Abstract
Salmonella Typhi differs from most other salmonellae in that it causes a life-threatening systemic infection known as typhoid fever1. The molecular bases for its unique clinical presentation are unknown2. Here we found that in an animal model, the systemic administration of typhoid toxin, a unique virulence factor of S. Typhi, reproduces many of the acute symptoms of typhoid fever. We identified specific carbohydrate moieties on specific surface glycoproteins that serve as receptors for typhoid toxin, which explains its broad cell target specificity. We present the atomic structure of typhoid toxin, which shows an unprecedented A2B5 organization with two covalently-linked A subunits non-covalently-associated to a pentameric B subunit. The structure provides insight into the toxin’s receptor-binding specificity and delivery mechanisms and reveals how the activities of two powerful toxins have been coopted into a single, unique toxin that can induce many of the symptoms characteristic of typhoid fever. These findings may lead to the development of potentially life-saving therapeutics against typhoid fever.
Keywords: Salmonella typhi, typhoid fever, bacterial toxins, Podocalyxin, CD45, bacterial pathogenesis, glycobiology, AB5 toxins
Salmonella enterica serovar Typhi (S. Typhi), the cause of typhoid fever, results in more than 200,000 annual deaths1,3,4. Unlike other Salmonella serovars, which typically cause self-limiting gastroenteritis, S. Typhi causes a systemic, life-threatening disease1. The genome of S. Typhi contains a dearth of unique virulence factors that are not found in non-typhoidal serovars, and the molecular bases for its unique virulence properties and clinical presentation are unknown5. One of the few S. Typhi-specific factors that have been shown to directly impact its interaction with host cells is an AB-type toxin dubbed typhoid toxin6–8. Unlike typical AB toxins9, typhoid toxin is composed of two A subunits, PltA and CdtB, which are homologs of the A subunits of the pertussis and cytolethal distending toxins, respectively7. Its single B subunit, PltB, is a homolog of one of the components of the heteropentameric B subunit of pertussis toxin. Although the cellular targets of the ADP-ribosyl transferase activity of PltA have not yet been identified, CdtB is a deoxyribonuclease that inflicts DNA-damage and induces cell cycle arrest6,10,11. S. Typhi produces typhoid toxin only within mammalian cells, and the toxin is then ferried to the extracellular environment by a unique transport mechanism that involves vesicle carrier intermediates7,8. Convalescent typhoid fever patients developed strong immune response to the toxin, indicating that typhoid toxin is produced during human infection12,13. However, the potential contribution of typhoid toxin to disease and pathogenesis is unknown.
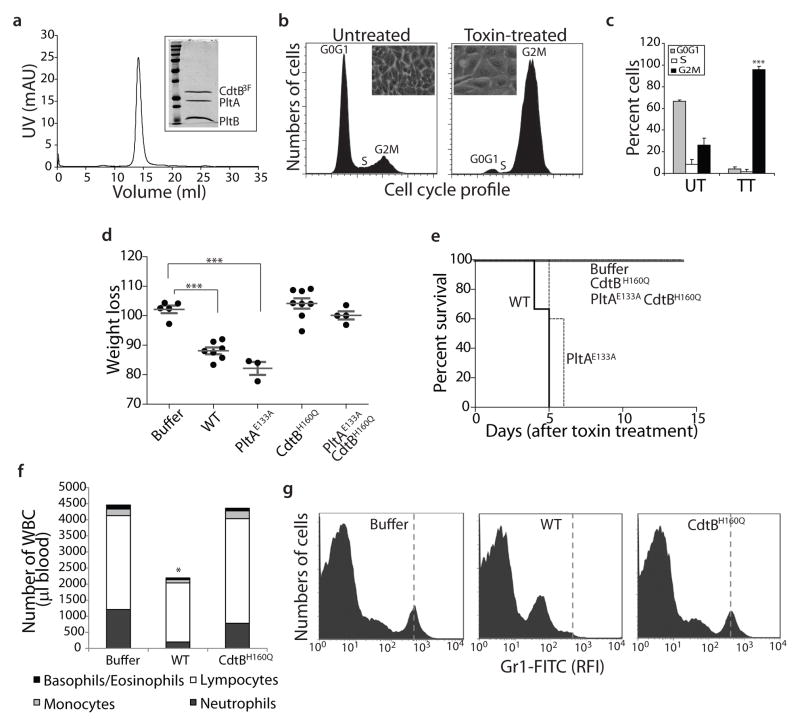
We found that systemic administration into mice of a highly purified preparation of biologically active typhoid toxin (Fig. 1a–c) caused many of the symptoms observed during the acute phase of typhoid fever1,14. Despite the lack of fever (Supplementary Fig. S1), the mice appeared lethargic showing clear signs of stupor and malaise (Supplementary Video S1), lost weight (Fig. 1d), and eventually died (Fig. 1e). The toxin-injected animals also showed a significant reduction in the number of circulating immune cells, resulting in the almost complete depletion of circulating neutrophils (Fig. 1f and 1g), a phenotype often observed during the acute phase of typhoid fever14. Consistent with this observation, typhoid toxin was able to intoxicate a broad range of cells in vitro, including several epithelial and immune cells (Supplementary Fig. S2). Although symptoms were observed in animals inoculated with a toxin carrying a catalytic mutant of PltA (Fig. 1d), no detectable symptoms were observed when animals were inoculated with a toxin carrying a catalytic mutant of CdtB (Fig. 1d–1g). Taken together, these results indicate that typhoid toxin, through its CdtB subunit, may contribute to the acute symptomatology observed during typhoid fever.
Figure 1.
Systemic administration of typhoid toxin causes symptoms observed during the acute phase of typhoid fever. a, Chromatographic profile of the typhoid toxin holotoxin used in the biological assays. The inset shows a Coomassie blue stained SDS-PAGE analysis of the peak fraction. b and c, Typhoid toxin induces cell cycle arrest in cultured cells. Human intestinal epithelial Henle-407 cells were left untreated, or treated with purified typhoid toxin and then analyzed by flow cytometry. The insets show representative light microscope images of mock or toxin treated cells and is representative of at least three independent experiments. c, Averages of cell cycle profiles from at least 3 independent experiments. Bar represents average ± standard deviation. ***, P < 0.001, compared to the number of cells in G2M of the control untreated group. UT: untreated; TT: toxin treated. d, Weight loss 5 days after intravenous administration of different typhoid toxin preparations. Lines are the mean ± standard error of the mean and represent the weight relative to the values before treatment. ***, P < 0.0001. e, Survival of animals receiving different typhoid toxin preparations. n= 3–5 animals per group. f and g, Circulating white blood cells were counted in a hematology analyzer after the indicated treatments (*, P < 0.05) (f). Alternatively, the number of neutrophils (vertical dashed line) in peripheral blood of animals treated as indicated was determined by flow cytometry. RFI, relative fluorescence intensity.
