Abstract
Bacterial meningitis is a serious health concern worldwide. Given that meningitis can be fatal and many meningitis cases occurred in high-poverty areas, a simple, low-cost, highly sensitive method is in great need for immediate and early diagnosis of meningitis. Herein, we report a versatile and cost-effective polydimethylsiloxane (PDMS)/paper hybrid microfluidic device integrated with loop-mediated isothermal amplification (LAMP) for the rapid, sensitive, and instrument-free detection of the main meningitis-causing bacteria, Neisseria meningitidis (N. meningitidis). The introduction of paper into the microfluidic device for LAMP reactions enables stable test results over a much longer period of time than a paper-free microfluidic system. This hybrid system also offers versatile functions, by providing not only on-site qualitative diagnostic analysis (i.e., a yes or no answer), but also confirmatory testing and quantitative analysis in laboratory settings. The limit of detection of N. meningitidis is about 3 copies per LAMP zone within 45 min, close to single-bacterium detection sensitivity. In addition, we have achieved simple pathogenic microorganism detection without a laborious sample preparation process and without the use of centrifuges. This low-cost hybrid microfluidic system provides a simple and highly sensitive approach for fast instrument-free diagnosis of N. meningitidis in resource-limited settings. This versatile PDMS/paper microfluidic platform has great potential for the point of care (POC) diagnosis of a wide range of infectious diseases, especially for developing nations.
The emergence of 335 infectious diseases that have been reported between 1940 and 2004 in the global human population has generated an extremely significant impact on global health and economies.1,2 Among various global infectious diseases, epidemic bacterial meningitis, a severe infection affecting the protective membranes covering the brain and spinal cord known as the meninges, is one of the most dangerous diseases due to its high morbidity and mortality. Meningitis is a contagious disease, which can become fatal in as early as 24 h after symptoms are noticed. According to the World Health Organization (WHO), “Worldwide, without epidemics one million cases of bacterial meningitis are estimated to occur and 200 000 of these die annually... Higher case-fatality rates (37–60%) have been reported in developing countries.”3 Additionally, many meningitis cases occurred in rural high-poverty areas, such as the so-called “meningitis belt” in Africa, where it remains an important and unresolved public health concern. Neisseria meningitidis (N. meningitidis), the etiologic agent of the meningococcal disease, is a leading cause of morbidity and mortality in children and young adults worldwide.4,5N. meningitidis is also the dominant etiologic bacterium in the African meningitis belt according to the bacteriologic and epidemiologic data collected over the past 30 years.6 Usually patients with meningitis share symptoms common to many febrile illnesses (e.g., influenza), which makes meningitis difficult to diagnose based on clinical symptoms alone. Because of the high fatality rate and damaging effects resulting from untreated meningitis in rural high-poverty areas, a simple, low-cost, highly sensitive methodology is in great need for the immediate and early diagnosis of meningitis.
There are several laboratory guidelines available from the Centers for Disease Control and Prevention (CDC) and the WHO for the diagnosis of meningitis. Currently, gram stain and bacterial culture appear to be the gold standard.7 However, both have to be done in a laboratory setting, and bacterial culture may take a few days. Although gram staining may aid in providing a fast identification after reaching the laboratory, it still has many limitations. (1) Gram stain has a lower detection rate for patients previously treated with antimicrobial therapy. (2) Its detection sensitivity is low. (3) It requires well-trained personnel, due to the fact that sometimes poor staining occurs.8
Recently, quantitative real-time polymerase chain reaction (qPCR),9,10 loop-mediated isothermal amplification (LAMP),11−15 and latex agglutination16 tests have been reported to provide rapid detection of bacterial meningitis. For example, there have been several reports on LAMP amplification methods for the clinical diagnosis of meningitis.11−15 However, these methods require specialized equipment in laboratories, such as qPCR thermocyclers (∼$60 000), turbidimeters, centrifuges, fluorescent microscopes, and so on, which render these methods incapable of rapid diagnosis of meningitis in the field or in low-resource settings.
Microfluidic lab-on-a-chip offers a unique opportunity for various biomedical applications due to a variety of advantages associated with miniaturization, integration, portability, and automation.17−22 It allows for significantly low reagent consumption, integrated processing and analysis of complex biological fluids with high efficiency and sensitivity in health care settings, as well as the possibility of rapid detection. The substrates of a microfluidic device can significantly affect many aspects of a microfluidic system from fabrication methods, cost, assay procedures, to detection. Various materials including Si,23 glass,24−28 PDMS,29−31 and paper32−35 have been used for microfluidic device fabrication. However, each substrate material has its own advantages and limitations. For instance, PDMS microfluidic devices are used extensively for biomedical applications, because of its moderate cost and ease of microfabrication (termed as soft lithography30) and transparent property for optical detection. However, PDMS devices often require complicated surface modification procedures to immobilize biosensors on a chip. Recently, paper-based microfluidic devices afford a new low-cost platform for different applications related to health care in low-resource settings.34−36 Paper-based devices however do not offer the high performance in flow control found in PDMS devices. Therefore, we previously developed a PDMS/paper hybrid microfluidic system, in which paper facilitated the integration of graphene oxide-based nanosensors on the chip, without any complicated surface treatment.37
Recently, microfluidic chips integrated with LAMP reactions have been developed for rapid pathogen detection including S. aureus, E. coli, Mycobacterium tuberculosis, and so on.31,38−41 These microfluidic chips showed potential for fast infectious disease diagnosis using miniaturized devices coupled with different detections such as colorimetric,39 absorbance,31,41 and electrochemical detections.40 For instance, Safavieh et al. developed a cassette-like device for colorimetric LAMP detection of bacteria.39 The limit of detection (LOD) for S. aureus was estimated at about 200 CFU/mL. Colorimetric detection is simple, but the sensitivity is not high, and it is challenging for quantitative analysis. Additionally, a microfluidic electrochemical assay using on-chip LAMP has been reported for rapid detection and quantitation of E. coli.40 It was demonstrated that the system could detect and quantify 24 CFU/mL of E. coli in 60 min using a linear sweep voltammetry method.
