Abstract
New data suggest that the global incidence of several types of fungal diseases have traditionally been under-documented. Of these, mortality caused by invasive fungal infections remains disturbingly high, equal to or exceeding deaths caused by drug-resistant tuberculosis and malaria. It is clear that basic research on new antifungal drugs, vaccines and diagnostic tools is needed. In this review, we focus upon antifungal drug discovery including in vitro assays, compound libraries and approaches to target identification. Genome mining has made it possible to identify fungal-specific targets; however, new compounds to these targets are apparently not in the antimicrobial pipeline. We suggest that ‘repurposing’ compounds (off patent) might be a more immediate starting point. Furthermore, we examine the dogma on antifungal discovery and suggest that a major thrust in technologies such as structural biology, homology modeling and virtual imaging is needed to drive discovery.
Keywords: antifungals, compound libraries, discovery, drug targets, repurposing
Global incidence of fungal diseases surpasses expectations
Recent data are of great significance to many of us whose research interest lies in pathogenic fungi of humans [1]. These data and relevant supportive information [2] indicate that fungal infections kill in excess of 1,300,000 people globally who have HIV/AIDS and other comorbidities. Mortality caused by these pathogens is now equal to drug-resistant Mycobacterium tuberculosis and exceeds malaria [1]. In the case of invasive fungal infections (IFIs), such as blood-borne candidiasis and invasive aspergillosis, these diseases mostly escape laboratory identification, which leads to a delay in therapeutic intervention or suggests the involvement of another pathogen. In the case of cryptococcal meningoencephalitis in HIV/AIDS patients, even with good diagnostic tests, the disease is still prevalent [1]. Often, symptoms are nonspecific, and the progression of the disease can be chronic. The levels of surveillance for fungal disease remain unacceptably low despite their human health consequences. Therapeutic interventions, including incorrect dosage, are common. IFIs only represent part of the magnitude of fungal diseases. The incidence of dermatophytosis, vaginal candidiasis, allergy and mycotoxicosis by far exceeds the frequency of IFI, yet the contribution of these diseases to morbidity is unchartered.
In the USA, the Generating Antibiotic Incentives Now (GAIN) Act (H.R. 2182) [2] constitutes a major effort to reduce the incidence of drug-resistant bacteria through new antimicrobial discovery. More recently, the list of drug-resistant bacteria has been expanded to include drug resistant, human pathogenic fungi. Advocacy through the Infectious Disease Society of America (IDSA), numerous grass-root groups and individuals, as well as congressional bipartisanship, was important in achieving this recognition [3]. The GAIN Act seeks to increase the commercial value of antibiotics by extending the term of exclusivity granted to innovator drugs by the US FDA. Following the GAIN Act, the FDA established a list of qualifying pathogens under the ‘Safety and Innovation Act’. Among the new additions to that list are Candida species and Aspergillus fumigatus due to their potential threats to public health. The FDA, in turn, will fast-track innovator drug delivery for all microbial pathogens on that same list [4]. Almost simultaneously, the CDC published a detailed description of each drug-resistant microbe including fluconazoleresistant C. glabrata [5]. Of importance, the incidence of fungal diseases and their resistance globally should command the attention of stakeholders to commit funds to new diagnostics, vaccines, and drug discovery. These areas of understrength will require even more basic science on the biology of these pathogens.
Resistance is associated with treatment failure
Is there a correlation of drug resistance with treatment failure? There are several reports that have addressed this issue by comparing susceptibilities of patient isolates to fluconazole, voriconazole and echinocandins [6–11]. Minimum inhibitory concentrations (MIC) of patient isolates (C. albicans, C tropicalis and C. parapsilosis) directly correlated with successful outcomes in patients with candidemia. Isolates were fluconazole-susceptible, susceptible-dose dependent or resistant. If the isolate was resistant, the chance of a successful outcome was reduced considerably with either fluconazole or voriconazole therapy [6]. Epidemiological cutoff values (ECVs) and clinical breakpoints (CBPs) have been partially established. If the pathogen is C. glabrata, a successful patient outcome is considerably lower. For example, the species-specific ECVs and CBPs for fluconazole are 2 µg/ml and 0.5–2 µg/ml for C. albicans, and 2 µg/ml for C. tropicalis and C. parapsilosis, while that of C. glabrata is 32 µg/ml [6]. These data indicate that C. glabrata isolates are often the most refractory to antifungal interventions. Similarly, the response rates of patients with candidemia or oropharyngeal episodes caused by Candida species correlated with dose:MIC ratios [9].
In summary, the high incidence and mortality of IFIs as well as the morbidity caused by the less life-threatening mucosal and superficial diseases is now established. The emergence of antifungal drug resistance, either inherent or acquired, is especially common in non-Candida species. New concepts in drug discovery may need to be discussed. In this review, we highlight approaches to antifungal drug discovery, arguing that improved diagnostics must accompany new drug discovery. Our review is divided into the following sections: in vitro susceptibility assay screens; compound libraries; target discovery; natural products (NPs); and synergy.
In vitro susceptibility assays
The Clinical and laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) represent two standards for in vitro susceptibility testing of select pathogenic fungi. Both methods use broth microdilutions, but with some differences in protocols [6–14]. However, there is a near-complete harmonization of assays. Of equal importance, interpretative breakpoints for CLSI and EUCAST are very similar for measurements of Candida species to azoles and echinocandins and azoles to Aspergillus species [10] (reviewed in [6]). We will not describe method details, but, instead refer readers to the published papers, which include breakpoint numbers and differences in assay procedures for CLSI and EUCAST [6–14]. Methodologies for both are easily done in the laboratory and there are now three susceptibility assays products that are approved by the FDA (Sensititre YeastOne, the Vitek 2 yeast susceptibility test and the Etest). Opposite of the standard in vitro methods, Butts and Krysan [15] suggest a high-throughput assay that identifies compounds with fungicidal activity against Candida species and C. neoformans. In this method, a luciferase-tagged adenylate kinase (AK) is utilized, which upon cell lysis by a test compound, causes a release of tagged AK which is measured by luminescence. This assay could very well be useful for screening large compound libraries.
Compound libraries
Compound libraries are of four types. First, existing, synthetic or semisynthetic compound libraries with known activities against nonmicrobial diseases (e.g., cancers) can be screened for antifungal activity (Table 1). This type of discovery is referred to as ‘repurposing’. An advantage here is that a minimal level of toxicity of ‘repurposed’ compounds may have already been established. A second type of library is compounds that are derived from newly synthesized synthetic scaffolds (Table 1) [16]. The activity of a specific scaffold is determined followed by lead optimization to identify the ‘most’ active synthetic derivative(s) of that scaffold. The third library is an outcome of tailoring of current clinically used antifungals. This approach is seen much more often nowadays since the cost of development is lower than with compounds to new targets. There are many examples of tailoring among the antibacterial drugs, penicillin being one of the most tailored as semisynthetics. However, certainly the postfluconazole triazole antifungals also represent tailored derivatives. This type of discovery is done to improve current compounds, that is, fluconazole but with the same exhaustive scrutiny of clinical trials using the tailored compound. In the case of newer triazoles, most remain fungistatic and cross-resistance among triazoles still can occur along with drug–drug toxicity. The fourth library is of NPs (Table 1), interest of which has increased over the past decade, mainly because of the slow development of synthetic compounds. We address these types of compound libraries and lead optimization below.
Table 1.
Compound libraries are represented as new synthetics, repurposed, natural products, or tailored and development is indicated as drug discovery steps.
| Drug discovery step | Compound libraries | |||
|---|---|---|---|---|
| New synthetics | Repurposing | Natural products | Tailoring | |
| Purification | − | − | + | − |
| Lead optimization | + | ± | + | + |
| In vitro cytotoxicity assays | + | ± | + | + |
| PK/PD | + | ± | + | ± |
| Antifungal target identification | + | + | + | − |
| Efficacy in vivo, survival and tissue loads | + | + | + | + |
+: A need to complete each specific step; ±: Sufficient information may be available; −: Not required; PK/PD: Pharmacokinetic/pharmacodynamics (the temporal effects of a drug administration on the body and microbes).
