Abstract
Programmed necrosis or necroptosis is controlled by the action of two serine/threonine kinases, receptor interacting protein kinase 1 (RIP1) and RIP3. The phosphorylation of RIP1 and RIP3 is critical for assembly of the necrosome, an amyloid-like complex that initiates transmission of the pro-necrotic signal. Here, we used site-directed mutagenesis to systematically examine the effects of putative phosphor acceptor sites on RIP1 and RIP3 on TNF-induced programmed necrosis. We found that mutation of individual serine residues in the kinase domain of RIP1 had little effects on RIP1 kinase activity and TNF-induced programmed necrosis. Surprisingly, alanine substitution of Ser89 enhanced RIP1 kinase activity and TNF-induced programmed necrosis without affecting RIP1-RIP3 necrosome formation. This indicates that Ser89 is an inhibitory phospho-acceptor site that can dampen the pro-necrotic function of RIP1. In addition, we show that a phosphor-mimetic mutant of RIP3, S204D, led to programmed necrosis that was refractory to RIP1 siRNA and insensitive to necrostatin-1 inhibition. Our results show that programmed necrosis is regulated by positive and inhibitory phosphorylation events.
Keywords: programmed necrosis, necroptosis, RIP1, RIPK1, RIP3, RIPK3, TNF
Introduction
Programmed necrosis/necroptosis is an inflammatory form of cell death with important functions in certain viral infections and trauma-induced tissue injury. TNF and related death ligands are strong inducers of programmed necrosis. Induction of programmed necrosis requires the assembly of the amyloid-like RIP1-RIP3 necrosome [1, 2]. The necrosome recruits and activates downstream RIP3 substrates such as the mixed lineage kinase domain-like (MLKL) and phosphoglycerate mutase family member 5 (Pgam5) to effect programmed necrosis [3–5]. Necrosome assembly and activation is controlled by three major types of post-translational modifications: caspase 8-mediated proteolytic cleavage, protein ubiquitination, and phosphorylation [6]. Upon TNF signaling, RIP1 bound to the TNFR-1 complex undergoes polyubiquitination. Polyubiquitinated RIP1 binds NEMO and other adaptors that are important for NF-κB signaling and is sterically restricted from engaging cytoplasmic death inducing factors including RIP3 [7]. The deubiquitinase CYLD promotes programmed necrosis by facilitating removal of the ubiquitin chains on RIP1 [8, 9].
Caspase-8 and its upstream adaptor Fadd are crucial for caspase-dependent apoptosis induced by TNF-like death cytokines. They inhibit programmed necrosis by cleavage and inactivation of essential necrosis mediators including RIP1, RIP3 and CYLD [10–12]. When caspase activity is inhibited, such as that in mice with genetic deletion of caspase-8 or its upstream adaptor Fadd, extensive necrosis ensued, giving rise to extensive injury-induced tissue inflammation and embryonic lethality [13–19].
The kinase function of RIP1 and RIP3 is essential for assembly and activation of the necrosome [20–22]. RIP1 is generally believed to be the apical kinase that phosphorylates RIP3 to initiate necrosis signaling. Consistent with this notion, necrostatin-1 (Nec-1), a RIP1 inhibitor [23], blocks necrosis-specific RIP1 and RIP3 phosphorylation [20]. However, neither RIP1 nor RIP3 was phosphorylated under necrosis inducing conditions in RIP3−/− cells [20]. These results raise the possibility that RIP3 might in fact act upstream to promote RIP1 activation. Identifying the key phosphor acceptor sites on RIP1 and RIP3 that regulate their activities will help clarify the hierarchical relationship between RIP1 and RIP3 in the necrosis signaling pathway. To this end, several serine residues in the kinase domains of RIP1 and RIP3 have been identified [3, 21, 23]. However, their roles in programmed necrosis have not been carefully investigated. It is also not known if phosphatases might counter necrosis by inhibiting the function of the RIP kinases.
In this report, we show that the previously identified phosphorylation sites on RIP1 had minimal effects on RIP1-dependent programmed necrosis. Instead, we identified Ser89 as a novel regulatory serine residue on RIP1. Alanine substitution at Ser89 resulted in hyperactive RIP1 kinase activity and increased TNF-induced programmed necrosis. However, necrosome assembly was not altered. We further show that cells expressing the phosphor-mimetic, gain-of-function RIP3 mutant S204D underwent programmed necrosis that was partially independent of RIP1. These results indicate that RIP1 activity is regulated by positive and inhibitory phosphorylation. Moreover, phosphorylation of RIP3 at Ser204 is likely a critical mechanism by which RIP1 promotes necrosis signaling.
