Abstract
Minicircle DNA vectors consisting of a circular expression cassette devoid of the bacterial plasmid DNA backbone provides several advantages including sustained transgene expression in quiescent cells/tissues. Their use has been limited by labor-intensive production. We report on a strategy for making multiple genetic modifications in E.coli to construct a producer strain that stably expresses a set of inducible minicircle-assembly enzymes, the øC31-integrase and I-SceI homing-endonuclease. This bacterial strain is capable of producing highly purified minicircle yields in the same time frame as routine plasmid DNA. It is now feasible for minicircle DNA vectors to replace routine plasmids in mammalian transgene expression studies.
Minicircle episomal DNA vectors have been used for years in preclinical gene transfer research because of their 10–1000-fold enhancement in long-term transgene expression in quiescent tissues in vivo1, 2 and in vitro3. The mechanism of enhanced transgene expression achieved by minicircle is not fully established but likely the result of elimination of the plasmid backbone and associated plasmid backbone DNA induced heterochromatin formation4 and/or transfected cell death triggered by inflammation due to CpG responses when used in combination with lipid delivery carriers5.
The limitation in the use of minicircles has been their time consuming and labor intensive production. In our previous minicircle production schemes (Figure 1a), the minicircle producer plasmid contained a transgene expression cassette flanked with attB and attP, a set of inducible enzyme genes (a homing endonuclease I-SceI gene and two copies of the øC31 integrase gene) and an I-SceI recognition site6. The attB and attP sites are the bacterial and phage attachment sites of øC31 integrase, and the øC31 and I-SceI genes are regulated by the L-arabinose-inducible araCBAD system. Minicircle DNA is generation by recombination between the attB and attP sites, while I-SceI initiated the destruction of the plasmid DNA backbone circle by cutting through the engineered I-SceI site (Figure 1a). While the yields were about 1 mg of minicircle DNA from 1 liter of overnight culture, the preparations still contained about 3 to 15% of the input minicircle producer plasmid plus the plasmid backbone circle as contaminants. Including CsCl equilibrium gradient centrifugation to remove these unwanted DNAs, the production procedure is four labor-intensive days longer than routine plasmid production protocols.
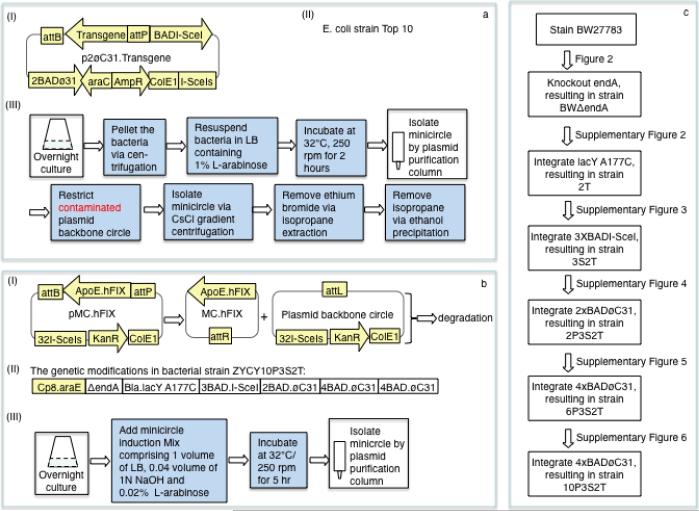
Figure 1. Comparison of the new and old minicircle systems.