The discovery of potential antifungal drugs is achieved by using compound libraries against an array of pathogenic fungal species. Compound libraries are either of public domain or privately owned. Some compound ‘providers’ are quite competitive in regard to their usage and require objectives in line with their focus. In the public domain, among the opportunities for the utilization of libraries is the NIH Libraries Program, which is designed to provide access for investigators of drug discovery, including antimicrobial discovery. This library can also be a rich source of compounds for chemical probes to study gene function (The Molecular Libraries Small Molecule Repository [MLSMR]) [17]. NIH also maintains facilities at other locations. One such facility that emphasizes antimicrobial searches is The University of New Mexico Center for Molecular Discovery (UNMCMD) [18]. Flow cytometric, cell-based multiplex, high-throughput screening (HTS) is used for the identification of active compounds. Antimicrobial drug efflux inhibitor discovery and substrate assays are one area of interest at UNMCMD. Collaborations are possible through the NIH and the Defense Threat Reduction Agency (DTRA). The National Screening Laboratory for the Regional Centers of Excellence in Biodefense and Emerging Infectious Disease (NSRB) (also known as the ICCB-Longwood Screening Facility) in Harvard Medical School is another option. Another is at the NIH as part of the National Center for Advancing Translational Sciences (NCATS). Other libraries are mentioned below.
Synthetics against nonfungal human diseases: ‘repurposing’
The use of compounds with established applications but with unexplored antifungal activity is referred to as ‘repurposing’ known medications [15,19–21]. Examples are listed in Table 2. To this end, we have recently engaged the NIH/NCI Developmental Therapeutics Program [22] to advance our program in antifungal discovery. The NCI/Developmental Therapeutics Program (DTP) repository maintains approximately >140,000 small molecules and NPs for nonclinical research usage. These compounds have most often been submitted to the NCI/DTP by other investigators for evaluation. A large number have demonstrated activity against human diseases such as cancers. A web-based ordering procedure is available [17]. We have screened 2600 compounds from the DTP for activity against a panel of pathogenic fungi, including many isolates with antifungal drug resistance. From this library, we have identified several with MIC values of 0.1 µg/ml (described below). Of importance, a number of closely related compounds that lack activity was also identified which provides lead optimization opportunities. Based upon their reasonable activity, we have initiated mechanism of action (MOA) studies (discussed hereafter). We next describe data using one of these compounds.
Table 2.
Repurposed compounds, their targets and their activities.
| Compound reference |
Mammalian target | Fungal target | Activity against fungi |
Ref. |
|---|---|---|---|---|
| Thiosemicarbazone | Mutated p53 tumor | Unknown | Broad-spectrum protein synthesis inhibitor | [Sun N, Li D, Groutas W, Calderone R, Unpublished Data] |
| Tamoxifen | Estrogen receptor agonist | Calmodulin (partial) | Reduced kidney burden in C. albicans | [15,21] |
| BQM | Anticancer | Unknown | Broad spectrum; reverses MDR1 resistance by accumulating in cells | [23] |
| Triethylperazine Sertaline | Antianxiety Antipsychotic Anticancer | Membrane vesicle | Synergizes fluconazole | [24] |
| Tosedostat | Amino peptidase | Unknown | Comparable with antifungal drugs | [25] |
BQM: Bis(1,6-a:5’,6’-g) quinolizinium-8-methyl-salt.
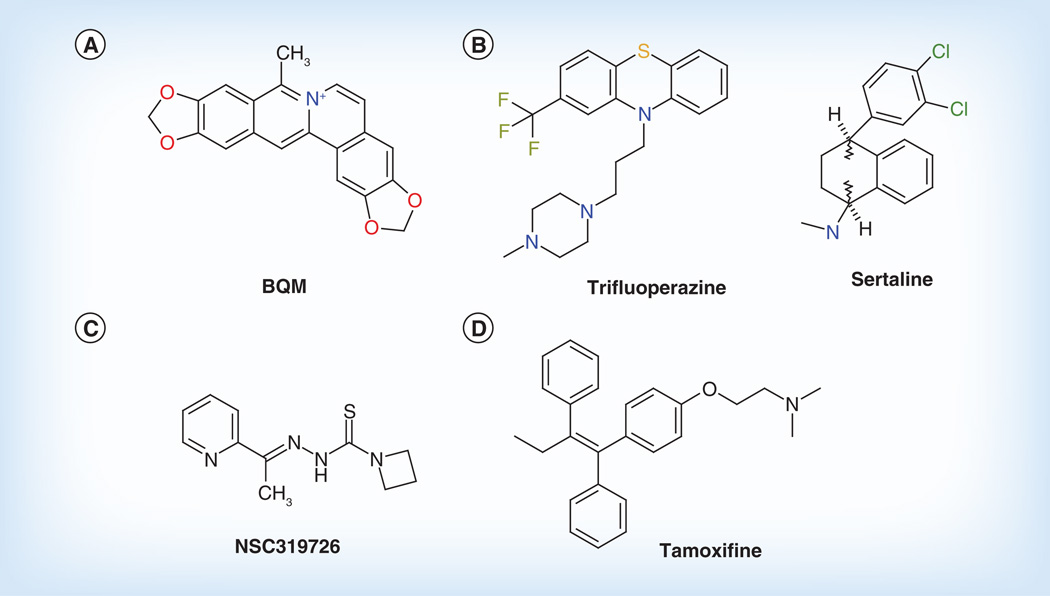
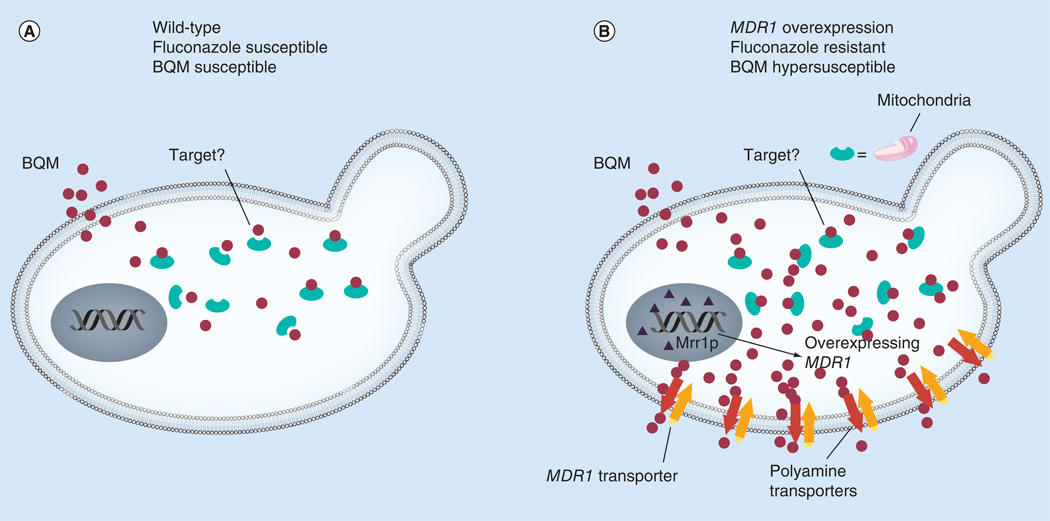
From the NIH–NCI library, we recently identified a novel antifungal small molecule, bis(1,6-a:5’,6’-g) quinolizinium-8-methyl-salt (BQM) [26,27], that has potent and broad antifungal activity (Figure 1A) [23]. While doing preliminary screening, we noticed that patient and laboratory-constructed isolates of C. albicans that were resistant to fluconazole because of MDR1 overexpression were hypersusceptible to BQM compared with parental or mdr1 null or mrr1 null mutants [2,23]. MRR1 is a known regulator of MDR1 overexpression and fluconazole resistance [2,24]. Of importance, BQM was also active against an MDR-overexpressed isolate of A. fumigatus. BQM activity of C. albicans was due to its facilitated intracellular accumulation only in MDR1-overexpressing isolates (Figure 2 & Table 2). To understand this observation at the molecular level, by microarray analyses, we found that a family of polyamine transporters was also upregulated in MDR1-resistant cells compared with non-MDR1 overexpressed cells. The BQM effect was reversed by either substrates for polyamine transporters or in a strain lacking a key enzyme (serine/threonine protein kinase, Ptk2) of the polyamine synthetic pathway [23].
