Abstract
Pseudomonas aeruginosa PGPR2 is a mung bean rhizosphere strain that produces secondary metabolites and hydrolytic enzymes contributing to excellent antifungal activity against Macrophomina phaseolina, one of the prevalent fungal pathogens of mung bean. Genome sequencing was performed using the Ion Torrent Personal Genome Machine generating 1,354,732 reads (6,772,433 sequenced bases) achieving ~25-fold coverage of the genome. Reference genome assembly using MIRA 3.4.0 yielded 198 contigs. The draft genome of PGPR2 encoded 6803 open reading frames, of which 5314 were genes with predicted functions, 1489 were genes of known functions, and 80 were RNA-coding genes. Strain specific and core genes of P. aeruginosa PGPR2 that are relevant to rhizospheric habitat were identified by pangenome analysis. Genes involved in plant growth promoting function such as synthesis of ACC deaminase, indole-3-acetic acid, trehalose, mineral scavenging siderophores, hydrogen cyanide, chitinases, acyl homoserine lactones, acetoin, 2,3-butanediol, and phytases were identified. In addition, niche-specific genes such as phosphate solubilising 3-phytase, adhesins, pathway-specific transcriptional regulators, a diguanylate cyclase involved in cellulose synthesis, a receptor for ferrienterochelin, a DEAD/DEAH-box helicase involved in stress tolerance, chemotaxis/motility determinants, an HtpX protease, and enzymes involved in the production of a chromanone derivative with potent antifungal activity were identified.
1. Introduction
Pseudomonas aeruginosa is one of the prevalent organisms present in diverse natural environments. It is an aquatic and soil bacterium that can infect a range of hosts including humans in an opportunistic manner [1]. Though they are specially noted for their pathogenicity, the recent research and genomic studies have thrown light into the unique property of these organisms to produce a wide array of secondary metabolites with multiple potential biotechnological applications. Pseudomonas aeruginosa has a large number of paralogous groups (distinct gene families), indicating that its genome has evolved through genetic expansion [2]. It has developed adaptation and competitive fitness in a diverse range of ecological niches. P. aeruginosa is known to produce bioactive compounds showing antagonistic activity against plant pathogens. P. aeruginosa strains such as 7NSK2 [3], PNA1 [4], NJ-15 [5], and PUPa3 [6] have been reported to have plant growth promoting ability and biocontrol activity against phytopathogens. Pseudomonas aeruginosa PGPR2 was isolated from the rhizosphere of mung bean plant with the ability to promote plant growth. This strain showed efficient antagonistic activity against Macrophomina phaseolina, the causative agent of root rot diseases [7].
Though genome sequences of several strains of medically relevant P. aeruginosa have been reported, the genome of only one agriculturally relevant strain, P. aeruginosa M18, has been reported until now. Thus, in order to define the differences in the genome structures between P. aeruginosa strains from nosocomial and rhizosphere niches, the whole genome of the strain PGPR2 was sequenced and compared with those of previously sequenced medically relevant strains. In this communication we report the genomic regions that are evolutionarily preserved and varied between P. aeruginosa PGPR2 and other medically relevant strains. We have comprehensively compared the PGPR2 draft genome with the genomes of six other strains (M18, DK2, LESB58, PA7, PAO1, and UCBPP-PA14). We report the core and niche-specific genome organization in this ubiquitous species.
2. Materials and Methods
2.1. Bacterial Growth and DNA Extraction
A single colony of P. aeruginosa PGPR2 grown on Luria-Bertani (LB) agar was inoculated into 5 mL of LB broth and grown overnight with agitation at 30°C. Bacterial cells were collected by centrifugation and the genomic DNA was isolated using the Qiagen DNeasy kit following the manufacturer's protocol for Gram negative bacteria (Qiagen, Hilden, Germany). The quantity, quality, and integrity of the isolated DNA were verified by spectrophotometry (NanoDrop products, Wilmington, DE, USA) and gel electrophoresis.
2.2. Whole Genome Sequencing and Assembly
Genomic libraries were generated using 1 μg of genomic DNA using the Ion Xpress Plus fragment library kit (Life technologies, NY, USA). Briefly, genomic DNA was enzymatically sheared and the quality and quantity were analyzed on a bioanalyzer (Multina, Shimadzu, Japan). Adapters were ligated to the sheared DNA fragments and the fragments were size-selected on 2% E-gel (Lifetechnologies). The size selected library (150 to 300 bp) was used for template preparation on an Ion one-touch automated template preparation system using the Ion One-touch Template 200 kit. Template positive ion sphere particles (ISPs) were enriched using the Ion one touch ES system, loaded onto an Ion 316 chip, and sequenced using the Ion sequencing 200 kit on Ion Torrent Personnel Genome machine. The MIRA assembler v 3.4.1 was used to generate a reference genome assembly of P. aeruginosa PGPR2 using P. aeruginosa DK2 genome (NC_018080) as template.
The template sequence was removed after alignment and the unaligned reads were extracted for de novo assembly using MIRA v 3.4.1 [8]. The resulting contigs of high quality and appreciable length were added to the assembly. The final draft genome was viewed and edited, when required, using Staden Package version 2.0 [9].