Despite the attractiveness of these on-chip LAMP systems for rapid pathogen detection,31,38−41 there are multiple limitations that impose restriction on their applications in low-resource settings. (1) Some microfluidic devices require complicated fabrication procedures because of microvalves and patterned electrodes used in those devices,38,41 which increase the device cost as well. (2) Complicated assay procedures are also obstacles to point-of-care analysis in low-resource settings, such as magnetic bead-based assays that require multiple steps for bacterial cell lysis, DNA denaturization, DNA hybridization on beads, and washing steps.41 Cassette-like devices demand a significant amount of manual micropipetting because of the lack of reagent delivery channels.39 (3) Sensitivity is not very high (e.g., LODs of 270 DNA copies38 or 200 CFU/mL39), especially for colorimetric detection.39 None of the aforementioned instrument-free detection systems has achieved a LOD down to 3 DNA copies. (4) Most systems still rely on fairly expensive and bulky detectors (e.g., potentiostats,40 spectrophotometers,31,41 and turbidimeters42) and other supporting equipment (e.g., pumps41 and water baths31,38) that are not commonly available in resource-poor settings. Conventional DNA extraction procedures that usually require the use of centrifuges are also not suitable for limited-resource settings.40
As far as we know, no low-cost microfluidic POC devices have been reported for the rapid diagnosis of meningitis in low-resource settings. Herein, we report a novel, versatile, and cost-effective PDMS/paper hybrid microfluidic platform for the rapid, sensitive, and instrument-free detection of the main meningitis-causing bacterium, Neisseria meningitidis. Chromatography paper used in this hybrid system serves as a 3D substrate for the prestorage of DNA primers for subsequent LAMP reactions to improve the detection sensitivity. The detection of N. meningitidis is highly sensitive, with a limit of detection about 3 DNA copies. To our best knowledge, this might be the lowest LOD from previously reported on-chip LAMP systems for instrument-free pathogenic microorganism detection. This hybrid microfluidic platform has versatile functions. First, a rapid qualitative detection (i.e., giving a yes or no answer) of N. meningitidis can be achieved within 45 min without using any specialized instruments. Results can be visualized by the naked eye. These features make the microfluidic POC platform capable of quick preliminary diagnosis of meningitis in the field or other resource-limited settings. Furthermore, on-chip LAMP products can be readily collected for confirmatory diagnosis or quantitative analysis of meningitis in a laboratory setting.
Experimental Section
Chemicals and Materials
LAMP detection and DNA preparation: The LAMP primers (Integrated DNA Technologies, Coralville, IA) for the target ctrA gene sequence12 from Neisseria meningitidis are shown in Supporting Information Table S-1. Loopamp DNA amplification kit and Loopamp fluorescence detection reagent (calcein) were purchased from Eiken Co. Ltd., Japan. DNA isolation kit containing a lysis buffer ATL and a ready-to-use proteinase K solution and LAMP product purification kit were purchased from Qiagen (Valencia, CA).
Microfluidic platform fabrication: Polydimethylsiloxane (PDMS, Sylgard 184) was obtained from Dow Corning (Midland, MI); Whatman#1 chromatography paper and Epoxy glue were purchased from Sigma (St. Louis, MO) and ITW Devcon (Danvers, MA), respectively.
All other chemicals were purchased from Sigma (St. Louis, MO) and used without further purification, unless stated otherwise. Unless otherwise noted, all solutions were prepared with ultrapure Milli-Q water (18.2 MΩ cm) from a Millipore Milli-Q system (Bedford, MA).
Microfluidic Platform Design and Fabrication
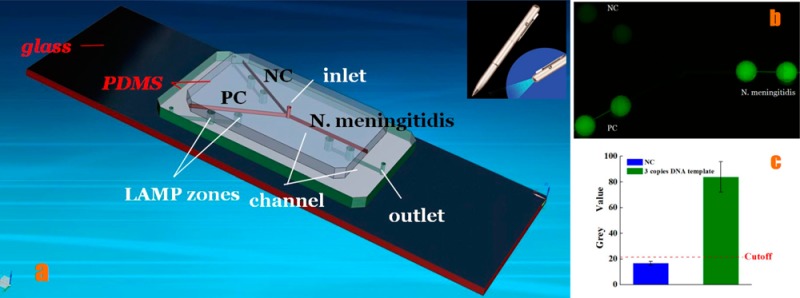
As shown in Figure 1, the microfluidic device comprises three layers, two PDMS layers on the top of a glass slide. The top layer is the PDMS layer used for reagent delivery, including three microchannels (width 100 μm, depth 100 μm) on the bottom side of the PDMS layer, and one inlet reservoir (diameter 1.0 mm, depth 1.5 mm). The middle PDMS layer consists of 6 wells as LAMP zones (diameter 2.0 mm), 3 outlet reservoirs (diameter 1.0 mm), and microchannels (width 100 μm, depth 100 μm) at the bottom side of the middle PDMS layer to connect to outlet reservoirs. The bottom layer is a glass slide mainly for structure support. Different LAMP zones were used for negative control (NC), positive control (PC), and N. meningitidis detection, respectively. Both omission of LAMP primers and omission of DNA template for negative control were tested and showed the same results. Thus, omission of LAMP primers was adopted as the main negative control herein. PC template DNA and its primer mix (PM) were provided by the Loopamp DNA amplification kit.
Figure 1.
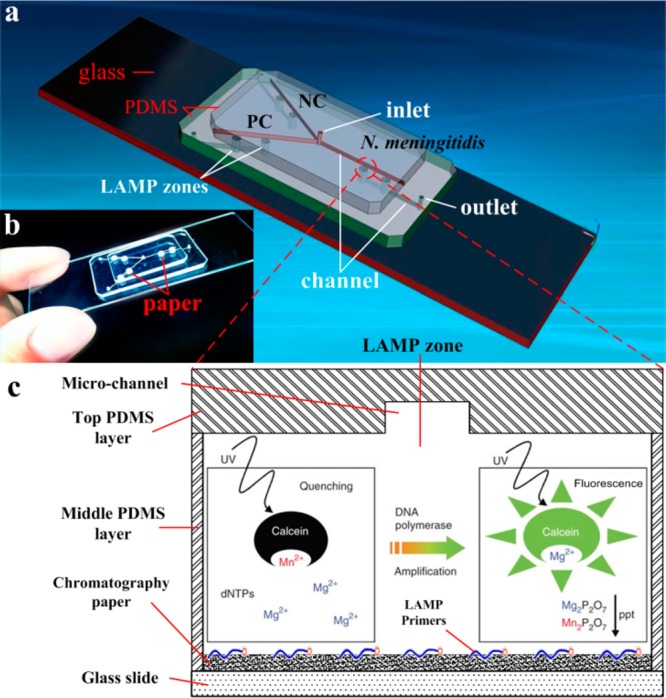
Chip layout of the PDMS/paper hybrid microfluidic device. (a) 3D illustration of the schematic of the chip layout. The chip consists of one top PDMS layer, one middle PDMS layer, and one glass slide for reagent delivery, LAMP reaction, and structure support, respectively. A chromatography paper disk is situated inside each LAMP zone to preload LAMP primers. (b) A photograph of the hybrid microfluidic device for infectious disease diagnosis. (c) A cross-section view of the LAMP zone illustrating the principle of the LAMP detection.