Figure 1. Repurposed drugs.
Examples of compounds that have applications to human diseases but also have antifungal activity alone or synergize with compounds such as fluconazole. The compounds shown have (A, C & D) anticancer or (B) antidepressant, antipsychotic activity.
BQM: Bis(1,6-a:5’,6’-g) quinolizinium-8-methyl-salt.
Structures for tamoxifen and sertraline taken from [15].
Figure 2. The activity of bis(1,6-a:5’,6’-g) quinolizinium-8-methyl-salt is associated with its accumulation in MDR1-resistant Candida albicans.
(A) A wild-type cell is shown that is susceptible to fluconazole and BQM. BQM is taken up by cells and binds to an unknown target. (B) In an MDR1-overexpressed and fluconazole-resistant cell, BQM accumulates in the cytoplasm much more than in a susceptible cell. Consequently, cells are both fluconazole susceptible and hypersusceptible to BQM. In MDR1-overexpressed cells, MRR1, a positive regulator of MDR1, accounts for the overexpression of MDR1. Both the mrr1Δ and mdr1Δ are susceptible to fluconazole but not hypersusceptible to BQM (data not shown). In addition to MDR1 upregulation (B), other transporters such as those of the polyamine family (many are regulated by Mrr1p) are also overexpressed in MDR1 fluconazole-resistant cells. BQM accumulation is reduced by substrates (spermidine) that likely compete with BQM for uptake by polyamine transporters or in a transporter regulator is deleted in cells [13].
BQM: Bis(1,6-a:5’,6’-g) quinolizinium-8-methyl-salt.
A different approach that exploits the well-known synthetic lethal phenotype of yeast has been explored by using a combination of compounds, referred to as ‘syncretic combinations’ (combination of different, seemingly contraindicated chemicals) [25,28–29]. In this case compounds were sought that potentiate fluconazole activity. In vitro screens were established against a panel of three fungal pathogens (C. albicans, C. neoformans and C. gattii) and S. cerevisiae with fluconazole and a library (Pestwick library) of 1120 ‘off-patent’ and other bioactive agents [25]. At least two of 148 compounds, trifluoperazine and sertraline, were each synergistic with fluconazole (Figure 1B & Table 2). Both compounds are antipsychotics/antidepressants. Microarray hybridization identified membrane organization and vesicle-mediated transport as putative target sites of both compounds.
Recently, we have initiated studies with another NCI compound (NSC319726, a thiosemicarbazone) and have established its activity in synergy experiments with several triazoles and caspofungin (Figure 1C) [Sun N, Li D, Groutas W, Calderone R, Unpublished Data]. The mode of action of this compound is not completely understood, but it is related to an inhibition of ribosome biogenesis and protein synthesis.
Breger et al. [30] utilized assembled multiple compound libraries against human diseases such as amyotrophic lateral sclerosis (ALS), Huntington’s disease and hereditary diseases. A total of 1266 compounds were screened against Caenorhadbditis elegans (round worms) infected with C. albicans. Of these, 15 prolonged survival of C. elegans and caused defective in vivo filamentation.
Following the same tactic of ‘repurposing,’ Stylianou et al. recently used the ENZO Life Sciences, Inc. and the Finland Institute of Molecular Medicine (FIMM) oncology collection library for in vitro susceptibility assays against Candida species using the EUCAST guidelines [28]. Of the 26 drugs that showed activity against C. albicans, 14 were off-target either previously reported (seven) or not previously reported (seven). Tosedostat, an aminopetidase cancer inhibitor, had broad activity among Candida species including C. glabrata (Table 2). Other repurposed anticancer compounds that had antifungal activity were identified by this same group.
Other active compounds include tamoxifine and toremifene, both of which are estrogen receptor agonists (Figure 1D) [15,21]. Tamoxifine has efficacy in an animal model of candidiasis [15,21] while toremifene has synergy with either caspofungin or amphotericin B (AmpB) against C. albicans or C. glabrata [29].
New synthetic antifungal compounds
In addition to the use of the DTP library as a source of compounds, we have screened a group of synthetic, low-molecular-weight compounds that were provided to the laboratory through an ongoing collaboration [16,31]. Scaffolds such as the amino acid-derived 1,2-benzisothiazolinone (BZT; BD-I-186) scaffold were screened against pathogens that included Candida species Cryptococcus neoformans, Aspergillus fumigatus and several dermatophytic fungi [16,31]. Lead optimization of the BZT scaffold identified structure–activity relationships (SARs) that established the importance of a heterocyclic ring, a methyl group and a phenyl ring for optimal antifungal activity. Four lead-optimized compounds had broad antifungal activity against all species mentioned above. These compounds have no structural similarity to existing antifungal drugs [31]. All compounds caused synergy with fluconazole, had fungicidal concentrations that were similar to micafungin and nearly that of AmpB, had effective time-kill (fungicidal concentrations) and minimal in vitro toxicity against human cell lines (3 of 4 compounds) [16].
One of the compounds (BD-I-186) displayed no toxicity in vitro and had very good activity against several fungal pathogens mentioned above (Table 2) [16] [Gay-Andrieu F, Groutas W, Li D, Alpha M, Calderone R, Unpublished Data]. Using the S. cerevisiae 6000+ heterozygous mutant library, which allows for the identification of haploinsufficiency or reduced fitness mutants, mentioned below in detail, we have demonstrated that this compound appears to target the kinetochore complex of S. cerevisiae [Gay-Andrieu F, Groutas W, Li D, Alpha M, Calderone R, Unpublished Data]. This complex is essential for the attachment of chromosomes to spindle fibers during nuclear division.
A similar approach was recently used to identify novel antifungal compounds from a Novartis compound archive using the S. cerevisiae haploinsufficiency profiling (HIP) assay [32]. Preliminary screens led to the identification of three compounds with MIC values of 0.25–24 µg/ml against C. albicans but with limited activity against A. fumigatus. Azole-resistant mutants were also resistant to these compounds. IC50 concentrations against mammalian cells varied among the three compounds but with reasonable low levels of cytotoxicity. The active compounds are nonazoles, but treated cells had features of an Erg11p target (sterol 14-alpha demethylase; CYP51/ERG11). Both studies clearly show that the S. cerevisiae HIP and other supportive profiling are useful ways to isolate new compounds while also enriching for information on drug target identification.
Natural products
Long appreciated as a rich source of compounds for antimicrobial drug discovery, the popularity of NPs in this research arena has waxed and waned over the past many decades. The echinocandins, first described from Aspergillus nidulans, provides us with confidence for the potential of NPs [33]. An important group of semisynthetic compounds derived from fermentation cultures of Sordaria araneosa (a terrestrial Ascomycetous fungus) are the sordarins (diterpene glycosides), inhibitors of protein synthesis and specifically elongation factor-2 of protein synthesis [33]. Both types of NPs are described in outstanding reviews on NPs [33,34]. Parameters of drug discovery of NP are shown in Table 1.
Roemer et al. and Lopez et al. [34,35] describe new efforts in NPs as antifungals that attempt to circumvent established dogma that has impeded discovery. This same group describes an elaborate protocol to purify active NP compounds and apply fitness-test (FT) profiling that resulted in the isolation of many NP with active components characterized [34].
The antifungal activity of NPs against bio-films produced by Candida species was recently described [36]. Active concentrations varied depending upon the Candida species that formed the biofilm. Most of the NPs listed were extracts and not purified derivatives.
