Abstract
Background
Schneckenbecken dysplasia (SBD) is an autosomal recessive lethal skeletal dysplasia that is classified into the severe spondylodysplastic dysplasias (SSDD) group in the international nosology for skeletal dysplasias. The radiological hallmark of SBD is the snaillike configuration of the hypoplastic iliac bone. SLC35D1 (solute carrier-35D1) is a nucleotide-sugar transporter involved in proteoglycan synthesis. Recently, based on human and mouse genetic studies, we showed that loss-of-function mutations of the SLC35D1 gene (SLC35D1) cause SBD.
Object
To explore further the range of SLC35D1 mutations in SBD and elucidate whether SLC35D1 mutations cause other skeletal dysplasias that belong to the SSDD group.
Methods and results
We searched for SLC35D1 mutations in five families with SBD and 15 patients with other SSDD group diseases, including achodrogenesis type 1A, spondylometaphyseal dysplasia Sedaghatian type and fibrochondrogenesis. We identified four novel mutations, c.319C>T (p.R107X), IVS4+3A>G, a 4959-bp deletion causing the removal of exon 7 (p.R178fsX15), and c.193A>C (p. T65P), in three SBD families. Exon trapping assay showed IVS4+3A>G caused skipping of exon 4 and a frameshift (p.L109fsX18). Yeast complementation assay showed the T65P mutant protein lost the transporter activity of nucleotide sugars. Therefore, all these mutations result in loss of function. No SLC35D1 mutations were identified in all patients with other SSDD group diseases.
Conclusion
Our findings suggest that SLC35D1 loss-of-function mutations result consistently in SBD and are exclusive to SBD.
Schneckenbecken dysplasia (SBD; OMIM 269250) is an autosomal recessive perinatally lethal skeletal dysplasia.1–3 The German term “Schneckenbecken” refers to the distinctive, snail-like appearance of the ilia due to medial bone projection from the inner iliac margin. Other skeletal hallmarks of SBD include thoracic hypoplasia, severe flattening of the vertebral bodies with absent ossification of the posterior parts of the vertebral bodies, and short thick long bones. Histological findings in SBD include large, round, and centrally located chondrocyte nuclei, scarce extracellular matrix, and an absence of columnar alignment of proliferating chondrocytes in the growth plate.
SBD belongs to the “severe spondylodysplastic dysplasias (SSDD)” group in the international nosology for skeletal dysplasias,4 which includes achodrogenesis type 1A (ACG1A; OMIM 200600), spondylometaphyseal dysplasia (SMD) Sedaghatian type (OMIM 250220), and fibrochondrogenesis (FCG; OMIM 228520) (supplemental table 1). ACG1A shows more poorly ossified vertebral bodies, more hypoplastic ilia with protruded medial margins and arched lower margins, and misshapen, stellate tubular bones.5 SMD Sedaghatian type exhibits better ossified vertebral bodies, less hypoplastic ilia, and milder tubular bone shortening. The medial projection of the ilia is very subtle, while the metaphyseal cupping and irregularity are conspicuous.6 FCG is a disorder most similar to SBD, particularly in its absent ossification of the posterior parts of the vertebral bodies; however, the iliac medial projection is less conspicuous and the tubular bone shortening is more pronounced with bulbous metaphyses.7 The aetiology of these disorders remains unknown except for a part of SMD Sedaghatian type.8
Recently, we reported mutations of the solute carrier-35 D1 (SLC35D1) gene (SLC35D1; OMIM 610804) in SBD—that is, three premature stop codon mutations that result in complete loss of SLC35D1 function in two typical SBD patients.9 This was the first report of identification of causative mutations for SBD and the first known gene for a disease in the SSDD group. SLC35D1 is the nucleotide sugar transporter (NST) expressed in the endoplasmic reticulum (ER) and involved in proteoglycan (PG) synthesis.9–12 SLC35D1 transports UDP-GalNAc and UDP-GlcUA, the substrates used to synthesise CS, from the cytoplasm into the ER lumen.9–12 We also reported that the Slc35d1 deficient mouse develops a lethal form of severe short limbed dwarfism very similar to SBD.9 The mice had defective chondroitin sulfate (CS) biosynthesis on the core protein of cartilage PGs. The CS chain in the mice was estimated to be half the length of the normal CS chain and it seemed to be reduced in number.
We showed that loss-of-function mutations of SLC35D1 result in SBD phenotype; however, phenotypic consequences of other types of SLC35D1 mutations remain elusive. It is intriguing to consider whether other types of SLC35D1 mutations cause other skeletal dysplasias, particularly other disorders belonging to the SSDD group, because a phenotypic similarity often reflects a causal similarity in the skeletal dysplasias.4 To address this question and to explore further the range of SLC35D1 mutations in SBD, we searched for SLC35D1 mutations in 20 families with SSDD, including five SBD, eight SMD Sedaghatian type, six ACG1A, and one FCG. Four novel loss-of-function mutations of SLC35D1 were identified in the three SBD families, but no mutations were found in all patients with other SSDD.
PATIENTS AND METHODS
Collection of patients
SSDD cases were recruited through International Skeletal Dysplasia Registry (ISDR) (http://www.csmc.edu/), European Skeletal Dysplasia Network (ESDN) (http://www.esdn.org/), and Japanese Skeletal Dysplasia Consortium (JSDC) (http://www.riken.jp/lab-www/OA-team/JSDC/). Clinical and radiographic phenotypes of the patients were evaluated by the experts of the organisations and reviewed by authors (AS, SU, GN). The diagnosis of these disorders was based on the whole patterns of skeletal abnormalities, but not on single signs alone or in combination. The core radiological findings are listed in supplemental table 1. A total of 20 SSDD cases—that is, five SBD, eight SMD Sedaghatian type, six ACG1A, and one FCG—were recruited for this study.
Clinical reports
The clinical and radiological manifestations of five patients diagnosed as SBD are shown in table 1.
Table 1.