2.3. Genome Annotation and Comparative Genome Analysis
The PGPR2 genome was annotated using the Rapid Annotation using Subsystems Technology (RAST) server [10]. The annotated genome was compared with other related and distant genomes maintained in the SEED Viewer environment. Blastp was used to find homologs of selected PGPR2 sequences in all the annotated proteins of the P. aeruginosa strains, M18, DK2, LESB58, PAO1, UCBPP-PA14, and PA7. Ribosomal RNA and transfer RNA genes were predicted by RNAmmer v1.2 [11] and tRNAScan-SE [12], respectively. A complete set of open reading frames predicted to encode proteins was identified using GLIMMER [13]. Genomes/contigs were aligned with each other using Mummer v 3.20 [14] and Mauve v 2.3.1 [15]. InterProScan was used to identify conserved domains in selected sequences [16]. A comprehensive genome comparison was performed across seven P. aeruginosa strains using Gview server and the results were visualized using the WebAct Tool [17]. DNAplotter [18] was used to construct a P. aeruginosa PGPR2 genome atlas while CRISPR repeats were identified using the CRISPR finder [19]. The CVtree tool [20] was used to perform phylogenetic analysis of PGPR2 in comparison with other P. aeruginosa genomes (M18, PAO1, DK2, LESB58, PA7, and UCBPP-PA14). Strain specific regions on the P. aeruginosa PGPR2 genome were detected using Panseq server with default parameter [21]. The metabolic pathways of the strain PGPR2 were constructed and compared with those of other strains using KAAS (KEGG Automatic Annotation Server) [22] and the MetaCyc database [23].
3. Results and Discussion
3.1. Genome Features
The mung bean rhizosphere isolate PGPR2 showed efficient plant growth promoting activity and antagonistic activity against Macrophomina phaseolina, a common fungal plant pathogen. In this study, we report the whole genome sequence of strain PGPR2 and the genetic factors responsible for plant growth promoting properties. The draft genome of P. aeruginosa PGPR2 was 6772433 bp long comprising 198 contigs (Genbank accession number: ASQO00000000). The P. aeruginosa PGPR2 genome contains 6803 predicted open reading frames (ORFs), of which 80 were RNA encoding genes, 5314 were protein encoding genes (PEGs) with predicted functions, and 1489 were PEGs with unknown functions. The average GC content of the PGPR2 genome was 66%, which is consistent with previously reported P. aeruginosa genomes. Approximately, 86.9% of the total PGPR2 genome was found to be coding regions. The P. aeruginosa PGPR2 genome is graphically represented in Figure 1 while the genomic features are summarized in Table 1.
Figure 1.
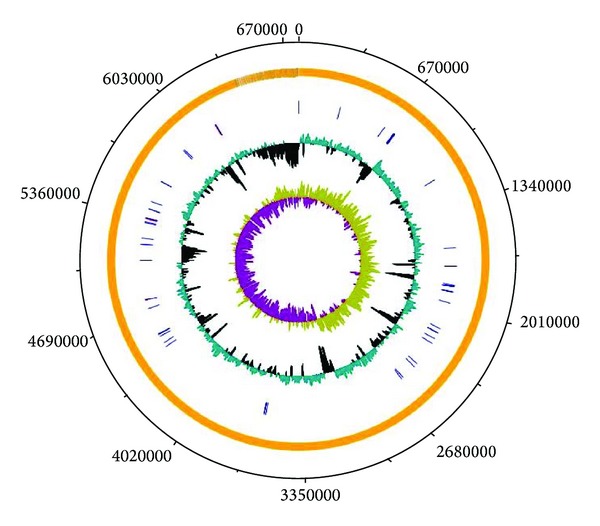
Graphical map of the P. aeruginosa PGPR2 draft genome. From the outside to the inside: open reading frames, rRNA operons, and tRNAs are shown in yellow, red, and blue, respectively. G+C content plot and GC skew (purple: negative values, olive: positive values) are also shown.
Table 1.
Features of the P. aeruginosa PGPR2 genome in comparison with P. aeruginosa strains from other ecological niches.
| Niche | PGPR2 | LESB58 | PA7 | PAO1 | UCBPP-PA14 | DK2 | M18 |
|---|---|---|---|---|---|---|---|
| Mung bean rhizosphere # |
Cystic fibrosis patient # |
Cystic fibrosis patient # |
Human burn wound # |
Human burn wound # |
Cystic fibrosis patient # |
Sweet melon rhizosphere # |
|
| Total number of genes | 6883 | 6061 | 6369 | 5697 | 5977 | 5959 | 5770 |
| PEGs | 6803 | 5925 | 6286 | 5566 | 5892 | 5884 | 5684 |
| 5S rRNA | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 16S rRNA | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| 23S rRNA | 4 | 4 | 4 | 4 | 4 | 4 | 4 |
| tRNA genes | 68 | 67 | 63 | 63 | 59 | 64 | 61 |
| misc_RNA | 0 | 21 | 0 | 4 | 0 | 0 | 5 |
| Predicted genes | 5314 | 4172 | 3638 | 3248 | 3583 | 3972 | 4116 |
| Hypothetical genes | 1489 | 1753 | 2648 | 2318 | 2309 | 1912 | 1568 |
| CRISPR repeats | 3 | 1 | 2 | 1 | 2 | 3 | 3 |
# Count.
3.2. Unique Genomic Regions of P. aeruginosa PGPR2 Relevant to Its Rhizospheric Habitat
Plant growth-promoting rhizobacteria have the ability to scavenge mineral nutrients, fix atmospheric nitrogen, solubilize soil phosphorus, and synthesize regulators of plant metabolism [24]. The strain PGPR2 was shown to exhibit effective plant growth promoting properties towards the mung bean plant in both in vivo and in vitro conditions. This strain also had a strong suppressive effect on the growth of Macrophomina phaseolina. Earlier, we have reported a secondary metabolite (3, 4-dihydroxy-N-methyl-4-(4-oxochroman-2-yl) butanamide) and a secreted protease responsible for the antifungal activity [7, 25]. Among several reported strains, P. aeruginosa M18 strain was the only reported genome with plant growth promotion and biocontrol activity [26]. Analysis of the genome sequence of PGPR2 revealed several features that reflect this bacterium highlighting its plant growth promoting ability (Figure 2), a few of which are discussed in detail.
Figure 2.

