SUMMARY
DNA damage in Arabidopsis thaliana seedlings results in upregulation of hundreds of genes. One of the earliest and highest levels of induction is displayed by a previously uncharacterized gene that we have termed X-ray induced 1 (XRI1). Analysis of plants carrying a null xri1 allele revealed two distinct requirements for this gene in plant fertility. XRI1 was important for the post-meiotic stages of pollen development, leading to inviability of xri− pollen and abnormal segregation of the mutant allele in heterozygous xri1+/− plants. In addition, XRI1 was essential for male and female meiosis, as indicated by the complete sterility of homozygous xri1 mutants due to extensive chromosome fragmentation visible in meiocytes. Abolition of programmed DNA double-strand breaks in a spo11-1 mutant background failed to rescue the DNA fragmentation of xri1 mutants, suggesting that XRI1 functions at an earlier stage than SPO11-1 does. Yeast two-hybrid studies identified an interaction between XRI1 and a novel component of the Arabidopsis MND1/AHP2 complex, indicating possible requirements for XRI1 in meiotic DNA repair.
Keywords: Arabidopsis, DNA damage response, DNA repair, homologous recombination, meiosis
INTRODUCTION
DNA in the cell is continuously being damaged by a combination of endogenous and environmental factors. This can lead to DNA replication failure, transcriptional blocks, mutagenesis and cell death (Sancar et al., 2004). DNA double-strand breaks (DSBs) are one of the most serious forms of DNA damage, and a single DSB can result in the loss of large amounts of genetic information and cell death. Genomic integrity in all organisms is maintained by the activities of DNA repair pathways, and mutants deficient in DNA repair display hypersensitivity to DNA damage, an increased frequency of mutation, and, in animals, predisposition to cancer. Elevated levels of DNA damage elicit a DNA damage response, including activation of repair processes, although the nature of this response differs between organisms. The plant DNA damage response includes transcriptional upregulation of hundreds of genes encoding proteins involved in DNA repair, DNA synthesis and cell-cycle control (Culligan et al., 2006; Ricaud et al., 2007). Gene induction may be rapid, as typified by AtRAD51, which shows increased transcript levels within 30 min post-irradiation (Garcia et al., 2003). The Arabidopsis DNA damage response is regulated by the phosphatidyl inositol kinase-like kinase AtATM (ataxia telangiectasia mutated), with a small contribution from the related protein kinase ATR (ATM and RAD3-related kinase) (Culligan et al., 2006). In plants, activation of ATM and ATR kinases results in histone H2AX phosphorylation and activation of cell-cycle checkpoints (Friesner et al., 2005; Culligan et al., 2006).
DSBs are repaired by non-homologous end-joining (NHEJ), independently of DNA sequence. This is the major pathway in vegetative plant cells, as illustrated by the hypersensitivity of Arabidopsis NHEJ mutants to X-rays and radio-mimetic drugs (Bray and West, 2005). Alternatively, DSBs can be repaired by homologous recombination (HR), which is dependent on availability of an intact copy of the damaged region as a template for repair. Phenotypic analysis of mutant plants suggests that HR is a minor DSB repair pathway in Arabidopsis vegetative cells, although HR-deficient plants do display hypersensitivity to DNA cross-linking agents that cause stalling or collapse of replication forks (Bleuyard et al., 2005). Collapsed replication forks form a DSB end, which then acts as a substrate for the HR repair machinery to restore the replication fork structure (Shrivastav et al., 2008).
HR also plays an important role in meiosis, during which programmed DSBs are induced by the protein SPO11, a homologue of the archaebacterial topoisomerase subunit Top6A (Bergerat et al., 1997; Keeney et al., 1997). Repair of these breaks by HR facilitates the pairing of homologous chromosomes; the absence of SPO11 activity causes chromosomes to segregate randomly in meiotic anaphase I as univalents (Grelon et al., 2001). HR mutants fail to repair SPO11-induced DSBs, resulting in extensive chromosome fragmentation in meiocytes and complete sterility. In addition to the core HR factors required for DSB repair in all cells, meiosis requires specific components, including DMC1, the meiotic homologue of RAD51 (Couteau et al., 1999). RAD51/DMC1-mediated repair of meiotic DSBs is dependent on MND1, which forms a complex with AHP2, the Arabidopsis HOP2 homologue (Schommer et al., 2003; Kerzendorfer et al., 2006; Vignard et al., 2007). In humans, the HOP2/MND1 complex promotes the strand-invasion activity of RAD51 and DMC1 (Petukhova et al., 2005). Successful completion of meiosis produces haploid spores, which subsequently undergo further mitotic cell divisions to generate male and female gametes.
Here we report the characterization of a novel gene termed X-RAY INDUCIBLE 1 (XRI1), which displays high levels of transcriptional induction in response to DSBs in Arabidopsis in an ATM-dependent pathway. Phenotypic analysis of xri1 mutants indicates that this gene plays an essential role in meiotic progression, with mutant plants displaying severely disrupted meiosis resulting in extensive chromosome fragmentation. The chromosome fragmentation was independent of SPO11-induced DSBs, indicating important functions for XRI1 in pre-meiotic S phase. XRI1 interacts with the recombination machinery, forming a complex in vitro and in yeast two-hybrid assays with an MND1 interacting protein (MIP1). In addition to the meiotic phenotype, male gametophyte development required de novo XRI1 synthesis, with xri1− mutant pollen arresting at the bicellular stage, leading to embryo abortion and loss of the mutant allele through the male line. Our results identify XRI1 as a novel DNA repair factor that is essential for both meiosis and male gametogenesis in plants.
RESULTS AND DISCUSSION
Microarray analysis of Arabidopsis seedlings exposed to gamma rays identified a number of uncharacterized genes that were upregulated in response to DNA damage and had potential roles in DNA repair, DNA damage signalling and cell-cycle control (Culligan et al., 2006). In the present study, we aimed to elucidate the molecular function of a gene that displays rapid and high transcriptional induction in response to X-rays, X-RAY INDUCED 1 (XRI1, AGI code At5g48720).