Results
Ser161 controls sensitivity of RIP1 to necrostatin-1
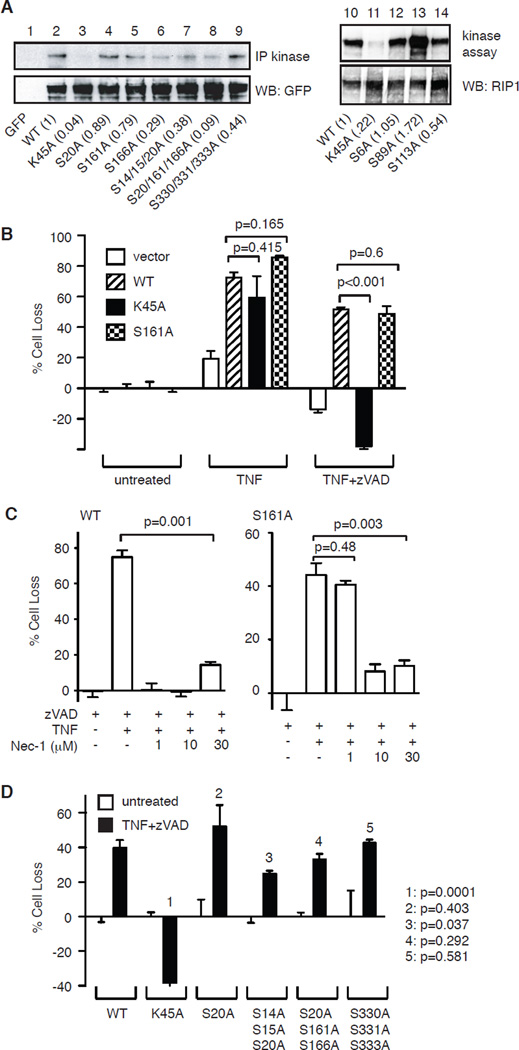
During programmed necrosis, RIP1 and RIP3 form filamentous punctate structures that exhibit amyloid-like properties. Amyloid assembly requires the “RIP homotypic interaction motif” (RHIM) that is present in both RIP1 and RIP3 [6]. Interestingly, expression of truncated RIP1 lacking the N-terminal kinase domain, but not full-length RIP1, resulted in spontaneous clustering of RIP1 into amyloid fibrils [2]. These results suggest that the kinase domain may mask the RHIM to prevent it from forming amyloid filaments. Upon activation, the negative charge generated from phosphorylation of the kinase domain may relieve this inhibition to permit RHIM-RHIM interaction and amyloid assembly. Indeed, the kinase domain of RIP1 was reported to be heavily phosphorylated at multiple serine and threonine residues [23]. In particular, structural modeling of RIP1 on B-Raf suggested that phosphorylation of Ser161 might regulate the pro-necrotic kinase activity of RIP1 [23]. We tested the function of Ser161 and found that alanine substitution only reduced RIP1 kinase activity by about 20% (Fig. 1A, compare lanes 2 and 5). We previously showed that reconstitution of wild type RIP1 into RIP1-deficient Jurkat cells stably expressing TNFR-2 restored TNF-induced programmed necrosis and enhanced TNF-induced apoptosis [24]. Using this system, we found that S161A-RIP1 enhanced TNF-induced apoptosis and restored programmed necrosis to similar levels achieved by wild type RIP1 (Fig. 1B). As control, kinase inactive RIP1 (K45A) only enhanced TNF-induced apoptosis, but did not restore TNF-induced programmed necrosis (Fig. 1B). Therefore, despite the minor reduction in kinase activity, S161A-RIP1 is functionally competent to mediate TNF-induced programmed necrosis.
Figure 1. The effect of serine phosphorylation on RIP1 kinase activity.
(A) Effect of alanine substitutions of reported phosphorylation sites on RIP1 kinase activity. HEK293T cells were transfected with the indicated GFP-tagged RIP1 plasmids. The in vitro kinase activity was determined using RIP1 autophosphorylation as readout. The numbers in parentheses represent kinase activity normalized to expression level of RIP1-GFP on bottom panel. (B) Ser161 does not control the pro-necrotic function of RIP1. RIP1-deficient Jurkat cells were transfected with the indicated RIP1-GFP plasmids or empty GFP vector. Transfected GFP+ cells were tested for TNF-induced apoptosis or TNF and zVAD-induced programmed necrosis. (C) Ser161 controls sensitivity to Nec-1 inhibition. RIP1-deficient Jurkat cells stably expressing wild type RIP1-GFP or S161A-RIP1-GFP were treated with TNF, zVAD, and the indicated doses of Nec-1. (D) RIP1-deficient Jurkat cells were transfected with the indicated RIP1-GFP plasmids. Transfected GFP+ cells were tested for TNF and zVAD-induced programmed necrosis. p values were calculated by comparing WT transfectants with the indicated mutants using Student’s t test.
Because Ser161 resides in a potential regulatory loop that contacts the RIP1 inhibitor necrostatin-1 (Nec-1) [25], we asked if this mutation might instead affect the sensitivity of RIP1 to Nec-1. We generated RIP1-deficient Jurkat cells that stably express WT RIP1-GFP or S161A-RIP1-GFP. Dose titration showed that TNF-induced programmed necrosis was inhibited by 1 µM of Nec-1 in cells expressing wild type RIP1, but not S161A-RIP1 (Fig. 1C). In contrast, higher doses of Nec-1 inhibited programmed necrosis in both cell types. Hence, Ser161 was not critical for the pro-necrotic function of RIP1. Rather, it contributes to Nec-1 mediated inhibition of RIP1 kinase activity.
