Hybrid nanomaterial systems comprising different functional components have begun to receive significant attention in recent years.1–9 The goal of such multicomponent systems is primarily to combine the functionalities of their constituents, which are typically metallic nanoparticles and semiconducting and/or magnetic quantum dots (QDs). They may also display emergent properties that cannot be realized in homogeneous systems as a result of specific interactions between the components. To date, several different approaches have been taken to assemble multicomponent systems, including colloidal assembly,2,6,9 thermal decomposition,1 selective growth4,9 and DNA-mediated assembly,3,8 including the formation of 3D superlattices.7
The distinctive programmability and versatility of DNA make it particularly attractive for coupling heterogeneous nanostructures. Indeed, binary complexes of Au nanoparticles (AuNPs) and QDs bound by double-stranded DNA have displayed unique photonic and optoelectronic properties that can be tuned by varying the length of the DNA duplex linker between them.8,10 Thus, DNA-based assembly opens up tremendous possibilities for material design and new applications. However, the binding schemes used to date for this purpose rely on DNA duplexes, which lack mechanically rigidity. Furthermore, additional steps (e.g., gel electrophoresis) are required to manage the ratio between the particles. These issues will become even more difficult to manage with increasing heterogeneity and number of particles.
In this work, we demonstrate, for the first time, the heterogeneous organization of semiconducting QDs and metallic nanoparticles on a single DNA origami11 template. The rigidity of the origami scaffold, along with the specific spatial addressability through defined staple binding sites provides extremely precise control over the inter-particle spacing. We also show that heterogeneous binding on DNA origami can be accomplished using distinct reaction chemistry, and we optimize the conditions for both.
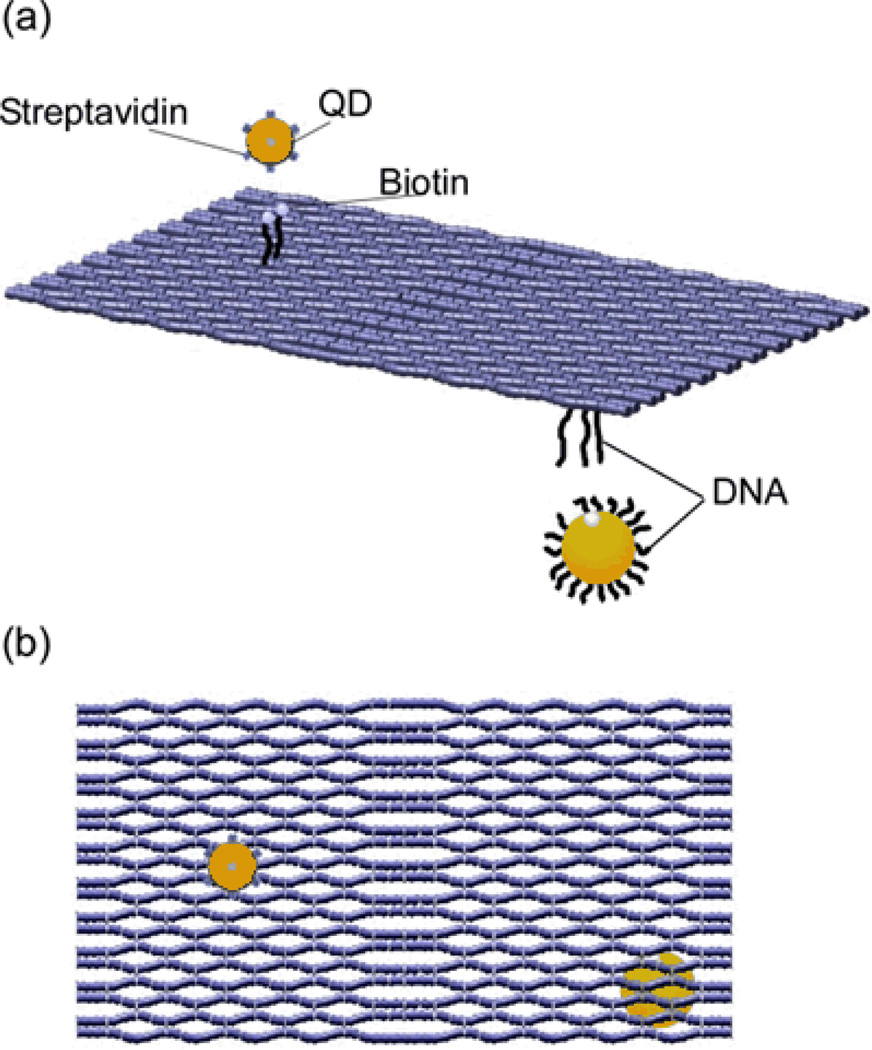
DNA origami scaffolds11 are a general platform for DNA nanotechnology. These structures are formed from a long single-stranded M13mp18 genomic (M13) DNA which is folded into nearly any predefined forms with the help of short staple strands. Owing to the unique sequence of each staple strand, DNA origami is a fully addressable nanostructure that has been used as a molecular breadboard to spatially organize single nanoparticles.12–16 Figure 1 shows our strategy for organizing both the quantum dot and the gold nanoparticle on opposite sides of a DNA origami structure. We used a rectangular DNA origami structure, ~120 nm × 60 nm on a side. We install two types of anchors. One anchor consists of three DNA sticky-ends (15 adenines) protruding from selected staple strands on one side of the origami to capture ssDNA-coated 10 nm AuNPs via DNA hybridization. The second anchor consists of two biotinylated DNA staple strands on the 5’ end with 6 random oligonucleotides protruding from the opposite side of the origami template to capture streptavidin-coated CdSe QDs (the core size is ~ 5 nm12,15) through the streptavidin–biotin interaction. These two anchors allow us to organize different types of differentially functionalized nanoscale objects by selective binding at the corresponding locations. It is also important to note that this enables the formation of sandwich-like three-dimensional DNA origami nanostructures. This strategy not only proves the functionality and versatility of DNA origami scaffold as a two-dimensional structure, but also offers flexibility in varying the inter-particle distance without regard to steric hindrance effects that can arise when closely spaced particles are placed on the same side.