Molecular cloning of XRI1
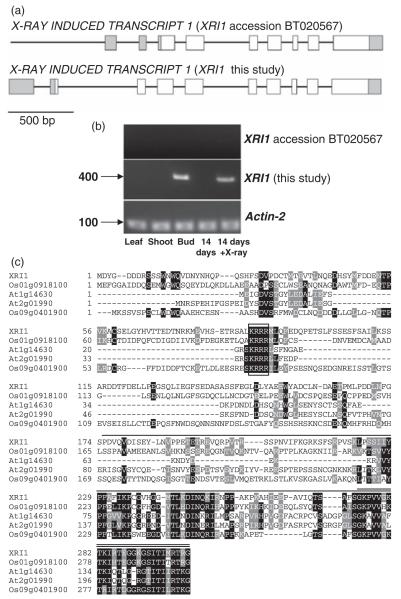
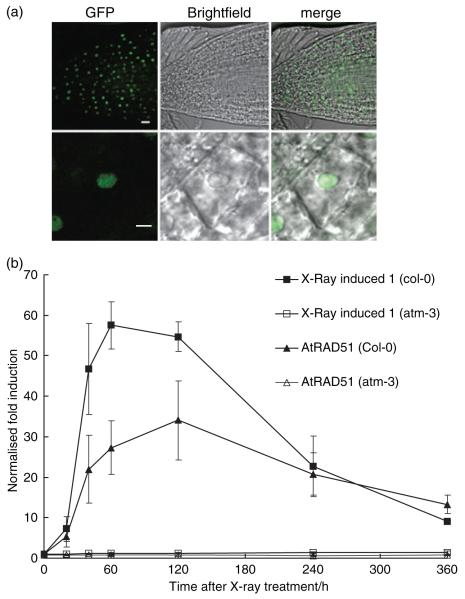
High-throughput cDNA cloning identified an 801 bp XRI1 coding sequence (Genbank accession number BT020567) and a 1066 bp XRI1 cDNA clone that included two untranslated 5′ exons (Genbank accession number NM_124249) (Figure 1a) (Yamada et al., 2003). However, we were unable to detect the 1066 bp XRI1 transcript in RT-PCR experiments despite using similar source materials, suggesting that the transcript was expressed at very low levels, is only expressed in very specific growth conditions, or represents an artefact. The predominant transcript in our studies was initiated from an upstream transcriptional start site relative to XRI1 accession BT020567 (Figure 1a). The resulting 1229 bp cDNA, identified using 5′ and 3′ RACE PCR, had a single untranslated first exon followed by a second exon upstream of XRI1 accession BT020567, and encoded a protein of 300 amino acids with no significant sequence similarity beyond the plant kingdom (Table S1). The primary sequence contained a consensus nuclear localization sequence (KRRR), which is conserved between the XRI1-like sequences identified in rice and Arabidopsis (Figure 1c). These sequences showed weak but significant sequence similarity to XRI1 (At2g01990 expect value 10−10; At1g14630 expect value 10−7). However, the sequence similarity was largely confined to the C-terminal domains of these proteins, and there was no evidence of X-ray induction of either the At2g01990 or At1g14630 transcripts. The nuclear localization of XRI1 was confirmed using confocal analysis of a GFP–XRI1 fusion protein expressed under the control of the CaMV 35S promoter and visualized in root tips of 10-day-old seedlings (Figure 2a). GFP fluorescence was localized exclusively to the nucleus, with evidence for nucleolar exclusion in some cells.
Figure 1.
Sequence analysis of X-RAY INDUCED 1 (XRI1).
(a) Schematic of the XRI1 short and XRI1 long transcripts. Exons are denoted by boxes, with filled boxes representing untranslated regions and empty boxes coding regions; introns are represented by a line.
(b) RT-PCR analysis of XRI1 accession BT020567 and the XRI1 transcript identified in the present study. RNA from leaf, silique, bud, untreated (14d) or X-ray-treated (14d + X-ray) 2-week-old seedlings was analysed by RT-PCR using a primer specific for the 5′ UTR of each cDNA as indicated. Control RT-PCR was performed using primers specific to ACTIN2.
(c) ClustalX sequence alignment of XRI1 and similar sequences from Arabidopsis (At) and O. sativa (Os). Aligned residues that are identical are shaded black and residues that are similar are shaded grey. A conserved nuclear localization sequence is boxed. The XRI1 fragment used in yeast two-hybrid analysis is indicated by a single line above the sequence. The C-terminal domain of XRI1 that is sufficient for interaction with MIP is indicated by a double line above the sequence.
Figure 2.
Expression of X-RAY INDUCED 1.
(a) Subcellular localization of an smrs–GFP–XRI1 fusion protein analysed by laser scanning confocal microscopy. Nuclear localization of XRI1 was observed in the root tip and at higher magnification in a root cell. Scale bar = 5 μm.
(b) Real-time RT-PCR analysis of XRI1 and AtRAD51 expression after 10 Gy X-ray irradiation. Fold induction of XRI1 (■) and AtRAD51 (▲) transcripts was measured at 1 h time points in wild-type (closed squares/triangles) and atm-3 (open squares/triangles) following X-ray treatment. The transcript levels were normalized against the ACTIN2 transcript levels in each sample. Error bars represent the standard error of the mean of three biological replicates.
XRI1 induction is dependent on DNA damage signalling by AtATM
The induction of XRI1 by ionizing radiation (IR) was further characterized using real-time RT-PCR analysis. Arabidopsis seedlings were subjected to 10 Gy X-rays, and XRI1 transcript levels were analysed in the period following irradiation. Whilst XRI1 accession BT020567 was undetectable, the longer XRI1 transcript was induced in response to X-rays (Figure 2b). The X-ray-induced transcriptional response was rapid, with transcript levels increasing after 20 min and peaking at 1–2 h post-irradiation at levels approximately 55-fold greater than in untreated plants, before declining over the next few hours. This induction was completely dependent on ATM, as atatm-3 mutants fail to display a significant increase in XRI1 transcript levels after irradiation (Figure 2b) (Culligan et al., 2006; Ricaud et al., 2007). The pattern of XRI1 induction closely resembled that of AtRAD51, and inspection of the publicly available microarray data (Zimmermann et al., 2004) indicated that this response was specific to reagents that cause DSBs, rather than a generic stress response (data not shown).
Isolation and complementation of an xri1 mutant
Analysis of the sequence-tagged T-DNA mutant database identified a line in the GABI-KAT collection (Rosso et al., 2003) that contains a T-DNA in the third exon of the XRI1 gene (Figure 3a). The T-DNA insertion used to generate these lines carries a CaMV 35S promoter adjacent to the right border for use in activation tagging experiments (Figure 3b). This led to the possibility that the 3′ end of the XRI1 transcript might be overexpressed in this line. However, RT-PCR analysis of homozygous xri1-1 mutants did not detect XRI1 expression downstream of the insertion site. PCR and sequence analysis revealed a 432 bp truncation of the T-DNA right border that resulted in deletion of most of the 537 bp CaMV 35S promoter (Figure 3b), explaining the absence of ectopically expressed 3′ XRI1 transcript in this line. Re-introduction of the wild-type XRI1 gene into the xri1-1 mutant background restored expression of XRI1 (Figure 3c), and complemented the xri1-1 mutant phenotypes (Figures 4 and 5).
Figure 3.
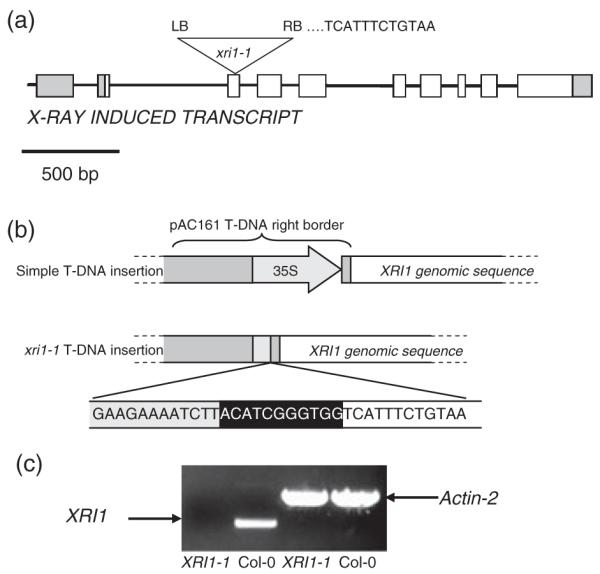
Isolation of x-ray induced 1 mutant lines.
