Abstract
The alveolates are composed of three major lineages, the ciliates, dinoflagellates, and apicomplexans. Together these ‘protist’ taxa play key roles in primary production and ecology, as well as in illness of humans and other animals. The interface between the dinoflagellate and apicomplexan clades has been an area of recent discovery, blurring the distinction between these two clades. Moreover, phylogenetic analysis has yet to determine the position of basal dinoflagellate clades hence the deepest branches of the dinoflagellate tree currently remain unresolved. Large-scale mRNA sequencing was applied to 11 species of dinoflagellates, including strains of the syndinean genera Hematodinium and Amoebophrya, parasites of crustaceans and dinoflagellates, respectively, to optimize and update the dinoflagellate tree. From the transcriptome-scale data a total of 73 ribosomal protein-coding genes were selected for phylogeny. After individual gene orthology assessment, the genes were concatenated into a >15,000 amino acid alignment with 76 taxa from dinoflagellates, apicomplexans, ciliates, and the outgroup heterokonts. Overall the tree was well resolved and supported, when the data was subsampled with gblocks or constraint trees were tested with the approximately unbiased test. The deepest branches of the dinoflagellate tree can now be resolved with strong support, and provides a clearer view of the evolution of the distinctive traits of dinoflagellates.
Keywords: Dinoflagellate, Alveolate, Heterokont, Apicomplexan, Ribosomal protein
1. Introduction
Alveolates include three major lineages, the ciliates, dinoflagellates and apicomplexans (Gajadhar et al., 1991; Taylor, 1999; Burki et al., 2007). The dinoflagellates are notable primary producers, especially in marine environments, and the apicomplexans are known as parasites, particularly the malaria-causing genus Plasmodium, which infects 149 million to 274 million and kills 537,000–907,000 individuals annually (WHO, 2010). The third alveolate group, the ciliates, is most notable for the diversity of their habitats and unusual cell biology including dual nuclei, one germinal and the other somatic. The alveolates, in turn, are most closely related to the heterokonts, a very diverse group ranging from members of the human gut flora, plant pathogens, to the photosynthetic diatoms and the giant kelps (Baldauf, 2003; Burki et al., 2007; Parfrey et al., 2010).
Within the alveolates, the dinoflagellates and apicomplexans are more closely related, and the area between them has been one of recent discovery which confounds simple interpretations of the evolution of these groups (Okamoto et al., 2012; Leander et al., 2003). For example, while parasitic apicomplexans are known to harbor a non-photosynthetic plastid, the discovery of Chromera velia demonstrates that photosynthetic members of the apicomplexan lineage are extant (Moore et al., 2008; Keeling, 2013). Meanwhile, in the dinoflagellates, best known as free-living photosynthetic autotrophs, the non-photosynthetic oyster parasite Perkinsus marinus diverges from the base of the dinoflagellate lineage (Bachvaroff et al., 2011; Reece et al., 1997; Saldarriaga et al., 2003). Clearly both C. velia and P. marinus have the potential to independently lose or gain characteristics, but at the simplest level the lifestyles of the deepest branching members of the apicomplexan and dinoflagellate clades strongly contrast with the more familiar members of these lineages.
Simultaneously with the description of new species between apicomplexans and dinoflagellates has been the discovery of an astonishing breadth and abundance of sequences attributable to ‘marine alveolates’ from marine environmental clone libraries. At a first approximation many of these sequences are placed with known syndinean dinoflagellates in phylogenies, although the raw abundance of such sequences (>1000 in GenBank) dwarfs the tens of sequences attributed to described syndinean species or genera (Bachvaroff et al., 2012). The relationships between Marine Alveolate clades I–VIII are not resolved. Indeed, all Marine Alveolate clades may not be syndinean dinoflagellates or parasites, although certainly clades I, II and IV contain syndinean taxa (Bachvaroff et al., 2012; Coats and Bachvaroff, 2012; Harada et al., 2007; Skovgaard et al., 2005, 2009). In the present study we define two major lineages of dinoflagellates, the syndineans and the core dinoflagellates (Hoppenrath and Leander, 2010; Okamoto et al., 2012). The term core dinoflagellate is used in preference to what were formally called the dinokaryotes, since recent studies have cast doubt on the synapomorphies of the dinokaryon (Gornik et al., 2012; Sano and Kato, 2009).
The dinokaryotic state lacks a strict definition, but could be defined as a nucleus with chromosomes condensed throughout the cell cycle, a very low basic protein:DNA ratio, and lack of bulk DNA packaging into nucleosomes. Together these characteristics produce an ‘arched fibrillar’ appearance of the DNA in transmission electron micrographs of dinokaryote chromosomes (Taylor, 1989). Also, these features appear to be correlated with a high degree of gene duplication (Bachvaroff and Place, 2008; Bachvaroff et al., 2009; Shoguchi et al., 2013).
The outlying species Oxyrrhis marina has features reminiscent of dinokaryotes including high DNA content, ‘conspicuously banded’ chromosomes, and multiple gene copies (Sano and Kato, 2009). In recent reviews on the evolution of the dinokaryon O. marina is placed just outside of the core dinoflagellates (Saldarriaga et al., 2004; Wisecaver and Hackett, 2011). Such a placement, however, disagrees with other taxonomic treatments that place O. marina outside of both the syndineans and core dinoflagellates based on cell morphology and flagellar arrangement (Adl et al., 2005; Fensome et al., 1993). Clearly independent phylogenetic assessment of O. marina is warranted to resolve this discordance.
Well-defined relationships between P. marinus, O. marina, syndineans and core dinoflagellates are essential to interpreting the evolution of distinctive characters found in dinoflagellates, notably the state of the dinokaryon. In pursuit of this used ‘next-generation’ sequencing data acquired from two syndineans and their dinoflagellate hosts and another 8 novel datasets from core dinoflagellate taxa, in combination with existing data from P. marinus and O. marina, to develop basic taxon sampling for the dinoflagellate clade.
Here a specific category of protein-coding genes, the ribosomal proteins, was used to create a phylogeny of the dinoflagellate lineage, other alveolates and heterokonts. These proteins contribute the protein fraction of the ribosome (Nakao et al., 2004). Given that the rRNA may be the most commonly used sequence for nuclear molecular phylogeny, a logical progression would be to use ribosome associated proteins where orthology assignment and horizontal gene transfer may be less of a problem. There are approximately 70+ ribosomal genes in eukaryotes, with some diversification into gene families (Nakao et al., 2004). Many of these genes are quite small, conserved, and highly expressed, providing an easily obtainable fraction of orthologous genes for phylogeny, particularly from EST datasets already available in GenBank. For the core dinoflagellates many genes for ribosomal proteins exist as multiple duplicated gene copies, however most of the differences between gene copies are synonymous, and thus amino acid translations were used (Bachvaroff et al., 2009; Bachvaroff and Place, 2008; Kim et al., 2011).