Figure 1.
Schematic drawings of the heterogeneous assembly of CdSe QDs and AuNPs on opposite sides of DNA origami scaffold. (a) Streptavidin coated CdSe QDs were attached to biotin-modified DNA anchors on one side, while ssDNA-wrapped 10 nm AuNPs were assembled to another side through DNA hybridization. (b) Schematic showing the binding position of QDs and AuNPs on DNA origami scaffold.
To organize the nanoparticles on the DNA origami scaffold, the DNA origami template was first annealed11, and then purified to remove the extra helper strands (see experimental section). We optimize the binding yield of AuNPs to the DNA origami template by varying the annealing protocols. First, the binding efficiency of was optimized as a function of annealing temperature. Freshly purified DNA origami rectangles were mixed with a 50 nM solution of 10 nm AuNPs at an elevated temperature, then allowed to slowly cool down to room temperature in a 2-liter water bath over the course of 18 hours. Three different starting temperatures were used: 47 °C, 37 °C and room temperature i.e., no change). We used atomic force microscopy to gauge the results of each protocol. We found that the binding efficiency of 10 nm AuNPs is strongly dependent on the annealing conditions, increasing from 6% at room temperature to 98% at 47 °C (see Figure S1). We hypothesize that the dependence on annealing conditions is a consequence of the increased diffusion kinetics for both the DNA origami and the AuNPs, which increases the contact probability and hybridization affinity between them, consistent with ref.
The second attachment reaction involves the attachment of streptavidin-coated QDs to the DNA origami. This has been well-studied at room temperature.12,15 Figure S2 shows an AFM image of the assembly that resulted from mixing the 50 nM solution of QDs and freshly purified DNA origami after incubation at room temperature over 18 hours. The QDs are selectively bound to the designed anchor position with high binding efficiency, indicating the room temperature is sufficient for streptavidin-biotin interaction, consistent with previous work.12,15
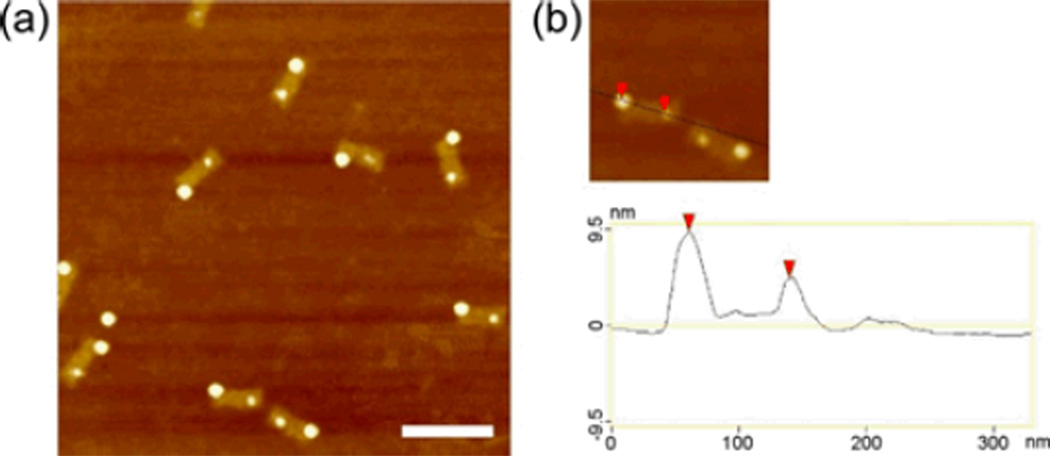
Based on the conditions found for the separate binding of the AuNPs and QDs to the origami scaffold, we tried a two-step annealing procedure for binding both: First, a mixture of rectangular origami with AuNPs was annealed from 47 °C to RT. Then the DNA origami/AuNP product was mixed with QDs and maintained at room temperature for 18 hours. The result, as measured by AFM, is shown in Fig. 2a. We use AuNPs and QDs of different size, so they can be distinguished in AFM height scans (Fig. 2b). The average center-to-center space between two particles is ~75 nm, consistent with the design of the staple binding sites (this distance was chosen so that the AuNP and QD can be clearly resolved in the AFM).