(a) Schematic showing the position of the T-DNA insertion in the xri1-1 line. The T-DNA insertion site in the XRI1 gene is shown. The first bases of the XRI sequence from the right border (RB) of xri1-1 are indicated.
(b) Schematic showing truncation of the pAC161 right border in the xri1-1 line.
(c) RT-PCR analysis of XRI expression in the xri1-1 homozygous mutant. The cDNA was amplified using primers designed to the 3′ end of XRI1, and ACTIN2 primers were used as a control for cDNA synthesis.
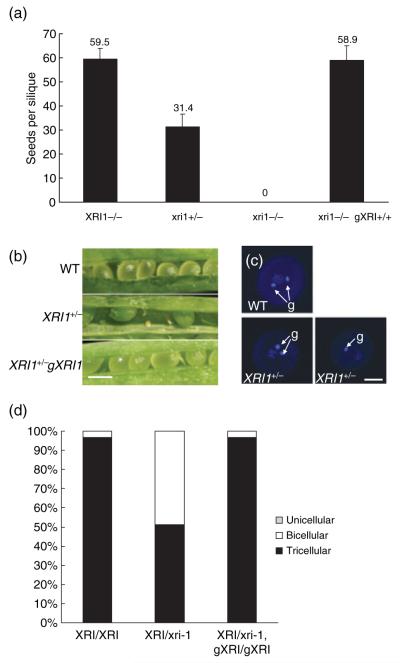
Figure 4.
Embryo lethality in x-ray induced 1-1 heterozygous mutants.
(a) xri1-1 heterozygotes have reduced seed set, with approximately half the number of seeds per silique relative to wild-type plants. This defect is complemented in plants homozygous for an XRI1 genomic transgene. Homozygous xri1-1 plants are sterile.
(b) Dissected siliques reveal the presence of aborted embryos (arrowed) in heterozygotes, which are not found in wild-type plants or plants complemented with the wild-type XRI1 gene. Scale bar = 1 mm.
(c) Confocal laser scanning microscopy images of DAPI-stained pollen from wild-type (WT) and heterozygous xri1-1+/− plants. WT pollen is tricellular, with two generative nuclei (arrowed) and a vegetative nucleus. Pollen from heterozygous xri1-1 is an approximately equal mix of tricellular and bicellular pollen, which has only one generative nucleus. Scale bar = 10 μm.
(d) Quantification of uni-, bi- and tricellular pollen frequencies in wild-type, xri−/+ heterozygotes and complemented xri−/+ heterozygotes.
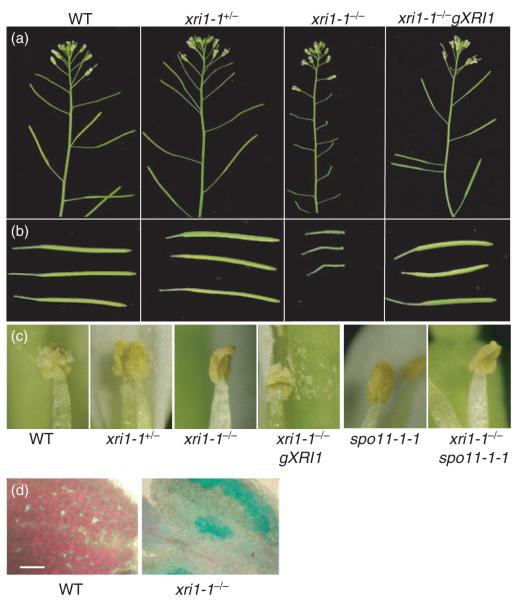
Figure 5.
Sterility of x-ray induced 1-1 mutants is rescued by complementation with the wild-type XRI1 gene.
(a) Inflorescences from wild-type, xri1-1+/− heterozygotes, xri1-1−/− mutants and complemented xri1-1−/− gXRI1 mutants. Scale bar = 1 cm.
(b) Siliques, showing very short siliques of xri1-1 mutants. Scale bar = 1 cm.
(c) Mature anthers, showing that no pollen is released from xri1-1 mutant anthers. Scale bar = 1 mm.
(d) Alexander staining of anthers before dehiscence. The protoplasm of wild-type pollen stained purple, whereas non-viable pollen in the homozygous xri1-1 anthers stained green due to the absence of developed protoplasm. Scale bar = 50 μm.
XRI1 is required for male gametophyte development
Southern analysis indicated that xri1-1 plants contain a single T-DNA insertion at a single locus, which would be expected to segregate 1:2:1 homozygous mutant:heterozygous:wild-type (data not shown). However, the observed segregation, both in terms of the sulphadiazine resistance gene and PCR genotyping, was found to be 0.11:1.19:1 (n = 223). This ratio is closer to 0:1:1 homozygous mutant:heterozygous:wild-type, as would result from loss of the mutant allele through either the male or female germline. Siliques from heterozygous plants contained a substantial number of aborted embryos, leading to a reduction in the number of seeds from a mean of 59.5 ± 0.4 per silique in wild-type plants to 31.4 ± 5.1 in xri1-1+/− plants (Figure 4a,b). Arabidopsis seed development requires a double fertilization event to form a viable embryo and endosperm tissue. Arabidopsis pollen contains two generative cells, one of which fertilizes the egg cell to form the zygote, whereas the second fertilizes the central cell of the ovule leading to endosperm development. Analysis of pollen from xri1-1+/− plants revealed that loss of the mutant allele was associated with defects in pollen development. DAPI staining revealed that 48.7% of pollen was arrested at the bicellular stage, with a vegetative cell nucleus but only a single generative cell nucleus rather than the two generative cell nuclei observed in both wild-type and the remaining half of the xri1-1+/− pollen (Figure 4c). These levels of bicellular pollen were far greater than those observed in wild-type plants (3.3%). Alexander staining suggested that the bicellular pollen was viable, but the presence of a single generative cell nucleus means that xri1-1− pollen would be incapable of double fertilization, leading to embryo lethality and failure to transmit the xri1-1 allele.