NPs such as berberine and isoquinoline alkaloids extracted from plants such as Berberis aquifolium (Oregon grape) and Berberis vulgaris (barberry) have been used therapeutically for a variety of infectious diseases and as anticancer and anti-inflammatory compounds [37]. Berberine has been shown to act synergistically with fluconazole against C. albicans in vitro [37,38]. Time-killing experiments demonstrated that the synergistic activity was berberine but not fluconazole concentration-dependent [37]. Berberine accumulated in treated cells and caused a cell cycle arrest and decreases in cell cycle gene transcription. The compound also worked in synergy with fluconazole against fluconazole-resistant strains.
Among the many other NPs are the antimicrobial peptides (AMPs) and proteins that for the most part induce apoptosis in treated cells. This topic has been recently reviewed [39]. The features of AMPs include: they are produced by a diverse range of organisms including plants, bacteria, amphibians, insects and humans; the antifungal spectrum may be AMP-specific and is diverse; most cause damage to cell membranes and cell death by apoptosis but mechanisms of action are complex characterized by changes in membrane potential and permeabilization, ion channel activation, hyperpolarization and activation of signal cascades; the Bacillus amyloliquefaciens AMP inhibits cell wall glucan synthesis; in human saliva, the AMPs include the nonapoptosis defensins such as the histatins; and the iron-binding compound lactoferrin can act synergistically with existing antibiotics.
Lead optimization of compounds
Hits identified via HTS, structure-based drug design [40] and/or fragment-based methodologies [41] are initially evaluated by embarking on a hit-to-lead optimization campaign that is intended to confirm the structural identity, activity and selectivity of a hit using relevant biochemical and cell-based assays. In addition, the synthetic tractability, physicochemical properties (chemical robustness, solubility, etc.) are assessed and, finally, a preliminary evaluation of the SAR landscape around the structure of the hit series (or hit) is conducted [42]. The outcome of these activities is the identification of a lead series (or lead compound) that can be further optimized. The lead compound can be a repurposed drug that needs to be optimized for a new indication.
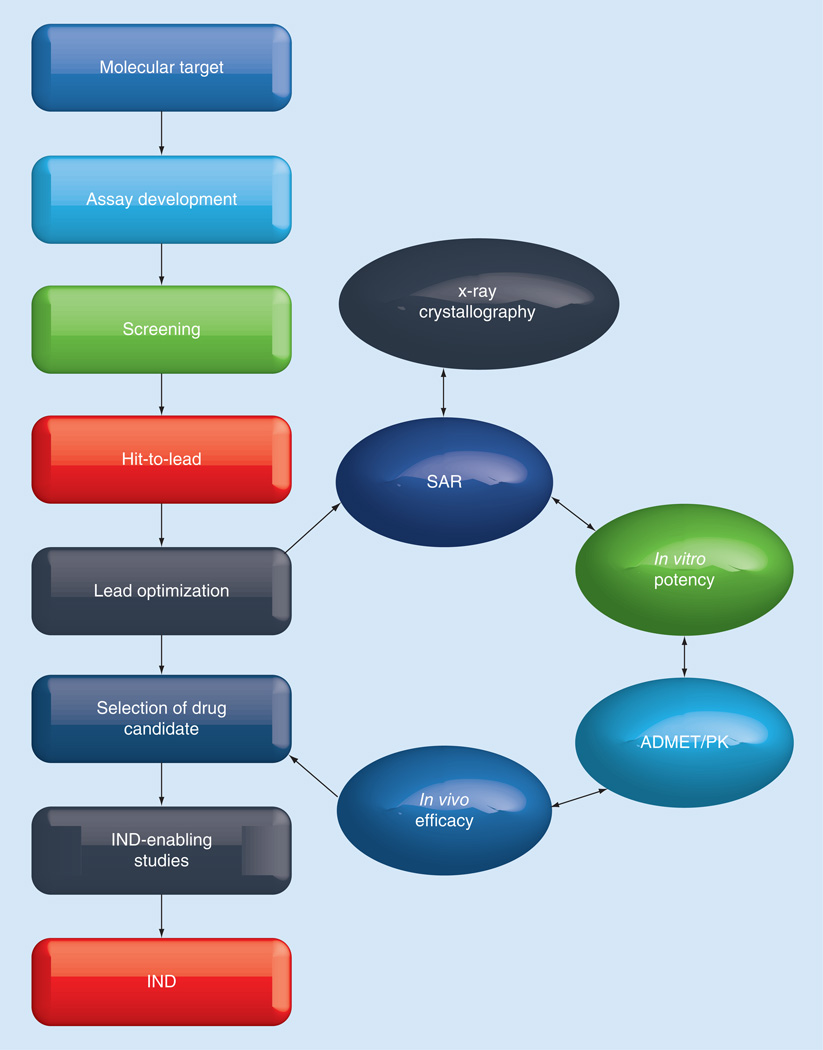
An important component of any drug development program (Figure 3) is lead optimization, since this ultimately results in the selection of a drug candidate suitable for further advancement. These endeavors entail the concurrent optimization of activity and absorption, distribution, metabolism, elimination and toxicity (ADMET)/pharmacokinetic (PK) characteristics through an integrated and iterative process involving SAR, x-ray crystallographic studies, in vitro screening and MOA studies, as well as demonstration of proof of concept using an appropriate animal model [43]. ADMET studies include assessment and optimization of cellular permeability, metabolic stability, and plasma protein binding. In addition, toxicological profiling studies (cytotoxicity, CYP450 inhibition or induction, hERG channel inhibition and genotoxicity) are conducted early on, since toxicity is a significant cause of attrition during drug development [44–46]. Lead candidates are further prioritized based on the results of in vivo PK studies, including oral bioavailability. Many early drug candidates fail to advance because of poor ADMET characteristics, consequently, activity (antifungal activity, for instance) and ADMET/PK properties are optimized in parallel. Thus, continuous assessment of ADMET/PK and rational feedback of the ADMET/PK data into the iterative medicinal chemistry design is of paramount importance.
Figure 3. General drug development path and lead optimization components.
ADMET: Absorption, distribution, metabolism, elimination and toxicity; IND: Investigational new drug; PK: Pharmacokinetic; SAR: Structure–activity relationships.
New target discovery
The current therapeutic antifungals target either ergosterol synthesis (azoles), cell wall β-1,3-glucan synthesis (echinocandins) or bind to membrane ergosterol perturbing membrane functions (polyenes and AmpB). However, these targets account for a minor component of suggested targets of the entire genome of most fungi. While the assumption is that approximately 8% of S. cerevisiae genome (508 proteins) can be exploited as drug targets, the number of potential proteins in pathogenic fungi should be much higher [47]. Therefore ‘genome mining’ is a reasonable approach to find new targets by comparing the genetic differences between fungal and mammalian cells as well as among fungal species with the hope that the targets are broadly represented in fungal pathogens. There are questions raised about whether any compound is active against such a structurally and physiologically diverse group of pathogens. It is also clear that while the azoles continue to be remodeled, the gains in efficacy are not necessarily substantial.
We now discuss approaches to target discovery with emphasis on requirements as well as the advantages/disadvantages of each (Table 3).
Table 3.
Approaches to target discovery, with requirements and advantages and disadvantages of each method.
| HIP | GRACE | Structural biology |
|---|---|---|
| Requirements | ||
| Agar plate application of a mutant library, or PCR-based technology | PCR- and microarray-based technology | Cloning, expression, protein purification, crystallization of apo and ligand bound fungal target complexes, x-ray diffraction data collection, phasing, 3D structure determination, model building, refinement and modeling of ligand into the electron density maps |
| Advantages | ||
| Total genome coverage Agar plate screens are not costly Batch culture screens by PCR Activity of compound means penetration into cells | HIP profiling in batch culture HTS | Resolution of overall 3D structure of human and fungal proteins Insight and knowledge into the key differences in substrate binding and active site regions of human and fungal homologs allowing to design fungal-specific drugs Allows for rapid and efficient optimization of lead compounds to achieve potency, selectivity and desired pharmacological properties Availability of synchrotron beam lines for data collection have significantly improved the speed and quality of data collection and eliminated the need for in-house x-ray source, reducing the costs HTS |
| Disadvantages | ||
| Tedious, transfer of cells unequal may miss gene targets not in S. cerevisiae Slow growth of some mutants Contamination Not HTS Identifying target in clustered genes | Complex methodologies Haploid pathogens not useful for HIP screens High cost | Membrane proteins that may be important antifungal drug targets can be difficult to express and crystallize Some proteins may require extensive manipulation to obtain high resolution diffracting quality crystals |
GRACE: Gene replacement and conditional expression; HIP: Haploinsufficiency profiling; HTS: High-throughput screening.
