Abstract
Patients with newly diagnosed AML (n=360) including 137 (38%) with normal karyotype (NK) were evaluated. Overall, 60 (16.6%) patients including 46 of the 137 (33.5%) NK patients had NPM1 mutation at baseline. Thirty nine patients (30 NK) had available NPM1 status at the time of CR and all (100%) were negative for mutated NPM1. Among the patients with mutated NPM1 at baseline, 10/39 overall (25%) and 7/30 NK (23%) patients relapsed. NPM1 status was available for 8 patients (6 with NK) at the time of relapse. Among them, 7/8 overall (87%) and 5/6 NK (83%) patients had mutated NPM1, while 1/8 overall (12%) and 1/6 NK (16%) patients remained NPM1 wild type. Among the 300 patients (including 91 with NK) with wild type NPM1 at diagnosis, none acquired a mutated NPM1 clone, either at CR or at relapse. We conclude that mutated NPM1 is a stable and reliable prognostic marker in AML and can be used to assess MRD.
Keywords: AML, NPM1 mutations, minimal residual disease, MRD
INTRODUCTION
The clinical course and response to therapy in patients with acute myeloid leukemia (AML) is largely dictated by the presence or absence of specific genomic aberrations and mutations.[1–3] Relapse continues to be a major cause of failure to achieve long term disease-free survival using available treatment strategies.[4] Recently, several groups have identified a number of recurring mutations in patients with de novo AML.[5–7] The suitability of these mutations as a marker of minimal residual disease (MRD) is being studied.[7, 8] Molecular markers that are reliably stable during the disease course and clonal evolution are sought after as markers for MRD detection. Mutations in Nucleophosmin-1 (NPM1) gene have been described in about 35% of adult patients with de novo AML and 45–60% of patients with a normal karyotype (NK).[9] Mutations in exon 12 are considered to be the most frequent mutations in AML (>50%) patients with normal karyotype (NK).[9] Since NPM1 mutation is considered as a founder mutation in AML leukemogenesis, its prognostic impact has been evaluated by a number of groups.[9–15] Furthermore, the most recent WHO classification of myeloid neoplasms considers NPM1 mutated AML as a separate entity.[16–18] Among patients with NK AML, the presence of NPM1 mutations predicts for a higher likelihood of achieving complete remission (CR), lower relapse rates and better overall outcomes. [10] [19–22] NPM1 is a phosphoprotein encoded by a gene on chromosome 5. It is thought to play a role in various intracellular processes [23] such as ribosomal assembly, shuttling or transport,[24, 25] DNA repair, stress responses and protection against P53 induced apoptosis.[26, 27] The cytoplasmic localization of the mutant NPM1 is considered to be the key event in inducing intracellular signaling pathways, although this localization is not always concordant with the presence of the mutations.[28] NPM1 mutations have also been shown to induce CD4 and CD8 T cell responses and are being explored as an immunotherapeutic target.[29] Cup like nuclei are recognized as a common feature of the AML blasts from patients with AML and NPM1 and/or FLT3 mutations.[14]
Different techniques have been used to analyze NPM1 mutations including RNA or DNA based real time quantitative polymerase chain reaction (RQ-PCR), [8] imaging flow cytometry, [30] and next generation sequencing.[31] Few prior studies have examined the role of NPM1 mutations as markers for MRD assessment. [8, 32, 33] In these reports, the investigators examined the presence of NPM1 mutations in paired samples at the diagnosis, and at the time of relapse. [8, 32–34] Kronke et al reported that among 245 patients with AML aged < 60 years, early detection of relapse was possible in patients with >200 NPM1 mutation/104 ABL copies (n=36) as assessed by real time PCR.[8] However, 9% of the relapsed samples did not contain a mutant NPM1 clone in this study. In another study of paired samples (at diagnosis and at relapse) from 84 patients with NPM1 mutated AML, NPM1 was found to be expressed at high levels (2 log range) at the time of relapse and was a stable MRD marker.[32] Two other studies have suggested that NPM1 mutations may not reliably recur at the time of relapse.[19, 20]
We have conducted this study to assess the prognostic significance of NPM1 mutations in patients with AML at the diagnosis, at CR and at the time of relapse in our single institute database.
PATIENTS AND METHODS
Population studied
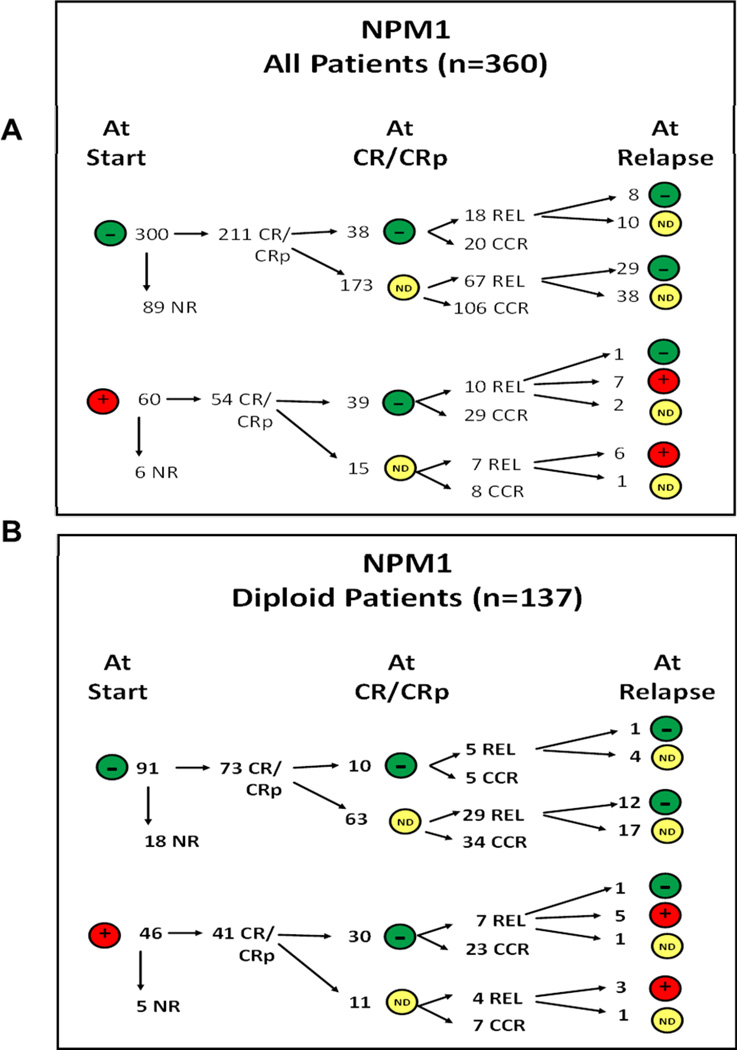
We conducted a retrospective analysis of patients (n=360, NK; n=137) with newly diagnosed AML who underwent testing for NPM1 status and who were treated at our institution between 2008 and 2012 (patients with acute promyelocytic leukemia were excluded). NPM1 mutations were detected in 60 (16.6%) patients and were undetectable in the other 300. All patients were treated on frontline induction protocols and had bone marrow biopsy and/or aspiration, cytogenetic, and molecular studies at the time of diagnosis. Cytogenetic and molecular studies at complete remission (CR) and relapse were performed at the discretion of the treating physician. All patients signed an informed consent for participation and the trials were conducted in accordance with the Declaration of Helsinki. All studies have been approved by the Institutional Review Board of the University of Texas - MD Anderson Cancer Center. CR and relapse were defined as described previously.[35] The available bone marrow samples at diagnosis, CR and first relapse were reviewed for the presence of NPM1 mutated clones (Figure 1A–B). A subset of NPM1 mutated patients were FLT3-ITD positive n=24 (41%) and were also analyzed for time to event variables.