Figure 2.
(a) AFM image of heterogeneous assembly of QDs and AuNPs on rectangle DNA origami template. A two- step annealing process was used: AuNPs self-assembled to origami template from 47 °C to RT over 18 hr, then the product of AuNPs/origami was incubated with QDs at RT keeping another 18hr. The scale bar is 200 nm. (b) Cross-section profile analysis showing the height difference between QDs and AuNPs. (Note: Due to the strong adhesion of the DNA origami to the mica substrate, some deformation may occur; it is therefore difficult to discern in the AFM on which side of the scaffold a particle is located.)
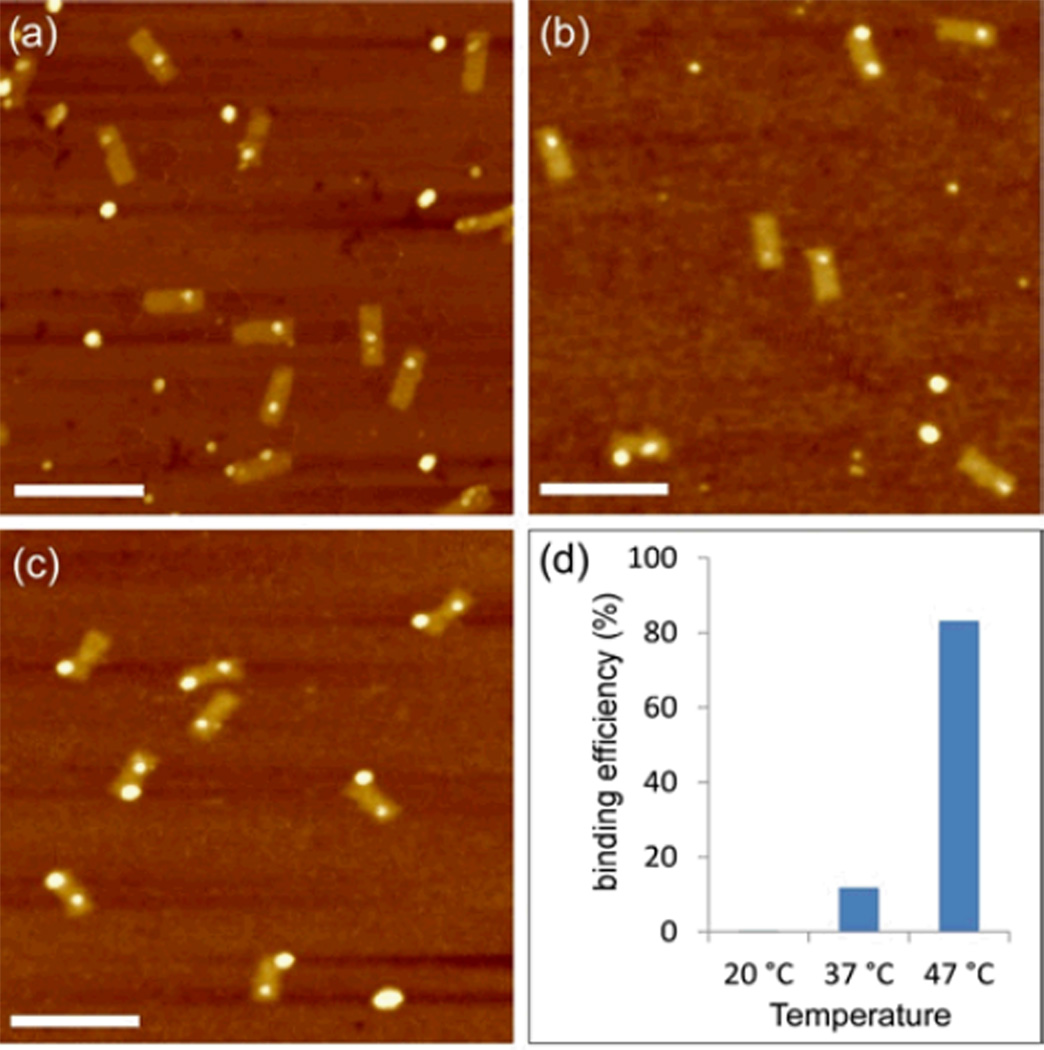
In order to simplify the process and reduce overall processing time, we found a set of experimental conditions that support multiple heterogeneous particle assembly in a single step, i.e., a process in which the DNA origami template, AuNPs and QDs are directly mixed together in the same ratio of two-step annealing. Three different annealing temperatures were tested. (1) When the mixture was annealed at room temperature, almost no AuNPs attached to the origami, as can be clearly seen from Figure 3a. (2) Increasing the annealing temperature to 37 °C resulted in an increase of the binding yield of both QDs and AuNPs to ~12% (Figure. 3b). (3) Using a slow anneal from 47 °C to RT, increased the yield of heterogeneous binding further up to 83% (Figures. 3c and S3). This is slightly lower than we found with the two-step annealing process, indicating that the mixture of two different moieties can interfere with the binding efficiency of each.
Figure 3.
Single-step heterogeneous assembly of QDs and AuNPs on DNA origami template as a function of annealing temperature. (a) The mixture of QDs, AuNPs and DNA origami was annealed at room temperature (25 °C) over 18 hr. (b) Annealed from 37 °C to RT over 18 hr. (c) Annealed from 47 °C to RT over 18 hr. The scale bar is 200 nm. (d) Binding efficiency of QDs and AuNPs on the both side of DNA origami template. These values were calculated for both particles bound to the origami scaffold at the same time.
Interestingly, when only one species was bound to the origami, it could be either a single AuNP or a single QD, as shown in Figure. 3c, i.e., there was no particular preference for one or the other. Clearly, the low heterogeneous binding is mainly restricted by the attachment efficiency of AuNPs at 25°C and 37 °C annealing process, which is lower than the single AuNPs binding. Further research is needed to fully understand the heterogeneous assembly mechanism. However, from this experiment, we demonstrate that heterogeneous assembly of multiple types of nano-objects on DNA origami can be convenient and time saving by the optimization of the reaction conditions.
