Abstract
p53 Inducible gene 6 (PIG6) encodes mitochondrial proline dehydrogenase (PRODH) and is up-regulated several fold upon p53 activation. Proline dehydrogenase is proposed to generate radicals that contribute to cancer cell apoptosis. However, there are at least 10 mitochondrial sites that can produce superoxide and/or H2O2, and it is unclear whether proline dehydrogenase generates these species directly, or instead drives production by other sites. Amongst six cancer cell lines, ZR75-30 human breast cancer cells had the highest basal proline dehydrogenase levels, and mitochondria isolated from ZR75-30 cells consumed oxygen and produced H2O2 with proline as sole substrate. Insects use proline oxidation to fuel flight, and mitochondria isolated from Drosophila melanogaster were even more active with proline as sole substrate than ZR75-30 mitochondria. Using mitochondria from these two models we identified the sites involved in formation of superoxide/H2O2 during proline oxidation. In mitochondria from Drosophila the main sites were respiratory complexes I and II. In mitochondria from ZR75-30 breast cancer cells the main sites were complex I and the oxoglutarate dehydrogenase complex. Even with combinations of substrates and respiratory chain inhibitors designed to minimize the contributions of other sites and maximize any superoxide/H2O2 production from proline dehydrogenase itself, there was no significant direct contribution of proline dehydrogenase to the observed H2O2 production. Thus proline oxidation by proline dehydrogenase drives superoxide/H2O2 production, but it does so mainly or exclusively by providing anaplerotic carbon for other mitochondrial dehydrogenases and not by producing superoxide/H2O2 directly.
Keywords: Proline dehydrogenase (PRODH), Cancer cell mitochondria, Drosophila, Electron transport chain, Reactive oxygen species, Superoxide, Hydrogen peroxide
Abbreviations: PIG6, proline dehydrogenase inducible gene 6; PRODH, proline dehydrogenase; GDH, glutamate dehydrogenase; OGDH, 2-oxoglutarate dehydrogenase complex; SCS, succinyl-CoA synthase; AT, aminotransferase; ROS, reactive oxygen species; P5C, Δ1-pyrroline-5-carboxylate; Oxa, oxaloacetate; Asp, asparate; TCA, tricarboxylic acid; A5, atpenin A5; GSA, glutamic semi-aldehyde; oAB, o-aminobenzaldehyde; IF, flavin of complex I; IIF, flavin of complex II; IIIQo, quinone binding site on the outer/cytosolic face of complex III; OF, Flavin of the oxoglutarate dehydrogenase complex
Graphical abstract
Highlights
-
•
Proline dehydrogenase is thought to produce reactive oxygen species (ROS) in cancer cells and to promote apoptosis.
-
•
Isolated mitochondria from Drosophila melanogaster and from a human breast cancer cell line oxidize proline producing superoxide/H2O2 at measurable rates.
-
•
Proline oxidation drives superoxide/H2O2 production indirectly at other sites and it is unlikely that proline dehydrogenase produces superoxide/H2O2 itself.
-
•
In Drosophila, superoxide/H2O2 arises from sites IF and IIF (the flavin sites from complexes I and II, respectively).
-
•
In the breast cancer cell line the main sites are IF and OF (from the oxoglutarate dehydrogenase complex).
Introduction
A hallmark of cancer cells is their ability to overcome signaling pathways that trigger cell death [1]. Programmed cell death and cell cycle arrest are partly controlled by p53 expression [2,3], therefore p53 in tumor cells is either mutated or sequestered in the cytoplasm by its negative modulator MDM2 [4]. Apoptosis induced by p53 activation is preceded by the expression of PIGs (p53 induced genes) [5]. One of the most prominent is PIG6, which encodes the mitochondrial enzyme proline dehydrogenase (PRODH) [5]. In most tumor cells proline dehydrogenase levels are low [6,7]. Proline dehydrogenase overexpression in certain cancer cell lines is associated with apoptosis through mechanisms involving proline-dependent mitochondrial production of reactive oxygen species (ROS) [6,8–10]. Since proline dehydrogenase overexpression is also associated with reduced tumor formation in vivo, it has been suggested as a direct mediator of p53-induced tumor suppression [7]. Paradoxically, some cancer cells may use proline oxidation to drive proliferation and protection against stress, and inhibition of proline dehydrogenase in such proline-addicted cells may be therapeutically useful [7,11–13].
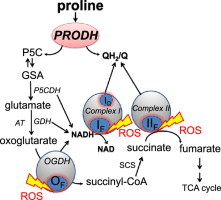
Cancer cells utilize a variety of metabolic pathways to support the high energy demands of proliferation and biosynthesis [14]. Recently, proline catabolism has gained attention as one such pathway, particularly under nutrient stress [11,12,15]. Proline and hydroxyproline comprise ~20% of the amino acids in collagen [16], the main constituent of the extracellular matrix. Collagen may be degraded to provide amino acids and energy for growing and proliferating cells. In eukaryotes, proline is catabolized to glutamate in the mitochondrial matrix through proline dehydrogenase and Δ1-pyrroline-5-carboxylate (P5C) dehydrogenase (Fig. 1a). Proline dehydrogenase oxidizes proline to P5C and reduces flavin adenine dinucleotide (FAD) to FADH2. In vitro, FAD is regenerated using artificial electron acceptors [17]. The natural electron acceptor is unknown but may be ubiquinone in the mitochondrial respiratory chain [18]. P5C dehydrogenase then oxidizes P5C to glutamate and reduces NAD+ to NADH+H+. The glutamate is metabolized to 2-oxoglutarate, succinate and other tricarboxylic acid intermediates; oxoglutarate and succinate levels have been shown to change upon proline dehydrogenase expression [7]. Thus proline oxidation feeds electrons into the electron transport chain at multiple sites: (1) at ubiquinone from succinate dehydrogenase and probably proline dehydrogenase, and (2) at NAD from the dehydrogenases for P5C, 2-oxoglutarate and glutamate (Fig. 1a). The two enzymes involved in the oxidation of proline to glutamate are shared amongst different organisms. Therefore, proline catabolic pathway was evolutionarily conserved between microorganisms, insects and mammals [11,17,18].
Fig. 1.
