Club cells are infected by influenza virus, survive acute infection, and establish a proinflammatory environment that contributes to lung pathology. Depletion of club cells reduces lung tissue damage associated with flu infection.
Abstract
Respiratory infection of influenza A virus (IAV) is frequently characterized by extensive immunopathology and proinflammatory signaling that can persist after virus clearance. In this report, we identify cells that become infected, but survive, acute influenza virus infection. We demonstrate that these cells, known as club cells, elicit a robust transcriptional response to virus infection, show increased interferon stimulation, and induce high levels of proinflammatory cytokines after successful viral clearance. Specific depletion of these surviving cells leads to a reduction in lung tissue damage associated with IAV infection. We propose a model in which infected, surviving club cells establish a proinflammatory environment aimed at controlling virus levels, but at the same time contribute to lung pathology.
Influenza A virus (IAV) is a seasonal pathogen with the capacity to cause devastating pandemics. IAV infects a variety of cells within the respiratory tract, including ciliated epithelial cells, type I and II alveolar cells, and immune cells (Matrosovich et al., 2004; Manicassamy et al., 2010; Shieh et al., 2010; Langlois et al., 2012; Smed-Sörensen et al., 2012). Classically, IAV-infected cells are tracked through detection of virus-derived products or reporters (e.g., virus RNA or protein), all of which have short half-lives and are therefore incapable of defining infected cell types in the long-term. Ultimately, acute IAV infections are resolved within 2 wk post-infection (Carrat et al., 2008).
Infected cells are eliminated through two major mechanisms, apoptosis/necrosis driven by virus replication (Sanders et al., 2011; Yatim and Albert, 2011) or clearance mediated through the innate and adaptive arms of the immune system (Zinkernagel and Doherty, 1979; Eichelberger et al., 1991; Julkunen et al., 2001; Takeuchi and Akira, 2009). Clearance of IAV infections can come at the cost of aberrant immune-mediated disease (Damjanovic et al., 2012). Therefore, a balance between virus clearance and immune-mediated tissue damage is important for recovery from IAV infections.
In this study, we define the long-term fate of virus-infected cells within the lung through an IAV expressing Cre recombinase and transgenic reporter mice (Nagy, 2000). This experimental model system allows for the indelible labeling of virus-infected cells, even at time points well after replication has ceased and virus has been cleared. Surprisingly, despite a potent viral lytic phase and generation of antiviral immune responses, we demonstrate that a small population of cells that were infected by IAV persist after virus clearance. Furthermore, using a combination of next-generation mRNA sequencing and flow cytometry, we determine that infected long-term surviving cells were comprised mainly of a single cell lineage, club cells (formerly termed Clara cells; Winkelmann and Noack, 2010), and that these cells have heightened interferon stimulated gene (ISG) levels. Specific depletion of surviving cells results in increased pulmonary pathology, suggesting a proinflammatory role in recovery. This study provides evidence of cellular survival from acute virus infection and details new cellular mechanisms of immunopathology.