Characteristics of the Schneckenbecken dysplasia patients
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | |
|---|---|---|---|---|---|
| Clinical information | |||||
| Ethnic origin | Caucasian | Turkish | Turkish | Turkish | Brazilian |
| Consanguinity | (−) | + | + | + | (−) |
| Gestational age (weeks) | 18 | 22 | 20 | 20 | 28 |
| Radiological manifestations | |||||
| Handle bar clavicle | + | + | + | + | + |
| Bell shaped thorax | + | + | + | + | + |
| Retardation of the vertebral body ossification | Very severe | Severe | Very severe | Very severe | Moderate |
| Ossification of the posterior arch | Relatively preserved | Relatively preserved | Relatively preserved | Poor | Well preserved |
| Interpediculate distance narrowing | + | + | + | + | + |
| Sacral ossification | Relatively preserved | Relatively preserved | Relatively preserved | Poor | Well preserved |
| Schneckenbecken sign | Prominent | Prominent | Prominent | Prominent | Mild |
| Pubic ossification | + | + | + | (−) | + |
| Metaphyseal flaring | + | + | + | + | + |
| Tarsal ossification | (−) | + | (−) | (−) | + |
| Angular deformity of the humerus | (−) | (−) | (−) | (−) | + |
Patient 1
Clinical information of this patient has been recorded in a previous report1 as Patient 2 of Family A. Briefly, the infant girl was a product of A non-consanguineous marriage between healthy Caucasian parents (the mother: Spanish-North African, the father: Portuguese). The family history was unremarkable for short stature or genetic disease. Prenatal ultrasound at 16 weeks gestation revealed a short-limb fetus and the pregnancy was terminated at 18 weeks. She had a severe form of short-limb short-stature with a relatively large head and prominent abdomen. Radiographic examination showed characteristic appearance of the spine and the iliac. Precocious ossification at the ischium was apparent.
Patient 2
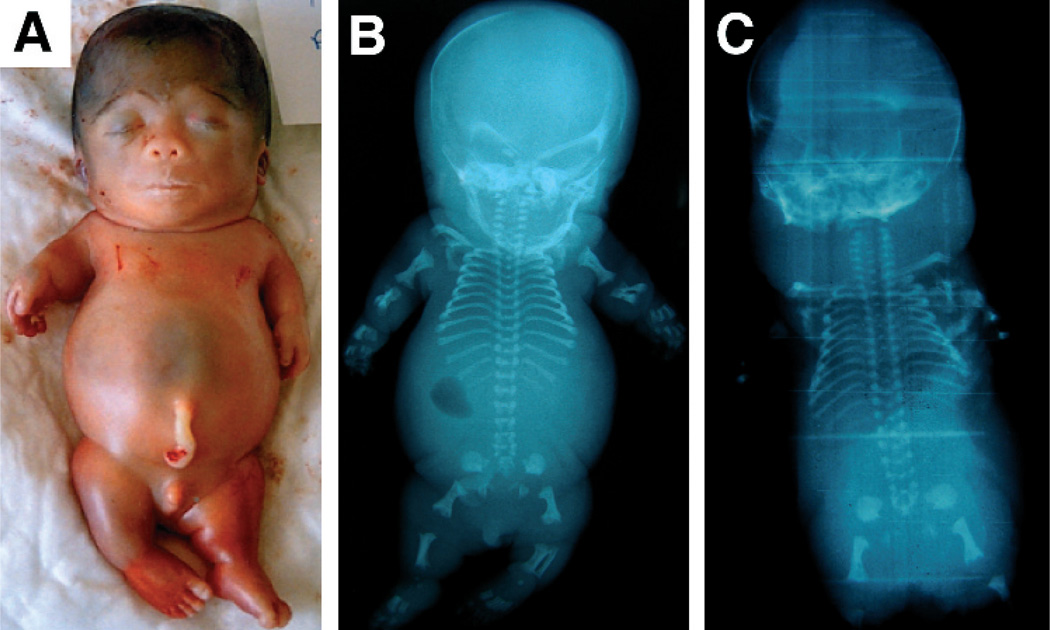
The parents of this fetus were first degree cousins of Turkish origin. The mother was a G3P1: one healthy boy and one stillbirth with short limbs (no radiographs or DNA available). On prenatal ultrasound at 22 weeks gestation, this fetus was identified as having a short neck with redundant nuchal skin, narrow thorax, scoliosis, and short limbs, and thus the pregnancy was interrupted. Postmortem investigations were limited to external examination and radiographs. The fetus had a small nose with anteverted nares (fig 1A). There was generalised micromelia but no polydactyly. The thorax was small and the abdomen protuberant. Radiographs revealed platyspondyly with pedicle width exceeding that of the vertebral bodies (fig 1B). The chest was narrow and bell shaped. The Scheneckenbecken sign was present in both iliac wings. All long bones were short and stocky with some metaphyseal flaring. Tarsal ossification was advanced.
Figure 1.
Schneckenbecken dysplasia patients with SLC35D1 mutations. (A) Clinical photograph of Patient 2. Note generalised micromelia, narrow thorax, and protuberant abdomen. (B) Radiograph of patient 2. Note platyspondyly, thoracic hypoplasia, and snail-like appearance of ilia. (C) Radiograph of patient 3. Similar to patient 2, but slightly more severe phenotype with wafer thin vertebral bodies.
Patient 3
The parents of this fetus were also first degree cousins of Turkish origin. The mother was a G4P2SA1: one healthy girl, one boy who died at 18 months of age without clear diagnosis, and one spontaneous abortion. This fetus was identified on 18 weeks ultrasound as having hydrops and short limbs. Thanatophoric dysplasia was suspected and the pregnancy was interrupted at 20 weeks. On inspection, there was massive hydrops and cystic hygroma. The facial features resembled those of patient 2 with small nose and anteverted nares. There was marked micromelia. The chest was narrow and short with a protuberant abdomen. Radiographs revealed features similar to those of patient 2 but slightly more severe with wafer thin vertebral bodies (fig 1C).
