Abstract
In humans over 15% of X-linked genes have been shown to ‘escape’ from X-chromosome inactivation (XCI): they continue to be expressed to some extent from the inactive X chromosome. Mono-allelic expression is anticipated within a cell for genes subject to XCI, but random XCI usually results in expression of both alleles in a cell population. Using a study of allelic expression from cultured lymphoblasts and fibroblasts, many of which showed substantial skewing of XCI, we recently reported that the expression of genes lies on a contiunuum between those that are subject to inactivation, and those that escape. We now review allelic expression studies from mouse, and discuss the variability in escape seen in both humans and mice in genic expression levels, between X chromosomes and between tissues. We also discuss current knowledge of the heterochromatic features, DNA elements and three-dimensional topology of the inactive X that contribute to the balance of expression from the otherwise inactive X chromosome.
Keywords: allelic imbalance, boundary elements, dosage compensation, epigenetic marks, RNA-seq, waystations, XIST
Introduction
The inactivation of almost a thousand genes on one of the two essentially identical X chromosomes in female nuclei is perhaps the most striking example known of epigenetic gene silencing; however, not all of the genes on the inactive X chromosome (Xi) are silenced. In humans, approximately 15% of X-linked genes escape from X-chromosome inactivation (XCI) and are likely principal contributors to the phenotypes of humans with X-chromosome aneuploidies. In mice, a smaller percentage (perhaps 3%) of genes escape from XCI, consistent with the viability and limited phenotypic consequences for female sex chromosome monosomy (reviewed in [1]). It has, however, been argued that only a subset of X-linked genes, in particular those that are involved in large protein complexes, need to be dosage compensated [2]; and similarly, a lack of dosage compensation for those genes that escape from XCI may only have phenotypic consequences for a subset of genes. In general, there is strong synteny of the content of the X chromosome amongst eutheria, although the pseudoautosomal regions (PARs), which continue to recombine between the X and Y chromosomes, differ between species [3]. To date, it is not known why human and mouse differ so substantially in the number of genes that escape from XCI.
Escape from XCI is not an absolute with either full or no expression from the Xi; rather, in both humans and mice, variability has been shown in the extent of expression, as well as differences between X chromosomes and between tissues as to which genes are expressed. Therefore, escape from XCI will not only result in differences in expression between males and females, but also between tissues and between females, which could have important implications for disease predispositions between men and women, or amongst women. Furthermore, XCI has long served as a paradigm for heterochromatin formation, and understanding how genes escape from XCI can inform our understanding of how silencing spreads, and how certain regions of the X chromosome evade inactivation.
The Xi is facultative heterochromatin; as in the case of classical position effect variegation, the dogma has been that inactivation spreads along the chromosome. The ability of silencing to spread incompletely into autosomal material translocated to the X chromosome led to the description of an X inactivation centre (XIC) from which inactivation initiated, and waystations – booster elements that support and extend the capacity for spread along the X chromosome [4]. A candidate for the initiating factor in the XIC is the long non-coding RNA XIST, and long interspersed nuclear elements (LINEs) have been proposed as potential waystations. Characterisation of cis-regulatory elements in humans has been hindered by the lack of a tractable developmental model system, as there is considerable epigenetic instability in human female embryonic stem (ES) cells that have often already undergone XCI (e.g. [5]). On the other hand, in female mouse ES cells, one X chromosome undergoes XCI during differentiation [6], thus providing a system to study XCI in culture as well as in mouse models.
The fundamental mechanisms of XCI and the marks of an Xi appear to be very similar between mice and humans. However, in addition to the differing number of genes that escape from XCI, there are a few key divergences in the XIC regions, including the regulatory function of the Xist antagoniser Tsix [7, 8], and the timing of XCI and presence of imprinted XCI in extraembryonic tissues (reviewed in [9]). Furthermore, another potential source of variability is the long non-coding RNA termed XACT, which transiently coats the active X chromosome (Xa) but has only been identified in humans and chimpanzees [10]. Despite these differences, the success of using human XIST transgenes to recapitulate XCI in the mouse [11, 12], and the recent demonstration that escape from XCI of human genes is possible from the mouse X chromosome [13], have illustrated the importance of manipulation of mouse models for understanding human XCI. The ability to follow the initial steps of XCI in mouse has been crucial in identifying the cascade of events during XCI, including recent examinations of the chromosome topology. These have revealed new insights into Xi chromatin and epigenetic interactions, an important step towards understanding the process of silencing – and escape.
Human genes that escape from XCI correlate with evolutionary history of the sex chromosomes
XCI achieves dosage equivalence between the two X chromosomes in females and the single X in males, and thus the need for XCI is believed to have been driven by the loss of genes from the Y chromosome, which also provided pressure to up-regulate expression levels from the Xa [14]. Genes in the PAR are prime examples of genes that escape from XCI [15]; however, evidence that a significant portion of human X-linked genes might escape from XCI accumulated as more genes outside the PAR1 were identified that were expressed from the Xi (e.g. STS [16]). In 2005, Carrel and Willard published the first large ‘survey’ of the XCI status of 634 X-linked human transcripts using two different methods [17]. The first method relied upon rodent/human somatic cell hybrids that retained either a human Xa or Xi. The second method examined the expression of polymorphisms from each X chromosome in human cell lines that had non-random or clonal XCI. In a female with clonal XCI, a gene that is subject to XCI will have mono-allelic expression whereas a gene that escapes from XCI will show bi-allelic expression proportional to the degree of escape. Cell lines with clonal XCI were obtained by examining fibroblasts from females with structurally abnormal X chromosomes, although other studies have used carriers of X-linked disease, selection, or clonal expansion from a single cell to achieve the same purpose. Escape from XCI and the level of Xi expression was shown to be strongly correlated with X and Y chromosome homology that in turn is related to the length of time since the genes on the X and Y chromosomes diverged [3, 17, 18].
Mouse RNA-seq studies demonstrate variability in escape from XCI
The growing plethora of genome-wide studies has provided further opportunities to identify genes that escape from XCI. Mouse RNA-seq studies in cells with non-random XCI have validated that mice have fewer genes that escape from XCI than humans and have laid the foundation for expanding RNA-seq analysis into humans [19–22]. As shown in Table 1, 31 X-linked mouse genes have shown some evidence for escape from XCI (defined as either greater than 10% expression from the Xi or bi-allelic expression) in brain, neural stem cell and/or kidney cell lines. Thirteen genes (highlighted in bold in Table 1) showed escape from XCI across the majority of studies. Variation between studies may be caused by several factors including: differences in genic Xi expression levels; differences between X chromosomes, differences between tissues and/or differences between females (illustrated in Fig. 1).
Table 1.