RESULTS AND DISCUSSION
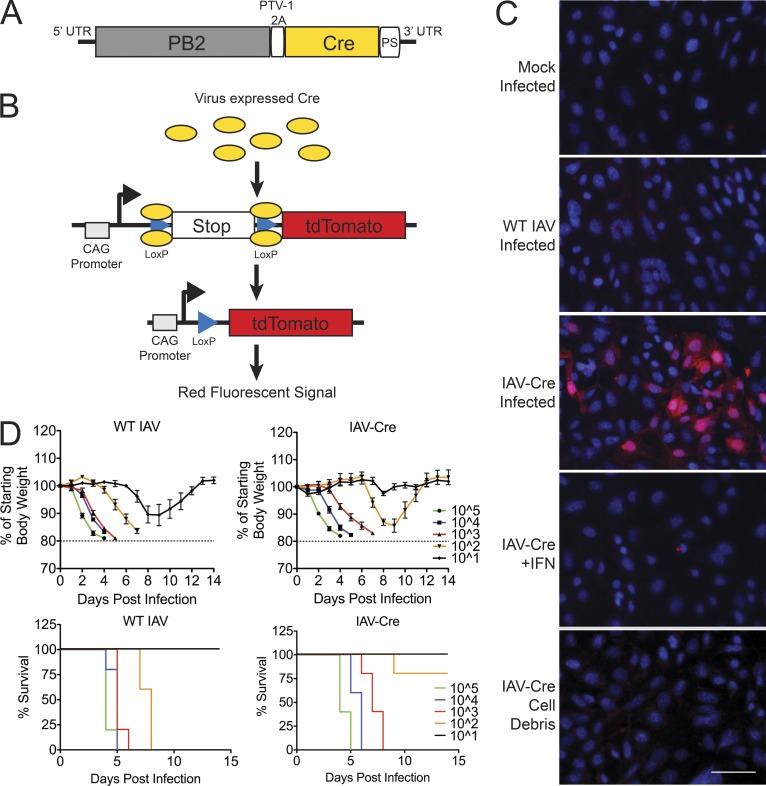
To identify and characterize cells that are productively infected by IAV but go on to survive infection, we generated an H1N1 strain (A/Puerto Rico/8/1934) expressing the bacteriophage protein Cre recombinase after a PTV-1 self-cleavage site with a glycine-serine linker (Kim et al., 2011) on the viral PB2 protein (Fig. 1 A). By infecting mice harboring the appropriate transgenic fluorescent reporter cassette, the expression of Cre leads to the excision of a stop cassette (Madisen et al., 2010). After the stop element is removed, the cells will constitutively express the red fluorescent protein tdTomato (Fig. 1 B). Because the host cell harbors the tdTomato expression cassette, the cells continue to express the reporter protein even if viral replication is stalled or eliminated.
Figure 1.
Generation of influenza A virus expressing Cre recombinase. (A) Schematic showing insertion of Cre recombinase (Cre) downstream of a PTV-1 2A site at the 3′ end of PB2 segment. (B) Model depicting Cre mediated excision of tdTomato reporter stop cassette. (C) Lung fibroblast generated from ROSA26 tdTomato lox-stop mice were mock infected or infected with WT IAV or IAV-Cre at an MOI of 5 (top three panels). Reporter fibroblasts were treated with 100 U of IFN-α/β for 6 h and infected with IAV-Cre at MOI of 5 (fourth panel). MDCK cells were infected with IAV Cre at an MOI of 5 for 24 h. Cell debris were treated with anti-HA antibodies for 30 min and placed on reporter fibroblast (bottom). All images were taken 36 hpi. Bar, 50 µm. Data are representative of two independent experiments. (D) WT C57BL/6 mice were infected with WT IAV (left) or IAV-Cre (right) at the indicated doses and monitored for morbidity and mortality. Calculated LD50 values are ∼50 PFU for WT IAV and ∼240PFU for IAV-Cre. n = 5 mice per group. The experiment was performed once.
To characterize the system, we performed ex vivo experiments on mouse lung fibroblasts isolated from the transgenic tdTomato reporter animals. Wild-type IAV or mock-infected fibroblasts failed to express tdTomato; however, upon infection with IAV-Cre, we observe red fluorescence (Fig. 1 C). To demonstrate that viral replication is required to activate the reporter, we pretreated cells with IFN-α/β and infected with IAV-Cre. Under these conditions, we observed no red signal, indicating that viral RNA replication and protein expression are required (Fig. 1 C). Finally, to determine if phagocytosis of infected cellular extract was sufficient for tdTomato expression, we applied lysed cell debris from IAV-Cre infections in the presence of a neutralizing antibody but found no evidence for fluorescence (Fig. 1 C). Collectively, these data suggest that activation of the tdTomato cellular reporter requires active viral replication.
