Abstract
Different brain components can evolve in a coordinated fashion or they can show divergent evolutionary trajectories according to a mosaic pattern of variation. Understanding the relationship between these brain evolutionary patterns, which are not mutually exclusive, can be informed by the examination of intraspecific variation. Our study evaluates patterns of brain anatomical covariation in chimpanzees and humans to infer their influence on brain evolution in the hominin clade. We show that chimpanzee and human brains have a modular structure that may have facilitated mosaic evolution from their last common ancestor. Spatially adjacent regions covary with one another to the strongest degree and separated regions are more independent from each other, which might be related to a predominance of local association connectivity. Despite the undoubted importance of developmental and functional factors in determining brain morphology, we find that these constraints are subordinate to the primary effect of local spatial interactions.
Introduction
Macroevolutionary studies of neuroanatomy tend to show coordinated variation of different brain structures that are related to developmental constraints1. Particular selective pressures, however, have been shown to exert effects on specific regions that result in a mosaic-like pattern of evolution, with different structures showing divergent evolutionary trajectories2–5. Brain evolutionary changes will tend to be concerted when different structures are highly integrated due to pleiotropy, genetic linkage or epigenetic processes6, which can constraint patterns of brain reorganization. Alternatively, mosaic evolution will be more likely when different brain regions are relatively independent from one another, thus giving rise to distinct modules. Modular architectures are expected to facilitate evolution because different traits can respond to selection in diverging ways7. Although this relative interdependence of different brain components has a macroevolutionary result, it also has a microevolutionary origin and thus requires an intraspecific context to be addressed.
Cranial and craniofacial integration and modularity have been extensively investigated in studies of human evolutionary anatomy6,8–10. Most of these studies, however, lack a critical comparison of the extent to which craniofacial variation is influenced by brain variation. Unlike studies of cranial evolution that can include hominin fossil species, because brain tissue does not fossilize, inferences about hominin brain evolution must rely on the comparison of extant species, i.e. chimpanzees and humans. Comparative analyses of extant species can help to identify aspects of brain organization that were present before the divergence of the hominin-panin lineages and those that have diverged across panin and hominin evolution, supplementing information that can be gleaned from endocranial evolution11–17.
The field of neuroscience has tended to analyze patterns of covariation, which are relevant to the study of brain networks, from a different perspective and using a different set of methodological tools18 than those used in anthropology and evolutionary biology. Elements within the same network are expected to show coordinated variation, whereas different networks are likely to represent different modules. Studies of structural and functional brain networks are common in the neuroimaging literature19,20 but, in many cases, they overlook evolutionary history. Additionally, the methodological advance of neuroimaging approaches in which many network analyses are based has progressed more rapidly than the capacity to infer biologically relevant patterns from these techniques21. For this reason, it is especially important to study brain variation in the context of general evolutionary theory, using the same theoretical framework and methodological tools employed in the analysis of other structures and organisms.
Geometric morphometric techniques provide a suitable methodological framework to evaluate these questions by combining some of the strengths of evolutionary biology and neuroscience approaches. These methods offer a suitable way to evaluate correlated variation among brain regions and provide a different perspective than that offered by scaling analyses of the size of neural structures1–5. Geometric morphometric methods allow for a multivariate assessment of covariance structure in which the spatial relationships between anatomically homologous locations are maintained across the different steps of the analysis22. In this way, not only the size of the different structures defined by homologous landmarks can be incorporated into the analysis of covariation, but also their position and orientation within the whole brain and with respect to each other. This allows for a much more complete analysis of covariation patterns, as, once overall size-related modifications are accounted for, different structures can show correlated variation that is not directly reflected in size increases or decreases.
In this study, we used a 3D geometric morphometric framework to analyze and compare intraspecific patterns of covariation between brain regions based on in vivo magnetic resonance imaging (MRI) scans of chimpanzee and human brains. We used both a hypothesis-free and a hypothesis-based approach, in which we compared three competing models based on different biological assumptions regarding the factors driving anatomical covariation: evolutionary and developmental factors, structural factors, or functional factors. Our results show that chimpanzee and human brains are characterized by a very weak level of integration. The structural model is the only one that shows significant modularity and, within this model, correlated variation is especially low between spatially separated regions. The observed modular organization, which is based predominantly on spatial proximity, may allow for relatively independent variation of non-adjacent regions, thus facilitating mosaic evolution. We suggest this architecture to have permitted the sequential acquisition along hominin evolution of traits that are typical of modern human brains.
Results
Hypothesis-free assessment of integration
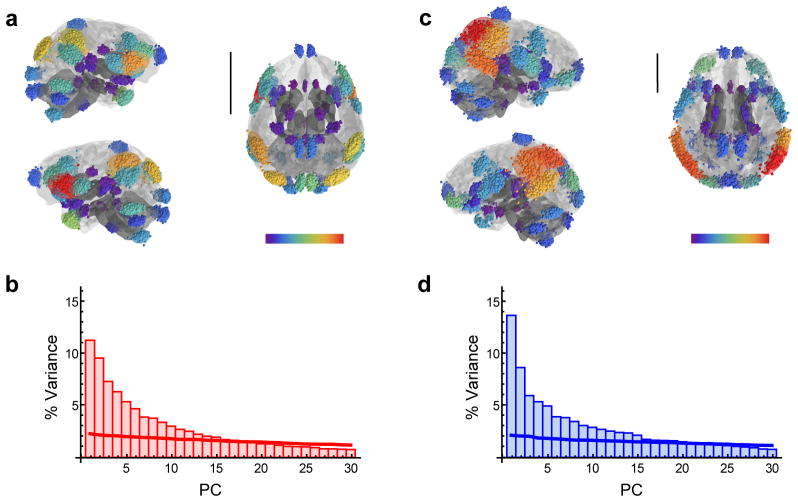
The distribution of variance across the different principal components (PCs) of shape variables corresponding to all landmarks (Fig. 1, Table 1) was used to obtain a general assessment of the degree of integration of chimpanzee and human brains before evaluating biological models of modularity (Fig. 2, Supplementary Table 1). This distribution demonstrates that no single PC accounts for a substantial proportion of variance in the samples (Fig. 3). On the contrary, different PCs account for low and similar proportions of variance both for chimpanzees and humans, with the first PC accounting for no more than 15% of variance. This points to a weakly integrated structure overall in both species.