(a) An earlier version of the minicircle production system. (aI) Structure of the previous minicircle producer plasmid. BAD and araC, the promoter and the repressor gene of the inducible L–arabinose-araC.BAD system; øC31, bacteriophage øC31 integrase gene; attB and attP, the bacterial and phage attachment sites of the øC31 integrase; I-SceI, I-SceI homing endonuclease gene; I-SceIs, the I-SceI recognition site; AmpR, ampicillin resistance gene; ColE1, DNA replication origin. (aII) E. coli strain Top10 Invitrogen (Carlsbad, California) original strain used to produce minicircle. (aIII) Flowchart showing the minicircle production protocol. Each box represents a major step and the darkened boxes represent the steps required in addition to a routine plasmid production protocol. (b) The new minicircle system. (bI) Diagram of new minicircle parental plasmids and its conversion to minicircle DNA. pMC.hFIX, minicrcle producer plasmid; hFIX, human factor IX; sApoE, promoter/enhancer as described previously2; KanR, kanamycin resistance gene. Upon L-arabinose induction, øc31 is expressed to mediate the formation of minicircle and plasmid backbone circle and I-SceI to induce the destruction of plasmid backbone circle. (bII) The genetic modifications of the minicircle producing bacterial strain ZYCY10P3S2T. 10P3S2T = (1) 10 copies of BAD.øC31 cassette, which were integrated in 3 loci of the bacterial genome: 2 tandem copies at the ΔendA locus (Supplementary Figure 4c), and 4 copies at the araD (Supplementary Figure 5a) and galK (Supplementary Figure 6a) each; (2) 3 tandem copies of BAD.I-SceI cassette, which were integrated at UMU locus (Supplementary Figure 3a) and (3) 2 constitutively expressing L-arabinose transporter genes: one was araE gene driven by an artificial promoter cp8, which presented in strain BW2778310; the other was the bla-lacY A177C cassette, which was integrated at the lacY locus (Supplementary Figure 2); bla, beta-galactosidase gene promoter; lacY A177C, the missense mutant of lacY gene. (bIII) Flowchart showing the new minicircle production protocol. (c) Stepwise genetic modification of the bacterial genome to make the current ZYCY10P3S2T strain.
Other groups have made minicircle DNA vectors using different recombinases such as the bacteriophage Lambda integrase7 or Cre recombinase8. These approaches have limitations due to lower yields, high contamination, or the need to use expensive and labor intensive techniques to isolate minicircle from bacterial lysates9.
The scheme showing the steps used in designing a new system to allow for a simple, rapid and inexpensive procedure for production of a high quality form of this DNA vector is shown in Figure 1b,c. We reasoned that the impurity plasmid DNAs in our earlier minicircle production schemes (Figure 1a) were largely the result of an “all-or-none” phenomenon10, such that in a subset of bacterial cells, the L-arabinose transporter AraE is not expressed, resulting in the failure of øC31 integrase-directed formation of minicircle and I-SceI-mediated degradation of the minicircle producer plasmid. To overcome this limitation, we replaced the Top10 cells with the BW27783 bacterial strain10, in which the araE gene was driven by the constitutive promoter, cp8. When used in combination with previous minicircle producer plasmids, the conversion to minicircle DNA was more complete even with the addition of a very small amount (0.001%) of L-arabinose to the culture (Figure 2a). There was variable but substantial plasmid DNA degradation in these preparations (Figure 2a&c). We hypothesized this was due to the presence of Endonuclease A, which was confirmed when we knocked the gene out (Figure 2c), resulting in strain BWΔendA (Figure 2b, Supplementary Figure 1).
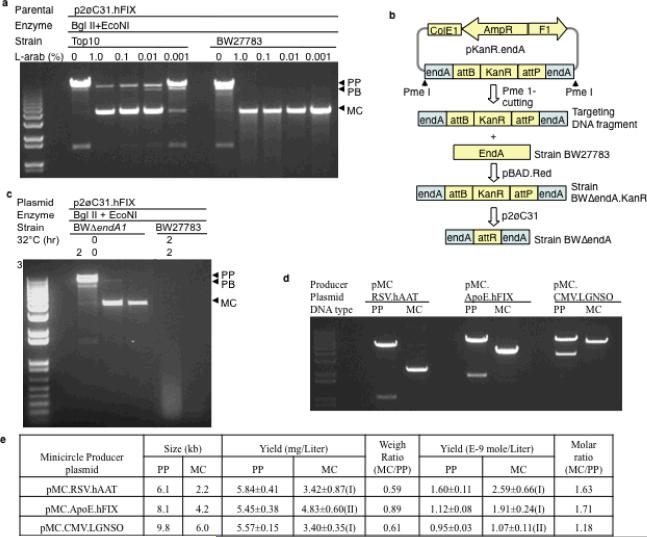
Figure 2. Improvement in minicircle quality and quantity by the new manufacturing system.