Diagrammatic representations of various unique and core genomic regions of PGPR2 strain relevant to its rhizospheric habitat. Predicted and annotated gene sequences were analyzed for similarity with the NCBI database followed by assignment of each gene into KEGG pathway. Based on individual analysis results of the KEGG pathway, proposed biochemical pathways were constructed which demonstrated characteristic features related to plant growth promotion. Unique genes are indicated by green color.
3.3. 3-Phytase
A 930 bp ORF (Genbank accession number ASQO01000075.1, region 1144 : 215) coding for 3-phytase was identified in PGPR2 with no homologs in other P. aeruginosa strains. Sequence analysis of this ORF revealed maximum similarity to 3-phytase from Pseudomonas protegens CHAO with 68% identity using BlastP analysis. Rhizospheric bacteria can benefit plant growth by solubilization of inorganic phosphates. This is achieved by conversion of insoluble phosphorus (P) to an accessible form via several enzymes. Nonspecific phosphatases are known to perform dephosphorylation of phosphoester or phosphoanhydride bonds in organic matter [27]. Phytases on the other hand specifically release P from phytic acid [28]. Phosphonatases and C-P Lyases perform C-P cleavage in organophosphonates [29]. All other analyzed P. aeruginosa strains do not produce this enzyme including P. aeruginosa M18, a reported PGPR strain.
3.4. TonB-Dependent Receptor and Hemagglutinin
TonB-dependent receptors (TBDRs) allow Gram-negative bacteria to uptake scarce resources from competitive environments with very high affinity. Earlier reports on TBDRs focused on the uptake of siderophore-iron complexes but recent studies have showed that the spectrum of ligands that can be scavenged includes sugars, vitamins, heme, and other nonferrous cations [30]. One of the TBDRs identified in PGPR2 was found to be unique to this strain. A total of 24 TBDRs were identified from the genome of P. aeruginosa PGPR2. BlastP analysis revealed a strain specific region (2514 bp; Genbank accession number ASQO01000014.1, regions 3109 : 596) contains a unique TBDR, which showed 98% identity with a TBDR of Pseudomonas nitroreducens. Filamentous hemagglutinins are involved in plant attachment and root adhesion of plant associated bacteria thereby indirectly responsible for plant growth promotion [31]. Bacterial attachment to the root surface by various adhesins is an important trait required for competitive colonization. The genome of P. aeruginosa PGPR2 encoded adhesin/hemagglutinin protein (2484 bp; Genbank accession number ASQO01000065.1, region 4016 : 1533), possibly involved in the initial adhesion to mung bean roots. This region was found to share 73% identity to the adhesin/hemagglutinin gene of Pseudomonas protegens CHA0.
3.5. LysR Family Transcriptional Regulator PA2877
Apart from the global regulatory systems, the pathway-specific regulators are responsible for the transcriptional activation of the secondary metabolite biosynthetic operons. LysR family response regulators have been shown to be involved in the positive regulation of antifungal metabolite production [32]. PGPR2 strain encoded a LysR family transcriptional regulator (909 bp; Genbank accession number ASQO01000026.1, regions 2019 : 1111), which showed maximum homology with a LysR family regulator from Achromobacter xylosoxidans (68% identity) in BlastP analysis. Pfam domain analysis identified a 60-amino acid helix-turn-helix domain (PF00126.22), which is known to be involved in DNA binding and 209-amino acid as LysR substrate binding domain (PF03466.15).
3.6. Diguanylate Cyclase
Diguanylate cyclases usually possess nucleotide cyclase activity and aid in cyclic-di-GMP formation. Cyclic-di-GMP signaling is used by many bacteria to regulate vital functions such as biofilm formation [33]. Since biofilm formation is an important survival strategy for many bacteria, the synthesis and degradation of cyclic-di-GMP is tightly regulated by diguanylate cyclases, which contain domains with conserved GGDEF and EAL sequence motifs. Most sequenced bacterial genomes contain several GGDEF motif containing proteins. The GGDEF domain is usually responsible for diguanylate cyclase activity and also for cellulose synthase activity. Cellulose fibres in well-documented PGPR (Agrobacterium sp. and Rhizobium sp.) help in anchoring of bacteria on plant surfaces, thus facilitating permanent colonization and establishment of biofilm-like structures on the root surface [34]. PGPR2 contained a unique region (1437 bp; Genbank accession number ASQO01000004.1, regions 2981 : 4417) encoding a diguanylate cyclase with the characteristic “GGDEF” domain. This gene shared 78% identity with a homolog from Pseudomonas putida using BlastP analysis.
3.7. UxpB Protein
The genome of PGPR2 harbours a strain specific region (1623 bp; Genbank accession number ASQO01000028.1, regions 3325 : 1703) coding for a UxpB homolog. This region shared 88% identity with the Pseudomonas denitrificans ATCC 13867 genome using BlantN analysis and TblastN showed homology with the same organism but with 51% identity. Earlier reports have demonstrated that UxpB and its homologs are induced under inorganic phosphate (Pi) limitation and thus enable the utilization of various organic phosphate sources. Expression of this gene helps the plants in accessing the scarce nutrients that are normally difficult to obtain, while limiting their concentration in the niche for competing pathogens [35].
3.8. Alpha/Beta-Hydrolase Fold Family Enzyme
A strain specific region (Genbank accession number ASQO01000004.1, regions 1492 : 2373) encoding a 293-amino acid protein, homologous to a hydrolytic enzyme containing an alpha/beta-hydrolase fold from Pseudomonas sp. GM25 with 92% identity in BlastP analysis was identified. Earlier studies have shown that members of the alpha/beta-hydrolase fold family are capable of cleaving the lactone ring of acylhomoserine lactones [36]. N-Acylhomoserine lactones (AHLs) are signaling molecules in quorum-sensing (QS) systems that regulate virulence, disease incidence, and establishment in pathogenic bacteria. The ability to cleave these signals (quorum quenching) and disrupt communication in the rhizosphere will help PGPR2 to nullify the advantage of population in competing pathogens.
