Abstract
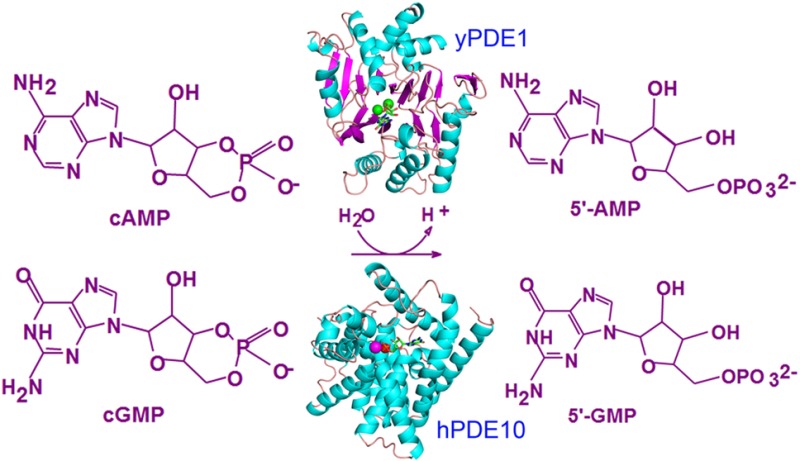
Cyclic nucleotide phosphodiesterases (PDEs) decompose second messengers cAMP and cGMP that play critical roles in many physiological processes. PDE1 of Saccharomyces cerevisiae has been subcloned and expressed in Escherichia coli. Recombinant yPDE1 has a KM of 110 μM and a kcat of 16.9 s–1 for cAMP and a KM of 105 μM and a kcat of 11.8 s–1 for cGMP. Thus, the specificity constant (kcat/KMcAMP)/(kcat/KMcGMP) of 1.4 indicates a dual specificity of yPDE1 for hydrolysis of both cAMP and cGMP. The crystal structures of unliganded yPDE1 and its complex with GMP at 1.31 Å resolution reveal a new structural folding that is different from those of human PDEs but is partially similar to that of some other metalloenzymes such as metallo-β-lactamase. In spite of their different structures and divalent metals, yPDE1 and human PDEs may share a common mechanism for hydrolysis of cAMP and cGMP.
Efficient integration of extracellular and intracellular signals is required to maintain adaptive cellular functions. The signaling systems of cyclic adenosine and guanosine 3′,5′-monophosphate (cAMP and cGMP, respectively) represent the earliest identified signaling pathways in the regulation of a vast number of critical physiological processes, including visual transduction, cell proliferation and differentiation, gene expression, inflammation, apoptosis, steroidogenesis, insulin secretion, glycogen synthesis, glycogenolysis, lipogenesis, and lipolysis.1−3 Cyclic nucleotide phosphodiesterases (PDEs) play important roles in these physiological processes with their ability to decompose cellular cAMP and cGMP.4−6 Inhibitors of human PDEs have been widely studied as therapeutics for the treatment of diseases, such as central nerve system diseases and cardiomyocyte hypertrophy.7−9 A well-known example of this drug class is the PDE5 inhibitor sildenafil (Viagra) that has been approved for the treatment of male erectile dysfunction and pulmonary hypertension.10,11
The superfamily of PDE can be categorized into two main classes of nonhomologous proteins that have been known to specifically hydrolyze cAMP and cGMP, although a third class of PDE was mentioned but not fully characterized.12 All PDEs found in mammals and flies belong to class I, while yeast and protozoans contain classes I and II PDEs. The conserved catalytic domain of class I PDEs consists of ∼300 amino acids and contains two divalent metal ions, typically zinc and magnesium/manganese, to facilitate the catalysis.13 The yeast genome contains two types of PDEs: yPDE1 and yPDE2. Yeast PDE2 is a member of the class I PDE superfamily and shares a similar folding of its catalytic domain with human PDEs, whereas yPDE1 belongs to the class II PDE superfamily and uses two zinc ions for its catalysis.14 The sequence alignment suggests that yPDE1 has a structural folding different from that of class I PDEs but shares a partially common pattern with other metalloproteins such as metallo-β-lactamase (MBL).15 Because no three-dimensional structure is available for any member of class II PDEs, how yPDE1 is evolutionarily related to class I PDEs and other metalloenzymes remains a puzzle.
The signaling of cAMP in yeast has been shown to play critical roles in physiological processes, including metabolism, cell wall biosynthesis, cell growth, and mating.16−23 Overexpression of the PDE2 gene enhances the tolerance of yeast to oxidative and ethanol stress for survival.24−26 Both yPDE1 and yPDE2 appear to participate in the cAMP signaling pathway, although yPDE2 shows high affinity with a KM of 0.17–1.0 μM for cAMP,27−30 and yPDE1 has a low affinity with a KM of 100–150 μM.16,31,32 In comparison with numerous publications about the roles of cAMP in the physiological processes of yeast, the cGMP signaling pathway has not been well studied, although it was reported that cGMP activated cAMP-dependent protein kinase of Pichia pastoris yeast.33 There is only one report showing that yPDE1 from Candida albicans has very weak cGMP activity with a KM of 250 μM and a Vmax of 0.044 μmol mg–1 min–1, in comparison with a KM of 490 μM and a Vmax of 1.17 μmol mg–1 min–1 for cAMP.34 Thus, physiological roles of the cGMP signaling pathway in yeast remain illusive. This paper reports the enzymatic properties of full length yPDE1 and shows that yPDE1 has a dual activity on hydrolysis of both cAMP and cGMP with similar enzymatic efficacies. The crystal structure of yPDE1 at 1.31 Å resolution reveals a new topological folding that is different from those of human PDEs.