(a) Strain BW27783 produced minicircle (MC) with enhanced purity. Minicircles were produced according to the protocol described previously6. p2øC3.hFIX, minicircle producer plasmid (PP); BglII+EcoN1, two restriction enzymes used to restrict MC before electrophoresis; L-arab(%), percent of L-arabinose in the minicircle induction reaction; PB, plasmid backbone circle. (b) Strategy for inactivation of endA. pKanR.endA, the plasmid used to generate the Pme1-restricted targeting DNA fragment; KanR, kanamycin-resistance gene; attB and attP, the bacterial and phage attachment sites of bacteriophage øC31 integrase; boxed endA, PCR-generated 329- and 754-bp end A fragments; pBAD.Red, a plasmid expressing the bacteriophage Lambda homology recombination complex (Red) under the control of araC.BAD (BAD); p2øC31, a complementing plasmid encoding two copies of BAD.øC31 gene, one copy of BAD.I-SceI gene and one I-SceI site; BWΔendA, a strain derived from BW27783 with the endA interrupted. (c) Minicircle DNA integrity before and after disruption of the endA gene. Before disrupting endA, we observed repeatedly large variations in the degree of plasmid degradation as demonstrated in Figures 2a&c. Because the Endonuclease A is a membrane-bound enzyme, it was possible that its membrane release and activation varied during plasmid preparation. 32°C and 37°C, the incubation temperature; hr., hours; all reactions contained 1% L-arabinose. (d) Quality of the minicircle determined by gel analyses. Minicircle was made according to the simplified protocol outlined in Figure 1b and Supplementary Figure 9, DNAs were restricted before electrophoresis; (e) Yield of minicircle producer plasmids and minicircle vector DNAs. The yield was derived from triplicate 400-ml overnight cultures; PP, Minicircle producer plasmid; MC, minicircle; Wilcoxon rank sum test comparing the yield of minicircle and its minicircle producer plasmid: (I), p<0.05; (II), p>0.05. We used the following formula to convert the yield from mg/L to mol/L: mol/L=[yield (mg/L)×10E-3 gram/L]/[size (kb)×1000 ×330×2 gram/mol], where 330 is the average molecular weight of dNTP. The minicircle producer plasmid, pMC.RSV-hAAT is schematically illustrated in Supplementary Figure 4d, while the pMC.CMV.LGNSO was described in details in the Constructs section of the Online Methods.
Trace amounts of impurity DNAs became visible after knocking-out the endonuclease A gene (Figure 2c), suggesting that there was still a lack of complete recombination after L-arabinose induction. Thus, we engineered the bacterial cells to simultaneously express a second L-arabinose transporter, LacY A177C11. Its wildtype counterpart LacY is a glucose transporter, which gains an L-arabinose transporter function with the described missense mutation (Supplementary Figures 1 and 2). While the contaminating DNAs were further reduced, they were not eliminated (data not shown).
To eliminate any possible contamination of øC31 integrase and I-SceI genes from the small amount of parental plasmids used to generate minicircle DNAs, we relocated both the øC31 integrase and I-SceI genes from the minicircle producer plasmid to the bacterial genome. Accordingly, we set forth to integrate three copies of the BAD.I-SceI cassette using the bacteriophage Lambda Red homologous recombination system12 and created the strain 3S2T (Supplementary Figure 3a, Supplementary Figure 1). We found that the integrated BAD.I-SceI was fully functional as evident by the efficient destruction of a plasmid with 8 consecutive I-SceI sites (Supplementary Figure 3b&c) after L-arabinose induction.
Integration of the BAD.øC31 gene was less straightforward. First, the BAD.øC31 cassette could not be integrated using our original Red-system without destroying the integrated BAD.I-SceI gene. Second, we attempted to integrate the BAD.øC31 cassette from a plasmid through øC31 integrase-mediated recombination between the attB site in the plasmid and attP pre-integrated into the genome. However, this integrant was unstable (data not shown), most likely because of a reverse reaction mediated by leaky øC31 integrase expression in concert with an uncharacterized cofactor13. Third, because many more copies of the BAD.øC31 were needed in order to efficiently generate minicircles, we designed a strategy using øC31 integrase and a second recombinase. After making the desired integrant by the øC31 integrase-mediated reaction, the resulting attL site was removed by the second recombinase, eliminating the possibility of the reverse excision reaction (Supplementary Figures 4a, b&c, 5a&e, 6a&d).