We next characterized the virulence of IAV-Cre in vivo to ensure that the pathogenesis of this recombinant virus was maintained. To this end, C57BL/6 mice were infected with either wild-type IAV or IAV-Cre at a range of infectious doses, and body weight and survival were monitored over time (Fig. 1 D). Excitingly, despite the insertion of Cre recombinase, intranasal inoculation of IAV-Cre was sufficient to induce morbidity in a manner comparable to the parental IAV stain (Fig. 1 D). Furthermore, mortality upon infection with IAV-Cre was only mildly attenuated when compared with the parental strain, with the median lethal dose (LD50) shifting from ∼50 to ∼240 PFU (Fig. 1 D). These data suggest that IAV-Cre retains the pathogenic properties associated with IAV disease and justifies its utilization as a tool to probe for cells that survive replicating virus in vivo.
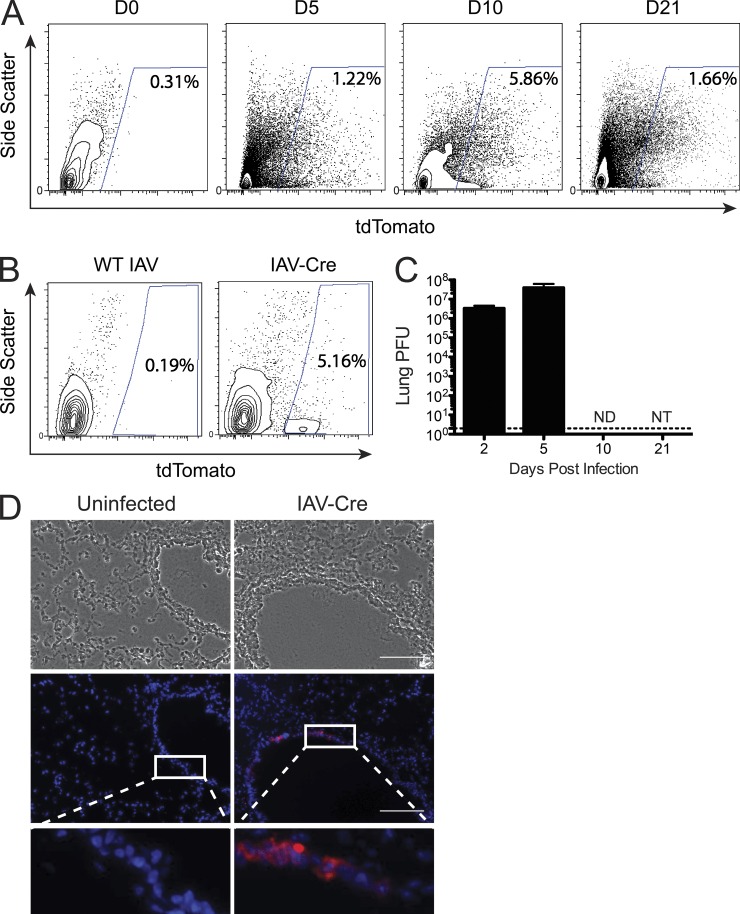
Although IAV infection and replication generally result in the induction of cell death, here we chose to determine whether any cell types could successfully clear virus and survive. We therefore infected tdTomato reporter mice with IAV-Cre and assessed the presence of reporter-positive cells using flow cytometry at various times after infection (Fig. 2 A). While no tdTomato+ cells were identified in uninfected mice or reporter mice infected with WT PR8 virus (Fig. 2, A and B), we observed a population of reporter-positive cells during active viral replication (5 d post-infection) as expected. Surprisingly, we also observed a population of tdTomato+ cells at 10 and 21 d post-infection (Fig. 2 A), time points that exceed the physiological course of IAV replication (Eichelberger et al., 1991; Carrat et al., 2008).
Figure 2.
IAV-Cre demonstrates long-term cell survival in vivo. (A) tdTomato reporter mice were infected with 500PFU of IAV-Cre and live CD45− cells were analyzed for tdTomato expression by FACS. Values represent percent live, CD45− tdTomato+ cells. Data are representative of five independent experiments. (B) The presence of surviving tdTomato+ cells was measured 10 d post-infection from mice infected with WT IAV or IAV-Cre. (C) Mice were infected with IAV-Cre and lungs were removed at indicated time points and virus titers from total lung homogenates were assessed by plaque assay. Dotted line represents the limit of detection. ND, not detected; NT, not titered. n = 3 mice per time point. Data are representative of two independent experiments. (D) Mice were infected as in A and lungs were harvested at 10 d post-infection. Frozen lung sections were counter stained with DAPI and analyzed for tdTomato expression. Insets are enlargements of the white-boxed areas. Bars, 100 µm. Data are representative of two independent experiments.