Summary of mouse genes that escape from XCI based on RNA-seq data
| Mouse gene name | Brain cells [19] | Neural stem cells [21] | Embryonic kidney cells [20] | Trophoblast stem cells [22] | Human gene name | Hybrid escape [17] |
|---|---|---|---|---|---|---|
| Shroom4 | – | 0% | 69%musc. | 1% | SHROOM4 | 0/9 |
| Clcn5 | 7% | 0% | – | 0% | CLCN5 | 0/9 |
| Syp | 5% | 0% | – | – | SYP | 2/9 |
| Timm17b | 28% | 0% | – | 1% | TIMM17B | 0/9 |
| Gm4984 | 11%musc. | – | – | – | – | – |
| 1810030O07Rik | 3% | 0% | 13%musc. | 0% | CXorf38 | 8/9 |
| Ddx3x | 20% | 0% | 71%musc. | – | DDX3X | 9/9 |
| Kdm6aa | 46% | 75% | 87%musc. | 9% | KDM6A | 9/9 |
| Utp14a | 10% | 50% | – | 16% | UTP14A | 3/9 |
| 6720401G13Rik | – | 0% | 130%musc. | – | – | – |
| Cdr1 | 9% | 0% | – | – | – | – |
| Hmgb3 | 4% | 0% | – | 0% | HMGB3 | 0/9 |
| Bgn | 6% | 0% | 25%musc. | – | BGN | 0/6 |
| Eif2s3x | 51% | 100% | 76%musc. | 28% | EIF2S3 | 9/9 |
| Med12 | 4% | 0% | 1% | – | MED12 | 0/9 |
| Taf1 | 6% | 0% | 0% | 37% | TAF1 | 0/5 |
| Chic1 | 5% | 0% | – | – | CHIC1 | 0/9 |
| Enoxb | 27% | – | – | 46% | JPX | – |
| 5530601H04Rik | 31% | 100% | – | 12%musc. | – | – |
| 2610029G23Rik | – | 50% | 77%musc. | 11%musc. | CXorf26 | 0/9 |
| Itm2a | 13%cast. | 0% | – | 5% | ITM2A | 5/9 |
| Wbp5 | 22%musc. | 0% | 0% | – | WBP5 | 4/9 |
| Gnl3l | 6% | – | – | 0% | GNL3L | 0/9 |
| Hsd17b10 | 3% | 0% | 3% | 0% | HSF17B10 | 1/9 |
| Kdm5cc | 29% | 75% | 43%musc. | 16% | KDM5C | 9/9 |
| Sat1 | 8% | 0% | – | 0% | SAT1 | 1/9 |
| Car5b | – | 0% | 41%musc. | 14%cast. | CA5B | 9/9 |
| Trappc2 | 3% | 0% | – | 6% | TRAPPC2 | 9/9 |
| BC022960 | – | – | 50%musc. | – | – | – |
| Mid1 | 52% | – | 184%musc. | 50%cast. | MID1 | 1/9 |
| G530011O06Rik | – | 33% | – | – | – | – |
| Trophoblast specific escape | – | – | – | 8 escape 7 escapemusc.1 escapecast | ||
| Total genes examined | 263 | 268 | 135 | 369 |
All studies examined both reciprocal crosses except for Yang et al. [20] in which only the B6 X chromosome was the Xi. The average percent Xi expression relative to the Xa is given for each examined gene in Wu et al. [19], Yang et al. [20] and Calabrese et al. [22]. For Li et al. [21], the percent of bi-allelic cell lines is given. Genes highlighted in bold suggest escape from XCI in the majority of studies. Superscripts of ‘musc.’ and ‘cast.’ indicate that escape from XCI was only observed in one mouse strain. Corresponding human homologues and the XCI status established in Carrel and Willard [17] are shown to the far right.
Kdm6a is also known as Utx.
Enox is also known as Jpx and 2010000I03Rik.
Kdm5c is also known as Jarid1c and Smcx.
Figure 1.
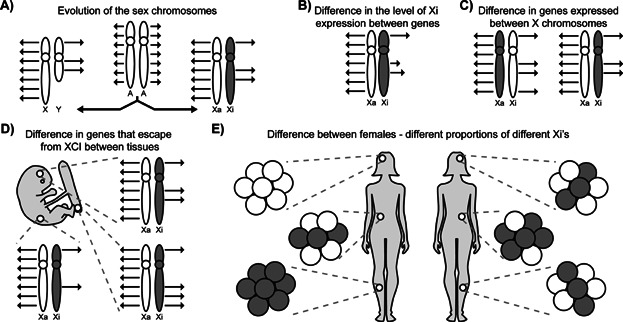
Variability in dosage compensation on the X chromosome. A: The X and Y chromosomes evolved from an ancestral pair of autosomes. After their divergence, expression of X-linked genes from the Xa was increased, and the majority of genes were silenced on the Xi. Note that only a subset of genes is illustrated in all figures. B: Not all genes that escape from XCI show the same level of expression. Longer arrows denote higher Xi expression but expression is still not equal to the Xa. C: A gene may escape from XCI on certain X chromosomes but not others. To simplify, variable Xi expression levels are not shown in parts (C) and (D). D: Escape from XCI may occur only in certain tissues for some genes. E: The level of skewing of XCI, as illustrated by grey and white circles, differs between females and tissues. As escape from inactivation can differ between X chromosomes (shown in C), skewing will alter the overall expression level.
A continuum of expression from the Xi, from zero to a maximum of approximately 70% of the level seen from the Xa was observed in humans [23], and similarly in Table 1, expression levels varied from the Xi in mice. It is not known whether all cells had Xi expression at the same level, or whether only a subset of cells showed full Xi expression while other cells showed none. Previous studies using RNA FISH suggested that even a consistent gene that escapes from XCI, Kdm5c, only showed expression from both X chromosomes in a small subset of cells [24]; while single-cell RT-PCR analysis of other genes in humans has demonstrated that expression from the Xi was present in all cells but highly varied [25].
These mouse RNA-seq studies used interspecies crosses to maximise the number of polymorphisms that could be examined, so it was possible to identify strain-specific escape from XCI, which is an example of the inter-chromosomal variability observed in escape from XCI between females (Fig. 1C). Seven mouse genes showed evidence for strain-specific escape, suggesting that escape from XCI may be influenced by features of the X chromosome present in one strain but not the other. In humans, Carrel and Willard [17] observed variability between females, and our survey suggested that approximately 13% of genes would variably escape in some but not all females and that an additional 10% of genes may show variable escape due to population differences in XCI [23].
Unlike the other mouse RNA-seq studies that examined somatic tissues, Calabrese et al. [22] examined trophoblast stem cells, which showed evidence for escape from XCI for an additional 22 genes. The higher degree of escape from XCI in trophoblast stem cells could be considered an extreme example of differences in XCI based on tissue (Fig. 1D), but also raises the question of whether escape from XCI is truly escape or if escape is rather reactivation; trophoblast has been reported to be more epigenetically plastic [26]. In the case of Kdm5c, there is initial silencing, but then rapid reactivation on the Xi [27], whereas Otc has been shown to reactivate slowly with age [28]. In humans, approximately 9% of genes show tissue-specific XCI [23], but little is known about early, or age-related reactivation.
Caveats in defining the variability in escape from XCI
Escape from XCI is also known to differ between females as a result of the above differences between X chromosomes and tissues, but with the added complexity of skewing of XCI (Fig. 1E). While mouse crosses can be undertaken with particular alleles to bias XCI towards the inactivation of a specific X chromosome (e.g. [29]), most human females have random XCI in all tissues including blood [30]. While new high throughput methodologies allow for the XCI status of more genes to be examined in an increasing number of tissues, studies examining non-clonal samples must take into account the level of skewing of XCI within each sample, which has not been done in all cases [31]. Studies using DNA methylation (DNAm) [32], which is acquired by the Xi but low to absent on the Xa, do not require clonal populations; however, studies of activity [23], or absence of active marks such as RNA polymerase II [33] must either use proven clonal cell lines or adjust for the extent of skewing of XCI.
