Abstract
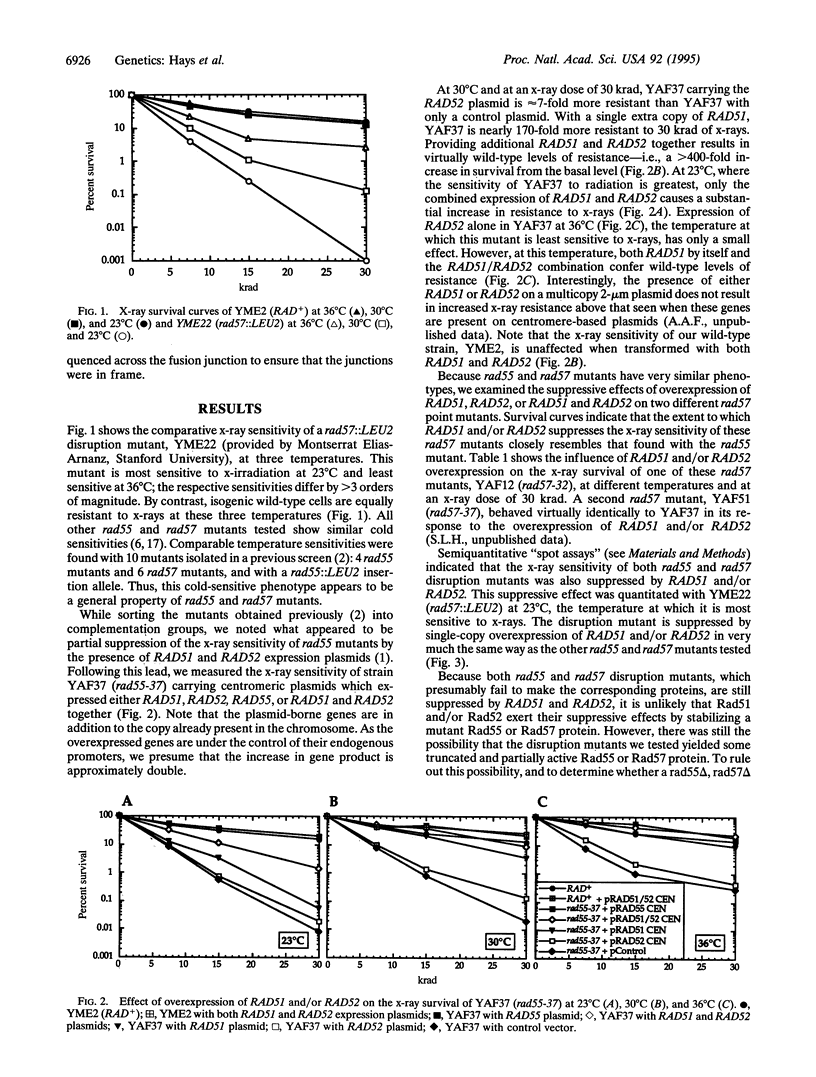
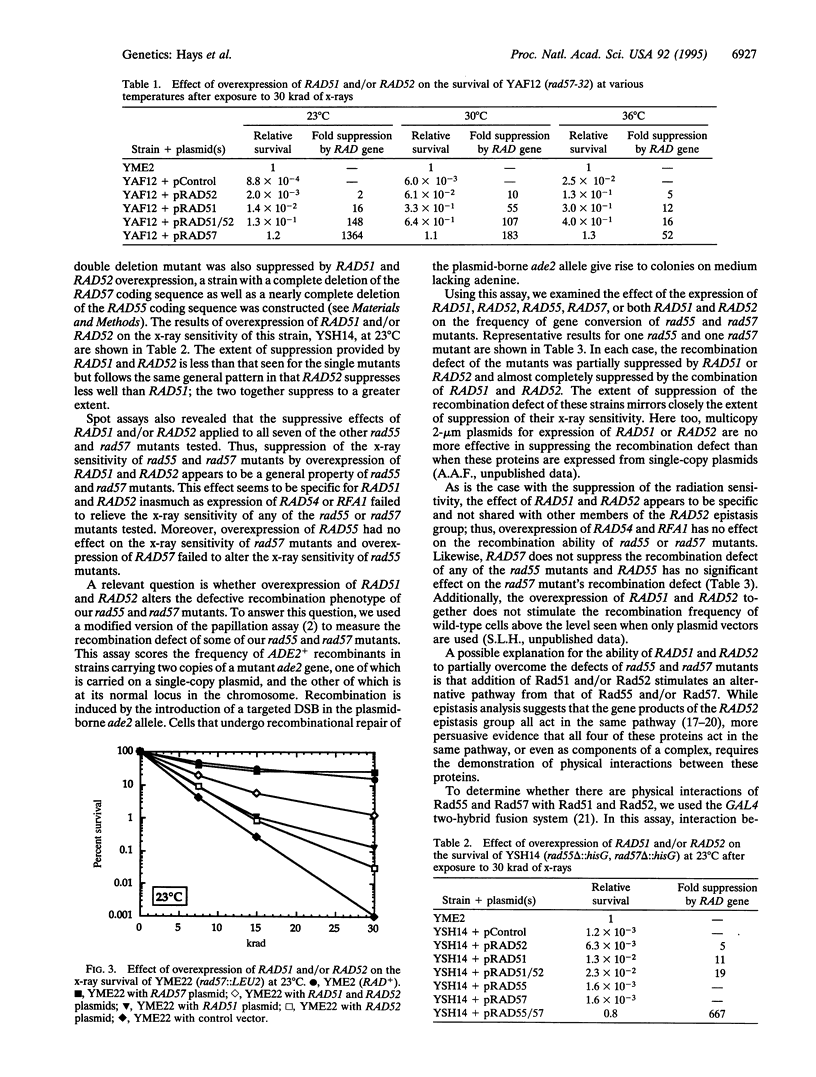
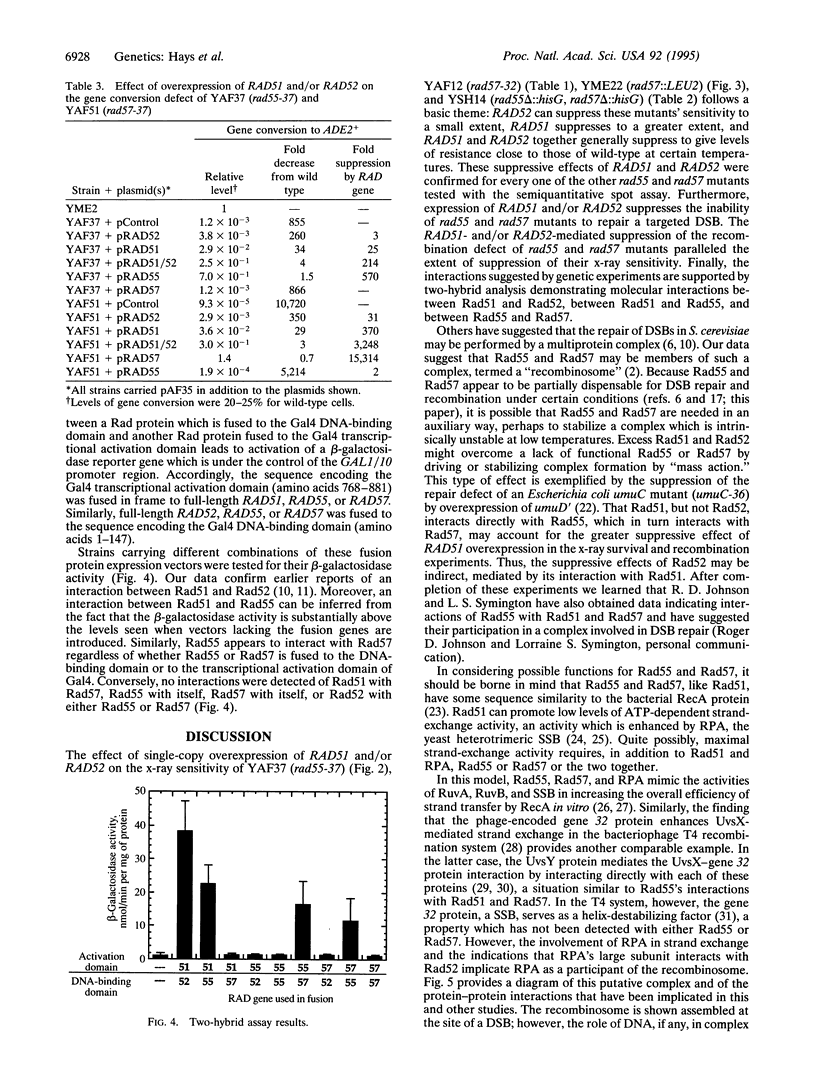
The repair of DNA double-strand breaks in Saccharomyces cerevisiae requires genes of the RAD52 epistasis group, of which RAD55 and RAD57 are members. Here, we show that the x-ray sensitivity of rad55 and rad57 mutant strains is suppressible by overexpression of RAD51 or RAD52. Virtually complete suppression is provided by the simultaneous overexpression of RAD51 and RAD52. This suppression occurs at 23 degrees C, where these mutants are more sensitive to x-rays, as well as at 30 degrees C and 36 degrees C. In addition, a recombination defect of rad55 and rad57 mutants is similarly suppressed. Direct in vivo interactions between the Rad51 and Rad55 proteins, and between Rad55 and Rad57, have also been identified by using the two-hybrid system. These results indicate that these four proteins constitute part of a complex, a "recombinosome," to effect the recombinational repair of double-strand breaks.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Cao L., Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987 Aug;116(4):541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E., Thresher R., Griffith J. D., Kolodner R. D. Characterization of DNA-binding and strand-exchange stimulation properties of y-RPA, a yeast single-strand-DNA-binding protein. J Mol Biol. 1992 Sep 5;227(1):54–71. doi: 10.1016/0022-2836(92)90681-9. [DOI] [PubMed] [Google Scholar]