To formally demonstrate that the observed tdTomato+ cells were not a reflection of prolonged replication of IAV-Cre, we determined the amount of recoverable virus from lungs over the same time course as the FACS experiments. While high titers were recovered at 2 and 5 post-infection, no virus was detected at day 10 post-infection (Fig. 2 C). To identify where in the lung architecture these surviving cells exist, we infected reporter animals and collected lungs for histological analysis at 10 d post-infection. Upon examination of the lung sections, tdTomato+ cells were only found in the epithelial layer of larger airway spaces (bronchi), never in the alveoli (Fig. 2 D).
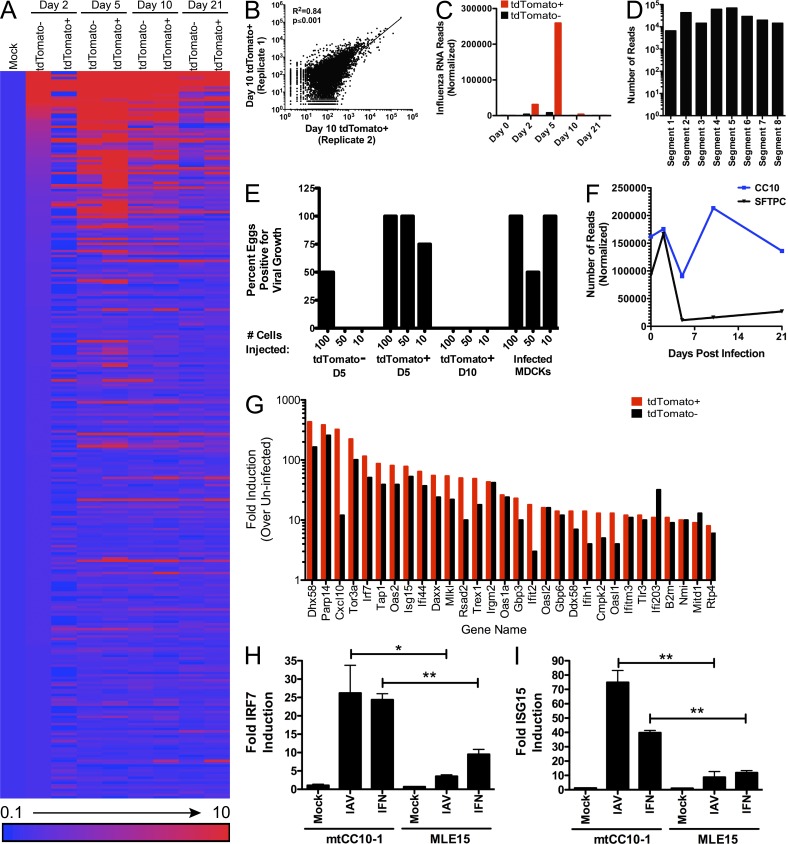
In an effort to better characterize and identify the surviving cells type(s), we profiled the transcriptome of tdTomato+ cells. To this end, reporter mice were infected with IAV-Cre and over a time course we performed fluorescence-activated cell sorting (FACS). At each time point (days 0, 2, 5, 10, 21 post-infection), we separated live, CD45−, tdTomato+ and negative cells and collected RNA from the different populations. Next-generation RNA-seq analysis was then performed to define the transcriptional profiles of each population. Transcriptome analyses of these two cellular cohorts demonstrated that despite deriving from comparable environments, there were significant differences in the gene expression profiles (Fig. 3 A). Results from replicate tdTomato+ cell isolation and RNA-seq experiments were highly reproducible (Fig. 3 B). Thus, cells that will go on to survive infection have a distinct transcriptional profile relative to uninfected cells.
Figure 3.