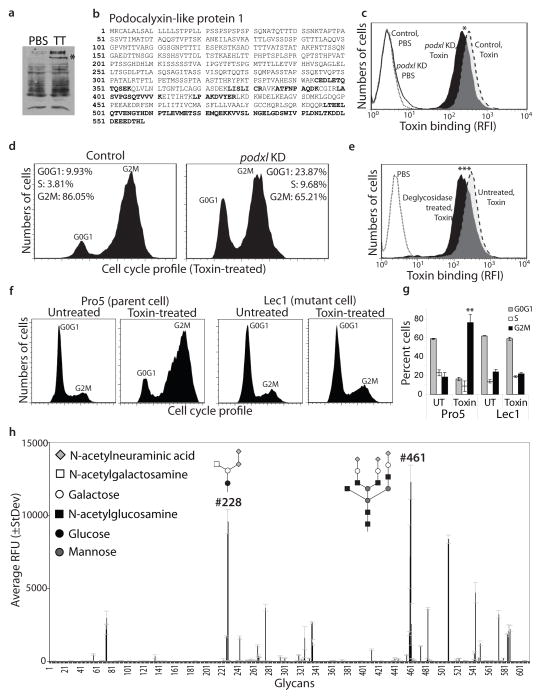
To identify typhoid toxin’s cellular receptor(s) we used a highly purified biologically active typhoid toxin preparation to affinity purify biotin-labeled host-cell-surface interacting proteins. We found by LC-MS/MS analyses of interacting proteins that in human epithelial cells, typhoid toxin interacts with Podocalyxin-like protein 1 (PODXL) (Fig. 2a and 2b), a member of the CD34 sialomucin protein family, which localizes to the apical side of epithelial cells and is also expressed in vascular endothelial cells15. Consistent with its potential role as a toxin receptor, shRNA-mediated depletion of PODXL resulted in a significant reduction in toxin binding (Fig. 2c and Supplementary Fig. S3) and toxin-mediated cell cycle arrest (Fig. 2d). Since we found that typhoid toxin is capable of intoxicating a broad range of cells (Supplementary Fig. S2), we examined the interaction of typhoid toxin with surface proteins of other cell lines. We found that in macrophages as well as in T and B cells typhoid toxin interacts with receptor tyrosine phosphatase C, also known as CD45 (Supplementary Fig S4), which is ubiquitously expressed in hematopoietic cells other than erythrocytes and platelets16. These results suggest that typhoid toxin may engage different receptors in different cells. The identified typhoid toxin-interacting proteins, however, are all glycosylated suggesting that typhoid toxin may interact with these different surface proteins through common carbohydrate moieties. Consistent with this hypothesis we found that removal of surface glycans significantly reduced typhoid toxin binding to cultured cells (Fig. 2e). Furthermore, a cell line lacking all complex and hybrid N-glycans on glycoproteins due to a mutation in N-acetylglucosaminyltransferase I17,18 was more resistant to typhoid toxin than its parent wild type cell line (Fig. 2f and 2g and Supplementary Fig. S5). These results indicate that a sugar moiety(s) on surface glycoproteins serves as a receptor for typhoid toxin.
Figure 2.
Typhoid toxin recognizes terminally sialylated glycans on surface glycoproteins. a and b, Henle-407 cell surface proteins were biotinlyated, co-immunoprecipitated with purified typhoid toxin (TT), and analyzed by SDS-PAGE (a) and LC-MS/MS. The peptides from Podocalyxin like protein 1 (PODXL) (indicated by an asterix in a) identified by LC-MS/MS are indicated in bold (b). c and d, PODXL-depleted (by an specifically targeted siRNA) and control cells were treated with fluorescently-labeled typhoid toxin and toxin binding was evaluated by flow cytometry (c) (* P=0.024 for three independent determinations). Alternatively, siRNA-depleted and control cells were treated with typhoid toxin and toxicity was evaluated by cell cycle analysis. e, Henle-407 cells were treated with a mixture of glycosidases and the ability of treated and control cells to bind fluorescently-labeled toxin was subsequently evaluated by flow cytometry (***, P < 0.001 from three independent experiments). f and g, The N-acetylglucosaminyltransferase I-deficient (Lec1) and its parent (Pro5) cell lines were treated with typhoid toxin and toxicity was evaluated by cell cycle analysis (f). The quantification of the cell cycle profiles is shown in (g). Bar represents average ± standard deviation of at least three independent determinations. **, P < 0.01, compared to the number of Pro5 cells in G2M. h, Glycan array analysis of typhoid toxin binding. Values represent the average relative fluorescence unit (RFU). The X axis depicts the glycan numbers. The structure of the most relevant glycans is shown.
To identify the glycans recognized by typhoid toxin we probed 610 glycans arrayed on a solid surface19 for binding to biologically active, highly purified, fluorescently labeled typhoid toxin. This analysis revealed a complex binding pattern involving 4 main glycan groups (Fig. 2h and Supplementary Tables S1 and S2). The first group, which is most commonly present in complex N-linked glycans, is represented by sialylated tri- or bi-antennary glycans (e. g. glycans #461, #483, and #482) with one or all of the branches terminally sialyated (note: glycan numbers correspond to the nomenclature used by the Consortium for Functional Glycomics, http://www.functionalglycomics.org). This group includes glycans with both Neu5Acα2-3 (e. g. #483) and Neu5Acα2-6 (e. g. #482) terminal linkages. However, typhoid toxin likely binds preferentially to Neu5Acα2-3 terminal linkages since glycan #483 showed stronger binding than glycan #482, which only differ in their terminal linkages (Fig. 2h and Supplementary Tables S1 and S2). Furthermore, typhoid toxin likely preferentially binds tri-antennary over bi-antennary compounds, since glycan #461 exhibited the highest binding affinity. In addition, the toxin may also bind preferentially the type 2-N-acetyllactosamine unit (Galβ1-4GlcNAc), present in glycan #461, over type 1-N-acetyllactosamine unit (Galβ1-3GlcNAc), present in the very similar tri-antennary N-glycan #474, which showed lower binding affinity (Fig. 2h and Supplementary Tables S1 and S2). The second group consists of non-sialylated tri- or bi-antennary glycans also commonly found in complex N-linked glycans. Overall this group exhibited lower binding affinity than sialylated glycans, implying a preference of typhoid toxin for the terminal sialic acid modification. The third group consists of glycans commonly found in glycolipids, mostly as gangliosides. Among these glycans typhoid toxin likely prefers Neu5Acα 2-8 Neu5Ac disialosides, present in ganglioside GD220 (glycan #228), over Neu5Ac monosialoside, present in GM3 (glycan #263), which did not bind the toxin. These results suggest that in certain cells, typhoid toxin may also be able to use glycolipids as a receptor. Consistent with this hypothesis, typhoid toxin was able to intoxicate an N-acetylglucosaminyltransferase I-deficient cell line although only when applied at high concentrations (Supplementary Fig. S6). The fourth group is defined by glycans commonly found on O-glycans such as glycan #243, which shares the canonical structure of mucin type O-GalNAcylated glycan. Taken together, these results indicate that typhoid toxin can use as receptors a broad range of glycans preferentially on surface glycoproteins but also, although less efficiently, on surface glycolipids, providing a mechanistic explanation for its broad cell target specificity.