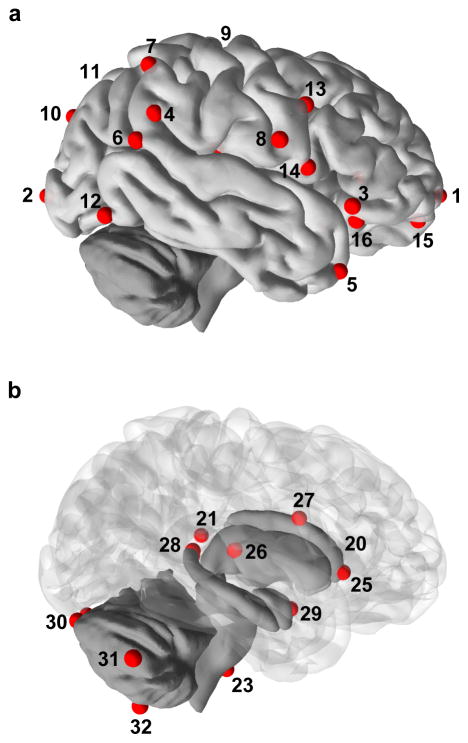
Figure 1. Landmarks used in this study.
Cortical (a) and subcortical landmarks (b) are represented on a human brain. Landmarks 17, 18 and 19 (insular landmarks), 22 and 24 are not represented. Note that the definition of landmarks 10, 11, 15, and 16 is not exactly the same in chimpanzees and humans.
Table 1.
Definition of anatomical landmarks
| Landmark | Definition |
|---|---|
| 1 | Frontal pole |
| 2 | Occipital pole |
| 3 | Anterior end of the Sylvian fissure (defined on the pars orbitalis in humans) |
| 4 | Posterior end of the Sylvian fissure (following the main course of the fissure when the terminal segment is divided) |
| 5 | Anterior end of the superior temporal sulcus (close to the temporal pole) |
| 6 | Inflection point between the horizontal segment and the ascending segment of the superior temporal sulcus |
| 7 | Most posterior and superior point of the superior temporal sulcus (following the anterior course of the sulcus when there are more than one ascending terminal branches) |
| 8 | Inferior termination of the central sulcus |
| 9 | Superior termination of the central sulcus (intersection between the central sulcus and the midline) |
| 10 | In chimpanzees: intersection between the intraparietal sulcus and the lunate sulcus In humans: intersection between the intraparietal sulcus and the transverse occipital sulcus |
| 11 | In chimpanzees: intersection of the lunate sulcus with the midline In humans: intersection of the parieto-occipital sulcus with the midline |
| 12 | Occipital notch |
| 13 | Intersection of the inferior frontal sulcus with the precentral sulcus |
| 14 | Inferior end of the precentral sulcus |
| 15 | In chimpanzees: superior end of the fronto-orbital sulcus In humans: anterior end of the latero-orbital sulcus |
| 16 | In chimpanzees: inferior end of the fronto-orbital sulcus In humans: posterior end of the latero-orbital sulcus |
| 17 | Intersection between the superior circular insular sulcus and the inferior circular insular sulcus |
| 18 | Intersection between the superior circular insular sulcus and the orbito-insular sulcus |
| 19 | Intersection between the inferior circular insular sulcus and the orbito-insular sulcus |
| 20* | Centroid of the genu of the corpus callosum |
| 21* | Centroid of the splenium of the corpus callosum |
| 22* | Superior aspect of the pons |
| 23* | Inferior aspect of the pons |
| 24* | Point where superior cerebellar peduncules meet |
| 25 | Most anterior point of the caudate nucleus |
| 26 | Most posterior point of the putamen nucleus |
| 27 | Most superior and central point of the caudate nucleus |
| 28 | Most superior point of the hippocampus |
| 29 | Centroid of the anterior aspect of the amygdala |
| 30 | Most posterior point of the cerebellum |
| 31 | Most lateral point of the cerebellum |
| 32 | Most inferior point of the cerebellum |
Unilateral landmarks
Figure 2. Modularity models.
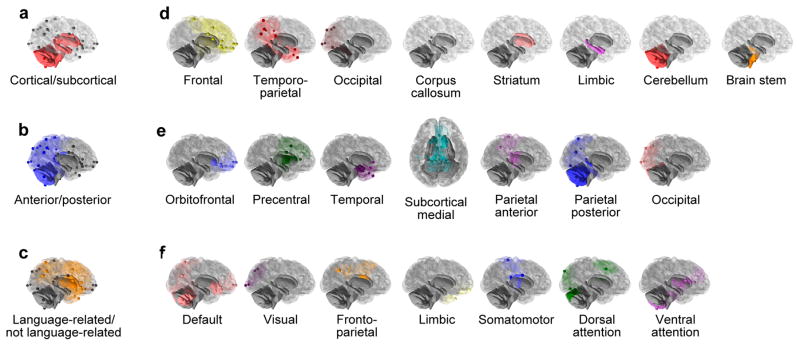
(a) large-scale EvoDevo model; (b) large-scale structural model; (c) large-scale functional model; (d) fine-scale EvoDevo model; (e) fine-scale structural model; (f) fine-scale functional model. Similarly colored modules are not equivalent across the three models. Note that the limbic module is not equivalent in the EvoDevo model and in the functional model. Note as well that our parcellation in the fine-scale functional model does not match exactly those in refs 25, 62, and 63 because our parcellation uses an interpolation based on the anatomical landmarks that are represented in each network.
Figure 3. Hypothesis-free evaluation of integration.
(a) Procrustes superimposed landmarks corresponding to the chimpanzee sample (n=189) before symmetrizing configurations of landmarks; (b) scree plot showing the percentage of variance (% Variance) explained by each principal component (PC) in the chimpanzee sample (red); (c) Procrustes superimposed landmarks corresponding to the human sample (n=189) before symmetrizing configurations of landmarks; (d) scree plot showing the distribution of variance across different principal components in the human sample (blue). In (a) and (c) landmarks have been Procrustes-superimposed and later translated to the original space of the individual represented as a brain surface model. Scale bars are 5 cm. Landmarks are color-coded to represent dispersion around their location in the mean or consensus shape of each sample (red represents high values and purple represent low values). This dispersion is highest at the inferior extreme of the left precentral sulcus in chimpanzees and at the most posterior extreme of the right superior temporal sulcus in humans. In (b) and (d) solid lines represent the scree plots obtained from a random model of variation. Only the first 30 principal components are represented.
The difference between the proportion of variance explained by the first and second principal components is slightly higher in humans than in chimpanzees, thus pointing to a slightly higher degree of integration in humans. These differences, however, are moderate between both species, as revealed by covariance matrix correlations between chimpanzees and humans of 0.68 (matrix permutation test with landmark permutation: P<0.001, which leads to a rejection of the null hypothesis of dissimilarity). This value demonstrates that covariance matrices are significantly similar in both species. Theoretical and comparative neuroanatomical studies suggest that allometric effects of increased brain size may lead to a higher degree of modularity in the neocortex of large brains since communication is more efficient across small areas of the cortex than between distant areas23. In this context, it is notable that human brains show a slightly higher level of integration than chimpanzees. Although it is tempting to speculate about a more distributed organization of brain networks in humans, differences between chimpanzees and humans implied in these results are minor and it is not clear that the distribution of variance we observe for the whole brain in this analysis can be interpreted to reflect the organization of functional networks, as it is described in the following section.