Patient 4
This fetus was the product of Turkish parents who denied consanguinity, yet came from the same small village. The mother was a G7P1 at referral. She had a history of recurrent first trimester fetal loss and had been immunised for Rhesus incompatibility on every intervention. Her fifth and sixth pregnancies were complicated with hydrops. The fetus was terminated 20 weeks gestation due to detection of short limbs, redundant nuchal skin and scalp oedema on prenatal ultrasound. The karyotype was normal. A severely hydropic fetus with a swollen face with a bipartite nose, thick lips, narrow thorax and micromelic limbs were noted at postmortem examination. The autopsy revealed placental hydropic changes, pulmonary hypoplasia and accessory spleen. Radiological manifestations were similar to those of patients 3 except for absence of pubic ossification and poorly ossified sacrum (supplemental fig 1A). Ossification of the posterior neural arches was very severely retarded (supplemental fig 1B).
Patient 5
The parents of this fetus were non-consanguineous Brazilian origin. The mother was a G2P2SA0: one healthy boy. This fetus was identified on prenatal ultrasound as having short limbs and very narrow thorax (<P5). The newborn died shortly after birth at 28 weeks gestation, weighting 1020 g. The facial features revealed a small nose and anteverted nares with a median groove. The palate was normal. There was marked micromelia without polydactyly. The chest was narrow and short with a protuberant abdomen. Autopsy showed no internal malformations. Radiological examination showed platyspondyly and Schneckenbecken sign (supplemental fig 1C). However, these were much milder than those of other cases. In addition, angular deformity of the humerus was somewhat distinctive (supplemental fig 1D).
Mutation screening
Genomic DNA was extracted by standard procedures. The exon sequence of SLC35D1 with its flanking intron sequence was amplified by polymerase chain reaction (PCR) from genomic DNA and PCR products were sequenced directly by using an ABI Prism 3700 automated sequencer (PE Biosystems, Foster City, California, USA). PCR primer sequence is available on request.
Exon trapping assay
A genomic region encompassing exons 4–5 of SLC35D1 in patient 1 was amplified by PCR using primer set: D1-exon 4_F (5′-TCTCTCGAGTTGGGTGTTTGGGTGTCAAA-3′) and D1-exon 5_R (5′-TTGGCGGCCGCAGAGAGAGGGTTGCTTAAAC-3′). PCR products were digested with XhoI and NotI, and cloned into an exon trapping vector (Mo Bi Tec, Goettingen, Germany). Exon trapping vectors were transfected into HEK293 cells. Total RNAs were isolated using SV Total RNA Isolation System (Promega, Madison, Wisconsin, USA) 24 h after transfection. Reverse transcriptase (RT) PCR was performed using primer sets supplied with the exon trapping assay kit and PCR products were cloned and sequenced.
Construction of yeast expression vectors
SLC35D1 cDNA with the c.193A>C mutation was obtained by a PCR based mutagenesis. pcDNA3 vector (Invitrogen, Carlsbad, California, USA) containing wild type SLC35D1 cDNA tagged with an HA epitope at the N-terminus was PCR amplified by the primer set: T65P_F (5′-CTCCCCAATTACAGATTTCCCTCCT-3′) and T65P_R (5′-CACGCTCTTATTCACCACCA-3′). The blunt ended PCR generated DNA fragment was ligated and used to transform Escherichia coli, JM109. Plasmid DNA was prepared from the transformant culture using HiSpeed Plasimd Midi Kit (Qiagen, Valencia, California, USA). The mutation was confirmed by DNA sequencing. The cDNA fragments with wild type and mutant SLC35D1 were subcloned into a yeast expression vector, pYEX-BESN,13 derivative of YEX-BX (Clontech, Palo Alto, California, USA).
Nucleotide sugar transport assay
The assay was performed with a heterologous expression system in Saccharomyces cerevisiae as previously described. Briefly, expression vectors encoding wild type or mutant SLC35D1 cDNA was transfected into S cerevisiae YPH500. Microsome fractions were prepared from the transformants, and nucleotide sugar transporter activity was measured by comparing the amount of radiolabelled nucleotide sugar incorporated into positive and negative microsomes.
Immunoblotting
Proteins in membrane vesicles were separated by electrophoresis on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (Amersham Biosciences, Piscataway, New Jersey, USA). The HA tagged proteins were detected with anti-HA monoclonal antibody conjugated with peroxidase, 3F10 (Roche Diagnostics, Basel, Switzerland), at 50 mU/ml. Chemiluminescent signals were detected using ECL pus western blotting detection reagents (Amersham).