2. Materials and methods
The photosynthetic dinoflagellates were cultured under autotrophic growth conditions (Table 1). The two parasitic dinoflagellates from the genus Amoebophrya used free-living photosynthetic hosts. One was grown on Karlodinium veneficum, and the second on Akashiwo sanguinea and so are referred to here as Amoebophrya sp. ex. Karlodinium veneficum and Amoebophrya sp. ex Akashiwo sanguinea (Gunderson et al., 1999, 2002). Host cultures of ∼10,000 hosts ml−1 were inoculated with ∼100,000 parasite dinospores ml−1. After incubation for 48–72 h, parasite dinospores were isolated from remaining hosts using nucleopore (Whatman, Piscataway, NJ) filters (5 μm for dinospores produced from K. veneficum host, and 8 μm for dinospores from A. sanguinea host) (Coats and Park, 2002; Park et al., 2002). Parasite cells were pelleted by centrifugation at 10,000g for 10 min. Total RNA was isolated using the RNAqueous kit (Ambion, Grand Island, NY) with LiCl precipitation as recommended by the manufacturer. The RNA quality was assessed on the Experion system (BioRad, Hercules, CA). Illumina (San Diego, CA) sequencing was performed by Macrogen with paired end reads of 76 or 100 bases (Table 1). The sequence data were assembled using Trinity for most datasets (Grabherr et al., 2011) or Abyss (Simpson et al., 2009). The choice of assembly program was arbitrary although Trinity required larger memory space computers and longer run times than Abyss. Hematodinium sp. ex Nephops norvegicus was cultured, its RNA extracted, sequenced and assembled as described in Jackson et al. 2012.
Table 1.
Culture and strain information for novel sequences used in this study.
| Binomial | Strain | Assembler | Temperature | Media | Sequencing type | Number of paired reads (millions) |
|---|---|---|---|---|---|---|
| Akashiwo sanguinea | Trinity | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 104a | |
| Amoebophrya sp. ex A. sanguinea | Trinity | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 104a | |
| Amoebophrya sp. ex K. veneficum | Trinity | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 76a | |
| Amphidinium carterae | CCMP1314 | Trinity | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 30 |
| Crypthecodinium cohnii | ATCC30340 | Newbler | 454 | |||
| Hematodinium sp. ex Nephrops norvegicus | Trinity, MIRA | 12 | NS/FCS | 454/Illumina 100 base paired end | ||
| Karlodinium veneficum | CCMP1974 | Trinity | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 76a |
| Polarella glacialis | CCMP1383 | Trinity | 4 | L1 @ 15 ppt | Illumina 76 base paired end | 34 |
| Prorocentrum sp. | CCMP3122 | Abyss | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 55 |
| Prorocentrum hoffmannianum | CCMP683 | Abyss | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 50 |
| Prorocentrum micans | CCMP1589 | Abyss | 20 | L1 @ 15 ppt | Illumina 100 base paired end | 50 |
Combined sequencing of host and parasite.
2.1. Assembling orthologous genes
A non-composite strategy was used in this study. Data from individual studies, strains and species were treated as individual taxa. Sequences were downloaded from GenBank using the species-specific taxonomic identifier from refseq, nr, or db_est as appropriate (Supplemental Table T1) and formatted into blast databases, with one database for each species. Similarly, in-house assembled datasets were formatted into blast databases. All species within the heterokonts and apicomplexans with >1000 EST sequences in db_est or a comparable sized nucleotide dataset in the nr database were used.
Sequences for Nannochloropsis gaditana were downloaded from http://nannochloropsis.genomeprojectsolutions-databases.com. Sequences from recent publications based on 454 or Illumina sequencing of RNA from dinoflagellates were also downloaded and formatted into local databases for Symbiodinium spp. (Bayer et al., 2012), Alexandrium tamarense (Moustafa et al., 2010) and Lingulodinium polyedrum (Roy and Morse, 2012).
A manually curated set of Perkinsus marinus ribosomal proteins was used as a reference query against the individual species' blast databases. Sequences were gleaned from the individual taxa by combining blast search (using the ncbi blast+ suite) with sequence retrieval and translation as necessary using perl scripts. For the sequences from the two Amoebophrya host–parasite cultures a total of 10 sequences (if available) from each host–parasite system were harvested. The increased depth for host–parasite datasets was used to ensure both host and parasite versions of each gene were collected. For nucleotide sequences from nr or db_est, or the autotrophic dinoflagellates three sequences were collected (if available) followed by translation into amino acids using the blast hit reading frame and the appropriate genetic code. Amongst the study organisms the ciliates use an alternate translation of the codons TAA and TAG which are translated as Glutamine (Caron and Meyer, 1985). Thus for Ichthyopthiriius multifiliis, Anophryoides haemophila, Entodinium caudatum and Miamiensis avidis the ciliate genetic code was used. For Tetrahymena thermophila and Paramecium tetraurelia the NCBI ref_seq protein database was used so translation was not necessary (see Supplemental Table T1). For amino acid sequences from ref_seq only the best hit was retained. The resulting amino acid sequences were then aligned with clustalo version 1.0.3 (Sievers et al., 2011) using the full iteration option. The sequences were then inspected in Mesquite version 2.74 to confirm start and stop sites, and to combine overlapping partial or frame-shifted sequences into a single consensus sequence. The manual curation step often found potential contaminant sequences from parasitic species, but these sequences were retained for individual gene analysis. No attempt was made at this stage to assign sequences to host or parasite, although partial or fragmentary sequences were removed.
2.2. Phylogenetic analysis
Individual gene alignments were used in a preliminary analysis to screen for contaminants, particularly from EST datasets for parasitic species and to confirm orthology. For these preliminary analyses RAxML (Stamatakis, 2006) with the JTT model with Gamma correction was used with 100 bootstrap replicates. For some organellar ribosomal genes, manual annotation of mitochondrial, plastid and bacterial (contaminant) clades was required.
Selected individual alignments were then concatenated and the optimal amino acid substitution matrix for the concatenated dataset was determined by comparing the full suite of substitution matrices. A second round of individual gene analysis was then done using the optimal LG model. In this analysis any redundant gene copies from a single species were eliminated. Two criteria were used to remove duplicate gene copies: first the longest sequences were retained, deleting any fragmentary gene copies. Secondly, amongst the full-length sequences for an individual taxon the least divergent gene copy based on the phylogeny, was retained. Putative contaminant host sequences from EST datasets of parasitic taxa were identified based on aberrant placement in the single gene phylogenies and removed after confirmation by blast searches against the nr database. The host and parasite assignments of the sequences from Amoebophrya-infected dinoflagellates were made based on phylogeny. The simple heuristic of AT bias for Amoebophrya sp. ex K. veneficum was also used to confirm parasite sequences (Bachvaroff et al., 2009).
The dataset was also tested for putative contamination or cross contamination of paralogs by searching for identical gi numbers within the dataset. This process revealed that two copies of genes from the rpl7 gene family in Alveolates aligning to the same copy from the outgroup heterokonts. This was resolved by removing the duplicate heterokont sequences from the alignment (see below). The program gblocks was used to trim the dataset using default parameters except for the ‘with-half’ gaps option (Castresana, 2000). This program was designed to scan for contiguous blocks of aligned sequence and remove poorly aligned regions.
A total of 73 ribosomal proteins were concatenated of which five were mitochondrial ribosomal proteins. For core dinoflagellates 16 taxa were sampled, with five additional taxa including three syndinean parasites. Outgroups included 22 apicomplexans, eight ciliates and 25 heterokonts for a total of 76 taxa. For the final analysis of concatenated datasets the optimal LG model for amino acid substitution with Gamma correction and 500 bootstrap replicates was used.