A chromatography paper disk (diameter 2.0 mm) cut by a laser cutter (Epilog Zing 16, Golden, CO) was placed inside each LAMP zone, as a 3D storage substrate for LAMP primers.
On-Chip LAMP Procedures
After the LAMP mix was prepared in a biosafety cabinet, the LAMP reaction mix was introduced to the biochip from the inlet reservoir to fill different LAMP zones. After the inlet and outlets reservoirs were sealed with Epoxy, the microfluidic device was placed on a heating film at 63 °C for 45 min for LAMP reactions, followed by the termination of LAMP reactions at 80 °C for 2 min. The heating film was controlled by an inexpensive proportional–integral–derivative (PID)-based temperature controller devised by our laboratory.
After LAMP reactions, a portable UV pen light was applied to shine LAMP products. The generated fluorescence was captured by a cellular phone camera (e.g., iPhone 5), and the images were processed with the NIH software ImageJ. Results were further confirmed by a high-sensitivity Nikon Ti-E fluorescence microscope (Melville, NY) that was equipped with a motorized stage and a cooled CCD camera to measure the fluorescence intensities, using appropriate FITC optical filters (Ex = 495 nm; Em = 520 nm) for calcein and Cy3 optical filters (Ex = 550 nm; Em = 570 nm) for Cy3-labeled primers.
Centrifuge-Free Detection of Microorganisms
Artificial cerebrospinal fluid (ACSF) was prepared according to previously published protocol.43 Centrifuge-free assay procedures for microorganisms in ACSF were as follows: First, a couple of bacterial colonies were picked from a pure culture using a sterile loop and solubilized in a 20 μL ACSF buffer to prepare the pathogen/ACSF mixture. Next, 2 μL of this mixture was added into 18 μL of bacterial lysis buffer that contained 50 mM Tris buffer (pH 7.5), 4 M urea, and 0.1% triton, and incubated at room temperature for 10 min. At last, 2 μL of the pathogen/pathogen mixture lysate was used in the LAMP reaction mixture for on-chip LAMP reaction, as performed in the aforementioned on-chip LAMP procedures.
Results and Discussion
PDMS/Paper Hybrid Microfluidic Device
Because different chip substrates have their own advantages and limitations, we previously developed a PDMS/paper hybrid microfluidic system for one-step pathogen detection, in which paper facilitated the integration of graphene oxide-based nanosensors on the chip.37 Although this method can directly measure microorganisms without complicated sample preparation due to the use of aptamers, the sensitivity is generally not as high as DNA-amplification-based methods. In this work, chromatography paper is placed in LAMP zones to form another PDMS/paper hybrid microfluidic device (Figure 1), in which paper serves as the 3D substrate for preloading DNA primers for subsequent LAMP reactions to improve detection sensitivity. Paper is a highly porous material, which renders it as an ideal 3D storage substrate for interaction-based assays.44 Paper’s 3D microstructures can also facilitate uniform reagent distribution. It can be seen from Figure 2a that when fluorescently labeled primers were initially loaded into LAMP zones, primers were uniformly distributed in LAMP zones either with paper or without paper inside. However, when devices were placed in a vacuum desiccator to dry, a necessary step to make a ready-to-use POC device in this work, primers in paper-free LAMP zones accumulated on the edge of the LAMP zones, while primers in LAMP zones with paper inside were still uniformly distributed (Figure 2b).
Figure 2.
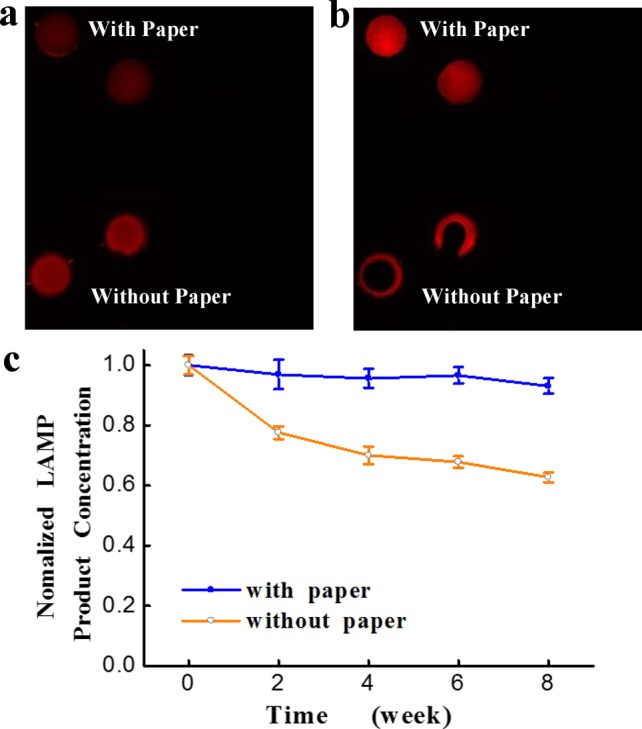
(a,b) Fluorescence images of preloaded Cy3-labeled primers (10.4 μM) in LAMP zones either with or without paper inside. When LAMP zones were dry, primers in paper-free LAMP zones accumulated on the edge of the LAMP zones, while primers in LAMP zones with paper inside were still uniformly distributed. (c) On-chip LAMP performance comparison between biochips with paper inside and without paper inside over a period of 2 months. Nucleic acid concentration of N. meningitidis LAMP products from LAMP zones with paper inside and without paper inside was measured to evaluate on-chip LAMP performance between these two different kinds of biochips. DNA concentration was normalized for convenient comparison.