Experimental Procedures
Subcloning and Protein Purification of Yeast PDE1
The cDNA of full length yPDE1 (residues 1–369) from bakers’ yeast Saccharomyces cerevisiae was subcloned into vector pET28a for expression. Oligonucleotide primers containing the NheI and EcoRI sites were designed and synthesized for amplification of yPDE1 by polymerase chain reaction. Amplified yPDE1 cDNA and expression vector pET28a were separately digested by restriction enzymes NheI and EcoRI, purified on an agarose gel, and then ligated by T4 DNA ligase. The resultant pET28-yPDE1 plasmid was confirmed by DNA sequencing and then transferred into Escherichia coli strain BL21 (CodonPlus) for expression. When the cells were grown in 2×YT culture medium at 37 °C to an OD600 of 0.7, 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to induce the overexpression of wild-type yPDE1 at 15 °C for 2 days.
The selenomethionyl mutant of yPDE1 was expressed in M9 minimal medium (6 g of Na2HPO4, 3 g of KH2PO4, 1 g of NH4Cl, 0.5 g of NaCl, 2 mM MgSO4, 4 g of glucose, 100 mg of ampicillin, 50 mg of selenomethionine, 100 mg of lysine, 100 mg of threonine, 100 mg of phenylalanine, 50 mg of leucine, 50 mg of isoleucine, and 50 mg of valine per liter). A colony of E. coli transferred with pET28-yPDE1 was grown in 10 mL of 2×YT medium at 37 °C overnight, centrifuged at 5000 rpm at room temperature, and then resuspended in 500 mL of prewarmed M9 medium at 37 °C. After the cells had been cultured at 37 °C to an OD600 of 0.4–0.5, 0.1 mM IPTG was added to induce the overexpression at 12 °C for ∼2 days.
The harvested cells were suspended in an extraction buffer of 20 mM Tris base (pH 8.0), 0.3 M NaCl, 15 mM imidazole, and 1 mM β-mercaptoethanol (β-ME) and lysed by being passed once through a Nano DeBee homogenizer. After centrifugation, the supernatant was loaded onto a 2.5 cm column containing 10 mL of Ni-nitriloacetic acid (NTA) agarose (QIAGEN). The column was washed with two buffers [20 mM Tris base (pH 8.0), 15 mM imidazole, and 1 mM β-ME with 50 or 300 mM NaCl], and then yPDE1 was eluted with 20 mM Tris base (pH 8.0), 50 mM NaCl, 150 mM imidazole, and 1 mM β-ME. After digestion with thrombin at room temperature for 2 h, the digested sample was loaded on a Q-Sepharose column (2.5 cm × 8 cm). The column was washed with ∼100 mL of 20 mM Tris base (pH 7.5), 50 mM NaCl, 1 mM β-ME, and 1 mM EDTA and eluted with the same buffer supplemented with 200 mM NaCl. The yPDE1 protein was finally purified with a Superdex 200 column using a buffer of 20 mM Tris base (pH 7.5), 1 mM β-ME, 1 mM EDTA, and 50 mM NaCl. The combined yPDE1 fractions were concentrated to 6–10 mg/mL and stored in −80 °C for use.
Enzymatic Assay
The enzymatic activity was assayed using [3H]cAMP or [3H]cGMP as a substrate, as previously reported for human PDE.35 Briefly, yPDE1 was incubated in a reaction mixture of 50 mM Tris-HCl (pH 7.5) and [3H]cAMP or [3H]cGMP (20–40K cpm/assay) at room temperature for 15 min. The reaction was terminated by the addition of 0.2 M ZnSO4. The reaction product [3H]AMP or [3H]GMP was precipitated out by addition of 0.2 N Ba(OH)2, while unreacted [3H]cAMP or [3H]cGMP remained in the supernatant. Radioactivity in the supernatant was measured by a liquid scintillation counter. The enzymatic properties were analyzed by the steady state kinetics. The Michaelis–Menten equation was used to obtain KM, Vmax, and kcat by nonlinear regression and also by an Eadie–Hofstee plot.36
The yPDE1 activity was tested in a buffer of 50 mM Tris-HCl (pH 7.5) and also this plain buffer with 10 mM MgCl2, 2 mM MnCl2, 0.01 mM ZnCl2, 1 mM EDTA, or 1 mM DTT. In addition, the yPDE1 activity was assayed after dialysis against a buffer of 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 1 mM EDTA, three times (1 h, 2 h, and overnight).
Crystallization, Structure Determination, and Comparison
The crystal of native yPDE1 (6 mg/mL) was grown by the hanging drop method against a buffer of 20 mM HEPES (pH 7.5), 5% PEG3350, 40 mM lithium sulfate, and 12% 2-methyl-2,4-pentanediol (MPD) at room temperature, or a similar buffer but with 15% glycerol and 20% MPD instead of 12% MPD. The native yPDE1 crystals diffracted to 1.31 Å resolution and are in space group C2221 with the following cell dimensions: a = 74 Å, b = 85 Å, and c = 130 Å (Table 1). The selenomethionyl yPDE1 (11 mg/mL) was crystallized at room temperature by the hanging drop method against a well buffer of 6% PEG3350, 30 mM sodium citrate (pH 5.6), 60 mM ammonium acetate, 75–100 mM ammonium sulfate, and 3% glycerol. The Se-yPDE1 crystals are in space group P21 with the following cell dimensions: a = 68 Å, b = 57 Å, c = 102 Å, and β = 106.7°. The yPDE1–GMP complex was prepared by soaking the native yPDE1 crystal in the crystallization buffer with 50 mM cGMP at room temperature for 2 days.