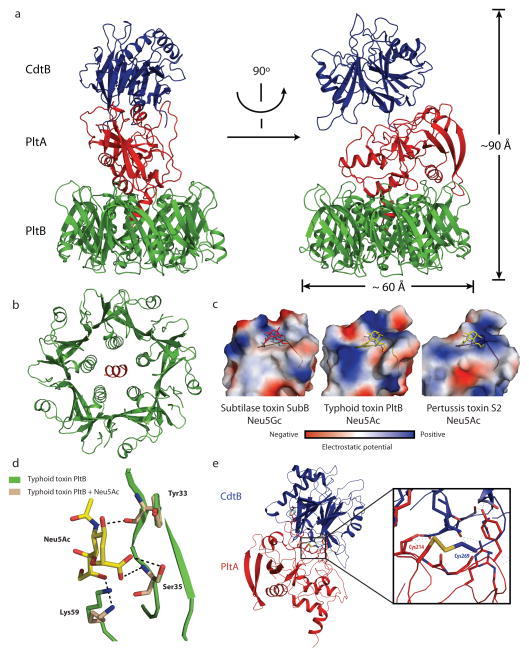
To uncover the organization of typhoid toxin, we solved its crystal structure to 2.4Å resolution. The structure showed a complex of 5 PltB molecules and one molecule each of PltA and CdtB (Fig. 3a and Table S3), which is consistent with the stoichiometry observed by size exclusion chromatography combined with dynamic light scattering and quantitative amino acid composition analysis (Supplementary Fig. S7). This indicates an unprecedented A2B5 organization for typhoid toxin, which is in contrast to other known AB5 toxins that have only one A subunit9,21. The pyramid shaped complex has a height of ~90Å and a maximum width of ~60Å (Fig. 3a), with CdtB located at the vertex, connected by PltA to a pentameric disc at the base of the pyramid made of 5 monomers of PltB (Fig. 3a and 3b). The tandem linear arrangement of the enzymatic subunits PltA and CdtB dictates that there are no interactions between CdtB and PltB. As predicted by the amino acid sequence similarities, the PltA and CdtB subunits exhibit a very similar structure to the pertussis toxin S1 (and other ADP ribosyl transferases)22 and to the CdtB subunit of Cytolethal distending toxin23. PltA aligns very well to the pertussis toxin S1 domain with a root-mean-squared deviation (rmsd) of 2.168 Å over 140 Cα atoms (with 31% sequence identity) (Supplementary Fig. S8). The positions of the conserved catalytic residues (Glu133 in typhoid toxin and Glu129 in pertussis toxin S1), as well as the disulfide bonds (Cys56-Cys207 in typhoid toxin and Cys41-Cys201 in pertussis toxin S1) overlap almost completely (Supplementary Fig. S8). The latter indicates that, similarly to the pertussis toxin S1 subunit 24, PltA would have to be reduced to allow the access of NAD and its putative substrates to the active site, and consequently, a reducing activating step must be necessary prior to contacting its host cell target(s). Typhoid toxin CdtB aligns very well to the Haemophilus ducreyi CdtB with an rmsd of 0.947 Å over 206 Cα atoms (with 52% sequence identity) (Supplementary Fig. S9). The positions of the conserved catalytic residues in Typhoid toxin’s CdtB overlap almost completely with those of its homolog in H. ducreyi.
Figure 3.
The crystal structure of typhoid toxin depicts a unique architecture. a, Two views of the overall structure of the typhoid holotoxin complex shown as a ribbon cartoon and related by 90° rotation about a vertical axis. CdtB, PltA and PltB are shown in blue, red, and green, respectively. b, Bottom view of the channel formed by the PltB pentamer (in green), depicting the PltA C-terminal α-helix (in red) within it. c, Surface charge distribution of the predicted sugar-binding pockets of different B subunit homologs of the indicated AB5 toxins (SubB for Subtilase and S2 for Pertussis toxins). A highly conserved serine residue critical for sugar binding is indicated within the sugar-binding pocket. The sugars N-glycolylneuraminic acid (within SubB) and N-acetylneuraminic acid (within typhoid and pertussis toxins) are shown. d, Molecular modeling of N-acetylneuraminic acid within the typhoid toxin binding pocket. Critical residues engaged in this interaction are shown. e, Atomic interface between CdtB and PltA. The inset shows a detailed view of a critical disulfide bond between PltA Cys214 and CdtB Cys269.
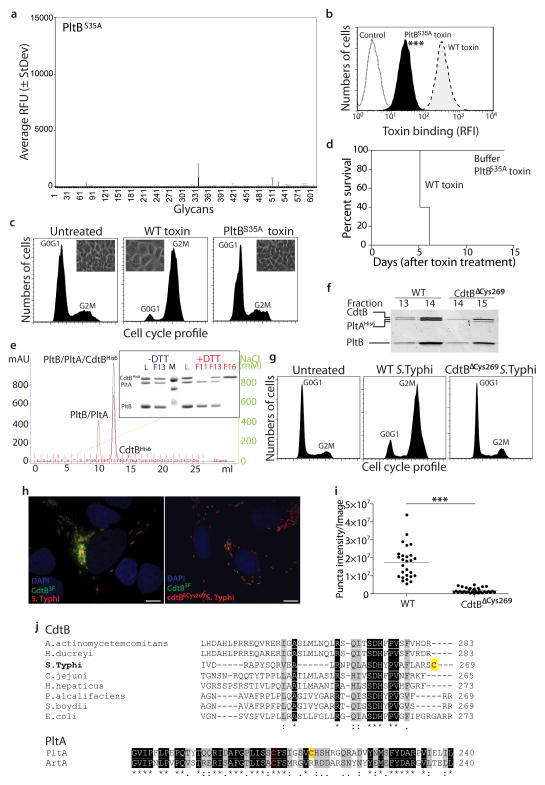
Similar to the B subunits of other AB5 toxins9, the PltB oligomer is arranged as a pentamer with a central channel that is lined by 5 helixes (Fig. 3b). The PltB protomer shows a typical oligosaccharide-binding fold on the side of the pentamer (Fig. 3c and Supplementary Fig. S10), a location similar to that of toxins that preferentially bind glycoproteins but different from those that preferentially bind glycolipids, which have the sugar-binding pockets on the membrane-proximal face of the toxin9. These findings are consistent with the observation that typhoid toxin preferentially binds glycans present on surface glycoproteins over those present on glycolipids (supplementary Table S1 and S2). The monomer of typhoid holotoxin PltB aligns very well with SubB, the subtilase cytotoxin B subunit, with an rmsd of ~0.5 Å over 97 Cα atoms (with 50% sequence identity) (Supplementary Fig. S10). Of note, the position of the conserved putative sugar-binding residue Ser35 overlap very well (Fig. 3c and Supplementary Fig. S10). The predicted sugar-binding pocket in PltB is not as deep and appears more extended than in SubB, which also differs in surface charge distribution (Fig. 3c). These differences may account for the significantly different binding specificities exhibited by these two toxins25. We also compared PltB to the pertussis toxin S2 B subunit26. Although their overall amino sequence similarity is low, the structures can be aligned very well around their sugar-binding domains with an rmsd of 1.752 Å over 80 Cα atoms (Supplementary Fig. S10). Residues in pertussis toxin S2 involved in sugar binding (Tyr102, Ser104, Arg125) are well conserved in PltB (Tyr33, Ser35, Lys59) (Fig. 3d), and the charge distribution and architecture of the sugar-binding pockets are similar (Fig. 3c). This is consistent with the observation that, despite overall less conservation, these two B subunits share sugar-binding specificity. For example, several of the glycans that bind typhoid toxin possess an Neu5Ac moiety at their terminal end, a determinant that also binds pertussis toxin27. Structural modeling of Neu5Ac bound to PltB predicts almost identical interaction to those observed in the atomic structure of pertussis toxin