Bicellular pollen is also observed in plants that are heterozygous for a mutation in the Arabidopsis cdc2 homologue CDKA;1, and the ratio of tricellular to bicellular pollen in the cdka-1+/− heterozygote (51.3% tricellular, 48.7% bicellular) is comparable to that in xri1-1+/− (Iwakawa et al., 2006). As observed with cdka;1 mutants, the xri-1+/− pollen defect was not fully penetrant; 1 in 20 seedlings from an xri1-1+/− plant were homozygous for the xri1-1 allele, indicating that approximately 10% of the xri1-1 pollen are tricellular and capable of producing viable embryos by double fertilization. The leaky nature of the xri1-1 phenotype in the male gametophyte is consistent with residual levels of XRI1 protein or transcript being sufficient to permit normal pollen development in approximately 10% of haploid mutants. Heterozygous xri1-1+/− plants that were homozygous for the complementation construct carrying the wild-type XRI1 gene displayed normal Mendelian inheritance of the mutant xri1-1 allele, and pollen was largely tricellular as seen in wild-type anthers (Figure 4d). This complementation demonstrated that the inviability of pollen and embryo abortion in xri1-1 heterozygotes originated from lack of a functional XRI1 gene in the developing male gametophyte. These results point to an essential role for XRI1 in pollen development.
Transmission of the xri1-1 allele was not affected in the female germline, as determined by segregation of the allele and crossing experiments. Using wild-type pollen to fertilize xri1 heterozygotes resulted in a ratio of wild-type to heterozygous mutant progeny of 1:0.92 (n = 238), which is not significantly different from the expected ratio of 1:1 (χ2 test, P > 0.05). This could reflect either a specific requirement for XRI1 in male gametogenesis, or differences in male and female gametophyte development (Wilson and Yang, 2004). On completion of meiosis in pollen mother cells, haploid spores undergo a mitotic division to produce the vegetative cell nucleus and a generative cell nucleus. Before the second mitotic division, the generative cell nucleus undergoes cellularization, whereby it is physically isolated from the surrounding vegetative cell and contains very little cytoplasm (McCormick, 2004). This is in contrast to female gametogenesis, in which cellularization occurs at completion of the mitotic divisions (Wilson and Yang, 2004). Analysis of xri1-1 mutants is consistent with a requirement for de novo XRI1 expression for mitotic division of the generative cell nucleus and the development of mature tricellular pollen grains.
xri1-1 mutants do not complete male and female meiosis
Homozygous xri1-1 mutants were indistinguishable from wild-type plants during germination, seedling establishment and subsequent vegetative growth up to the point of flowering. However, in comparison to wild-type plants, xri1-1 displayed prolonged flowering and developed very short siliques that were devoid of seeds (Figure 5a,b). The sterility of xri1-1 null mutants was complemented by the wild-type XRI1 gene, resulting in wild-type levels of seeds (58.8 ± 6.2 per silique) in these lines (Figure 4a).
The nature of the sterility of homozygous xri1-1 mutants was further analysed by out-crossing. Pollen from wild-type plants was unable to fertilize xri1-1 mutants. This indicates an essential role for XRI1 in female gametogenesis, with xri1-1 plants unable to produce viable ovules. Similarly, homozygous xri1-1 mutant pollen was unable to fertilize wild-type plants in out-crossing experiments. Male sterility is consistent with the requirement for XRI1 in male gametogenesis as observed in heterozygous xri1-1+/− plants. Inspection of anthers found that xri1-1 mutants released no mature pollen grains, in contrast to xri1-1+/− and wild-type plants (Figure 5c). Alexander staining of anthers revealed that pollen from homozygous xri1-1 plants was non-viable, staining green in comparison to the strong purple stain of wild-type pollen (Figure 5d). Although the bicellular pollen of xri1-1+/− appears to be viable by Alexander staining, there remains the possibility that pollen development is more severely affected in homozygous mutants where XRI1 is absent as opposed to the reduced levels found in xri1-1+/− plants. Alternatively, underlying meiotic defects in xri1-1 plants could be responsible for pollen inviability in Alexander staining. These two possibilities (meiotic failure versus defects later in gametophyte development) could also apply to the observed female sterility, even though female gametophyte development is unaffected in heterozygous plants. This is because the defects observed in male gametogenesis may be attributed to the early cellularization of pollen cells, whereas female levels of XRI1 could be sufficient for completion of embryo sac development in the heterozygous xri1-1+/− background. The presence of meiotic defects in xri1-1 plants was therefore investigated by cytological analysis of megaspore mother cells and pollen mother cells.
Severe DNA fragmentation during meiosis in xri1-1 mutants
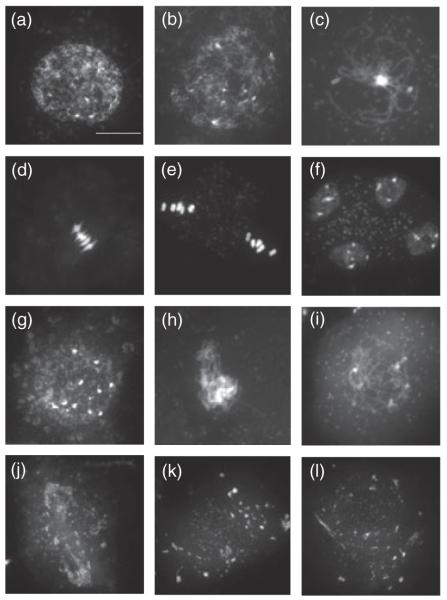
Wild-type meiosis in pollen mother cells consists of two rounds of chromosome segregation, preceded by a single S phase, which results in haploid meiotic products (Figure 6a–f). During prophase I, homologous chromosomes align, synapse and recombine before they are separated in anaphase I (meiosis I). The sister chromatid products of DNA replication are separated in the second round of division (meiosis II). Prophase I is sub-divided into several stages. During leptotene, the chromosomes start to condense and appear as thin thread-like structures (Figure 6a,b). This stage is followed by zygotene, with chromosomes further condensing and partial pairing between homologues. In pachytene, pairing of homologous chromosomes is complete and synapsis is established (Figure 6c). At the diakinesis stage of prophase I, wild-type chromosomes are identified as pairs of highly condensed chromosome bivalents held together by chiasmata (crossovers). At meta-phase I, the five bivalents are aligned on the equator prior to segregation of the homologous chromosomes to the poles (Figure 6d). During metaphase II (Figure 6e), the chromosomes align and the sister chromatids are separated at anaphase II. Finally, tetrads consisting of four sets of five chromatids are seen, resulting from completion of the second round of meiotic segregation (Figure 6f).
Figure 6.
Progression through male meiosis in wild-type plants and homozygous x-ray induced 1-1 mutants. The pollen mother cells are stained with DAPI.
(a–f) Wild-type (WT) meiosis is shown in the stages of early leptotene/leptotene (a, b), pachytene (c), metaphase I (d), metaphase II (e) and a tetrad of meiotic products (f) after the completion of meiosis.
(g–l) Homozygous xri1-1 pollen mother cells at early leptotene/leptotene (g, h), later asynaptic prophase (i), anaphase I (j), anaphase II (k) and polyad (l). Cells in (j), (k) and (l) show extensive chromosome fragmentation. Scale bar = 10 μm.