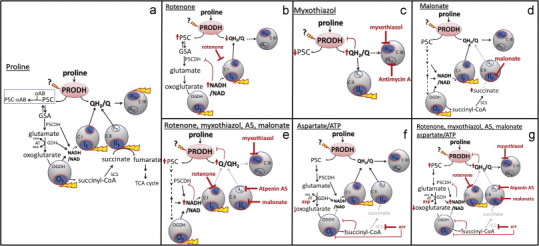
Metabolic pathways and sites of superoxide/H2O2 production during mitochondrial proline oxidation. Electron transport chain complexes I, II, and III and the 2-oxoglutarate dehydrogenase complex (OGDH) are shown as gray circles; sites that can generate superoxide and/or H2O2 as blue ovals; sites that may produce superoxide/H2O2 in particular conditions by yellow tags; sites of inhibition by red blunted arrows. (a) Control conditions. Proline dehydrogenase (PRODH) oxidizes proline to Δ1-pyrroline-5-carboxylate (P5C), directly reducing ubiquinone (Q) to ubiquinol (QH2) and indirectly reducing NAD to NADH through the dehydrogenases of P5C (P5CDH), glutamate (GDH) and 2-oxoglutarate (OGDH). P5C spontaneously forms glutamate semi-aldehyde (GSA) and its formation can be monitored colorimetrically with o-aminobenzaldehyde (oAB) (dotted rectangle). Sites active in the presence of: (b) rotenone to inhibit IQ; (c) myxothiazol or antimycin A to inhibit complex III; (d) malonate to inhibit IIF; (e) rotenone, myxothiazol, malonate and A5, leaving only site IF (and OF) active to produce ROS; (f) aspartate and ATP to inhibit OGDH [22]; (g) rotenone, myxothiazol, malonate, A5, aspartate and ATP to inhibit superoxide/H2O2 production from IQ, IIIQo, IIF and (partially) OF, leaving only IF and proline dehydrogenase active to produce ROS. Here we examine whether proline dehydrogenase can generate superoxide/H2O2, as indicated by “?”. Oxa, oxaloacetate; asp, asparate; AT, aminotransferase; SCS, succinyl-CoA synthase; TCA, tricarboxylic acid; A5, atpenin A5.
Overexpression of proline dehydrogenase in different cancers increases ROS production and apoptosis [8,9,19]. In vitro, proline dehydrogenases from different microorganisms can reduce oxygen directly, but 16–2500 times more slowly than artificial electron acceptors [17,20]. There are at least ten sites linked to mitochondrial electron transport that can also produce superoxide and/or H2O2 [21–23], and it is unclear whether proline dehydrogenase generates these species directly, or indirectly by providing substrate to other sites. The mitochondrial sites with the greatest maximum capacities for superoxide/H2O2 production are the outer quinone-binding site of respiratory complex III (site IIIQo), the flavin of complex II (site IIF), the quinone-binding site of complex I (site IQ) and the E3 flavin of the 2-oxoglutarate dehydrogenase (site OF), pyruvate dehydrogenase (site PF) and branched-chain 2-oxoacid dehydrogenase complexes (site BF) [22–24].
Here, we use mitochondria isolated from Drosophila and the breast cancer cell line ZR75-30 to identify the sites involved in superoxide/H2O2 production during proline oxidation. We show that sites IF, IIF and OF dominate the observed H2O2 production. Proline dehydrogenase itself may not generate superoxide/H2O2, but proline oxidation does so indirectly from other sites.
Materials and methods
Flies
Drosophila melanogaster strain w1118 were raised on standard lab food [25]. 10–15 Day old adult males were used.
Cells
All cells were from ATCC. ZR75-30, SJSA-1 and DU4475 cells were cultured in RPMI, MCF7 and RKO cells in DMEM, and PC3 cells in DMEM/F-12+GlutaMAX. All media were supplemented with 10% (v/v) fetal bovine serum (Gibco).
Western blotting
Cells were harvested at ~70% confluence, washed with ice-cold PBS, lysed, and sonicated. Protein (15 µg, determined using BCA (Thermo Scientific)) was separated on 4–12% NUPAGE gels using 1× MES buffer (Invitrogen). Anti-PRODH 68 kDa (Santa Cruz: 376,401) was used at 1:1000 dilution. Anti β-actin (Santa Cruz: 47,778) was used at 1:1000 dilution. Chemiluminescence was normalized to β-actin.
Mitochondria
~200 Flies were homogenized and mitochondria were isolated [26]. The final mitochondrial pellet was resuspended in KHE (120 mM KCl, 3 mM HEPES, 5 mM KH2PO4, pH 7.2) and kept on ice. Protein was determined by Bradford assay (Biorad).
ZR75-30 cells were harvested from 2 to 4 500 cm2 dishes at ~90% confluence, washed three times in ice-cold STE (250 mM sucrose, 5 mM Tris–HCl, 2 mM EGTA, pH 7.4), scraped off, suspended in 50 ml STE and centrifuged for 10 min at 500g. The pellet was resuspended in STE supplemented with 1% (w/v) fatty acid-free bovine serum albumin and homogenized (up to 20 strokes). The suspension was brought to 50 ml and centrifuged for 5 min at 1000g. The supernatant was centrifuged for 10 min at 11,000g. The pellet was resuspended in STE and centrifuged for 10 min at 10,000g. The mitochondrial pellet was resuspended in STE and kept on ice. Protein was determined by Bradford assay (Biorad).
Respiration
Oxygen consumption was measured at 37 °C for ZR75-30 mitochondria and 25 °C for Drosophila mitochondria using an XF24 extracellular flux analyzer (Seahorse Bioscience) [27]. 10 µg Mitochondrial protein in 20 µl was added to each well and centrifuged for 15 min at 2000g. Volume was brought to 500 µl with KHE containing 5 mM K2HPO4, 2 mM MgCl2, 0.3% (w/v) fatty acid-free bovine serum albumin (KHEPMB) and 1 mM ADP. Proline was added from port A and the “phosphorylating” rate of oxygen consumption was determined.
Proline oxidation to P5C
Mitochondria (0.1 mg protein/ml) were incubated for 10 min in KHE containing 1 mM ADP, 5 µM carbonylcyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP), 2.5 µg/ml oligomycin and 5 mM o-aminobenzaldehyde (oAB). 5 mM Proline was added ±4 µM rotenone or 2 µM myxothiazol. At 30 min reaction was stopped by adding 0.5% (v/v) trichloroacetic acid. pH was brought to 7.1 using KOH. Tubes were centrifuged for 10 min at 10,000g and the supernatants were transferred to new tubes containing an extra 5 mM oAB to fully develop the assay [28]. A440 of the P5C–oAB complex was measured and the concentration was calculated using ϵM=2580. A440 without mitochondria or proline was ~14% of the standard value.
Superoxide/H2O2
H2O2 was detected using 5 U/ml horseradish peroxidase to catalyze the oxidation of 50 µM Amplex UltraRed (Molecular Probes) by H2O2 to its fluorescent product. Exogenous superoxide dismutase (25 U/ml) converted any superoxide to H2O2. Fly mitochondria (0.075 mg mitochondrial protein/ml) were suspended in KHE at 25 °C with 1 mM ADP, 2.5 µg/ml oligomycin, 5 µM FCCP, and the detection system. Fluorescence was monitored using a microplate reader (Pherastar, BMG Labtech) at λexcitation=540 nm, λemission=590 nm. ZR75-30 mitochondria (0.15 mg protein/ml) were resuspended in KHEPMB with 1 mM ADP plus 2.5 µg/ml oligomycin. Fluorescence was recorded using a Varian, Cary Eclipse spectrofluorimeter (λexcitation=560 nm, λemission=590 nm) with constant stirring at 37 °C. Fluorescence was calibrated using H2O2 standards under the exact conditions of the assay.