Ser89 phosphorylation limits RIP1 kinase activity
The lack of effect of S161A-RIP1 on programmed necrosis prompted us to evaluate if other reported phospho-serine residues in the kinase domain might regulate RIP1 function. We introduced alanine substitutions at other previously reported phospho-acceptor sites and found that the majority of the single site mutants as well as many compound mutations had little effects on RIP1 kinase activity (Fig. 1A, compare lanes 1–9, 12 and 14). Moreover, expression of these mutants in RIP1-deficient Jurkat cells restored TNF-induced programmed necrosis to similar levels achieved by wild type RIP1 (Fig. 1D). These results show that individually, the previously identified phosphorylation sites on RIP1 have minimal role in programmed necrosis.
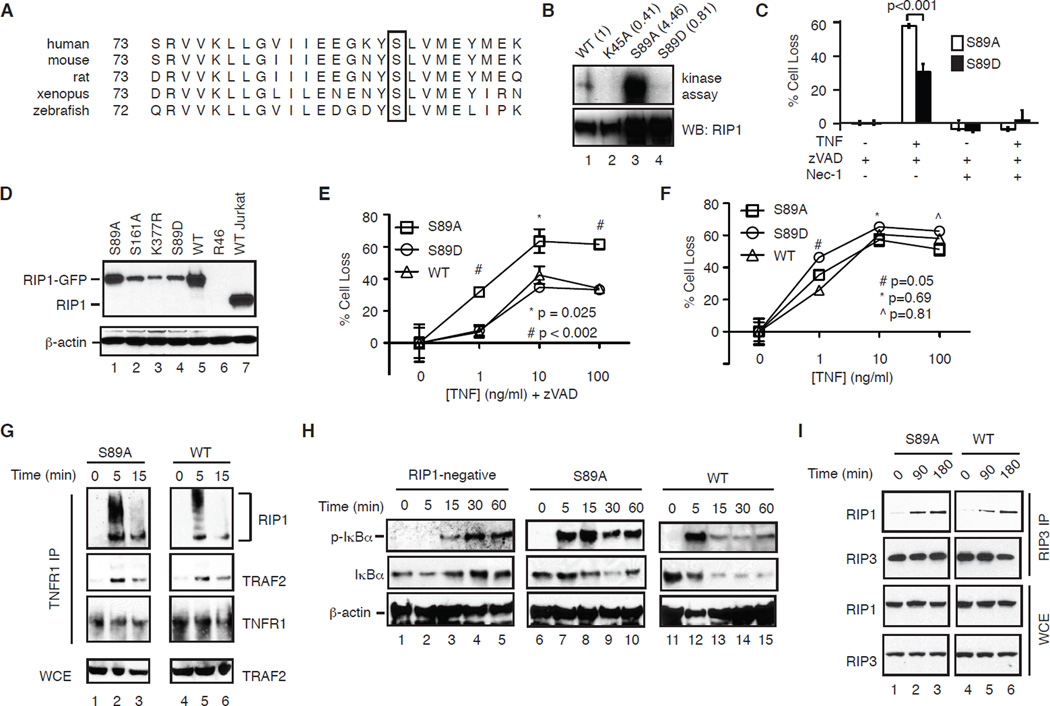
During our analysis, we noticed that S89A-RIP1 was hyperactive compared with wild type RIP1 and other RIP1 mutants in in vitro kinase assay (Fig. 1A, lane 13). Ser89 in the kinase domain of RIP1 is conserved in divergent species including xenopus and zebrafish (Fig. 2A). We suspected that phosphorylation of Ser89 might inhibit RIP1 kinase function and programmed necrosis. To test our hypothesis, we introduced aspartic acid substitution to mimic the negative charge generated from phosphorylation. In contrast to S89A-RIP1, S89D-RIP1 showed reduced kinase activity compared to WT RIP1 (Fig. 2B, compare lanes 1–4). Transient expression into RIP1-deficient Jurkat cells revealed that S89A-RIP1 restored programmed necrosis to a higher level compared to that by S89D-RIP1 (Fig. 2C). To further elucidate the mechanism by which Ser89 regulates RIP1 function, we generated RIP1-deficient Jurkat cells that stably express WT RIP1, S89A-RIP1 or S89D-RIP1 (Fig. 2D). Importantly, Jurkat cells reconstituted with S89A-RIP1 restored TNF-induced programmed necrosis to a higher level than that achieved by S89D-RIP1 or WT RIP1 (Fig. 2E). Despite slightly lower expression level, cells expressing S89D-RIP1 responded similarly to TNF-induced programmed necrosis compared with WT RIP1 expressing cells (Fig. 2E). This suggests that the reduced kinase activity in S89D-RIP1 was sufficient to initiate programmed necrosis (Fig. 2B, lane 4). In contrast, TNF-induced apoptosis was comparable in all cell lines (Fig. 2F). These results strongly implicate that phosphorylation of Ser89 inhibits RIP1 kinase activity to limit RIP1-dependent programmed necrosis.
Figure 2. S89A-RIP1 promotes necrosome assembly and programmed necrosis.