The progression of male meiosis was severely impaired in homozygous xri1-1 mutants (Figure 6g–l) The meiocytes at early leptotene/leptotene (Figure 6g,h) appeared to be normal. Homozygous xri1-1 meiocytes that had progressed further through prophase I, as indicated by the condensation of the chromosomes, appeared to fail to synapse (Figure 6i). We did not see any equivalent stages of late prophase or metaphase I chromosomes, but observed meiocytes at anaphase I (Figure 6j) and anaphase II (Figure 6k) that displayed extensive fragmentation, and, on the completion of meiosis, xri1-1 formed polyads (Figure 6l) that contained unequal amounts of fragmented chromosomal material. These results identified the cause of male sterility in homozygous xri1-1 mutants as a severe disruption of meiosis.
To determine whether female meiosis was disrupted as observed for male meiosis, cytological analysis was performed on megaspore mother cells. Embryo sac mother cells prepared from homozygous xri1-1 mutants were similar in appearance at leptotene to wild-type, with chromosomes visible as unsynapsed threads with areas of bright DAPI staining corresponding to the centromeres and nucleolar organizing regions. No further wild-type-like stages of prophase I were identified in the xri1-1 mutants. Instead the embryo sac mother cells that had progressed further through prophase I had chromosomes that had a ‘clumped’ appearance and did not condense to form the same thread-like structures observed in wild-type spreads. There was no evidence of synapsis in xri1-1 female meiosis, and the meiocytes displayed extensive chromosome fragmentation (data not shown). These results of cytological analysis of meiosis of embryo sac mother cells confirmed that female sterility in homozygous xri1-1 mutants is due to an early defect in female meiosis that is similar to that of male meiosis.
Meiotic DNA fragmentation in xri1-1 does not depend on SPO11-1
Programmed DSBs are induced early in meiosis by SPO11-1/SPO11-2 proteins (Grelon et al., 2001; Stacey et al., 2006). These DSBs form a substrate for the HR-mediated repair that is required for chromosome pairing during prophase I. Mutants in HR factors show extensive chromosome fragmentation resulting from an inability to repair SPO11-induced DSBs. This is illustrated by the fact that mutations in SPO11-1 are epistatic to mutations in genes needed for HR repair (e.g. atrad51), with atrad51 spo11-1 double mutants having spo11-1 levels of fertility. The rescue of atrad51 by spo11-1 is confirmed at the cytological level, with atrad51 spo11-1 double mutants losing the chromosome fragmentation observed in atrad51 mutants (Li et al., 2004). To test whether the fragmentation observed in homozygous xri1-1 mutants was due to failure to repair SPO11-induced DSBs, we analysed the progression of male meiosis in an xri1-1 spo11-1 double mutant.
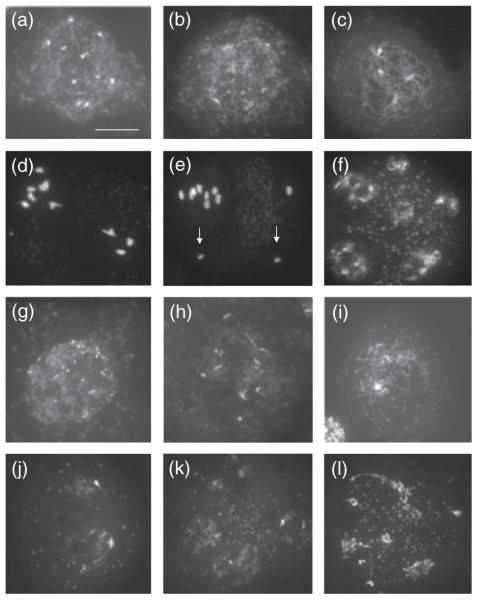
Meiosis in spo11-1 pollen mother cells (Figure 7a–h) appeared similar to that in wild-type plants during early prophase I, although synapsed cells were not seen (Figure 7a–c). At metaphase I (Figure 7d), the chromosomes were always seen as ten univalents, and segregated unevenly at anaphase I leading to unbalanced tetrads (Figure 7f). Cytological analysis of meiotic progression in spo11-1-1 xri1-1 (Figure 7g–l) revealed that double mutant pollen mother cells closely resembled those seen during meiosis of homozygous xri1-1 single mutants. The failure of the spo11-1-1 mutation to rescue xri1-1 meiotic chromosome fragmentation was also reflected in the complete sterility of spo11-1-1 xri1-1 double mutants compared to spo11-1-1 mutants, which produce approximately two seeds per silique (Grelon et al., 2001), and the absence of pollen in spo11-1-1 xri1-1 anthers, which was not observed in spo11-1-1 plants (Figure 5c).
Figure 7.
Progression through male meiosis in spo11-1-1 and spo11-1-1 xri1-1 double homozygotes. The pollen mother cells are stained with DAPI.
(a–f) Homozygous spo11-1-1 showing early leptotene/leptotene (a, b), later asynaptic prophase (c), metaphase I with 10 univalents (d), anaphase II (e) with premature sister chromatid separation shown by arrows, and polyad (f).
(g–l) Pollen mother cells from spo11-1-1 xri1-1 double homozygotes are shown at early leptotene/leptotene (g, h), later asynaptic prophase (i), anaphase I (j), anaphase II (k) and polyad (l). The cells in (j), (k) and (l) show extensive chromosome fragmentation. Scale bar = 10 μm.
A role for XRI1 in pre-meiotic S phase?
These results illustrate that meiosis is severely disrupted in xri1-1 mutants. The failure of the spo11-1-1 mutation to rescue the chromosome fragmentation suggests that the function of XRI1 may be related to preventing the accumulation of DSBs that arise during pre-meiotic DNA replication. The observations do not exclude the possibility that XRI1 has an additional role during subsequent meiotic DNA repair, a scenario consistent with the physical interaction of XRI1 with a novel component of the MND1/HOP2 recombination complex (see below).
Pre-meiotic S phase is a specialized instance of DNA replication, and has a longer duration than mitotic S phase (Armstrong and Jones, 2003). While many of the components of the DNA replication machinery are the same as those of mitotic cells, meiosis-specific factors are also required, which in plants includes the DNA replication factors CDC45 and MEI1; disruption of CDC45 or MEI1 in Arabidopsis results in SPO11-independent DNA fragmentation and sterility, indicating a non-redundant role in pre-meiotic S phase (Grelon et al., 2003; Stevens et al., 2004). The MEI1 protein shows sequence similarity to yeast CUT5/DPB11, which is required in the initiation of DNA replication (Tanaka et al., 2007; Moore and Aves, 2008) and the human homologue TOPBP1. Altering the activities of CDC45 or DPB11 in yeast was sufficient to overcome the requirement for CDK activity to initiate DNA replication (Tanaka et al., 2007). Interestingly, DPB11 (and potentially MEI1) is also required for HR-mediated repair of DNA damage (Grelon et al., 2003; Ogiwara et al., 2006).