Using the above approach, we generated our first bacterial host containing two copies of the BAD.øC31 inserted into the deleted endA site (ΔendA), called the 2P3S2T strain (Supplementary Figure 4a, b&c; Supplementary Figure 1). We found that øC31 integrase and I-SceI mediated the formation of minicircle, and destruction of the unrecombined minicircle producer plasmid and plasmid backbone circle in a minicircle producer plasmid encoding a 2.2-kb transgene and 32 tandem copies of the I-SceI site in the plasmid backbone, respectively (Supplementary Figure 4d&e). However, the minicircle yield was low, suggesting that most of the minicircle producer plasmid was destroyed by I-SceI prior to minicircle formation, and that additional øC31 integrase activity was needed to mediate greater minicircle formation.
To do this, we used another targeting plasmid to integrate 4 copies of the BAD.øC31 cassette into the araD locus (Supplementary Figures 1 and 5), and 4 additional copies into the galK gene (Supplementary Figures 1 and 6), resulting in strains 6P3S2T and ZYCY10P3S2T carrying 6 and 10 copies of BAD.øC31, respectively. These two new strains, in concert with the minicircle producer plasmid described earlier (Supplementary Figure 4d), rendered three improvements over the previous system as outlined in Figure 1a6: First, the procedure is greatly simplified, and when compared to a routine plasmid preparation requires only an additional temperature change and 5-hour incubation after addition of L-arabinose (Figure 1b). Second, the yield of three minicircles, with a size range of 2.2 to 6.0 kb, was 3.4 to 4.8 mg/1000-ml of overnight culture making it ~3 to 5-times higher (Figure 2e) than our previous minicircle producing system (Figure 1a). In addition, when compared with the previous minicircle production protocol there was 10-times less contaminating plasmid DNAs ranging from 0.4% to 1.5% of the input minicircle preparation as determined by qPCR (see Online Methods qPCR section)6. On a molar scale, the yield of minicircle was 20 to 70% higher than the parental plasmid (Figure 2e). Third, the cost of minicircle production is now similar to a standard plasmid.
The improvements made in the production along with the superior expression profiles obtained from minicircle DNA vectors now makes it feasible that they be considered for replacement of plasmid DNAs in mammalian expression studies.
During our quest to develop an optimized minicircle production system, we developed a new bacterial genome modification strategy. The enhancements include: (1) the use of a circular integrating plasmid instead of linear DNA, allowing repeated integration of the same or different DNA sequences of up to 10-kb in selected targets (Supplementary Figure 4b, Supplementary Figures 5a, 6a); (2) Inclusion of a second recombinase allowing selective removal of unwanted sequences such as the hybrid sequences responsible for the reverse recombinase reaction, along with the useless or harmful plasmid backbone DNAs, from the genome (Figures 4b, Supplementary Figures 5a, 6a); and (3) the use of the TPin/9attB.9attP recombination system (Supplementary Figures 5a, 6a), eliminating the inherent problems with the FLP/FRT system where the uncontrolled reaction between the substrate and product FRTs makes repeated integration virtually impossible. While these bacterial gene modifications would be difficult to achieve by using conventional homologous recombination methods, there are other technologies using dual site-specific recombinases for targeted integration of circular DNA into the bacterial genome and creation of a marker-less bacterial strain14. However, the novelty to our approach is that by strategic design we were able to remove one of the two hybrids, (ie., the attL/attR and 9attL/9attR), leaving a stable integrant. Recent studies have shown that repeated gene sequences inserted into the bacterial genome are stable for at least 80 generations15 Our new integrating system will broaden its application in genetic modifications of both pro-and eukaryotic genomes.
Online Methods
Materials
Bacterial strain BW27783 was a gift of Jay D. Keasling of University of California at Berkley10. Luria-Bertani Broth (LB) powder was purchased from MP Biomedicals (Solon, Ohio) and Terrific Broth (TB) powder from Invitrogen (Carlsbad, California). Plasmid DNA purification kits were from Qiagen (Valencia, California).