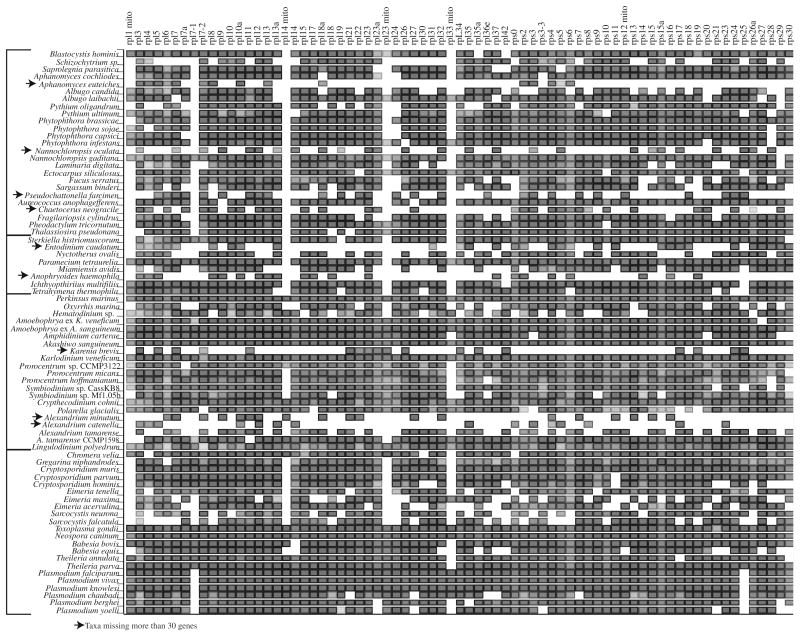
Near-complete gene sampling was available for many species (Fig. 1; Supplemental Table T1; Supplemental Fig. S1). For the 76 taxa in this study a mean of 59 genes were present, although excluding the nine taxa with poor coverage increased the average to 65 genes per taxon. The largest alignment was composed of 76 taxa and 15,487 amino acid sites (Treebase Accession 13997). Three additional datasets were constructed by subsampling the first dataset using either gblocks to remove sites or reducing taxa with missing data, or both. The second dataset had all taxa, but gblocks reduced the alignment to 10,127 sites. In the third dataset the 9 OTUs with the most missing data (30 or more genes absent, 67 taxa remained) were removed and the alignment was not trimmed with gblocks (15,392 sites). Finally, the fourth dataset was the result of gblocks trimming of the third dataset to 9597 sites, combining taxon and site reduction.
Fig. 1.
A representation of the alignment used for phylogenetic analysis. Along the y axis are the taxa used in the study in the same order as in the tree, with the genes along the x axis. The genes were sorted by name with the large ribosomal subunit associated genes on the left and the small subunit genes on the right. Nuclear encoded mitochondrial targeted genes are noted with the abbreviation ‘mito’. The fill color is proportional to the fraction of aligned characters, with darker colors representing more aligned characters. The nine taxa missing more than 30 genes are highlighted with arrows.
A set of minimally constrained trees were constructed for Approximately Unbiased (AU) testing (Table 2), and the most likely tree compatible with the constraint was constructed with RAxML (Shimodaira, 2000). The CONSEL package was used to calculate AU test values based on the site likelihoods from RAxML (Shimodaira and Hasegawa, 2001).
Table 2.
Results of AU tests.
| Dataset 1 | Dataset 2 | Dataset 3 | Dataset 4 | Constraint | ||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||
| Δ ln | AU | Δ ln | AU | Δ ln | AU | Δ ln | AU | |
| 0 | 0.819 | 0 | 0.859 | 0 | 0.779 | 0 | 0.721 | Best unconstrained tree |
| 55.6 | 0.004 | 54.7 | 0.005 | 54.3 | 0.008 | 51.2 | 0.008 | (P. marinus, O. marinus) |
| 234.9 | 0.001 | 210.8 | 8.00E–55 | 227.6 | 1.00E–62 | 216.2 | 7.00E–71 | (P. marinus, Syndineans) |
| 258.7 | 0.001 | 230.1 | 1.00E–10 | 254 | 0.001 | 233.7 | 1.00E–04 | (P. marinus) |
| 100.3 | 0.061 | 85.1 | 2.00E–04 | 99.2 | 4.00E–04 | 85.9 | 3.00E–04 | (O. marinus, Syndineans) |
| 100.7 | 0.067 | 83.9 | 7.00E–05 | 104.3 | 1.00E–39 | 89 | 3.00E–04 | (O. marinus, Core dinoflagellates) |
| 100.7 | 0.068 | 83.9 | 7.00E–05 | 104.3 | 1.00E–68 | 89 | 3.00E–04 | P. marinus (Syndineans (O. marinus and Core dinoflagellates)) |
| 196.1 | 3.00E–35 | 150 | 0.004 | 191 | 9.00E–108 | 150.1 | 4.00E–115 | (O. marinus, Hematodinium sp.) |
| 334.9 | 5.00E–157 | 268.4 | 0.001 | 330.4 | 2.00E–05 | 267.7 | 5.00E–67 | (P. marinus, Hematodinium sp.) |
| 122.4 | 2.00E–108 | 94.3 | 0.077 | 121.5 | 3.00E–04 | 85 | 2.00E–05 | (Hematodinium sp., Core dinoflagellates) |
| 40.4 | 0.183 | 21.6 | 0.306 | 46.6 | 0.135 | 27.2 | 0.279 | (C. velia, all dinoflagellates) |
| 21.3 | 0.369 | 12.9 | 0.474 | 25.3 | 0.292 | 25.1 | 0.247 | (G. niphandrodes (Apicomplexans)) |
| 260.9 | 7.00E–05 | 196.6 | 2.00E–05 | 255.1 | 0.001 | 203 | 2.00E–16 | (Cryptosporidium (Apicomplexans)) |
| 2.5 | 0.619 | 14.1 | 0.322 | 2.8 | 0.567 | 8.2 | 0.426 | (P. falciparum (P. knowlesi, P. vivax) (P. yoelli, P. berghei)) |
| 81.5 | 0.002 | 37.1 | 0.074 | 79.4 | 0.016 | 49.9 | 0.083 | (Diatoms (other photosynthetic heterokonts)) |
| 23.8 | 0.258 | 0 | 0.859 | 15 | 0.327 | 0 | 0.59 | (Nannochloropsis, Phaeophytes) |
3. Results
3.1. Orthology assignment
One rpl7 gene seems to have been duplicated in the alveolates when compared to the heterokonts. For this gene, independent assembly of alignments with rpl7-1 and rpl7-2 from P. marinus collected identical sequences for heterokonts with both P. marinus genes. When analyzed as single genes both rpl7-1 and 7-2 placed heterokont versions as the outgroup to alveolates. When both genes were combined in a single alignment rpl7-1 and rpl7-2 formed two distinct clades in alveolates, independently recapitulating the (Ciliate (Apicomplexan, Dinoflagellate)) phylogeny. For purposes of concatenation, the rpl7-1 gene copies were more closely related to heterokonts on the combined tree, and so heterokonts were removed from the rpl7-2 alignment.