Protein interaction studies associate XRI1 with homologous recombination
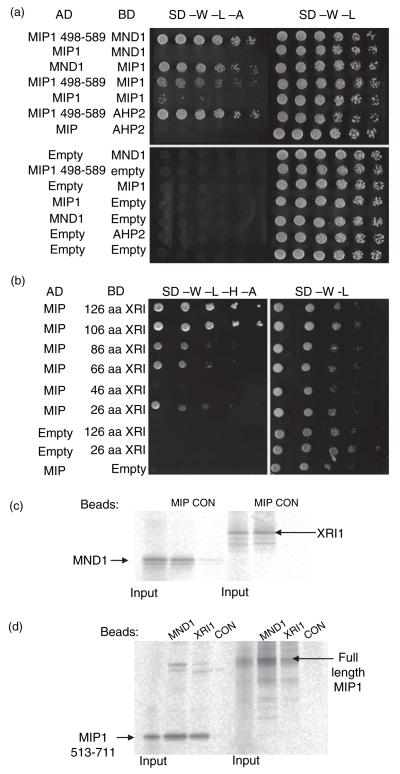
To further characterize the potential roles of XRI1 in DNA replication and/or repair, we performed a yeast two-hybrid screen for proteins that interact with the C-terminal region of XRI1, as full-length XRI1 had intrinsic transcriptional activation activity. Several putative interactors were identified and further tested to verify the yeast two-hybrid results. Of these, one was confirmed by several additional approaches (below), and encoded a protein previously identified in another yeast two-hybrid screen as a protein partner of the meiotic HR factor AtMND1 (Figure 8a). AtMND1-interacting protein 1 (MIP1, At1g32530) has a conserved structural maintenance of chromosomes (SMC) domain, typical of chromosome segregation ATPases (Jessberger, 2002), and a C-terminal RING finger domain, implicated in protein–protein interactions (Figure S1). The AtMND1 interaction screen identified a MIP1 fragment towards the C-terminal region (498–589) as sufficient for At-MND1 binding. This region of MIP also interacted with AHP2 (the Arabidopsis HOP2 homologue), but not with the GAL4 DNA binding domain alone (Figure 8a). These results suggest that MIP1 forms multiple interactions with the AHP2/AtMND1 complex in Arabidopsis.
Figure 8.
Interactions between XRI1, MIP1 and AtMND1.
(a) Yeast two-hybrid analysis showing interaction between MIP1, AtMND1 and AHP2. Yeast grown in liquid culture was plated in a series of fivefold dilutions onto plates selecting for activation and DNA binding domain plasmids (synthetic defined media lacking tryptophan and leucine, SD-W-L) or selecting for plasmids and activation of the ADE2 and HIS3 reporter genes (synthetic defined media lacking tryptophan, leucine, higtidine and adenine, SD-W-L-H-A).
(b) Yeast two-hybrid interaction between a deletion series of XRI1 and the MIP1 C-terminal fragment identified in library screening.
(c) In vitro interaction between radio-labelled AtMND1 or radio-labelled XRI1 and full-length MIP1 immobilized on paramagnetic beads. Control beads were pre-treated with bacterial cell lysate before incubation with the radio-labelled proteins.
(d) In vitro interaction between radio-labelled full-length or C-terminal MIP1 and AtMND1, XRI1 or control beads as indicated.
The C-terminal domain (amino acids 513–711) of MIP1 was sufficient for interaction with XRI1. This region included the RING finger domain and overlapped with the AHP2/AtMND1 interaction domain. Further analysis of XRI1 identified the C-terminal 26 amino acids as sufficient for MIP1 interaction in yeast two-hybrid analysis (Figure 8b). This is the most highly conserved region shared between the three XRI1-like sequences found in Arabidopsis and the two found in rice. Full-length XRI1, expressed as a GAL4 activation domain (AD) fusion, failed to interact with an MIP1–GAL4 DNA binding domain (DB) fusion in yeast two-hybrid analysis, which may reflect conformational or steric constraints that prevent binding of the proteins in this orientation. In vitro studies verified the interactions between MIP1 and both MND1 and XRI1. MND1 and XRI1 bound to MIP1-coated beads, with only trace levels of MND1 binding to Escherichia coli lysate-coated control beads (Figure 8c). Similarly, full-length MIP1 and its C-terminal region bound to MND1- or XRI1-coated beads, but not to controls (Figure 8d). Interaction between XRI1 and MIP was also observed in co-immunoprecipitation experiments using either in vitro translated proteins or extracts of yeast expressing XRI1–GAL4DB and either MIP1–GAL4AD or GAL4AD alone (Figure S2). These data provide further evidence that XRI1 interacts with MIP, a protein that binds MND1, which is required for meiotic HR in eukaryotes.
In mitotic cells, HR is mediated by the conserved RAD52 epistasis group that was first characterized in yeast (Symington, 2002). However, HR-mediated repair in plants may differ from that of yeast, as AtMND1 may be involved in DNA repair in vegetative tissues (Domenichini et al., 2006; Kerzendorfer et al., 2006). HR has multiple roles during DNA replication as it is required for the repair of collapsed replication forks and in post-replication repair pathways (Gangavarapu et al., 2007; Hanawalt, 2007). The links between HR and DNA replication provide an explanation for the expression patterns and mutant phenotypes of Arabidopsis XRI1. A role for XRI1 in DNA replication is indicated by the SPO11-1-independent chromosome fragmentation in meiocytes that is displayed by plants deficient in XRI1, while X-ray induction of XRI1 and interaction with the AtMND1-interacting protein indicate a connection with HR. XRI1 therefore provides a novel link between DNA replication and repair in plants, and, like CDKA;1, is an essential requirement for meiosis and pollen mitosis.
EXPERIMENTAL PROCEDURES
Plant material and growth conditions
Arabidopsis (Col-0) plants were raised in growth chambers under 70% humidity, with 16 h light/8 h dark cycles at 20 °C. Arabidopsis line 075E02 was obtained from GABI-KAT (Rosso et al., 2003), and Col-0 and atatm-3 mutants (SALK_089805) were obtained from the Nottingham Arabidopsis Stock Centre. Agrobacterium-mediated plant transformation was performed as described previously (Clough and Bent, 1998).
Cytological procedures
Mutant and wild-type pollen mother cells were examined by fluorescence microscopy in DAPI-stained spreads as described by Armstrong et al. (2001). Female meiosis was examined in DAPI-stained spreads of embryo sac mother cells as described by Armstrong et al. (2001).