Constructs
Plasmid pKanR.endA (Figure 2b; Supplementary Figure 7) was made by inserting the following DNA elements: the kanamycin-resistance gene (KanR) flanked by the bacterial (attB) and phage (attP) attachment sites of the Streptomyce bacteriophage øC31 integrase and PCR-generated 329- and 734-bp fragments of endonuclease A gene (endA) into the pBlueScript (Stretagene, La Jolla, California) plasmid. The plasmid p2øC31 (Figures 2b; Supplementary Figures 1, 2b,3a and 7) was made by removing the human factor IX (hFIX) transgene and flanking attB and attP sites from p2øC31.hFIX6, the BAD.I-SceI cassette and I-SceI site were retained. To make the plasmid p3BAD.I-SceIg (Supplementary Figure 3a), the araC repressor gene together with 3 tandem copies of BAD.I-SceI gene were inserted downstream of the attB site of the plasmid pKanR.endA (Figure 2b), followed by replacement of endA with the 737- and 647-bp UMU fragment generated by PCR. The plasmid pBS.8I-SceIs (Supplementary Figure 3b; Supplementary Figure 7) was constructed by inserting 8 consecutive copies of the 18-bp I-SceI site into pBlueScript. The plasmids, pBAD.Red (Figure 2b; Supplementary Figures 1, 2a&c, 3a, 4a, 5a, 6a and 7)16 and pcI587.FLP (Supplementary Figures 1, 4a&b and 7)17, both carry the temperature-sensitive A101 origin of replication, were obtained from the E. coli Genetic Resource Center of Yale University. Plasmid pFRT.KanR.attB (Supplementary Figures 1, 4a and 5) was generated by inserting the attB site and the KanR flanked with FRT sites of flipase into pBlueScript using DNA oligonucleotides. Plasmid p2øC31.attP.FRT; (Supplementary Figures 1, 4b and 7) was constructed using our previous minicircle producing plasmid p2øC31.hFIX6 as starting material. The sApoE.hFIX and BAD.I-SceI cassettes and the I-SceI site were eliminated, the ColE1 origin and the AmpR were replaced with the KanR, the FRT and attP sites and the plasmid replication origin R6K plus Zeocin resistence gene (Zeo) derived from the plasmid pCpG-mcs (InvivoGen-San Diego, California). The plasmid pMC.hFIX (Figure 2a; Supplementary Figures 1 and 7) was made by stepwise replacement of the AmpR and the F1 origin in the plasmid pBS.8I-SceI (Supplementary Figures 1, 3b and 7) with KanR and sApoE.hFIX and the flanking attB and attP sites from the plasmid p2øC31.hFIX6, followed by the insertion of an additional 24 consecutive I-SceI recognition sites. The plasmid pMC.CMV.LGNSO (Figure 2d) was made by replacing the attB.RSV.hAAT.attP fragment in the plasmid pMC.RSV.hAAT (Supplementary Figures 1, 4d and 7) with the fragment flanked with the attB and attP derived from the plasmid p2øC31.LGNSO as described previously3. The plasmid placY.TetR (Supplementary Figures 1, 2a, and 7) was made by inserting the TetR sequence from pACYC184 (New England Biolabs, Inc., Beverly Massachusetts) and flanked with PCR-generated 425- and 227-bp of the z and a lactose operon genes, respectively. Plasmid pbla.lacY A177C (Supplementary Figures 1, 2a and 7) was made by inserting the beta-lactosidase gene promoter (bla) derived from pBlueScript fused with the lacY A177C cDNA derived from plasmid placY A177C obtained from John E Cronan at the University of Illinois11. Plasmid p9attP.TetR.attB (Supplementary Figures 3a, 4a and 5) was made by replacing the z and a fragments in the plasmid placY.TetR (Supplementary Figures 2a and 5) with the attachment site (9attP) from bacteriophage TP901-118 and attB. Plasmid p4øC31.attP.9attB (Supplementary Figures 1, 5a, 4a and 7) was generated by replacing the FRT and ZEO.R6K sequences of p2øC31.attP.FRT (Supplementary Figures 1, 4b and 7) with the bacterial attachment site 9attB of bacteriophage TP901-1 and A101 from plasmid pBAD.Red16, followed by insertion of 2 additional copies of the BAD.øC31 sequence.