bound to Neu5Ac (Figure 3c and 3d and Supplementary Fig. S11). Consistent with the structural predictions, a mutation in the sugar-binding pocket of PltB (PltBSer35A) abrogated the ability of typhoid toxin to bind glycans in glycoarray (Fig. 4a and Table S4) and surface plasmon resonance assays (Supplementary Fig. S12), and the ability of the toxin to bind (Fig. 4b) and intoxicate (Fig. 4c) cultured cells or to cause symptoms when systemically applied to mice (Fig. 4d). These results are also consistent with the notion that there is a single carbohydrate-binding domain in typhoid toxin since the mutant abrogated binding to all carbohydrates. Surface plasmon resonance assays also indicated that on average at least for the glycan tested (GD2) each PltB pentamer binds 2.5 sugar molecules with an affinity of ~ 1.2 mM (supplementary Fig. S12). However, the glycoarray analysis predicts that the binding affinity is likely to be higher for more complex glycans.
Figure 4.
Structure-function analysis of typhoid toxin. a–d, Fluorescently-labeled typhoid toxin containing PltBSer35A was tested for its binding to glycans (a) and to cultured cells (b) (see Fig. 2 for details) (***, P < 0.001 from at least three independent determinations). Alternatively, toxicity was assayed by flow cytometric cell cycle analysis of toxin-treated cells (at least three independent experiments) (c), or by systemic administration to mice (n=3 to 5 mice) (d). e–i, The typhoid toxin complex was analyzed by ion exchange chromatography before (blue) and after (red) treatment with DTT (L: loading control; M: molecular weight markers; F: chromatographic fraction) (e). Inset shows SDS-PAGE analyzes of the indicated fractions (e). f, A toxin preparation obtained from a bacterial strain expressing CdtBΔcys269 was analyzed by gel filtration chromatography and compared to wild-type toxin (the experiment was repeated two times). While wild-type holotoxin eluted in fractions 13 and 14, toxin obtained from a bacterial strain encoding CdtBΔcys269 eluted in fractions 14 and 15 due to the lack of CdtB. g–i, Henle-407 cells were infected with S. Typhi strains encoding FLAG-tagged CdtB or CdtBΔcys269 and cells were examined for toxicity by flow cytometry (g). Alternatively, cells were fixed, stained with anti FLAG antibody, and the amount of puncta staining, which represent CdtB in typhoid toxin export carriers8, were determined by immunofluorescence analysis (h and i). Bar represents average of puncta-associated flurorescence intensity (at least 100 cells were analyzed in three independent experiments).***, P < 0.0001, Scale bar:10 μm. j, ClustalW amino acid sequence comparison of CdtB and PltA homologs. Conserved cysteines are shown in red while unique cysteines are indicagted with a yellow shade.
The interaction of PltA with the PltB oligomer occurs largely through its carboxy terminus, which buries 1,657 Å2 and has a ΔiG of −17.7 kcal/mol. A critical element in this interaction is a short helix at the carboxyterminal end of PltA, which inserts into the hydrophobic lumen of the PltB channel (Fig. 3b and Supplementary Fig. S13) and stabilizes the complex by critical interactions mediated by Pro234, Val235, Ile236, Leu238, Ile239, and Leu240 in PltA.
The most salient feature of the interface between PltA and CdtB is a disulfide bond between Cys214 of PltA and Cys269 of CdtB (Fig. 3e). The rest of the interface between the two A subunits is unremarkable burying only 950 Å2 with a ΔiG of only −5.2 kcal/mol, suggesting that the disulfide bound is essential for the integrity of the holotoxin. Consistent with this hypothesis, subjecting the typhoid toxin to reducing conditions resulted in the dissociation of CdtB from the PltA-PltB complex (Fig. 4e). Furthermore, introduction of a mutation in Cys269 of CdtB destabilized the holotoxin complex in vitro (Fig. 4f), and resulted in a loss of CdtB-dependent toxicity in S. Typhi-infected cells (Fig. 4g and Supplementary Fig. S14). This mutation also prevented the assembly of CdtB into its vesicle carrier intermediates that can be visualized as fluorescent puncta in S. Typhi-infected cells (Fig. 4h and 4i), despite the fact that the mutant was expressed at equivalent levels to those of wild type (Supplementary Fig. S15). These results indicate that the CdtBCys269/PltACys214 disulfide bond is critical for the assembly of typhoid toxin holotoxin in the periplasm of the bacteria. Remarkably, even though the sequence conservation between typhoid toxin’s CdtB and that of its cytolethal distending toxin homologs is very high, the Cys269 is unique to Typhoid toxin’s CdtB (Fig. 4j). Likewise, although all close homologs of PltA including ArtA, a closely related mono ADP ribosyl transferase from S. Typhimurium28, have only two Cys that form an intramolecular disulfide bond, PltA is unique in that it has a third Cys (Cys214) to pair with S. Typhi’s CdtB (Fig. 4j). Therefore typhoid toxin’s CdtB has been specifically adapted for its tethering to the PltA/PltB complex by the evolution of uniquely positioned Cys residues in both PltA and CdtB. The covalent linkage of CdtB to the PltA/PltB complex by a disulfide bond ensures that CdtB and PltA are simultaneously delivered to the same target cell. Furthermore, this arrangement would ensure that, after endocytosis and retrograde transport to its place of translocation (most likely the endoplasmic reticulum), PltA and CdtB would be freed from one another upon reduction of the disulfide bond by local reductases thus allowing them to become competent for translocation to the cytosol and delivery to their place of action.