NAD(P)H redox state
NAD(P)H was measured using 0.3 mg fly mitochondrial protein/ml at 25 °C in KHE plus 1 mM ADP, 2.5 µg/ml oligomycin and 5 µM FCCP, or 0.3 mg ZR75-30 mitochondrial protein/ml at 37 °C in KHEPMB plus 1 mM ADP and 2.5 µg/ml oligomycin. The reduction state of endogenous NAD(P)H was determined by autofluorescence using the Pherastar microplate reader at λexcitation=340 nm, λemission=460 nm. NAD(P)H was assumed to be 0% reduced after 5 min without added substrate, then rotenone was added. With Drosophila mitochondria, rotenone increased the reduction of NAD(P)H to ~80% due to endogenous substrates. Addition of proline further increased NAD(P)H reduction to 98%. The 100% reduced level was established with saturating substrate (e.g. 5 mM malate, glutamate and proline) and 4 µM rotenone. Intermediate values were determined as % reduced NAD(P)H relative to the 0% and 100% values.
Cytochrome b566 reduction state
The reduction state of endogenous cytochrome b566 was measured using 0.75 mg mitochondrial protein/ml in KHE plus 2.5 µg/ml oligomycin and 1 mM ADP with constant stirring at 25 °C in an Olis DW 2-dual wavelength spectrophotometer at A566−A575 nanometers [29,30]. Cytochrome b566 was assumed to be 0% reduced after 5 min without added substrate, then 4 µM rotenone, 5 mM malonate and 2 µM antimycin A were added to report reduction caused by endogenous substrates. 5 mM Proline was subsequently added and %cytochrome b566 reduction was determined relative to the 0% and 100% values. Cytochrome b566 was assumed to be 100% reduced after addition of 5 mM succinate plus 5 mM glycerolphosphate.
Curve fitting
Data in Figs. 3e and 4d were fitted by nonlinear regression to an exponential, yielding the parameter values in Eq. (1) for Drosophila mitochondria
| (1) |
Fig. 3.
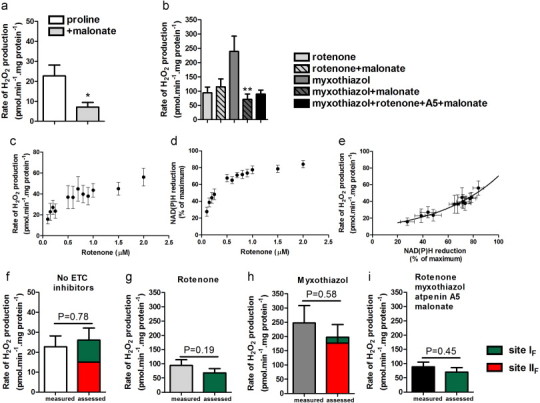
Superoxide/H2O2 production at 25 °C during proline oxidation by Drosophila mitochondria. (a,b) H2O2 production with proline plus different inhibitors. (c) Dependence of superoxide/H2O2 production driven by endogenous substrates on rotenone concentration. (d) Dependence of %NAD(P)H reduction on rotenone concentration. (e) Relationship between the rate of superoxide production from site IF and %NAD(P)H reduction, replotted from (c) and (d). (f–i) Measured and predicted rates of superoxide/H2O2 production during oxidation of proline with inhibitors as shown. Left-hand bars represent the measured rates from panels (a) and (b). Colored bars represent predicted rates for site IF (green) and site IIF (red). *p<0.05; **p<0.01 by unpaired one-tailed t-test (by Welch’s t-test assuming unequal variances in f–i). Additions where shown were 5 mM proline, 5 mM malonate, 4 µM rotenone, 2 µM myxothiazol, 1 µM atpenin A5.
Fig. 4.
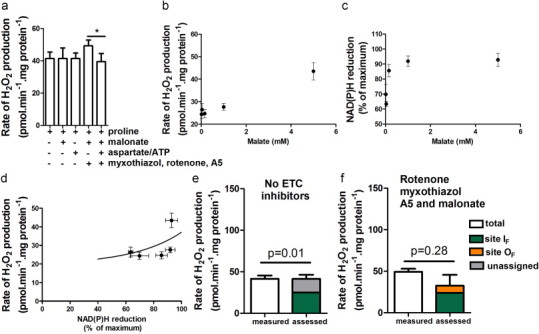
Sites involved in superoxide/H2O2 production at 37 °C during proline oxidation by ZR75-30 mitochondria. (a) H2O2 production. (b–d) Relationship between the rate of superoxide production by site IF and the reduction state of NAD(P)H in the presence of different concentrations of malate plus rotenone, ATP and aspartate. (b) Dependence of H2O2 production rate on malate concentration. (c) Dependence of %NAD(P)H reduction on malate concentration. (d) Relationship between the rate of superoxide production from site IF and %NAD(P)H reduction, replotted from (b) and (c). (e,f) Measured and predicted rates of superoxide/H2O2 production in the presence of proline (e) in the absence of electron transport chain (ETC) inhibitors and (f) in the presence of inhibitors as shown. White bars represent rates from (a). Colored bars represent predicted rates for sites IF (green) and OF (orange) and unassigned rates (gray). *p<0.05 by paired one-tailed t-test (by Welch’s t-test assuming unequal variances in (e), and (f). In (e) p=0.01 when comparing the measured rate to the rate predicted from site IF (green). Additions where indicated were 5 mM proline, 5 mM malonate, 2.5 mM ATP, 1.5 mM aspartate, 2 µM myxothiazol, 4 µM rotenone, and1 µM atpenin A5.
and Eq. 2 for ZR75-30 mitochondria:
| (2) |
where rate is in pmol H2O2 min−1 mg protein−1. S.E.M. values for predicted rates of H2O2 production in Figs 3f–i and 4e,f were calculated using error propagation [31].
Statistics
All data are means±S.E.M (n≥3). Differences between two conditions were analyzed by Student’s t-test or (where the variance was assumed to be unequal, when comparing measured and predicted values) Welch’s t-test [31]. One-way ANOVA with Dunnett’s post-hoc test was used to compare multiple conditions. p<0.05 was considered significant (Prism).