Surviving cells have an altered transcriptional profile and represent a unique cellular lineage. Live, CD45− tdTomato+, and tdTomato− cells were sorted at the indicated time points and transcriptional profile assessed by mRNA-seq. (A) The 300 most differentially expressed transcripts mapping to the mouse genome were ranked by magnitude of induction (at 2 d post-infection) and represented as fold change relative to mock-infected lung cells. Data from 2, 5, 10, and 21 d post-infection are represented. (B) Replicate sequencing experiments of 10 d post-infection tdTomato+ samples were plotted against each other to demonstrate reproducibility. Data are representative of all time points analyzed. R2 indicates the coefficient of determination and the p-value indicates the significance of the association between the two variables. (C) Transcripts from tdTomato+ and tdTomato− cells that mapped to IAV mRNA were analyzed at the indicated time points. (D) Transcripts that mapped to each of the eight IAV segments were analyzed from tdTomato+ cells at 5 d post-infection. (E) FACS sorted cells from tdTomato+ or tdTomato− populations at the indicated time points were injected into embryonated chicken eggs. 48 h after injection, the detection of amplified virus (via hemagglutination assay) was scored as positive. (F) mRNA reads were analyzed for cell type–specific signatures, Cc10 and Sftpc at the indicated time points. Data are representative of two independent experiments. (G) The fold induction of interferon stimulated genes in tdTomato+ and tdTomato− cells at 5 d post-infection were ranked and plotted by largest change in tdTomato+ cells. (H) Club or mouse lung epithelial cell lines infected with IAV (MOI = 1) or treated with IFN-α/β and analyzed for Irf7 expression. (I) Cells were infected/treated as in H and analyzed for Isg15 expression. Data in H and I are representative of two independent experiments. Significance is based on an unpaired Student’s t test. *, P ≤ 0.05; **, P ≤ 0.001.
Within the RNA-seq data, we assessed IAV-specific mRNA reads. In agreement with data in Fig. 2 C, we observed high levels of viral mRNA at early time points (days 2 and 5 post-infection), but viral mRNA is lost from tdTomato+ cells by day 10 post-infection (Fig. 3 C). Recovered viral mRNA reads from day 5 infected cells mapped to all 8 viral segments (Fig. 3 D) indicating that activation of the reporter is unlikely to be a reflection of defective interfering particle entry but instead competent viral infection and replication. To further demonstrate that tdTomato+ cells were the result of productive viral infection, we injected FACS-purified tdTomato+ cells from day 5 post-infection animals into embryonated chicken eggs, for which 11/12 eggs became positive for virus (Fig. 3 E). Conversely, eggs injected with tdTomato+ cells from 10 d post-infection animals failed to induce viral amplification (Fig. 3 E). Together, these data argue that the observed tdTomato+ cells are the result of cells surviving and successfully clearing a productive IAV infection.
In an effort to define the cell lineage capable of surviving a productive IAV infection, we analyzed the RNA-seq data for cell type-specific markers over a 21-d time period. At early time points during active IAV replication, tdTomato+ cells demonstrate high levels of Sftpc and Cc10 (Fig. 3 F), markers specific for type II alveolar cells and club cells, respectively (Glasser et al., 1990; Hackett et al., 1992; Kalina et al., 1992; Daly et al., 1997). Interestingly, time points as early as day 5 post-infection show almost a complete loss of Sftpc in tdTomato+ cells while the levels of Cc10 are maintained (Fig. 3 F). Furthermore, with the exception of Cc10, no cell-specific marker was detected in appreciable amounts in the surviving cell population implicating club cells as the long-term cell survivors of IAV infection (Table S1). This hypothesis is further supported by data demonstrating that club cells are restricted to the larger airways of the respiratory tract, the same physiological location where tdTomato+ expression was observed (Fig. 2 D).
We next turned our attention to understanding the mechanism by which tdTomato+ cells resisted the cytopathic effects of IAV. The mammalian response to infection is mediated by the type I IFN (IFN-I) response and the transcriptional up-regulation of a barrage of antiviral genes (Bowie and Unterholzner, 2008). Strikingly, we observed an increase in magnitude of ISGs in tdTomato+ cells relative to tdTomato− cells in the same lungs (Fig. 3 G). Furthermore, this enhanced antiviral transcriptional signature persisted for the duration of the time course (Table S1). Given the capacity of ISGs to block IAV replication (Fig. 1 C), we postulate that this transcriptional signature is responsible for allowing cellular survival and clearance of viral replication.