In addition, we also investigated and compared the on-chip LAMP performance of the devices with paper inside and without paper inside over a period of 2 months at room temperature, as shown in Figure 2c. We precoated LAMP primers of N. meningitidis in LAMP zones with paper inside or without paper inside. The two kinds of ready-to-use microfluidic chips were stored at the same conditions in dark. LAMP reactions were performed within 2, 4, 6, and 8 weeks, respectively. LAMP products were quantified by using Nanodrop after each LAMP reaction to evaluate the on-chip LAMP performance. As shown in Figure 2c, the nucleic acid concentration of LAMP product from LAMP zones without paper inside kept decreasing through the whole experimental period. A sharp decrease within the first 2 weeks was observed. At week 8, the on-chip LAMP performance decreased by ∼40%, which implied that the devices without paper inside were not able to provide consistent results over a period of time. Such devices without paper inside needed to be used right away. On the contrary, the on-chip LAMP performance from the device with paper inside remained stable within 2 months. Only a slight decrease (less than 6%) over time was observed. Therefore, we concluded that the introduction of paper in this hybrid microfluidic biochip as a primer storage substrate also enabled stable on-chip LAMP performance and longer shelf life than those without paper inside. This is mainly because (1) highly interwoven paper fibers can provide DNA primers a 3D protection matrix from harsh environmental elements, and thus are commonly used to collect samples for forensic DNA analysis.45,46 (2) DNA primers can be physically adsorbed onto paper fibers, avoiding DNA loss in the air in the form of aerosols.
On-Chip LAMP Detection of N. meningitidis Using Purified DNA
To offer high-sensitivity detection, DNA amplification is usually required. Although PCR is the most commonly used DNA amplification method, it requires a carefully controlled sequence of heating and cooling cyclers. The fabrication of heaters and temperature sensors on a chip is complicated.47 In contrast, the LAMP method that utilizes the Bacillus stearothermophilus DNA polymerase, a thermally stable enzyme with high strand displacement ability over the template–primer complex,48,49 is a simple, rapid, specific, and cost-effective nucleic acid amplification method when compared to PCR.50 This isothermal LAMP DNA amplification technique allows nucleic acid amplification to be carried out under thermally constant conditions, eliminating the use of expensive instrumentation (e.g., thermal cyclers) for stringent thermal cycling as in conventional PCR, or complicated and costly microfabrication of heating elements on a chip for on-chip PCR.47
The feasibility of the PDMS/paper hybrid microfluidic platform for N. meningitidis detection was first tested by using purified DNA of N. meningitidis. The N. meningitidis DNA template was isolated and purified from bacterial culture.
A 26 μL LAMP reaction mix was introduced through the inlet reservoir into different sample test, positive control (PC), and negative control (NC) LAMP zones, where the specific LAMP primers for the target N. meningitidis and PC DNA were preloaded, respectively. A notable feature of the device design is that the LAMP reaction wells in the middle PDMS layer were independent without connections in the same layer, which can effectively prevent cross-talk among the sample test, PC, and NC LAMP zones during the LAMP reactions.
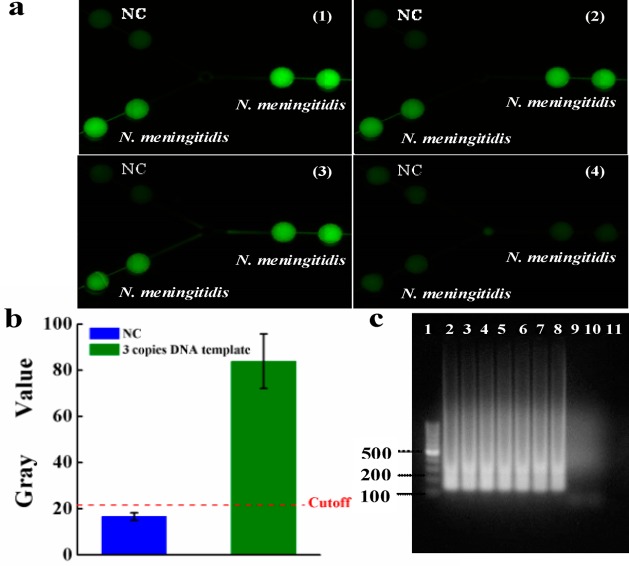
During the LAMP amplification process, it was observed that a magnesium pyrophosphate precipitate was formed as a turbid byproduct of the nucleic acid amplification process.51 This precipitate forms only when the targeted DNA is present in the LAMP amplification process, such that the presence of the pyrophosphate can serve as an indicator of the presence of a pathogen’s target DNA by turbidity detection. Turbidity however is challenging for high-sensitivity visual detection. Thus, turbidity detection usually requires a turbidimeter.42 Actually, visual confirmation by the naked eye can be achieved by the addition of a mixture of calcein in the presence of manganese ions. The fluorescence of calcein is quenched by manganese ions before LAMP amplification. When the amplification reaction proceeds, the manganese–calcein complex is deprived of manganese ions by generated pyrophosphate, which results in the emission of fluorescence under UV light (Figure 1c).50 As such, clinical diagnosis of the pathogen N. meningitidis can be achieved by visual confirmation of the green color with a portable UV light pen, as shown in Figure 3. The fluorescence generated was captured by a cellular phone camera, as used in Figure 3a. It was observed that the N. meningitidis sample and PC showed bright green fluorescence under a portable UV light pen, while NC only showed weak background. To quantify the difference of the fluorescence between N. meningitidis and NC, the gray value, an indication of the brightness of a pixel, of fluorescence images was processed by the software ImageJ, as shown in Figure 3c. The cutoff value was determined as 3-fold standard deviations of the mean gray value of the negative controls on the basis of the negative control. Figure 3c clearly shows the difference between NC and PC and N. meningitidis.
Figure 3.
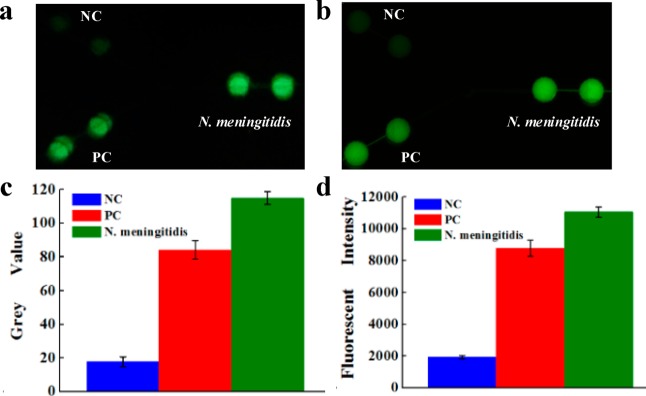
On-chip LAMP detection of N. meningitidis using purified DNA by a portable UV light pen (a) and fluorescence microscopy (b). Strong fluorescence was observed in N. meningitidis and PC LAMP zones, but not in NC zones. (c) Gray value of the LAMP products measured by ImageJ; (d) fluorescent intensity of the LAMP products measured by fluorescence microscope. The purified DNA template used was 3 × 106 copies per LAMP zone.