3.2. Phylogenies
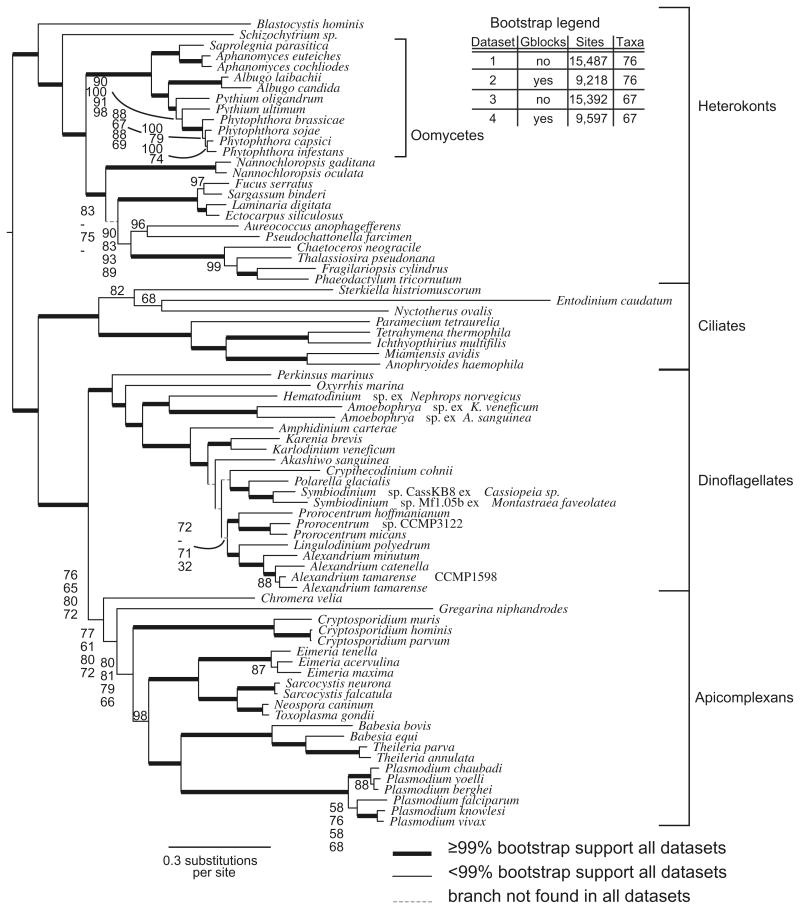
All phylogenies generated with the four datasets showed the alveolates as a monophyletic lineage with respect to the heterokonts. Within the alveolates, the apicomplexans and dinoflagellates were more closely related to each other than to the ciliates. In all four datasets, most branches consistently had 100% bootstrap support and the overall topology was mostly consistent (Fig. 2).
Fig. 2.
The most likely tree found by RAxML using the LG amino acid substitution matrix with gamma correction and 500 rapid bootstraps. Bold branches were found in 100% of bootstrap replicates. The dataset was partitioned both by using gblocks and by removing the 9 taxa with fewer aligned characters as shown on the legend in the figure. For branches with variable support in different partitions of the alignment (>5% difference in bootstrap values) the values are shown for all four different alignments.
3.2.1. Dinoflagellate phylogeny
Dinoflagellates resolved as a monophyletic clade grouped with the apicomplexans and then ciliates, all with good bootstrap support, and consistent with the accepted relationship of Alveolata. Within the broad dinoflagellate lineage, Perkinsus marinus was followed by Oxyrrhis marina, then the three syndinean taxa Hematodinium sp ex. Nephrops norvegicus, Amoebophrya sp. ex Karlodinium veneficum, and Amoebophrya sp. ex Akashiwo sanguinea formed one clade, and the 16 core dinoflagellates formed a second clade. Alternate topologies of P. marinus, O. marina, the syndineans, and core dinoflagellates were all rejected by the AU test (Table 2).
Consistent and well-supported features of the core dinoflagellate clade included the divergence of A. carterae from the base of the clade, and the monophyly of the suessialeans (Polarella glacialis and Symbiodinium sp.), Kareniaceae (Karenia brevis and Karlodinium veneficum), the genus Prorocentrum (3 spp.), and the gonyaulacoids composed of Alexandrium (3 spp.) together with Lingulodinium polyedrum.
Within the core dinoflagellates many branches were poorly supported or inconsistently found using the four different datasets, although the dataset with all taxa and gblocks trimming differed the most (dataset 2). For example, Prorocentrum and the gonyaulacoids (Alexandrium with Lingulodinium polyedrum) formed two well-supported monophyletic clades in all datasets. These two clades were grouped together with poor (32–72%) bootstrap support in three of the four datasets, but not in the dataset with all taxa and gblocks trimming. In this dataset, the gonyaulacoids were placed between Crypthecodinium cohnii and the remaining dinoflagellates with poor bootstrap support. Similarly, C. cohnii was placed with the suessaleans in three of the four datasets, albeit with poor support (up to 59%). The haptophyte-pigmented Kareniaceae (Karlodinium veneficum and Karenia brevis) were found between A. carterae and Akashiwo sanguinea, again with poor support (60–74%) in three of four datasets.
3.2.2. Apicomplexan phylogeny
Within the apicomplexans, Chromera velia was the first branching taxon with poor to moderate bootstrap support (71–86%), followed by Gregarina niphandrodes on a long branch, again with variable and reduced bootstrap support (67–85%). The AU test did not reject placement of C. velia with the dinoflagellate lineage, or a topology where G. niphandrodes was constrained outside of the apicomplexans including C. velia. The remaining features of the tree were consistently well supported except for the relationship of Eimeria acervulina with Eimeria maxima (85–87%), and the placement of Plasmodium falciparum with Plasmodium knowlesi and Plasmodium vivax (52–69%). The non-gregarine apicomplexans were well supported and well resolved with Cryptosporidium (3 spp.), followed by the Eimeria, Sarcocystis, and Toxoplasma clade next, and finally the clade composed of Babesia (2 spp.) plus Theileria (2 spp) next to the Plasmodium clade (6 spp.). Interestingly, the genus Theileria was embedded within the Babesia clade. Within Plasmodium the subgenus Vinckeia (P. chabaudi, yoelli, and berghei) was well supported, although the branch grouping P. yoelli with P. berghei varied from 81% to 96%. Intriguingly, P. falciparum (representing subgenus Laveriana) was placed with the subgenus Plasmodium (P. knowlesi and P. vivax), although the alternate topology of P. falciparum as the basal member of the Plasmodium clade was not rejected by the AU test.
3.2.3. Outgroup phylogeny
The heterokonts could be divided into two major lineages, the photosynthetic heterokonts and the oomycetes, with Schizochytrium sp. and Blastocystis hominis outside of these two lineages. Within heterokonts one branch was inconsistent with different analyses, with the deep-branching Nannochloropsis spp. placed with the phaeophytes when gblocks was used. This alternate topology was not rejected by the AU test. In addition gblocks trimming reduced bootstrap support by about 20% for two branches within the Phytophthora spp. clade, while increasing support for the deeper branch with Pythium ultimum.
4. Discussion
With advances in data availability, ‘genome-scale’ or phylogenomic studies have become possible. For species for which genome data remain unavailable due to genome size or other constraints, phylogenies can still be generated using a subset of the transcriptome data. Some phylogenetic studies employ a composite strategy where data from multiple species or studies are combined to represent specific groups (Burki et al., 2007; Parfrey et al., 2010), whereas here the data from individual studies, strains and species were treated as individual taxa. Ribosomal proteins are particularly accessible because these genes are well conserved, allowing for easy identification, homology assessment, and alignment. For these highly expressed and often short genes (<200 amino acids), full-length coding sequences can be gleaned from even small scale Sanger sequencing EST surveys. Within the dinoflagellates, and across the tree as a whole, gene sampling was very robust, with most taxa represented by 59 genes even when including relatively poorly sampled taxa. These short gene sequences provide relatively little phylogenetic resolution when analyzed individually, but can be concatenated to alleviate alignment length as a limiting factor. Even with the sharp increase in high-throughput sequencing that has occurred in recent years, relatively few taxa have been sequenced when compared to PCR-based single-gene datasets, and taxon sampling is certainly an area that could be improved. The data reported here for the dinoflagellate clade includes 11 novel datasets combined with a further 10 dinoflagellates from previously published surveys.
The tree agrees with generally accepted phylogeny where dinoflagellates grouped with apicomplexans, followed by ciliates and then heterokonts, a topology consistently recovered in multiple studies using both rRNA and protein-coding genes (Bachvaroff et al., 2011; Baldauf, 2003; Burki et al., 2007; Parfrey et al., 2010). Almost all of the branches on the tree are consistent and well supported even when the large alignment was trimmed into three subsampled datasets.