To determine if club cells are more responsive to IFN-I, as suggested by in vivo RNA-seq data, we treated a murine club cell line (mtCC10-1; Magdaleno et al., 1997) and a lung epithelial cell line (MLE-15; Wikenheiser et al., 1993) with either IFN-I or IAV (Fig. 3, H and I). In agreement with the in vivo transcriptome analyses, club cells had significantly higher expression of ISGs—including the master regulator of the response—Irf7 (Honda et al., 2005). Although the relationship between ISG up-regulation and cellular survival is only correlative, we favor the hypothesis that the robust IFN-I response in club cells permits a subpopulation of cells to successfully clear infection despite active virus replication.
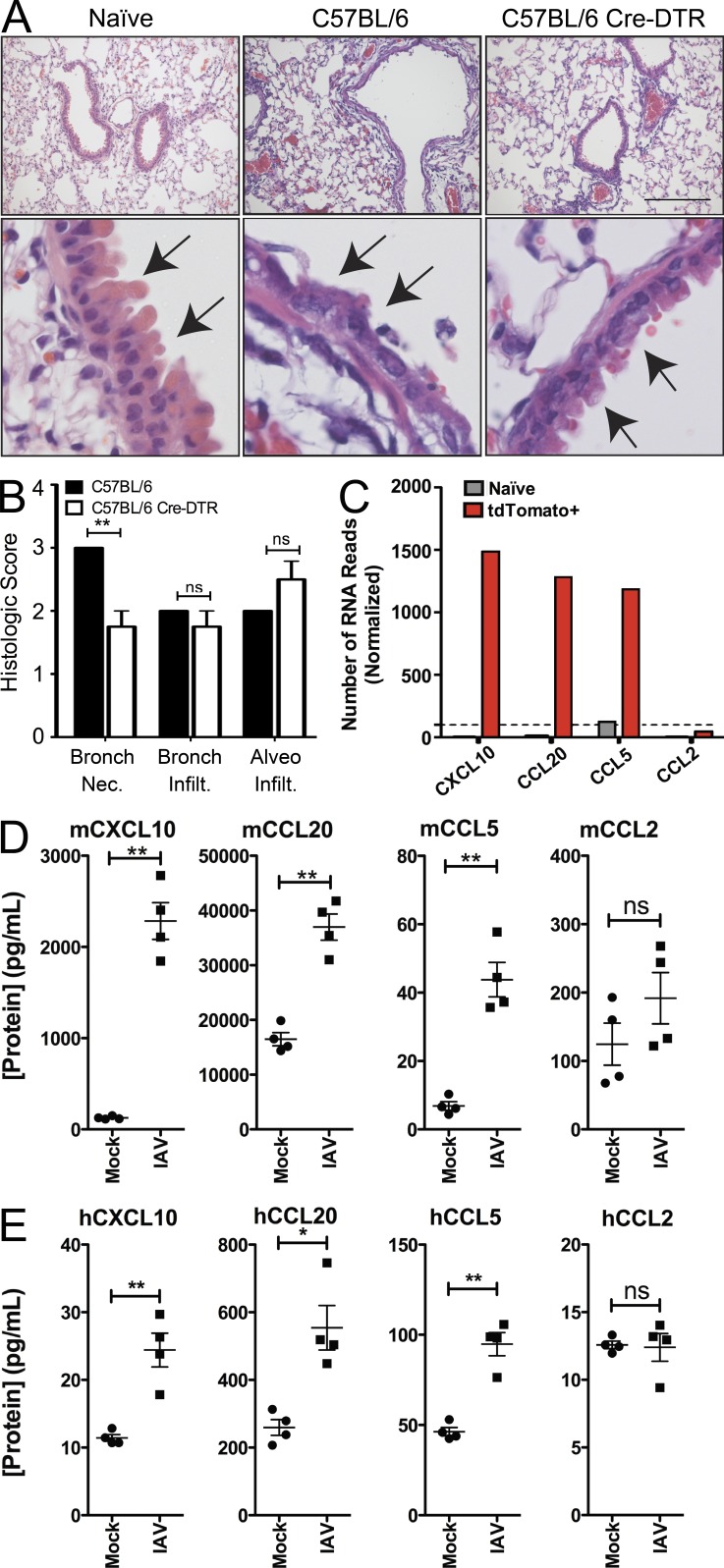
Finally, we wanted to ascertain the function of club cells as they relate to the pathogenesis of IAV infection. To define a role for surviving cells, we used another Cre-responsive transgenic mouse strain, one in which instead of turning on tdTomato expression, the diphtheria toxin receptor is expressed (Buch et al., 2005). By using these mice, we can administer diphtheria toxin and selectively deplete only those cells that have been infected by IAV. To this end, we infected wild-type or the diphtheria receptor transgenic mice with 500 PFU of IAV-Cre as in previous studies with the tdTomato reporter mice (Fig. 2). After allowing the virus to replicate for 5 d, we administered diphtheria toxin—a time where diphtheria toxin receptor expression should be limited to long-term survivors of viral infection (Fig. 3 F). At 10 d post-infection (5 d after diphtheria toxin administration), lung sections were processed for histological analysis. Sections were then analyzed and scored by an independent pathologist. Counterintuitively, there was a noticeable improvement in lung pathology under conditions where surviving cells were depleted (Fig. 4 A). Although the overall cell infiltration scores were not significantly different between the treatment groups, there was a significant reduction in bronchiolar epithelial pathology, the exact localization of club cells (Fig. 4 B). Specifically, the epithelium in nondepleted mice was observed to frequently be necrotic and attenuated in a segmental manner that affected most of the