Haploinsufficiency & homozygous profiling with mutant libraries
An approach to identifying gene targets is compound screens of mutant libraries, described below. The prerequisite for the screens is the availability of fungal mutant libraries. The choices of libraries are those of Saccharomyces cerevisiae that cover its entire genome or partial mutant libraries such as those described below for C. albicans and Aspergillus fumigatus.
The S. cerevisiae libraries
The lead-optimized compounds described above that were derived from the amino acid-derived BZT scaffold also inhibited the growth of S. cerevisiae, which can be a pathogen in immunocompromised patients [16,47]. Thus, screening yeast mutant libraries is quite useful to gain information on targets and MOA of compounds. The homozygous profiling (HOP) library contains approximately 4700 null homozygotes (YSC1056), while the haploinsufficiency profiling (HIP) library contains 6000+ mutants lacking one allele (YSC1055 Heterozygous Collection; Thermo Scientific). For the HOP null library, mutants that are resistant to the test compound may reflect the target gene of interest or perhaps indicate that the deleted gene is part of a pathway required for target activity in wild-type cells. With the heterozygous mutant library, our objective was to screen for hypersusceptibility (reduced ‘fitness’) after compound treatment compared with a parental strain and untreated mutants. Those mutants with reduce fitness due to the loss of one allele were selected after two rounds of screening the entire 6000+ library. Mutants that displayed a reduced ‘fitness’ were then verified by tube dilution assays to obtain MIC values. Hypersensitive mutants were always compared with untreated mutants. Clustering of genes to functional classes was done using the Functional Specification (FunSpec) database [48].
Two of the four active compounds with the broadest activity against the fungal pathogens were screened with the S. cerevisiae mutant libraries [16] [Gay-Andrieu F, Groutas W, Alpha M, Li D, Calderone R, Unpublished Data]. One of those compounds (DFD-VI-15) apparently targets mitochondria proteins and may have possible limitations since some toxicity was noted using in vitro assays with human cell lines [16]. We have not pursued animal studies with this compound.
We routinely use 96-well microtiter plates to grow each mutant, and a pin applicator that allows direct transfer of 96 mutant strains from an overnight-grown inoculum of each mutant to large agar plates containing a compound. Assays can also be performed in batch cultures since each mutant is bar-coded. PCR-based methods allow for determinations of fitness or growth in the presence of a compound. The advantages of HIP include the comprehensive analysis of the entire genome. Virtually every encoded gene mutant is assayed against compounds by screening for a reduced fitness in all mutants. Additionally, the approach is applicable to any compound that is able to enter yeast cells or other target organisms, and of importance, yields a cluster of potential targets, which can be functionally grouped using algorithms such as FunSpec as described above. With the agar plate method, there are disadvantages, the most important of which is that the procedure is quite time consuming. Contamination can be a problem and reproducibility is difficult to achieve without much practice. With regard to reproducibility, if the transfer volume of cells is not equal for some mutants, then assumed growth reductions in that mutant due to compound may be erroneous. Therefore, all mutants must be grown in the absence of compound. One does not find a single mutant per compound that is hypersensitive, and more often 5–15 mutants with hypersensitivity may be identified. With BD-I-186, we were fortunate enough to see that most all of these mutants were associated with loss of genes required for kinetochore and cell division functions, as mentioned above. Our interpretation is that this cluster of genes represents a global pathway that is affected by the compound.
Pathogen mutant libraries
HIP has also been applied to a heterozygous collection of deletion strains of C. albicans [49]. Each deletion strain had unique barcodes in the up- and down-stream regions of the deleted genes allowing compound screens to be done in batch cultures. The growth inhibition readout was determined against compounds using PCR amplification and DNA microarray to identify hypersensitive mutants by barcode PCR. In addition to the C. albicans library just described, a library of 3633 C. albicans tagged, heterozygous transposon disruption mutants were constructed to do drug-induced HIP [50]. The compounds for this screening were obtained from ChemDiv, Inc. Of interest, one compound targeted a C. albicans-specific target, Tfp1p [50].
Gene replacement and conditional expression (GRACE) is a time-proven method to identify growth-essential genes (Table 2) [48–53]. In diploid C. albicans, one of the two alleles is deleted and the second is placed under conditional induction/repression. Repression of the second allele thus allows one to identify genes essential for growth. A. fumigatus is haploid, however, so establishing GRACE requires additional maneuvering [52]. Comparative genomics has yielded information on conserved essential genes in fungal pathogens [51,53–54]. A total of 57 growth-essential genes were identified in C. albicans and Aspergillus fumigatus and six other fungal pathogens. Ten of these genes were found in all pathogens (Paracocidioides species, Blastomyces dermatitidis, Coccidioides immitis, Histoplasma capsulatum and C. neoformans) and absent in the human genome. Among the conserved targets thought to be potential targets were those critical to processes such as redox homeostasis, cell metabolism and biogenesis, protein transport cell wall biogenesis/degradation, ergosterol biosynthesis and pH response regulators [51]. As far as can be determined, compound screens of these targets have not been pursued.
C. albicans unique proteome targets were also identified. Orthologs conserved in both human cells and S. cerevisiae were eliminated from further evaluation [53]. From 14,633 proteins retrieved from the US National Center for Biotechnology Information (NCBI), 1618 were unique to C. albicans, of which 42 were known functionally.
Synergy
Over four decades ago, parenteral AmpB therapeutic was exclusively used as intervention against IFIs [55]. Toxicity to patients was inevitable but reduced when a combination of AmpB with 5-fluorocytosine (5-FC) was used, which also provided an improvement in clinical outcome. As 5-FC selected for resistant isolates, its use as a singular was fairly rapidly abandoned. Synergy was used to explain the combination effect of both drugs. Synergy continues to be discussed in the current literature. Much of this literature is on in vitro testing but with little information on efficacy in clinical trials [56,57]. A benefit of synergy would be to reduce resistance of isolates as we have described for one of our compounds, and there are several recent reports to this effect cited below.
Antifungal drug resistance is associated with one or more of the following mechanisms: strains overexpress efflux pumps, such as Cdr1p, Cdr2p and Mdr1p; have point mutations in the drug target protein (Erg11p or Fks1p) such that triazoles or echinocandins do not bind; or there is overexpression of target genes [6,58–60].
As described above, one of the rationales behind new drug discovery is to overcome the resistance to current antifungals such as the triazoles and echinocandins by reversing drug resistance. If so, the sustained use of those drugs that select for resistant pathogens is possible if synergy existed with another compound that countered selection. In addition to BQM, which has hyperactivity to MDR1-overexpressed strains [23], there are other examples of compounds that reverse resistance. For example, in vivo inhibitors of the calcineurin pathway reversed echinocandin and azole resistance, and Hsp90 inhibitors against A. fumigatus have been reported [61]. Also, decreased genetic expression of Hsp90 reduced virulence of A. fumigatus and promoter modification of Hsp90 resulted in hypersensitivity to caspofungin [62] in C. glabrata [63]. Calcineurin inhibitors are also active against C. albicans as well as C. neoformans [64,65].
Conclusion & future perspective
The recent 2012 review on the global incidence and mortality of fungal infections [1] should serve as an impetus for a greater commitment of funds to support outstanding basic and translational research of fungal pathogens. However, there are speed bumps along the way to new drug discovery. They include: the market size and, hence, profit, is believed too small and new drug development is too expensive; current NIH support has dwindled in part because of the sequestration that has forced huge budget downfalls; remodeling of current drugs is cheaper; new antimicrobials are used for 2–3 weeks to cure and chronic diseases may require use for perhaps one’s life; and lastly at least in the case of antibacterial drugs, eventually resistance will develop to these new drugs. Therefore, there is an uphill battle to justify new antimicrobial (and antifungal in particular) development.