The results were further confirmed by high-sensitivity fluorescence microscopy (see Figure 3b and d). Similar to that observed in Figure 3a, strong fluorescence was observed in N. meningitidis and PC LAMP zones, but not in NC zones. The fluorescence intensity of the N. meningitidis LAMP products was about 6 times higher than that of the NC.
Besides, the multiple-layer biochip is designed in a way to render versatile functions for the N. meningitidis diagnosis. In addition to on-chip LAMP detection, different LAMP products can be collected separately for further confirmatory tests using conventional gel electrophoresis and quantitative analysis, as demonstrated in Supporting Information Figures S-1 and 6 in laboratory settings, respectively. When the NC LAMP mix and the N. meningitidis LAMP mix in two PCR tubes were placed under UV light before LAMP reactions, neither tube showed notable fluorescence (Supporting Information Figure S-1a). However, after on-chip LAMP reaction, similar to on-chip visual detection as shown in Figure 3a, the collected N. meningitidis LAMP products in a PCR tube showed bright green fluorescence under portable UV light (Supporting Information Figure S-1b). Conversely, the NC mixture had no difference after LAMP reaction. The obvious difference between N. meningitidis tests and NC could be seen even by the naked eye. Subsequently, the results were further confirmed by conventional gel electrophoresis of extracted LAMP products, as shown in Supporting Information Figure S-1c, the comparison of LAMP products in lane 2 with the DNA sizing ladder in lane 1 verified the success of the on-chip LAMP. As expected, no DNA bands were observed in NC in lane 3.
Specificity Test
As reported previously, LAMP is a reliable method for pathogen diagnosis with high specificity.52,53 We tested the specificity of our method among N. meningitidis, Giardia lamblia (Giardia), and Bordetella pertussis (B. pertussis) (data not shown), and among three common types of meningitis-causing bacteria, N. meningitidis, Haemophilus influenzae (H. influenzae), and Streptococcus pneumoniae (S. pneumoniae) (See Figure 4). Both tests showed high specificity of our approach. For instance, signals from LAMP zones with H. influenzae and Streptococcus pneumoniae (S. pneumoniae) DNA templates were observed to be similar to NC in Figure 4, in contrast to bright fluorescence from N. meningitidis, which confirmed the high specificity of our approach in the detection of N. meningitidis.
Figure 4.
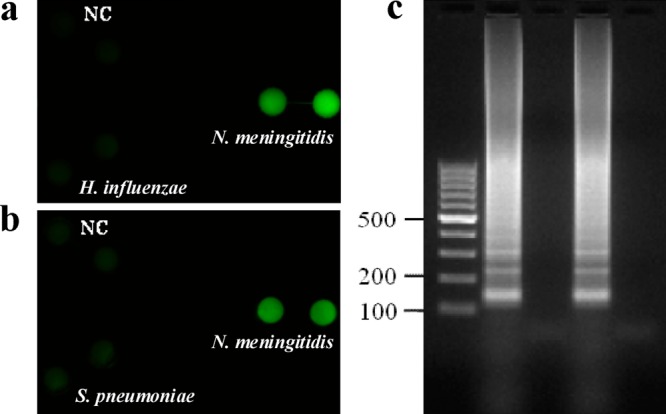
Specificity study among N. meningitidis, H. influenzae, and S. pneumoniae. Fluorescence images of on-chip LAMP products to test specificity between N. meningitidis and H. influenzae (a) and specificity between N. meningitidis and S. pneumoniae (b). Only the LAMP zones with N. meningitidis template DNA showed bright fluorescent signal, whereas LAMP zones loaded with H. influenzae and S. pneumoniae template DNA showed signal similar to that of NC. (c) Gel electrophoresis of the on-chip LAMP products for confirmatory analysis. Lane 1, 100 bp marker; lanes 2 and 3, products of N. meningitidis and H. influenzae LAMP zones from (a); lanes 4 and 5, LAMP products of N. meningitidis and S. pneumoniae LAMP zones from (b).
Instrument-Free Detection of Pathogenic Microorganisms
The detection discussed above was carried out by using a purified DNA template. There have also been reports on rapid PCR detection for clinic samples of pathogenic bacteria by using special lysis reagents without the inhibition of PCR.47,54−57 Yet for LAMP DNA amplification, traditional sample preparation procedures such as DNA isolation and purification are needed.12,13 These procedures are time-consuming, and require the use of centrifuges that however usually do not exist in the field, making them not suitable for POC detection in resource-poor settings.
In this work, we have developed a simple microfluidic approach for pathogenic microorganism detection (not just pathogen DNA). This approach combines a simple bacteria lysis procedure with on-chip LAMP detection, without using any centrifuge and prepurified DNA templates. Because the examination of cerebrospinal fluids (CSF) is a cornerstone of current meningitis diagnosis, we spiked N. meningitidis bacteria in artificial cerebrospinal fluid (ACSF) to mimic the real clinic samples for on-chip LAMP detection.
The first step is to discover a simple method to lysis N. meningitidis microorganisms in a resource-limited setting. More importantly, the method should be fully compatible to the subsequent LAMP reactions (i.e., without inhibition to LAMP reactions). We tried various lysis buffers including some common commercial buffers used for PCR (e.g., Buffet ATL from Qiagen, MagNA Pure Bacteria Lysis Buffer from Roche Applied Science), but found only this lysis buffer (50 mM Tris buffer (pH 7.5), 4 M urea, and 0.1% triton) is compatible to LAMP reactions, while others completely inhibited LAMP reaction (see gel electrophoresis results in Figure S-2 of the Supporting Information). This lysis buffer can be adapted for other microorganism detection using LAMP without problems of inhibition.
Upon the success of microorganism lysis without the use of any centrifuges, the lysate without any further preparation was used directly for the on-chip LAMP detection of N. meningitidis microorganisms in ACSF. The results in Figure 5 showed that the LAMP products of the spiked ACSF sample could still produce strong fluorescence as purified DNA samples under portable UV light and fluorescence microscope. Figure 5c and d showed good discrimination among a spiked N. meningitidis sample, PC, and NC. Gel electrophoresis of on-chip LAMP products from pathogen/ACSF sample further confirmed the success of the LAMP reaction (Figure 5e), and the successful detection of N. meningitidis.