Results
Tumor cells generally have very low endogenous levels of proline dehydrogenase [6,32], complicating evaluation of its direct production of superoxide/H2O2. Basal levels of proline dehydrogenase in different tumor cell lines were analyzed by Western blot. Levels in five lines were low relative to the breast cancer cell line ZR75-30 (Fig. 2a). Mitochondria isolated from ZR75-30 cells had measurable proline-dependent respiration in the presence of ADP (Fig. 2b, gray bar) and measurable proline-dependent H2O2 production when several other sites of superoxide/H2O2 production were inhibited (Fig. 2c, gray bar). However, to investigate superoxide/H2O2 production from proline dehydrogenase itself we first examined insect flight muscle, which is rich in proline dehydrogenase because proline is a major fuel used for flight [33]. Mitochondria prepared from the fruit fly Drosophila melanogaster come mostly from flight muscle, and their H2O2 production has been previously characterized [26]. Their proline-dependent respiration rates at 25 °C were 4-fold higher than those of mitochondria from ZR75-30 cells at 37 °C (Fig. 2b, white bar) and their proline-dependent H2O2 production rates were more than double (Fig. 2c, white bar). The rate differences would be even more marked if temperature corrected. Therefore, we first characterized proline-dependent H2O2 production in Drosophila mitochondria before doing so in mitochondria isolated from ZR75-30 cells.
Fig. 2.
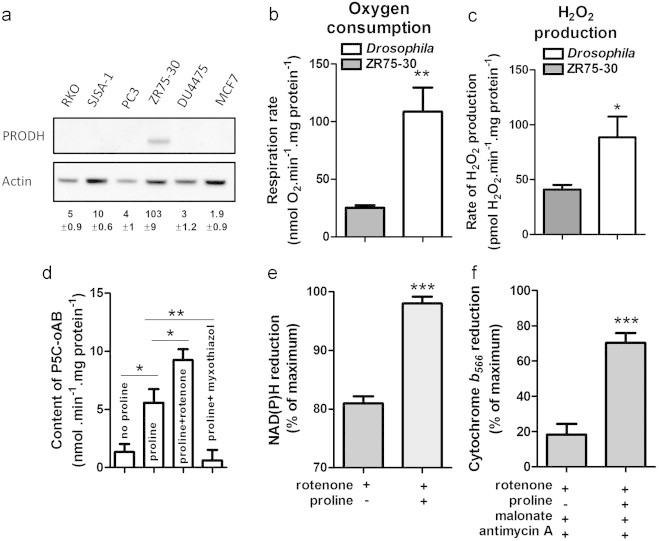
Proline dehydrogenase levels in cancer cells and effects of proline oxidation in mitochondria isolated from ZR75-30 cells and Drosophila. (a) Western blot analysis of proline dehydrogenase (PRODH) levels in different cancer cell lines. Numbers are PRODH intensities normalized to actin. (b) ADP-stimulated oxygen consumption by ZR75-30 and Drosophila mitochondria oxidizing proline. (c) H2O2 production by ZR75-30 and Drosophila mitochondria oxidizing proline in the presence of rotenone, myxothiazol, atpenin A5 and malonate. (d) Δ1-pyrroline-5-carboxylate (P5C) in Drosophila mitochondria after oxidation of proline for 20 min. (e) NAD(P)H and (f) cytochrome b566 reduction state in Drosophila mitochondria. *p<0.05; **p<0.01; ***p<0.001 by unpaired t-test (one-way ANOVA with Dunnett’s post-test in d). Additions where indicated were 5 mM proline, 4 µM rotenone, 5 mM malonate, 2 µM antimycin A, 2 µM myxothiazol, and 1 µM atpenin A5.
Proline dehydrogenase is a ubiquinone reductase
The crystal structures and kinetics of proline dehydrogenase from different microorganisms are well characterized [17,34–36]. Less is known about proline dehydrogenases from eukaryotes, particularly humans [37]. Proline oxidation by proline dehydrogenase can be monitored when the product P5C forms a colored adduct with o-aminobenzaldehyde (oAB) (Fig. 1a) [38]. P5C–oAB formation, in the presence of different inhibitors of the electron transport chain, was used to define the endogenous electron acceptor used by proline dehydrogenase (Figs. 1 and 2d). Drosophila mitochondria were incubated with 1 mM ADP (allosterically stimulates proline dehydrogenase [39]), 2.5 µg/ml Oligomycin (inhibits ADP consumption) and 5 µM FCCP (promotes rapid substrate utilization). In this condition (Fig. 1a), proline induced formation of P5C (Fig. 2d, proline). In the absence of exogenous proline P5C accumulation was indistinguishable from zero (Fig. 2d, no proline); therefore endogenous proline levels are low.
NADH is formed during proline oxidation (Fig. 1a). In the presence of rotenone to block NADH reoxidation (Fig. 1b) proline addition increased the reduction level of NADH (Fig. 2e). High NADH/NAD inhibits NAD-linked dehydrogenases. If proline dehydrogenase uses NAD as its electron acceptor, rotenone should slow formation of P5C–oAB, whereas if P5C dehydrogenase uses NAD as electron acceptor and proline dehydrogenase does not, P5C–oAB levels will increase as P5C consumption through P5C dehydrogenase is prevented. Fig. 2d shows that rotenone increased P5C-oAB formation, indicating that proline dehydrogenase in situ does not use NAD directly as its electron acceptor.
Myxothiazol and antimycin A inhibit complex III and block reoxidation of ubiquinol (QH2) formed during proline oxidation (Fig. 1c). Addition of proline in the presence of rotenone, malonate (complex II inhibitor) and antimycin A increased Q-pool reduction, reported by the reduction state of cytochrome b566[29] (Fig. 2f), indicating that proline oxidation reduces ubiquinone directly (and not only through complex I). High QH2/Q inhibits ubiquinone-linked dehydrogenases. If proline dehydrogenase uses ubiquinone as its electron acceptor, complex III inhibitors should slow adduct formation. Addition of myxothiazol decreased P5C accumulation (Fig. 2d), showing that proline dehydrogenase uses ubiquinone as its electron acceptor in intact mitochondria.
H2O2 production during proline oxidation by uncoupled Drosophila mitochondria
Fig. 3a shows that in the absence of inhibitors of electron transport, 5 mM proline induced H2O2 production by uncoupled Drosophila mitochondria. Proline oxidation can increase ubiquinol and NADH levels (Fig. 2e,f) and generate oxoglutarate and succinate [7,40]. Therefore, several different sites could be active during proline oxidation, including proline dehydrogenase but also sites IF, IQ, IIF, IIIQo and OF (Fig. 1a). To test the contribution of site IIF, complex II was inhibited by malonate [41] (Fig. 1d). H2O2 production decreased by ~70% (Fig. 3a, +malonate), showing that site IIF was probably responsible for much of the H2O2 produced during proline oxidation under these conditions.
To maximize any production of superoxide/H2O2 from proline dehydrogenase itself, we added proline and manipulated electron transport flow with inhibitors. First, complex I was inhibited with rotenone, thereby increasing NADH/NAD (Fig. 2e, Table 1), slowing P5C dehydrogenase, raising P5C concentration (Fig. 2d, proline+rotenone) and stalling proline dehydrogenase (Fig. 1b). The rate of H2O2 generation (Fig. 3b, first bar) increased >4-fold compared to Fig. 3a. However, under this condition sites IF and OF will be active [22] and may contribute to the signal. Site IIF was inactive, since malonate did not decrease the rate (Fig. 3b, second bar).