We have discussed escape from XCI in humans and mice; however, escape from XCI is observed in other species. There are genes for which escape from XCI is conserved across species; but in general, it appears that rodents have fewer genes that escape from XCI than other species examined [24] although there are also genes that escape XCI only in primates (RPS4X, [34]). Characterizing the variability of escape from inactivation, whether between species, individuals, tissues or genes, will inform our understanding of how inactivation spreads to most, but not all, of the genes on the 160 Mb X chromosome.
“Escape” genes differ from “subject” genes in their associated chromatin features
In addition to observing a continuum of expression from the Xi, examination of allelic imbalance of histone modifications demonstrated a gradation of histone modifications on genes subject to or escaping from XCI [23]. More and more studies have revealed that chromatin modifications not only differ between the Xa and Xi, but that genes that escape from XCI show different patterns than genes subject to XCI on the Xi (see Table 2). The ability of inducible transgenes of XIST/Xist to trigger inactivation in humans [35] and mice [36], suggests that the XIST RNA is the initiating signal for the assembly of the facultative heterochromatin on the Xi, and a lack of Xist RNA has been noted on some genes that escape from XCI [37–39]. Whether this deficiency reflects an initial lack of spreading to these areas or whether it is actively cleared or passively lost at a later point in time remains unanswered, as there are suggestions that the initial spread of Xist and silencing may be more extensive [27, 40].
Table 2.
Summary of chromatin marks distinguishing genes subject to and escaping from XCI
| Feature | Subject (mouse and human) | Escape (mouse) | Escape (human) |
|---|---|---|---|
| XIST/Xist coating | Present [96–99] | Depleted Kdm5c, Kdm6a [37] a [38, 39] b | |
| H3K4 methylation | Depleted [100–103] | Present [45] b | Present UBA1 [46] a [23, 45] b |
| H3 and H4 acetylation | Depleted [100, 104, 105] | Present Kdm5c, Eif2s3x [76] a [104] b | Present EIF2SI3 [76] a XIST, ZFX, KDM5C [101] a UBA1 [46] a [23] b |
| H3K27 methylation | Present [103, 106] | Depleted [20] b | Depleted UBA1 [46] a [23] b |
| H2A ubiquitination | Present [107–109] | ||
| H3K9 methylation | Present [100, 101, 110–112] | Depleted UBA1 [46] a | |
| H4K20 methylation | Present [113] | Depleted UBA1 [46] a | |
| MacroH2A | Present [43, 114–116] | Depleted Xist, Eif2s3x, Kdm5c, Mid1 [47] a | |
| Replication timing | Late [48] | Early [49, 50] b | |
| DNAm of CpG islands in promoter region | Present [117–120] | Depleted Kdm5c [76] a | Depleted MIC2 [121] a; TIMP1 [122] a UBA1, PCTK1 [123] a [32, 60] b |
Features seen in genes subject to XCI are consistent between mouse and human, and are grouped together; while features associated with genes that escape from XCI are separate for mouse and human as several of the marks remain untested in one or the other species. References for escape features are specified based on the number of escape genes/domains studied (see footnote). Genes studied for features conferring XCI are not indicated. Alternate gene names are listed in Table 1, in addition UBA1 is also known as UBE1.
Studies examined specific genes (<5) that escape from XCI, gene names are listed.
Studies examined a group or domain of genes (>5) that escape from XCI.
Other modifications found to be associated with the Xi include the loss of active chromatin marks such as acetylation of histones and H3K4me3, as well as the gain of repressive histone marks including the polycomb repressive complex (PRC)-1-mediated H2A ubiquitination, H3K9me2/3 and the PRC2-mediated H3K27me3 (reviewed in [41]). Genomic regions of H3K27me3 and XIST were reported to recruit SMCHD1, which in turn interacts with the HP1-binding protein HBiX1 from the distinct H3K9me3 domain, bridging the two domains and working together to create the compact Xi structure [42], which can then recruit further marks such as DNAm. In addition, enrichment of the variant histones MacroH2A1 and MacroH2A2 was noted on the Xi [43], and other histone variants showed varying levels of enrichment or depletion [44]. Analysed genes that escape from XCI appear to have euchromatic histone features, while demonstrating depletion in the repressive marks [20, 23, 45, 46] and loss of MacroH2A recruitment compared to subject domains [47].
Late replication was an early feature associated with the Xi [48], and recent genome-wide assessment of replication timing has demonstrated that replication of the Xi occurs in an unstructured fashion in which origins fire randomly and replication finishes quickly despite its lagging start [49]. There was a significant exception to the randomness seen in an ∼8 Mb region on the distal short arm of the X chromosome, which replicated relatively early and with similar timing and structure for the homologous chromosome pairs in both females (Xa and Xi) and males (Xa and Y) [49]. Interestingly, this region contains the PAR1, which is actively expressed on both the Xi and Y, as well as a cluster of genes that escape from XCI in females [17]. In agreement with these findings, early studies of replication timing and transcriptional activity on the X chromosome showed evidence of earlier replication of putative escape regions [50], or of reactivated genes on the Xi (e.g. [51]), although the Xa and Xi have been shown to use the same origins of replication [52]. Advances to these high-resolution chromosome-wide methods will allow the closer examination of the replication timing of individual genes that escape from XCI.
DNA methylation is an indicator of XCI status in eutherians
DNAm was another epigenetic mark proposed quite early to be associated with XCI [53], and unlike the majority of CpG islands in the genome, CpG islands at the promoters of genes subject to XCI are heavily methylated on the Xi, with the exception of genes that escape from XCI, which tend to be hypomethylated (e.g. [54]). Interestingly, the CpG island promoters of X-linked genes are biased against the strongest class of CpG island promoters, which were generally associated with broadly expressed housekeeping genes and strong GC skew, suggesting that a high GC content was protective against DNAm. However, no correlation between CpG island promoter class and the ability of a gene to escape from XCI was observed [55].
In contrast to mice and humans, the Xi of marsupials lacks expression of an Xist homologue and DNAm, with only one X-linked gene, Rsx, identified as differentially methylated between the Xa and Xi [56]. Rsx was recently described as a large non-coding, repeat-rich gene sharing the Xist-like property of coating the Xi, and is capable of silencing in cis when integrated into mouse ES cells [57]. RNA-seq revealed that ∼14% of genes escape from XCI in marsupials, but the corresponding hypomethylation of the Xi means that DNAm is not a distinguishing factor between genes subject to and escaping from XCI in this species. However, a significant decrease in repressive H3K27me3 at genes that escape from XCI suggested that histone modifications still correlate with marsupial XCI [56], and may highlight an important difference in escape mechanisms between species.
DNAm has generally been considered to be a late event in XCI, and is often cited as a maintenance mechanism to lock in silencing; closer analyses of several X-linked CpG islands across the chromosome revealed that DNAm is actually established in two separate waves [58]. In addition to the Smchd1-dependent pathway that is acquired gradually over an extended period [59], Smchd1-independent DNAm occurs more rapidly after onset of XCI [58]. Given that XCI marks such as DNAm are currently being used as tests for predicting the Xi status of an X-linked gene (e.g. [60]), timing of their recruitment to the Xi as well as the mechanism of their establishment are important processes to be kept in mind.