Figure 5.
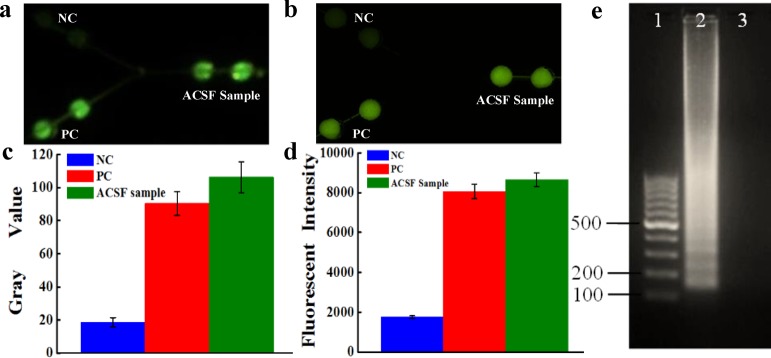
Instrument-free detection of N. meningitidis microorganisms. Fluorescence images of on-chip LAMP detection of N. meningitidis in ACSF under portable UV light (a) and by fluorescence microscopy (b). (c) Gray value of the LAMP products from pathogen/ACSF mixtures measured by fluorescence microscope. (d) Fluorescent intensity of the LAMP products from pathogen/ACSF mixtures measured by fluorescence microscope. (e) Gel electrophoresis of the on-chip LAMP products of N. meningitidis in ACSF. Lanes 1–3: 100 bp marker; LAMP products of N. meningitidis in ACSF; NC.
The instrument-free pathogen detection was very simple and fully compatible with LAMP reactions, without using any equipment. The success of this instrument-free LAMP detection using the microfluidic device is significant because it indicates that real samples could be directly used for the on-chip LAMP reaction without any laborious and time-consuming DNA isolation or purification procedures.
Calibration Curve
The visual LAMP fluorescence detection is a simple method for rapid pathogen detection, from which a yes or no qualitative answer for POC detection can be achieved quickly based on the fluorescence of the LAMP products. During our experiments, it was found that the fluorescence intensity was not directly proportional to pathogen concentrations, indicating the fluorescence intensity of calcein is not suitable for quantitative analysis. Although the qualitative analysis was sufficient for most diseases diagnosis, we developed an indirect method for quantitative analysis of N. meningitidis, based on the versatile functions of the microfluidic chip. Before on-chip LAMP reactions, the DNA concentrations of a series of 10-fold diluted pathogen/ACSF lysate solutions ranging from 6 × 106 to 6 were quantified to obtain initial DNA copy numbers per LAMP zone by Nanodrop. After LAMP amplifications in the microfluidic platform, because microwells for samples PC and NC were separated from each other in the middle PDMS layer, LAMP products could be simply collected, purified, and quantified. Thus, the calibration was generated by plotting the DNA LAMP product concentration against initial DNA copy numbers, as shown in Figure 6. It can be seen that the product DNA concentration is directly proportional to the log of the initial template DNA numbers, with an R2 of 0.98. Therefore, by measuring the nucleic acid concentration of the LAMP products from unknown N. meningitidis samples, we can calculate the amount of the bacteria in unknown samples, and thus estimate the seriousness of infection.
Figure 6.
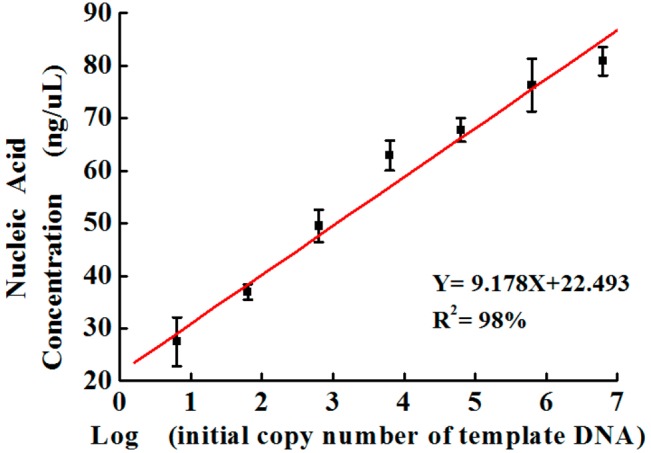
Calibration curve of nucleic acid concentration of the LAMP products (after LAMP amplification) versus the initial copy number of template DNA (before LAMP amplification) of N. meningitidis in ACSF.
Limit of Detection (LOD)
By using a series of 10-fold diluted N. meningitidis DNA template solutions whose DNA copy numbers were determined by Nanodrop, the limit of detection was studied. The top left LAMP zones of the device were used for NC. All other LAMP zones were used for N. meningitidis DNA detection. The initial DNA copy numbers of the template (before LAMP) ranged from 3 × 106, 3 × 105, 3 × 104, ... to 3 × 10–2 per LAMP zone. After LAMP reactions, it was observed from Figure 7a that even the initial DNA templates were as low as 3 copies per LAMP zone; the on-chip LAMP products still exhibited strong fluorescence. However, when the initial DNA templates were less than one copy, the fluorescence of the LAMP zones was as dim as the NC. On the basis of 3-fold standard deviations of the mean gray value of the negative controls on the basis of the negative control, we calculated the gray value of the cutoff line for N. meningitidis as 21.5, as shown with the dashed line in Figure 7b. The gray value of the LAMP product from 3 copies of initial DNA template was much higher than that of the cutoff line. This was further confirmed by off-chip gel electrophoresis of all LAMP products, as shown in Figure 7c. Therefore, the LOD of the microfluidic approach was estimated to be ∼3 DNA copies (or 7.4 fg), which is close to single-bacterium detection sensitivity. This LOD was even 3-fold as low as that of the conventional real-time PCR method for N. meningitidis detection (9 copies/reaction).58
Figure 7.