Table 1.
NAD(P)H reduction and predicted rates of superoxide production by site IF in Drosophila mitochondria.
| Substrates and inhibitors | NAD(P)H reduction(% of maximum) | Predicted rate of superoxide |
|---|---|---|
| Production from site IF | ||
| (pmol min−1 mg protein−1) | ||
| Proline (Fig. 3a) | 5.5±1.9 | 11.0±1.0 |
| Proline, malonate (Fig. 3a) | 2.6±1.8 | 10.3±0.6 |
| Proline, rotenone (Fig. 3b) | 98.0±1.0 | 67.8±14.7 |
| Proline, myxothiazol (Fig. 3b) | 37.9±5.6 | 20.8±8.7 |
| Proline, myxothiazol, malonate (Fig. 3b) | 20.8±4.9** | 17.7±5.8 |
| Proline, myxothiazol, rotenone, A5, malonate (Fig. 3b) | 100 | 70.4±15.4 |
p=0.007 comparing proline, myxothiazol and proline, myxothiazol and malonate (paired t-test, one-tailed).
Next, myxothiazol was added to inhibit complex III, thereby reducing ubiquinone (Fig. 2f) and stalling proline dehydrogenase (Fig. 1c). This gave high rates of H2O2 production (Fig. 3b, third bar). However, NAD was 38% reduced (Table 1), so site IF might be active. Furthermore, addition of malonate strongly decreased the signal (Fig. 3b, fourth bar). Therefore, during proline oxidation plus myxothiazol most superoxide/H2O2 production arises from complex II at site IIF.
When sites IQ, IIF and IIIQo were all inhibited by addition of rotenone, myxothiazol, malonate and atpenin A5 (inhibits site IIQ) (Fig. 1e), a measurable rate remained (Fig. 3b, fifth bar). Thus, after inhibiting most of the sites predicted to be active during proline oxidation, some site was still able to produce superoxide/H2O2 at a measurable rate.
Rates of superoxide/H2O2 production from specific sites during proline oxidation by Drosophila mitochondria
Complex I produces superoxide through sites IF and I Q[42]. Site IQ can be blocked by rotenone, but there are no suitable inhibitors of site IF. However, the rate of superoxide production from site IF is uniquely related to NADH redox state [31,43]. Fig. 3c shows the rate of H2O2 generation driven by endogenous substrates during titration with rotenone in Drosophila mitochondria. This signal was assumed to be from site IF since all other putative sites were presumably oxidized and inactive; it was not from proline dehydrogenase itself, since any endogenous proline was insufficient to drive significant proline dehydrogenase activity (Fig. 2d, no proline). Fig. 3d shows the corresponding NAD(P)H reduction state. Fig. 3e replots the rate of H2O2 production as a function of NAD(P)H reduction. This curve was used to assess the contribution of site IF at a given NAD(P)H reduction state.
We then measured NAD(P)H during proline oxidation. Table 1 shows the NAD(P)H levels and assessed rates from site IF under the conditions of Fig. 3a and b. Fig. 3(f–i) shows the contribution of site IF to the rates observed in Fig. 3a and b.
The contributions of other sites were assessed using inhibitors of electron transport after correcting for any alterations in NAD(P)H and the rate of site I F[31]. Fig. 3(f–i) shows that under the conditions of Fig. 3a without malonate, sites IF (green) and IIF (red) fully accounted for the measured H2O2 production; there was no evidence for any direct contribution of site OF or proline dehydrogenase.
Under the conditions of Fig. 3b plus rotenone (Fig. 1b), NAD was ~98% reduced (Table 1). Fig. 3g shows that under this condition site IF fully accounted for the measured rate of H2O2 production; there was no evidence for any direct contribution of proline dehydrogenase. Similarly, Fig. 3h shows that when proline was oxidized in the presence of myxothiazol (Fig. 1c) H2O2 production was dominated by site IIF, a small fraction was attributed to site IF, and there was no evidence for any direct contribution of proline dehydrogenase.
To maximize any superoxide/H2O2 production by proline dehydrogenase itself, the enzyme was pharmacologically isolated by blocking sites IQ, IIF and IIIQo with rotenone, malonate, atpenin A5 and myxothiazol (Figs. 1e and 3b, fifth bar). Fig. 3i shows that even under these conditions H2O2 production was dominated by site IF, which was not inactivated under these conditions; there was no evidence for any direct contribution of proline dehydrogenase.
Thus, Drosophila mitochondria oxidizing proline through proline dehydrogenase generated superoxide/H2O2 at substantial rates from other sites, but there was no evidence of superoxide/H2O2 production from proline dehydrogenase itself even under conditions designed to maximize such production.
Mitochondria isolated from the breast cancer cell line ZR75-30 produce H2O2 during proline oxidation
To investigate whether proline dehydrogenase generates superoxide/H2O2 directly or indirectly in cancer cell mitochondria, we repeated the analysis using mitochondria from ZR75-30 human breast cancer cells.
Fig. 4a (proline) shows that ZR75-30 mitochondria produced 41.5±4.0 pmol H2O2 min−1 mg protein−1 when oxidizing proline in the absence of inhibitors of electron transport. Under these conditions sites IF, IQ, IIF, IIIQo and OF might be active (Fig. 1a). However, inhibiting sites IQ, IIF, IIIQo and OF did not decrease the rate of H2O2 production (Fig. 4a). Specifically, blocking site IIF using malonate did not decrease the rate of H2O2 production (Fig. 4a, proline+malonate). The 2-oxoglutarate dehydrogenase complex is an important source of superoxide/H2O2 in rat skeletal muscle mitochondria [22], and in cancer cells there is a positive relationship between proline dehydrogenase levels and 2-oxoglutarate concentration [7], so site OF might produce superoxide/H2O2 during proline oxidation (Fig. 1a). To decrease its activity, aspartate and ATP were added (Figs 1f and 4a) (aspartate removes 2-oxoglutarate by transamination; ATP inhibits the 2-oxoglutarate complex directly and by increasing succinyl-CoA levels [22]). Fig. 4a (proline+aspartate/ATP) shows that H2O2 production was not decreased, although this might be because aspartate/ATP failed to suppress site OF when 2-oxoglutarate was produced by proline metabolism (Fig. 1a). However, H2O2 production during proline oxidation was sensitive to aspartate/ATP when sites IQ, IIIQo and IIF were inhibited by rotenone, myxothiazol, atpenin A5 and malonate, respectively (Figs. 1e and 4a, fourth and fifth bars), so site OF was active under this specific condition, and aspartate/ATP was a valid test of the contribution of site OF in the initial condition (Fig. 4a, proline).