For the pursuit of the translational science of drug discovery, investigators must rely on extensive collaborations with medicinal chemists, investigators with knowledge of the entire discovery process and/or structural biologists. The availability of compound libraries and HTP facilities is also essential. The initial lead person is the expert in fungal diseases and the basic sciences of these pathogens with knowledge of the need for new antifungals. In Table 1, the pathway to discovery is shown as sequential steps in the process. Of most importance along the way to discovery is the in vitro and/or in vivo toxicity to the compound.
The dogma on new antimicrobial discovery may be in need of subtle change. For example, can we really find that antifungal bullet that will kill all pathogens? The existing armamentarium suggests otherwise. Fluconazole is ineffective against A. fumigatus, the echinocandins are of low utility against C. neoformans since this fungus lacks β-1,3 cell wall glucan, and the best choice for treatment of emerging diseases caused by mold fungi is unresolved. Do we need another triazole? Perhaps we have gotten as much out of a family of compounds as is possible with only minimal increments in advantages. However, new imidazole and triazole derivatives are in development and appear promising [66].
Other interventions such as efungumab, a monoclonal antibody that targets HSP 90, is suggested in combination with other antifungals [67].
Repurposing offers nearly immediate use since much of the groundwork on toxicity problems is completed. Some of the published repurposed compounds are described in Table 2. Of these, an interesting pursuit in anticancer research is the p53 tumor suppressor protein, which is critical to maintenance of normal cell growth. Mutations in that protein result in growth of cancer cells. One of several experimental anticancer drugs (NSC319726, NCI-NIH) that have been developed binds only to the mutated p53 and rescues the function of p53 [68,69]. Importantly, the same compound we have been following also has great antifungal activity; we are resolving the target of this compound in fungi since we cannot identify a p53 ortholog in fungi (Figure 1C & Table 2). We advocate for ‘repurposing’ as at least part of the direction we should be following with the complimentary application of HIP or microarray/RNA sequencing to find drugs and targets.
The genomics era remains important to antifungal drug discovery; there are exceptional laboratory groups that have used chemogenetics to identify numerous fungal targets that obey the specificity issues that are so critical to drug discovery. The payoff of this effort towards the development of drugs to these targets is not upon us yet, but this type of direction should still be encouraged.
Equally important is an emphasis on structural biology and structural genomics in antifungal drug discovery (Table 3). This approach is especially new to fungal drug discovery. Mabanglo et al. [70] report on the crystal structure of the Aspergillus fumigatus protein farnesyltransferase AFFTase, a protein critical for growth and virulence of C. albicans, C. neoformans and A. fumigatus. Substrate binding to the AFFTase monitored by high-resolution structures resulted in the identification of structural differences compared with the human FTase that correlated with differences in inhibitor binding (ED5 and tipifarnib) between the two proteins that could be critical to antifungal drug discovery. Equally important, the S. cerevisiae tryptophanyl-tRNA synthetase (sTrp-RS), which is required for tryptophan activation during translation, has binding differences compared with orthologous human proteins. As described above for the FTase of pathogens, there is promise of specific inhibitors [71]. In fact, inhibitors of LeuRS and TrpRS are now being evaluated for treatment against bacterial pathogens (summarized in [71]).
In addition to using 3D x-ray crystal structures of enzyme:ligand complexes for antifungal drug discovery, one can also use pharmacophore modeling methods. This method can be used as a standalone or in combination with experimental structural methods. Pharmacophore modeling is a computational method that makes use of 3D structural information of the drug targets determined either by experimental methods (x-ray or NMR) or using homology-based modeling to understand how chemically and structurally diverse set of ligands (inhibitors and/or activators) bind and interact within a common binding and/or active of the receptor molecule. This method can be used as a virtual HTS tool to identify novel hits and for optimization of hits to improve the potency and ‘drug-like’ properties to discover safe and efficacious lead drug candidates [72].
Items on the horizon
Emerging pathogens such as Candida nonalbicans species that have multiple drug resistance including to AmpB are reported [73,74]. Their prevalence in patients globally is probably underestimated. A very provocative hypothesis has been suggested that nonantifungal drug interventions may in fact select for commensal fungi [75]. As one of many examples put forth, steroid therapy may select for steroid-tolerant commensal fungi. In addition to evaluating patient isolates that are resistant to antifungal drugs, it may be time to measure not only antifungal susceptibilities of patient isolates, but also isolate susceptibilities to the complex drug regiments that are used to treat these same patients. Metalloenzyme inhibitors, a glycosylphosphatidylinositiol biosynthesis inhibitor and a new β-1,3 glucan inhibitor may offer promise as new antifungal therapeutics [76].
Executive summary.
Incidence of fungal infections is much higher than previously acknowledged
The invasive fungal infections (IFIs) are most likely underdiagnosed and underappreciated for their frequency.
At least some of the IFIs are inappropriately treated, meaning improper length, dosage or delay in treatment.
Treatment failure may be due to inherent, acquired or both types of resistance of Candida species and other fungal pathogens.
Complete intervention to recurrent diseases (vulvovaginal candidiasis, chronic dermatophytosis or oral candidiasis) is not fully mapped.
A change in dogma of antifungal discovery may be needed
Is it possible to identify a ‘super’ drug that can cure all infections?
Should we direct new therapies to specific fungi or high-impact diseases such as invasive cryptococcosis, candidiasis and aspergillosis?
Is ‘repurposing’ compounds as antifungals (off-patent) that currently cure only nonfungal diseases useful?
Should synergy and natural products be emphasized more in discussions of new interventions? Efungumab (monoclonal antibody) used in combination with cytokines has been proposed.
Problems in screening of natural products extracts
The laborious chemistry and isolation methods to identify active compounds.
Already known natural products purified compounds may be part of the analysis and have to be ‘dereplicated’ to distinguish others of interest.
Is continued remodeling of triazoles needed?
New-generation triazoles are more broad spectrum than fluconazole.
The newer triazoles remain fungistatic and therefore resistance may remain among isolates.
Cross-resistance is still a problem.
Drug–drug toxicities still occur. Toxicity can be managed but only by using other nontriazole compounds.
Acknowledgments
This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Footnotes
Financial & competing interests disclosure
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1. Brown GD, Denning DW, Gow NA, et al. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4(165):165rv13. doi: 10.1126/scitranslmed.3004404. •• Up-to-date summary of fungal disease incidence.
- 2.H.R. 2182 (112th): Generating Antibiotic Incentives Now Act of 2011. www.govtrack.us/congress/bills/112/hr2182.
- 3.Statements for 10 × ‘20 Initiative. www.idsociety.org/IDSA_Statements_for_10x20_Initiative. [Google Scholar]
- 4.Proposed Rules from the US FDA. www.gpo.gov/fdsys/pkg/FR-2013-2006-2012/pdf/2013-13865.pdf.
- 5.Antibiotic Resistance Threats in the United States, 2013. www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-2508.pdf. [PubMed]
- 6. Pfaller M. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 2012;125(1 Suppl.):S3–S13. doi: 10.1016/j.amjmed.2011.11.001. •• Addresses standardized testing, mechanisms of resistance and clinical correlates of resistance and patient outcome.