Many of the features tested so far have correlated well with expression data, as in mouse, particularly mouse ES cells, it has been possible to determine when different features become visible on the Xi, and whether they are lacking in genes that escape from XCI. There is redundancy as well as co-operativity between many of the marks of XCI, since removal of a single silencing feature does not lead to complete reactivation of the entire X chromosome as demonstrated in several studies [61–66]. Understanding the interplay of heterochromatin formation on the Xi and the synergy of marks would provide insight into the silencing pathways, and perhaps offer clues to the right combination of marks needed to tip the scale to favour escape from XCI.
DNA sequences associated with escape from XCI
The consistent ability of some genes to avoid the silencing and associated heterochromatic features of XCI suggests that the genes' neighbourhood is an important contributor to expression. The analysis of spread of silencing into autosomal material had suggested that there might be waystations, which Lyon proposed to be LINEs [67]. Recently, using high-density arrays to profile DNAm as a mark of silencing, spread into autosomal material was examined at a high resolution. Cotton et al. found an enrichment of L1s within 100 kb of genes subject to XCI; however the enrichment of Alu elements in gene regions escaping from XCI was more significant [68]. A similar study of six different translocations also observed an enrichment of Alu, as well as simple repeats, around genes that escape from XCI [69], and that L1 and L2 classes of LINEs were enriched around genes subject to XCI in most translocations, the most consistently enriched short motif being derived from recently active L1 elements. An enrichment for the younger L1 subclasses on the X chromosome in general – and around genes known to escape from XCI – had been noted earlier [70]. Using a larger catalogue of genes that escape from XCI [17] an enrichment for Alu and short ACG/CGT motifs was observed around genes that escape from XCI, while a number of repeat features were enriched around genes that were subject to XCI, with L1 and L2 having the greatest discriminatory power to predict whether a gene was subject to XCI [71]. In mice, comparison of a gene that is X-linked in Mus spretus and autosomal in Mus musculus demonstrated enrichment of an AT motif on the X-linked version that was subject to XCI [72]. Several mouse protein coding genes which escape from XCI have been found to be located in close proximity to long non-coding RNAs, which escape from XCI suggesting that long non-coding RNAs may also play a role in determining escape from XCI [73]. A subset of young L1 elements were shown to be expressed during XCI in differentiating mouse ES cells, and enriched at the boundaries of escape regions, where they were suggested to be facilitating the spread of XCI [74].
The assessment of the DNA sequences flanking genes that resist inactivation has given clues to the potential identity of putative waystations. In contrast, recurrent integration of a BAC containing a mouse gene that escaped from XCI provided evidence supporting intrinsic elements that promote expression from the otherwise silent Xi [75]. Comparisons of human and mouse regions escaping XCI have suggested a role for CTCF in boundary regions between escape and subject [76], although it has generally been considered that these boundaries are preventing spread of silencing, rather than spread of escape. Interestingly, a deletion of a boundary in the transgene that demonstrated intrinsic escape from XCI led to spread of escape into adjacent genes [77], suggesting that boundary elements may also block spread of euchromatin.
While these studies have shown consistent correlation of elements, notably L1/2 with subject and Alu with escape, there has not been clear identification of a waystation or other element. Overall, we need to dig deeper into the structure of the X chromosome and the process of XCI to understand what differs between the genes that are consistently subject to XCI and those that escape from XCI to various extents; but we will need to bear in mind that the genomic distributions of many DNA elements are likely not independent of each other.
There is a step-wise spreading of Xist RNA across the X
Genes that escape from XCI have been noted to be depleted in Xist [37], raising the question of how Xist RNA spread occurs temporally and spatially. In order to address how the Xist RNA ‘coats’ the ∼150 Mb chromosome researchers have utilised biotinylated antisense probes to isolate DNA regions interacting with the noncoding RNA. Engreitz et al. [39] showed that the initial localisation of Xist was to inactive genes in gene-rich domains that were in close proximity to the Xist gene as determined by chromosome conformation studies [78]. The regions with the highest Xist enrichment also had higher H3K27me3, and as XCI proceeded the Xist localisation spread to active gene-rich regions. Intriguingly, while the initial localisation was not silencing-dependent, the spread to actively transcribed regions required a transcriptionally competent Xist RNA [39]. The elegance of these studies demonstrated the power of using mouse ES cells in which transgenes and knockouts can be created and followed over the initial stages of XCI. Interestingly, only four of the 13 consistent genes that escape from XCI from Table 1 were found on the list of 53 genes noted to be depleted for Xist, suggesting that lack of Xist interaction is not the sole source of escape from XCI.
With a related capture approach, Simon et al. [38] observed a two-step spread of Xist. Once again the first localisation was detectable at gene-rich islands, before spreading to the intervening gene-poor regions [38]. Depletion of Xist from the gene bodies of genes that escape from XCI was observed, while the earliest domains to recruit Xist were active gene-rich domains, and as generally seen for active regions, correlated with an enrichment for short interspersed nuclear elements (SINEs) and reduction in LINEs, early replication and DNase I hypersensitivity. A modest enrichment for chromosome conformation-defined contacts was again observed. Spreading of XCI was also suggested to occur through a hierarchy of PRC2 binding sites. Allele-specific PRC2 binding was examined during ES cell differentiation and ∼150 initial strong sites of enrichment were observed [79], which were concentrated at bivalent ES cell domains coinciding with CpG islands. Interestingly, these strong PRC2 sites did not definitively correlate with Xist interaction [38], although the subsequent spread to ∼400 PRC2 sites did show a correlation with Xist binding. A clear correlation was observed across differentiation between PRC2 and H3K27me3 enrichment [38], with targeting of H3K27me3 to active regions, and subsequent loss of H3K4me3 being more important in the maintenance of XCI in somatic cells [80]. While direct interaction of the Xist repeat A region with PRC2 has been reported [81], more recently spatial separation between Xist and PRC2 has been observed along the Xi [82], and Jarid2 has been implicated as a mediator of PRC2 recruitment by Xist [83].
Initial tethering of Xist has been shown to involve binding of both the RNA and the DNA by the transcription factor YY1 [84]. From this foothold, the Xist RNA spreads to target closely interacting gene-rich regions and recruits a hierarchy of PRC2 sites that in turn establishes H3K27me3 and silencing, followed by spread of Xist and H3K27me3 more fully across the Xi. An unsolved question is how Xist itself manages to avoid being silenced. The spreading of Xist leads to one of the earliest events in XCI, the creation of a silent nuclear compartment, depleted of RNA polymerase II and transcription. This inner compartment of the Xi was predicted to consist mainly of silent repeats both in mice [85] and in humans [86], with genes located more to the periphery; however upon silencing genes appeared to be drawn into this condensed Xist-dense core. Recently, repeat-rich stable nuclear RNAs have been found to be associated with the scaffold of euchromatic chromosomes [87]. Such RNAs are excluded from the Xi; however, they are reminiscent of the XACT RNA association with the Xa early in development [10], potentially demonstrating an interplay between non-coding RNAs, chromatin remodelling and the three-dimensional topology of the chromosome.
Conformation of the Xi differs from the Xa
Chromosomes have long been seen to have different locations in the nucleus, and the Xi is a striking example, being preferentially found at the nuclear or nucleolar periphery [88, 89]. Targeting of the Xi, or even Xic-containing transgenes to the SNF-rich perinucleolar region was critical for the maintenance of silencing [88]. Recent advances in molecular approaches to dissect the subchromosomal three-dimensional structure of DNA within the nucleus by examining the intranuclear conformation of chromosomes involve capturing contacts between distant regions of the genome with a cross-linking technique. Interrogation of these contacts has been accomplished through a variety of chromosome conformation approaches (reviewed in [90, 91]) revealing a separation into topological domains of different sizes.