To determine the contribution of site IF in ZR75-30 mitochondria oxidizing proline (Fig. 4a, proline), we measured NAD(P)H. Fig. 4b and c shows the dependence of H2O2 production and NAD(P)H reduction levels on the concentration of added malate in the presence of rotenone, aspartate and ATP to suppress other sites [22]. Fig. 4d shows the relationship between the rate of superoxide/H2O2 production from site IF and NAD(P)H reduction. This relationship was used to assess the rates of superoxide/H2O2 production from site IF during proline oxidation by ZR75-30 mitochondria in Fig. 4e and f. Table 2 shows the levels of NAD(P)H reduction and the rates of superoxide/H2O2 production from site IF during proline oxidation.
Table 2.
NAD(P)H reduction and predicted rates of superoxide production by site IF in ZR75-30 mitochondria.
| Substrates and inhibitors | NAD(P)H reduction | Predicted rate of superoxide |
|---|---|---|
| (% of maximum) | production from site IF | |
| (pmol min−1 mg protein−1) | ||
| Proline | 59.5 ± 8.0 | 24.9 ± 2.8 |
| Proline, aspartate and ATP | 60.0 ± 5.0 | 25.0 ± 2.65 |
| Proline, rotenone, myxothiazol, malonate, A5 | 51.9 ± 1.4 | 23.8 ± 2.1 |
| Proline, rotenone, myxothiazol, malonate, A5, Aspartate and ATP, | 41.3 ± 3.9* | 22.8 ± 1.8 |
Conditions are described in Fig. 4a.
p=0.04 vs proline, rotenone, myxthiazol, malonate and A5 (paired t-test, one-tailed).
With proline alone (Fig. 1a), site IF was responsible for 60% of measured H2O2 production (Fig. 4e). Under this condition the measured rate was significantly different from the rate assessed for site IF (p=0.01, white bar vs site IF green) suggesting that another site was active. The remaining 40% may be from site OF (because suppression of site OF by aspartate/ATP may be incomplete with proline as substrate) or from other sites, including proline dehydrogenase itself. If proline dehydrogenase did produce superoxide/H2O2 directly under this condition, the rates were low, <16±7 pmol H2O2 min−1 mg protein−1 (Fig. 4e, unassigned gray).
When proline dehydrogenase was pharmacologically isolated by inhibiting sites IQ, IIF, IIQ and IIIQo with rotenone, malonate, atpenin A5 and myxothiazol (Fig. 1g), site IF produced 50% of the H2O2 (Fig. 4f). Another 20% was sensitive to aspartate and ATP, suggesting that it came from site OF. The sum of the rates from sites IF and OF was not significantly different from the measured rate, so there was no evidence for any direct contribution of proline dehydrogenase to H2O2 production. If proline dehydrogenase did contribute under this condition, its contribution was small, <17±13 pmol min−1 mg protein−1.
Thus, ZR75-30 mitochondria oxidizing proline through proline dehydrogenase generated superoxide/H2O2 at measurable rates from other sites, but there was little evidence of superoxide/H2O2 production from proline dehydrogenase itself even under conditions designed to maximize such production.
Discussion
The role of proline dehydrogenase as a direct source of superoxide or H2O2 has not been explored in situ, although proline oxidation generates ROS in cells [6,8]. Because proline oxidation feeds electrons into multiple sites of the citric acid cycle and electron transport chain, it has been unclear whether this ROS arises directly from proline dehydrogenase or from downstream mitochondrial sites. Purified proline dehydrogenases from humans and different microorganisms react directly with molecular oxygen in the absence of endogenous electron acceptors to produce superoxide/H2O2[17,20,44], supporting the view that proline dehydrogenase produces mitochondrial ROS and promotes apoptosis in cancer cells [6,10]. However, under certain circumstances including nutrient and oxidative stress and hypoxia, proline and proline dehydrogenase may exert protective effects by acting as an alternative energy source [11–13]. In this context, depletion of proline dehydrogenase is detrimental [44].
It has been challenging to understand the roles of proline and proline dehydrogenase in cellular metabolism and cancer, and whether ROS production directly by the enzyme is responsible for triggering apoptosis in cancer cells. To address this issue, we measured the rates of production of superoxide/H2O2 from different candidate sites during proline oxidation in isolated mitochondria in situ.
Initially, Drosophila mitochondria were chosen as a model because they have abundant proline dehydrogenase and oxidize proline faster than cancer cell mitochondria (Fig. 2b). In insects proline is used as the source of carbon to power flight [33]. We confirmed that the initial acceptor for electrons from proline dehydrogenase in situ is ubiquinone, not NAD (Fig. 2d–f). The sites involved in superoxide/H2O2 production during proline oxidation under a variety of conditions were quantified. Although proline oxidation caused substantial rates of superoxide/H2O2 production from other sites, specifically sites IIF in complex II and IF in complex I (Fig. 3f), there was no significant unassigned rate and no evidence of superoxide/H2O2 production from proline dehydrogenase itself even under conditions designed to maximize such production (Fig. 3i).
Next, the breast cancer line ZR75-30 was chosen as a model to investigate the role of proline dehydrogenase in cancer cells because of its high basal amounts of proline dehydrogenase relative to other cancer cell lines tested (Fig. 2a). Mitochondria from ZR75-30 cells consumed oxygen and produced H2O2 with proline as substrate (Fig. 2b and c). The sites involved in superoxide/H2O2 production during proline oxidation under a variety of conditions were quantified. In the absence of inhibitors of electron transport, most of the measured H2O2 production was from site IF. However, a significant proportion was unassigned, and may have arisen from site OF in the 2-oxoglutarate dehydrogenase complex or from proline dehydrogenase (Fig. 4e). Under conditions designed to maximize any contribution from proline dehydrogenase itself (Fig. 4f), proline oxidation caused measurable superoxide/H2O2 production from sites IF and OF. There was no significant unassigned rate and no evidence of superoxide/H2O2 production from proline dehydrogenase itself. Together the results in the two systems show that proline oxidation by isolated mitochondria generates significant superoxide/H2O2 from downstream sites, specifically IF, IIF and OF, but there was little evidence for superoxide/H2O2 production from any other site, and no evidence for any superoxide/H2O2 production by proline dehydrogenase itself.
Our conclusion that proline dehydrogenase is not a significant source of superoxide/H2O2 in mitochondria contrasts with the observations cited above that the isolated enzyme can generate ROS. This might be because production by the enzyme itself is insufficient to register in the context of other much more active mitochondrial sites. Alternatively, proline dehydrogenase in situ may have different radical production properties than the purified enzyme. Evidence comes from kinetic analyses showing that the Km of proline dehydrogenase for proline depends greatly on the electron acceptor [17,18,20,45]. Ubiquinone may suppress the ability of the enzyme to donate electrons directly to oxygen in situ and in vivo.
We conclude that proline dehydrogenase does not directly produce superoxide or H2O2 at significant rates in flight muscle or cancer cell mitochondria. Instead, proline oxidation feeds electrons into the respiratory chain directly through ubiquinone reduction and indirectly through further metabolism, and triggers superoxide/H2O2 production from other downstream sites. This does not preclude proline dehydrogenase from playing important roles in cancer cell metabolism, but it is not itself a significant source of superoxide/H2O2.