Table 1. Data Collection and Structural Refinement Statistics.
| unliganded yPDE1 | yPDE1–GMP | selenomethionyl yPDE1 | |
|---|---|---|---|
| Data Collection | |||
| space group | C2221 | C2221 | P21 |
| unit cell | |||
| a (Å) | 73.5 | 73.7 | 68.2 |
| b (Å) | 85.2 | 85.2 | 56.9 |
| c (Å) | 130.6 | 130.8 | 102.3 |
| β (deg) | 106.7 | ||
| wavelength (Å) | 1.075 | 1.075 | 0.97920 |
| resolution (Å) | 50–1.31 (1.33–1.31) | 50–1.31 (1.33–1.31) | 50–2.3 (2.38–2.3) |
| total no. of measurements | 1272718 | 1398700 | 241585 |
| no. of unique reflections | 96740 | 98268 | 31820 |
| completeness (%) | 98.4 (79.8) | 99.3 (88.0) | 95.1 (96.1) |
| average I/σ | 12.6 (4.9) | 10.1 (2.3) | 10.3 (3.5) |
| Rmerge | 0.084 (0.55) | 0.064 (0.81) | 0.095 (0.501) |
| Structural Refinement | |||
| R-factor/Rfree | 0.172/0.184 | 0.169/0.180 | |
| no. of reflections | 91848/4844 (5%) | 93309/4910 (5%) | |
| resolution (Å) | 15–1.31 | 15–1.31 | |
| rmsd | |||
| bond lengths (Å) | 0.008 | 0.008 | |
| bond angles (deg) | 1.2 | 1.3 | |
| average B factor (Å2) (no. of atoms) | |||
| protein | 18.4 (2914) | 18.7 (2914) | |
| MPD | 15.2 (8) | 19.7 (8) | |
| Zn | 13.6 (2) | 13.2 (2) | |
| SO4 | 17.8 (5) | ||
| GMP | 35.9 (23) | ||
| water | 25.3 (245) | 24.7 (215) | |
The single-wavelength anomalous diffraction (SAD) data for selenomethionyl yPDE1 were collected at the k-edge of Se (λ = 0.97920), and the data of native yPDE1 and its complex with cGMP were collected at λ = 1.075 on beamline X29 of the Brookhaven National Laboratory (Table 1). All the data were processed with HKL.37 Three Se sites were found for the SAD data with SHELX and used to phase the structure with PHENIX.38 Improvement of the SAD phases yielded a figure of merit of 0.66 for the data at 2.3 Å resolution, and the Fo map clearly revealed the trace of the yPDE1 structure. The atomic model was built with O39 and refined with REFMAC.40
The yPDE1 structure was compared with structures in the Protein Data Bank by the online program Dali (http://www.ebi.ac.uk/Tools/structure/dalilite). The results were ranked by Dali’s Z score and rmsd.41 A structure with a Z score of >2.0 is assumed thought to be partially similar with that of yPDE1, and a higher Z score indicates better similarity.
Results
Yeast PDE1 Hydrolyzes both cAMP and cGMP
The native bakers’ yeast yPDE1 was shown to contain two zinc ions per molecule by an atomic absorption spectrometer.16 In a plain assay buffer without divalent metals, our recombinant yPDE1 shows similar enzymatic efficacy for hydrolysis of cAMP and cGMP: KM of 110 μM and kcat of 16.9 s–1 for cAMP and KM of 105 μM and kcat of 11.8 s–1 for cGMP (Table 2). Thus, the enzymatic efficacy kcat/KM of yPDE1 is 0.153 and 0.113 for cAMP and cGMP, respectively. The specificity constant (kcat/KMcAMP)/(kcat/KMcGMP) of 1.4 suggests a dual activity of yPDE1 on hydrolysis of both cAMP and cGMP, with a slightly better efficacy on cAMP than cGMP. Our KM of yPDE1 for cAMP is comparable with the early report of KM values of 100–120 μM, but our yPDE1 enzyme is much more active, as shown by a Vmax at least 20-fold higher than those of the previously reported proteins.16,31,32
Table 2. Kinetics of Yeast PDE1 (1–369).