Evaluation of modularity and comparison between models
Given the previous results indicating weak integration in chimpanzee and human brains, which is a necessary but not sufficient prerequisite for a modular structure, we evaluated three competing models of modularity that are based on different biological assumptions concerning the causes of anatomical parcellation (Fig. 2, Supplementary Table 1). Each of these models was evaluated at a large scale (i.e., using only two different modules) and at a fine scale (i.e., using a larger number of modules corresponding to fine-grained systems). 1. An EvoDevo model evaluated covariation between brain structures with different developmental origins and evolutionary trajectories. 2. A structural model analyzed modularity based on spatial proximity under the assumption that anatomically adjacent regions covary to a stronger degree than regions separated by long distances. 3. A functional model evaluated modularity among different functional areas under the assumption that anatomical variation might reflect underlying functional connectivity. This model evaluated covariation between regions functionally involved in language processing and production in humans and regions without that functional involvement24 (large-scale model), or between modules that were defined based on resting-state functional connectivity25 (fine-scale model).
Only the structural model showed significantly higher modularity than random partitions of the sample for chimpanzees and humans, and for the large-scale and fine-scale models (Fig. 4, Table 2). Several studies have demonstrated that modularity is usually influenced by developmental origin26, which, in this case, corresponds to embryological segmentation of brain regions. In our analyses, evolutionary and developmental factors, however, seem to play a minor role in driving patterns of covariation within each species when compared with spatial proximity (Table 2). Similarly, functional networks do not show a major role in determining patterns of correlated variation in comparison with structural factors. Most cerebral shape changes evaluated in our study describe variation in the relative position of sulci and gyri that, in the end, are related to the expansion or reduction of certain regions. A few studies have tried to connect brain macro- and microstructure in humans27,28, chimpanzees29 and other primates30, but more conclusive associations await larger sample sizes and a multivariate approach to understanding intraspecific variation. Sulcal variation has been demonstrated to be a reasonable predictor of cytoarchitecture for primary and secondary regions such as visual, somatosensory and motor areas28, but it appears to be less reliable to identify higher order cognitive areas in both chimpanzees and humans28,31. In any case, sulcal and gyral variation has an indirect relevance as a macromorphological manifestation of functionally key microstructural changes.
Figure 4. Hypothesis-based evaluation of modularity in the large-scale models.
(a) EvoDevo model in chimpanzees; (b) structural model in chimpanzees; (c) functional model in chimpanzees; (d) EvoDevo model in humans; (e) structural model in humans; (f) functional model in humans. For (a), (b), (c), (d), (e) and (f), arrows represent the actual RV coefficient obtained for the corresponding model, and histograms represent the distribution of RV coefficients yielded by random partitions of landmarks from which the P-values discussed in the comparison of the models are calculated. Scale bars are 5 cm.
Table 2.
RV coefficients and P-values (based on permutation tests) obtained for the three modularity models at a large and fine scale.
| Scale of the model | Species | EvoDevo model | Structural model | Functional model |
|---|---|---|---|---|
| Large-scale | Chimpanzee | RV=0.35 P=0.310 |
RV=0.26 P<0.001 |
RV=0.32 P=0.093 |
| Large-scale | Human | RV=0.29 P=0.406 |
RV=0.20 P<0.001 |
RV=0.25 P=0.107 |
| Fine-scale | Chimpanzee | RV=0.13 P=0.351 |
RV=0.13 P=0.005 |
RV=0.13 P=0.386 |
| Fine-scale | Human | RV=0.11 P=0.938 |
RV=0.10 P=0.001 |
RV=0.09 P=0.098 |
Sample size is 189 in all cases. P-values indicate the proportion of 10000 random models with a higher level of modularity (lower RV coefficient) than the evaluated biological model.
Among the large-scale structural models that we tested, only a model separating anterior from posterior landmarks shows significant modularity for both species (Table 3). Models separating medial from lateral landmarks, and superior from inferior landmarks do not show significant levels of modularity. A spatial distance-based model of modularity is a default expectation for most biological systems32, but anterior-posterior partitions of landmarks show much clearer parcellation than other partitions (Table 3). Other features of anatomical variation have been demonstrated to show a rostro-caudal gradient in primate (including human) brains33, a morphological continuum that is likely related to gene expression gradients34. Similarly, numbers of neurons in the neocortex show a rostro-caudal gradient in humans27 and nonhuman primates35. This rostro-caudal partition is partially related to developmental factors such as the timing of neurogenesis, as demonstrated by the separation in our EvoDevo model of a frontal, temporo-parietal and occipital regions, a parcellation that is consistent with evolutionary and developmental regionalization of the neocortex36. However, more local factors appear to drive anatomical covariation, as the EvoDevo model does not show significant modularity in either species.
Table 3.
RV coefficients and P-values (based on permutation tests) obtained for different structural divisions of landmarks.
| Anterior-Posterior | Medial-Lateral | Superior-Inferior | |
|---|---|---|---|
| Chimpanzee | RV=0.26 P<0.001 |
RV=0.31 P=0.268 |
RV=0.32 P=0.232 |
| Human | RV=0.20 P<0.001 |
RV=0.27 P=0.424 |
RV=0.23 P=0.072 |
Sample size is 189 in all cases. P-values indicate the proportion of 10000 random models with a higher level of modularity (lower RV coefficient) than the evaluated biological model.