This study provides unique insight into typhoid toxin, a critical virulence factor of S. Typhi, revealing unprecedented features for a bacterial exotoxin and providing significant information on the pathogenesis of typhoid fever. Although human experimentation will be required to confirm the relevance of these findings, the potential implication of typhoid toxin in the development of symptoms during the acute, life threatening phase of typhoid fever may facilitate the development of potentially life-saving therapeutics against this disease.
Material and Methods
Bacterial Strains, mammalian cells, and growth conditions
The wild-type Salmonella enterica serovar Typhi strain ISP2825 has been described previously1. A derivative of this strain encoding a FLAG-epitope tagged CdtB has been previously described2. Other S. Typhi mutant strains were constructed by allelic exchange following previously described procedures3. Site directed mutations and epitope tagging was carried out following standard recombinant DNA procedures. For all infection experiments, S. Typhi strains were grown at 37°C in 2 ml LB broth containing 0.3 M NaCl to an OD600 of ~0.9 after inoculation from overnight cultures at a dilution of 1:50. Cultured cells and culture medium used in these studies were as follows: Henle-407 (human intestinal epithelial cells): DMEM + 10% BCS; Jurkat (human T lymphocytes): RPMI1640 + 10% FBS; Ramos (human B lymphocytes): RPMI1640 + 10% FBS; THP1 (human monocytic cells): RPMI1640 + 10% FBS + 0.05 mM β-mercaptoethanol; Raw (mouse monocytes/macrophages): DMEM + 10% FBS; NIH3T3 (mouse embryonic fibroblasts): DMEM + 10% BCS; COS1 (monkey kidney fibroblasts): DMEM +10% BCS; CHO (Chinese hamster ovary epithelial cells): Ham’s F12 + 10% FBS; MDCK (canine kidney epithelial cells): MEM + 10% FBS; Lec-1 (N-acetylglucosaminyltransferase I mutant) and parent cell Pro-5: alpha MEM with ribonucleosides and deoxyribonucleosides + 10% FBS. All mammalian cells were originally obtained from American Type Culture Collection and cultured at 37 °C under an atmosphere of 5% CO2.
Typhoid toxin expression and purification
pltB, pltA, and cdtB (wild type or mutant alleles, and 3 x FLAG or 6 x His tagged at the carboxy terminus of CdtB or internally in the case of PltA, as indicated) were cloned as a single operon in either a low copy plasmid derived from pWSK1294, or in pET28a+ (Novagen). E. coli strains carrying the different plasmids were grown to an OD600 of 0.6–0.7 at 37°C, expression of typhoid toxin was subsequently induced by the addition of 0.5 mM IPTG, and induced cultures were incubated overnight at 30 °C. Bacterial cells were pelleted by centrifugation, and bacterial cells were resuspended in a buffer containing 200 mM Tris-HCl (pH 7.5), 20% sucrose, 1 mM EDTA, 2 mg/ml lysozyme, and 1x protease inhibitors (2 ml per each gram of wet cell pellet weight), incubated for 5 min at RT, mixed with dH2O (3ml per each gram of wet cell pellet weight) by inversion, and incubated for additional 10 min on ice. A crude periplasmic protein fraction was obtained by centrifugation at 4,000 x g for 15 min at RT, and used as a source of typhoid toxin for affinity chromatography purification using M2 Flag affinity gel (Sigma). The periplamic fraction containing typhoid holotoxin was incubated for 3 hrs at 4 °C with M2 agarose beads packed on a 10 ml column (Bio-rad), washed with ~20 bed volume of PBS, and eluted twice with PBS containing 3xFlag peptide (Sigma; 150 ng/μl). Partially purified holotoxin was applied on a superdex 200 size-exclusion chromatography (Tris-HCl buffer, 15–50 mM, in a pH range of 7.6–8.0 supplemented with 150 mM NaCl) to complete its purification. His-epitope-tagged typhoid toxin was purified as follows. E. coli cultures were resuspended in a buffer containing 15 mM Tris-HCl, pH8.0, 150 mM NaCl, 2 mg/ml lysozyme, 10 μg/ml DNase, and 1x protease inhibitor cocktail, lysed by passing them through a French press three times, pelleted, and affinity-purified using a Nickel-resin (Qiagen) according to the vendor’s recommendation. The eluates were diluted in 20 mM MES, pH6.0 buffer and loaded onto a Hitrap ion-exchange column. Fractions from the ion-exchange chromatography were monitored on SDS-PAGE, concentrated, and further purified by using a superdex 200 column. Final fractions were examined for purity on a 15% SDS-PAGE.
Mammalian cell intoxication assay
Cell cycle arrest after typhoid toxin intoxication was examined by Flow cytometry as previously described2. Briefly, after treatment with 3 x FLAG-tagged typhoid toxin or bacterial infection for different times (as indicated), cells were trypsinized, harvested, washed, and fixed overnight at −20°C in ~70% ethanol/PBS. Fixed cells were washed with PBS and resuspended in 500 μl of PBS containing 50 μg/ml propidium iodide (PI), 0.1 mg/ml RNase A, and 0.05% Triton X-100. After incubation for 40 min at 37°C, cells were washed with PBS, resuspended in 500 μl PBS, filtered, and analyzed by a flow cytometry. DNA contents of cells was determined using Flowjo (Treestar).
Light-Scattering Size Exclusion Chromatography and Amino Acid Composition Analysis
Light-Scattering Size Exclusion Chromatography (SEC-LS) and amino acid composition analysis were carried out at the Keck Biotechnology Resource Laboratory at the Yale University School of Medicine (http://medicine.yale.edu/keck/index.aspx). For SEC-LS analysis, the toxin was purified in 50 mM Hepes buffer containing 150 mM NaCl and run on a Superdex 200 column equipped with a light scattering detector using a same Hepes buffer. For amino acid composition analysis, typhoid toxin was resolved on a 15% SDS-PAGE gel, stained with coomassie brilliant blue, and the three individual bands were excised and used for amino acid composition analysis.
Mouse intoxication experiments
All animal experiments were conducted according to protocols approved by Yale University’s Institutional Animal Care and Use Committee. Age- and sex-matched 7–14 weeks old C57BL/6 mice were randomly allocated in each group and numbers of animals per each group were estimated to obtain statistically significant data after a pilot experiment to get a sense of the potential variability. Groups of mice were intravenously injected with 100 μl solutions containing either TBS buffer alone or 10 μg of each of the purified holotoxin preparations (endotoxin free). His-tagged wild type, PltA catalytic mutant (PltAE133A), CdtB catalytic mutant (CdtBH160Q), double catalytic mutant (PltAE133ACdtBH160Q), and PltBS35A mutant holotoxins were purified as indicated above. Changes in behavior, weight, and temperature of the toxin-injected mice as well as their survival were closely monitored during the duration of the experiment. To minimize bias, blind end-point assessment was applied to examine the behavior of the animals after toxin inoculation.