Analysis of chromosome conformation around the Xic in undifferentiated and differentiated mouse ES cells revealed topologically associating domains (TADs), which aligned with H3K27me3 and H3K9me2 [92]. Boundaries between alternate topological domains were enriched for CTCF, housekeeping genes, tRNAs and SINEs [78]. Within the Xic region, the boundaries were often observed to contain CTCF, however CTCF sites are also seen within TADs, implicating additional features for functionality of boundaries, the deletion of which resulted in disruption of a TAD [92]. In differentiated cells, global organisation into TADs continued on the Xa, but the Xi lost most long-range contacts. This more random organisation of the Xi was consistent with the limited interactions observed with an allele-specific chromosome conformation approach [93]; however the genes that escaped from XCI also participated in long range contacts with each other. Such contacts suggested that 10 more genes might escape from XCI; however only two of these were found in the additional studies reviewed in Table 1 [22]. While such extensive studies have not been performed in humans, analysis of the spread of XCI into autosomal material showed that genes that were subject to (or escaped from) XCI clustered within TADs, and genes subject to XCI were more likely to be found in regions that have PRC2 and H3K27me3 marks normally on non-rearranged chromosomes [68].
Since Lyon first suggested that genes with Y homologues would escape from XCI in 1962, more complete maps of the genes that escape from XCI have been generated (Fig. 2A), and many of the players in the process of XCI and marks that are assembled onto silent genes have been identified (Fig. 2B). Models have theorised that there would be waystations, boundary and escape elements involved in XCI (Fig. 2C; reviewed in [94]), and to date, multiple elements have been correlated with either genes that escape from or are subject to XCI. Elucidation of the three-dimensional structure of the Xi has yielded new insights into the spread of XCI and the interactions between genes that escape from XCI (Fig. 2D). Despite the continuing progress in generating a comprehensive model of XCI, the lack of a single feature whose presence or absence is necessary for a gene to escape from XCI suggests that the variability that we observe is a reflection of multiple contributions to each gene or domain's activity (see Fig. 2E). Given the complexity of the silencing process, and the reported independence of many of the features from each other (e.g. [82, 95]), or from silencing [41], it would not be surprising that a gene's expression from the Xi is impacted by more than one of the features we have discussed in this review.
Figure 2.
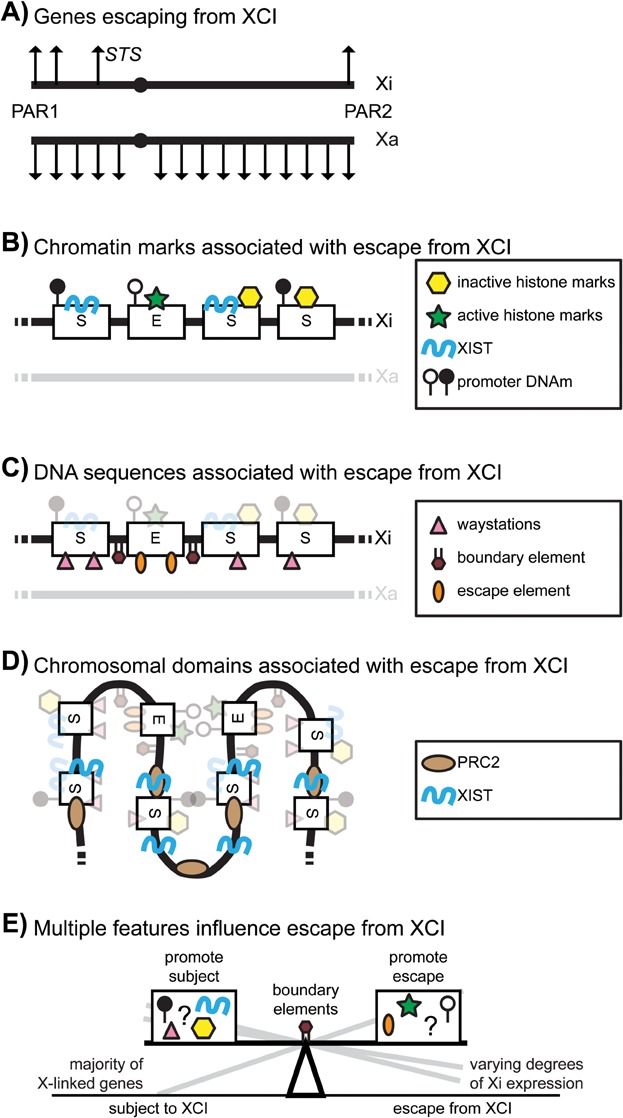
Features contributing to escape from XCI. A: STS was the first non-PAR gene found to escape from XCI in humans, and maps of genes escaping XCI show that genes with the least divergence from the Y are most likely to escape from XCI. B: Genes, which escape from XCI differ with respect to inactive (yellow hexagons) and active (green stars) chromatin marks as well as the presence of XIST RNA (blue wavy line) and promoter DNAm (white lollipops = unmethylated, black lollipops = methylated). C: DNA sequences such as waystations (pink triangles), escape elements (orange ovals) and boundary elements (maroon hexagons) have been hypothesised to account for genes that are subject to and escape from XCI. D: The three dimensional structure of the Xi appears to bring together genes that escape from XCI and to involve XIST (blue wavy lines) and PRC2 (tan ovals) in the spread of XCI. E: Together all the above features influence whether a gene is subject to, or can escape from, XCI. There does not appear to be a definitive set of features that cause a gene to escape from XCI, rather, it is likely a combination of multiple features that determines the degree to which escape occurs.
Conclusions and prospects
Improvements to the catalogues of genes that escape from XCI are still possible as genome-wide methodologies increase the depth of coverage. With these improved catalogues of XCI status, we may be able to refine the correlations with specific features. Overall, an approach to test specific regions will substantially further our understanding of the process of escape from XCI. A promising approach in recent studies has been the integration of over 100 kb sized BAC constructs to delineate the smallest regions necessary for silencing or escape. These have involved both multiple integrations of one construct into different locations [75] or integration of multiple constructs into one site on the X chromosome [13]. Similar to the spread onto autosomes, these studies are not as biased by the evolutionary history of the X chromosome; however they have the advantage of still studying the spread of silencing on the Xi. Mechanistic studies in humans have been limited because of the lack of a developmental model; however, a recent report of a human XIST transgene in an induced pluripotent stem cell may provide a means around that challenge [35]. New methodologies may therefore tip the balance towards elucidation of the complementary roles that DNA sequences, chromatin modifications and chromosomal domains play in the variable expression from the Xi in both humans and mice.
Acknowledgments
Research in the Brown lab is funded by CIHR (MOP-13690 and MOP-119586). We thank Christine Yang and members of the Brown Lab for helpful discussions and Bing Ge, Nicholas Light and Veronique Adoue of the Pastinen lab, coauthors on our recent study.