(A) Sequence alignment showing conservation of Ser89 in RIP1 from different species. (B) The indicated RIP1-GFP mutants were expressed and purified from HEK293T cells and subjected to in vitro kinase. (C) RIP1-deficient Jurkat cells were transiently transfected with the indicated RIP1-GFP plasmids and tested for TNF and zVAD-induced programmed necrosis. (D) Expression of RIP1-GFP in different clones of RIP1-deficient Jurkat cells. (E) S89A-RIP1 cells exhibited higher level of TNF and zVAD-induced programmed necrosis. (F) TNF-induced apoptosis in Jurkat cells expressing S89A, S89D or WT RIP1. (G) Similar assembly of TNFR-1 signaling complex in WT and S89A-RIP1 Jurkat cells. (H) TNF-induced phosphorylation of IκBα in parental RIP1-deficient Jurkat and clones expressing different RIP1. (I) Assembly of the RIP1-RIP3 necrosome is not affected in the S89A-RIP1 Jurkat cells.
Ser89 does not affect assembly of the RIP1-RIP3 necrosome
NF-κB activation protects cells against the cytotoxic effects of death cytokines. RIP1 polyubiquitination at the TNFR-1 complex has been shown to inhibit cell survival through NF-κB dependent and independent mechanisms [7]. We compared RIP1 ubiquitination in TNFR-1 complexes and found that RIP1 polyubiquitin chains were somewhat shorter in S89A-RIP1 cells compared with those in WT RIP1 cells (Fig. 2G, compare lanes 2 and 5). This led us to further examine if NF-κB activation was impaired. We confirmed that RIP1-deficient Jurkat cells were defective in early, but not late IκBα phosphorylation (Fig. 2H, lanes 1–5). Cells reconstituted with either WT or S89A-RIP1 fully restored this early IκBα phosphorylation deficiency (Fig. 2H, compare lanes 2, 7 and 12). In fact, IκBα phosphorylation was more robust in S89A-RIP1 cells at all time points tested (Fig. 2H). Moreover, we did not observe detectable level of p100 in Jurkat cells (data not shown), indicating that non-canonical NF-κB activation also did not contribute to the increased necrosis in S89A-RIP1 expressing cells. Assembly of the RIP1-RIP3 necrosome is critical for programmed necrosis [2]. We found that formation of the RIP1-RIP3 necrosome moderately increased at 90 minutes post-TNF stimulation. However, the overall level of RIP1-RIP3 necrosome was not greatly altered by 3 hours after TNF stimulation (Fig. 2I, compare lanes 1–3 with 4–6). Hence, enhanced programmed necrosis in S89A-RIP1 cells was not due to reduced NF-κB activation or necrosome assembly. Rather, these results suggest that phosphorylation of Ser89 inhibits the kinase activity of RIP1 at a step subsequent to necrosome assembly.
Ser204 regulates the pro-necrotic function of RIP3
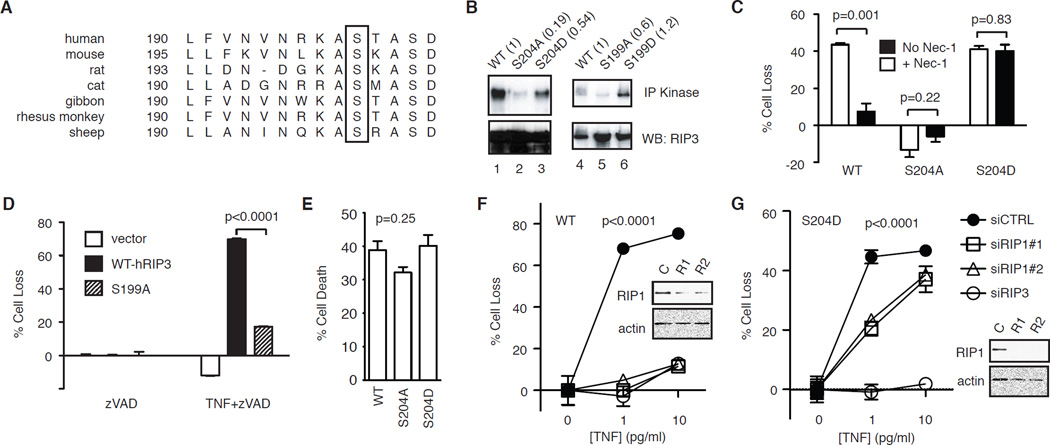
Similar to RIP1, RIP3 is also phosphorylated during programmed necrosis. Sequence alignment revealed that Ser204 in the kinase domain of mouse RIP3 (Ser199 for human RIP3) is highly conserved among different species (Fig. 3A). Previous work shows that S199A-RIP3 mutant was unable to undergo necrosis-dependent phosphorylation [21]. However, it was not clear if that was due to the mutation causing protein misfolding or abolition of an important phospho-acceptor site. We confirmed that alanine substitution at Ser204 (S199A for human RIP3 and S204A for mouse RIP3) abolished in vitro kinase activity of RIP3 (Fig. 3B, lanes 2 and 5). By contrast, aspartic acid substitution mutants (S204D for mouse RIP3 and S199D for human RIP3) retained in vitro kinase activity (Fig. 3B, lane 3 and 6). When introduced into RIP3−/− fibroblasts, S204D-RIP3, but not S204A-RIP3, restored TNF-induced programmed necrosis to level similar to that achieved by wild type RIP3 (Fig. 3C). Similar results were obtained with human RIP3 mutants (Fig. 3D). Consistent with the fact that RIP3 does not participate in death receptor-induced apoptosis, TNF and cycloheximide induced apoptosis was not affected by mutation of Ser204 (Fig. 3E).