RESULTS
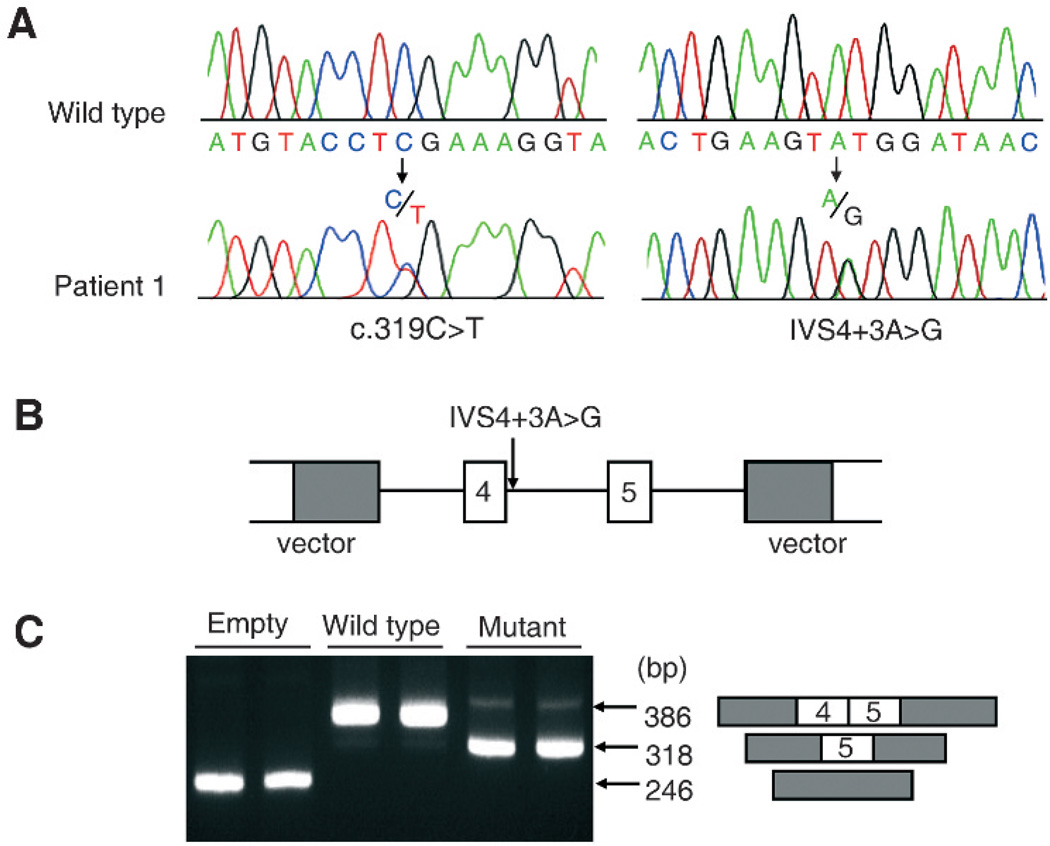
A heterozygous nonsense mutation, c.319C>T (p.R107X), and a splice donor site mutation, IVS4+3A>G, were identified in SBD patient 1 (fig 2A). Sequencing of subcloned DNAs revealed that patient 1 is a compound heterozygote for the mutations. These mutations were not found in 52 Caucasian-American controls or in public sequence variation databases. The score of IVS4+3G (mutation) by a splice donor site predictor was clearly lower than that of IVS4+3A (wild type) (0.13 vs 0.02), suggesting that the mutation caused abnormal splicing. To verify this hypothesis, we performed the exon trapping assay and found that the mutation did indeed cause skipping of exon 4 (fig 2B, C). The skipping results in a frame-shift that generates a truncated protein, p.L109fsX18.
Figure 2.
Identification of compound heterozygous mutations for c.319C>T (p.R107X) and IVS4+3A>G in patient 1. (A) The mutations. (B,C) Exon trapping assay to evaluate the effect of IVS4+3A>G on RNA splicing. (B) The SCL35D1 genomic regions encompassing exons 4–5, corresponding to wild type (IVS4+3A) and mutant (IVS4+3G) sequences, were cloned into an exon trapping vector. (C) The reverse transcriptase polymerase chain reaction (RT-PCR) products following transfection of exon trapping vectors into HEK293 cells. Note that the wild type vector trapped exons 4–5 but the mutant vector caused skipping of exon 4.
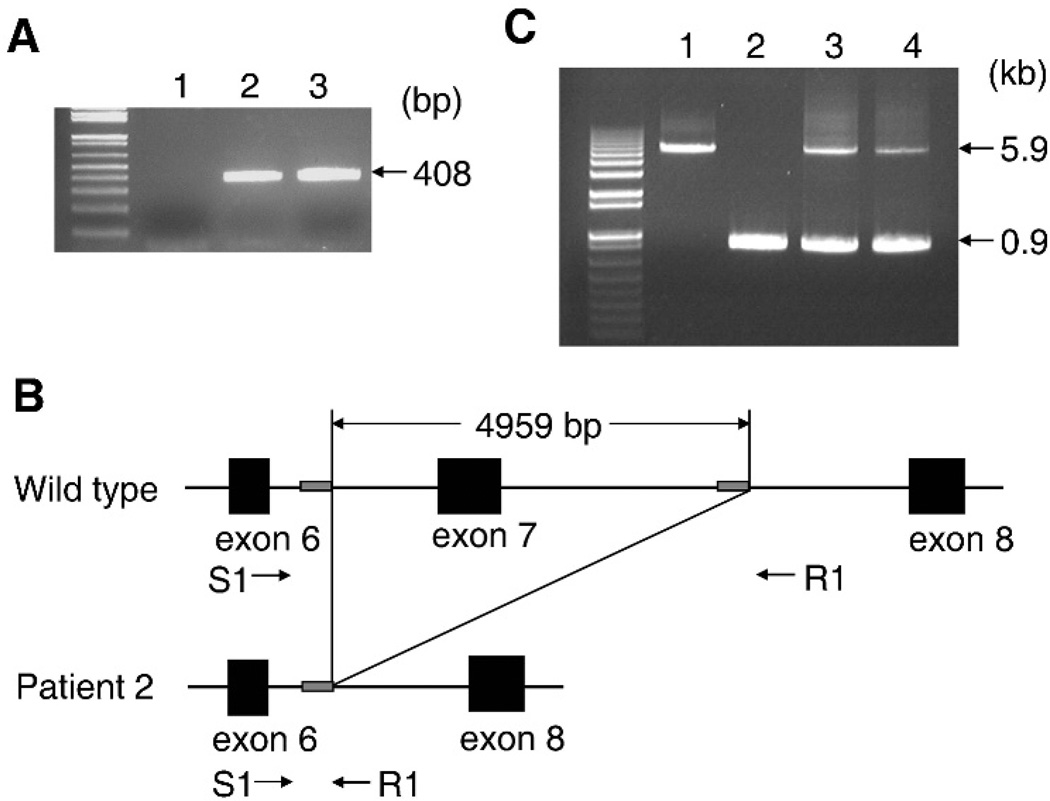
When using the genomic DNA from patient 2, the DNA fragment encompassing exon 7 of SLC35D1 could not be amplified, suggesting a homozygous deletion that includes exon 7 (fig 3A). To confirm this hypothesis, we designed a PCR primer set (S1 and R1) for amplifying a 5.9 kb genomic region encompassing exon 7 (fig 3B). A DNA fragment of an unexpected size (0.9 kb) was generated from patient 2’s DNA whereas a fragment of the expected size (5.9 kb) was generated from a wild-type DNA (fig 3C). Sequence comparison between the DNA fragments indicated the presence of a 4959 bp deletion causing complete removal of exon 7 (fig 3B). This analysis also revealed the presence of two 11 bp direct repeats (CTTGGCCTCCC) flanking the break points. One repeat was retained, whereas the other was lost in the deletion process, strongly suggesting that the deletion was generated by the mechanism of slipped mispairing during DNA replication.17,18 This deletion mutation IVS6+730_IVS7+3171del4959 results in skipping of exon 7 in cDNA (c.534_636del103) that produces a premature stop codon (p.R178fsX15). As expected, both parents of patient 2 were heterozygous carriers of the mutation (fig 3C).