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- Bates H., Bridges B. A., Woodgate R. Mutagenic DNA repair in Escherichia coli, XX. Overproduction of UmuD' protein results in suppression of the umuC36 mutation in excision defective bacteria. Mutat Res. 1991 Sep-Oct;250(1-2):199–204. doi: 10.1016/0027-5107(91)90176-o. [DOI] [PubMed] [Google Scholar]
- Donovan J. W., Milne G. T., Weaver D. T. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 1994 Nov 1;8(21):2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989 Jul 20;340(6230):245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Firmenich A. A., Elias-Arnanz M., Berg P. A novel allele of Saccharomyces cerevisiae RFA1 that is deficient in recombination and repair and suppressible by RAD52. Mol Cell Biol. 1995 Mar;15(3):1620–1631. doi: 10.1128/mcb.15.3.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game J. C. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993 Apr;4(2):73–83. [PubMed] [Google Scholar]
- Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993 Nov 19;75(4):805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Yonesaki T. The characterization of a complex of three bacteriophage T4 recombination proteins, uvsX protein, uvsY protein, and gene 32 protein, on single-stranded DNA. J Biol Chem. 1991 Mar 15;266(8):4883–4888. [PubMed] [Google Scholar]
- Jiang H., Giedroc D., Kodadek T. The role of protein-protein interactions in the assembly of the presynaptic filament for T4 homologous recombination. J Biol Chem. 1993 Apr 15;268(11):7904–7911. [PubMed] [Google Scholar]
- Kodadek T. The role of the bacteriophage T4 gene 32 protein in homologous pairing. J Biol Chem. 1990 Dec 5;265(34):20966–20969. [PubMed] [Google Scholar]