Figure 3. Ser204 critically regulates RIP3 function.
(A) Sequence alignment of RIP3 reveals conservation of Ser204 (in mouse RIP3) among different species (box). (B) Effects of alanine and aspartic acid substitution of Ser204 on mRIP3 and Ser199 on hRIP3 kinase activity. The indicated RIP3-GFP fusion proteins were expressed in 293T cells were subjected to in vitro kinase assay. The numbers in parentheses represent the normalized kinase activity compared to wild type RIP3. (C) RIP3−/− fibroblasts stably expressing the indicated RIP3-GFP fusion proteins were treated with TNF and zVAD to induce programmed necrosis, with or without 1 µM Nec-1. (D) The indicated RIP3-GFP plasmids were transfected into RIP3−/− fibroblasts and tested for TNF and zVAD-induced programmed necrosis. (E) RIP3−/− fibroblasts stably transfected with the indicated RIP3-GFP plasmids were tested for TNF and cycloheximide (CHX) induced apoptosis. One-way ANOVA analysis was performed to compare the three different groups. (F-G) RIP3−/− fibroblasts stably expressing the indicated WT or S204D RIP3-GFP fusion protein were transfected with the indicated siRNAs. TNF and zVAD-induced programmed necrosis was determined. Western blots show the level of silencing by the RIP siRNAs. Two-way ANOVA analysis was performed.
Although wild type and S204D-RIP3 restored TNF-induced programmed necrosis to similar levels, we found that cells expressing S204D-RIP3 were not protected from TNF-induced programmed necrosis by Nec-1. In contrast, cells transfected with WT RIP3 were completely protected by Nec-1 (Fig. 3C). This surprising result indicates that RIP1 kinase function is not essential for S204D-RIP3-mediated programmed necrosis. To further elucidate the mechanism of programmed necrosis in S204D-RIP3 cells, we transfected siRNAs against RIP1 into wild type RIP3 and S204D-RIP3 expressing fibroblasts. We found that WT RIP3 expressing cells were completely protected from TNF-induced programmed necrosis by two different RIP1 siRNAs despite incomplete silencing of RIP1 expression (Fig. 3F). By contrast, RIP1 siRNAs only partially protected cells expressing S204D-RIP3 from TNF-induced programmed necrosis, despite more efficient silencing of RIP1 expression (Fig. 3G). RIP3 siRNA equally protected both cell types from programmed necrosis (Fig. 3F–G). These results strongly suggest that a primary function of RIP1 in programmed necrosis is to directly or indirectly facilitate RIP3 phosphorylation at Ser204. However, RIP1 phosphorylation of RIP3 at other sites or other substrates is required for full activation of programmed necrosis.
Ser232 regulates RIP3 function in programmed necrosis, but not its kinase activity
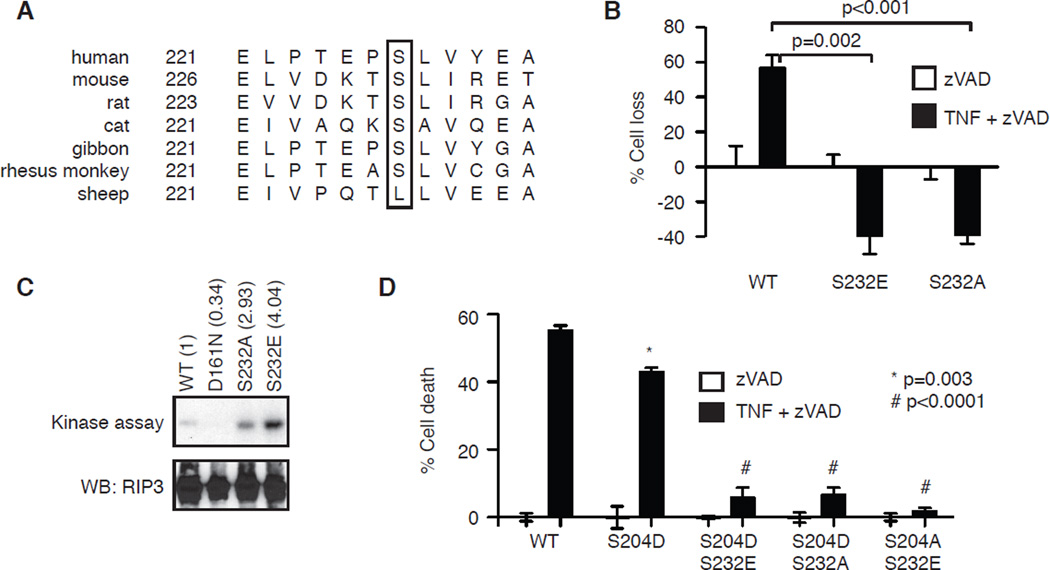
Besides Ser204, phosphorylation of Ser232 (Ser227 in human RIP3) was recently implicated to promote RIP3 function [3]. Specifically, S227A-hRIP3 and S232A-mRIP3 were unable to bind MLKL [3, 26]. As such, phosphorylation of Ser232 was thought to be critical for recruitment of MLKL. However, sequence alignment revealed that this serine residue is not conserved in sheep RIP3 (Fig. 4A). This observation prompted us to re-evaluate Ser232 phosphorylation in programmed necrosis. Consistent with previous reports, S232A-RIP3 was unable to restore TNF and zVAD-induced programmed necrosis in RIP3−/− fibroblasts (Fig. 4B). Surprisingly, the phosphomimetic mutant S232E-RIP3 also failed to restore programmed necrosis in RIP3−/− fibroblasts (Fig. 4B). In vitro kinase assay showed that S232A-RIP3 and S232E-RIP3 retained kinase activity (Fig. 4C). These results show that Ser232 phosphorylation dose not control RIP3 kinase activity and that the negative charge from Ser232 phosphorylation is not sufficient for MLKL binding and programmed necrosis. We reasoned that mutations at Ser232 might simply disrupt the binding surface for MLKL. To test this hypothesis, we introduced S232A or S232E into the gain-of-function S204D-RIP3 mutant and generated RIP3−/− fibroblasts that stably expressed these double mutants. Consistent with our hypothesis, both S204D/S232A-RIP3 and S204D/S232E-RIP3 failed to restore TNF and zVAD-induced programmed necrosis in RIP3−/− fibroblasts (Fig. 4D). Hence, we conclude that the binding surface of RIP3 for MLKL is highly sensitive to perturbations and that the mere presence of negative charge is not sufficient to mediate binding to MLKL and programmed necrosis.