airways. Rarely, a few normal segments were found at the terminal bronchioles. In contrast, the bronchiolar epithelium in mice where surviving club cells were depleted showed only minor necrotic lesions and less of the airway epithelium was affected. Frequently, the terminal bronchioles were completely unaffected (Fig. 4, A and B). Although the interpretation of these data are complicated by the possible contribution of surviving nonclub cells, the data show that surviving cells do significantly contribute to the lung pathology in the larger airways after IAV infection.
Figure 4.
Surviving cells contribute to IAV-induced immunopathology. Wild-type (C57BL/6) or diphtheria toxin receptor Cre inducible (C57BL/6 Cre-DTR) mice were infected with 500 PFU of IAV-Cre. 5 d post-infection, mice were administered diphtheria toxin to deplete surviving club cells. At 10 d post-infection (5 d after toxin depletion), lungs were collected, and lung pathology was assessed. (A) Representative images of lung sections from uninfected mice, C57BL/6 mice infected and toxin treated (nondepleted), and C57BL/6 Cre-DTR mice infected and toxin treated (depleted for surviving club cells). Enlargements (bottom) show a reduction in bronchiolar epithelial necrosis when surviving club cells are depleted, arrows indicate the epithelial cell layer. Bar, 200 µm. (B) Independent pathologist scoring of virus induced bronchiolar epithelial cell necrosis (Bronch. Nec.), bronchiolar infiltration (Bronch. Infilt.), and alveolar infiltration (Aveo. Infilt.). Data are representative of two independent experiments. (C) RNA-seq reads of chemokine transcripts highly induced in tdTomato+ surviving cells relative to uninfected mice. The dashed line indicates the reproducible level of detection for RNA transcripts based on Fig 3 B. (D) The murine club cell line mtCC10-1 was mock or IAV-infected at an MOI of 2 for 24 h. The concentration of the secreted chemokines in the cellular supernatant is shown. (E) The human club cell line H441 was mock or IAV infected at an MOI of 2 for 24 h. The concentrations of the secreted chemokines in the cellular supernatant are indicated. Data in D and E are representative of two independent experiments. Significance is based on an unpaired Student’s t test. *, P ≤ 0.05; **, P ≤ 0.001.