Nucleic acid purification, amplification and cloning
DNA procedures and bacterial manipulations used established protocols (Sambrook et al., 1989). RNA was isolated from above-ground tissues of flowering Arabidopsis using the SV total RNA isolation kit (Promega; http://www.promega.com/) according to the manufacturer’s instructions. XRI1 was cloned by RT-PCR using Superscript II reverse transcriptase (Invitrogen; http://www.invitrogen.com/) for cDNA synthesis followed by amplification using iPROOF (Bio-Rad; http://www.bio-rad.com/). RACE PCR was performed using a FirstChoice RNA ligase-mediated RACE kit (Ambion, http://www.ambion.com), according to the manufacturer’s instructions. PCR products were cloned using a TOPO-TA cloning kit and E. coli TOP10 cells (Invitrogen), and plasmid DNA was prepared using spin columns (Qiagen; http://www.qiagen.com/) prior to fluorescent dye-terminator DNA sequencing (Perkin Elmer, http://www.perkinelmer.com). To determine the subcellular localization of XRI1, the XRI1 cDNA was cloned into pCB1300 containing soluble modified red-shifted (smrs)–GFP (Davis and Vierstra, 1998) under the control of the 35S promoter to create XRI1–smrs–GFP, and used for Agrobacterium-mediated plant transformation. Confocal analysis for detection of GFP fluorescence was performed as described previously (Sunderland et al., 2006). Real-time PCR analysis was performed on an iCycler thermocycler (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad) and primers At5g48720_f (GCTACCTGATGACTTAAACTTTGGTTC) and At5g48720_r (CATTTGGAGAAGATCGAGTCACAG) for XRI1, and qPCR_ACTf (CTCAGGTATCGCTGACCGTATGAG) and qPCR_ACTr (CTTGGAGATCCACATCTGCTGGAATG) for ACTIN2 (At1g49240). AtRAD51 (At5G20850) was amplified using primers rad51RTf (GTTCTTGAGAAGTCTTCAAGAAGTTAG) and rad51RTr (GCTGAACCATCTACTTGCGCAACTAC). The XRI1 and RAD51 transcript levels were normalized against those for ACTIN2. Complementation of the xri1-1 mutation was performed using a genomic clone of chromosome 5 region 19774598–19779380. The complementation construct consisted of a 4782 bp XbaI/SpeI fragment of BAC clone JAtY55H08 cloned into the XbaI site of the pCB1300 binary vector, and this was used to transform heterozygous xri1-1 plants according to standard protocols (Fields and Song, 1989; Durfee et al., 1993).
Overexpression of XRI1, MND1 and MIP1
Protein expression and interaction studies were performed as described previously (Waterworth et al., 2007). Full-length AtMND1 and MIP1 cDNAs were cloned into separate pET32EkLIC vectors (Novagen, http://www.merckbiosciences.co.uk) and XRI1 was cloned into pET30EkLIC, generating C-terminal 6× His-tagged proteins. These constructs were used to transform aliquots of E. coli BL21(DE3)pLysS. Expression was induced using 1 mm IPTG (Promega) once cells had reached an absorbance at 600 nm of 0.4, and growth at 37°C was continued for a further 3 h. Bacteria were recovered by centrifugation at 5000 g for 10 min at 22°C, re-suspended in Bugbuster (Novagen), and lysed by freezing and thawing. Nucleic acids were removed by benzonase treatment (25 U ml−1, Novagen) at 37°C for 15 min, and the extract was cleared by centrifugation at 25 000 g for 30 min at 4°C. As XRI1 and MND1 were soluble, lysate containing the His-tagged proteins [or from untransformed BL21(DE3) controls] was applied to 20 μl of nickel-coated paramagnetic beads (Promega), and washed five times for 1 min with 1 ml fresh binding buffer (100 mm HEPES pH 7.4, 200 mm NaCl, 100 mm imidazole, 0.1% v/v Triton X-100), then resuspended in 100 μl binding buffer and used immediately for in vitro interaction studies. MIP1 was insoluble under various induction conditions, and on-bead renaturation was therefore used. Following the same induction protocol as for XRI1 and MND1, pelleted MIP1 was solubilized in denaturing buffer (100 mm HEPES pH 7.4, 8 m urea, 200 mm NaCl, 10 mm imidazole, 0.1% Triton X-100), and centrifuged at 25 000 g for 10 min at 25°C to remove particulates. The supernatant was applied to 20 μl paramagnetic beads (Promega), washed five times in denaturing buffer, and resuspended in 100 μl denaturing buffer. Renaturation buffer (100 mm HEPES pH 7.4, 200 mm NaCl, 10 mm imidazole, 0.1% Triton X-100) was added slowly to the beads to reduce the urea content to 0.5 m, at which point the beads were recovered and equilibrated with binding buffer as described above.
Yeast two-hybrid analysis
Two-hybrid analysis was performed as described previously (Fields and Song, 1989; Durfee et al., 1993). XRI1 cDNA encoding amino acid residues 175–300 was cloned into plasmid pGBKT7 to create a GAL4DB fusion protein for use in yeast two-hybrid screening. RNA was isolated from two-week-old Arabidopsis seedlings 2 h after exposure to 10 Gy X-rays, and mixed with an equal quantity of RNA isolated from buds and flowers at various stages of development. RT-PCR and in vivo cloning were used to generate an activation domain library and perform the library screening using the Match-maker kit according to the manufacturer’s instructions (Clontech; http://www.clontech.com/). Transformants were plated directly onto selective media lacking tryptophan, leucine and histidine, and supplemented with 2.5 mm 3-amino-1,2,4-triazole. Colonies growing after one week were re-plated onto selective media that also lacked adenine, before analysis by PCR and sequencing. Interactions were verified by plasmid isolation and re-transformation into the yeast reporter strain AH109 (MATa, trp1-901, leu2-3, 112, ura3-52, his3-200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3, GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1) as described previously (Soni et al., 1993).
Preparation and microscopic analysis of plant tissues
Alexander staining was performed on anthers according to published protocols (Johnson-Brousseau and McCormick, 2004). DAPI (4′,6-diamidino-2-phenylindole) staining of pollen grains was performed as described by Park et al. (1998), and viewed on a Zeiss LSM 510 META Axiovert 200M inverted confocal microscope. Preparation of embryo sac and pollen mother cells was as described previously (Armstrong et al., 2001), except cytohelicase was omitted from the digestion mix and the digestion time was extended to 75 min.
ACKNOWLEDGEMENTS
We thank Mathilde Grelon (Station de Génétique et Amélioration des Plantes, INRA, Versailles) for the kind gift of spo11-1-1. We acknowledge the financial support of the UK Biotechnology and Biological Sciences Research Council to C.E.W. (grant numbers BBSSC200412599 and JF20608) and S.J.A. (grant numbers BBSSC200512932 and BBF0028581). Funding from the Royal Society to C.E.W. is also gratefully acknowledged. This work was furthermore supported by the Austrian Science Foundation (grant P18036 to P.S.) and the Austrian Academy of Sciences (APART fellowship for P.S.).
Footnotes
SUPPORTING INFORMATION
Additional Supporting Information may be found in the online version of this article:
Figure S1. Sequence analysis of MND1-INTERACTING PROTEIN 1 (MIP1).
Figure S2. Interaction between XRI1 and MIP1 by co-immunoprecipitation of in vitro-transcribed and translated epitope-tagged proteins, and in yeast lysates.