Figure 4. Ser232 does not regulate RIP3 function in programmed necrosis.
(A) Sequence alignment of RIP3 reveals that Ser232 (in mouse RIP3) is not conserved in sheep RIP3 (box). (B) RIP3−/− fibroblasts were transiently transfected with the indicated GFP-tagged RIP3. Programmed necrosis was induced by TNF and zVAD. Cell death was determined in the GFP+ population. (C) The indicated RIP3-GFP fusion proteins were expressed in 293T cells and subjected to in vitro kinase assay. Autophosphorylation of RIP3 was revealed. (D) RIP3−/− fibroblasts stably expressing the indicated RIP3-GFP. Programmed necrosis was induced by TNF and zVAD. p values represent comparison between WT and mutant clones using Student’s t test.
Discussion
In this study, we show that Ser89 in RIP1 and Ser204 in RIP3 play critical roles in programmed necrosis signaling. Neither of these residues was identified as phospho-acceptor sites in mass spectroscopic analyses [3, 23, 26]. This might be due to the fact that only a small fraction of the total cellular pool of RIP1 and RIP3 is involved in TNF signaling and that the active RIP1-RIP3 complex is sequestered in detergent insoluble compartment [2]. Our mutagenesis studies revealed several important mechanistic insights into the control of programmed necrosis. First, the S89A-RIP1 mutant revealed that programmed necrosis could be regulated by positive as well as inhibitory phosphorylation. Presently, we do not know the identity of the kinase that mediates this phosphorylation. However, we can rule out PKA, PKC and Jnk as the kinase that phosphorylates RIP1 at this site (data not shown). Conversely, dephosphorylation of Ser89 may be an important regulatory event for TNF-induced programmed necrosis. To this end, it is noteworthy that the recently identified Pgam5 promotes TNF-induced necrosis by acting as a phosphatase [5]. In addition, the phosphatase WIP1 has been shown to negatively regulate NF-κB signaling [27, 28]. It will be interesting to determine if Pgam5 or other phosphatases have a similar regulatory role in promoting RIP1 dephosphorylation and necrosis. Second, our results suggest that a key function of RIP1 is to facilitate RIP3 phosphorylation at Ser204. Our results are consistent with recent observations that RIP3-dependent, but RIP1-independent necrosis could occur under certain conditions [29, 30]. Interestingly, RIP3 was not phosphorylated by RIP1 in in vitro kinase assays [20]. These discrepant results raise the possibility that a third kinase may be involved in RIP3 activation. Finally, our results show that the mere presence of negative charge at Ser232 is not sufficient to mediate necrosis signaling. Rather, phosphorylation of T231/Ser232 likely creates a conformation-sensitive binding surface that is conducive for recruitment of downstream factors such as MLKL [26].
Methods
Reagents
Antibodies used are: GFP (Roche), RIP1, TRAF2, β-actin (BD Pharmingen), p-IκBα and total IκBα (Cell Signaling), TNFR-1 (R&D Systems and Abcam) and RIP3 (ProSci). Jurkat cells and fibroblasts were grown in RPMI1640 and DMEM media respectively, supplemented with 10% fetal bovine serum (Invitrogen). GFP-tagged RIP1 and RIP3 have been described [20]. All mutants were generated by Quikchange site directed mutagenesis (Stratagene) on the GFP-tagged RIP1 and RIP3 backbone. The RIP1 clones were also tagged at the N-termini with HA. The mutations and sequence integrity were confirmed by DNA sequencing.