LOD investigation. (a) Fluorescence images of LAMP products using a series of 10-fold diluted N. meningitidis DNA template solutions ranging from (1)–(4): 3 × 102, 3 × 101, 3 × 100, 3 × 10–1 DNA copies per LAMP zone. The on-chip LAMP products still exhibited strong fluorescence even though the initial DNA templates were as low as 3 copies per LAMP zone. (b) Gray values of the image of (a)-3 for LAMP products from 3 copies DNA template. The dotted line is the calculated gray value (21.5) of the cutoff line for N. meningitidis detection based on 3 times SD of negative controls. (c) Gel electrophoresis of on-chip LAMP products using a series of diluted DNA template solutions. Lanes 1–11: 100 bp marker, 3 × 106, 3 × 105, 3 × 104, ...3 × 100, 3 × 10–1, 3 × 10–2 DNA copies of the template per LAMP zone, NC.
Conclusion
We have developed a versatile PDMS/paper hybrid microfluidic platform for rapid and sensitive detection of N. meningitidis. Because of the integrated LAMP DNA amplification on the chip, the limit of detection of ∼3 DNA copies of N. meningitidis has been achieved within 45 min, overcoming lengthy assay time and low-sensitivity issues in conventional methods for the diagnosis of meningitis. This hybrid microfluidic platform incorporates the advantages of high performance in liquid control from PDMS and of high porosity from paper for preloading LAMP primers.
The function of this hybrid microfluidic system is versatile. (1) Its on-chip LAMP detection based on calcein under portable UV light does not require any bulky specialized equipment without the use of any centrifuges and cumbersome procedures for DNA isolation and purification. The instrument-free detection makes the microfluidic system highly capable for the diagnosis of meningitis in the field or in other resource-limited settings. (2) The design of the microfluidic biochip allows on-chip LAMP products to be readily extracted for more confirmatory tests (e.g., gel electrophoresis) and quantitative analysis based on the calibration curve, as demonstrated in this work. This feature is suitable to the in-depth analysis and study of patient samples in clinical laboratory settings. Combining features (1) and (2) can provide a comprehensive examination of patient samples in different settings. For instance, after an initial qualitative assay of a patient sample in the field or resource-limited settings, the sample tested by the biochip can be sent back to a clinical laboratory for further confirmatory tests or quantitative analysis to examine the disease seriousness of infection. Moreover, by designing and changing different primers specific to other infectious diseases, this microfluidic platform can have great potential in quick and early diagnosis of a broad range of other infectious diseases, such as whooping cough, malaria, H1N1, and severe acute respiratory syndrome (SARS), especially for developing nations. By scaling up channels and LAMP zones, our method can be used for high-throughput screening of different infectious diseases.
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health (NIH) under Award Number SC2GM105584. Financial support from the IDR Program at the University of Texas at El Paso (UTEP), University of Texas (UT) System for the STARS award, and Multidisciplinary Research Award Program (MRAP) from College of Science at UTEP is also gratefully acknowledged. We would like to express special thanks to Dr. Siddhartha Das’s group for providing Giardia DNA and the staff of the Genomic Analysis Core Facility of The University of Texas at El Paso. This core is supported by Grant G12MD007592 from NIMHD.
Supporting Information Available
Detailed information on the Experimental Section, some figures for confirmatory analysis, and bacterial cell lysis. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Jones K. E.; Patel N. G.; Levy M. A.; Storeygard A.; Balk D.; Gittleman J. L.; Daszak P. Nature 2008, 451, 990–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M.; Folkers G. K.; Fauci A. S. Nature 2004, 430, 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo D.WHO Manual, 2nd ed.; 2011. [Google Scholar]
- Goldacre M. J.; Roberts S. E.; Yeates D. BMJ [Br. Med. J.] 2003, 327, 596–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyderman R. S.; Ben-Shlomo Y.; Brennan C. A.; Somerset M. Arch. Dis. Child. 2004, 89, 1064–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc LaForce F.; Ravenscroft N.; Djingarey M.; Viviani S. Vaccine 2009, 27, B13–B19. [DOI] [PubMed] [Google Scholar]
- Brouwer M. C.; Tunkel A. R.; van de Beek D. Clin. Microbiol. Rev. 2010, 23, 467–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman M. I.; Tolford S.; Harper M. B.. Pediatr. Infect. Dis. J. 2008, 27, 309–313310.1097/INF.1090b1013e31815f31853ba. [DOI] [PubMed] [Google Scholar]
- Poppert S.; Essig A.; Stoehr B.; Steingruber A.; Wirths B.; Juretschko S.; Reischl U.; Wellinghausen N. J. Clin. Microbiol. 2005, 43, 3390–3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson D. C.; Louie L.; Louie M.; Simor A. E. J. Clin. Microbiol. 2003, 41, 3851–3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagdev K. J.; Kashyap R. S.; Parida M. M.; Kapgate R. C.; Purohit H. J.; Taori G. M.; Daginawala H. F. J. Clin. Microbiol. 2011, 49, 1861–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna J. P.; Fairley D. J.; Shields M. D.; Cosby S. L.; Wyatt D. E.; McCaughey C.; Coyle P. V. Diagn. Microbiol. Infect. Dis. 2011, 69, 137–144. [DOI] [PubMed] [Google Scholar]