4.1. Improved phylogeny of basal dinoflagellates
The central objective of this study was to improve the understanding of dinoflagellate phylogeny and the novel data collection includes both syndineans and core dinoflagellates. Within the dinoflagellate lineage the present tree provides a well-resolved framework at the foundation of the clade, with significantly increased support relative to previous studies for relationships between P. marinus, O. marina, syndineans, and core dinoflagellates (Bachvaroff et al., 2011; Saldarriaga et al., 2003). Oxyrrhis marina is a clear outgroup to the syndineans and core dinoflagellates. In contrast to a previous smaller scale ribosomal protein phylogeny (Bachvaroff et al., 2011), the AU tests with a longer alignment and more taxa reject alternate topologies at the base of the dinoflagellate clade. The AU test rejects placing O. marina with the core dinoflagellates, or P. marinus with the syndineans. Interestingly, the AU test did not reject an alternate topology placing Chromera velia with dinoflagellates. Further, the phylogeny resolves ambiguous rRNA phylogenies of syndineans and core dinoflagellates. Syndinean rRNA sequences, mostly from environmental clone libraries, did not resolve the branching order of the different ‘Marine Alveolate Group’ (MAG) clades I–VIII and the core dinoflagellates (Bachvaroff et al., 2012; Guillou et al., 2008). Here, the syndineans representing Marine Alveolate clades II (Amoebophrya) and IV (Hematodinium) form a distinct monophyletic lineage sister to the core dinoflagellates.
Previous core dinoflagellate trees based on rRNA suffer from poor bootstrap support and conflict between analytic methods, a result that is completely consistent with the results from the ribosomal proteins presented here (Bråte et al., 2010; Coats et al., 2010; Saldarriaga et al., 2001; Shalchian-Tabrizi et al., 2006; Jørgensen et al., 2004; Hoppenrath and Leander, 2010; Logares et al., 2007; Okamoto et al., 2012; Murray et al., 2005). The most strongly supported and consistent features within the core dinoflagellates in the present study have also been seen in rRNA phylogenies. For example the gonyaulacoids (Alexandrium and Gonyaulax) are monophyletic with both rRNA and ribosomal protein trees. The Suessiales including Symbiodinium sp. and Polarella glacialis are found in both the rRNA and the protein-coding gene trees. Monophyly of the Kareniaceae is often poorly supported in the rRNA phylogenies cited above, which is surprising because these dinoflagellates share a tertiary plastid of haptophyte origin (Tengs et al., 2000). The two Kareniaceae are monophyletic with good support in the present analysis. Similarly the morphologically distinct genus Prorocentrum, although not recovered on SSU trees is monophyletic in this study. The divergence of A. carterae from the remaining core dinoflagellates was a feature first found with LSU trees (Jørgensen et al., 2004) and reinforced by protein-coding genes (Zhang et al., 2007). Within the core dinoflagellates the position of A. carterae suggests thecate dinoflagellates arose from an athecate ancestor (Orr et al., 2012), although multiple groups of thecate and athecate dinoflagellates were not sampled here. (Bachvaroff et al., 2009; Bachvaroff and Place, 2008; Kim et al., 2011).
4.2. Different permutations of the data
Gblocks prefilters rapidly changing or poorly sampled sites and excludes them from the analysis, but likelihood parameters for site-to-site rate variation account for rapidly changing sites. In the context of this study, gblocks trimmed alignments did not consistently improve bootstrap support, with values shifted up in some parts of the tree and down in others. Within genera Pythium and Phytopthera, previously shown to be paraphyletic (Villa et al., 2006; Runge et al., 2011), large changes in bootstrap values occurred with gblocks trimming. The short branches between the four Phytopthora spp. had much lower bootstrap support with gblocks trimming, although the next branch with Pythium ultimum had increased support. This result suggests more variable or poorly sampled regions help distinguish the subtly different Phytopthora spp. but may introduce inconsistency in bootstrap support at the next deeper branch.
This becomes important within the core dinoflagellates where the combination of the gonyaulacoids and Prorocentrum spp. is very sensitive to the gblocks trimming. Only datasets without gblocks trimming moderately supported this clade (69–72%) and when gblocks was used the support fell, or the branch was not observed. The sites removed by gblocks may be important for resolving core dinoflagellate relationships, but in the global context of the alignment such sites were excluded by gblocks. An additional confounding factor is the gene duplication and potential for sequencing error in EST datasets, which can mask true orthologous gene copies (Kim et al., 2011) For example, the two datasets for Alexandrium tamarense were collected by traditional EST and 454 methods from the same strain, yet they appear to be distinct taxa on the tree. Likely this represents a combination of sequencing, assembly, and alignment error potentially masking the most similar or identical gene copies and instead substituting slightly variant gene copies. Similarly, two strains of Symbiodinium are distinct on the phylogeny. Thus, while additional taxon sampling is an obvious next step in resolving the core dinoflagellate topology, methods to account for subtly variant copies will also be required, particularly when comparing closely related taxa (Bachvaroff and Place, 2008; Kim et al., 2011). Even more distinct were the differences between the two Amoebophrya strains, where previous studies of rRNA previously suggested multiple species might be present (Gunderson et al., 2002), and gene duplication is not as common as in core dinoflagellates (Bachvaroff et al., 2009).
4.3. Additional novel features of the phylogeny
Despite many points of agreement, there are several points on the tree that differ from previous phylogenies. For example within the heterokonts, the Khakista group combining diatoms, Aureococcus anophagefferens, and Pseudochattonella farcimen, are relatively well supported (81–95%) (Edvardsen et al., 2007). This result contrasts strongly with the study of Riisberg et al. 2009 where the diatoms were at the base of the heterokonts.
A controversial portion of the tree within the apicomplexans would be the placement of Plasmodium falciparum within the subgenus Plasmodium, albeit with poor bootstrap support. In previous studies with deeper taxon sampling but shorter alignments, P. falciparum was placed outside of the two subgenera Plasmodium (P. knowlesi, and P. vivax) and Vinkeia (P. chabaudi, P. yoelli, and P. berghei) (Martinsen et al., 2008; Duval et al., 2010). However, in this tree P. falciparum, representing the Laveriana subgenus is found within subgenus Plasmodium, implying a closer relationship of P. falciparum with the other primate malarias.
4.4. Conclusions
There are two parallel developments that clarify character evolution in the dinoflagellate lineage. First, broader taxon sampling and longer alignments have improved the resolution and support for molecular phylogenies. Second, the actual character states of many species have been more precisely described. For example, the previous analysis of 21 ribosomal proteins from dinoflagellates was not able to clearly place O. marina, P. marinus, and Amoebophrya sp. in relation to the outgroup and core dinoflagellates (Bachvaroff et al., 2011), whereas with the addition of both more taxa and longer alignments now these branches are clearly resolved.