Glossary
- DNAm
DNA methylation
- ES cells
embryonic stem cells
- LINE
long interspersed nuclear element
- PAR
pseudoautosomal region
- PRC
Polycomb Repressive Complex
- SINE
short interspersed nuclear element
- TAD
topologically associating domain
- Xa
active X chromosome
- XCI
X-chromosome inactivation
- Xi
inactive X chromosome
- XIC
X inactivation centre
References
- 1.Berletch JB, Yang F, Disteche CM. Escape from X inactivation in mice and humans. Genome Biol. 2010;11:213. doi: 10.1186/gb-2010-11-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, et al. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci USA. 2012;109:5346–51. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ross MT, Grafham DV, Coffey AJ, Scherer S, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gartler SM, Riggs AD. Mammalian X-chromosome inactivation. Ann Rev Genet. 1983;17:155–90. doi: 10.1146/annurev.ge.17.120183.001103. [DOI] [PubMed] [Google Scholar]
- 5.Lengner CJ, Gimelbrant AA, Erwin JA, Cheng AW, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell. 2010;141:872–83. doi: 10.1016/j.cell.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Rastan S, Robertson EJ. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–88. [PubMed] [Google Scholar]
- 7.Lee JT, Davidow LS, Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation center. Nat Genet. 1999;21:400–4. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 8.Migeon BR, Lee CH, Chowdury AK, Carpenter H. Species differences in TSIX/Tsix reveal the roles of these genes in X-chromosome inactivation. Am J Hum Genet. 2002;71:286–93. doi: 10.1086/341605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont C, Gribnau J. Different flavors of X-chromosome inactivation in mammals. Curr Opin Cell Biol. 2013;25:314–21. doi: 10.1016/j.ceb.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Vallot C, Huret C, Lesecque Y, Resch A, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet. 2013;45:239–41. doi: 10.1038/ng.2530. [DOI] [PubMed] [Google Scholar]
- 11.Migeon BR, Kazi E, Haisley-Royster C, Hu J, et al. Human X inactivation center induces random X chromosome inactivation in male transgenic mice. Genomics. 1999;59:113–21. doi: 10.1006/geno.1999.5861. [DOI] [PubMed] [Google Scholar]
- 12.Heard E, Mongelard F, Arnaud D, Chureau C, et al. Human XIST yeast artificial chromosome transgenes show partial X inactivation center function in mouse embryonic stem cells. Proc Natl Acad Sci USA. 1999;96:6841–6. doi: 10.1073/pnas.96.12.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C, McLeod AJ, Cotton AM, de Leeuw CN, et al. Targeting of over 1.5 Mb of human DNA into the mouse X chromosome reveals presence of cis-acting regulators of epigenetic silencing. Genetics. 2012;192:1281–93. doi: 10.1534/genetics.112.143743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- 15.Lyon MF. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet. 1962;14:135–45. [PMC free article] [PubMed] [Google Scholar]
- 16.Mohandas T, Sparkes RS, Hellkuhl B, Grzeschik KH, et al. Expression of an X-linked gene from an inactive human X chromosome in mouse-human hybrid cells: futher evidence for the noninactivation of the steroid sulfatase locus in man. Proc Natl Acad Sci USA. 1977;77:6759–63. doi: 10.1073/pnas.77.11.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 18.Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–7. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Luo J, Yu H, Rattner A, et al. Cellular resolution maps of x chromosome inactivation: implications for neural development, function, and disease. Neuron. 2014;81:103–19. doi: 10.1016/j.neuron.2013.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–22. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li SM, Valo Z, Wang J, Gao H, et al. Transcriptome-wide survey of mouse CNS-derived cells reveals monoallelic expression within novel gene families. PLoS One. 2012;7:e31751. doi: 10.1371/journal.pone.0031751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese JM, Sun W, Song L, Mugford JW, et al. Site-specific silencing of regulatory elements as a mechanism of X inactivation. Cell. 2012;151:951–63. doi: 10.1016/j.cell.2012.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cotton AM, Ge B, Light N, Adoue V, et al. Analysis of expressed SNPs identifies variable extents of expression from the human inactive X chromosome. Genome Biol. 2013;14:R122. doi: 10.1186/gb-2013-14-11-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al Nadaf S, Deakin JE, Gilbert C, Robinson TJ, et al. A cross-species comparison of escape from X inactivation in Eutheria: implications for evolution of X chromosome inactivation. Chromosoma. 2012;121:71–8. doi: 10.1007/s00412-011-0343-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrel L, Willard HF. Heterogeneous gene expression from the inactive X chromosome: an X-linked gene that escapes X inactivation in some human cell lines but is inactivated in others. Proc Natl Acad Sci USA. 1999;96:7364–9. doi: 10.1073/pnas.96.13.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dubois A, Deuve JL, Navarro P, Merzouk S, et al. Spontaneous reactivation of clusters of x-linked genes is associated with the plasticity of X-inactivation in mouse trophoblast stem cells. Stem Cells. 2014;32:377–90. doi: 10.1002/stem.1557. [DOI] [PubMed] [Google Scholar]
- 27.Lingenfelter PA, Adler DA, Poslinski D, Thomas S, et al. Escape from X inactivation of Smcx is preceded by silencing during mouse development. Nat Genet. 1998;18:212–3. doi: 10.1038/ng0398-212. [DOI] [PubMed] [Google Scholar]
- 28.Brown S, Rastan S. Age-related reactivation of an X-linked gene close to the inactivation centre in the mouse. Genet Res. 1988;52:151–4. doi: 10.1017/s0016672300027531. [DOI] [PubMed] [Google Scholar]
- 29.Kalantry S, Purushothaman S, Bowen RB, Starmer J, et al. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–51. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amos-Landgraf JM, Cottle A, Plenge RM, Friez M, et al. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am J Hum Genet. 2006;79:493–9. doi: 10.1086/507565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Castillo-Morales A, Jiang M, Zhu Y, et al. Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol. 2013;30:2588–601. doi: 10.1093/molbev/mst148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sharp AJ, Stathaki E, Migliavacca E, Brahmachary M, et al. DNA methylation profiles of human active and inactive X chromosomes. Genome Res. 2011;21:1592–600. doi: 10.1101/gr.112680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kucera KS, Reddy TE, Pauli F, Gertz J, et al. Allele-specific distribution of RNA polymerase II on female X chromosomes. Hum Mol Genet. 2011;20:3964–73. doi: 10.1093/hmg/ddr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jegalian K, Page DC. A proposed path by which genes common to mammalian X and Y chromosomes evolve to become X inactivated. Nature. 1998;394:776–80. doi: 10.1038/29522. [DOI] [PubMed] [Google Scholar]
- 35.Jiang J, Jing Y, Cost GJ, Chiang JC, et al. Translating dosage compensation to trisomy 21. Nature. 2013;500:296–300. doi: 10.1038/nature12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- 37.Murakami K, Ohhira T, Oshiro E, Qi D, et al. Identification of the chromatin regions coated by non-coding Xist RNA. Cytogenet Genome Res. 2009;125:19–25. doi: 10.1159/000207514. [DOI] [PubMed] [Google Scholar]
- 38.Simon MD, Pinter SF, Fang R, Sarma K, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504:465–9. doi: 10.1038/nature12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341:1237973. doi: 10.1126/science.1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hall LL, Clemson CM, Byron M, Wydner K, et al. Unbalanced X;autosome translocations provide evidence for sequence specificity in the association of XIST RNA with chromatin. Hum Mol Genet. 2002;11:3157–65. doi: 10.1093/hmg/11.25.3157. [DOI] [PubMed] [Google Scholar]