Figure 3.
Identification of the homozygous 4959-bp deletion encompassing exon 7 of SLC35D1 in patient 2. (A) Electrophoretogram for the shorter PCR product encompassing exon 7 of SLC35D1. Lane 1: patient 2; lanes 2, 3: wild-type controls. The predicted size of the PCR product from the wild-type allele is 408 bp. The expected sized band is missing in patient 2. (B) Schematic representation of the genomic region containing the deletion and the location of the primer pair (S1 and R1) used for its identification. The deletion was flanked by 11 bp direct repeats (CTTGGCCTCCC) (grey box). (C) Electrophoretogram for the longer PCR product encompassing exon 7 of SLC35D1. Lane 1: control; lane 2: patient 2; lane 3: the father; lane 4: the mother. The primer pair in (B) was used. The predicted size of the PCR product from the wild-type allele is 5.9 kb and that from the deletion allele is 0.9 kb.
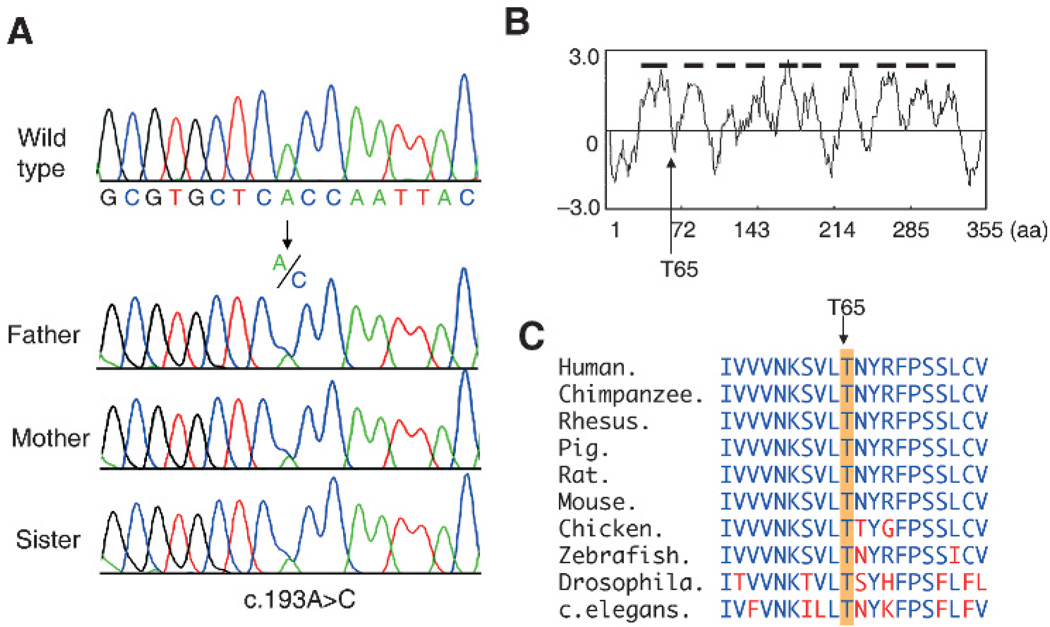
Since biological material of patient 3 was unavailable, SLC35D1 mutations were searched for in the consanguineous parents and an unaffected sister. A heterozygous missense mutation (c.193A>C), which results in the substitution of conserved threonine to proline in the first intracellular domain of SLC35D1 (p.T65P), was identified in the three relatives (fig 4A,B). This mutation was not found in 100 ethnically matched Turkish controls, 66 Caucasian-American controls, or in public sequence variation databases. The T65 amino acid residue was conserved among various species from human to Caenorhabditis elegans (C elegans) (fig 4C).
Figure 4.
Identification of the missense mutation (c.193A>C, p.T65P) in family members of patient 3. (A) Heterozygous c.193A>C mutation in the family members. (B) Hydrophobicity plot of the human SLC35D1 protein and the position of T65. The plot was based on a calculation using the hydrophobicity values of Kyte and Doolittle. The x axis represents the amino acid position within the SLC35D1, and the y axis represents the hydrophobicity values of the amino acids. SLC35D1 is predicted to have 10 transmembrane domains (thick horizontal bars). T65 is predicted to be located in the first intra-ER domain. (C) Comparison of the amino acid sequences around T65 of SLC35D1 among various species. Amino acids identical to human SLC35D1 are represented by blue letters and those not identical by red letters. The location of T65 is indicated by an arrow and orange shading. T65 is highly conserved.