Nuclear characters were used to divide the dinoflagellates two major clades, the syndineans and core dinoflagellates (Adl et al., 2005). The core dinoflagellates share aberrant nuclear characters including high DNA content, numerous chromosomes condensed during interphase, and large-scale gene duplication. The syndineans have been considered more like typical eukaryotes because of reduced chromosome number and no obvious gene amplification (Bachvaroff et al., 2009; Ris and Kubai, 1974). Syndinean nuclei also stain with alkaline fast green, suggesting larger basic protein to DNA ratios than core dinoflagellates (Cachon and Cachon, 1987). These features were explicit or implicit evidence that syndinean nuclei were more like typical eukaryotes forming an outgroup to the core dinoflagellates. Rooted in this manner, the monophyletic core dinoflagellates (often termed dinokaryotes), appeared to share a suite of derived nuclear characters (Adl et al., 2005; Saldarriaga et al., 2004; Wisecaver and Hackett, 2011; Lin, 2011). The character state of the Oxyrrhis marina nucleus, however, was more ambiguous, with a large genome (55.8 pg), duplicated genes, and alkaline fast green positive chromatin, deviating from the typical eukaryotic state and perhaps representing an intermediate dinokaryotic state (Sano and Kato, 2009). Our ribosomal protein phylogenies now firmly place O. marina outside of both the syndineans and core dinoflagellates, suggesting that remodeling of the nucleus commenced early in dinoflagellate radiation prior to the emergence of O. marina. Further, recent discovery of a specific protein (DVNP) that has apparently displaced major histone functions in dinoflagellates was shown to be present in the syndinean Hematodinium sp., the core dinoflagellates, and O. marina, but it is absent in P. marinus (Gornik et al., 2012). In Hematodinium bulk nucleosomal packing is absent, chromosomes are substantially condensed, and the DNA content is relatively large (4.5 pg, although less than O. marinus and many of the core dinoflagellates). Similarly, in the dinospore stage of Amoebophrya sp. ex A. sanguinea partially condensed interphase chromosomes were observed (Miller et al., 2012). The combined evidence suggests that major changes observed in the nuclei of dinoflagellates are shared between O. marina, syndineans and core dinoflagellates. This new interpretation is, in a sense, a return to the arguments of Chatton, where he determined the dinoflagellate affinity of many syndinean parasites based on nuclear and mitotic characteristics (Chatton, 1920).
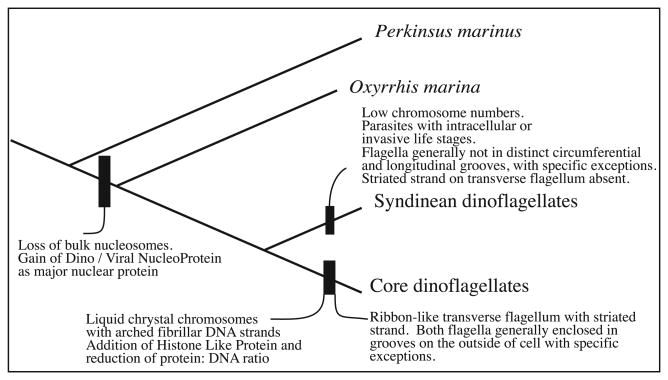
Remaining potential synapomorphies for core dinoflagellates include more detailed features, such as liquid crystal ‘arched fibrillar’ chromosomes, although this character is only strictly observable by transmission electron microscopy the refringent chromosomes are often visible with light microscopy (Bütschli, 1880) (Fig. 3). Perhaps arched fibrillar chromosome arrangement coincides with the histone-like proteins representing an additional protein substitution unique to core dinoflagellates beyond the DVNP shared by both core and syndinean dinoflagellates (Vernet et al. 1990, Chudnovsky et al. 2002).
Fig. 3.
A putative evolutionary scheme for the evolution of dinoflagellate characters. Evolution of nuclear and morphologic characters are described based on the ribosomal protein phylogeny.
The diagnostic dinokont morphology of the core dinoflagellate clade is based on the helical transverse and undulating trailing flagella both typically partly enclosed in grooves on the outside of the cell, although the genus Prorocentrum lacks such grooves. Oxyrrhis marina and most syndineans lack obvious grooves for flagella (Coats, 1988; Coats et al. 2012; Miller et al., 2012; Dodge and Crawford, 1971), although in the syndinean genera Ichthyodinium and Syndinium grooves are present in some life stages (Skovgaard et al., 2005, 2009). In core dinoflagellates the transverse flagellum is usually widened into a ribbon shape with an additional membrane-enclosed fiber called the striated strand (Dodge and Greuet, 1987). This type of transverse flagellum has not been observed in syndineans (Cachon and Cachon, 1970; Miller et al., 2012), although clearly more detailed investigations are required.
Supplementary Material
Acknowledgments
The authors would like to acknowledge D. Wayne Coats for generous support, useful discussion, and the Amoebophrya cultures. This work was funded by a NSF Assembling the Tree of Life Grant (EF-0629624) and an Australian Research Council Discovery Grant (DP1093395).
Footnotes
Appendix A. Supplementary material: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.ympev.2013.10.007.
References
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor FJR. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–451. doi: 10.1111/j.1550-7408.2005.00053.x. [DOI] [PubMed] [Google Scholar]
- Bachvaroff TR, Place AR. From stop to start: tandem gene arrangement, copy number and trans-splicing sites in the dinoflagellate Amphidinium carterae. PLoS One. 2008;3:e2929. doi: 10.1371/journal.pone.0002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvaroff TR, Place AR, Coats DW. expressed sequence tags from Amoebophrya sp. infecting Karlodinium veneficum: comparing host and parasite sequences. J Eukaryot Microbiol. 2009;56:531–541. doi: 10.1111/j.1550-7408.2009.00433.x. [DOI] [PubMed] [Google Scholar]
- Bachvaroff TR, Handy SM, Place AR, Delwiche CF. Alveolate phylogeny using ribosomal proteins. J Eukaryot Microbiol. 2011;58:223–233. doi: 10.1111/j.1550-7408.2011.00555.x. [DOI] [PubMed] [Google Scholar]
- Bachvaroff TR, Kim S, Guillou L, Delwiche CF, Coats DW. Diversity of the syndinian genus Euduboscquella based on single cell PCR. Appl Environ Microbiol. 2012 doi: 10.1128/AEM.06678-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf S. The deep roots of eukaryotes. Science. 2003;5626:1703–1706. doi: 10.1126/science.1085544. [DOI] [PubMed] [Google Scholar]
- Bayer T, Aranda M, Sunagawa S, Yum LK, Desalvo MK, Lindquist E, Coffroth MA, Voolstra CR, Medina M. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One. 2012;7:e35269. doi: 10.1371/journal.pone.0035269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bråte J, Logares R, Berney C, Ree DK, Klaveness D, Jakobsen KS, Shalchian-Tabrizi K. Freshwater Perkinsea and marine–freshwater colonizations revealed by pyrosequencing and phylogeny of environmental rDNA. ISME J. 2010;4:1144–1153. doi: 10.1038/ismej.2010.39. [DOI] [PubMed] [Google Scholar]
- Bütschli O Protozoa. Dr H G Bronns Klassen und Ordnungen des Tierreichs. Vol. 1. C.F. Winter'sche Verlagshandlung; Leipzig: 1880. pp. 906–1029. [Google Scholar]
- Burki F, Shalchian-Tabrizi K, Minge M, Skjaevelenad A, Nikolaev S, Jakobsen KS, Pawlowski J. Phylogenomics reshuffles the eukaryotic supergroups. PLoS One. 2007:e790. doi: 10.1371/journal.pone.0000790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon J, Cachon M. Ultrastructure des Amoebophryidae (Péridiniens Duboscquodinida) II Systèmes atractophoriens et microtubulaires; leur intervention dans la mitose. Protistologica. 1970;6:57–70. [Google Scholar]