Peripheral blood leukocyte preparation, immunostaining, and flow cytometry analysis
Peripheral blood samples of typhoid toxin treated and control mice were collected into heparinized tubes, incubated with 1 ml ACK lysis buffer (BioWhittaker) (to remove red blood cells), incubated for 5 min on ice, washed with 2 ml PBS, and centrifuged to collect peripheral blood leukocytes (PBLs). After a repetition of the red blood cell removal step, PBLs were used for immunostaining as described below. After washing, the cells were immediately incubated for 30 min on ice with 100 μl of anti-mouse Ly-6G (Gr-1) antibody conjugated with FITC (eBioscience, cat. # 11-5931-81). The cells were then washed with 2 ml FACS buffer (PBS, 0.16% BSA), resuspended in 100 μl FACS fixation buffer (PBS, 1% paraformaldehyde, 1% FCS), and used for flow cytometric analyses on a FACSCalibur flow cytometer (BD Biosciences). Alternatively, blood samples collected by heart puncture 4 days after toxin treatment were analyzed in a Hemavet 950FS hematology analyszer (Drew Scientific).
Identification of typhoid toxin interacting, biotin-labeled host cell surface proteins
Cultured cells (Henle, Jurkat, Ramos, or THP1 at ~50% confluency) were washed with PBS, treated with PBS containing ~100 μg/ml of Sulfo-NHS-SS-Biotin (Thermo) for 30 min at RT, and subsequently washed 3 times with 50 mM Tris-HCl pH8.0/150 mM NaCl buffer to quench and to remove extra biotin reagent. After additional washings (3 times with PBS), cells were resuspended in a lysis buffer containing 50 mM Tris-HCl, pH7.4, 150 mM NaCl, 1% NP-40, and 1x protease inhibitor, incubated for 20 min on ice, and homogenized by passing a 26-G needle ~ 20 times. After removal of cellular debris by centrifugation, the supernatants were mixed with 10 μg purified FLAG-tagged typhoid toxin and incubated for 3 hrs at 4 °C. Anti-FLAG antibody-containing agarose beads were added, incubated for additional 1 hr at 4 °C, washed 5 times with PBS, SDS-PAGE sample buffer was added, boiled, and run on SDS-PAGE. The gels were transferred to nitrocellulose membranes, blocked with TBS containing 5% BSA, incubated overnight at 4 °C with Streptavidin-HRP (1:5000) in TBS/1% BSA, washed with TBST, and developed with ECL substrates (Pierce). To identify the typhoid toxin-interacting proteins by LC-MS/MS, equivalently obtained samples were run in parallel, stained with Coomassie blue, and gel regions corresponding to the molecular weight of typhoid-toxin interacting proteins identified by western blot analysis were excised and processed for LC-MS/MS as previously described5. Briefly, gel slices were destained in destaining buffer [50 mM NH4HCO3. 50% Acetonitrile (ACN)], and dehydrated with ACN. Disulfide bonds were reduced by incubating the samples with NH4HCO3 containing 10 mM DTT and alkylated by incubating them with 55 mM 2-iodoacetamide in 100 mM NH4HCO3 buffer for 20 min at RT. Gel pieces were dehydrated, trypsin-digested overnight, extracted, run on an LTQ-Velos Mass Spectrometer, and spectra analyzed with Mascot (Matrixscience). As negative controls, equivalently processed samples obtained with two irrelevant baits (GST-3xFlag and InvC-3xFlag) were used.
Oregon Green 488 typhoid toxin labeling
Purified wild type and PltBS35A mutant typhoid toxin preparations were fluorescently labeled with Oregon Green (OG)-488 dye (Invitrogen) according to the vendor’s recommendation. OG-488 dye has a succinimidyl ester moiety that reacts with primary amines of proteins to form stable dye-protein conjugates. Purified toxin preparations (1 mg/ml) were incubated with reactive dye in 500 μl of 100 mM bicarbonate buffer for 1 hr at RT, and applied to a size exclusion chromatography column provided by the vendor to separate the dye-protein conjugates from free dye. Degree of labeling was determined by measuring the absorbance of the conjugate solution at 280 nm and 496 nm, which yielded comparable toxin labeling for both toxin preparations (4.4:1 and 4.36:1 dye/holotoxin ratios for wild type and and PltBS35A mutant toxin, respectively). A predicted extinction coefficient of 191,400 M−1 cm−1 was used to calculate the dye/toxin ratio.
Typhoid toxin binding assay
Cells were harvested by trypsinization, washed with HBSS, resuspended in 100 μl HBSS containing 0.2 μg of Oregon Green-488 (Invitrogen)-labeled purified wild type or mutant toxin preparations. Cells were incubated in the presence of the labeled toxin preparations for 30 min at room temperature, washed with PBS, resuspended in 100 μl PBS containing 1% paraformaldehyde, and analyzed by flow cytometry. When indicated, Henle-407 cells were treated for 2 hrs with 10 μl deglycosidase mix (NEB) in 2 ml HBSS prior to processing for toxin-binding assays as indicated above.
Amino acid sequence alignment
ClustalW amino acid sequence comparison analyses of CdtB and PltA homologs was carried out using the following sequences (Genbank entry numbers): CdtB homologs from Shigella boydii (AAU88264.1), Providencia alcalifaciens (BAL72684.1), Helicobacter hepaticus (AAF19158.1), Haemophilus ducreyi (NP_873398.1), E. coli (BAH72965.1), Campylobacter jejuni (AAS01598.1), and Aggregatibacter actinomycetemcomitans (AAC70898.1). The PltA homolog used in the alignment was the highly related S. Typhimurium DT104 ArtA (BAE20153.1).
Generation of PODXL-depleted cell lines
RNA interference vector pSUPER-H1 (Oligoengine) was used to generate a plasmid expressing an shRNA construct targeted to podxl. Oligos including a target region for podxl 5′-GATCCCCGGACAAATGGGATGAACTATTCAAGAGATAGTTCATCCCATTTGTCCTTTTTC and 5′-TCGAGAAAAAGGACAAATGGGATGAACTATCTCTTGAATAGTTCATCCCATTTGTCCGGG were annealed to form double-stranded DNA and cloned into the BglII and HindIII sites of pSUPER-H1 vector. Henle-407 cells were transfected with this plasmid using Lipofectamine 2000 (Invitrogen) and puromycin-resistant stable-transfected cell lines were screened for PODXL expression by real time-PCR using a podxl-specific primer set. The primer sequences were as follows: 5′-ACCGGGGACTACAACCCTG (sense) and 5′-TGTGGTGTTAGGTTTAGCTGTG (antisense) for podxl and 5′-GATTACTGCTCTGGCTCCTAGC (sense) and 5′-GACTCATCGTACTCCTGCTTGC (antisense) for β-actin.