Figure 1.
(A-B). Schema describing the NPM1 status at CR1 and first relapse. Panel A shows the numbers of patients overall and panel B shows number of patients (NK alone). ND indicates NPM1 mutation testing not done. CR/CCR/CRp indicates complete remission with without platelet recovery. One patient who remained negative at relapse had an extramedullary relapse.
Detection of NPM1 Mutations
NPM1 mutation status was determined from DNA from unsorted bone marrow (BM) aspirate samples by a PCR-based method at baseline, remission, and relapse samples, when available. Genomic DNA from bone marrow samples was isolated using the Autopure extractor (QIAGEN/Gentra, Valencia, CA). Mutations in exon 12 of NPM1 were assessed by a DNA-based semi-quantitative polymerase chain reaction capillary electrophoresis (PCR-CE) assay with analytical sensitivity of approximately 2.5%. FLT3-ITD mutations were also assessed using the same method.
Statistical analysis
Differences among variables were evaluated by the chi-square test and Mann- Whitney test for categorical and continuous variables, respectively. Survival curves were calculated for overall survival (OS; defines as time from diagnosis to death), event free survival (EFS; defined as time from diagnosis to time of treatment failure, relapse or death), complete remission duration (CRD; defined as time from documentation of CR to first relapse) according to the Kaplan-Meier method and compared by use of a 2 sided log rank test. All P values were two sided and values less than 0.05 were considered to be significant. Statistica (version 9) software was used for statistical analysis.
RESULTS
Patient characteristics
Data from 360 previously untreated patients with AML, who had available NPM1 analysis on their BM at the time of initial diagnosis, was collected. The characteristics of the patients (both overall and those with NK) are shown in Tables 1 and 3. Median age was 60 years (range 19 – 83 years). Median WBC count at diagnosis was 5.2 × 109/L and was similar in NPM1 mutated and wild-type patients. Patients with mutated NPM1 exhibited significantly higher BM blasts percentage as compared to patients with wild-type NPM1 (Median 62% vs. 40% respectively; p<0.001) and significantly lower % of CD34+ cells (Median 0.7 vs. 72.5% respectively; p<0.001). NPM1 mutations were present with similar frequency in men and women.
Table 1.
Patient characteristics (Overall population)
| Parameter | NPM1 WT | NPM1 MUT | Total | P value |
|---|---|---|---|---|
| N | 300 | 60 | 360 | |
| Age | 60 (19–83) | 60 (23–80) | 60 (19–83) | 0.58 |
| WBC (K/µL) | 5.0 (0.1–191.0) | 6.3 (0.1–228.5) | 5.2 (0–228.5) | 0.22 |
| BM Blasts | 40 (0–95) | 62 (12–95) | 42 (0–95) | <0.001 |
| CD34+ | 72.5 (0–99.8) | 0.7 (0–94.8) | 54.9 (0–54.9) | <0.001 |
| Female n (%) | 137 (46) | 29 (48) | 166 (46) | |
| Male n (%) | 163 (54) | 31 (52) | 194 (54) | 0.7 |
| Diagnosis: | ||||
| AML | 209 (70) | 33 (55) | 242 (67) | |
| AMML | 46 (15) | 17 (28) | 63 (18) | |
| AMoL | 26 (9) | 8 (13) | 34 (9) | 0.07 |
| AEL | 16 (5) | 2 (3) | 18 (5) | |
| Mega | 3 (1) | 0 | 3 (1) | |
| AML type: | ||||
| De novo | 210 (58) | 52 (87) | 262 (73) | |
| Secondary | 90 (42) | 8 (13) | 98 (27) | 0.008 |
| FLT3 Status | ||||
| FLT3 WT | 249 (85) | 34 (59) | 283 (81) | |
| FLT3 MUT | 43 (15) | 24 (41) | 67 (19) | <0.001 |
| ND | 8 | 2 | 10 | |
| RAS Status | ||||
| RAS WT | 236 (85) | 43 (81) | 279 (84) | |
| RAS MUT | 43 (15) | 10 (19) | 53 (16) | 0.53 |
| ND | 21 | 7 | 28 | |
| Karyotype: | ||||
| CBF [t(8;21) & inv(16)] | 41 (15) | 2 (3) | 43 (12) | |
| Normal | 91 (30) | 46 (77) | 137 (38) | |
| Miscellaneous | 88 (29) | 8 (13) | 96 (27) | <0.001 |
| −5/−7 | 73 (24) | 1 (2) | 74 (21) | |
| IM/ND | 7 (2) | 3 (5) | 10 (3) |
Legends – WT: wild type; MUT: mutant; WBC: white blood count; BM: bone marrow; ND: not done; IM: insufficient metaphases
Table 3.