3.9. Chemotaxis/Motility
Two unique regions (712 bp and 723 bp; Genbank accession number ASQO01000138.1, regions 1559 : 846, 2311 : 1589) from PGPR2 strain were identified that showed homology to corresponding homologs from Pseudomonas sp. UW4 in BlastP analysis with 95 and 85% identity, respectively. They possessed highly conserved domains and were annotated as MotB-related protein essential for chemotaxis and methyl-accepting chemotaxis protein (MCPs). Nearly, all motile bacteria have genes encoding the signal transduction pathway for chemotaxis. Earlier studies reported that motility and chemotaxis are important traits for root colonization [37] and a requisite for improved biocontrol activity against the pathogenic fungi. Methyl-accepting chemotaxis proteins (MCPs) are responsible for the detection of ligands, the binding of which initiates a chemotaxis signalling cascade thereby leading to root directed motility.
3.10. DEAD/DEAH-Box Helicase
DEAD-box helicases play a key role in tolerance to various abiotic stresses including temperature, light, oxygen, and salt stress. Earlier studies suggested that genes belonging to the DEAD-box helicase family are induced by salinity and function as a typical helicase to unwind DNA and RNA [38]. A PGPR2 genomic region (1587 bp; Genbank accession number ASQO01000097.1, region 3154 : 1568) was identified as DEAD/DEAH-box helicase and shared 81% identity with a helicase from Pseudomonas stutzeri A1501 in BlastP analysis.
3.11. Receptor for Ferrienterochelin
A unique region (1271 bp, Genbank accession number ASQO01000059.1, regions 111 : 1382) coding for ferrienterochelin receptor—FepA was identified on the PGPR2 genome. FepA is an 81,000-dalton protein in the E. coli outer membrane that functions in the initial step of iron uptake by binding ferrienterochelin [39]. Receptors for siderophores such as ferrienterochelin are necessary for survival in the rhizosphere by competition. The ability to uptake self siderophores and siderophores that are secreted by other competing bacteria provides an advantage for PGPR2 in the rhizosphere. This may subsequently lead to depletion of iron, leading to suppression of pathogen growth in the immediate vicinity of the plant.
3.12. Antifungal Proteases
Nearly 60 loci on the PGPR2 genome were annotated as different types of proteases. Their molecular weight ranged from 4000 to 100,000 Da. Among 60 sequences only two proteases showed molecular weight in the range 30 to 34 kDa. Earlier, we have identified and characterized a proteolytic enzyme with a molecular weight of ~33 kDa. This protein exhibited efficient antifungal activity. Peptide mass fingerprinting results showed that the mass value matched with the amino acid sequence of protease HtpX of Pseudomonas aeruginosa strain PA7. The two proteases of interest on the PGPR2 genome (molecular weights of 31592.92 and 34389.2 Da) showed considerable sequence homology with a probable protease HtpX homolog and a putative cysteine protease, respectively. A genomic region (876 bp, ASQO01000001.1, regions 2206178_2205303) was found to encode a homolog of the protease HtpX of Pseudomonas aeruginosa strain PGPR2.
3.13. Antifungal Secondary Metabolites from PGPR2
Genes belonging to metabolic pathways involved in antifungal activity were identified from the genome of PGPR2. These metabolites include phenazine, pyocyanin, paerucumarin, pyoverdine, pyochelin, hydrogen cyanide, AHLs, rhamnolipids, phenylacetic acid, and macrolide antibiotics. In addition, we have demonstrated earlier that 3, 4-dihydroxy-N-methyl-4-(4-oxochroman-2-yl) butanamide, a secondary metabolite synthesized by PGPR2, showed antagonism against Macrophomina phaseolina [7]. Based on the survey of the PGPR2 genome putative biosynthetic pathway was identified. One molecule of acetyl CoA can condense with five molecules of malonyl CoA, which in turn can be obtained by the action of acetyl CoA carboxylase with biotin as the cofactor (Equation 1), to give a hexa keto compound (Equation 2), which can then be cyclised to give different chromone intermediates (Equation 3) which is represented in Figure 3. The basic chromanone skeleton of 3, 4-dihydroxy-N-methyl-4-(4-oxochroman-2-yl) butanamide could have been generated by a similar reaction. However, the mode of dehydroxylation of the aromatic ring of the constructed chromanone unit and the route by which the side chain moiety is obtained and gets itself attached to the 2-position of chromanone ring are still unclear.
Figure 3.
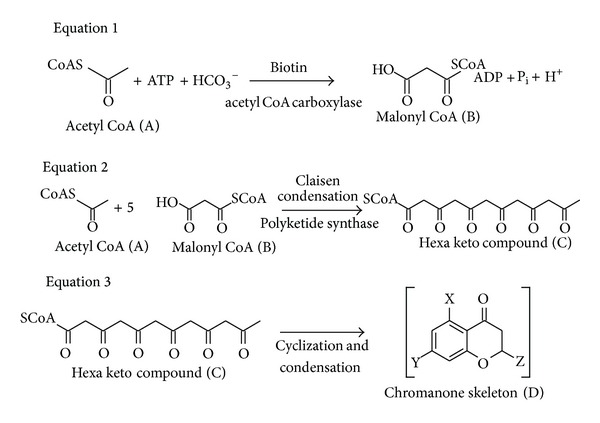
Pathway proposed for production of the antifungal metabolite, 3, 4-dihydroxy-N-methyl-4-(4-oxochroman-2-yl) butanamide by PGPR2. Equation 1: Acetyl CoA carboxylase catalyzed the formation of malonyl CoA from acetyl CoA; Equation 2: Polyketide synthase catalyzed the formation of hexa keto compound by condensation of a molecule of acetyl CoA with five molecules of malonyl CoA; Equation 3: The hexa keto compound undergoes cyclization and condensation reactions to form different chromanone intermediates.