Organization of structural modules
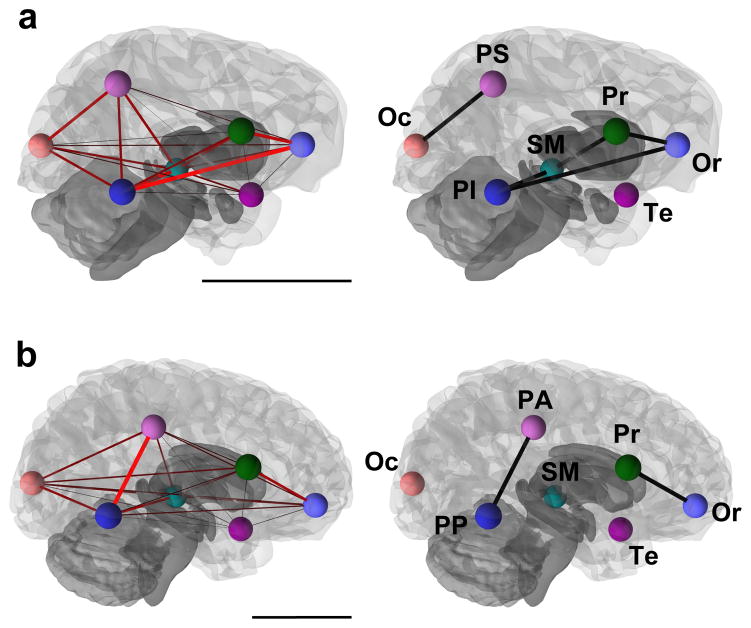
When covariation between the different modules demarcated in the fine-scale structural model is evaluated through pairwise comparisons, it can be observed that modules are not completely independent from each other in spite of the significant degree of modularity observed for this partition of landmarks (Supplementary Tables 2 and 3). On the contrary, all the modules covary significantly with each other with a strength that is influenced by spatial proximity. This is represented in graphical models, where significant edges with high bootstrap support are mostly those grouping spatially adjacent modules (Fig. 5). Graphical models show the separation of anterior versus posterior areas in both species. In humans, the anterior and posterior parts of the brain are more clearly regionalized than in chimpanzees. The anterior complex in humans includes the orbitofrontal and precentral modules, and the posterior one includes both parietal submodules. In chimpanzees, the posterior complex appears to be more clearly spatially defined than the anterior one. The posterior complex includes the occipital and parietal superior modules, whereas the anterior complex includes both frontal modules, but also the subcortical medial and parietal inferior modules. These association patterns might indicate an inverse relationship between distance and strength of covariation. However, although this trend exists in both species, it is not significant (Supplementary Fig. 1).
Figure 5. Graphical models showing pairwise correlations between structural modules.
(a) chimpanzees; (b) humans. In (a) and (b) the model to the left shows pairwise RV coefficients between all modules, with edge thicknesses and color intensity proportional to pairwise RV values (red thick edges represent the highest RV values). Models to the right include only edges showing significant partial correlations between structural modules. Graphical models are superimposed on 3D models of brain surface. Colored spheres represent the centroid of each structural module (Or: orbitofrontal; Pr: precentral; Te: temporal; SM: subcortical medial; PS: parietal superior; PI: parietal inferior; PA: parietal anterior; PP: parietal posterior; Oc: occipital). (a) Chimpanzees: two complexes are identified, a posterior one including the parietal superior and occipital modules, and an anterior complex that includes frontal modules, as well as subcortical medial and inferior parietal modules. (b) Humans: Two complexes are identified, an anterior one formed by both frontal modules and a posterior one formed by both parietal modules. Scale bars are 5 cm.
It is important to note that our results are based only on patterns of correlated variation in the position of the anatomically homologous landmarks that we have examined. However, potential concerns about different scaling methods of other commonly evaluated variables (such as volumetric measures), which can have a strong impact on results, are taken into account in our geometric morphometric approach, as landmark configurations (and, hence, brain morphologies represented by them) are scaled at the initial steps of the analysis. In fact, our results have some similarities with the ones obtained when using variables that are more typically evaluated in neuroscience, such as regional volume, surface area or cortical thickness (Supplementary Methods and Supplementary Table 4). It has to be noted, nonetheless, that results based on these different variables are not expected to match because they have different genetic, developmental, and evolutionary underpinnings37. As the landmark-based model, the volume-, area-, and thickness-based models show a predominance of significant edges between modules that are physically adjacent (Fig. 5 and Supplementary Fig. 2). The volume-based model shows the clearest similarity to the landmark-based model, especially in the low number of significant partial correlations and in the separation of an anterior and a more posterior complex. Surface area- and cortical thickness-based models, however, do not show a clear parcellation of anterior and posterior regions.
Discussion
Intraspecific analyses of correlated variation provide a fundamental way to evaluate the problem of concerted versus mosaic evolution of the brain because macroevolutionary patterns of covariation are grounded in the variation that is observed within species. Intraspecific studies can help elucidate whether different regions of the brain are more or less prone to evolve in a coordinated or independent way. Our observation of modular variation in chimpanzee and human brains is in line with reports of independent variation and regulation between brain components based on quantitative genetic analyses of mice38. Our results, however, shift the focus from relative independence of developmentally distinct regions to relative independence of spatially separated regions. This distance-dependent modular organization might be a particularity of great ape and human brains due to their large size and high morphological complexity as compared with other mammalian brains. The spatial relationships among ontogenetically related structures, which would be initially expected to show the highest relative independence, ultimately become altered due to the high morphological and connectional complexity of the large and gyrified brains of chimpanzees and humans. For example, prefrontal and occipital regions of the cerebral cortex are spatially separated along the rostro-caudal axis even though they share a common developmental origin from the dorsal telencephalon. Our results show that the frontal cortical region covaries most with other frontal regions, and that the occipital neocortical areas covary with the posterior region of the cerebellum, which is derived from the rhombencephalon, in spite of their different developmental and evolutionary trajectories. A similar lack of cortical-subcortical independent modularity has been described using a similar methodological approach in midsagital sections of human brains, where a model of spatially contiguous parietal landmarks has been suggested to show the highest independence from the rest of the brain39.
These results highlight the important influence of basic local spatial interactions in constraining brain morphological variation. These interactions can include biophysical properties of axonal connections, which are likely to cause correlated changes in the areas that they connect40, regardless of whether they are cortico-cortical or cortico-subcortical connections. Landmarks included within each structural module in our study are mostly those located on adjacent sulci and gyri. Therefore, the observation of distance-dependent modularity might be related to the presence of short association fibers (also called U-fibers) between neighboring cortical areas, which have been proposed to be increased in human brains41. It has to be noted that similar (and probably homologous) U-fibers have been described in the rhesus macaque, and that these fibers are probably highly conserved during primate brain evolution42, thus leading to the similar patterns of covariation observed in chimpanzees and humans. The structural model of modularity, however, includes within certain modules regions of the brain outside of the neocortex that are not connected by short association fibers. A potential factor causing correlated variation between adjacent regions not connected by U-fibers and even with different developmental origins is spatial packing, which can have a role in causing covariation between neighboring regions of the occipital cortex and the cerebellum. In this regard, our definition of spatial packing is slightly more general than the classically studied relationship between basicranial flexion and encephalization43, but it makes reference to the physical constraints imposed by the limited available space within the braincase on large brains.