Characteristics of patients with NPM1 mutations and normal karyotype
| Diploid | NPM1 WT | NPM1 MUT | Total | P value |
|---|---|---|---|---|
| N | 91 | 46 | 137 | |
| Age | 61 (20–81) | 59 (23–80) | 61 (20–81) | 0.11 |
| WBC (K/µL) | 2.4 (0.5–132.3) | 6.3 (1.1–100.2) | 3.8 (0.5–132.3) | 0.04 |
| BM Blasts | 36 (0–93) | 59 (12–95) | 44 (0–95) | <0.001 |
| CD34+ | 79 (0–99) | 1(0–64) | 31 (0–100) | <0.001 |
| Female n (%) | 37 (41) | 25 (54) | 62 (45) | |
| Male n (%) | 54 (59) | 21 (46) | 75 (55) | 0.13 |
| Diagnosis: | ||||
| AML | 71 (78) | 24 (52) | 95 (69) | |
| AMML | 8 (9) | 14 (30) | 22 (16) | |
| AMoL | 3 (3) | 7 (15) | 10 (7) | <0.001 |
| AEL | 7 (8) | 1 (2) | 8 (6) | |
| Mega | 2 (2) | 0 | 2 (1) | |
| AML Type: | ||||
| De novo | 69 (76) | 42 (91) | 111 (81) | |
| Secondary | 22 (24) | 4 (9) | 26 (19) | 0.029 |
| FLT3 Status: | ||||
| FLT3 WT | 69 (76) | 23 (51) | 92 (68) | |
| FLT3 MUT | 22 (24) | 22 (49) | 44 (32) | 0.004 |
| ND | 1 | 1 | ||
| RAS Status: | ||||
| RAS WT | 77 (89) | 34 (81) | 111 (86) | |
| RAS MUT | 10 (11) | 8 (19) | 18 14) | 0.29 |
| ND | 4 | 4 | 8 |
Legends – WT: wild type; MUT: mutant; WBC: white blood count; BM: bone marrow; ND: not done
Cytogenetics was normal in 137 (38%) patients of whom 46 (33%) had mutated NPM1 as compared to wild type NPM1 in 91 (66%) patients. Patients with NPM1 mutations had a lower proportion of other cytogenetic abnormalities as compared to patients with wild type NPM1. Overall, 60 of 360 (16.6%) patients including the previously mentioned 46 of the 137 (33%) NK patients had mutated NPM1 at the baseline. RAS mutations were equally present among the two groups. 262 patients (72%) had de novo AML, and 98 (27%) secondary or therapy-related AML. Secondary leukemia was more common in the NPM1 wild type (30%) than in the NPM1 mutated (13%) category (p=0.008). NPM1 mutated patients had higher proportions of FLT3-ITD positive cases as compared to NPM1 wild type including in NK (n=24; 41% and n=22; 49% in NK group respectively).
Prognostic significance of NPM1 mutations
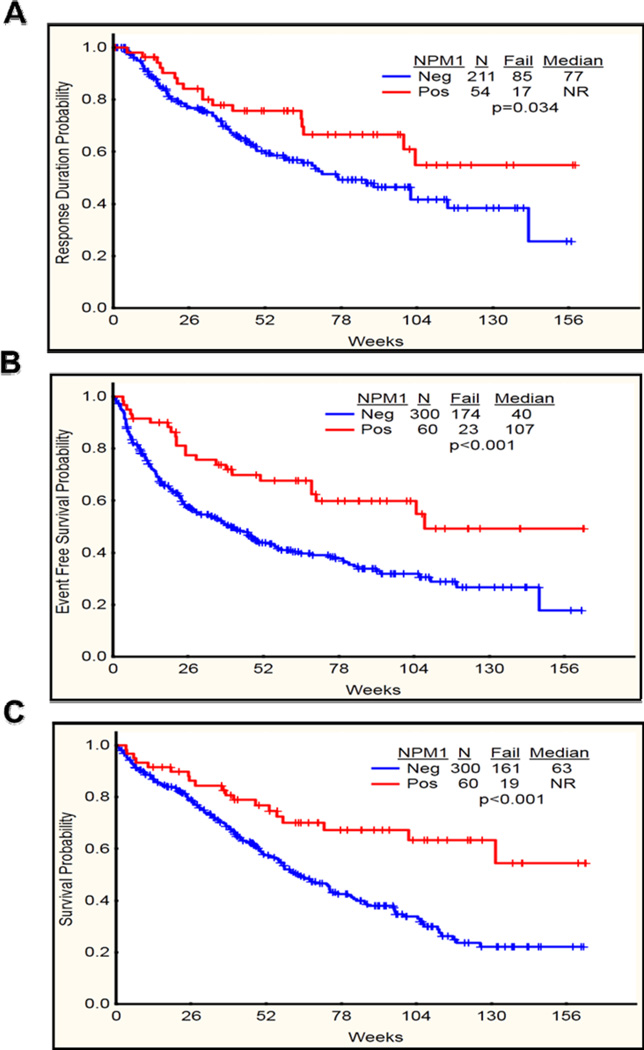
Overall, median follow up of the patients was 61 weeks (Range 0–163) while in NK it was 70 weeks (3–163 weeks). Patients with mutated NPM1 had a significantly longer complete remission duration (CRD) (P=0.03) and a higher proportion n=54 (90%) achieved complete remission as compared to NPM1 wild type patients (P=0.002) (Table 2 and Figure 2A). Only 6 of 95 (6%) patients who did not achieve CR had mutated NPM1. Both event-free survival (EFS) and overall survival (OS) were significantly longer in patients with NPM1 mutation (P <0.001 and 0.001 respectively) (Figure 2B–C). When analyzed by age, in patients < 60 years (n=175), OS, EFS and response rates were significantly superior in NPM1 mutated subgroup (p=0.001, 0.007, 0.02 respectively). Among patients ≥ 60 years (n=185) EFS and response rates were also significantly higher in the NPM1 mutated subgroup (p=0.008, 0.03 respectively) (Table 2).