| cAMP |
cGMP |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| enzyme | ion in assay buffer | KM (μM) | Vmax (μmol mg–1 min–1) | kcat (s–1) | kcat/KM (s–1/μM) | KM (μM) | Vmax (μmol mg–1 min–1) | kcat (s–1) | kcat/KM (s–1/μM) | (kcat/KMcAMP)/(kcat/KMcGMP) |
| yPDE1 | 10 mM MgCl2 | 98.8 ± 8.5 | 30.1 ± 0.7 | 21.0 ± 1.0 | 0.213 | 96.8 ± 8.1 | 21.5 ± 0.5 | 15.1 ± 1.5 | 0.156 | 1.4 |
| yPDE1 | 0.01 mM ZnCl2 | 115.7 ± 7.4 | 24.8 ± 0.4 | 17.4 ± 1.1 | 0.150 | 104.8 ± 13.7 | 15.4 ± 0.5 | 10.8 ± 0.2 | 0.103 | 1.5 |
| yPDE1 | 2 mM MnCl2 | 117.5 ± 16.8 | 16.8 ± 0.6 | 11.8 ± 0.9 | 0.100 | 191.3 ± 32.3 | 20.4 ± 1.3 | 14.3 ± 2.0 | 0.075 | 1.3 |
| yPDE1 | no ion | 110.1 ± 14.2 | 24.1 ± 0.8 | 16.9 ± 1.3 | 0.153 | 104.7 ± 10.2 | 16.9 ± 0.4 | 11.8 ± 1.0 | 0.113 | 1.4 |
| yPDE1a | no ion | 81.2 ± 7.4 | 20.5 ± 0.5 | 14.3 ± 0.4 | 0.176 | 147.4 ± 20.0 | 17.2 ± 0.7 | 12.1 ± 1.6 | 0.082 | 2.1 |
| yPDE1a | no ion with 1 mM EDTA | 77.3 ± 12.0 | 15.8 ± 0.6 | 11.1 ± 1.5 | 0.144 | 153.9 ± 23.2 | 16.7 ± 0.7 | 11.7 ± 2.0 | 0.076 | 1.9 |
yPDE1 protein was dialyzed three times against a buffer of 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, and 1 mM EDTA (1 h, 2 h, and overnight).
Because human PDEs require magnesium or manganese for maximization of their activities, a few types of divalent metal ions were added to the assay buffer to test if they promote the activity of yPDE1. In summary, addition of 10 μM ZnCl2, 2 mM MnCl2, or 10 mM MgCl2 to the assay buffer did not significantly promote or inhibit the catalytic activities of our recombinant yPDE1 upon hydrolysis of cAMP and cGMP (Table 2). These results are consistent with the early observations that yPDE1 from S. cerevisiae or C. albicans was not sensitive to the divalent metals.31,34 In addition, the dialysis of native yPDE1 against 1 mM EDTA overnight did not change the catalytic efficacy (kcat/KM) for cAMP but slightly decreased kcat/KM for cGMP (Table 2), implying that the dialysis was not efficient for the extraction of the zinc ions that coordinate with four yPDE1 residues. On the other hand, DTT significantly inhibits our yPDE1 activity, in contrast to the fact that DTT promotes the activity of human PDEs.
Architecture of the yPDE1 Structure
The 369 amino acids of yPDE1 are folded into 16 β-stands, 10 α-helices, and two 310-helices (Figure 1A). The 16 β-strands can be divided into two sheets that form a sandwich and a hydrophobic core of the molecule. The center of the molecular core looks like a distorted β-barrel and is flanked by the helices. Two molecules of yPDE1 are tightly associated into a dimer (Figure 1B,C). For the massive interactions between two monomers, we assume that the dimeric form of yPDE1 is a biologic unit for its function. This argument is supported by the gel filtration experiment in which the recombinant yPDE1 protein appears to be a dimer in the solution (data not shown) and also by the structural assembly in which two monomers jointly form the active site of yPDE1 (Figure 1).
Figure 1.

Structure of yPDE1. (A) Ribbon diagram of monomeric yPDE1. The linker between H1 and B4 (residues 55–63) was disordered. (B) Dimer of yPDE1. GMP is shown as sticks. Zincs are represented by red spheres. (C) Surface presentation of the yPDE1 dimer. Molecule A (top) is colored as follows: white for carbon, blue for nitrogen, and red for oxygen. Molecule B is colored cyan.
The active site pocket of yPDE1 can be divided into three portions. The bottom part of the pocket is composed of residues His128, His130, Asp132, His133, His213, Asp244, and His326, which are entirely contributed by molecule A of the dimer. Residues Glu289, Tyr292, His294, and Lys328 of molecule A constitute one wall of the catalytic pocket, while Pro142, Tyr145, Trp175, Pro176, and Leu178 of molecule B form another wall of the pocket (Figure 2A). Two divalent metal ions sit in the bottom of the pocket, each of which forms four coordinations with His128, His130, His213, and Asp244, or with Asp132, His133, Asp244, and His326 of the same molecule of the dimer (Figure 2B). In addition, a sulfate ion was found to coordinate with the two zinc ions in the structure of unliganded yPDE1. This sulfate must come from the excess Li2SO4 (40 mM) in the crystallization buffer, and its occupancy is supported by the clear electron density and a small B factor of 17.8 Å2 comparable with the average value of 18.4 Å2 for protein atoms. Also, a water molecule or hydroxide ion bridges both metal ions, to form an octahedral configuration of each ion (Figure 2B). Because zinc was identified by atomic absorption spectrometry,31 two zinc ions were assigned for the structural refinement without further verification. This assignment is supported by the comparable B factors of the zinc ions (13.6 Å2) with the overall average B factor of 18.4 Å2 for protein atoms (Table 1).
Figure 2.

Active site of yPDE1. (A) GMP (yellow sticks) binding pocket that is constituted with residues from both molecules A (top left, colored red, blue, and white) and B (bottom right, colored cyan). Dotted lines represent the coordination with zinc ions or hydrogen bonds between GMP and yPDE1. (B) Six coordinations of each zinc ion in an octahedral configuration. (C) Ribbon diagram of GMP binding. The blue mesh is the electron density of the Fo – Fc map that was calculated from the structure with omission of GMP and contoured at 2.5σ.