Network analyses of the human skull have shown the importance of spatial relationships and proximity to account for the modular structure of correlated shape changes44. These and our results, however, contrast with the pattern of pervasive genetic integration described for human skulls, which has been proposed to drive the evolution of cranial shape10. Due to the reciprocal interaction between cerebral and cranial development45, related patterns of covariation can be expected between the brain and the skull. One possible explanation for the apparent difference is that our analyses of pairwise comparisons actually do demonstrate that all modules covary significantly with each other (Supplementary Tables 2 and 3). Nonetheless, the strength of this covariation differs across different modules, with the highest values corresponding to spatially adjacent regions and the lowest values to distant regions (Fig. 5). These results are supportive of spatially distant brain areas varying more independently from each other than adjacent areas, but not of completely independent variation. Additionally, the relationship between genetic and phenotypic integration is not entirely clear46. It has been previously demonstrated that fluctuating asymmetry, which can be indicative of developmental plasticity and a proxy for non-genetic variance, accounts for a substantial proportion of morphological variance in chimpanzee and, even more so, in human brains47. In this light, it is important to note that the human cortical surface shows a clear proximity-dependent hierarchical organization when genetic correlations are studied, but a less clear structure based on phenotypic correlations48. One of the main differences reported between phenotypic and genetic correlations is allocated at the posterior perisylvian area, where a very asymmetric pattern is observed in humans for phenotypic correlations, but not for genetic correlations48. This observation is consistent with previous reports of increased fluctuating asymmetry of postsylvian regions in humans47. Environmental effects are known to modify the phenotype of brain morphology, which is initiated by genetic correlations. Non-genetic influences can give rise to a more modular pattern of variation than expected solely on the basis of genetics. If the adult brain phenotype is affected by environmental interactions, which are heavily dependent on social and cultural influences in humans, it is possible that different brain regions respond in a relatively independent way to non-genetic factors, thus giving rise to the observed modular pattern of phenotypic variation.
One of the most consistent results in our study is the general similarity in brain modular structure between chimpanzees and humans (Figs 3, 4, 5; Table 2 and 3), a result that strikingly contrasts with reported differences in craniofacial integration49. Similar patterns of brain modularity are especially relevant because anatomical differences in chimpanzee and human brains are clear47,50, but they seem to be built upon fundamental symplesiomorphic structural interactions that were probably present in the hominin-panin last common ancestor. The predominance of modularity based on spatial proximity may permit evolutionary flexibility, allowing for anatomical and functional differences that have arisen between chimpanzee and human brains. Studies of the mammalian skull have demonstrated that Homo and Pan show the lowest levels of integration and constraints and the highest levels of evolutionary flexibility not only across primates, but also across most mammals51, a result that is compatible with our results showing a modular architecture in chimpanzee and human brains. This modular organization might allow for increased responsiveness of the brain to particular selective pressures51, facilitating mosaic evolution.
Mosaic variation is indeed observed in the hominin fossil record through the sequential appearance of traits considered to be typical of modern human brains. A reorganization of the occipital areas, represented in fossil endocasts as a change in the position of the lunate sulcus marking a reduction of the primary visual cortex, appears to be one of the first acquisitions during hominin brain evolution, perhaps already present in A. afarensis and A. africanus11 (but see ref. 14). Significant orbitofrontal reorganization has been suggested to be present in A. sediba52, and is likely to have occurred during the transition between Australopithecus and early Homo13. Both occipital and orbitofrontal reorganization are observed in the hominin fossil record before a substantial increase in brain size (and a likely related increase in gyrification) is observed in H. erectus. Both Neanderthals and modern humans show substantial frontal broadening53, so it can be parsimoniously assumed that this trait originated before the divergence of both species. An anterior projection of the temporal poles9, as well as a general globularization of the brain involving both a frontal bulging and a parietal reorganization16,17,54, have been proposed to be recent acquisitions in human evolution, only observed in H. sapiens. Other traits that are not visible in endocasts are likely to have evolved as well in a sequential, mosaic fashion, facilitated by the absence of a general, large-scale integration. Our results provide an explanation through which the interspecific patterns of variation observed in the hominin fossil record can be explained on the basis of intraspecific variation in brain anatomy.
Methods
Material
A sample of 189 chimpanzees (Pan troglodytes; 117 females, 72 males; age rage 6–53 years old) and 189 humans (Homo sapiens; 113 females, 76 males; age range 18–60 years old) was studied through in vivo MRI scans. The number of human subjects was chosen to match the number of available chimpanzee scans. Chimpanzees housed at the Yerkes National Primate Research Center (YNPRC) in Atlanta, GE, and at the University of Texas MD Anderson Cancer Center (UTMDACC) in Bastrop, TX, were scanned using a 3T scanner (Siemens Trio, Siemens Medical Solutions, Malvern, USA) or a 1.5T scanner (Phillips, Model 51, Philips Medical Systems, N.A., Bothell, Washington, USA). Scanning procedures in chimpanzees were approved by the Institutional Animal Care and Use Committees at YNPRC and UTMDACC, and also followed the guidelines of the Institute of Medicine on the use of chimpanzees in research. Chimpanzees showed a variety of hand preferences for different tasks. Human scans were obtained from the OASIS database55, selecting only healthy (nondemented) and moderately aged individuals (always younger than 60 years old). All the scans stored in this database belong to right-handed individuals and were acquired with a 1.5T Vision scanner (Siemens, Erlangen, Germany). Informed consent for all participants was obtained in accordance with guidelines of the Washington University Human Studies Committee in the context of the OASIS project. Technical details of human MRI acquisition, as well as population information, is provided in ref. 55, whereas the equivalent information for chimpanzee MRI scans is provided in ref. 56.
Geometric morphometrics and modularity models
Three-dimensional surface models of brain hemispheres for both species were reconstructed using BrainVISA software57. Our analyses were based on a set of 19 bilateral cortical landmarks and 13 subcortical landmarks (5 unilateral and 8 bilateral) defined after refs. 47 and 50 (Fig. 1, Table 1). Although landmark-based geometric morphometric techniques are commonly used in evolutionary and paleontological studies (and, therefore, in the analysis of skeletal remains), these techniques are readily usable in soft tissues, such as brains, as long as homologous locations can be defined and identified. This is the case for the set of landmarks used in our study, which were defined using major sulci in the cortical surface and clearly defined anatomical subdivisions. Cortical landmarks were digitized on 3D reconstructions of the brain surface using IDAV Landmark Editor58, whereas subcortical landmarks were digitized on triplanar views of the MRI scans using MIPAV software59. Cortical and subcortical landmarks were later merged so as to obtain one single landmark configuration for each individual. Non-shape information corresponding to position, size and orientation was removed using a Procrustes superimposition60. Unlike other neuroimaging approaches, no further registration was performed in order to retain all shape variance in the sample, which is the focus of our study.