Table 2.
Prognostic significance of NPM1 mutations (Overall population)
| Parameter | NPM1 WT | NPM1 MUT | Total | P value |
|---|---|---|---|---|
| CR/CRp | 211 (70) | 54 (90) | 265 (74) | 0.002 |
| NR | 89 (20) | 6 (10) | 95 (26) | |
| CRD (wk) | 77 | NR | 0.034 | |
| OS (wk) (N=360) | 63 | NR | <0.001 | |
| OS (wk) (<60 yrs) (N=175) | 82 | NR | <0.001 | |
| OS (wk) (≥60 yrs) (N=185) | 49 | 73 | 0.064 | |
| EFS (wk) | 40 | 107 | 0.001 | |
| EFS (wk) (<60 yrs) | 69 | NR | 0.007 | |
| EFS (wk) (≥60 yrs) | 27 | 105 | 0.008 |
Legends – WT: wild type; MUT: mutant; CR: complete response; CRp: complete response without platelet recovery; NR: no response; CRD: CR duration; OS: overall survival; EFS: event-free survival; wk: week
Figure 2.
(A-C). Complete remission duration (CRD), event free survival (EFS) and overall survival (OS) in NPM1 mutated (Positive) vs NPM1 wild type (Negative) among all the patients (n=360). A) CRD significantly longer in NPM1 mutated as compared to NPM1 wild type (P=0.034). B) EFS significantly longer in NPM1 mutated as compared to NPM1 wild type (P<0.001) C) OS significantly longer in NPM1 mutated as compared to NPM1 wild type (P<0.001).
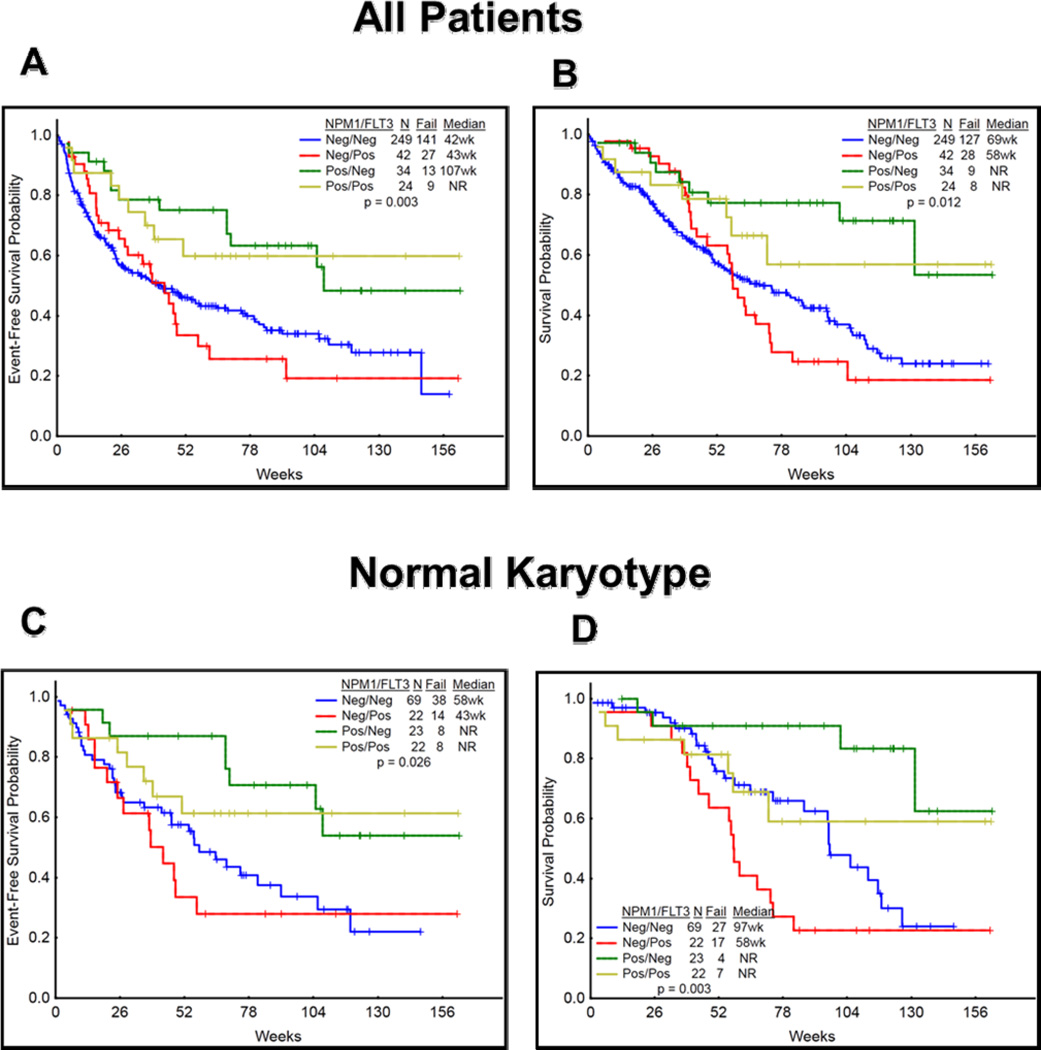
Due to a significant association between NPM1 mutated patients with FLT3-ITD positivity, we have further analyzed the EFS and OS among NPM1+/FLT3-ITD+, NPM1+/FLT3-ITD-, NPM1−/FLT3-ITD+, and NPM1−/FLT3-ITD- subgroups (Figure 3A–D). Patients who were NPM1−/FLT3-ITD+ had significantly inferior EFS and OS as compared to other categories including in patients with normal karyotype.
Figure 3.
(A-D). Event free survival (EFS) and overall survival (OS) among NPM1+/FLT3+, NPM1+/FLT3−, NPM1−/FLT3+, and NPM1−/FLT3− subgroups is shown (A–D). A) EFS was significantly inferior in NPM1−/FLT3+ mutated as compared to other subgroups (P=0.003) B) OS significantly inferior in NPM1−/FLT3+ mutated as compared to other subgroups (P=0.012). Similar results were seen in patients with normal karyotype (EFS: P=0.02; OS: P=0.003) in various subgroups. NPM1+/FLT3− have significantly better outcomes.