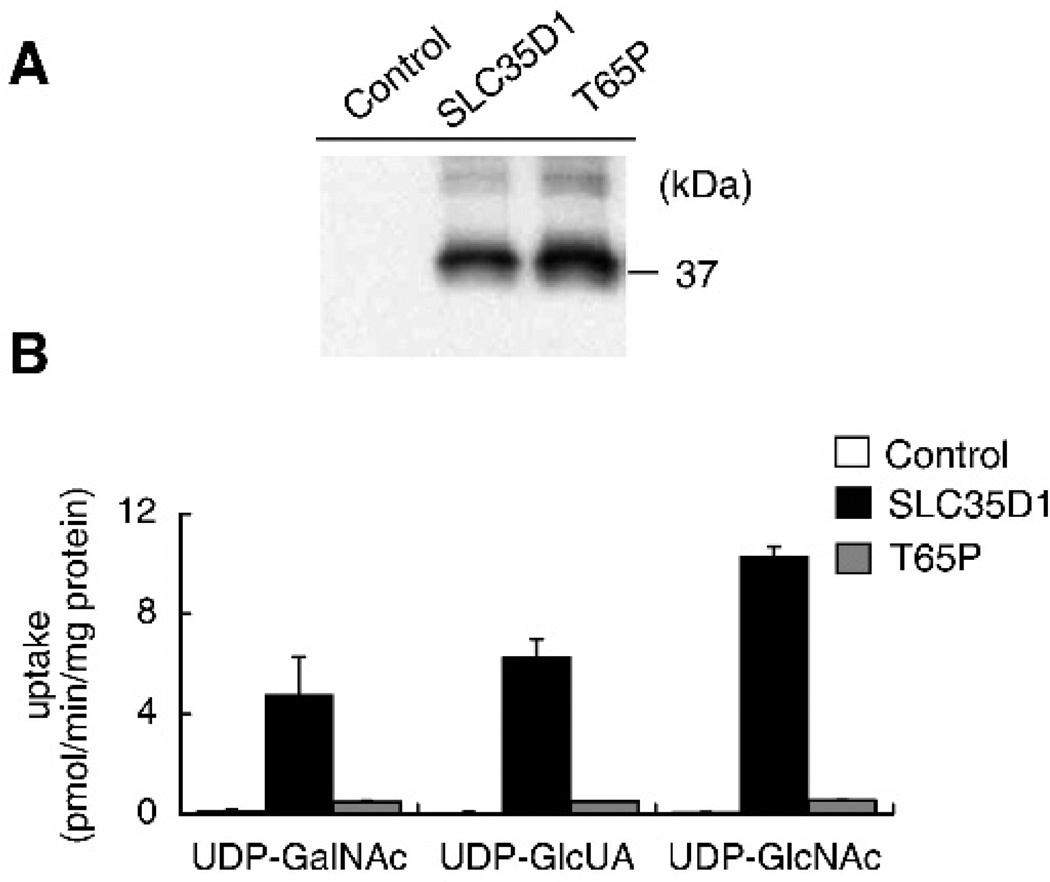
We evaluated the NST activity of the T65P mutant protein by using a yeast complementation assay.12,14,15 Western blot analysis confirmed expression of the mutant and wild-type SLC35D1 in microsome fractions prepared from transfected yeast cells (fig 5A). SLC35D1 expressing microsomes showed higher incorporation of UDP-GalNAc, UDP-GlcUA, and UDP-N- acetylglucosamine (UDP-GlcNAc) relative to non-expressing control (fig 5B). The incorporation levels of nucleotide sugar in T65P-expressing microsomes were reduced to one tenth for UDP-GalNAc, one twelfth for UDP-GlcUA, and one nineteenth for UDP-GlcNAc, compared to those in SLC35D1 expressing microsomes. Therefore, we can conclude that p.T65P mutant results in severely diminished NST activity and that the T65 residue is a critical amino acid for NST activity of SLC35D1.
Figure 5.
Nucleotide sugar transporter (NST) activity of T65P mutant protein. (A) Western blot analysis for the wild-type and mutant SLC35D1 proteins in yeast microsomes. (B) Activity of the mutant protein. The uptake of nucleotide sugars by microsomes prepared from the yeast cells transfected with empty vector, SLC35D1 or T65P mutant expression vector. The incorporation of nucleotide sugars for 1 min at 30uC per mg protein of microsomes was illustrated. Each value is the mean (SD) of duplicate experiments. Note that the NST activity of T65P mutant was drastically reduced.
No SLC35D1 mutations were identified in SBD patients 4 and 5 and in a total of 15 patients with other SSDD group diseases.
The radiographic features common to the three cases caused by SLC35D1 mutations were: (1) handle bar clavicle, (2) bell shaped thorax, (3) interpediculate distance narrowing, (4) metaphyseal flaring, (5) severe retardation of the vertebral body ossification, (6) relatively preserved ossification of the posterior arch, (7) relatively preserved sacral ossification, (8) pubic ossification, and (9) lack of angular deformity of the humerus (table 1). The first four features are also found in the two SLC35D1 mutation negative SBD cases. However, some of the other features were absent in the mutation negative cases. In patient 4, ossification was poor in the posterior arch and sacrum and was absent in pubis. In patient 5, retardation of the vertebral body ossification was moderate, and ossification of the posterior arch and the sacrum was well preserved. The angular deformity of the humerus was present.
DISCUSSION
We identified four novel types of SLC35D1 loss-of-function mutations, in three typical SBD families. This is the second report of identification of loss-of-function mutations and the fist report of a missense mutation of SLC35D1 in SBD patients. Unfortunately, DNA from patient 3 was unavailable; yet, the facts of a heterozygous missense mutation in both parents and an unaffected sister, absence of the mutation in ethnically matched controls, and the experimental evidence of impaired transporter activity of the mutant protein are sufficient to consider that the missense mutation is responsible for the SBD phenotype in the patient. Patient 3 is most likely a homozygote for c.193A>C (p. T65P), although the possibility of a compound heterozygote for c.193A>C and a de novo mutation cannot be denied.
