillus thuringiensis Cry1Ac Toxin. Plos Genetics 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter S. W. et al. Parallel Evolution of Bacillus thuringiensis Toxin Resistance in Lepidoptera. Genetics 189, 675–U814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atsumi S. et al. Single amino acid mutation in an ATP-binding cassette transporter gene causes resistance to Bt toxin Cry1Ab in the silkworm, Bombyx mori. Proc Natl Acad Sci U S A 109, E1591–1598 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Early warning of cotton bollworm resistance associated with intensive planting of Bt cotton in China. PLoS One 6, e22874 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. et al. Diverse genetic basis of field-evolved resistance to Bt cotton in cotton bollworm from China. Proc Natl Acad Sci U S A 109, 10275–10280 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L. et al. Dominant resistance to Bt cotton and minor cross-resistance to Bt toxin Cry2Ab in cotton bollworm from China. Evol Appl 6, 1222–1235 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S. et al. The ATP-binding cassette transporter subfamily C member 2 in Bombyx mori larvae is a functional receptor for Cry toxins from Bacillus thuringiensis. FEBS J 280, 1782–1794 (2013). [DOI] [PubMed] [Google Scholar]
- Heckel D. G. Learning the ABCs of Bt: ABC transporters and insect resistance to Bacillus thuringiensis provide clues to a crucial step in toxin mode of action. Pestic Biochem Physiol 104, 103–110 (2012). [Google Scholar]
- Fabrick J. A., Mathew L. G., Tabashnik B. E. & Li X. Insertion of an intact CR1 retrotransposon in a cadherin gene linked with Bt resistance in the pink bollworm, Pectinophora gossypiella. Insect Mol Biol 20, 651–665 (2011). [DOI] [PubMed] [Google Scholar]
- Zhang H., Tang M., Yang F., Yang Y. & Wu Y. DNA-based screening for an intracellular cadherin mutation conferring non-recessive Cry1Ac resistance in field populations of Helicoverpa armigera. Pestic Biochem Physiol 107, 148–152 (2013). [DOI] [PubMed] [Google Scholar]
- Fabrick J. A. et al. Alternative splicing and highly variable cadherin transcripts associated with field-evolved resistance of pink bollworm to bt cotton in India. PLoS One 9, e97900 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Wu S., Yang Y., Tabashnik B. E. & Wu Y. Non-recessive Bt toxin resistance conferred by an intracellular cadherin mutation in field-selected populations of cotton bollworm. PLoS One 7, e53418 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yu L. & Wu Y. Disruption of a cadherin gene associated with resistance to Cry1Ac {delta}-endotoxin of Bacillus thuringiensis in Helicoverpa armigera. Appl Environ Microbiol 71, 948–954 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caccia S. et al. Binding Site Alteration Is Responsible for Field-Isolated Resistance to Bacillus thuringiensis Cry2A Insecticidal Proteins in Two Helicoverpa Species. Plos One 5 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma G. et al. Is the mature endotoxin Cry1Ac from Bacillus thuringiensis inactivated by a coagulation reaction in the gut lumen of resistant, Helicoverpa armigera larvae? Insect Biochem Mol Biol 35, 729–739 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang S. et al. Mutation of an aminopeptidase N gene is associated with Helicoverpa armigera resistance to Bacillus thuringiensis Cry1Ac toxin. Insect Biochem Mol Biol 39, 421–429 (2009). [DOI] [PubMed] [Google Scholar]
- Gunning R. V., Dang H. T., Kemp F. C., Nicholson I. C. & Moores G. D. New resistance mechanism in Helicoverpa armigera threatens transgenic crops expressing Bacillus thuringiensis Cry1Ac toxin. Appl Enviro Microbiol 71, 2558–2563 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal R. et al. Resistance of Helicoverpa armigera to Cry1Ac toxin from Bacillus thuringiensis is due to improper processing of the protoxin. Biochem J 419, 309–316 (2009). [DOI] [PubMed] [Google Scholar]
- Liang G. M. et al. Changes of inheritance mode and fitness in Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae) along with its resistance evolution to Cry1Ac toxin. J Invertebr Pathol 97, 142–149 (2008). [DOI] [PubMed] [Google Scholar]
- Liang G. M., Tan W. J. & Guo Y. Y. An improvement in the technique of artificial rearing cotton bollworm. Plant Protec 25, 15–17 (1999). [Google Scholar]
- Wu K. & Guo Y. Changes in susceptibility to conventional insecticides of a Cry1Ac-selected population of Helicoverpa armigera (Hubner) (Lepidoptera: Noctuidae). Pest Manag Sci 60, 680–684 (2004). [DOI] [PubMed] [Google Scholar]
- Cao G., Zhang L., Liang G., Li X. & Wu K. Involvement of nonbinding site proteinases in the development of resistance of Helicoverpa armigera (Lepidoptera: Noctuidae) to Cry1Ac. J Econ Entomol 106, 2514–2521 (2013). [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E. et al. Inheritance of resistance to Bt toxin crylac in a field-derived strain of pink bollworm (Lepidoptera: Gelechiidae). J Econ Entomol 95, 1018–1026 (2002). [DOI] [PubMed] [Google Scholar]
- Tabashnik B. E. et al. Control of resistant pink bollworm (Pectinophora gossypiella) by transgenic cotton that produces Bacillus thuringiensis toxin Cry2Ab. Appl Environ Microbiol 68, 3790–3794 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Tang Q. Y. & Zhang C. X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci 20, 254–260 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information