- Cachon J, Cachon M. Parasitic dinoflagellates. In: Taylor FJR, editor. The Biology of Dinoflagellates. Vol. 21. Blackwell Scientific Publications; Oxford: 1987. pp. 571–610. [Google Scholar]
- Caron F, Meyer E. Does Paramecium primaurelia use a different genetic code in its macronucleus? Nature. 1985;314:185–188. doi: 10.1038/314185a0. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chatton E. Les Peridiniens parasites. Morphologie, reproduction, ethologie. Arch Zool Exp Gen. 1920;59:1–475. [Google Scholar]
- Coats DW. Duboscquella cachoni n. sp., a parasitic dinoflagellate lethal to its tintinine host Eutintinnus pectinis. J Protozool. 1988;35:607–617. [Google Scholar]
- Coats DW, Bachvaroff TR, Delwiche CF. Revision of the Duboscquellidae with description of Euduboscquella crenulata n. g., n. sp. (Dinoflagellata, Syndinea), an intracellular parasite of the ciliate Favella panamensis Kofoid and Campbell, 1929. J Eukaryot Microbiol. 2012;59 doi: 10.1111/j.1550-7408.2011.00588.x. [DOI] [PubMed] [Google Scholar]
- Coats DW, Park MG. Parasitism of photosynthetic dinoflagellates by three strains of Amoebophyra (Dinophyta): parasite survival, infectivity, generation time, and host specificity. J Phycol. 2002;38:520–528. [Google Scholar]
- Coats DW, Kim S, Bachvaroff TR, Handy SM, Delwiche CF. Tintinnophagus acutus n.g., n. sp. (Phylum Dinoflagellata), an Ectoparasite of the Ciliate Tintinnopsis cylindrica Daday 1887, and its relationship to Duboscquodinium collini Grassse 1952. J Eukaryot Microbiol. 2010;57:468–482. doi: 10.1111/j.1550-7408.2010.00504.x. [DOI] [PubMed] [Google Scholar]
- Chudnovsky Y, Li JF, Rizzo PJ, Hastings JW. Cloning, expression, and characterization of a histone-like protein from the marine dinoflagellate Lingulodinium polyedrum (Dinophyceae) J Phycol. 2002;38:543–550. [Google Scholar]
- Dodge JD, Crawford RM. Fine structure of the dinoflagellate Oxyrrhis marina. I. The general structure of the cell. Protistologica. 1971;7:295–304. [Google Scholar]
- Dodge JD, Greuet C. Dinoflagellate ultrastructure and complex organelles. In: Taylor FJR, editor. The Biology of Dinoflagellates. Blackwell Scientific Publications; Oxford, UK: 1987. pp. 92–142. [Google Scholar]
- Duval L, Fourment M, Nerrienet E, Rousset D, Sadeuh SA, Goodman SM, Andriaholinirina NV, Randrianarivelojosia M, Paul RE, Robert V, Ayala FJ, Ariey F. African apes as reservoirs of Plasmodium falciparum and the origin and diversification of the Laverania subgenus. Proc Natl Acad Sci USA. 2010;107:10561–10566. doi: 10.1073/pnas.1005435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardsen B, Eikrem W, Shalchian-Tabrizi K, Riisberg I, Johnsen G, Naustvoll L, Throndsen J. Verrucophora farcimen gen. et sp nov (Dictyochophyceae, Heterokonta) – a bloom-forming ichthyotoxic flagellate from the Skagerrak, Norway. J Phycol. 2007;43:1054–1070. [Google Scholar]
- Fensome RA, Taylor FJR, Norris G, Sarjeant WAS, Wharton DI, Williams GL. A classification of living and fossil dinoflagellates. Micropaleontology (Special publication number 7) 1993 [Google Scholar]
- Gajadhar AA, Marquardt WC, Hall R, Gunderson J, Ariztia-Carmona EV, Sogin ML. Ribosomal RNA sequences of Sarcocystis muris, Theileria annulata and Crypthecodinium cohnii reveal evolutionary relationships among apicomplexans, dinoflagellates, and ciliates. Mol Biochem Parasitol. 1991;45:147–154. doi: 10.1016/0166-6851(91)90036-6. [DOI] [PubMed] [Google Scholar]
- Gornik SG, Ford KL, Mulhern TD, Bacic A, McFadden GI, Waller RF. Loss of nucleosomal DNA condensation coincides with appearance of a novel nuclear protein in dinoflagellates. Current Biology. 2012;22:2303–2312. doi: 10.1016/j.cub.2012.10.036. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Viprey M, Chambouvet A, Welsh RM, Kirkham AR, Massana R, Scanlan DJ, Worden AZ. Widespread occurrence and genetic diversity of marine parasitoids belonging to Syndiniales (Alveolata) Environ Microbiol. 2008;10:3349–3365. doi: 10.1111/j.1462-2920.2008.01731.x. [DOI] [PubMed] [Google Scholar]
- Gunderson JH, Goss SH, Coats DW. The phylogenetic position of Amoebophrya sp. infecting Gymnodinium sanguineum. J Eukaryot Microbiol. 1999;46:194–197. doi: 10.1111/j.1550-7408.1999.tb04603.x. [DOI] [PubMed] [Google Scholar]
- Gunderson JH, John SA, Boman WC, Coats DW. Multiple strains of the parasitic dinoflagellate Amoebophrya exist in Chesapeake Bay. J Eukaryot Microbiol. 2002;49:469–474. doi: 10.1111/j.1550-7408.2002.tb00230.x. [DOI] [PubMed] [Google Scholar]
- Harada A, Ohtsuka S, Horiguchi T. Species of the parasitic genus Duboscquella are members of the enigmatic Marine Alveolate Group I. Protist. 2007;158:337–347. doi: 10.1016/j.protis.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Hoppenrath M, Leander BS. Dinoflagellate phylogeny as inferred from heat shock protein 90 and ribosomal gene sequences. PLoS One. 2010;5:e13220. doi: 10.1371/journal.pone.0013220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CJ, Gornik SG, Waller RF. The mitochondrial genome of the basal dinoflagellate Hematodinium sp.: Character evolution within the highly derived mitochondrial genomes of dinoflagellates. Genome Biology and Evolution. 2012;4:59–72. doi: 10.1093/gbe/evr122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen MF, Murray S, Daugbjerg N. Amphidinium revisited. I. Redefinition of Amphidinium (Dinophyceae) based on cladistic and molecular phylogenetic analysis. J Phycol. 2004;40:351–365. [Google Scholar]
- Keeling PJ. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol. 2013;64:583–607. doi: 10.1146/annurev-arplant-050312-120144. [DOI] [PubMed] [Google Scholar]
- Kim S, Bachvaroff TR, Handy SM, Delwiche CF. Dynamics of actin evolution in dinoflagellates. Mol Biol Evol. 2011;28:1469–1480. doi: 10.1093/molbev/msq332. [DOI] [PubMed] [Google Scholar]
- Leander BS, Kuvardina ON, Aleshin VV, Mylnikov AP, Keeling PJ. Molecular phylogeny and surface morphology of Colpodella edax (Alveolata): insights into the phagotrophic ancestry of apicomplexans. J Eukaryot Microbiol. 2003;50:334–340. doi: 10.1111/j.1550-7408.2003.tb00145.x. [DOI] [PubMed] [Google Scholar]
- Lin S. Genomic understanding of dinoflagellates. Res Microbiol. 2011;162:55–569. doi: 10.1016/j.resmic.2011.04.006. [DOI] [PubMed] [Google Scholar]