Landmark configurations were symmetrized by averaging the original and mirrored configurations of landmarks for each brain (Supplementary Data 1). Principal components analyses (PCA) of superimposed Procrustes coordinates were used for a preliminary hypothesis-free assessment of the degree of integration in our samples. Highly integrated structures are characterized by one leading PC explaining most variance in the sample, whereas non-integrated structures show a regular distribution of variance across different PCs61. We evaluated modular variation via three competing models based on different biological assumptions (Fig. 2, Supplementary Table 1), evaluated at a large-scale (i.e., including only two modules) and at a fine-scale (i.e., including a larger number of modules).
An EvoDevo model evaluated covariation between brain structures with different developmental origins and evolutionary trajectories. In the large-scale case, this model evaluated covariation between cortical and subcortical landmarks. In the fine-scale case, this model evaluated covariation between eight different modules: frontal (cortical), temporo-parietal (cortical), occipital (cortical), corpus callosum, striatum, limbic system, pons and cerebellum. The parcellation of cortical landmarks into three different modules (frontal, temporo-parietal, and occipital) is mainly based on their different evolutionary trajectories.
A structural model studied modularity based on spatial proximity. In the large-scale case, this model quantified covariation between rostral and caudal sets of landmarks (corresponding roughly to landmarks located anteriorly or posteriorly to the central sulcus, Fig. 2 and Supplementary Table 1). Alternative structural models separating medial from lateral, and superior from inferior partitions of landmarks were also evaluated. In the fine-scale case, this model included seven modules defined using spatial proximity: orbitofrontal, precentral, temporal, subcortical medial, parietal and occipital. The parietal module was separated into an anterior and a posterior submodules in humans, and into a superior and an inferior submodules in chimpanzees due to the different spatial relationships among landmarks in both species.
A functional model evaluated modularity among areas with different functional involvement. In the large-scale case, this model evaluated covariation between regions involved in language processing and production versus the remaining brain regions. Regions within the language network were selected as those showing positron emission tomography (PET) or functional magnetic resonance imaging (fMRI) activation for specific language related tasks24. In the fine scale case, modules were defined using models of cortical parcellation based on functional connectivity25. This parcellation has been also mapped onto the cerebellum62 and the striatum63. Landmarks in other locations (corpus callosum, pons, hippocampus and amygdala) were not included in this model. In order to design this model of modularity, each landmark was assigned to the functional network to which that anatomical location belonged in the initial parcellation of the cortex25, striatum63 and cerebellum62, although extensive variability in the anatomical location of these functional networks has been described64. Functional models were defined using information based on humans because equivalent information about functional parcellation in the chimpanzee brain is not available, although some studies of intrinsic connectivity networks have revealed striking similarities between humans, chimpanzees, baboons, and capuchins65. Nonetheless, this human-based definition of the model may be involved in the higher level of modularity observed for humans in the fine-scale functional model. A similar result might be expected for the large-scale functional model, which is based on the definition of language-related areas in humans24, but, in that case, both chimpanzees and humans show similar, marginally significant results.
Quantification of integration and modularity
Correlated variation between modules was evaluated using the RV coefficient66. This approach measures the strength of correlated changes in the position of landmarks regardless of the direction of those changes. The RV coefficient is described as a multivariate analogue to the squared correlation coefficient, for which a value of 0 indicates complete modularity and a value of 1 indicates complete integration. These extreme values, however, are orientative, and the significance of the evaluated patterns of modularity was assessed using permutation tests. P-values for modularity models were calculated as the proportion of 10000 random partitions of landmarks (which observe bilateral symmetry) showing higher modularity (lower RV value) than the evaluated model. For models including more than two modules, a multi-set RV coefficient was used66.
Because size, age and sex have a quantitatively very minor effect on the evaluated morphologies47 (Supplementary Tables 5 and 6), we did not correct shape variables for allometry, age or sex. Similarly, significant although quantitatively very minor differences located mainly at occipital landmarks were observed between chimpanzee brains scanned with a 1.5T versus a 3T scanner. These differences have a quantitative value of less than half of the smallest inter-individual Procrustes distance observed in the sample (Supplementary Table 6). Even though these differences appear to be minor, we repeated our analyses within each of the samples to confirm that no scan-related bias impacts our results. These additional analyses showed that modularity patterns observed in chimpanzees are consistent regardless of the type of scanner (Supplementary Table 7).
Models showing significant modularity are not necessarily formed by modules that are completely independent from each other. Therefore, RV coefficients between each pair of modules were calculated for the structural model. Significance was evaluated using permutation tests for the null hypothesis of complete independence between modules. Pairwise RV coefficients were calculated using a common Procrustes superimposition for all the modules. The geometry of those patterns of covariation was graphically depicted using a graphical modelling approach, which has been extensively used in the study of brain networks19. Graphical models were obtained with a modification of the approach presented in ref. 67 based on between-module pairwise RV coefficients. These coefficients were used to calculate partial correlations, which measure the correlation of two variables conditioned on all other variables, and their significance was assessed using the edge exclusion deviance criterion67. Consensus graphical models were obtained to account for the variability in estimating RV coefficients. Only edges showing significant partial correlations (P<0.05) in more than 90% of 1000 bootstrap resamplings of the original chimpanzee and human populations were represented. Analyses were performed in MorphoJ68 and in Mathematica using the Morphometrics package written by P.D. Polly69 and the Modularity package written by P.D. Polly and A. Goswami70.
Supplementary Material
Acknowledgments
We are grateful to David Sánchez-Martín for his continuous assistance with technical questions. The Open Access Series of Imaging Studies (OASIS) project, from which we have obtained human MRI scans, is supported by grants P50 AG05681, P01 AG03991, R01 AG021910, P50 MH071616, U24 RR021382 and R01 MH56584. This work was supported by National Science Foundation Grants BCS-0824531; National Institutes of Health Grants NS-42867; and James S. McDonnell Foundation Grants 22002078, and 220020293.