Immunoprecipitations and Western blotting
Ten million Jurkat cells were resuspended in 1-ml of medium, pretreated with 50 mM zVAD-fmk, and stimulated with 100 ng/ml TNF for the indicated times. After washing with cold PBS, cells were lysed with 150 mM NaCl, 0.2% NP-40, 1 mM EDTA, 3 mM NaF, 1 mM b-glycerophosphate, 1 mM sodium orthovanadate, 10% glycerol supplemented with protease and phosphatase inhibitor cocktails (Sigma and Roche). After pre-clearing with sepharose 6B beads (Sigma), lysates were incubated with TNFR-1 antibody (R&D Systems) or RIP3 antibody (ProSci) for overnight. The resulting immune complexes were washed and resolved on SDS-PAGE. For p-IκBα Western blot, 10 ng/ml of TNF was used.
In vitro kinase assays
HEK293T cells were transfected with different GFP-tagged RIP1 or RIP3 mutants using Fugene HD (Promega) as per manufacturer’s instructions. Twenty-four hours post-transfections, cells were washed with PBS, lysed in buffer containing 150 mM NaCl, 20 mM Tris-Cl [pH7.5], 1% NP-40, 1 mM EDTA, 1 mM sodium orthovanadate, 3 mM NaF, 2 mM β-glycerophosphate and supplemented with Complete protease inhibitor cocktail (Roche). The lysates were incubated with anti-GFP antibody (Roche) and Dynabead protein G magnetic beads for 4 hours to overnight. The immune complex was washed and resuspended in kinase buffer (20 mM β-glycerophosphate, 20 mM MgCl2, 20 mM MnCl2, 1 mM EDTA, 100 µM cold ATP, 10 µCi γ-32P-ATP). After incubation at 30°C for 30 minutes, the complex was boiled and resolved by SDS-PAGE. Kinase activity was determined by autophosphorylation and radiography.
Generation of stable Jurkat and fibroblasts
RIP1-deficient Jurkat cells stably transfected with TNFR-2 were previously described [24]. Wild type and mutant RIP1 (HA-tagged at the N-terminus and GFP-tagged at the C-terminus) were transfected to these TNFR2+ RIP1-deficient Jurkat cells along with pEF6/Myc-HisA (Invitrogen, for blasticidin selection) by electroporation as described before [31]. For fibroblasts, RIP3−/− fibroblasts were transfected with RIP3 tagged with GFP at the C-terminus with Fugene HD. Clones were selected with 1 mg/ml geneticin (Gibco/Invitrogen). Clones with comparable expression were selected for experiments.
Cell death assay
Fifty thousand Jurkat cells were pre-treated with 20 µM zVAD-fmk (Enzo Life) for 1 hour before treatment with 100 ng/ml recombinant human TNF (Biosource). Cell death was measured after 16 hours by flow cytometry using uptake of propidium iodide (PI) as indicator of cell death. For fibroblasts, cells were pre-treated with 20 µM zVAD-fmk for 1 hour before 100 ng/ml (or otherwise indicated) of mouse TNF was added. Cell death was measured 18 hours later by flow cytometry and PI uptake or by MTS assay (Promega) as per manufacturer’s instructions. Percentage cell loss was calculated using the formula: % Cell Loss = (1-(number of live cells in treated sample/number of live cells in control sample)) × 100%. Results shown are mean +/− SEM of triplicates of at least two representative experiments. Student’s t test or two-way ANOVA analysis was used for statistical analysis.
Summary statement.
This study shows that RIP1 is regulated by positive as well as inhibitory phosphorylation. It also shows that gain-of-function RIP3 mutant can partially bypass the requirement of RIP1 to induce programmed necrosis.
Acknowledgement
We thank Kenta Moriwaki for critical reading of the manuscript. This work was supported by NIH grants (AI083497 and AI088502). F.K-M.C. is a member of the UMass DERC (DK32520).
Abbreviations
- RIP1/RIPK1
receptor interacting protein kinase 1
- RIP3/RIPK3
receptor interacting protein kinase 3
- MLKL
mixed lineage kinase domain-like
- Pgam5
phosphoglycerate mutase family member 5
Footnotes
Author contributions
T.M. and Y.S.C. performed the experiments. F.K-M.C. designed and supervised the research and wrote the paper with input from T.M and Y.S.C.