Insertional inactivation of the endonuclease A gene in BW27783 (Figure 2b)
This was achieved by Red-mediated homologous recombination between a linear DNA and the endA gene. To do this, BW27783 cells were transformed with pBAD.Red. Cells from one transformed colony were used to make competent cells according to the protocol of Datsenko et al.16. Briefly, the cells were cultured in 25-ml of low salt LB containing ampicillin (50 μg/ml) and 1% L-arabinose and incubated at 32°C with shaking at 250 rpm until the OD600 was 0.5. The competent cells were immediately transformed with 50-ng of the Pme1-restriced targeting DNA fragment prepared from pKanR.endA, cultured at 32°C for 1 hour, spread onto a plate containing Kanamycin (Kan, 25μg/ml) and incubated at 43°C overnight. To select the KanR+ colonies free of pBAD.Red, 8 KanR+ -colonies were cultured in 500-μl LB with Kan at 43°C for 30 minutes and 1 μl each was loaded onto the antibiotic-free, Kan+, and Amp+ plates, which were incubated at 43°C overnight. To remove the KanR gene in the strain BWΔendA.KanR, competent cells were prepared from one KanR-colony, transformed with p2øC31 and spread onto a Amp+ plate; subsequently, cells from eight p2øC31-transformed colonies were cultured each in 500-μl antibiotic-free LB containing 1% L-arabinose at 32°C for 2 hours to induce the øC31 integrase-mediated recombination between the attB and attP flanking the KanR, followed by 4 additional hours at 37°C to induce I-SceI-mediated destruction of p2øC31. The cells were spread onto an antibiotic-free plate. Cells from 8 colonies were cultured in 500-μl antibiotic-free LB at 37°C for 30 minutes, and 1-μl each was loaded onto the antibiotic-free, Kan+ and Amp+ plates to select the AmpR−/KanR− colonies. This resulted in the BWΔendA strain. The disruption of the endA gene was confirmed by DNA sequencing using locus-specific PCR products generated from selected colonies.
Integration of the 2nd L-arabinose transporter lacY A177C and 3 copies of the BAD.I-SceI gene (Supplementary Figure 2)
Replacement of the y gene (lacY) of the lactose operon with the mutant lacY A177C (Supplementary Figure 2) and integration of 3 copies of the I-SceI gene into the UMU locus (Supplementary Figure 3) were achieved by the same Red-mediated homology recombination protocol as described for the endA gene (Figure 2b). To integrate the lacY A177C, however, knockout of the wild-type lacY was performed before knocking in the lacY A177C. Both were accomplished using the same Red-mediated homologous recombination. We used a DNA fragment containing TetR flanked with fragments of the z and a genes to knockout lacY, and selected the intermediate, BWΔendA.TetR on the Tet (6 μg/ml)-plate. This allowed the selection of the next integrant using differential KanR/TetR selection (Supplementary Figures 2a, 5a and 6a).
Integration of the BAD.øC31 cassettes
Three experiments were conducted to integrate 10 copies of BAD.øC31 into the bacterial genome: 2, 4, and 4 copies into the ΔendA locus (Supplementary Figure 4), araD gene (Supplementary Figure 5), and galK gene (Supplementary Figure 6), respectively. All three integration events were achieved by øC31 integrase-mediated site-specific recombination between the attB or attP site in an integrating plasmid, and the attB or attP in the targeted genomic sites as described for the endA gene disruption (legend of Figure 2b).