- 7.Clancy CJ, Yu VL, Morris AJ, et al. Fluconazole MIC and the fluconazole dose/MIC ratio correlate with therapeutic response among patients with candidemia. Antimicrob. Agents Chemother. 2005;49:3171–3177. doi: 10.1128/AAC.49.8.3171-3177.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rex JH, Pfaller MA, Galgiani JN, et al. Development of interpretive breakpoints for antifungal susceptibility testing: conceptual framework and analysis of in vitro-in vivo correlation data for fluconazole, itraconazole, and candida infections. Subcommittee on Antifungal Susceptibility Testing of the National Committee for Clinical Laboratory Standards. Clin. Infect. Dis. 1997;24(2):235–247. doi: 10.1093/clinids/24.2.235. [DOI] [PubMed] [Google Scholar]
- 9.Rodríguez-Tudela JL, Almirante B, Rodríguez-Pardo D, et al. Correlation of the MIC and dose/MIC ratio of fluconazole to the therapeutic response of patients with mucosal candidiasis and candidemia. Antimicrob. Agents Chemother. 1997;51:3599–3604. doi: 10.1128/AAC.00296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaller MA, Diekema DJ, Ostrosky-Zeichner L, et al. Correlation of MIC with outcome for Candida species tested against caspofungin, anidulafungin, and micafungin: analysis and proposal for interpretive MIC breakpoints. J. Clin. Microbiol. 2007;46:2620–2629. doi: 10.1128/JCM.00566-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alexander BD, Johnson MD, Pfeiffer CD, et al. Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with the presence of FKS mutations and elevated minimum inhibitory concentrations. Clin. Infect. Dis. 2013;56:1724–1732. doi: 10.1093/cid/cit136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cuenca-Estrella M, Rodriguez-Tudela J. The current role of the reference procedures by CLSI and EUCAST in the detection of resistance to antifungals in vitro. Expert Rev. Antinfect. Ther. 2010;8:267–276. doi: 10.1586/eri.10.2. [DOI] [PubMed] [Google Scholar]
- 13.Pfaller M, Castanheira M, Diekema D, et al. Comparison of European Committee of antimicrobial susceptibility testing (EUCAST) and Etest methods with the CLSI broth microdilution method fo echinocandin susceptibility testing of Candida species. J. Clin. Microbiol. 2010;48:1592–1599. doi: 10.1128/JCM.02445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfaller M, Boyken L, Hollis R, et al. Comparison of the broth microdilution methods of the European Committee on Antimicrobial Susceptibility Testing and the Clinical Laboratory Standards Institute for testing itraconzaole, posaconazole, and voriconazole against Aspergillus isolates. J. Clin. Microbiol. 2011;49:1110–1112. doi: 10.1128/JCM.02432-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Butts A, Krysan D. Antifungal drug discovery: something old and something new. PLoS Pathog. 2012;8:e1002870. doi: 10.1371/journal.ppat.1002870. • Very good review of repurposing compounds.
- 16.Alex D, Gay-Andrieu F, May J, et al. Amino acid derived 1,2-benzisothiazolinone derivatives as novel small molecule inhibitors: identification of potential genetic targets. Antimicrob. Agents Chemother. 2012;56:4630–4639. doi: 10.1128/AAC.00477-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molecular Libraries Program – Overview. http://mli.nih.gov/mli/mlp-overview.
- 18.UNM School of Medicine, Department of Pathology, Translational Antimicrobial Discovery. http://pathology.unm.edu/research/faculty-laboratories/translational-antimicrobial-discovery/index.html. [Google Scholar]
- 19.Ericson E, Gebbia M, Heisler L, et al. Off-target effects of psychoactive drugs revealed by genome-wide assays in yeast. PLoS Genet. 2008;4(8):e1000151. doi: 10.1371/journal.pgen.1000151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giaever G, Flaherty P, Kumm J, et al. Chemogenomic profiling: identifying the functional interactions of small molecules. Proc. Natl Acad. Sci. USA. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dolan K, Montgomery S, Buchheit B, et al. Antifungal action of tamoxifen: in vitro and in vivo activities and mechanistic characterization. Antimicrob. Agents Chemother. 2009;53:3337–3346. doi: 10.1128/AAC.01564-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NIH/NCI Developmental Therapeutics Program. http://dtp.nci.nih.gov. [PubMed]
- 23.Sun N, Li D, Fonzi W, et al. Multidrug resistant transporter Mdr1p mediated uptake of a novel antifungal compound. Antimicrob. Agents Chemother. 2013;57:5931–5939. doi: 10.1128/AAC.01504-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lohberger A, Coste A, Sanglard D. Distinct roles of the drug resistance transcription factors TAC1, MRR1 and UPC2 from Candida albicans in virulence. Eukaryot. Cell. 2013;3(1):127–142. doi: 10.1128/EC.00245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spitzer M, Griffiths E, Blakely K, et al. Cross-species discovery of syncretic drug combinations that potentiate the antifungal fluconazole. Mol. Systems Biol. 2011;7:499–522. doi: 10.1038/msb.2011.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calderone R, Alex D, Groutas W. US61/793080. 2013 [Google Scholar]
- 27.Cook N. US5623463. 2002 [Google Scholar]
- 28.Stylianou M, Kulessky E, Lopes JP, et al. Antifungal application of non-antifungal drugs. Antimicrob. Agents Chemother. 2013;58(2):1087–1013. doi: 10.1128/AAC.01087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delattin N, DeBrucker K, VanDamme K, Moert E, Marchand A. Repurposing as a means to increase the activity of amphotericin B and caspofungin. J. Antimicrob. Chemother. 2013;69(4):1035–1044. doi: 10.1093/jac/dkt449. [DOI] [PubMed] [Google Scholar]
- 30.Breger J, Fuchs BB, Aperis G, et al. Antifungal chemical compounds identified using a C. alegans pathogenicity assay. PLoS Pathog. 2007;3:e18. doi: 10.1371/journal.ppat.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dou D, Alex D, Du B, et al. Antifungal activity of a series of 1,2-benzisothiazol-3(2H)-one derivatives. Bioorg. Med. Chem. 2011;19:5782–5787. doi: 10.1016/j.bmc.2011.08.029. [DOI] [PubMed] [Google Scholar]
- 32.Hoepfner D, Karkaree S, Helliwell S, et al. An integrated approach for identification and target validation of antifungal compounds active against Erg11p. Antimicrob. Agents Chemother. 2012;56:4233–4240. doi: 10.1128/AAC.06332-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DiSanto R. Natural products as antifungal agents against clinically relevant pathogens. Nat. Prod. Rep. 2010;27:1084–1098. doi: 10.1039/b914961a. •• Must-read for those interested in natural product discovery.
- 34.Roemer T, Xu D, Singh S, et al. Confronting the challenges of natural product based-antifungal discovery. Chem. Biol. 2011;18(2):148–164. doi: 10.1016/j.chembiol.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 35.Lopez A, Parsons A, Nislow C, et al. Chemical-genetic approaches for exploring the mode of action of natural products. Prog. Drug Res. 2008;237:239–271. doi: 10.1007/978-3-7643-8595-8_5. [DOI] [PubMed] [Google Scholar]
- 36.Sardi J, Scorzoni L, Bernardi T, et al. Candida species: current epidemiology, pathogenicity, biofilm formation, natural antifungal products and new therapeutic options. J. Med. Microbiol. 2013;62(Pt 1):10–24. doi: 10.1099/jmm.0.045054-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang L, Chang JJ, Zhang SL, et al. Synthesis and bioactive evaluation of novel hybrids of metronidazole and berberine as new types of antimicrobial agents and their transportation behavior by human serum albumin. Bioorgan. Med. Chem. 2013;21(14):4158–4169. doi: 10.1016/j.bmc.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Li D, Zhang D, Quan H, et al. Fluconazole assists berberine to kill fluconazole-resistant Candida albicans. Antimicrob Agents Chemother. 2013;57:6016–6027. doi: 10.1128/AAC.00499-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeBruker K, Cammue P, Thevissen K. Apoptosis-inducing antifungal peptides and proteins. Biochem. Soc. Trans. 2011;39:1527–1532. doi: 10.1042/BST0391527. [DOI] [PubMed] [Google Scholar]
- 40.Maddaford SP. A medicinal chemistry perspective on structure-based drug design and development. Methods Mol. Biol. 2012;841:351–381. doi: 10.1007/978-1-61779-520-6_15. [DOI] [PubMed] [Google Scholar]
- 41.Caliandro R, Belviso DB, Aresta BM, et al. Protein crystallography and fragment-based drug design. Future Med. Chem. 2013;5:1121–1140. doi: 10.4155/fmc.13.84. [DOI] [PubMed] [Google Scholar]
- 42.Gillespie P, Goodnow R. The hit-to-lead process in drug discovery. Ann. Rep. Med. Chem. 2004;39:293–304. [Google Scholar]
- 43.Meanwell NA. Improving drug candidates by design: a focus on physicochemical properties as a means of improving compound disposition and safety. Chem. Res. Toxicol. 2011;24:1420–1456. doi: 10.1021/tx200211v. [DOI] [PubMed] [Google Scholar]
- 44.Leeson PD, Empfield JR. Reducing the risk of drug attrition associated with physicochemical properties. Ann. Rep. Med. Chem. 2010;45:394–407. [Google Scholar]
- 45.Waring MJ. Lipophilicity in drug discovery. Expert Opin. Drug Discov. 2010;5:235–248. doi: 10.1517/17460441003605098. [DOI] [PubMed] [Google Scholar]
- 46.Pellegatti M. Preclinical in vivo ADME studies in drug development: a critical review. Expert Opin. Drug Metab. Toxicol. 2012;8:161–172. doi: 10.1517/17425255.2012.652084. [DOI] [PubMed] [Google Scholar]
- 47.Hopkin A, Groom C. The druggable genome. Nat. Rev. Drug Dis. 2002;1:727–730. doi: 10.1038/nrd892. [DOI] [PubMed] [Google Scholar]
- 48.FunSpec. http://funspec.med.utoronto.ca. [Google Scholar]
- 49. Xu D, Jiang B, Ketela T, et al. Genome-wide fitness test and mechanisms of action studies on inhibitory compounds in Candida albicans. PLoS Pathog. 2007;3:835–848. doi: 10.1371/journal.ppat.0030092. •• Good description of fitness experiments.