- Lovett S. T., Mortimer R. K. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics. 1987 Aug;116(4):547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of RAD55 to prokaryotic RecA and other RecA-like proteins. Gene. 1994 May 3;142(1):103–106. doi: 10.1016/0378-1119(94)90362-x. [DOI] [PubMed] [Google Scholar]
- MORTIMER R. K. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radiat Res. 1958 Sep;9(3):312–326. [PubMed] [Google Scholar]
- Milne G. T., Weaver D. T. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993 Sep;7(9):1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- Nakai S., Mortimer R. K. Studies on the genetic mechanism of radiation-induced mitotic segregation in yeast. Mol Gen Genet. 1969;103(4):329–338. doi: 10.1007/BF00383483. [DOI] [PubMed] [Google Scholar]
- Rattray A. J., Symington L. S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995 Jan;139(1):45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHERAGA H. A., NEMETHY G., STEINBERG I. Z. The contribution of hydrophobic bonds to the thermal stability of protein conformations. J Biol Chem. 1962 Aug;237:2506–2508. [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994 Aug 26;265(5176):1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Tsaneva I. R., Illing G., Lloyd R. G., West S. C. Purification and properties of the RuvA and RuvB proteins of Escherichia coli. Mol Gen Genet. 1992 Oct;235(1):1–10. doi: 10.1007/BF00286175. [DOI] [PubMed] [Google Scholar]
- Tsaneva I. R., Müller B., West S. C. ATP-dependent branch migration of Holliday junctions promoted by the RuvA and RuvB proteins of E. coli. Cell. 1992 Jun 26;69(7):1171–1180. doi: 10.1016/0092-8674(92)90638-s. [DOI] [PubMed] [Google Scholar]