3.14. Other Features of P. aeruginosa PGPR2 Genome Relevant to Its Plant Growth Promoting Ability
3.14.1. ACC Deaminase Synthesis
In addition to PGPR2, the genomes of M18, LesB58, PAO1, PA7, DK2, and UCBPP-PA14 contained a putative 1-aminocyclopropane-1-carboxylate (ACC) deaminase which showed 100% homology in BlastP analysis (899 bp, Genbank accession number ASQO01000001.1, regions 1920268 : 1921167). P. aeruginosa strains containing 1-aminocyclopropane-1-carboxylate deaminase (ACCD) are capable of growing on ACC as a nitrogen source. These organisms when colonized on the surface of plant roots break down the ACC exuded from the plant to ammonia and α-ketobutyrate. This in turn is advantageous for the plant as these organisms sequester ACC thereby lowering the level of the stress hormone ethylene in plants [40].
3.14.2. Siderophores Synthesis
The PGPR2 genome encoded a number of genes involved in iron acquisition such as the siderophore pyoverdine, one of the major siderophores found in all fluorescent pseudomonads. In addition, all the genomes contain the gene coding for TonB. This protein spans the periplasm and is anchored to the cytoplasmic membrane interacting with receptors in the outer membrane to facilitate the uptake of iron-siderophore complexes. Thus, the production of active siderophores could contribute to potent biocontrol property against soil-borne pathogens. Earlier reports have suggested that P. fluorescens Pf-5 and P. aeruginosa M18 contained genes (pch) encoding a secondary siderophore pyochelin, which has antifungal activity [41]. The pyochelin biosynthetic gene cluster (pchCEFGR) was identified using BlastP analysis in all chosen P. aeruginosa strains (Table 2). Interestingly, the strain PGPR2 contained the complete pyoverdine (Pvd) biosynthetic gene cluster (pvdJAEGXSLHIONMQPY), whereas one or more of these genes were missing in other genomes (Table 2). The ability of bacteria to produce multiple siderophores benefits them, as they may function in different environments, making them more competitive against other organisms in the same niche.
Table 2.
Genes associated with pyoverdine synthesis in P. aeruginosa strains.
| Gene | Function | PGPR2 | M18 | DK2 | LESB58 | UCBPP-PA14 | PAO1 | PA7 |
|---|---|---|---|---|---|---|---|---|
| pvdj | Pyoverdine sidechain nonribosomal peptide synthetase | + | + | + | + | + | + | + |
| pvdX | Hypothetical protein | + | + | + | + | + | + | + |
| pvdS | Sigma factor controlling pyoverdine biosynthesis | + | + | + | + | + | + | + |
| pvdL | Pyoverdine chromophore precursor synthetase | + | + | + | + | + | + | + |
| pvdH | L-2,4-diaminobutyrate: 2-oxoglutarate aminotransferase | + | + | + | + | + | + | + |
| pvdI | Pyoverdine sidechain nonribosomal peptide synthetase | + | + | + | + | + | + | + |
| pvdO | Pyoverdine responsive serine/threonine kinase | + | + | + | + | + | + | + |
| pvdN | Putative aminotransferase, class V | + | + | + | + | + | + | + |
| pvdM | Putative dipeptidase | + | + | + | + | + | + | + |
| pvdQ | Acyl-homoserine lactone acylase | + | + | + | + | + | + | + |
| pvdY | Hypothetical protein | + | + | + | − | − | − | + |
| pvdG | Thioesterase involved in nonribosomal peptide synthesis | + | + | + | + | + | + | − |
| pvdE | Pyoverdine ABC export system: ATPase and permease components | + | + | + | − | − | − | + |
| FpvB | Outer membrane ferripyoverdine receptor FpvB, for Type I pyoverdine | + | + | − | + | + | + | − |
| pvdP | Pyoverdine biosynthesis related protein | + | + | + | + | − | − | + |
| pvdA | L-ornithine 5-monooxygenase | + | − | + | + | + | + | + |
3.14.3. Indole Acetic Acid Production
Pseudomonads, fluorescent strains in particular, have been known to synthesize the phytohormone indole acetic acid (IAA). IAA is the most common and best characterized phytohormone. It has been estimated that 80% of bacteria isolated from the rhizosphere produce IAA [42]. However, the amount of IAA produced may vary significantly among strains. Based on the concentration, IAA can either stimulate or inhibit plant growth. The production of indole-3-acetic acid by rhizobacteria has been associated with plant growth promotion, especially root initiation and elongation. The role of bacterial IAA in suppression of charcoal rot disease of chickpea has been reported [43]. A genomic region containing an 837 bp CDS was found to have homologs in all chosen P. aeruginosa strains with 100% identity using BlastP analysis. This region was annotated as indole-3-glycerol phosphate synthase (ASQO01000001, regions 682275-683111), an enzyme responsible for the synthesis of IAA.
3.14.4. Trehalose Synthesis
Trehalose is a nonreducing disaccharide present in a wide variety of organisms. Certain species of plant, fungi, bacteria, and invertebrates are known as anhydrobionts due to the production of trehalose. This enables them to revive themselves in the presence of water in a few hours after being completely dehydrated for months or years [44]. Rodríguez-Salazar et al. [45] reported the effects of genetically engineered Azospirillum brasilense for trehalose biosynthesis conferred drought tolerance and also significantly increased leaf and root biomass of maize plant. The PGPR2 genome encoded a maltooligosyl trehalose synthase gene treY (accession number ASQO01000001, regions 3205742-3206398), Maltooligosyl trehalose hydrolase gene treZ, (accession number ASQO01000001, regions 3199525-3201276), and trehalose synthase gene treS (accession number ASQO01000001, regions 3219761-3216459), required for biosynthesis and accumulation of trehalose, a metabolite that is of key importance in conferring tolerance to low water environments. The homologs of these genes were found in all P. aeruginosa strains with 100% identity in BlastP analysis.