Table S1. BLASTP database search for proteins with sequence similarity to XRI1.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- Armstrong SJ, Jones GH. Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J. Exp. Bot. 2003;54:1–10. doi: 10.1093/jxb/erg034. [DOI] [PubMed] [Google Scholar]
- Armstrong SJ, Franklin FC, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J. Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, Savigny F, White CI. Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 2005;41:533–545. doi: 10.1111/j.1365-313X.2004.02318.x. [DOI] [PubMed] [Google Scholar]
- Bray CM, West CE. DNA repair mechanisms in plants: crucial sensors and effectors for the maintenance of genome integrity. New Phytol. 2005;168:511–528. doi: 10.1111/j.1469-8137.2005.01548.x. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Couteau F, Belzile F, Horlow C, Grandjean O, Vezon D, Doutriaux MP. Random chromosome segregation without meiotic arrest in both male and female meiocytes of a dmc1 mutant of Arabidopsis. Plant Cell. 1999;11:1623, 1634. doi: 10.1105/tpc.11.9.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culligan KM, Robertson CE, Foreman J, Doerner P, Britt AB. ATR and ATM play both distinct and additive roles in response to ionizing radiation. Plant J. 2006;48:947–961. doi: 10.1111/j.1365-313X.2006.02931.x. [DOI] [PubMed] [Google Scholar]
- Davis SJ, Vierstra RD. Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 1998;36:521–528. doi: 10.1023/a:1005991617182. [DOI] [PubMed] [Google Scholar]
- Domenichini S, Raynaud C, Ni D-A, Henry Y, Bergounioux C. Atmnd1-Δ1 is sensitive to gamma-irradiation and defective in meiotic DNA repair. DNA Repair. 2006;5:455–464. doi: 10.1016/j.dnarep.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Durfee T, Becherer K, Chen PL, Yeh SH, Yang Y, Kilburn AE, Lee WH, Elledge SJ. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein–protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Friesner JD, Liu B, Culligan K, Britt AB. Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell. 2005;16:2566–2576. doi: 10.1091/mbc.E04-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangavarapu V, Prakash S, Prakash L. Requirement of RAD52 group genes for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 2007;27:7758–7764. doi: 10.1128/MCB.01331-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Bruchet H, Camescasse D, Granier F, Bouchez D, Tissier A. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15:119–132. doi: 10.1105/tpc.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grelon M, Gendrot G, Vezon D, Pelletier G. The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 2003;35:465–475. doi: 10.1046/j.1365-313x.2003.01820.x. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC. Paradigms for the three rs: DNA replication, recombination, and repair. Mol. Cell. 2007;28:702, 707. doi: 10.1016/j.molcel.2007.11.014. [DOI] [PubMed] [Google Scholar]
- Iwakawa H, Shinmyo A, Sekine M. Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 2006;45:819–831. doi: 10.1111/j.1365-313X.2005.02643.x. [DOI] [PubMed] [Google Scholar]
- Jessberger R. The many functions of SMC proteins in chromosome dynamics. Nat. Rev. Mol. Cell Biol. 2002;3:767–778. doi: 10.1038/nrm930. [DOI] [PubMed] [Google Scholar]
- Johnson-Brousseau SA, McCormick S. A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically-expressed genes. Plant J. 2004;39:761–775. doi: 10.1111/j.1365-313X.2004.02147.x. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Kerzendorfer C, Vignard J, Pedrosa-Harand A, et al. The Arabidopsis thaliana MND1 homologue plays a key role in meiotic homologous pairing, synapsis and recombination. J. Cell Sci. 2006;119:2486–2496. doi: 10.1242/jcs.02967. [DOI] [PubMed] [Google Scholar]
- Li W, Chen C, Markmann-Mulisch U, Timofejeva L, Schmelzer E, Ma H, Reiss B. The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl Acad. Sci. USA. 2004;101:10596–10601. doi: 10.1073/pnas.0404110101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S. Control of male gametophyte development. Plant Cell. 2004;16:S142–S153. doi: 10.1105/tpc.016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K, Aves SJ. Mcml0 and DNA replication in fission yeast. SEB Exp. Biol. Ser. 2008;59:45–69. [PubMed] [Google Scholar]
- Ogiwara H, Ui A, Onoda F, Tada S, Enomoto T, Seki M. Dpb11, the budding yeast homolog of TopBP1, functions with the checkpoint clamp in recombination repair. Nucleic Acids Res. 2006;34:3389–3398. doi: 10.1093/nar/gkl411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SK, Howden R, Twell D. The Arabidopsis thaliana gametophytic mutation gemini pollen1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development. 1998;125:3789–3799. doi: 10.1242/dev.125.19.3789. [DOI] [PubMed] [Google Scholar]
- Petukhova GV, Pezza RJ, Vanevski F, Ploquin M, Masson JY, Camerini-Otero RD. The Hop2 and Mnd1 proteins act in concert with Rad51 and Dmc1 in meiotic recombination. Nat. Struct. Mol. Biol. 2005;12:449–453. doi: 10.1038/nsmb923. [DOI] [PubMed] [Google Scholar]
- Ricaud L, Proux C, Renou JP, Pichon O, Fochesato S, Ortet P, Montane MH. ATM-mediated transcriptional and developmental responses to gamma-rays in Arabidopsis. PLoS ONE. 2007;2:e430. doi: 10.1371/journal.pone.0000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Manniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- Schommer C, Beven A, Lawrenson T, Shaw P, Sablowski R. AHP2 is required for bivalent formation and for segregation of homologous chromosomes in Arabidopsis meiosis. Plant J. 2003;36:1–11. doi: 10.1046/j.1365-313x.2003.01850.x. [DOI] [PubMed] [Google Scholar]
- Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- Soni R, Carmichael JP, Murray JA. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr. Genet. 1993;24:455–459. doi: 10.1007/BF00351857. [DOI] [PubMed] [Google Scholar]
- Stacey NJ, Kuromori T, Azumi Y, Roberts G, Breuer C, Wada T, Maxwell A, Roberts K, Sugimoto-Shirasu K. Arabidopsis SPO11-2 functions with SPO11-1 in meiotic recombination. Plant J. 2006;48:206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- Stevens R, Grelon M, Vezon D, Oh J, Meyer P, Perennes C, Domenichini S, Bergounioux C. A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell. 2004;16:99–113. doi: 10.1105/tpc.016865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunderland PA, West CE, Waterworth WM, Bray CM. An evolutionarily conserved translation initiation mechanism regulates nuclear or mitochondrial targeting of DNA ligase 1 in Arabidopsis thaliana. Plant J. 2006;47:356–367. doi: 10.1111/j.1365-313X.2006.02791.x. [DOI] [PubMed] [Google Scholar]
- Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Vignard J, Siwiec T, Chelysheva L, Vrielynck N, Gonord F, Armstrong SJ, Schlogelhofer P, Mercier R. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 2007;3:1894–1906. doi: 10.1371/journal.pgen.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, Altun C, Armstrong SJ, Roberts N, Dean PJ, Young K, Weil CF, Bray CM, West CE. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 2007;52:41–52. doi: 10.1111/j.1365-313X.2007.03220.x. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Yang C. Plant gametogenesis: conservation and contrasts in development. Reproduction. 2004;128:483–492. doi: 10.1530/rep.1.00306. [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]