Binding of GMP to the yPDE1 Dimer
The unliganded yPDE1 crystals were soaked in 50 mM cGMP or 25 mM cAMP for a couple of days. Two data sets were collected from the soaked crystals to 1.31 Å resolution (Table 1). The Fo – Fc and 2Fo – Fc electron density maps reveal the definite binding and conformation of GMP in the cGMP-soaked crystal (Figure 2C). However, the structural refinement shows a B factor of 35.9 Å2 for GMP, which is significantly higher than the average B factor of 18.7 Å2 for protein atoms, suggesting its low occupancy. This should not be surprising because the product GMP is expected to weakly bind to yPDE1 on the basis of the low affinity of substrate cGMP (KM of 104 μM). On the other hand, the cAMP-soaked crystal shows some electron density at the binding pocket, but no unique conformation can be identified.
The phosphate oxygens of GMP chelate with two zinc ions, form two hydrogen bonds with the side chains of His294 and Lys328 of molecule A, and also closely contact zinc binding residues His130, Asp132, His213, Asp244, and His326 of molecule A of the dimer (Figure 2). The ribose of GMP forms a hydrogen bond with the side chain of Lys328 of molecule A, and van der Waals contacts with residues Glu289 and His324 of molecule A, and Pro142 and Pro176 of molecule B. The guanine of GMP takes an anti configuration and is located in a hydrophobic environment that is composed of Val217 and Tyr292 of molecule A and Tyr145, Pro176, and Leu178 of molecule B.
Structural Comparison of yPDE1 with Other Metalloproteins
The online program Dali41 lists 591 metalloproteins and/or subunits in the Protein Data Bank (PDB), which have Z scores of >2 and thus possess similar topological folding, including ribonuclease Z, tRNA Z, metallo-β-lactamase (MBL), protein PhnP, cleavage and polyadenylation specificity factor, RNase J, metal-dependent hydrolase, human 5′-exonuclease Apollo, coenzyme PQQ synthesis protein B, and human glyoxalase II, although some of the protein structures contain manganese or ferric ions, instead of zinc. Dali gives the best Z score of 24 to ribonuclease Z (PDB entry 2CBN, rmsd of 2.9 Å for 245 of 306 total residues). However, two zinc ions in ribonuclease Z are not directly involved in RNA binding but apparently serve as the structural metals to stabilize the three-dimensional structure. Thus, the structural similarity between yPDE1 and ribonuclease Z might just mean a scaffold support for zinc binding without a functional relationship. Also, on the top of the Dali list is a MBL-like protein from Gram-negative Brucella melitensis [PDB entry 3MD7, Z score of 21.4, rmsd of 2.6 Å for 231 of total 270 residues (Figure 3)], although it contains two manganese ions as the catalytic metals. In the structure of the MBL-like protein, the low occupancy of GMP at the Mn binding pocket was identified when the compound was cocrystallized with GMP-PNP.42 The orientation of GMP in the MBL-like protein is the opposite of that of GMP in yPDE1, while both of them have the common phosphate interactions with the divalent metals (Figure 3). Considering the fact that both MBL-like protein and yPDE1 require a dimer for catalysis and have identical metal binding residues, yPDE1 might have an evolutionary relationship with the MBL-like protein. In short, the structural comparison described above may suggest that the zinc ions in yPDE1 may stabilize and facilitate the proper folding of the yPDE1 structure, in addition to their main role as the catalytic ions.
Figure 3.

Structural comparison. (A) Superimposition of yPDE1 (cyan ribbons) over a metallo-β-lactamase-like protein from Gram-negative B. melitensis (green). Two zinc ions of yPDE1 are shown as red spheres, while two manganese ions of the bacterium are drawn as blue spheres. GMP (pink) is from yPDE1, and GMP in the bacterial structure (yellow) shows an opposite orientation. (B) Detailed view of GMP binding. The metal binding residues are identical in the two proteins. The yPDE1 residues are labeled.
Discussion
A Putative Mechanism for the Catalysis of yPDE1
It has been well established that hydrolysis of a phosphodiester bond by class I PDEs is accomplished via a nucleophilic attack of the bridging water or hydroxide ion.43,44 However, it is unknown whether yPDE1 hydrolyzes cAMP and cGMP with the same catalytic mechanism as human PDEs. Historically, an inversion of configuration at phosphorus was proposed for the stereochemistry of the cAMP hydrolysis by PDE in 1979.45 Later, the crystal structures of PDE443 and PDE9-cGMP44 revealed a hydroxide ion (or a water molecule) that bridges the divalent metal ions may serve as the nucleophile to attack the phosphorus. This hydroxide ion may be activated by an aspartic or glutamic acid (Asp318 or Glu230 in PDE4D2). After formation of an intermediate covalent bond between the hydroxide ion and the phosphorus, a histidine (His160 in PDE4D2) may donate a proton to complete the hydrolysis of the phosphodiester bond. This model is consistent with the mechanism of many other zinc enzymes that use an aspartic acid as a general base to activate a water or hydroxide ion.46
Yeast PDE1 contains two zinc ions as the catalytic metals, instead of zinc and magnesium or manganese in human PDEs. The crystal structure reveals a hydroxide ion or water molecule that bridges two zinc ions of yPDE1, in a pattern similar to that seen in the case of human PDEs. This hydroxide ion forms a hydrogen bond with Asp132 and may thus be activated by Asp132 to initiate the nucleophilic attack (Figures 2B and 4). After the reaction intermediate forms, His294 that forms a hydrogen bond with the phosphate group of GMP in the crystal structure of yPDE1-GMP may serve as the proton donor to complete the conversion of cGMP to GMP. Therefore, class II yPDE1 may share a common catalytic mechanism with class I PDEs for the hydrolysis of cAMP and cGMP, although they possess different structures and divalent metals.