3.14.5. Acetoin and Butanediol Production
Certain bacterial species ferment pyruvate to acetoin and 2,3-butanediol through 2-acetolactate. Pyruvate is converted into either lactate by the enzyme L-lactate dehydrogenase or D-lactate dehydrogenase or to 2-acetolactate by the enzyme α-acetolactate synthase (α-ALS) [46]. Genes coding for the enzymes involved in acetoin and 2,3-butanediol synthesis were detected using BlastP in the genome of PGPR2 (182 bp, accession number ASQO01000001, region 5377021-5376839). The conversion of pyruvate to lactate requires NADH and when the levels of NADH are limited, a majority of the pyruvate is converted to 2-acetolactate. Since 2-acetolactate is an unstable intermediate, it undergoes spontaneous decarboxylation in the presence of oxygen, producing diacetyl. Once diacetyl is formed, it can be converted to acetoin by the activity of the enzyme diacetyl reductase (755 bp, accession number ASQO01000016.1, regions 144-899) (also known as acetoin dehydrogenase). Under anaerobic conditions 2-acetolactate is converted directly to acetoin by α-acetolactate decarboxylase (α-ALD). P. aeruginosa strains depend on acetoin dehydrogenase for synthesis of acetoin rather than using α-acetolactate decarboxylase. The production of acetoin and 2,3-butanediol by plant growth promoting bacteria was reported to increase systemic disease resistance [47] and drought tolerance [48].
3.14.6. Hydrogen Cyanide Production
HCN, a secondary metabolite produced in Pseudomonas spp., is catalyzed by the membrane-bound enzyme HCN synthase, which forms HCN and CO2 from glycine [49]. The hcnABC genes encoding HCN synthase (2961 bp, accession number ASQO01000001.1, regions 3175436-3172475) were identified on a genomic fragment of P. aeruginosa PGPR2. A blastp search using the region as query identified homologs in all other strains of P. aeruginosa. Lanteigne et al. [50] isolated HCN producing Pseudomonas sp. LBUM300 and observed effective biological control activity against bacterial canker of tomato. HCN production in P. aeruginosa PGPR2 was verified as described earlier by Bakker and Schippers [51].
3.14.7. Chitinase Production n
Chitinases produced by Pseudomonas spp. can hydrolyze the β-1,4-linkages in chitin, an abundant N-acetyl-β-D-glucosamine polysaccharide which is an integral structural component of fungal cell walls. Thus, bacterial chitinases contribute to the biological control of fungal phytopathogens [52]. P. aeruginosa PGPR2 encodes a chitinase (1451 bp, ASQO01000001.1, region 2978066-2979517). Similarly, using BlastP, all the strains of P. aeruginosa were found to encode at least one chitinase.
3.14.8. Acyl Homoserine Lactone Synthesis
P. aeruginosa PGPR2 produce N-acyl-homoserine lactone (AHL), a well-known quorum sensing signalling molecule. Earlier studies have reported the involvement of these molecules in plant growth promotion and protection against salt stress [53]. As quorum sensing is known to modulate population dependent genes including virulence factors, it is necessary to study the means of interference with quorum sensing systems. Interruption of quorum sensing, known as quorum quenching, is usually accomplished by production of lactonases or acylases. Lactonases inactivate AHLs by hydrolyzing the ester bond of the lactone ring while acylases cleave the amide bond that connects the lactone moiety and acyl side-chain. The products of the enzymatic breakdown do not function as active signal molecules. As described earlier, the P. aeruginosa PGPR2 genome encodes pvdQ (2289 bp, ASQO01000001.1, regions 2872266-2872652) involved in the production of acyl-homoserine lactone acylase. Earlier studies have shown that the PvdQ prefers long c-length AHL molecules as a substrate [54]. The overexpression of PvdQ in P. aeruginosa strains is shown to inhibit the accumulation of signal molecules (3-oxo-C12-HSL and 2-heptyl-3-hydroxy-4(1H)-quinolone) and thereby decreases expression of several virulence factors such as elastase and pyocyanin that are important for P. aeruginosa pathogenicity in animals [55].
4. Conclusion
In this study, the genome of the P. aeruginosa strain PGPR2 was sequenced and analyzed. The draft genome PGPR2 possesses an array of genes encoding unique functions corresponding to its plant growth promotion and biocontrol abilities, besides sharing a common core genome to other sequenced strains of this species. Interestingly, the strain PGPR2 genome has acquired several genes needed for the rhizopheric environments from other organisms, which are not found in other medically relevant P. aeruginosa strains.