In the present and previous studies,9 we searched for SLC35D1 mutations in 10 SBD patients and identified the mutations in five patients with the common radiographic features. These results indicate that a variety of SLC35D1 mutations can cause SBD and that SLC35D1 mutations are responsible for SBD in patients of different ethnic backgrounds. The following features were common to the mutation positive cases (patients 1–3) and some of them were absent in the mutation negative cases (patients 4 and 5): severe retardation of the vertebral body ossification, relatively preserved ossification of the posterior arch and sacrum, preserved pubic ossification, and lack of angular deformity of the humerus (table 1). These features are also found in the two mutation positive cases that we previously identified.9 These findings would solidify the diagnostic criteria for SBD.
Formerly, SSDD group included achondrogenesis type 1B (ACG1B; OMIM 600972), which is now re-classified in the “sulfate transporter dysplasias” group.4 ACG1B is a particularly interesting disorder in the context of our study because of its clinical and radiographic phenotypic similarities to SBD and its pathogenesis related to impairment of a membrane sulfate transporter termed DTDST (diastrophic dysplasia sulfate transporter) affecting sulfation of GAG chains.19,20 ACG1B represents the most severe end in a group of skeletal dysplasias due to impaired DTDST ranging from the perinatally lethal ACGIB and atelosteogenesis type II (OMIM 256050), through to non-lethal diastrophic dysplasia, to mild autosomal recessive multiple epiphyseal dysplasia.19–21 It is also intriguing that despite widespread expression of DTDST, the phenotypes in the sulfation defects group manifest mainly in the cartilage. This fact may be explained by the particularly high rate of PG synthesis in chondrocytes, and thus the high requirement for sulfate.22 The SLC35D1 protein is also ubiquitously expressed and the phenotype of SBD is confined to the cartilage. Similar clinical and pathogenic stories may pertain to the SLC35D1 defect. Therefore, it will be interesting to investigate potential associations between partial function mutations of SLC35D1 and other non-lethal skeletal dysplasias.
The SLC35 family genes encode NSTs. The SLC35 family consists of at least 17 molecular species in human.11 Among the six subfamilies in the SLC35 classification, the SLC35D subfamily consists of three species, SLC35D1, D2, and D3. SLC35D2 is expressed in the Golgi membrane and mainly transports UDP-GlcNAc,23 while SLC35D3 function has not yet been clarified. Other members of the SLC35 gene family are potential candidates for skeletal dysplasias and other constitutional bone diseases. Indeed, a missense mutation in the bovine Slc35a3 gene, which encodes a UDP-GlcNAc transporter, is responsible for complex vertebral malformations.24 Therefore, mutation search of the SLC35 gene family may lead to the identification of new causative genes for skeletal dysplasias.
Supplementary Material
Key points.
-
►
We identified four novel SLC35D1 mutations in three Schneckenbecken dysplasia (SBD) families, including the first missense mutation (p.T65P).
-
►
All mutations result in loss of function.
-
►
No SLC35D1 mutations were identified in 15 patients with other severe spondylodysplastic dysplasias group diseases.
-
►
SBD with SLC35D1 mutations are defined radiolographically by several characteristics.
Acknowledgements
We are grateful to Dr N Ishida for providing materials to measure NST activity and to Dr S Tominaga for her help in SLC35D1 mutation search.
Funding: This project was supported by Grants-in-aid from the Ministry of Education, Culture, Sports and Science of Japan (Contract grant Nos. 19209049 and 20390408) and from Research on Child Health and Development (Contract grant Nos. H18-005 and 20-S-3).
Footnotes
Competing interests: None.
Patient consent: Obtained.
REFERENCES
- 1.Borochowitz Z, Jones KL, Silbey R, Adomian G, Lachman R, Rimoin DL. A distinct lethal neonatal chondrodysplasia with snail-like pelvis: Schneckenbecken dysplasia. Am J Med Genet. 1986;25:47–59. doi: 10.1002/ajmg.1320250107. [DOI] [PubMed] [Google Scholar]
- 2.Giedion A, Biedermann K, Briner J, Soler R, Spycher M. Case report 693. Skeletal Radiol. 1991;20:534–538. doi: 10.1007/BF00194254. [DOI] [PubMed] [Google Scholar]
- 3.Nikkels PG, Stigter RH, Knol IE, van der Harten HJ. Schneckenbecken dysplasia, radiology, and histology. Pediatr Radiol. 2001;31:27–30. doi: 10.1007/s002470000357. [DOI] [PubMed] [Google Scholar]
- 4.Superti-Furga A, Unger S. Nosology and classification of genetic skeletal disorders: 2006 revision. Am J Med Genet A. 2007;143:1–18. doi: 10.1002/ajmg.a.31483. [DOI] [PubMed] [Google Scholar]
- 5.Borochowitz Z, Lachman R, Adomian GE, Spear G, Jones K, Rimoin DL. Achondrogenesis type I: delineation of further heterogeneity and identification of two distinct subgroups. J Pediatr. 1988;112:23–31. doi: 10.1016/s0022-3476(88)80113-6. [DOI] [PubMed] [Google Scholar]
- 6.Sedaghatian MR. Congenital lethal metaphyseal chondrodysplasia: a newly recognized complex autosomal recessive disorder. Am J Med Genet. 1980;6:269–274. doi: 10.1002/ajmg.1320060403. [DOI] [PubMed] [Google Scholar]