Figure 4.
Proposed mechanism for yPDE1 catalysis. The catalysis may contain the following steps. (A) The bridging hydroxide ion is activated by Asp132 and attacks the phosphorus of cGMP. (B) Formation of the intermediate of the reaction. (C) His294 donates a proton to the intermediate. (D) Completion of the reaction. This mechanism is assumed to apply to the catalysis of cAMP by yPDE1, too.
Implication of the Physiological Roles of yPDE1
Although class II PDEs are found in a diverse number of species, including S. cerevisiae, C. albicans, Schizosaccharomyces pombe, Cryptococcus neoformans, Dictyostelium discoideum, Leishmania mexicana, Trypanosoma brucei, Vibrio fischeri, and Trypanosoma cruzi,47 their physiological roles remain unclear. The low affinity of cAMP for yPDE1 renders it nonfunctional at the usual physiological concentration of cAMP. However, several studies have shown critical roles of yPDE1 under extreme environments or stresses. Deletion of yPDE1, but not yPDE2, resulted in a much higher level of accumulation of cAMP upon addition of glucose or intracellular acidification, suggesting a specific role of yPDE1 on agonist-induced cAMP signaling.48 Also, deletion of PDE1 in C. albicans changed the level of expression of genes, stress responses, cell wall and membrane biogenesis, adherence, and virulence, suggesting irreplaceable roles of class II PDEs in stress, adhesion, and virulence.49 Furthermore, the genetic interactions of PDE1 with GPA2, a Gα protein controlling yeast differentiation,50 caused synthetic defects in growth, morphogenesis, and responses to some stresses, suggesting that yPDE1 and Gpa2 modulate agonist-induced cAMP signaling.47
While the cAMP signaling pathway of yPDE1 appears to play a secondary role in yeast physiological processes, the function of the cGMP signaling pathway in yeast has not been well characterized. Most studies focused on the cAMP signaling in which yPDE1 and yPDE2 are involved,16−32 but only a few papers about the function of the cGMP signaling in yeast have been published.33,34 Because only the cAMP activity of the four yPDE2 genes was reported, it is unknown if they also have cGMP hydrolysis activities. Our study reveals for the first time that yPDE1 is capable of hydrolyzing both cAMP and cGMP with similar efficacies. However, it remains to be elucidated whether the cGMP activity of yPDE1 plays an independent role in yeast physiological processes or an auxiliary function to the cAMP signaling pathway.
Acknowledgments
We thank beamline X29 of the National Synchrotron Light Source for collection of the diffraction data.
Glossary
Abbreviations
- PDEs
phosphodiesterases
- cGMP
cyclic guanosine 3′,5′-monophosphate
- cAMP
cyclic adenosine 3′,5′-monophosphate
- β-ME
β-mercaptoethanol
- IPTG
isopropyl β-d-1-thiogalactopyranoside
- rmsd
root-mean-square deviation.
Accession Codes
The coordinates and structural factors of unliganded yPDE1 and its GMP complex have been deposited as Protein Data Bank entries 4OJV and 4OJX.
Author Contributions
Y.T., W.C., and M.H. contributed equally to this work.
This work was supported in part by National Institutes of Health Grant GM59791 to H.K., the National Natural Science Foundation of China (31291944, to Y.W.), and the Offices of Biological and Environmental Research and of Basic Energy Sciences of the U.S. Department of Energy and the National Center for Research Resources of the National Institutes of Health (H.R.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Conti M.; Beavo J. A. (2007) Biochemistry and physiology of cyclic nucleotide phosphodiesterases: Essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76, 481–511. [DOI] [PubMed] [Google Scholar]
- Zaccolo M.; Movsesian M. A. (2007) cAMP and cGMP signaling crosstalk: Role of phosphodiesterases and implications for cardiac pathophysiology. Circ. Res. 100, 1569–1578. [DOI] [PubMed] [Google Scholar]
- O’Neill J. S.; Maywood E. S.; Chesham J. E.; Takahashi J. S.; Hastings M. H. (2008) cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320, 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A. T.; Beavo J. (2006) Cyclic nucleotide phosphodiesterases: Molecular regulation to clinical use. Pharmacol. Rev. 58, 488–520. [DOI] [PubMed] [Google Scholar]
- Omori K.; Kotera J. (2007) Overview of PDEs and their regulation. Circ. Res. 100, 309–327. [DOI] [PubMed] [Google Scholar]
- Maurice D. H.; Ke H.; Ahmad F.; Wang Y.; Chung J.; Manganiello V. C. (2014) Advances in targeting cyclic nucleotide phosphodiesterases. Nat. Rev. Drug Discovery 13, 290–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menniti F. S.; Faraci W. S.; Schmidt C. J. (2006) Phosphodiesterases in the CNS: Targets for drug development. Nat. Rev. Drug Discovery 5, 660–670. [DOI] [PubMed] [Google Scholar]