Acknowledgments
The work was financially supported by the Indian Council for Agricultural Research 217 (NBAIM/AMAAS/2007–2012/MG (5)/PG/BG/3), India. The central facilities from CAS, CEGS, UGC-NRCBS, and DBT-IPLS at MKU are gratefully acknowledged.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: lessons from a versatile opportunist. Microbes and Infection. 2000;2(9):1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 2.Stover CK, Pham XQ, Erwin AL, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 3.Buysens S, Heungens K, Poppe J, Hofte M. Involvement of pyochelin and pyoverdin in suppression of pythium-induced damping-off of tomato by Pseudomonas aeruginosa 7NSK2. Applied and Environmental Microbiology. 1996;62(3):865–871. doi: 10.1128/aem.62.3.865-871.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anjaiah V, Cornelis P, Koedam N. Effect of genotype and root colonization in biological control of fusarium wilts in pigeonpea and chickpea by Pseudomonas aeruginosa PNA1. Canadian Journal of Microbiology. 2003;49(2):85–91. doi: 10.1139/w03-011. [DOI] [PubMed] [Google Scholar]
- 5.Bano N, Musarrat J. Characterization of a new Pseudomonas aeruginosa strain NJ-15 as a potential biocontrol agent. Current Microbiology. 2003;46(5):324–328. doi: 10.1007/s00284-002-3857-8. [DOI] [PubMed] [Google Scholar]
- 6.Kumar RS, Ayyadurai N, Pandiaraja P, et al. Characterization of antifungal metabolite produced by a new strain Pseudomonas aeruginosa PUPa3 that exhibits broad-spectrum antifungal activity biofertilizing traits. Journal of Applied Microbiology. 2005;98(1):145–154. doi: 10.1111/j.1365-2672.2004.02435.x. [DOI] [PubMed] [Google Scholar]
- 7.Illakkiam D, Ponraj P, Shankar M, Muthusubramanian S, Rajendhran J, Gunasekaran P. Identification and structure elucidation of a novel antifungal compound produced by Pseudomonas aeruginosa PGPR2 against Macrophomina phaseolina . Applied Biochemistry and Biotechnology. 2013;171:2176–2185. doi: 10.1007/s12010-013-0469-7. [DOI] [PubMed] [Google Scholar]
- 8.Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. Computer science and biology. Proceedings of the German Conference on Bioinformatics; 1999; pp. 45–56. [Google Scholar]
- 9.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods in Molecular Biology. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]
- 10.Aziz RK, Bartels D, Best A, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9, article 75 doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lagesen K, Hallin P, Rødland EA, Stærfeldt H-H, Rognes T, Ussery DW. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Research. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25(5):955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Research. 1999;27(23):4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delcher AL, Phillippy A, Carlton J, Salzberg SL. Fast algorithms for large-scale genome alignment and comparison. Nucleic Acids Research. 2002;30(11):2478–2483. doi: 10.1093/nar/30.11.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Research. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quevillon E, Silventoinen V, Pillai S, et al. InterProScan: protein domains identifier. Nucleic Acids Research. 2005;33(2):W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. ACT: the artemis comparison tool. Bioinformatics. 2005;21(16):3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 18.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25(1):119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Research. 2007;35(2):W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z, Hao B. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Research. 2009;37(2):W174–W178. doi: 10.1093/nar/gkp278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laing C, Buchanan C, Taboada EN, et al. Pan-genome sequence analysis using Panseq: an online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinformatics. 2010;11, article 461 doi: 10.1186/1471-2105-11-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36(1):D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspi R, Altman T, Dreher K, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Research. 2012;40(1):D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashan Y, de-Bashan LE. Fresh-weight measurements of roots provide inaccurate estimates of the effects of plant growth-promoting bacteria on root growth: a critical examination. Soil Biology & Biochemistry. 2005;37(10):1795–1804. [Google Scholar]
- 25.Illakkiam D, Anuj NL, Ponraj P, Shankar M, Rajendhran J, Gunasekaran P. Proteolytic enzyme mediated antagonistic potential of Pseudomonas aeruginosa against Macrophomina phaseolina . Indian Journal of Experimental Biology. 2013;51:1024–1031. [PubMed] [Google Scholar]
- 26.Wu D, Ye J, Ou H, et al. Genomic analysis and temperature-dependent transcriptome profiles of the rhizosphere originating strain Pseudomonas aeruginosa M18. BMC Genomics. 2011;12, article 438 doi: 10.1186/1471-2164-12-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nannipieri P, Giagnoni L, Landi L, Renella G. Role of phosphatase enzymes in soil. In: Bunemann EK, Obreson A, Frossard E, editors. Phosphorus in Action. Berlin, Germany: Springer; 2011. pp. 215–243. [Google Scholar]
- 28.Richardson AE. Soil microorganisms and phosphorous availability. In: Pankhurst CE, Doube BM, Gupta VVSR, editors. Soil Biota: Management in Sustainable Farming Systems. Victoria, Australia: CSIRO; 1994. pp. 50–62. [Google Scholar]
- 29.Rodriguez H, Fraga R, Gonzalez T, Bashan Y Y. Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant and Soil. 2006;287:15–21. [Google Scholar]
- 30.Lim BL. TonB-dependent receptors in nitrogen-fixing nodulating bacteria. Microbes and Environments. 2010;25(2):67–74. doi: 10.1264/jsme2.me10102. [DOI] [PubMed] [Google Scholar]