Analysis for detecting both IDH1 and IDH2 mutations were performed at baseline in 103 patients. Among the 300 patients with wild type NPM1 at baseline 85 patients had testing for IDH1 and IDH2 mutations at baseline; 5 (6%) were IDH1 mutated and 80 (94%) were IDH1 wild type. 7 of 85 (8%) patients were IDH2 mutated and 78 (92%) were IDH2 wild type. On the other hand, among the 60 patients who were NPM1 mutated at baseline, 18 patients had testing for IDH1 and IDH2 mutations. Four (22%) had IDH1 mutation and fourteen (78%) were IDH1 wild type. One (5%) patient was IDH2 mutated and 17 (94%) were IDH2 wild type.
Characteristics and outcome of patients with normal karyotype (NK)
A total of one hundred and thirty seven patients with NK (normal karyotype) were evaluated. Table 3 shows the main characteristics of patients with NK with or without mutated NPM1. Median age was 61 years (range 20 – 81 years). Median WBC count at diagnosis was significantly higher in NPM1 mutated patients as compared to NPM1 wild type (Median WBC 6.3 vs 2.4 K/µL; P=0.04 respectively). Patients with NPM1 mutation exhibited significantly higher bone marrow (BM) blasts percentage as compared with NPM1 wild type cases (Median 59% vs. 36% respectively; p<0.001). Higher proportions of NPM1 mutated patients presented with acute monocytic (AMoL) and acute myelomonocytic leukemia (AMML) as compared to NPM1 wild type patients (P<0.001). Proportions of secondary leukemia was higher in NPM1 negative as compared to NPM1 mutated patients (P=0.02). As expected, FLT3-ITD mutations were higher in NPM1 mutated as compared to NPM1 wild type (P=0.004). [2, 11]
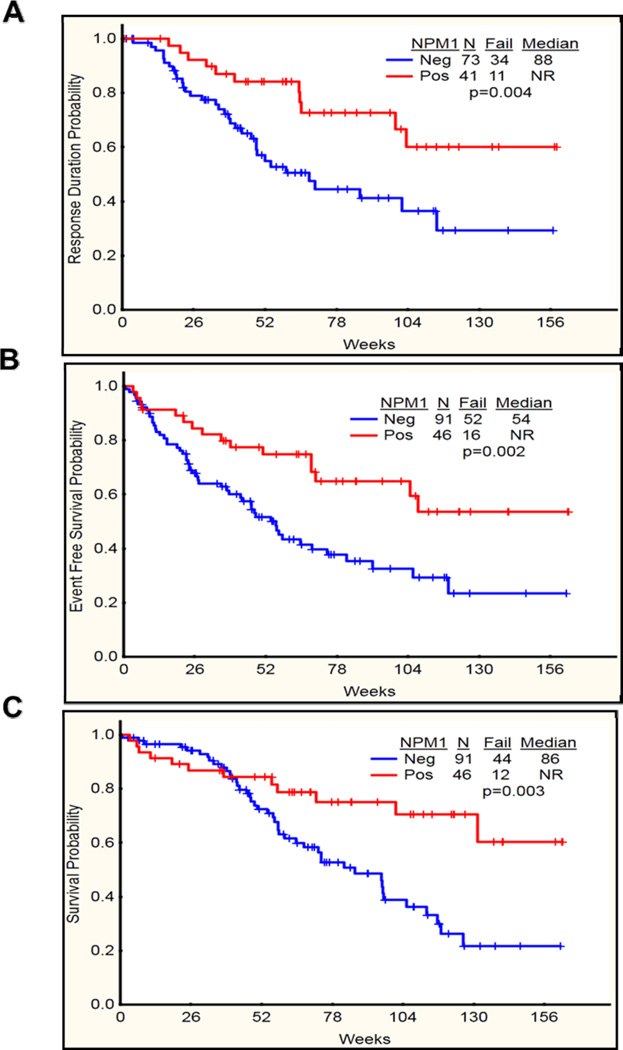
Among patients with NK, those with NPM1 mutations had a higher proportions (89%) achieving CR, as compared to those with wild type NPM1 (80%; P=NS) (Table 4). They also had a significantly longer CRD (P=0.004) (Figure 4A). Both EFS and OS were also significantly longer in patients with mutated NPM1 (P =0.003 and 0.002 respectively) (Figure 4B–C). When analyzed by age, in patients < 60 years (n=60), OS, EFS and CRD were significantly superior in NPM1 mutated subgroup (p=0.007, 0.007, 0.02 respectively), while among patients ≥ 60 years (n=77) none were significantly higher in the NPM1 mutated subgroup except that CRD showed a trend of being longer in NPM1 mutated as compared to NPM1 wild type (104 weeks vs 60 weeks respectively) (Table 2).
Table 4.
Prognostic significance of NPM1 mutations (normal karyotype)
| Parameter | NPM1 WT | NPM1 MUT | Total | P value |
|---|---|---|---|---|
| CR/CRp | 73 (80) | 41 (89) | 114 (83) | |
| NR | 18 (20) | 5 (11) | 23 (17) | 0.19 |
| CRD (wk) | 68 | NR | 0.004 | |
| OS (wk) (n=137) | 86 | NR | 0.003 | |
| OS (wk) (<60 yrs) (n=60) | 113 | NR | 0.007 | |
| OS (wk) (≥60 yrs) (n=77) | 74 | 101 | 0.3 | |
| EFS (wk) | 54 | NR | 0.002 | |
| EFS (wk) (<60 yrs) | 58 | NR | 0.007 | |
| EFS (wk) (≥60 yrs) | 54 | 105 | 0.13 |
Legends – WT: wild type; MUT: mutant; CR: complete response; CRp: complete response without platelet recovery; NR: no response; CRD: CR duration; OS: overall survival; EFS: event-free survival; wk: week
Figure 4.