- Miller C. L.; Oikawa M.; Cai Y.; Wojtovich A. P.; Nagel D. J.; Xu X.; Xu H.; Florio V.; Rybalkin S. D.; Beavo J. A.; Chen Y. F.; Li J. D.; Blaxall B. C.; Abe J.; Yan C. (2009) Role of Ca2+/calmodulin-stimulated cyclic nucleotide phosphodiesterase 1 in mediating cardiomyocyte hypertrophy. Circ. Res. 105, 956–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipworth B. J. (2005) Phosphodiesterase-4 inhibitors for asthma and chronic obstructive pulmonary disease. Lancet 365, 167–175. [DOI] [PubMed] [Google Scholar]
- Galie N.; Ghofrani H. A.; Torbicki A.; Barst R. J.; Rubin L. J.; Badesch D.; Fleming T.; Parpia T.; Burgess G.; Branzi A.; Grimminger F.; Kurzyna M.; Simonneau G. (2005) Sildenafil citrate therapy for pulmonary arterial hypertension. N. Engl. J. Med. 353, 2148–2157. [DOI] [PubMed] [Google Scholar]
- Rotella D. P. (2002) Phosphodiesterase 5 inhibitors: Current status and potential applications. Nat. Rev. Drug Discovery 1, 674–682. [DOI] [PubMed] [Google Scholar]
- Beavo J. A., Houslay M. D., and Francis S. H. (2007) Cyclic nucleotide phosphodiesterase superfamily. In Cyclic nucleotide phosphodiesterases in health and disease (Beavo J. A., Francis S. H., and Houslay M. D., Eds.) pp 3–17, CRC Press, Boca Raton, FL. [Google Scholar]
- Ke H.; Wang H. (2007) Crystal structures of phosphodiesterases and implications on substrate specificity and inhibitor selectivity. Curr. Top. Med. Chem. 7, 391–403. [DOI] [PubMed] [Google Scholar]
- Wentzinger L., and Seebeck T. (2007) Protozoal phosphodiesterases. In Cyclic nucleotide phosphodiesterases in health and disease (Beavo J. A., Francis S. H., and Houslay M. D., Eds.) pp 277–300, CRC Press, Boca Raton, FL. [Google Scholar]
- Bebrone C. (2007) Metallo-β-lactamases (classification, activity, genetic organization, structure, zinc coordination) and their superfamily. Biochem. Pharmacol. 74, 1686–1701. [DOI] [PubMed] [Google Scholar]
- Londesborough J.; Suoranta K. (1983) The zinc-containing high Km cyclic nucleotide phosphodiesterase of bakers’ yeast. J. Biol. Chem. 258, 2966–2972. [PubMed] [Google Scholar]
- Thevelein J. M.; Cauwenberg L.; Colombo S.; De Winde J. H.; Donation M.; Dumortier F.; Kraakman L.; Lemaire K.; Ma P.; Nauwelaers D.; Rolland F.; Teunissen A.; Van Dijck P.; Versele M.; Wera S.; Winderickx J. (2000) Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast. Enzyme Microb. Technol. 26, 819–825. [DOI] [PubMed] [Google Scholar]
- Jung W. H.; Stateva L. (2003) The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology 149, 2961–2976. [DOI] [PubMed] [Google Scholar]
- Jung W. H.; Warn P.; Ragni E.; Popolo L.; Nunn C. D.; Turner M. P.; Stateva L. (2005) Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast 22, 285–294. [DOI] [PubMed] [Google Scholar]
- Bahn Y. S.; Staab J.; Sundstrom P. (2003) Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol. Microbiol. 50, 391–409. [DOI] [PubMed] [Google Scholar]
- Wilson D.; Tutulan-Cunita A.; Jung W. H.; Hauser N. C.; Hernandez R.; Williamson T.; Piekarska K.; Rupp S.; Young T.; Stateva L. (2007) Deletion of the high affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol. Microbiol. 65, 841–856. [DOI] [PubMed] [Google Scholar]
- Wilson D.; Fiori A.; Brucker K. D.; Dijck P. V.; Stateva L. (2010) Candida albicans Pde1p and Gpa2p comprise a regulatory module mediating agonist-induced cAMP signalling and environmental adaptation. Fungal Genet. Biol. 47, 742–752. [DOI] [PubMed] [Google Scholar]
- Barsoum E.; Rajaei N.; Aström S. U. (2011) RAS/cyclic AMP and transcription factor Msn2 regulate mating and mating-type switching in the yeast Kluyveromyces lactis. Eukaryotic Cell 10, 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J. I.; Grant C. M.; Dawes I. W. (2005) The high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae is the major determinant of cAMP levels in stationary phase: Involvement of different branches of the Ras–cyclic AMP pathway in stress responses. Biochem. Biophys. Res. Commun. 327, 311–319. [DOI] [PubMed] [Google Scholar]
- Drobna E.; Gazdag Z.; Culakova H.; Dzugasova V.; Gbelska Y.; Pesti M.; Subik J. (2012) Overexpression of the YAP1, PDE2, and STB3 genes enhances the tolerance of yeast to oxidative stress induced by 7-chlorotetrazolo[5,1-c]benzo[1,2,4]triazine. FEMS Yeast Res. 12, 958–968. [DOI] [PubMed] [Google Scholar]
- Avrahami-Moyal L.; Braun S.; Engelberg D. (2012) Overexpression of PDE2 or SSD1-V in Saccharomyces cerevisiae W303-1A strain renders it ethanol-tolerant. FEMS Yeast Res. 12, 447–455. [DOI] [PubMed] [Google Scholar]
- Suoranta K.; Londesborough J. (1984) Purification of intact and nicked forms of a zinc-containing, Mg2+-dependent, low Km cyclic AMP phosphodiesterase from bakers’ yeast. J. Biol. Chem. 259, 6964–6971. [PubMed] [Google Scholar]