Glycan array analysis
OG488-labeled wild type and PltBS35A mutant holotoxins were diluted to 180 μg/ml or 20 μg/ml and an aliquot (70 μl) was applied to separate microarray slides (version 5.1) at the Consortium for Functional Glycomics Protein-Glycan Interaction Core, at Emory University (http://www.functionalglycomics.org). The data are reported as average relative fluorescence units of four of six replicates (after removal of the highest and lowest values) for each glycan represented on the array. Glycans showing typhoid toxin binding activity (listed in Table S1) were selected considering a cut off value that was larger or equal to than 1% of the values obtained with the glycan showing the highest binding activity. Glycans showing a variation coefficient higher than 30% were eliminated from this group. In addition some specific glycans were eliminated from the group because they are physiologically irrelevant (glycan #509) or showed non-specific binding (glycans #335,336,523).
Surface plasmon resonance
Surface plasmon resonance analysis was carried out at the Keck Biotechnology Resource Laboratory at the Yale University School of Medicine (http://medicine.yale.edu/keck/index.aspx) using a BiaCore biosensor. Briefly, 50 μg/ml anti-M2 Flag antibody was immobilized on the surface of a chip by amine coupling. Purified wild type or PltBS35A (both FLAG tagged on the C-terminus of CdtB) mutant toxin preparations were applied to the chip followed by application of Ganglisoside GD2 glycan (Elicityl) at various concentrations.
Crystallization
The purification of 6 x His-tagged typhoid toxin used for crystallization is described above. Initial spare matrix crystallization trials of full-length holotoxin protein preparations (2 mg/ml) were carried out at the Yale University School of Medicine Structural Biology Core facility. After crystal optimization trials, full length typhoid toxin (4.5 mg/ml) crystals grew in ~ 3 weeks at room temperature using the hanging-drop vapour-diffusion method in a mix of 1 μl of protein with 1μl of reservoir solution consisting of 1.6 M sodium formate and 0.1 M sodium acetate, pH 4.5.
X-ray data collection and structure determination
X-ray data were collected to 2.4Å at the wavelength of 1.5418 Å on a Rigaku Homelab system at the Yale University Chemical and Biophysical Instrumentation Center (CBIC)(http://cbic.yale.edu). Data were integrated and scaled using the HKL-2000 package6. Further processing was performed with programs from the CCP4 suite7. The holotoxin structure was determined by molecular replacement using PHASER8 with the atomic coordinates of Chain B of H. ducreyi CDT (PDB ID, 1SR4)9, Chain A of pertussis toxin (PDB ID, 1PRT)10 and Subtilase cytotoxin B-subunit (PDB ID,3DWP)11 as the initial search model. To complete the model, manual building was carried out in COOT12. Figures were prepared using PyMol13. The structure refinement was done by PHENIX14. Ramachandran plot statistics (%): Most favorable: 90.1; additionally allowed: 8.8; generously allowed: 1.1; disallowed: 0.0. The data collection and refinement statistics are summarized in Supplementary Table S2.
Molecular docking
Molecular docking of Neu5Ac onto PltB was carried out with AutoDock Vina 15. Based on the available structural and functional information on pertussis and subtilase toxins as well as our own functional data indicating the importance of PltBSer35 in sugar binding (see Fig. 4 and Supplementary data), we chose to model the binding of Neu5Ac onto a pocket surrounding PltB Ser35 and consider several amino acid residues (Tyr33, Tyr34, Ser35 and Lys59) as flexible. The calculation yielded 20 possible models, of which the one with the highest ranking was selected as the most likely.
Fluorescence microscopy
Fluorescence microscopy analysis of typhoid toxin in S. Typhi infected cells was carried out as previously described2. Briefly, Henle-407 cells were seeded on coverslips placed within 24-well plates and cultured overnight. Cultured cells were infected with different strains of S. Typhi expressing FLAG-epitope tagged typhoid toxin with a multiplicity of infection of 20 for 1 hr. Infected cells were washed, treated for 1 hr with 100 μg/ml gentamicin to kill extracellular bacteria, washed again, and incubated for 24 hr in a cell culture medium containing 10 μg/ml gentamicin. Infected cells were washed with PBS, fixed with 4% paraformaldehyde for 10 min at RT, washed, and incubated for 30 min at RT with PBS containing 50 mM NH4Cl, 0.2% Triton X-100 and 3% BSA. Cells were then incubated for 30 min at RT with a primary antibody mixture of mouse anti-Flag M2 (Sigma; 1:4000; catalog # F3165) and rabbit anti-S. Typhi (Difco; 1:4000; catalog # 240993) in PBS containing 3% BSA, washed, incubated for 30 min at RT with a secondary antibody mixture of Alexa-488 anti-mouse (1:2000; catalog # A21121) and Alexa-594 anti-rabbit (1:2000; catalog # A11012) in PBS containing 3% BSA. Cells were washed, stained with DAPI (1:10,000), washed again, mounted, and viewed using a 100 x objective on a fluorescence microscope (Nikon TE2000). Puncta intensities of images were analyzed using ImageJ software as previously described16.
Statistics
The two-tailed student T-test was preformed to determine the statistical significance of experimental changes from control values. A p value of less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank members of the Galán laboratory for careful review of this manuscript. We thank Ewa Folta-Stogniew from the Biophysical Resource at the Keck Biotechnology Resource Laboratory, Yale University School of Medicine (supported by PHS Grant SI0RR026992-0110), for help with conducting the Surface Plasmon Resonance and light-scattering size exclusion chromatography (SEC-LS) assays. We also thank María Lara-Tejero and Xiaoyun Liu (Yale University) for Mass Spectrometry analysis, Jong M. Kim (Duke University) for help with Glycan array analysis software, and Ki-Wook Kim (Yale University) for help in animal inoculations. We thank Wuyi Meng (Yale University) for providing help with X-Ray diffraction data collection, Jiawei Wang and Chuangye Yan (Tsinghua University) for suggestions and providing help with structure refinement, and Xinqi Gong and Meng Ke (Tsinghua University) for help and suggestions with molecular docking. The glycan array analysis was carried out at the Consortium for Functional Glycomics Protein-Glycan Interaction Core, at Emory University, which is supported by PHS Grant GM098791. J.S. was supported in part by a grant from the Northeast Biodefense Center U54-AI057158 and this work was supported by NIAID Grant AI079022 to J. E. G. The atomic coordinates have been deposited in the RCSB Protein Data Bank (entry number 4K6L).
Footnotes
Authors contributions: J. S., X. G., and J. E. G. designed the studies and interpreted the results. J. S. and X. G carried out the experiments. J. S., X. G., and J. E. G. prepared the manuscript.
The authors declare no financial competing interests. Readers are welcome to comment on the on line version of the manuscript.