In summary, we have demonstrated, for the first time, the successful heterogeneous organization of quantum dots and gold nanoparticles with precisely controlled position on both sides of a DNA origami template. Both a two-step and single-step assembly process resulted in relatively high binding yields when the annealing process was optimized. This method has great promise, not only as a path toward the organization of more complex heteromaterial systems, but also as a powerful tool to explore interactions between these materials in a site-specific manner.
Experimental Section
Self-Assembly of rectangle DNA origami template
DNA origami template was formed according to Rohtermund (reference S1). M13 viral DNA and all the staple strands were mixed together at a 1:10 ratio, in a 1×TAE buffer solution containing 40 mM Tris-HCl, 20 mM acetic acid, 2 mM EDTA and 12.5 mM magnesium acetate. The DNA origami solution was slow cooled from 90 °C to 16 °C with PCR over 1.5h. The final concentration of M13mp18 DNA genome in the solution was 10 nM. DNA origami was then purified to remove the excess DNA helper strands using 100kDa MWCO centrifuge filters. The mixture of AuNPs with/or QD and DNA origami was annealed by incubating the sample in a 2-liter beaker containing heated water (47°C or 37°C) and letting it cool down slowly in an insulated (Styrofoam) box to room temperature over the course of 18 hours.
Preparation of DNA coated 10 nm AuNPs
The detailed procedure can be found in reference S2 with slight modification. Briefly, the 10 nm AuNPs were stabilized with adsorption of Bis(p-sulfonatophenyl)phenylphosphine dihydrate dipotassium (BSPP), then sodium chloride was added slowly to this mixture while stirring until the color changed from deep burgundy to light purple. The resulting mixture was centrifuged and supernatant was carefully removed. The concentrated AuNPs were resuspended in BSPP solution and concentration was estimated at ~520 nm. The thiol functionalized single stranded oligonucleotides (5’-TTTTTTTTTTTTTTT-S) were first reduced by TCEP in water, followed by purification using G25 column (GE Healthcare) to remove the small molecules. Then thiol modified oligonucleotides mixed with phosphinatd AuNPs at 100:1 ratio in 0.5×TBE buffer containing 50 mM NaCl for two days at room temperature. AuNP-DNA conjugates were washed with 0.5 × TBE buffer with 100kDa MWCO centrifuge filters to get rid of the extra oligonucleotides. The concentration of conjugates was estimated from the optical absorbance at ~520 nm.