- Logares R, Shalchian-Tabrizi K, Boltovskoy A, Rengefors K. Extensive dinoflagellate phylogenies indicate infrequent marine–freshwater transitions. Mol Phylogenet Evol. 2007;45:887–903. doi: 10.1016/j.ympev.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 2008;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Miller JJ, Delwiche CF, Coats DW. Ultrastructure of Amoebophrya sp. and its changes during the course of infection. Protist. 2012;163:720–745. doi: 10.1016/j.protis.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Moore RB, Obornik M, Janouskovec J, Chrudimsky T, Vancova M, Green DH, Wright SM, Davies NW, Bolch CJ, Heimann K, Slapeta J, Hoegh-Guldberg O, Logsdon JM, Carter DA. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Moustafa A, Evans AN, Kulis DM, Hackett JD, Erdner DL, Anderson DM, Bhattacharya D. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS One. 2010;5:e9688. doi: 10.1371/journal.pone.0009688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray S, Jørgensen MF, Ho SYW, Patterson DJ, Jermlin LS. Improving the analysis of dinoflagellate phylogeny based on rDNA. Protist. 2005;156:269–286. doi: 10.1016/j.protis.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Nakao A, Yoshihama M, Kenmochi N. RPG: the ribosomal protein gene database. Nucleic Acids Res. 2004;32:D168–D170. doi: 10.1093/nar/gkh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Horak A, Keeling PJ. Description of two species of early branching dinoflagellates, Psammosa pacifica n.g., n. sp. and P. atlantica n. sp. PLoS One. 2012;7:e34900. doi: 10.1371/journal.pone.0034900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr RJ, Murray SA, Stüken A, Rhodes L, Jakobsen KS. When naked became armored: an eight-gene phylogeny reveals monophyletic origin of theca in dinoflagellates. PLoS One. 2012;7:e50004. doi: 10.1371/journal.pone.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Grant J, Tekle YI, Lasek-Nesselquist E, Morrison HG, Sogin ML, Patterson DJ, Katz LA. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59:518–533. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MG, Cooney SK, Yih W, Coats DW. Effects of two strains of the parasitic dinoflagellate Amoebophrya on growth, photosynthesis, light absorption, and quantum yield of bloom-forming dinoflagellates. Mar Ecol Prog Ser. 2002;227:281–292. [Google Scholar]
- Reece KS, Siddall ME, Burreson EM, Graves JE. Phylogenetic analysis of Perkinsus based on actin gene sequences. J Parasitol. 1997;83:417–423. [PubMed] [Google Scholar]
- Riisberg I, Orr RJS, Kluge R, Shalchian-Tabrizi K, Bowers HA, Patil V, Edvardsen B, Jakobsen KS. Seven gene phylogeny of heterokonts. Protist. 2009;160:191–204. doi: 10.1016/j.protis.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Ris H, Kubai DF. An unusual mitotic mechanism in the parasitic protozoan Syndinium sp. J Cell Biol. 1974;60:702–720. doi: 10.1083/jcb.60.3.702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Morse D. A full suite of histone and histone modifying genes are transcribed in the dinoflagellate Lingulodinium. PLoS One. 2012;7:e34340. doi: 10.1371/journal.pone.0034340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge F, Telle S, Ploch S, Savory E, Day B, Sharma R, Thines M. The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in Phytophthora. IMA Fungus. 2011;2:163–171. doi: 10.5598/imafungus.2011.02.02.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldarriaga JF, Taylor FJ, Keeling PJ, Cavalier-Smith T. Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J Mol Evol. 2001;53:204–213. doi: 10.1007/s002390010210. [DOI] [PubMed] [Google Scholar]
- Saldarriaga JF, McEwan ML, Fast NM, Taylor FJR, Keeling PJ. Multiple protein phylogenies show that Oxyrrhis marina and Perkinsus marinus are early branches of the dinoflagellate lineage. IJSEM. 2003;53:355–365. doi: 10.1099/ijs.0.02328-0. [DOI] [PubMed] [Google Scholar]
- Saldarriaga JF, Taylor FJRM, Cavalier-Smith T, Menden-Deuer S, Keeling PJ. Molecular data and the evolutionary history of dinoflagellates. Eur J Protistol. 2004;40:85–111. [Google Scholar]
- Sano J, Kato K. Localization and copy number of the protein-coding genes actin, α-tubulin, and hsp90 in the nucleus of a primitive dinoflagellate, Oxyrrhis marina. Zool Sci. 2009;26:745–753. doi: 10.2108/zsj.26.745. [DOI] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Skanseng M, Ronquist F, Klaveness D, Bachvaroff TR, Delwiche CF, Botnen A, Tengs T, Jakobsen KS. Heterotachy processes in rhodophyte-derived second hand plastid genes: implications for addressing the origin and evolution of dinoflagellate plastids. Mol Biol Evol. 2006;23:1504–1515. doi: 10.1093/molbev/msl011. [DOI] [PubMed] [Google Scholar]
- Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst Biol. 2000;51 doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. CONSEL: for assessing the confidence of phylogenetic tree selection. Bioinformatics. 2001;17:1246–1247. doi: 10.1093/bioinformatics/17.12.1246. [DOI] [PubMed] [Google Scholar]
- Shoguchi E, Shinzato C, Kawashima T, Gyoja F, Mungpakdee S, Koyanagi R, Takeuchi T, Hisata K, Tanaka M, Fujiwara M, Hamada M, Seidi A, Fujie M, Usami T, Goto H, Yamasaki S, Arakaki N, Suzuki Y, Sugano S, Toyoda A, Kuroki Y, Fujiyama A, Medina M, Coffroth MA, Bhattacharya D, Satoh N. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol: CB. 2013;23:1399–1408. doi: 10.1016/j.cub.2013.05.062. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skovgaard A, Massana R, Balague V, Saiz E. Phylogenetic position of the copepod-infesting parasite Syndinium turbo (Dinoflagellata, Syndinea) Protist. 2005;156:413–423. doi: 10.1016/j.protis.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Skovgaard A, Meneses I, Angélico MM. Identifying the lethal fish egg parasite Ichthyodinium chabelardi as a member of Marine Alveolate Group I. Environ Microbiol. 2009;11:2030–2041. doi: 10.1111/j.1462-2920.2009.01924.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Taylor FJR. Phylum Dinoflagellata. In: Margulis L, Corliss JO, Melkonian M, Chapman DJ, editors. Handbook of Protoctista. Jones and Bartlett; Boston: 1989. pp. 419–437. [Google Scholar]
- Taylor FJR. Ultrastructure as a control for protistan molecular phylogeny. Am Nat. 1999;154:S125–S136. doi: 10.1086/303288. [DOI] [PubMed] [Google Scholar]
- Tengs T, Dahlberg OJ, Shalchian-Tabrizi K, Klaveness D, Rudi K, Delwiche CF, Jakobsen KS. Phylogenetic analyses indicate that the 19′ hexanoyloxyfucoxanthin-containing dinoflagellates have tertiary plastids of haptophyte origin. Mol Biol Evol. 2000;17:718–729. doi: 10.1093/oxfordjournals.molbev.a026350. [DOI] [PubMed] [Google Scholar]
- Vernet G, Sala-Rovira M, Maeder M, Jacques F, Herzog M. Basic nuclear proteins of the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta): two-dimensional electrophoresis and DNA-binding properties. Biochimica et Biophysica Acta. 1990;1048:281–289. doi: 10.1016/0167-4781(90)90068-d. [DOI] [PubMed] [Google Scholar]
- Villa NO, Kageyama K, Asano T, Suga H. Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and beta-tubulin gene sequences. Mycologia. 2006;98:410–422. doi: 10.3852/mycologia.98.3.410. [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Hackett JD. Dinoflagellate genome evolution. Annu Rev Microbiol. 2011;65:369–387. doi: 10.1146/annurev-micro-090110-102841. [DOI] [PubMed] [Google Scholar]
- World Health Organization. World Malaria Report. Geneva Switzerland: 2010. http://www.who.int/malaria/world_malaria_report_2010/en/index.html) [Google Scholar]
- Zhang H, Bhattacharya D, Lin S. A three-gene dinoflagellate phylogeny suggests monophyly of Prorocentrales and a basal position for Amphidinium and Heterocapsa. J Mol Evol. 2007;65:463–474. doi: 10.1007/s00239-007-9038-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.