References
- 1.Parry C, Hien TT, Dougan G, White N, Farrar J. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 2.Parkhill J, et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- 3.Butler T. Treatment of typhoid fever in the 21st century: promises and shortcomings. Clin Microbiol Infect. 2011;17:959–963. doi: 10.1111/j.1469-0691.2011.03552.x. [DOI] [PubMed] [Google Scholar]
- 4.Crump J, Mintz E. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabbagh S, Forest C, Lepage C, Leclerc J, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010;305:1–13. doi: 10.1111/j.1574-6968.2010.01904.x. [DOI] [PubMed] [Google Scholar]
- 6.Haghjoo E, Galan JE. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A. 2004;101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spano S, Ugalde JE, Galan JE. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe. 2008;3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 8.Spano S, Galan JE. A novel pathway for exotoxin delivery by an intracellular pathogen. Curr Opin Microbiol. 2008;11:15–20. doi: 10.1016/j.mib.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beddoe T, Paton A, Le Nours J, Rossjohn J, Paton J. Structure, biological functions and applications of the AB5 toxins. Trends Biochem Sci. 2010;35:411–418. doi: 10.1016/j.tibs.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lara-Tejero M, Galan JE. A bacterial toxin that controls cell cycle progression as a deoxyribonuclease I-like protein. Science. 2000;290:354–357. doi: 10.1126/science.290.5490.354. [DOI] [PubMed] [Google Scholar]
- 11.Lara-Tejero M, Galan JE. Cytolethal distending toxin: limited damage as a strategy to modulate cellular functions. Trends in microbiology. 2002;10:147–152. doi: 10.1016/s0966-842x(02)02316-8. [DOI] [PubMed] [Google Scholar]
- 12.Charles R, et al. Characterization of anti-Salmonella enterica serotype Typhi antibody responses in bacteremic Bangladeshi patients by an immunoaffinity proteomics-based technology. Clin Vaccine Immunol. 2010 doi: 10.1128/CVI.00104-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang L, et al. Immune profiling with a Salmonella Typhi antigen microarray identifies new diagnostic biomarkers of human typhoid. Sci Rep. 2013;3:1043. doi: 10.1038/srep01043. doi:1010.1038/srep01043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Connor B, Schwartz E. Typhoid and paratyphoid fever in travellers. Lancet Infect Dis. 2005;5:623–628. doi: 10.1016/S1473-3099(05)70239-5. [DOI] [PubMed] [Google Scholar]
- 15.Yu C, et al. A bipartite signal regulates the faithful delivery of apical domain marker podocalyxin/Gp135. Mol Biol Cell. 2007;18:1710–1722. doi: 10.1091/mbc.E06-07-0629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermiston M, Zikherman J, Zhu J. CD45, CD148, and Lyp/Pep: critical phosphatases regulating Src family kinase signaling networks in immune cells. Immunol Rev. 2009;228:288–311. doi: 10.1111/j.1600-065X.2008.00752.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R, Yang J, Larsen R, Stanley P. Cloning and expression of N-acetylglucosaminyltransferase I, the medial Golgi transferase that initiates complex N-linked carbohydrate formation. Proc Natl Acad Sci U S A. 1990;87:9948–9952. doi: 10.1073/pnas.87.24.9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanley P, Narasimhan S, Siminovitch L, Schachter H. Chinese hamster ovary cells selected for resistance to the cytotoxicity of phytohemagglutinin are deficient in a UDP-N-acetylglucosamine--glycoprotein N-acetylglucosaminyltransferase activity. Proc Natl Acad Sci U S A. 1975;72:3323–3327. doi: 10.1073/pnas.72.9.3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song X, Lasanajak Y, et al. Shotgun glycomics: a microarray strategy for functional glycomics. Nat Methods. 2011 Jan;8:2085–2090. doi: 10.1038/nmeth.1540. 2018(2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu R, Tsai Y, Ariga T, Yanagisawa M. Structures, biosynthesis, and functions of gangliosides--an overview. J Oleo Sci. 2011;60:537–544. doi: 10.5650/jos.60.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merritt E, Hol W. AB5 toxins. Curr Opin Struct Biol. 1995;5:165–171. doi: 10.1016/0959-440x(95)80071-9. [DOI] [PubMed] [Google Scholar]
- 22.Stein P, et al. The crystal structure of pertussis toxin. Structure. 1994;2:45–57. doi: 10.1016/s0969-2126(00)00007-1. [DOI] [PubMed] [Google Scholar]
- 23.Nesić D, Hsu Y, Stebbins C. Assembly and function of a bacterial genotoxin. Nature. 2004;429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 24.Locht C, Coutte L, Mielcarek N. The ins and outs of pertussis toxin. FEBS J. 2011;278:4668–4682. doi: 10.1111/j.1742-4658.2011.08237.x. [DOI] [PubMed] [Google Scholar]
- 25.Byres E, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008;456:648–652. doi: 10.1038/nature07428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein P, et al. Structure of a pertussis toxin-sugar complex as a model for receptor binding. Nat Struct Biol. 1994;1:591–596. doi: 10.1038/nsb0994-591. [DOI] [PubMed] [Google Scholar]
- 27.Millen S, Lewallen D, Herr A, Iyer S, Weiss A. Identification and characterization of the carbohydrate ligands recognized by pertussis toxin via a glycan microarray and surface plasmon resonance. Biochemistry. 2010;49:5954–5967. doi: 10.1021/bi100474z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saitoh M, et al. The artAB genes encode a putative ADP-ribosyltransferase toxin homologue associated with Salmonella enterica serovar Typhimurium DT104. Microbiology. 2005;151:3089–3096. doi: 10.1099/mic.0.27933-0. [DOI] [PubMed] [Google Scholar]
- 29.Galán JE, Curtiss R., III Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaniga K, Bossio JC, Galan JE. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
References
- 1.Galán JE, Curtiss R., III Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spano S, Ugalde JE, Galan JE. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe. 2008;3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Kaniga K, Bossio JC, Galan JE. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang RF, Kushner SR. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 5.Liu X, Gao B, Novik V, Galan JE. Quantitative Proteomics of Intracellular Campylobacter jejuni Reveals Metabolic Reprogramming. PLoS pathogens. 2012;8:e1002562. doi: 10.1371/journal.ppat.1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 7.Project CC. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 8.McCoy A, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nesić D, Hsu Y, Stebbins C. Assembly and function of a bacterial genotoxin. Nature. 2004;429:429–433. doi: 10.1038/nature02532. [DOI] [PubMed] [Google Scholar]
- 10.Stein P, et al. Structure of a pertussis toxin-sugar complex as a model for receptor binding. Nat Struct Biol. 1994;1:591–596. doi: 10.1038/nsb0994-591. [DOI] [PubMed] [Google Scholar]
- 11.Byres E, et al. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 2008;456:648–652. doi: 10.1038/nature07428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 13.DeLano WLS. The PyMOL Molecular Graphics System. 2002;2002 http://www.pymol.org. [Google Scholar]
- 14.Adams P, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. 2010;D66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of computational chemistry. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spano S, Liu X, Galan JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci U S A. 2011;108:18418–18423. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.