(A-C). Complete remission duration (CRD), event free survival (EFS) and overall survival (OS) in NPM1 mutated (Positive) vs NPM1 wild type (Negative) among patients with AML with NK karyotype (DK) (n=137). A) CRD significantly longer in NPM1 mutated as compared to NPM1 wild type (P=0.004). B) EFS significantly longer in NPM1 mutated as compared to NPM1 wild type (P=0.002) C) OS significantly longer in NPM1 mutated as compared to NPM1 wild type (P=0.003).
NPM1 mutations at CR and at relapse
Among the 300 patients (including 91 with NK) with wild type NPM1 at diagnosis, none acquired a mutated NPM1 clone, either at CR or at the time of relapse suggesting that NPM1 mutations are stable. Among the 60 patients who had NPM1 mutation at the time of diagnosis, 39 patients (including 30 with NK) had available NPM1 status at the time of CR and all (100%) were negative for the mutated clone (Figure 1A–B). Among these 39 patients who have achieved a NPM1 negative status at CR, 10/39 (25%) patients overall [7/30 (23%) NK patients] relapsed. NPM1 status was available for 8 patients overall including 6 with NK at the time of relapse. Among them, 7/8 (87%) overall [including 5/6 (83%) NK patients] had mutated NPM1, while 1/8 (12%) [1/6 (16%) NK patients] remained wild type for NPM1. This patient relapsed with extramedullary disease (leukemia cutis) without any BM involvement.
Seven patients relapsed with NPM1 mutated leukemia including 3 with acquired FLT3 mutation; 3 had wild type FLT3 and 1 in whom FLT3 mutational analysis was not done. Among these patients, the distribution of other mutations was N-RAS: 4/7 wild type and 3 not done, IDH1: 1/2 wild type and 1 mutated, IDH2: 2/2 wild type, CEBPA: 2/2 wild type and KIT: 2/2 wild type.
Among the 60 patients with mutated NPM1 at baseline, 17 relapsed. Of these 17 patients, only 13 had mutated NPM1 at the time of relapse. We compared the characteristics of these 17 patients with those NPM1 mutated patients who did not relapse (n=37)(Table 5). Among these 17 patients, 8 achieved a second CR, 6 were non-responders (NR) and 3 were not evaluable for response. Their median survival was 33 weeks (range 1–131 weeks).
Table 5.
Characteristics of NPM1 mutated patients (relapsed vs. those remaining in remission)
|
Remission N=37 |
Relapsed N=17 |
P value | |
|---|---|---|---|
| Diploid (n) | 30 | 11 | |
| Other (n) | 7 | 6 | 0.19 |
| FLT3+ | 15 | 6 | |
| FLT3− | 21 | 10 | 0.78 |
| RAS+ | 8 | 2 | |
| RAS− | 25 | 12 | 0.45 |
| P.S. | |||
| 0–1 | 33 | 16 | |
| 2–3 | 4 | 1 | 0.56 |
| Age (Yrs) | 57 (27–75) | 60 (23–77) | 0.54 |
| WBC (K/µL) | 6.2 (1.1–186.5) | 8.5 (1.6–228.5) | 0.32 |
DISCUSSION
In this study, we have examined and confirmed the prognostic relevance of NPM1 mutations in previously untreated patients with AML who were evaluated and treated at our institute. We have also examined (among patients with evaluable samples) the fate of NPM1 mutations by analyzing BM samples at diagnosis, remission and at relapse. There are some shortcomings in our study including lack of serial samples in all the patients studied, use of a semi quantitative low sensitivity technique rather than a more sensitive quantitative method for detecting NPM1 mutations, and lack of subtyping of NPM1 mutations. Although, this type of analysis has been reported by other groups in the literature, we believe that this study further confirms that NPM1 mutations are stable markers of the disease and, unlike FLT3 mutations, do not fluctuate during the disease course, in particular, at relapse.
Monitoring minimal residual disease (MRD) by detection of mutations present in the leukemic clone is being increasingly utilized for the management of patients with leukemia.[1, 36] Various techniques and markers have been studied to detect MRD.[37] NPM1 mutations are an ideal target for use in clinical practice as they are present in about 40–50% of patients with de novo AML.[11, 38] Other mutations such as FLT3-ITD have been evaluated but their applicability to common practice is controversial.[39] Here, we have shown that none of the 300 patients who were initially negative for mutated NPM1 were positive at the time of CR or became positive at the time of relapse (100% of patients with evaluable samples retained their initial NPM1 status). Furthermore, among the 39 evaluable patients with mutated NPM1 at diagnosis, all became negative at the time of CR and the majority (>85%) regained the mutated NPM1 clone at the time of relapse; one patient with only extramedullary relapse remained negative. Our findings are consistent with two other large studies which have evaluated paired samples of NPM1 mutated patients at the time of CR and relapse.[8, 32] However, two other studies have reported that NPM1 mutations do not always reappear at the time of relapse; although this occurred in only 2/17 and 2/27 relapsed patients in these studies. Furthermore, in one study, the lack of NPM1 mutated clone at relapse was potentially attributable to therapy and clonal evolution.[19, 20]. In addition, we also found that all of our patients (irrespective of whether they were originally mutated or wild type for NPM1) were negative for the mutant clone at the time of CR, further supporting the usefulness of NPM1 mutations as a marker for MRD.
The characteristics of our patients with mutated NPM1 and its prognostic relevance are consistent with other published studies.[2, 9–11, 13] In our experience, presence of mutated NPM1 predicted for significantly longer CRD, EFS, and OS in the overall population and in patients with normal karyotypes (NK) (Figures 2 and 4, respectively). Furthermore, we confirm the findings from other groups that NPM1+/FLT3 (ITD) patients have significantly inferiors EFS and OS as compared with other subgroups. Of note, however, is that in our study, the patients with mutated NPM1 constituted only 33% (46/137) of those with NK which is lower than what has been reported in a number of other published reports (45–50% NPM1 mutated in NK AML)[2, 9–11, 13] but consistent with several other studies,[20, 21, 40, 41] including a prior study from our center.[28] This difference could possibly be explained by a higher proportion of older patients in our cohort (185 older than 60 compared with 175 who were <60). This is consistent with the reports that the incidence of NPM1 mutations decreases with advancing age [42] with some exceptions as in the study by Becket et al.[43]
In our experience patients with mutated NPM1 have a favorable prognosis and mutated NPM1 is a stable and reliable marker which does not fluctuate and can detect relapse in patients with AML harboring the mutation. NPM1 status should be further evaluated for routine use as a potential MRD marker among the commonly available markers in management of patients with AML. Prospective studies with high sensitivity assay to detect NPM1 mutations should be designed to better define the dynamics of NPM1 mutations in predicting relapse, and to prospectively evaluate NPM1 in conjunction with other molecular markers to develop a prognostic model and molecular MRD panel in patients with AML.