Footnotes
Authors’ contributions
A.G.-R. and C.C.S. conceived of the study; W.D.H. collected and provided data; A.G.-R. designed the analyses, conducted data collection and analyses; A.G.-R., W.D.H. and C.C.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Finlay B, Darlington R. Linked regularities in the development and evolution of mammalian brains. Science. 1995;268:1578–1584. doi: 10.1126/science.7777856. [DOI] [PubMed] [Google Scholar]
- 2.Barton RA, Harvey PH. Mosaic evolution of brain structure in mammals. Nature. 2000;405:1055–1058. doi: 10.1038/35016580. [DOI] [PubMed] [Google Scholar]
- 3.De Winter W, Oxnard CE. Evolutionary radiations and convergences in the structural organization of mammalian brains. Nature. 2001;409:710–714. doi: 10.1038/35055547. [DOI] [PubMed] [Google Scholar]
- 4.Smaers JB, Soligo C. Brain reorganization, not relative brain size, primarily characterizes anthropoid brain evolution. Proc R Soc B. 2013;280:20130269. doi: 10.1098/rspb.2013.0269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oxnard CE. Brain evolution: Mammals, primates, chimpanzees, and humans. Int J Primatol. 2004;25:1127–1158. [Google Scholar]
- 6.Mitteroecker P, Bookstein F. The evolutionary role of modularity and integration in the hominoid cranium. Evolution. 2008;62:943–958. doi: 10.1111/j.1558-5646.2008.00321.x. [DOI] [PubMed] [Google Scholar]
- 7.Wagner GP. Homologues, natural kinds and the evolution of modularity. Am Zool. 1996;36:36–43. [Google Scholar]
- 8.Lieberman DE, Krovitz GE, McBratney-Owen B. Testing hypotheses about tinkering in the fossil record: the case of the human skull. J Exp Zool. 2004;302B:284–301. doi: 10.1002/jez.b.21004. [DOI] [PubMed] [Google Scholar]
- 9.Bastir M, et al. Evolution of the base of the brain in highly encephalized human species. Nat Commun. 2011;2:588. doi: 10.1038/ncomms1593. [DOI] [PubMed] [Google Scholar]
- 10.Martínez-Abadías N, et al. Pervasive genetic integration directs the evolution of human skull shape. Evolution. 2012;66:1010–1023. doi: 10.1111/j.1558-5646.2011.01496.x. [DOI] [PubMed] [Google Scholar]
- 11.Holloway RL. The human brain evolving: a personal retrospective. Ann Rev Anthropol. 2008;37:1–19. [Google Scholar]
- 12.Balzeau A, Gilissen E, Grimaud-Herve D. Shared pattern of endocranial shape asymmetries among great apes, anatomically modern humans, and fossil hominins. PLoS One. 2011;7:e29581. doi: 10.1371/journal.pone.0029581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Sousa A, Cunha E. In: Evolution of the Primate Brain. Hofman MA, Falk D, editors. Elsevier; 2012. pp. 293–322. [Google Scholar]
- 14.Falk D. In: Evolution of the Primate Brain. Hofman MA, Falk D, editors. Elsevier; 2012. pp. 255–272. [DOI] [PubMed] [Google Scholar]
- 15.Neubauer S, Gunz P, Hublin JJ. Endocranial shape changes during growth in chimpanzees and humans: a morphometric analysis of unique and shared aspects. J Hum Evol. 2010;59:555–566. doi: 10.1016/j.jhevol.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 16.Bruner E, Manzi G, Arsuaga JL. Encephalization and allometric trajectories in the genus Homo: evidence from the Neandertal and modern lineages. Proc Natl Acad Sci USA. 2003;100:15335–15340. doi: 10.1073/pnas.2536671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunz P, Neubauer S, Maureille B, Hublin JJ. Brain development after birth differs between Neanderthals and modern humans. Curr Biol. 2010;20:R921–R922. doi: 10.1016/j.cub.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Alexander-Bloch A, Giedd JN, Bullmore E. Imaging structural co-variance between human brain regions. Nat Rev Neurosci. 2013;14:322–336. doi: 10.1038/nrn3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 20.Meunier D, Lambiotte R, Bullmore ET. Modular and hierarchically modular organization of brain networks. Front Neurosci. 2010;4:200. doi: 10.3389/fnins.2010.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buckner RL, Krienen FM, Yeo BTT. Opportunities and limitations of intrinsic functional connectivity MRI. Nat Neurosci. 2013;16:832–837. doi: 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- 22.Zelditch ML, Swiderski DL, Sheets DH, Fink WL. Geometric Morphometrics for Biologists. Academic Press; 2004. [Google Scholar]
- 23.Ringo JL. Neuronal interconnection as a function of brain size. Brain Behav Evol. 1991;38:1–6. doi: 10.1159/000114375. [DOI] [PubMed] [Google Scholar]
- 24.Price CJ. A review and synthesis of the first 20 years of PET and fMRI studies of heard speech, spoken language and reading. NeuroImage. 2012;62:816–847. doi: 10.1016/j.neuroimage.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeo BT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klingenberg CP. Morphological integration and developmental modularity. Ann Rev Ecol Evol Syst. 2008;39:115–132. [Google Scholar]
- 27.Ribeiro PFM, et al. The human cerebral cortex is neither one nor many: neuronal distribution reveals two quantitatively different zones in the gray matter, three in the white matter, and explains local variations in cortical folding. Front Neuroanat. 2013;7:28. doi: 10.3389/fnana.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischl B, et al. Cortical folding patterns and predicting cytoarchitecture. Cereb Cortex. 2008;18:1973–1980. doi: 10.1093/cercor/bhm225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spocter MA, et al. Wernicke’s area homologue in chimpanzees (Pan troglodytes) and its relation to the appearance of modern human language. Proc R Soc B. 2010;277:2165–2174. doi: 10.1098/rspb.2010.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Collins CE, Airey DC, Young NA, Leitch DB, Kaas JH. Neuron densities vary across and within cortical areas in primates. Proc Natl Acad Sci USA. 2010;107:15927–15932. doi: 10.1073/pnas.1010356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherwood CC, Broadfield DC, Holloway RL, Gannon PJ, Hof PR. Variability of Broca’s area homologue in African great apes: implications for language evolution. Anat Rec. 2003;271:276–285. doi: 10.1002/ar.a.10046. [DOI] [PubMed] [Google Scholar]
- 32.Adams DC, Rohlf FJ, Slice DE. A field comes of age: geometric morphometrics in the 21st century. Hystrix. 2013;24:7–14. [Google Scholar]
- 33.Toro R. On the possible shapes of the brain. Evol Biol. 2012;39:600–612. [Google Scholar]