- 41.Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–53. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- 42.Nozawa RS, Nagao K, Igami KT, Shibata S, et al. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol. 2013;20:566–73. doi: 10.1038/nsmb.2532. [DOI] [PubMed] [Google Scholar]
- 43.Costanzi C, Pehrson JR. Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature. 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 44.Chadwick BP, Willard HF. Chromatin of the Barr body: histone and non-histone proteins associated with or excluded from the inactive X chromosome. Hum Mol Genet. 2003;12:2167–78. doi: 10.1093/hmg/ddg229. [DOI] [PubMed] [Google Scholar]
- 45.Khalil AM, Driscoll DJ. Trimethylation of histone H3 lysine 4 is an epigenetic mark at regions escaping mammalian X inactivation. Epigenetics. 2007;2:114–8. doi: 10.4161/epi.2.2.4612. [DOI] [PubMed] [Google Scholar]
- 46.Goto Y, Kimura H. Inactive X chromosome-specific histone H3 modifications and CpG hypomethylation flank a chromatin boundary between an X-inactivated and an escape gene. Nucleic Acids Res. 2009;37:7416–28. doi: 10.1093/nar/gkp860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Changolkar LN, Pehrson JR. macroH2A1 histone variants are depleted on active genes but concentrated on the inactive X chromosome. Mol Cell Biol. 2006;26:4410–20. doi: 10.1128/MCB.02258-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morishima A, Grumbach MM, Taylor JH. Asynchronous duplication of human chromosomes and the origin of sex chromatin. Proc Natl Acad Sci USA. 1962;48:756–63. doi: 10.1073/pnas.48.5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koren A, McCarroll SA. Random replication of the inactive X chromosome. Genome Res. 2014;24:64–9. doi: 10.1101/gr.161828.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schempp W, Meer B. Cytologic evidence for three human X-chromosomal segments escaping inactivation. Hum Genet. 1983;63:171–4. doi: 10.1007/BF00291539. [DOI] [PubMed] [Google Scholar]
- 51.Hansen RS, Canfield TK, Fjeld AD, Gartler SM. Role of late replication timing in the silencing of X-linked genes. Hum Mol Genet. 1996;5:1345–53. doi: 10.1093/hmg/5.9.1345. [DOI] [PubMed] [Google Scholar]
- 52.Cohen SM, Brylawski BP, Cordeiro-Stone M, Kaufman DG. Same origins of DNA replication function on the active and inactive human X chromosomes. J Cell Biochem. 2003;88:923–31. doi: 10.1002/jcb.10429. [DOI] [PubMed] [Google Scholar]
- 53.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 54.Weber M, Davies JJ, Wittig D, Oakeley EJ, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–62. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 55.Ginno PA, Lim YW, Lott PL, Korf I, et al. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013;23:1590–600. doi: 10.1101/gr.158436.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang X, Douglas KC, Vandeberg JL, Clark AG, et al. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 2014;24:70–83. doi: 10.1101/gr.161919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, et al. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487:254–8. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gendrel AV, Apedaile A, Coker H, Termanis A, et al. Smchd1-dependent and -independent pathways determine developmental dynamics of CpG island methylation on the inactive X chromosome. Dev Cell. 2012;23:265–79. doi: 10.1016/j.devcel.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, et al. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet. 2008;40:663–9. doi: 10.1038/ng.142. [DOI] [PubMed] [Google Scholar]
- 60.Cotton AM, Lam L, Affleck JG, Wilson IM, et al. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 2011;130:187–201. doi: 10.1007/s00439-011-1007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chadwick BP, Valley CM, Willard HF. Histone variant macroH2A contains two distinct macrochromatin domains capable of directing macroH2A to the inactive X chromosome. Nucleic Acids Res. 2001;29:2699–705. doi: 10.1093/nar/29.13.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science. 1981;211:393–6. doi: 10.1126/science.6164095. [DOI] [PubMed] [Google Scholar]
- 63.Csankovszki G, Nagy A, Jaenisch R. Synergism of Xist RNA, DNA methylation, and histone hypoacetylation in maintaining X chromosome inactivation. J Cell Biol. 2001;153:773–83. doi: 10.1083/jcb.153.4.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Csankovszki G, Panning B, Bates B, Pehrson JR, et al. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat Genet. 1999;22:323–4. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- 65.Gartler SM, Goldman MA. Reactivation of inactive X-linked genes. Dev Genet. 1994;15:504–14. doi: 10.1002/dvg.1020150609. [DOI] [PubMed] [Google Scholar]
- 66.Brown CJ, Willard HF. The human X inactivation center is not required for maintenance of X inactivation. Nature. 1994;368:154–6. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 67.Lyon MF. X-chromosome inactivation: a repeat hypothesis. Cytogenet Cell Genet. 1998;80:133–7. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- 68.Cotton AM, Chen CY, Lam LL, Wasserman WW, et al. Spread of X-chromosome inactivation into autosomal sequences: role for DNA elements, chromatin features and chromosomal domains. Hum Mol Genet. 2014;23:1211–23. doi: 10.1093/hmg/ddt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bala Tannan N, Brahmachary M, Garg P, Borel C, et al. DNA methylation profiling in X;autosome translocations supports a role for L1 repeats in the spread of X chromosome inactivation. Hum Mol Genet. 2014;23:1224–36. doi: 10.1093/hmg/ddt553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bailey JA, Carrel L, Chakravarti A, Eichler E. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis. Proc Natl Acad Sci USA. 2000;97:6634–9. doi: 10.1073/pnas.97.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang Z, Willard HF, Mukherjee S, Furey TS. Evidence of influence of genomic DNA sequence on human X chromosome inactivation. PLoS Comput Biol. 2006;2:e113. doi: 10.1371/journal.pcbi.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nguyen DK, Yang F, Kaul R, Alkan C, et al. Clcn4-2 genomic structure differs between the X locus in Mus spretus and the autosomal locus in Mus musculus: AT motif enrichment on the X. Genome Res. 2011;21:402–9. doi: 10.1101/gr.108563.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopes AM, Arnold-Croop SE, Amorim A, Carrel L. Clustered transcripts that escape X inactivation at mouse XqD. Mamm Genome. 2011;22:572–82. doi: 10.1007/s00335-011-9350-6. [DOI] [PubMed] [Google Scholar]
- 74.Chow JC, Ciaudo C, Fazzari MJ, Mise N, et al. LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell. 2010;141:956–69. doi: 10.1016/j.cell.2010.04.042. [DOI] [PubMed] [Google Scholar]