Authors’ contributions
Conception and design: MDB, RLSG, CLQ, CCB, GKS; Development of methodology: RLSG, CLQ, MDB; Acquisition of data: RLSG, DER; Analysis and interpretation of data: RLSG, CLQ, MDB; Writing, review, and/or revision of the manuscript: RLSG, MDB; Administrative, technical, or material support: MDB, CCB; Study supervision: MDB.
Conflict of interest
The funding sources had no role in study design; in collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Acknowledgments
We thank I.V. Perevoshchikova and A.A. Gerencser for fitting the IF calibration curve, and P. Kapahi for providing facilities for Drosophila work. This work was supported by The Brazilian Government through the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) e ao Conselho de Nacional de Desenvolvimento Científico e Tecnológico programa Ciências Sem Fronteiras (CNPq-CSF) and The Glenn Foundation (RLSG); National Institutes of Health grant TL1-AG032116 (CLQ); NIH/NCI grants R21-CA155679, R01-CA071468, and U24-CA14358 (CCB).
Contributor Information
Renata L.S. Goncalves, Email: rgoncalves@buckinstitute.org.
Daniel E. Rothschild, Email: drothman8@gmail.com.
Casey L. Quinlan, Email: casey.quinlan@pfizer.com.
Gary K. Scott, Email: gscott@buckinstitute.org.
Christopher C. Benz, Email: cbenz@buckinstitute.org.
Martin D. Brand, Email: mbrand@buckinstitute.org.
References
- 1.Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. 10647931 [DOI] [PubMed] [Google Scholar]
- 2.Shangary S., Wang S. Targeting the MDM2–p53 interaction for cancer therapy. Clinical Cancer Research: An Official Journal of the American Association for Cancer Research. 2008;14:5318–5324. doi: 10.1158/1078-0432.CCR-07-5136. 18765522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vousden K.H., Lu X. Live or let die: the cell’s response to p53. Nature Reviews. Cancer. 2002;2:594–604. doi: 10.1038/nrc864. 12154352 [DOI] [PubMed] [Google Scholar]
- 4.Vassilev L.T. MDM2 inhibitors for cancer therapy. Trends in Molecular Medicine. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. 17126603 [DOI] [PubMed] [Google Scholar]
- 5.Polyak K., Xia Y., Zweier J.L., Kinzler K.W., Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. 9305847 [DOI] [PubMed] [Google Scholar]
- 6.Maxwell S.A., Rivera A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. Journal of Biological Chemistry. 2003;278:9784–9789. doi: 10.1074/jbc.M210012200. 12514185 [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Borchert G.L., Donald S.P., Diwan B.A., Anver M., Phang J.M. Proline oxidase functions as a mitochondrial tumor suppressor in human cancers. Cancer Research. 2009;69:6414–6422. doi: 10.1158/0008-5472.CAN-09-1223. 19654292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donald S.P., Sun X.Y., Hu C.A., Yu J., Mei J.M., Valle D., Phang J.M. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Research. 2001;61:1810–1815. http://www.ncbi.nlm.nih.gov/pubmed/11280728 [PubMed] [Google Scholar]
- 9.Liu Y., Borchert G.L., Donald S.P., Surazynski A., Hu C.A., Weydert C.J., Oberley L.W., Phang J.M. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis. 2005;26:1335–1342. doi: 10.1093/carcin/bgi083. 15817612 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y., Borchert G.L., Surazynski A., Hu C.A., Phang J.M. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: The role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25:5640–5647. doi: 10.1038/sj.onc.1209564. 16619034 [DOI] [PubMed] [Google Scholar]
- 11.Pandhare J., Donald S.P., Cooper S.K., Phang J.M. Regulation and function of proline oxidase under nutrient stress. Journal of Cellular Biochemistry. 2009;107:759–768. doi: 10.1002/jcb.22174. 19415679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu W., Glunde K., Bhujwalla Z.M., Raman V., Sharma A., Phang J.M. Proline oxidase promotes tumor cell survival in hypoxic tumor microenvironments. Cancer Research. 2012;72:3677–3686. doi: 10.1158/0008-5472.CAN-12-0080. 22609800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Natarajan S.K., Zhu W., Liang X., Zhang L., Demers A.J., Zimmerman M.C., Simpson M.A., Becker D.F. Proline dehydrogenase is essential for proline protection against hydrogen peroxide-induced cell death. Free Radical Biology and Medicine. 2012;53:1181–1191. doi: 10.1016/j.freeradbiomed.2012.07.002. 22796327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Oliveira M.F., Amoêdo N.D., Rumjanek F.D. Energy and redox homeostasis in tumor cells. International Journal of Cell Biology. 2012;2012:593838. doi: 10.1155/2012/593838. 22693511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu W., Phang J.M. Proline dehydrogenase (oxidase), a mitochondrial tumor suppressor, and autophagy under the hypoxia microenvironment. Autophagy. 2012;8:1407–1409. doi: 10.4161/auto.21152. 22885468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bella J., Eaton M., Brodsky B., Berman H.M. Crystal and molecular structure of a collagen-like peptide at 1.9 A resolution. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
- 17.White T.A., Krishnan N., Becker D.F., Tanner J.J. Structure and kinetics of monofunctional proline dehydrogenase from Thermus thermophilus. Journal of Biological Chemistry. 2007;282:14316–14327. doi: 10.1074/jbc.M700912200. 17344208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moxley M.A., Tanner J.J., Becker D.F. Steady-state kinetic mechanism of the proline:ubiquinone oxidoreductase activity of proline utilization A (PutA) from Escherichia coli. Archives of Biochemistry and Biophysics. 2011;516:113–120. doi: 10.1016/j.abb.2011.10.011. 22040654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu C.A., Donald S.P., Yu J., Lin W.W., Liu Z., Steel G., Obie C., Valle D., Phang J.M. Overexpression of proline oxidase induces proline-dependent and mitochondria-mediated apoptosis. Molecular and Cellular Biochemistry. 2007;295:85–92. doi: 10.1007/s11010-006-9276-6. [DOI] [PubMed] [Google Scholar]
- 20.Krishnan N., Becker D.F. Oxygen reactivity of PutA from Helicobacter species and proline-linked oxidative stress. Journal of Bacteriology. 2006;188:1227–1235. doi: 10.1128/JB.188.4.1227-1235.2006. 16452403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brand M.D. The sites and topology of mitochondrial superoxide production. Experimental Gerontology. 2010;45:466–472. doi: 10.1016/j.exger.2010.01.003. 20064600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quinlan C.L., Goncalves R.L., Hey-Mogensen M., Yadava N., Bunik V.I., Brand M.D. The 2-oxoacid dehydrogenase complexes in mitochondria can produce superoxide/hydrogen peroxide at much higher rates than complex I. Journal of Biological Chemistry. 2014;289:8312–8325. doi: 10.1074/jbc.M113.545301. 24515115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hey-Mogensen M., Goncalves R.L., Orr A.L., Brand M.D. Production of superoxide/H2O2 by dihydroorotate dehydrogenase in rat skeletal muscle mitochondria. Free Radical Biology & Medicine. 2014;72:149–155. doi: 10.1016/j.freeradbiomed.2014.04.007. 24746616 [DOI] [PubMed] [Google Scholar]