- 34.Sansom SN, Livesey FJ. Gradients in the brain: the control of the development of form and function in the cerebral cortex. Cold Spring Harbor Perspect Biol. 2009;1:a002519. doi: 10.1101/cshperspect.a002519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charvet CJ, Cahalane DJ, Finlay BL. Systematic, cross-cortex variation in neuron numbers in rodents and primates. Cereb Cortex. 2013 doi: 10.1093/cercor/bht214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen CH, et al. Genetic influences on cortical regionalization in the human brain. Neuron. 2011;72:537–544. doi: 10.1016/j.neuron.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanabria-Diaz G, et al. Surface area and cortical thickness descriptors reveal different attributes of the structural human brain networks. NeuroImage. 2010;50:1497–1510. doi: 10.1016/j.neuroimage.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 38.Hager R, Lu L, Rosen GD, Williams RW. Genetic architecture supports mosaic brain evolution and independent brain–body size regulation. Nat Commun. 2012;3:1079. doi: 10.1038/ncomms2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bruner E, Martin-Loeches M, Colom R. Human midsagittal brain shape variation: patterns, allometry and integration. J Anat. 2010;216:589–599. doi: 10.1111/j.1469-7580.2010.01221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.VanEssen DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- 41.Schenker NM, Desgouttes AM, Semendeferi K. Neural connectivity and cortical substrates of cognition in hominoids. J Hum Evol. 2005;49:547–569. doi: 10.1016/j.jhevol.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 42.Oishi K, et al. Superficially located white matter structures commonly seen in the human and the macaque brain with diffusion tensor imaging. Brain connectivity. 2011;1:37–47. doi: 10.1089/brain.2011.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross CF, Ravosa MJ. Basicranial flexion, relative brain size, and facial kyphosis in nonhuman primates. Am J Phys Anthropol. 1993;91:305–324. doi: 10.1002/ajpa.1330910306. [DOI] [PubMed] [Google Scholar]
- 44.Esteve-Altava B, Marugán-Lobón J, Botella H, Bastir M, Rasskin-Gutman D. Grist for Riedl’s mill: A network model perspective on the integration and modularity of the human skull. J Exp Zool B. 2013;320:489–500. doi: 10.1002/jez.b.22524. [DOI] [PubMed] [Google Scholar]
- 45.Richtsmeier JT, et al. Phenotypic integration of neurocranium and brain. J Exp Zool B. 2006;306:360–378. doi: 10.1002/jez.b.21092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hallgrímsson B, et al. Deciphering the palimpsest: studying the relationship between morphological integration and phenotypic covariation. Evol Biol. 2009;36:355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gómez-Robles A, Hopkins WD, Sherwood CC. Increased morphological asymmetry, evolvability and plasticity in human brain evolution. Proc R Soc B. 2013;280:20130575. doi: 10.1098/rspb.2013.0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen CH, et al. Hierarchical genetic organization of human cortical surface area. Science. 2012;335:1634–1636. doi: 10.1126/science.1215330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neaux D, Guy F, Gilissen E, Coudyzer W, Ducrocq S. Covariation between midline cranial base, lateral basicranium, and face in modern humans and chimpanzees: a 3D geometric morphometric analysis. Anat Rec. 2013;296:568–579. doi: 10.1002/ar.22654. [DOI] [PubMed] [Google Scholar]
- 50.Aldridge K. Patterns of differences in brain morphology in humans as compared to extant apes. J Hum Evol. 2011;60:94–105. doi: 10.1016/j.jhevol.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marroig G, Shirai L, Porto A, de Oliveira F, De Conto V. The evolution of modularity in the mammalian skull II: evolutionary consequences. Evol Biol. 2009;36:136–148. [Google Scholar]
- 52.Carlson KJ, et al. The endocast of MH1, Australopithecus sediba. Science. 2011;333:1402–1407. doi: 10.1126/science.1203922. [DOI] [PubMed] [Google Scholar]
- 53.Bruner E, Holloway RL. A bivariate approach to the widening of the frontal lobes in the genus Homo. J Hum Evol. 2010;58:138–46. doi: 10.1016/j.jhevol.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 54.Lieberman DE, McBratney BM, Krovitz G. The evolution and development of cranial form in Homo sapiens. Proc Natl Acad Sci USA. 2002;99:1134–1139. doi: 10.1073/pnas.022440799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marcus DS, et al. Open access series of imaging studies (OASIS): cross-sectional MRI data in young, middle aged, nondemented, and demented older adults. J Cogn Neurosci. 2007;19:1498–1507. doi: 10.1162/jocn.2007.19.9.1498. [DOI] [PubMed] [Google Scholar]
- 56.Bogart SL, et al. Cortical sulci asymmetries in chimpanzees and macaques: a new look at an old idea. NeuroImage. 2012;61:533–541. doi: 10.1016/j.neuroimage.2012.03.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cointepas Y, Mangin JF, Garnero L, Poline JB, Benali H. BrainVISA: software platform for visualization and analysis of multi-modality brain data. NeuroImage. 2001;13:98. [Google Scholar]
- 58.Wiley DF, et al. Evolutionary morphing. Proc IEEE Vis Conf. 2005;2005:431–438. [Google Scholar]
- 59.McAuliffe MJ, et al. Medical Image Processing, Analysis and Visualization in clinical research. Proc. 14th IEEE Symposium. Computer-Based Medical Systems (CBMS 2001); 2001. pp. 381–386. [Google Scholar]
- 60.Rohlf FJ, Slice D. Extensions of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39:40–59. [Google Scholar]
- 61.Wagner GP. On the eigenvalue distribution of genetic and phenotypic dispersion matrices: Evidence for a nonrandom organization of quantitative character variation. J Math Biology. 1984;21:77–95. [Google Scholar]
- 62.Buckner RL, Krienen FM, Castellanos A, Diaz JC, Yeo BTT. The organization of the human cerebellum estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:2322–2345. doi: 10.1152/jn.00339.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi EY, Yeo BTT, Buckner RL. The organization of the human striatum estimated by intrinsic functional connectivity. J Neurophysiol. 2012;108:2242–2263. doi: 10.1152/jn.00270.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mueller S, et al. Individual variability in functional connectivity architecture of the human brain. Neuron. 2013;77:586–595. doi: 10.1016/j.neuron.2012.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wey H-Y, et al. Multi-region hemispheric specialization differentiates human from nonhuman primate brain function. Brain Struct Funct. 2013 doi: 10.1007/s00429-013-0620-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Klingenberg CP. Morphometric integration and modularity in configurations of landmarks: tools for evaluating a priori hypotheses. Evol Dev. 2009;11:405–421. doi: 10.1111/j.1525-142X.2009.00347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Magwene PM. New tools for studying integration and modularity. Evolution. 2001;55:1734–1745. doi: 10.1111/j.0014-3820.2001.tb00823.x. [DOI] [PubMed] [Google Scholar]
- 68.Klingenberg CP. MorphoJ: an integrated software package for geometric morphometrics. Mol Ecol Resour. 2011;11:353–357. doi: 10.1111/j.1755-0998.2010.02924.x. [DOI] [PubMed] [Google Scholar]
- 69.Polly PD. Geometric Morphometrics for Mathematica. Indiana University ScholarWorks; 2012. at https://scholarworks.iu.edu/dspace/handle/2022/14613. [Google Scholar]
- 70.Polly PD, Goswami A. Modularity for Mathematica. Indiana University ScholarWorks; 2010. at https://scholarworks.iu.edu/dspace/handle/2022/14541. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.