In an earlier attempt, we found that the integrated BAD.øC31 was lost soon after its integration. This is likely the result of a reverse reaction mediated by an uncharacterized excisionase or cofactor that worked in concert with øC31 integrase produced by the leaky expression from the integrated BAD.øC31. To stabilize the integrant, we designed a double recombination strategy. The øC31 integrase was used to mediate an integration event followed by FLP- or TPin-mediated recombination to remove the attL. This would eliminate the possibility of a reverse reaction between the attL and attR. For integration of the 2BAD.øC31 (Supplementary Figure 4), we used the Red-mediated homologous recombination strategy with a linear DNA containing an attB site, a KanR gene and 2 flanking FRTs for insertion into the ΔendA locus. To remove the KanR, cells from one 3S2T.KanR colony were transformed with pcI587.FLP, and cells from a transformed colony were grown in 5-ml of antibiotic-free LB at 42°C for 8 hours. This induced the expression of FLP to mediate the recombination between the two FRTs, resulting in the elimination of KanR followed by the loss of the temperature-sensitive pcI587.FLP. Subsequently, the cells named 3S2T.attB were transformed with p2øC31.attP.FRT. Cells from one transformed colony were grown in 2-ml of LB containing 1% L-arabinose at 32°C for 2 hours, which induced the expression of øC31 integrase to mediate the recombination between the attB in the ΔendA locus and the attP in the plasmid. The same FLP-mediated FRT-FRT recombination was conducted to mediate the removal of the attL and R6K.Zeo and KanR, resulting in the strain of 2P3S2T (Supplementary Figure 4b). The integrated 2BAD.øC31 was confirmed by DNA sequencing of the locus specific PCR product (Supplementary Figure 4c) and by demonstrating the function in mediating minicircle formation (Supplementary Figure 4e). Integration of the 4 copies of the BAD.øC31 gene into the araD (Supplementary Figure 5) and galK (Supplementary Figure 6) loci, respectively, was also mediated by øC31 integrase; however, the removal of the attL was mediated by bacteriophage TP901-1 integrase (TPin). To do this, we used the Red recombination system to integrate a DNA fragment encoding TetR flanked with an attB and a phage attachment site of TPin (9attP) into the araD or galK site and confirmed the integrant by DNA sequencing of the integrant-specific PCR product (Supplementary Figures 5b and 6b). We integrated p4øC31.attP.9attB into the modified araD or galK locus following the same procedure as used for integrating the p2øC31.attP.FRT (Supplementary Figure 4) with modifications. We selected the colonies with integrants using plates containing both Tet and Kan, transformed the selected intermediates 6P3S2T.KanR.TetR or 10P3S2T.KanR.TetR with plasmid pBAD.TPin, and screened the resulting colonies using triple antibiotic resistance (Tet, Kan and Amp). We grew the cells from one colony in 2-ml of antibiotic-free LB containing 1% L-arabinose, with shaking (250 rpm) at 43°C for 6- to 8-hour before spreading onto an antibiotic-free plate and incubated at 43°C overnight. L-arabinose induced expression of both recombinases, however, the incubation temperature allowed significant TPin integrase activity18 but little øC31 integrase activity, resulting in the TPin intgrase-mediated removal of attL and selection of colonies with the desired integrant, 6P3S2T (Supplementary Figure 5a&d) and ZYCY10P3S2T (Supplementary Figure 6a&c).
Minicircle production protocol
Our new minicircle producing system, the strain ZYCY10P3S2T plus the minicircle producer plasmid pMC.hFIX or pMC.RSV.hAAT, allowed for a greatly simplified minicircle production protocol (Figure 1b). On day one, we inoculated cells from one transformed colony in 5-ml of TB (pH 7.0) with Kan (50 μg/ml) and incubated at 37°C with shaking at 250 rpm. Later that day, we amplified the bacteria by combining 100-μl of culture to every 400-ml TB containing Kan (50 μg/ml) and continued incubation for 16 to 18 hours. For the yield comparison study (Figure 2e), a 400-ml overnight culture was used to prepare intact plasmid DNA. At the end of the culture period the OD600 was 3.5 to 4.2 with a pH of ~6.5. On day 2, we prepared a Minicircle Induction Mix comprising 400-ml fresh LB, 16-ml 1N sodium hydroxide and 0.4-ml 20% L-arabinose, and combined it with a 400-ml overnight culture, and incubated the culture at 32°C with shaking at 250 rpm for an additional 5 hours. We used Qiagen plasmid purification kits to isolate minicircle from bacterial lysates following the manufacturer's protocol with modifications. For every 400-ml overnight culture, we used a 2500 column and 100-ml each of buffers P1, P2 and P3 to ensure complete re-suspension and lysis of the bacteria and a high yield of minicircle DNA vector. A step-by-step protocol is provided in Supplementary Figure 8.
qPCR determination of impurity DNAs
The presence of impurity DNAs, the unrecombined minicircle producer plasmid and plasmid backbone circle, in the three minicircle preparations was determined as described in legends of Figure 2d&e by qPCR. To optimize DNA-polymerase activity, all the DNA templates were digested with AflII plus XhoI flanking the ColE1 origin before qPCR. The minicircle producer plasmid (Supplementary Figure 9) without a transgene expression cassette was used as standard. The PCR primers, 5'-TCCTGTTACCAGTGGCTGCT and 5'-AGTTCGGTGTAGGTCGTTCG, were specific for ColE1 DNA origin shared by all templates. qPCR was performed using the Qiagen Quant SYBR Green RT-PCR kit with a RCorbett system (Corbett). The program included a denaturing step of 94°C for 10 minutes, followed by 25 cycles at 94°C for 20 seconds, 55°C for 15 seconds, and 68°C for 20 seconds each.