- Kim D. W.; Kilgore P. E.; Kim E. J.; Kim S. A.; Anh D. D.; Dong B. Q.; Kim J. S.; Seki M. PLoS One 2012, 7, e42954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M.; Yamashita Y.; Torigoe H.; Tsuda H.; Sato S.; Maeno M. J. Clin. Microbiol. 2005, 43, 1581–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. W.; Kilgore P. E.; Kim E. J.; Kim S. A.; Anh D. D.; Seki M. J. Clin. Microbiol. 2011, 49, 3621–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann O.; Braun J. S.; Becker D.; Halle A.; Freyer D.; Dagand E.; Lehnardt S.; Weber J. R. J. Immunol. 2007, 178, 6476–6481. [DOI] [PubMed] [Google Scholar]
- Culbertson C. T.; Mickleburgh T. G.; Stewart-James S. A.; Sellens K. A.; Pressnall M. Anal. Chem. 2013, 86, 95–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. J.; Zhou Y.. Microfluidic Devices for Biomedical Applications; Woodhead Publishing: Cambridge, 2013. [Google Scholar]
- Li X. J.; Valadez A. V.; Zuo P.; Nie Z. Bioanalysis 2012, 4, 1509–1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.; Li P. C. H. Expert Rev. Clin. Pharmacol. 2010, 3, 267–280. [DOI] [PubMed] [Google Scholar]
- Salieb-Beugelaar G. B.; Simone G.; Arora A.; Philippi A.; Manz A. Anal. Chem. 2010, 82, 4848–4864. [DOI] [PubMed] [Google Scholar]
- Kovarik M. L.; Gach P. C.; Ornoff D. M.; Wang Y.; Balowski J.; Farrag L.; Allbritton N. L. Anal. Chem. 2011, 84, 516–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung A. J.; Kim D.; Erickson D. Lab Chip 2008, 8, 330–338. [DOI] [PubMed] [Google Scholar]
- Li X. J.; Chen Y. C.; Li P. C. H. Lab Chip 2011, 11, 1378–1384. [DOI] [PubMed] [Google Scholar]
- Li X. J.; Xue X.; Li P. C. H. Integr. Biol. 2009, 1, 90–98. [DOI] [PubMed] [Google Scholar]
- Li X. J.; Ling V.; Li P. C. H. Anal. Chem. 2008, 80, 4095–4102. [DOI] [PubMed] [Google Scholar]
- Li X. J.; Huang J.; Tibbits G. F.; Li P. C. H. Electrophoresis 2007, 28, 4723–4733. [DOI] [PubMed] [Google Scholar]
- Li X. J.; Li P. C. H. Anal. Chem. 2005, 77, 4315–4322. [DOI] [PubMed] [Google Scholar]
- Chen H.; Li X.; Wang L.; Li P. C. Talanta 2010, 81, 1203–1208. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Whitesides G. M. Annu. Rev. Mater. Sci. 1998, 1998, 153–184. [Google Scholar]
- Fang X.; Liu Y.; Kong J.; Jiang X. Anal. Chem. 2010, 82, 3002–3006. [DOI] [PubMed] [Google Scholar]
- Liu W.; Cassano C. L.; Xu X.; Fan Z. H. Anal. Chem. 2013, 85, 10270–10276. [DOI] [PubMed] [Google Scholar]
- Li X. J.; Nie Z. H.; Cheng C.-M.; Goodale A. B.; Whitesides G. M. Proc. Micro Total Anal. Syst. 2010, 14, 1487–1489. [Google Scholar]
- Nie Z.; Nijhuis C. A.; Gong J.; Chen X.; Kumachev A.; Martinez A. W.; Narovlyansky M.; Whitesides G. M. Lab Chip 2010, 10, 477–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. M.; Martinez A. W.; Gong J.; Mace C. R.; Phillips S. T.; Carrilho E.; Mirica K. A.; Whitesides G. M. Angew. Chem., Int. Ed. 2010, 49, 4771–4774. [DOI] [PubMed] [Google Scholar]
- Liu X. Y.; Mwangi M.; Li X. J.; O’Brien M.; Whitesides G. M. Lab Chip 2011, 11, 2189–2196. [DOI] [PubMed] [Google Scholar]
- Zuo P.; Li X.; Dominguez D. C.; Ye B.-C. Lab Chip 2013, 13, 3921–3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X.; Chen H.; Xu L.; Jiang X.; Wu W.; Kong J. Lab Chip 2012, 12, 1495–1499. [DOI] [PubMed] [Google Scholar]
- Safavieh M.; Ahmed M. U.; Sokullu E.; Ng A.; Braescu L.; Zourob M. Analyst 2014, 139, 482–487. [DOI] [PubMed] [Google Scholar]
- Safavieh M.; Ahmed M. U.; Tolba M.; Zourob M. Biosens. Bioelectron. 2012, 31, 523–528. [DOI] [PubMed] [Google Scholar]
- Wang C.-H.; Lien K.-Y.; Wu J.-J.; Lee G.-B. Lab Chip 2011, 11, 1521–1531. [DOI] [PubMed] [Google Scholar]
- Mori Y.; Kitao M.; Tomita N.; Notomi T. J. Biochem. Biophys. Methods 2004, 59, 145–157. [DOI] [PubMed] [Google Scholar]
- Cold Spring Harbor Protocols, 2011, pdb.rec065730.
- Martinez A. W. Bioanalysis 2011, 3, 2589–2592. [DOI] [PubMed] [Google Scholar]
- Smith L.; Burgoyne L. BMC Ecol. 2004, 4, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisomchit S.; Wichajarn R.; Janejai N.; Chareonsiriwatana W. Southeast Asian J. Trop Med. Public Health 2005, 36, 270–273. [PubMed] [Google Scholar]
- Liu P.; Li X.; Greenspoon S. A.; Scherer J. R.; Mathies R. A. Lab Chip 2011, 11, 1041–1048. [DOI] [PubMed] [Google Scholar]
- M S.; H S.; M E.-M. Dis. Aquat. Org. 2008, 81, 143–151. [DOI] [PubMed] [Google Scholar]
- Notomi T.; Okayama H.; Masubuchi H.; Yonekawa T.; Watanabe K.; Amino N.; Hase T. Nucleic Acids Res. 2000, 28, e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita N.; Mori Y.; Kanda H.; Notomi T. Nat. Protoc. 2008, 3, 877–882. [DOI] [PubMed] [Google Scholar]
- Mori Y.; Nagamine K.; Tomita N.; Notomi T. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [DOI] [PubMed] [Google Scholar]
- Sirichaisinthop J.; Buates S.; Watanabe R.; Han E. T.; Suktawonjaroenpon W.; Krasaesub S.; Takeo S.; Tsuboi T.; Sattabongkot J. Am. J. Trop. Med. Hygiene 2011, 85, 594–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöschl B.; Waneesorn J.; Thekisoe O.; Chutipongvivate S.; Panagiotis K. Am. J. Trop. Med. Hygiene 2010, 83, 56–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr K.; Harper R.; Linacre A. Plant Mol. Biol. Rep. 2001, 19, 367–371. [Google Scholar]
- Vickers I.; O’Flanagan D.; Cafferkey M.; Humphreys H. Eur. J. Clin. Microbiol. Infect. Dis. 2011, 30, 447–453. [DOI] [PubMed] [Google Scholar]
- Ke C.; Kelleher A.-M.; Berney H.; Sheehan M.; Mathewson A. Sens. Actuators, B 2007, 120, 538–544. [Google Scholar]
- Brewster J. D.; Paoli G. C. Anal. Biochem. 2013, 442, 107–109. [DOI] [PubMed] [Google Scholar]
- Wang X.; Theodore M. J.; Mair R.; Trujillo-Lopez E.; du Plessis M.; Wolter N.; Baughman A. L.; Hatcher C.; Vuong J.; Lott L. J. Clin. Microbiol. 2012, 50, 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.