- 31.Gottig N, Garavaglia BS, Garofalo CG, Orellano EG, Ottado J. A filamentous hemagglutinin -like protein of Xanthomonas axonopodis pv . citri , the phytopathogen responsible for citrus canker, is involved in bacterial virulence. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0004358.e4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim EJ, Kim JC, Choi GJ, et al. Forest soil metagenome gene cluster involved in antifungal activity expression in Escherichia coli. Applied and Environmental Microbiology. 2008;74:723–730. doi: 10.1128/AEM.01911-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newell PD, Yoshioka S, Hvorecny KL, Monds RD, O’Toole GA. Systematic analysis of diguanylate cyclases that promote biofilm formation by Pseudomonas fluorescens Pf0-1. Journal of Bacteriology. 2011;193(18):4685–4698. doi: 10.1128/JB.05483-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ausmees N, Mayer R, Weinhouse H, et al. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiology Letters. 2001;204(1):163–167. doi: 10.1111/j.1574-6968.2001.tb10880.x. [DOI] [PubMed] [Google Scholar]
- 35.Putker F, Tommassen-van Boxtel R, Stork M, Rodríguez-Herva JJ, Koster M, Tommassen J. The type II secretion system (Xcp) of Pseudomonas putida is active and involved in the secretion of phosphatases. Environmental Microbiology. 2013;15:2658–2671. doi: 10.1111/1462-2920.12115. [DOI] [PubMed] [Google Scholar]
- 36.Mei G, Yan X, Turak A, Luo Z, Zhang L. AidH, an alpha/beta-hydrolase fold family member from an ochrobactrum sp. strain, Is a novel N-acylhomoserine lactonase. Applied and Environmental Microbiology. 2010;76(15):4933–4942. doi: 10.1128/AEM.00477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oku S, Komatsu A, Tajima T, Nakashimada Y, Kato J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes and Environments. 2012;27(4):462–469. doi: 10.1264/jsme2.ME12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu HH, Liu J, Fan SL, et al. Molecular cloning and characterization of a salinity stress-induced gene encoding DEAD-box helicase from the halophyte Apocynum venetum . Journal of Experimental Botany. 2008;59(3):633–644. doi: 10.1093/jxb/erm355. [DOI] [PubMed] [Google Scholar]
- 39.Lundrigan MD, Kadner RJ. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. Homology among outer membrane receptors that interact with TonB . The Journal of Biological Chemistry. 1986;261(23):10797–10801. [PubMed] [Google Scholar]
- 40.Glick BR. The enhancement of plant growth by free-living bacteria. Canadian Journal of Microbiology. 1995;41(2):109–117. [Google Scholar]
- 41.Phoebe CHJ, Combie J, Albert FG, et al. Extremophilic organisms as an unexplored source of antifungal compounds. Journal of Antibiotics. 2001;54(1):56–65. doi: 10.7164/antibiotics.54.56. [DOI] [PubMed] [Google Scholar]
- 42.Patten CL, Glick BR. Bacterial biosynthesis of indole-3-acetic acid. Canadian Journal of Microbiology. 1996;42(3):207–220. doi: 10.1139/m96-032. [DOI] [PubMed] [Google Scholar]
- 43.Khare E, Arora NK. Effect of indole-3-acetic acid (IAA) produced by Pseudomonas aeruginosa in suppression of charcoal rot disease of chickpea. Current Microbiology. 2010;61(1):64–68. doi: 10.1007/s00284-009-9577-6. [DOI] [PubMed] [Google Scholar]
- 44.Paul MJ, Primavesi LF, Jhurreea D, Zhang Y. Trehalose metabolism and signaling. Annual Review of Plant Biology. 2008;59:417–441. doi: 10.1146/annurev.arplant.59.032607.092945. [DOI] [PubMed] [Google Scholar]
- 45.Rodríguez-Salazar J, Suárez R, Caballero-Mellado J, Iturriaga G. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiology Letters. 2009;296(1):52–59. doi: 10.1111/j.1574-6968.2009.01614.x. [DOI] [PubMed] [Google Scholar]
- 46.Blomqvist K, Nikkola M, Lehtovaara P, et al. Characterization of the genes of the 2,3-butanediol operons from Klebsiella terrigena and Enterobacter aerogenes . Journal of Bacteriology. 1993;175(5):1392–1404. doi: 10.1128/jb.175.5.1392-1404.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.S. M. Cho, B. R. Kang, S. H. Han, et al. 2R,3R-butanediol, a bacterial volatile produced by Pseudomonas chlororaphis O6, is involved in induction of systemic tolerance to drought in Arabidopsis thaliana . Molecular Plant-Microbe Interactions. 2008;21(8):1067–1075. doi: 10.1094/MPMI-21-8-1067. [DOI] [PubMed] [Google Scholar]
- 48.Han SH, Lee SJ, Moon JH, et al. GacS-dependent production of 2R, 3R-butanediol by Pseudomonas chlororaphis O6 is a major determinant for eliciting systemic resistance against Erwinia carotovora but not against Pseudomonas syringae pv. tabaci in tobacco. Molecular Plant-Microbe Interactions. 2006;19(8):924–930. doi: 10.1094/MPMI-19-0924. [DOI] [PubMed] [Google Scholar]
- 49.Laville J, Blumer C, von Schroetter C, et al. Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0. Journal of Bacteriology. 1998;180(12):3187–3196. doi: 10.1128/jb.180.12.3187-3196.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lanteigne C, Gadkar VJ, Wallon T, Novinscak A, Filion M. Production of DAPG and HCN by Pseudomonas sp. LBUM300 contributes to the biological control of bacterial canker of tomato. Phytopathology. 2012;102(10):967–973. doi: 10.1094/PHYTO-11-11-0312. [DOI] [PubMed] [Google Scholar]
- 51.Bakker AW, Schippers B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas SPP-mediated plant growth-stimulation. Soil Biology & Biochemistry. 1987;19(4):451–457. [Google Scholar]
- 52.Ramyasmruthi S, Pallavi O, Pallavi S, Tilak K, Srividya S. Chitinolytic and secondary metabolite producing Pseudomonas fluorescens isolated from Solanaceae rhizosphere effective against broad spectrum fungal phytopathogens. Asian Journal of Plant Science Research. 2012;2(1):16–24. [Google Scholar]
- 53.Barriuso J, Solano BR, Mañero FJG. Protection against pathogen and salt stress by four plant growth-promoting rhizobacteria isolated from Pinus sp. on Arabidopsis thaliana . Phytopathology. 2008;98(6):666–672. doi: 10.1094/PHYTO-98-6-0666. [DOI] [PubMed] [Google Scholar]
- 54.Bokhove M, Jimenez PN, Quax WJ, Dijkstra BW. The quorum-quenching N-acyl homoserine lactone acylase PvdQ is an Ntn-hydrolase with an unusual substrate-binding pocket. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(2):686–691. doi: 10.1073/pnas.0911839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sio CF, Otten LG, Cool RH, et al. Quorum quenching by an N-acyl-homoserine lactone acylase from Pseudomonas aeruginosa PAO1. Infection and Immunity. 2006;74(3):1673–1682. doi: 10.1128/IAI.74.3.1673-1682.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