- 24.Orr A.L., Quinlan C.L., Perevoshchikova I.V., Brand M.D. A refined analysis of superoxide production by mitochondrial sn-glycerol 3-phosphate dehydrogenase. Journal of Biological Chemistry. 2012;287:42921–42935. doi: 10.1074/jbc.M112.397828. 23124204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katewa S.D., Demontis F., Kolipinski M., Hubbard A., Gill M.S., Perrimon N., Melov S., Kapahi, P. Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metabolism. 2012;16:97–103. doi: 10.1016/j.cmet.2012.06.005. [Pubmed: 22768842] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miwa S., St-Pierre J., Partridge L., Brand M.D. Superoxide and hydrogen peroxide production by Drosophila mitochondria. Free Radical Biology and Medicine. 2003;35:938–948. doi: 10.1016/s0891-5849(03)00464-7. 14556858 [DOI] [PubMed] [Google Scholar]
- 27.Rogers G.W., Brand M.D., Petrosyan S., Ashok D., Elorza A.A., Ferrick D.A., Murphy A.N. High throughput microplate respiratory measurements using minimal quantities of isolated mitochondria. PloS One. 2011;6:e21746. doi: 10.1371/journal.pone.0021746. 21799747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mezl V.A., Knox W.E. Properties and analysis of a stable derivative of pyrroline-5-carboxylic acid for use in metabolic studies. Analytical Biochemistry. 1976;74:430–440. doi: 10.1016/0003-2697(76)90223-2. 962101 [DOI] [PubMed] [Google Scholar]
- 29.Quinlan C.L., Gerencser A.A., Treberg J.R., Brand M.D. The mechanism of superoxide production by the antimycin-inhibited mitochondrial Q-cycle. Journal of Biological Chemistry. 2011;286:31361–31372. doi: 10.1074/jbc.M111.267898. 21708945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crofts A.R., Meinhardt S.W., Jones K.R., Snozzi M. The role of the quinone pool in the cyclic electron-transfer chain of Rhodopseudomonas sphaeroides: a modified q-cycle mechanism. Biochimica et Biophysica Acta. 1983;723:202–218. doi: 10.1016/0005-2728(83)90120-2. 21494412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinlan C.L., Treberg J.R., Perevoshchikova I.V., Orr A.L., Brand M.D. Native rates of superoxide production from multiple sites in isolated mitochondria measured using endogenous reporters. Free Radical Biology and Medicine. 2012;53:1807–1817. doi: 10.1016/j.freeradbiomed.2012.08.015. 22940066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herzfeld A., Mezl V.A., Knox W.E. Enzymes metabolizing delta1-pyrroline-5-carboxylate in rat tissues. Biochemical Journal. 1977;166:95. doi: 10.1042/bj1660095. 103 901423] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scaraffia P.Y., Wells M.A. Proline can be utilized as an energy substrate during flight of Aedes aegypti females. Journal of Insect Physiology. 2003;49:591–601. doi: 10.1016/s0022-1910(03)00031-3. 12804719 [DOI] [PubMed] [Google Scholar]
- 34.Vinod M.P., Bellur P., Becker D.F. Electrochemical and functional characterization of the proline dehydrogenase domain of the PutA flavoprotein from Escherichia coli. Biochemistry. 2002;41:6525–6532. doi: 10.1021/bi025706f. 12009917 [DOI] [PubMed] [Google Scholar]
- 35.Sakuraba H., Takamatsu Y., Satomura T., Kawakami R., Ohshima T. Purification, characterization, and application of a novel dye-linked l-proline dehydrogenase from a hyperthermophilic archaeon, Thermococcus profundus. Applied and Environmental Microbiology. 2001;67:1470–1475. doi: 10.1128/AEM.67.4.1470-1475.2001. 11282592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanduragala S., Sanyal N., Liang X., Becker D.F. Purification and characterization of Put1p from Saccharomyces cerevisiae. Archives of Biochemistry and Biophysics. 2010;498:136–142. doi: 10.1016/j.abb.2010.04.020. 20450881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tallarita E., Pollegioni L., Servi S., Molla G. Expression in Escherichia coli of the catalytic domain of human proline oxidase. Protein Expression and Purification. 2012;82:345–351. doi: 10.1016/j.pep.2012.01.021. 22333530 [DOI] [PubMed] [Google Scholar]
- 38.McKnight J.A., Hird F.J. The oxidation of proline by mitochondrial preparations. Comparative Biochemistry and Physiology B: Comparative Biochemistry. 1986;85:289–294. doi: 10.1016/0305-0491(86)90002-7. 3780181 [DOI] [PubMed] [Google Scholar]
- 39.Hansford R.G., Sacktor B. The control of the oxidation of proline by isolated flight muscle mitochondria. Journal of Biological Chemistry. 1970;245:991–994. 5417269 [PubMed] [Google Scholar]
- 40.Bursell E. Oxidation of proline by sarcosomes of the tsetse fly, Glossina morsitans. Insect Biochemistry. 1976;6:159–167. [Google Scholar]
- 41.Quinlan C.L., Orr A.L., Perevoshchikova I.V., Treberg J.R., Ackrell B.A., Brand M.D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. Journal of Biological Chemistry. 2012;287:27255–27264. doi: 10.1074/jbc.M112.374629. 22689576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treberg J.R., Quinlan C.L., Brand M.D. Evidence for two sites of superoxide production by mitochondrial NADH-ubiquinone oxidoreductase (complex I) Journal of Biological Chemistry. 2011;286:27103–27110. doi: 10.1074/jbc.M111.252502. 21659507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kussmaul L., Hirst J. The mechanism of superoxide production by NADH:ubiquinone oxidoreductase (complex I) from bovine heart mitochondria. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:7607–7612. doi: 10.1073/pnas.0510977103. 16682634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krishnan N., Dickman M.B., Becker D.F. Proline modulates the intracellular redox environment and protects mammalian cells against oxidative stress. Free Radical Biology and Medicine. 2008;44:671–681. doi: 10.1016/j.freeradbiomed.2007.10.054. 18036351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baban B.A., Vinod M.P., Tanner J.J., Becker D.F. Probing a hydrogen bond pair and the FAD redox properties in the proline dehydrogenase domain of Escherichia coli PutA. Biochimica et Biophysica Acta. 2004;1701:49–59. doi: 10.1016/j.bbapap.2004.06.001. 15450175 [DOI] [PubMed] [Google Scholar]