Supplementary Material
Acknowledgments
The authors would like to thank Paul Valdmanis for critical review of the manuscript. This work was supported by NIH - HL064274 (MAK).
References
- 1.Chen ZY, He CY, Ehrhrdt A, Kay MA. Minicircle DNA vectors devoid of bacteial DNA result in persistent and high level transgene expression in vivo. Mol. Ther. 2003;8:495–450. doi: 10.1016/s1525-0016(03)00168-0. [DOI] [PubMed] [Google Scholar]
- 2.Huang M, et al. Novel minicircle vector for gene therapy in murine myocardial infarction. Circulation. 2009;120:S230–237. doi: 10.1161/CIRCULATIONAHA.108.841155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jia FJ, et al. A nonviral minicircle vector for deriving human iPS cells. Nature Methods. 2010 doi: 10.1038/nmeth.1426. published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riu E, Chen ZY, Xu H, He CY, Kay MA. Histone modifications are associated with the persistence or silencing of vector-mediated transgene expression in vivo. Molecular Therapy: the Journal of the American Society of Gene Therapy. 2007;15:1348–1355. doi: 10.1038/sj.mt.6300177. [DOI] [PubMed] [Google Scholar]
- 5.Tan Y, Li S, Pitt BR, Huang L. The inhibitory role of CpG immunostimulatory motifs in cationic lipid vector-mediated transgene expression in vivo. Human gene therapy. 1999;10:2153–2161. doi: 10.1089/10430349950017149. [DOI] [PubMed] [Google Scholar]
- 6.Chen ZY, He CY, Kay MA. Improved production and purification of minicircle DNA vector free of plasmid bacterial sequences and capable of persistent transgene expression in vivo. Human Gene Therapy. 2005;16:126–131. doi: 10.1089/hum.2005.16.126. [DOI] [PubMed] [Google Scholar]
- 7.Darquet AM, Cameron B, Wils P, Scherman D, Crouzet J. A new DNA vehicle for nonviral gene delivery: supercoiled minicircle. Gene Ther. 1997;4:1341–1349. doi: 10.1038/sj.gt.3300540. [DOI] [PubMed] [Google Scholar]
- 8.Bigger BW, et al. An araC-controlled bacterial cre expression system to produce DNA minicircle vectors for nuclear and mitochondrial gene therapy. J. Biol. Chem. 2001;276:23018–23027. doi: 10.1074/jbc.M010873200. [DOI] [PubMed] [Google Scholar]
- 9.Mayrhofer P, Blaesen M, Schleef M, Jechlinger W. Minicircle-DNA production by site specific recombination and protein-DNA interaction chromatography. The journal of gene medicine. 2008;10:1253–1269. doi: 10.1002/jgm.1243. [DOI] [PubMed] [Google Scholar]
- 10.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. Homogeneous expression of the P(BAD) promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology (Reading, England) 2001;147:3241–3247. doi: 10.1099/00221287-147-12-3241. [DOI] [PubMed] [Google Scholar]
- 11.Morgan-Kiss RM, Wadler C, Cronan JE., Jr. Long-term and homogeneous regulation of the Escherichia coli araBAD promoter by use of a lactose transporter of relaxed specificity. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:7373–7377. doi: 10.1073/pnas.122227599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorpe HM, Smith MC. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. U. S. A. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Minaeva NI, et al. Dual-In/Out strategy for genes integration into bacterial chromosome: a novel approach to step-by-step construction of plasmid-less marker-less recombinant E. coli strains with predesigned genome structure. BMC Biotechnology. 2008;8:63. doi: 10.1186/1472-6750-8-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sallam KI, Tamura N, Imoto N, Tamura T. New vector system for random, single-step integration of multiple copies of DNA into the Rohodococcus genome. Appl Environ Microbiol. 2010;76:2531–2539. doi: 10.1128/AEM.02131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 18.Stoll SM, Ginsburg DS, Calos MP. Phage TP901-1 site-specific integrase functions in human cells. Journal of bacteriology. 2002;184:3657–3663. doi: 10.1128/JB.184.13.3657-3663.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.