- Sass P.; Field J.; Nikawa J.; Toda T.; Wigler M. (1986) Cloning and characterization of the high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 83, 9303–9307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaeli T.; Bloom T. J.; Martins T.; Loughney K.; Ferguson K.; Riggs M.; Rodgers L.; Beavo J. A.; Wigler M. (1993) Isolation and characterization of a previously undetected human cAMP phosphodiesterase by complementation of cAMP phosphodiesterase-deficient Saccharomyces cerevisiae. J. Biol. Chem. 268, 12925–12932. [PubMed] [Google Scholar]
- Suoranta K.; Londesborough J. (1985) The specificity of yeast low-Km cyclic AMP phosphodiesterase towards free bivalent metal ions and the diastereoisomers of cyclic adenosine phosphorothioate. Biochem. J. 226, 897–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londesborough J.; Lukkari T. (1980) The pH and temperature dependence of the activity of the high K, Cyclic Nucleotide Phosphodiesterase of Bakers’ Yeast. J. Biol. Chem. 255, 9262–9267. [PubMed] [Google Scholar]
- Uno I.; Matsumotog K.; Ishikawa T. (1983) Characterization of a cyclic nucleotide phosphodiesterase-deficient mutant in yeast. J. Biol. Chem. 258, 3539–3542. [PubMed] [Google Scholar]
- Frajnt M.; Cytryñska M.; Jakubowicz T. (2003) The effect of cAMP and cGMP on the activity and substrate specificity of protein kinase A from methylotrophic yeast Pichia pastoris. Acta Biochim. Pol. 50, 1111–1118. [PubMed] [Google Scholar]
- Hoyer L. L.; Cieslinski L. B.; McLaughlin M. M.; Torphy T. J.; Shatzmanl A. R.; Livil G. P. (1994) A Candida albicans cyclic nucleotide phosphodiesterase: Cloning and expression in Saccharomyces cerevisiae and biochemical characterization of the recombinant enzyme. Microbiology 140, 1533–1542. [DOI] [PubMed] [Google Scholar]
- Wang H.; Liu Y.; Chen Y.; Robinson H.; Ke H. (2005) Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J. Biol. Chem. 280, 30949–30955. [DOI] [PubMed] [Google Scholar]
- Fersht A. (1999) The basic equations of enzyme kinetics. In Structure and mechanism in protein science, pp 103–132, Freeman and Company, New York. [Google Scholar]
- Otwinowski Z.; Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Grosse-Kunstleve R. W.; Hung L.-W.; Ioerger T. R.; McCoy A. J.; Moriarty N. W.; Read R. J.; Sacchettini J. C.; Sauter N. K.; Terwilliger T. C. (2002) PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D58, 1948–1954. [DOI] [PubMed] [Google Scholar]
- Jones T. A.; Zou J.-Y.; Cowan S. W.; Kjeldgaard M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Schomaker V.; Trueblood K. N. (1998) Correlation of internal torsional motion with overall molecular motion in crystals. Acta Crystallogr. B54, 507–514. [Google Scholar]
- Holm L.; Park J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics 16, 566–567. [DOI] [PubMed] [Google Scholar]
- Abendroth J.; Sankaran B.; Edwards T. E.; Gardberg A. S.; Dieterich S.; Bhandari J.; Napuli A. J.; Van Voorhis W. C.; Staker B. L.; Myler P. J.; Stewart L. J. (2011) BrabA.11339.a: Anomalous diffraction and ligand binding guide towards the elucidation of the function of a ‘putative β-lactamase-like protein’ from Brucella melitensis. Acta Crystallogr. F67, 1106–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huai Q.; Colicelli J.; Ke H. (2003) The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry 42, 13220–13226. [DOI] [PubMed] [Google Scholar]
- Liu S.; Mansour M. N.; Dillman K. S.; Perez J. R.; Danley D. E.; Aeed P. A.; Simons S. P.; Lemotte P. K.; Menniti F. S. (2008) Structural basis for the catalytic mechanism of human phosphodiesterase 9. Proc. Natl. Acad. Sci. U.S.A. 105, 13309–13314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M.; Eckstein F.; Hunneman D. H.; Haraniak J.; Kinas R. W.; Lesiak K.; Stec W. J. (1979) Stereochemistry of hydrolysis of adenosine 3′:5′-cyclic phosphorothioate by the cyclic phosphodiesterase from beef heart. J. Biol. Chem. 254, 9959–9961. [PubMed] [Google Scholar]
- Lipscomb W. N.; Sträter N. (1996) Recent Advances in Zinc Enzymology. Chem. Rev. 96, 2375–2433. [DOI] [PubMed] [Google Scholar]
- Wilson D.; Fiori A.; Brucker K. D.; Dijck P. V.; Stateva L. (2010) Candida albicans Pde1p and Gpa2p comprise a regulatory module mediating agonist-induced cAMP signalling and environmental adaptation. Fungal Genet. Biol. 47, 742–752. [DOI] [PubMed] [Google Scholar]
- Ma P.; Wera S.; Van Dijck P.; Thevelein J. M. (1999) The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol. Biol. Cell 10, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D.; Tutulan-Cunita A.; Jung W.; Hauser N. C.; Hernandez R.; Williamson T.; Piekarska K.; Rupp S.; Young T.; Stateva L. (2007) Deletion of the high-affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol. Microbiol. 65, 841–856. [DOI] [PubMed] [Google Scholar]
- Harashima T.; Heitman J. (2002) The Gα protein Gpa2 controls yeast differentiation by interacting with Kelch repeat proteins that mimic Gβ subunits. Mol. Cell 10, 163–173. [DOI] [PubMed] [Google Scholar]