Footnotes
AUTHORSHIP
P.J. and F.R. designed the research.
J.C., S.F., T.K., G.B., N.D., N.P., H.K., and F.R. contributed patient samples.
K.P. and R.L. performed the NPM1 mutational analysis.
P.J., S.P., and F.R. analyzed and interpreted the data.
P.J., O.B. and F.R. wrote the manuscript.
All authors reviewed and gave the final approval for the paper.
DISCLOSURES
Authors declare no relevant conflict of interest.
REFERENCES
- 1.Patel JP, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlenk RF, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18):1909–1918. doi: 10.1056/NEJMoa074306. [DOI] [PubMed] [Google Scholar]
- 3.Grossmann V, et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood. 2012;120(15):2963–2972. doi: 10.1182/blood-2012-03-419622. [DOI] [PubMed] [Google Scholar]
- 4.Estey EH. Acute myeloid leukemia: 2012 update on diagnosis, risk stratification, and management. Am J Hematol. 2012;87(1):89–99. doi: 10.1002/ajh.22246. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi S. Current findings for recurring mutations in acute myeloid leukemia. J Hematol Oncol. 2011;4:36. doi: 10.1186/1756-8722-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gianfelici V, Lahortiga I, Cools J. Chromosomal aberrations and fusion genes in myeloid malignancies. Expert Rev Hematol. 2012;5(4):381–393. doi: 10.1586/ehm.12.30. [DOI] [PubMed] [Google Scholar]
- 7.Wakita S, et al. Mutations of the epigenetics modifying gene (DNMT3a, TET2, IDH1/2) at diagnosis may induce FLT3-ITD at relapse in de novo acute myeloid leukemia. Leukemia. 2012 doi: 10.1038/leu.2012.317. [DOI] [PubMed] [Google Scholar]
- 8.Kronke J, et al. Monitoring of minimal residual disease in NPM1-mutated acute myeloid leukemia: a study from the German-Austrian acute myeloid leukemia study group. J Clin Oncol. 2011;29(19):2709–2716. doi: 10.1200/JCO.2011.35.0371. [DOI] [PubMed] [Google Scholar]
- 9.Falini B, et al. Cytoplasmic nucleophosmin in acute myelogenous leukemia with a normal karyotype. N Engl J Med. 2005;352(3):254–266. doi: 10.1056/NEJMoa041974. [DOI] [PubMed] [Google Scholar]
- 10.Schnittger S, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106(12):3733–3739. doi: 10.1182/blood-2005-06-2248. [DOI] [PubMed] [Google Scholar]
- 11.Thiede C, et al. Prevalence and prognostic impact of NPM1 mutations in 1485 adult patients with acute myeloid leukemia (AML) Blood. 2006;107(10):4011–4020. doi: 10.1182/blood-2005-08-3167. [DOI] [PubMed] [Google Scholar]
- 12.Dohner K, et al. Mutant nucleophosmin (NPM1) predicts favorable prognosis in younger adults with acute myeloid leukemia and normal cytogenetics: interaction with other gene mutations. Blood. 2005;106(12):3740–3746. doi: 10.1182/blood-2005-05-2164. [DOI] [PubMed] [Google Scholar]
- 13.Verhaak RG, et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood. 2005;106(12):3747–3754. doi: 10.1182/blood-2005-05-2168. [DOI] [PubMed] [Google Scholar]
- 14.Shamaa S, et al. Prognostic implications of NPM1 mutations and FLT3 internal tandem duplications in Egyptian patients with cytogenetically normal acute myeloid leukemia. Hematology. 2013 doi: 10.1179/1607845413Y.0000000085. [DOI] [PubMed] [Google Scholar]
- 15.Pratcorona M, et al. Favorable outcome of patients with acute myeloid leukemia harboring a low-allelic burden FLT3-ITD mutation and concomitant NPM1 mutation: relevance to post-remission therapy. Blood. 2013;121(14):2734–2738. doi: 10.1182/blood-2012-06-431122. [DOI] [PubMed] [Google Scholar]
- 16.Vardiman JW, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114(5):937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 17.Falini B, Sportoletti P, Martelli MP. Acute myeloid leukemia with mutated NPM1: diagnosis, prognosis and therapeutic perspectives. Curr Opin Oncol. 2009;21(6):573–581. doi: 10.1097/CCO.0b013e3283313dfa. [DOI] [PubMed] [Google Scholar]
- 18.Falini B, et al. Acute myeloid leukemia with mutated nucleophosmin (NPM1): is it a distinct entity? Blood. 2011;117(4):1109–1120. doi: 10.1182/blood-2010-08-299990. [DOI] [PubMed] [Google Scholar]
- 19.Papadaki C, et al. Monitoring minimal residual disease in acute myeloid leukaemia with NPM1 mutations by quantitative PCR: clonal evolution is a limiting factor. Br J Haematol. 2009;144(4):517–523. doi: 10.1111/j.1365-2141.2008.07488.x. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, et al. Clinical characteristics and prognostic implications of NPM1 mutations in acute myeloid leukemia. Blood. 2005;106(8):2854–2861. doi: 10.1182/blood-2005-04-1733. [DOI] [PubMed] [Google Scholar]
- 21.Kuzmanovic M, et al. Prognostic Impact of NPM1 Mutations in Serbian Adult Patients with Acute Myeloid Leukemia. Acta Haematol. 2012;128(4):203–212. doi: 10.1159/000339506. [DOI] [PubMed] [Google Scholar]
- 22.Boissel N, et al. Prevalence, clinical profile, and prognosis of NPM mutations in AML with normal karyotype. Blood. 2005;106(10):3618–3620. doi: 10.1182/blood-2005-05-2174. [DOI] [PubMed] [Google Scholar]