References
- 1.Moriwaki K, Chan FK. RIP3: a molecular switch for necrosis and inflammation. Genes Dev. 2013;27:1640–1649. doi: 10.1101/gad.223321.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A, Chan FK, Wu H. The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell. 2012;150:339–350. doi: 10.1016/j.cell.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X, Wang X. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 4.Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu ZG. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci U S A. 2012;109:5322–5327. doi: 10.1073/pnas.1200012109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z, Jiang H, Chen S, Du F, Wang X. The mitochondrial phosphatase PGAM5 functions at the convergence point of multiple necrotic death pathways. Cell. 2012;148:228–243. doi: 10.1016/j.cell.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 6.Chan FK. Fueling the flames: Mammalian programmed necrosis in inflammatory diseases. Cold Spring Harb Perspect Biol. 2012;4 doi: 10.1101/cshperspect.a008805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Donnell MA, Hase H, Legarda D, Ting AT. NEMO inhibits programmed necrosis in an NFkappaB-independent manner by restraining RIP1. Plos One. 2012;7:e41238. doi: 10.1371/journal.pone.0041238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hitomi J, Christofferson DE, Ng A, Yao J, Degterev A, Xavier RJ, Yuan J. Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell. 2008;135:1311–1323. doi: 10.1016/j.cell.2008.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanlangenakker N, Vanden Berghe T, Bogaert P, Laukens B, Zobel K, Deshayes K, Vucic D, Fulda S, Vandenabeele P, Bertrand MJ. cIAP1 and TAK1 protect cells from TNF-induced necrosis by preventing RIP1/RIP3-dependent reactive oxygen species production. Cell Death Differ. 2011;18:656–665. doi: 10.1038/cdd.2010.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin Y, Devin A, Rodriguez Y, Liu ZG. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999;13:2514–2526. doi: 10.1101/gad.13.19.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell Signal. 2007;19:2056–2067. doi: 10.1016/j.cellsig.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 12.O'Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Green DR, Ting AT. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol. 2011;13:1437–1442. doi: 10.1038/ncb2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kovalenko A, Kim JC, Kang TB, Rajput A, Bogdanov K, Dittrich-Breiholz O, Kracht M, Brenner O, Wallach D. Caspase-8 deficiency in epidermal keratinocytes triggers an inflammatory skin disease. Journal of Experimental Medicine. 2009;206:2161–2177. doi: 10.1084/jem.20090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M, Kirsch P, Sterner-Kock A, van Loo G, Pasparakis M. FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature. 2011;477:330–334. doi: 10.1038/nature10273. [DOI] [PubMed] [Google Scholar]
- 15.Bonnet MC, Preukschat D, Welz PS, van Loo G, Ermolaeva MA, Bloch W, Haase I, Pasparakis M. The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity. 2011;35:572–582. doi: 10.1016/j.immuni.2011.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Kang TB, Yang SH, Toth B, Kovalenko A, Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 17.Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J. Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature. 2011;471:373–376. doi: 10.1038/nature09878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature. 2011;471:368–372. doi: 10.1038/nature09857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature. 2011;471:363–367. doi: 10.1038/nature09852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell. 2009;137:1112–1123. doi: 10.1016/j.cell.2009.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He S, Wang L, Miao L, Du F, Zhao L, Wang X. Receptor Interacting Protein Kinase-3 Determines Cellular Necrotic Response to TNF-α. Cell. 2009;137:1100–1111. doi: 10.1016/j.cell.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 22.Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J. RIP3, an Energy Metabolism Regulator that Switches TNF-Induced Cell Death from Apoptosis to Necrosis. Science. 2009 doi: 10.1126/science.1172308. [DOI] [PubMed] [Google Scholar]
- 23.Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, Hedrick SM, Gerber SA, Lugovskoy A, Yuan J. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan FK, Shisler J, Bixby JG, Felices M, Zheng L, Appel M, Orenstein J, Moss B, Lenardo MJ. A role for tumor necrosis factor receptor-2 and receptorinteracting protein in programmed necrosis and antiviral responses. J Biol Chem. 2003;278:51613–51621. doi: 10.1074/jbc.M305633200. [DOI] [PubMed] [Google Scholar]
- 25.Xie T, Peng W, Liu Y, Yan C, Maki J, Degterev A, Yuan J, Shi Y. Structural Basis of RIP1 Inhibition by Necrostatins. Structure. 2013;21:493–499. doi: 10.1016/j.str.2013.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Chen W, Zhou Z, Li L, Zhong CQ, Zheng X, Wu X, Zhang Y, Ma H, Huang D, Li W, Xia Z, Han J. Diverse Sequence Determinants Control Human and Mouse Receptor Interacting Protein 3 (RIP3) and Mixed Lineage Kinase domain-Like (MLKL) Interaction in Necroptotic Signaling. Journal of Biological Chemistry. 2013 doi: 10.1074/jbc.M112.435545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe JM, Cha H, Yang Q, Fornace AJ., Jr Nuclear factor-kappaB (NFkappaB) is a novel positive transcriptional regulator of the oncogenic Wip1 phosphatase. Journal of Biological Chemistry. 2010;285:5249–5257. doi: 10.1074/jbc.M109.034579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chew J, Biswas S, Shreeram S, Humaidi M, Wong ET, Dhillion MK, Teo H, Hazra A, Fang CC, Lopez-Collazo E, Bulavin DV, Tergaonkar V. WIP1 phosphatase is a negative regulator of NF-kappaB signalling. Nat Cell Biol. 2009;11:659–666. doi: 10.1038/ncb1873. [DOI] [PubMed] [Google Scholar]
- 29.Cho Y, McQuade T, Zhang HB, Zhang JK, Chan FKM. RIP1- Dependent and Independent Effects of Necrostatin-1 in Necrosis and T Cell Activation. Plos One. 2011;6 doi: 10.1371/journal.pone.0023209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moujalled DM, Cook WD, Okamoto T, Murphy J, Lawlor KE, Vince JE, Vaux DL. TNF can activate RIPK3 and cause programmed necrosis in the absence of RIPK1. Cell Death Dis. 2013;4:e465. doi: 10.1038/cddis.2012.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan FK, Lenardo MJ. A crucial role for p80 TNF-R2 in amplifying p60 TNF-R1 apoptosis signals in T lymphocytes. Eur J Immunol. 2000;30:652–660. doi: 10.1002/1521-4141(200002)30:2<652::AID-IMMU652>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]