- 50.Oh J, Fung E, Schlecht U, et al. Gene annotation and drug target discovery in Candida albicans with a tagged transposon mutant collection. PLoS Pathog. 2010;6:e1001140. doi: 10.1371/journal.ppat.1001140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abadio A, Kioshima E, Teixeira M, et al. Comparative genomics allowed the identification of drug targets agains human fungal pathogens. BMC Genomics. 2011;12:75. doi: 10.1186/1471-2164-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hu W, Silllaots S, Lemieux S, et al. Essential gene identification and drug target priotization in Aspergillus fumigatus. PLoS Pathog. 2007;3:e24. doi: 10.1371/journal.ppat.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roemer T, Jiang B, Davison J, et al. Large-scale essential gene identification in andida albicans and applications to antifungal drug discovery. Mol. Microbiol. 2003;50:67–81. doi: 10.1046/j.1365-2958.2003.03697.x. [DOI] [PubMed] [Google Scholar]
- 54.Tripathi H, Lugman S, Meena A, et al. Genomic identification of potential targets unique to Candida albicans for the discovery of antifungal agent. Curr. Drug Targets. 2013;5(1):136–149. doi: 10.2174/138945011501140115112242. [DOI] [PubMed] [Google Scholar]
- 55.Ostrosky-Zeichner L, Casadevall A, Galgiani J, et al. An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 2010;9:719–727. doi: 10.1038/nrd3074. [DOI] [PubMed] [Google Scholar]
- 56.Andrusiak K, Piotrowski JS, Boone C. Chemical-genomic profiling: systematic analysis of the cellular targets of bioactive molecules. Bioorg. Med. Chem. 2012;20:1952–1960. doi: 10.1016/j.bmc.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Camps I. Combination therapy for invasive aspergillosis. Enferm. Infect. Microbiol. Clin. 2011;29(Suppl. 2):38–42. doi: 10.1016/S0213-005X(11)70008-8. [DOI] [PubMed] [Google Scholar]
- 58.Morschhauser J, Barker KS, Liu TT, et al. The transcription factor Mrr1p controls expression of the MDR1 efflux pump and mediates multidrug resistance in Candida albicans. PLoS Pathog. 2007;3:e164. doi: 10.1371/journal.ppat.0030164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cannon RD, Lamping E, Holmes AR, et al. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 2009;22:291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shapiro RS, Robbins N, Cowen LE. Regulatory circuitry governing fungal development, drug resistance, and disease. Microbiol. Mol. Biol. Rev. 2011;75:213–267. doi: 10.1128/MMBR.00045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lamoth F, Juvvadi P, Gehrke C, Steinbach W. In vitro activity of calcineurin and heat shock protein 90 inhibitors against Aspergillus fumigatus azole and echinocandin-resistant strains. Antimicrob. Agents Chemother. 2013;57(2):1035–1039. doi: 10.1128/AAC.01857-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lamoth F, Juvvadi P, Gehrke C, et al. Transcriptional activation of heat shock 90 mediated via a proximam promoter regions as trigger of caspofungin resistance in Aspergillus fumigatus. J. Infect. Dis. 2013;209(3):473–481. doi: 10.1093/infdis/jit530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Singh-Babak SD, Babak T, Diezmann S, et al. Global analysis of the evolution and mechanism of echinocandin resistance in Candida glabrata. PLoS Pathog. 2012;8:e1002718. doi: 10.1371/journal.ppat.1002718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Singh SD, Robbins N, Zaas AK, et al. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5:e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Steinbach WJ, Reedy JL, Cramer RA, Jr, et al. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat. Rev. Microbiol. 2007;5:418–430. doi: 10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]
- 66.Rezael Z, Khabnadideh S, Zomorodian K, et al. Design, synthesis and antifungal activity of some new imidazole and triazole derivatives. Arch. Pharm. Chem. Life Soc. 2011;344:658–665. doi: 10.1002/ardp.201000357. [DOI] [PubMed] [Google Scholar]
- 67.Karwa R, Wargo KA. Efungumab: a novel agent in the treatment of invasive candidiasis. Ann. Pharmacother. 2009;43:1818–1823. doi: 10.1345/aph.1M218. [DOI] [PubMed] [Google Scholar]
- 68.Yu X, Vazquez A, Levine A, Carpizo D. Allele-specific p53 mutant reactivation. Cancer Cell. 2012;21:614–625. doi: 10.1016/j.ccr.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Frezza C, Martins C. From tumor prevention to therapy: empowering p53 to fight back. Drug Res. Update. 2012;15:258–267. doi: 10.1016/j.drup.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Mabanglo M, Hast M, Lubock N, et al. Crystal structures of the fungal pathogen Aspergillus fumigatus protein farnesyltransferase complexed with substrates and inhibitors reveal features for antifungal drug design. Protein Sci. 2013;23(3):289–301. doi: 10.1002/pro.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Zhou M, Dong X, Shen N, et al. Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthease: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design. Nucleic Acids Res. 2010;38:3399–3413. doi: 10.1093/nar/gkp1254. • Structural biology and target discovery.
- 72.Laggner C, Schiefer C, Fiechtner B, Poles G, Hoffmann D, et al. Discovery of high-affinity ligands of σ1 receptor, ERG2, and emopamil binding protein by pharmacophore modeling and virtual screening. J. Med. Chem. 2005;48:4754–4764. doi: 10.1021/jm049073+. [DOI] [PubMed] [Google Scholar]
- 73.Oberoi JK, Wattal C, Goel N, Raveendran R, Datta S, Prasad K. Non albicans Candida species in blood stream infections in a tertiary care hospital. Indian J. Med. Res. 2012;136:997–1003. [PMC free article] [PubMed] [Google Scholar]
- 74.Kim M-N, Shin H, Sung H, et al. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin. Infect. Dis. 2009;48:57–61. doi: 10.1086/597108. [DOI] [PubMed] [Google Scholar]
- 75.Hull C, Purdy N. Non antifungal clinical drug interventions and human commensal fungi: what are we selecting? Future Microbiol. 2013;8:813–816. doi: 10.2217/fmb.13.45. [DOI] [PubMed] [Google Scholar]
- 76.Walker I. Interscience conference on antimicrobial agents and chemotherapy – 50th Annual Meeting – research on promising new agents: part 2. IDrugs. 2010;13:746–748. [PubMed] [Google Scholar]