- 23.Colombo E, Alcalay M, Pelicci PG. Nucleophosmin and its complex network: a possible therapeutic target in hematological diseases. Oncogene. 2011;30(23):2595–2609. doi: 10.1038/onc.2010.646. [DOI] [PubMed] [Google Scholar]
- 24.Borer RA, et al. Major nucleolar proteins shuttle between nucleus and cytoplasm. Cell. 1989;56(3):379–390. doi: 10.1016/0092-8674(89)90241-9. [DOI] [PubMed] [Google Scholar]
- 25.Federici L, Falini B. Nucleophosmin mutations in acute myeloid leukemia: A tale of protein unfolding and mislocalization. Protein Sci. 2013 doi: 10.1002/pro.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Colombo E, et al. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat Cell Biol. 2002;4(7):529–533. doi: 10.1038/ncb814. [DOI] [PubMed] [Google Scholar]
- 27.Bertwistle D, Sugimoto M, Sherr CJ. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol Cell Biol. 2004;24(3):985–996. doi: 10.1128/MCB.24.3.985-996.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konoplev S, et al. Cytoplasmic localization of nucleophosmin in bone marrow blasts of acute myeloid leukemia patients is not completely concordant with NPM1 mutation and is not predictive of prognosis. Cancer. 2009;115(20):4737–4744. doi: 10.1002/cncr.24543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greiner J, et al. Mutated regions of nucleophosmin 1 elicit both CD4(+) and CD8(+) T-cell responses in patients with acute myeloid leukemia. Blood. 2012;120(6):1282–1289. doi: 10.1182/blood-2011-11-394395. [DOI] [PubMed] [Google Scholar]
- 30.Grimwade L, et al. Detection of cytoplasmic nucleophosmin expression by imaging flow cytometry. Cytometry A. 2012;81(10):896–900. doi: 10.1002/cyto.a.22116. [DOI] [PubMed] [Google Scholar]
- 31.Thol F, et al. Next-generation sequencing for minimal residual disease monitoring in acute myeloid leukemia patients with FLT3-ITD or NPM1 mutations. Genes Chromosomes Cancer. 2012;51(7):689–695. doi: 10.1002/gcc.21955. [DOI] [PubMed] [Google Scholar]
- 32.Schnittger S, et al. Minimal residual disease levels assessed by NPM1 mutation-specific RQ-PCR provide important prognostic information in AML. Blood. 2009;114(11):2220–2231. doi: 10.1182/blood-2009-03-213389. [DOI] [PubMed] [Google Scholar]
- 33.Ommen HB, et al. Strikingly different molecular relapse kinetics in NPM1c, PML-RARA, RUNX1-RUNX1T1, and CBFB-MYH11 acute myeloid leukemias. Blood. 2010;115(2):198–205. doi: 10.1182/blood-2009-04-212530. [DOI] [PubMed] [Google Scholar]
- 34.Kristensen T, et al. NPM1 mutation is a stable marker for minimal residual disease monitoring in acute myeloid leukaemia patients with increased sensitivity compared to WT1 expression. Eur J Haematol. 2011;87(5):400–408. doi: 10.1111/j.1600-0609.2011.01673.x. [DOI] [PubMed] [Google Scholar]
- 35.Cheson BD, et al. Revised recommendations of the International Working Group for Diagnosis, Standardization of Response Criteria, Treatment Outcomes, and Reporting Standards for Therapeutic Trials in Acute Myeloid Leukemia. J Clin Oncol. 2003;21(24):4642–4649. doi: 10.1200/JCO.2003.04.036. [DOI] [PubMed] [Google Scholar]
- 36.Ravandi F, Jorgensen JL. Monitoring minimal residual disease in acute myeloid leukemia: ready for prime time? J Natl Compr Canc Netw. 2012;10(8):1029–1036. doi: 10.6004/jnccn.2012.0105. [DOI] [PubMed] [Google Scholar]
- 37.Buccisano F, et al. Prognostic and therapeutic implications of minimal residual disease detection in acute myeloid leukemia. Blood. 2012;119(2):332–341. doi: 10.1182/blood-2011-08-363291. [DOI] [PubMed] [Google Scholar]
- 38.Palmisano M, et al. NPM1 mutations are more stable than FLT3 mutations during the course of disease in patients with acute myeloid leukemia. Haematologica. 2007;92(9):1268–1269. doi: 10.3324/haematol.11202. [DOI] [PubMed] [Google Scholar]
- 39.Bacher U, et al. Prognostic relevance of FLT3-TKD mutations in AML: the combination matters--an analysis of 3082 patients. Blood. 2008;111(5):2527–2537. doi: 10.1182/blood-2007-05-091215. [DOI] [PubMed] [Google Scholar]
- 40.Boonthimat C, Thongnoppakhun W, Auewarakul CU. Nucleophosmin mutation in Southeast Asian acute myeloid leukemia: eight novel variants, FLT3 coexistence and prognostic impact of NPM1/FLT3 mutations. Haematologica. 2008;93(10):1565–1569. doi: 10.3324/haematol.12937. [DOI] [PubMed] [Google Scholar]
- 41.Chou WC, et al. Nucleophosmin mutations in de novo acute myeloid leukemia: the age-dependent incidences and the stability during disease evolution. Cancer Res. 2006;66(6):3310–3316. doi: 10.1158/0008-5472.CAN-05-4316. [DOI] [PubMed] [Google Scholar]
- 42.Schneider F, et al. Age-dependent frequencies of NPM1 mutations and FLT3-ITD in patients with normal karyotype AML (NK-AML) Ann Hematol. 2012;91(1):9–18. doi: 10.1007/s00277-011-1280-6. [DOI] [PubMed] [Google Scholar]
- 43.Becker H, et al. Favorable prognostic impact of NPM1 mutations in older patients with cytogenetically normal de novo acute myeloid leukemia and associated gene- and microRNA-expression signatures: a Cancer and Leukemia Group B study. J Clin Oncol. 2010;28(4):596–604. doi: 10.1200/JCO.2009.25.1496. [DOI] [PMC free article] [PubMed] [Google Scholar]