- 75.Li N, Carrel L. Escape from X chromosome inactivation is an intrinsic property of the Jarid1c locus. Proc Natl Acad Sci USA. 2008;105:17055–60. doi: 10.1073/pnas.0807765105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Filippova GN, Cheng MK, Moore JM, Truong JP, et al. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 77.Horvath LM, Li N, Carrel L. Deletion of an X-inactivation boundary disrupts adjacent gene silencing. PLoS Genet. 2013;9:e1003952. doi: 10.1371/journal.pgen.1003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dixon JR, Selvaraj S, Yue F, Kim A, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pinter SF, Sadreyev RI, Yildirim E, Jeon Y, et al. Spreading of X chromosome inactivation via a hierarchy of defined Polycomb stations. Genome Res. 2012;22:1864–76. doi: 10.1101/gr.133751.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sadreyev RI, Yildirim E, Pinter SF, Lee JT. Bimodal quantitative relationships between histone modifications for X-linked and autosomal loci. Proc Natl Acad Sci USA. 2013;110:6949–54. doi: 10.1073/pnas.1216449110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao J, Sun BK, Erwin JA, Song JJ, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–6. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cerase A, Smeets D, Tang YA, Gdula M, et al. Spatial separation of Xist RNA and polycomb proteins revealed by superresolution microscopy. Proc Natl Acad Sci USA. 2014;111:2235–40. doi: 10.1073/pnas.1312951111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.da Rocha ST, Boeva V, Escamilla-Del-Arenal M, Ancelin K, et al. Jarid2 is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell. 2014;53:301–16. doi: 10.1016/j.molcel.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 84.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell. 2011;146:119–33. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chaumeil J, Le Baccon P, Wutz A, Heard E. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev. 2006;20:2223–37. doi: 10.1101/gad.380906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Clemson CM, Hall LL, Byron M, McNeil J, et al. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci USA. 2006;103:7688–93. doi: 10.1073/pnas.0601069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hall LL, Carone DM, Gomez AV, Kolpa HJ, et al. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–19. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 89.Barr ML, Bertram EG. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676–7. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 90.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49:773–82. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nora EP, Dekker J, Heard E. Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods. BioEssays. 2013;35:818–28. doi: 10.1002/bies.201300040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Splinter E, de Wit E, Nora EP, Klous P, et al. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev. 2011;25:1371–83. doi: 10.1101/gad.633311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yang C, Chapman AG, Kelsey AD, Minks J, et al. X-chromosome inactivation: molecular mechanisms from the human perspective. Hum Genet. 2011;130:175–85. doi: 10.1007/s00439-011-0994-9. [DOI] [PubMed] [Google Scholar]
- 95.Tavares L, Dimitrova E, Oxley D, Webster J, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 2012;148:664–78. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 97.Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–75. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Penny GD, Kay GF, Sheardown SA, Rastan S, et al. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–7. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 99.Marahrens Y, Panning B, Dausman J, Strauss W, et al. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–66. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 100.Heard E, Rougelle C, Arnaud D, Avner P, et al. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–38. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- 101.Boggs BA, Cheung P, Heard E, Spector DL, et al. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nat Genet. 2002;30:73–6. doi: 10.1038/ng787. [DOI] [PubMed] [Google Scholar]
- 102.Goto Y, Gomez M, Brockdorff N, Feil R. Differential patterns of histone methylation and acetylation distinguish active and repressed alleles at X-linked genes. Cytogenet Genome Res. 2002;99:66–74. doi: 10.1159/000071576. [DOI] [PubMed] [Google Scholar]
- 103.Marks H, Chow JC, Denissov S, Francoijs KJ, et al. High-resolution analysis of epigenetic changes associated with X inactivation. Genome Res. 2009;19:1361–73. doi: 10.1101/gr.092643.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jeppesen P, Turner B. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–9. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 105.Keohane AM, O'Neill LP, Belyaev ND, Lavender JS, et al. X-inactivation and histone H4 acetylation in embryonic stem cells. Dev Biol. 1996;180:618–30. doi: 10.1006/dbio.1996.0333. [DOI] [PubMed] [Google Scholar]
- 106.Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–5. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- 107.de Napoles M, Mermoud JE, Wakao R, Tang YA, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 108.Fang J, Chen T, Chadwick B, Li E, et al. Ring1b-mediated H2A ubiquitination associates with inactive X chromosomes and is involved in initiation of X-inactivation. J Biol Chem. 2004;279:52812–5. doi: 10.1074/jbc.C400493200. [DOI] [PubMed] [Google Scholar]
- 109.Smith KP, Byron M, Clemson CM, Lawrence JB. Ubiquitinated proteins including uH2A on the human and mouse inactive X chromosome: enrichment in gene rich bands. Chromosoma. 2004;113:324–35. doi: 10.1007/s00412-004-0325-1. [DOI] [PubMed] [Google Scholar]
- 110.Mermoud JE, Popova B, Peters AH, Jenuwein T, et al. Histone H3 lysine 9 methylation occurs rapidly at the onset of random X chromosome inactivation. Curr Biol. 2002;12:247–51. doi: 10.1016/s0960-9822(02)00660-7. [DOI] [PubMed] [Google Scholar]
- 111.Peters AH, Mermoud JE, O'Carroll D, Pagani M, et al. Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet. 2002;30:77–80. doi: 10.1038/ng789. [DOI] [PubMed] [Google Scholar]
- 112.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA. 2004;101:17450–5. doi: 10.1073/pnas.0408021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kohlmaier A, Savarese F, Lachner M, Martens J, et al. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Perche P-Y, Vourc'h C, Konecny L, Souchier C, et al. Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr Biol. 2000;10:1531–4. doi: 10.1016/s0960-9822(00)00832-0. [DOI] [PubMed] [Google Scholar]
- 115.Rasmussen TP, Wutz A, Pehrson JR, Jaenisch R. Expression of Xist RNA is sufficient to initiate macrochromatin body formation. Chromosoma. 2001;110:411–20. doi: 10.1007/s004120100158. [DOI] [PubMed] [Google Scholar]
- 116.Mietton F, Sengupta AK, Molla A, Picchi G, et al. Weak but uniform enrichment of the histone variant macroH2A1 along the inactive X chromosome. Mol Cell Biol. 2009;29:150–6. doi: 10.1128/MCB.00997-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wolf SF, Jolly DJ, Lunnen KD, Friedmann T, et al. Methylation of the hypoxanthine phosphoribosyltransferase locus on the human X chromosome: implications for X-chromosome inactivation. Proc Natl Acad Sci USA. 1984;81:2806–10. doi: 10.1073/pnas.81.9.2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wolf SF, Dintzis S, Toniolo D, Persico G, et al. Complete concordance between glucose-6-phosphate dehydrogenase activity and hypomethylation of 3' CpG clusters: implications for X chromosome dosage compensation. Nucleic Acids Res. 1984;12:9333–48. doi: 10.1093/nar/12.24.9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hansen RS, Gartler SM. 5-Azacytidine-induced reactivation of the human X chromosome-linked PGK1 gene is associated with a large region of cytosine demethylation in the 5′ CpG island. Proc Natl Acad Sci USA. 1990;87:4174–8. doi: 10.1073/pnas.87.11.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cotton AM, Avila L, Penaherrera MS, Affleck JG, et al. Inactive X chromosome-specific reduction in placental DNA methylation. Hum Mol Genet. 2009;18:3544–52. doi: 10.1093/hmg/ddp299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Goodfellow PJ, Mondello C, Darling SM, Pym B, et al. Absence of methylation of a CpG-rich region at the 5′ end of the MIC2 gene on the active X, and inactive X, and the Y chromosome. Proc Natl Acad Sci USA. 1988;85:5605–9. doi: 10.1073/pnas.85.15.5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anderson CL, Brown CJ. Variability of X chromosome inactivation: effect on levels of TIMP1 RNA and role of DNA methylation. Hum Genet. 2002;110:271–8. doi: 10.1007/s00439-002-0676-8. [DOI] [PubMed] [Google Scholar]
- 123.Carrel L, Clemson CM, Dunn JM, Miller AP, et al. X inactivation analysis and DNA methylation studies of the ubiquitin activating enzyme E1 and PCTAIRE-1 genes in human and mouse. Hum Mol Genet. 1996;5:391–402. doi: 10.1093/hmg/5.3.391. [DOI] [PubMed] [Google Scholar]