- 7.Whitley CB, Langer LO, Jr, Ophoven J, Gilbert EF, Gonzalez CH, Mammel M, Coleman M, Rosemberg S, Rodriques CJ, Sibley R, Horton WA, Opitz JM, Gorlin RJ. Fibrochondrogenesis: lethal, autosomal recessive chondrodysplasia with distinctive cartilage histopathology. Am J Med Genet. 1984;19:265–275. doi: 10.1002/ajmg.1320190209. [DOI] [PubMed] [Google Scholar]
- 8.Nishimura G, Nakashima E, Hirose Y, Cole T, Cox P, Cohn DH, Rimoin DL, Lachman RS, Miyamoto Y, Kerr B, Unger S, Ohashi H, Superti-Furga A, Ikegawa S. The Shwachman-Bodian-Diamond syndrome gene mutations cause a neonatal form of spondylometaphysial dysplasia (SMD) resembling SMD Sedaghatian type. J Med Genet. 2007;44:e73. doi: 10.1136/jmg.2006.043869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiraoka S, Furuichi T, Nishimura G, Shibata S, Yanagishita M, Rimoin DL, Superti-Furga A, Nikkels PG, Ogawa M, Katsuyama K, Toyoda H, Kinoshita-Toyoda A, Ishida N, Isono K, Sanai Y, Cohn DH, Koseki H, Ikegawa S. Nucleotide-sugar transporter SLC35D1 is critical to chondroitin sulfate synthesis in cartilage and skeletal development in mouse and human. Nat Med. 2007;13:1363–1367. doi: 10.1038/nm1655. [DOI] [PubMed] [Google Scholar]
- 10.Muraoka M, Kawakita M, Ishida N. Molecular characterization of human UDP-glucuronic acid/UDP-N-acetylgalactosamine transporter, a novel nucleotide sugar transporter with dual substrate specificity. FEBS Lett. 2001;495:87–93. doi: 10.1016/s0014-5793(01)02358-4. [DOI] [PubMed] [Google Scholar]
- 11.Ishida N, Kawakita M. Molecular physiology and pathology of the nucleotide sugar transporter family (SLC35) Pflugers Arch-Eur J Phisiol. 2004;447:768–775. doi: 10.1007/s00424-003-1093-0. [DOI] [PubMed] [Google Scholar]
- 12.Muraoka M, Miki T, Ishida N, Hara T, Kawakita M. Variety of nucleotide sugar transporters with respect to the interaction with nucleoside mono- and diphosphates. J Biol Chem. 2007;282:24615–24622. doi: 10.1074/jbc.M611358200. [DOI] [PubMed] [Google Scholar]
- 13.Segawa H, Kawakita M, Ishida N. Human and drosophila UDP-galactose transporters transport UDP-N-acetylgalactosamine in addition to UDP-galactose. Eur J Biochem. 2002;269:128–138. doi: 10.1046/j.0014-2956.2001.02632.x. [DOI] [PubMed] [Google Scholar]
- 14.Ishida N, Yoshioka S, Chiba Y, Takeuchi M, Kawakita M. Molecular cloning and functional expression of the human Golgi UDP-N-acethylglucosamine transporter. J Biochem. 1999;126:68–77. doi: 10.1093/oxfordjournals.jbchem.a022437. [DOI] [PubMed] [Google Scholar]
- 15.Sun-Wada GH, Yoshioka S, Ishida N, Kawakita M. Functional expression of the human UDP-galactose transporters in the yeast Saccharomyces cerevisiae. J Biochem. 1998;123:912–917. doi: 10.1093/oxfordjournals.jbchem.a022024. [DOI] [PubMed] [Google Scholar]
- 16.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 17.Magnani C, Cremonesi L, Giunta A, Magnaghi P, Taramelli R, Ferrari M. Short direct repeats at the breakpoints of a nobel large deletion in the CFTR gene suggest a likely slipped mispairing mechanism. Hum Genet. 1996;98:102–108. doi: 10.1007/s004390050167. [DOI] [PubMed] [Google Scholar]
- 18.Tavassoli K, Eigel A, Horst J. A deletion/insertion leading to the generation of a direct repeat as a result of slipped mispairing and intragenic recombination in the factor VIII gene. Hum Genet. 1999;104:435–437. doi: 10.1007/s004390050981. [DOI] [PubMed] [Google Scholar]
- 19.Superti-Furga A, Hästbacka J, Wilcox WR, Cohn DH, van der Harten HJ, Rossi A, Blau N, Rimoin DL, Steinmann B, Lander ES, Gitzelmann R. Achondrogenesis type IB is caused by mutations in the diastrophic dysplasia sulphate transporter gene. Nat Genet. 1996;12:100–102. doi: 10.1038/ng0196-100. [DOI] [PubMed] [Google Scholar]
- 20.Rossi A, Superti-Furga A. Mutations in the diastrophic dysplasia sulfate transporter (DTDST) gene (SLC26A2): 22 novel mutations, mutation review, associated skeletal phenotypes, and diagnostic relevance. Hum Mut. 2001;17:159–171. doi: 10.1002/humu.1. [DOI] [PubMed] [Google Scholar]
- 21.Hästbacka J, Superti-Furga A, Wilcox WR, Rimoin DL, Cohn DH, Lander ES. Atelosteogenesis type II is caused by mutations in the diastrophic dysplasia sulfate transporter gene (DTDST): evidence for a phenotypic series involving three chondrodysplasias. Am J Hum Genet. 1996;58:255–262. [PMC free article] [PubMed] [Google Scholar]
- 22.Rossi A, Kaitila I, Wilcox WR, Rimoin DL, Steinmann B, Cetta G, Superti-Furga A. Proteoglycan sulfation in cartilage and cell cultures from patients with sulfate transporter chondrodysplasias: relationship to clinical severity and indications on the role of intracellular sulfate production. Matrix Biol. 1998;17:361–369. doi: 10.1016/s0945-053x(98)90088-9. [DOI] [PubMed] [Google Scholar]
- 23.Ishida N, Kuba T, Aoki K, Miyatake S, Kawakita M, Sanai Y. Identification and characterization of human Golgi nucleotide sugar transporter SLC35D2, a novel member of the SLC35 nucleotide sugar transporter family. Genomics. 2005;85:106–116. doi: 10.1016/j.ygeno.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 24.Thomsen B, Horn P, Panitz F, Bendixen E, Petersen AH, Holm LE, Nielsen VH, Agerholm JS, Arnbjerg J, Bendixen C. A missense mutation in the bovine SLC35A3 gene, encoding a UDP-N-acetylglucosamine transporter, causes complex vertebral malformation. Genome Res. 2006;16:97–105. doi: 10.1101/gr.3690506. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.