Characterization of AuNP-QD-DNA origami structure by atomic force microscopy
5 µl of sample solution were spotted onto freshly cleaved muscovite mica (Ted Pella inc) and absorbed for ~3 min. (Note, the physisorption of QDs on origami template will happen with long time incubation). To remove buffer salts, 20–30 µl of doubly distilled H2O was placed on the mica, the drop was wicked off and the sample was dried with compressed air. Atomic force imaging was done utilizing Nanoscope IV (Digital Instruments) tapping in air, with ultrasharp 14 series (NSC 14) tips purchased from MikroMasch (www.SPMTIPS.com).
Supplementary Material
Footnotes
We gratefully acknowledge financial support from the Office of Naval Research under Award N00014-09-1-1117. Additional support from the Nanoscale Science and Engineering Initiative of the National Science Foundation under NSF Award CHE-0641523 and from the New York State Office of Science, Technology, and Academic Research (NYSTAR) is also gratefully acknowledged.
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
References
- 1.Chio SH, Na HB, Park YI, An K, Kwon SG, Jang Y, Park MH, Moon J, Son JS, Song IC, Moon WK, Hyeon T. Journal of the American Chemical Society. 2008;130:15573–15580. doi: 10.1021/ja805311x. [DOI] [PubMed] [Google Scholar]
- 2.Dong AG, Chen J, Vora PM, Kikkawa JM, Murray CB. Nature. 2010;466:474–477. doi: 10.1038/nature09188. [DOI] [PubMed] [Google Scholar]
- 3.Fu A, Micheel CM, Cha J, Chang H, Yang H, Alivisatos AP. Journal of the American Chemical Society. 2004;126:10832–10833. doi: 10.1021/ja046747x. [DOI] [PubMed] [Google Scholar]
- 4.Mokari T, Rothenberg E, Popov I, Costi R, Banin U. Science. 2004;304:1787–1790. doi: 10.1126/science.1097830. [DOI] [PubMed] [Google Scholar]
- 5.Ratchford D, Shafiei F, Kim S, Gray SK, Li X. Nano letters. 2011;11:1049–1054. doi: 10.1021/nl103906f. [DOI] [PubMed] [Google Scholar]
- 6.Redl FX, Cho KS, Murray CB, O’Brien S. Nature. 2003;423:968–971. doi: 10.1038/nature01702. [DOI] [PubMed] [Google Scholar]
- 7.Sun D, Gang O. Journal of the American Chemical Society. 2011;133:5252–5254. doi: 10.1021/ja111542t. [DOI] [PubMed] [Google Scholar]
- 8.Wang Q, Wang H, Lin C, Sharma J, Zou S, Liu Y. Chemical communication. 2010;46:240–242. doi: 10.1039/b915712c. [DOI] [PubMed] [Google Scholar]
- 9.Zeng H, Sun SH. Adv Funct Mater. 2008;18:391–400. [Google Scholar]
- 10.Maye MM, Gang O, Cotlet M. Chemical communication. 2010;46:6111–6113. doi: 10.1039/c0cc00660b. [DOI] [PubMed] [Google Scholar]
- 11.Rothemund PWK. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 12.Bui H, Onodera C, Kidwell C, Tan Y, Graugnard E, Kuang W, Lee J, Knowlton WB, Yurke B, Hughes WL. Nano letter. 2010;10:3367–3372. doi: 10.1021/nl101079u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding B, Deng Z, Yan H, Cabrini S, Zuckermann RN, Bokor J. Journal of the American Chemical Society. 2010;132:3248–3249. doi: 10.1021/ja9101198. [DOI] [PubMed] [Google Scholar]
- 14.Hung AM, Micheel CM, Bozano LD, Osterbur LW, Wallraff GM, Cha JN. Nature Nanotech. 2010;5:121–126. doi: 10.1038/nnano.2009.450. [DOI] [PubMed] [Google Scholar]
- 15.Ko SH, Gallatin GM, Liddle JA. Adv Funct Mater. 2012;22:1015–1023. [Google Scholar]
- 16.Maune HT, Han SP, Barish RD, Bochrath M, Iii WA, Rothemund PW, Winfree E. Nature Nanotech. 2010;5:61–66. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.