To understand the mechanism of how surviving cells were negatively influencing the bronchiolar epithelium, we looked for highly induced transcripts that were also differentially expressed between tdTomato+ and tdTomato− cells. Many of the genes that fit these criteria were proinflammatory cytokines and chemokines, most strikingly: Cxcl10, Ccl20, and Ccl5 (Fig. 4 C). Interestingly, it does not appear that proinflammatory mediators are universally up-regulated, as the monocyte chemoattractant Ccl2 was not appreciably induced in surviving tdTomato+ cells (Fig. 4 C). To validate secretion of the chemokines identified from our in vivo RNA-seq data, we infected murine club cells in vitro and quantified CXCL10, CCL20, CCL5, and CCL2 protein levels in the supernatant (Fig. 4 D). In agreement with our RNA data, infection of club cells led to a significant increase of secreted CXCL10, CCL20, and CCL5 (Fig. 4 D). No significant changes were observed in CCL2 levels, as expected (Fig. 4 D).
Finally, we wanted to ensure that the club cell biology described here was not murine specific. We therefore infected the human club cell line H441 (Brower et al., 1986) with IAV and measured release of a panel of proinflammatory factors. Similar to mouse club cells, CXCL10, CCL20, and CCL5 were significantly up-regulated upon infection (Fig. 4 E). CCL2 was not reproducibly induced by viral infection as expected (Fig. 4 E). These data suggest that human club cells and murine club cells respond similarly during viral infection.
In summary, we demonstrate that despite IAV causing an acute and lytic infection, a subpopulation of lung epithelial cells survive viral replication. Long-term surviving cells were identified to be predominantly club cells that expressed a robust ISG response. Furthermore, surviving club cells demonstrate elevated levels of proinflammatory mediators relative to uninfected cells in the same lungs, which ultimately leads to increased lung pathology in the bronchi. We therefore propose a model in which club cells that are directly infected by IAV but survive that infection provide a local proinflammatory environment in the bronchi. Although club cells are likely critical for the initial up-regulation of an antiviral response, their prolonged activation appears to be detrimental to bronchus remodeling after the pathogen has been cleared. This finding is particularly relevant given that the significant mortality in the most severe influenza infections (recently with the 2009 swine H1N1 virus [Gao et al., 2013] and avian H5N1 viruses [de Jong et al., 2006]) is attributed to a sustained proinflammatory response after infection. Although it is unlikely that the small numbers of surviving club cells contribute significantly to the global cytokine profile of an infected host, the localized effect on bronchiolar epithelial layer integrity may be an important component of IAV-related immunopathology.
MATERIALS AND METHODS
Cell lines and in vitro infections.
293T, H441, and MDCK cells (American type Culture Collection) were used in this study. mtCC10-1 and MLE-15 cells were gifts from F. DeMayo (Baylor College of Medicine, Houston, TX) and J. Whitsett (University of Cincinnati, Cincinnati, OH), respectively. All cells were maintained in DMEM supplemented with 10% FCS, l-glutamine, and Pen/Strep with the exception of the H441 cells, which were maintained in RPMI-1640 and supplemented with 10% FCS and Pen/Strep. For infection, virus was diluted to the appropriate concentration in PBS supplemented with 3% BSA and used to infect cells for 1 h at 37°C, followed by replacement of culture media. Virus was titered on MDCK cells as previously described (Langlois et al., 2013).
Generation of Cre recombinase expressing influenza virus and virus quantification.
The plasmid encoding PB2-Cre in the pDZ vector, which expresses negative sense vRNA and positive sense mRNA. PB2 with silent mutations in the packaging signal was amplified from the previously published PB2-GLuc plasmid (Heaton et al., 2013a) with the following primers: forward 5′-CTCCGAAGTTGGGGGGGAGCGAAAGCAGG-3′ and reverse 5′-GTTAATAGCCATACGGATCCTCTTAG-3′. We synthesized a sequence encoding a PTV-1 2A site, a codon-optimized Cre recombinase with an N-terminal SV40 NLS and a duplicated PB2 packaging signal. The Cre region was amplified with: forward 5′-ATCCGTATGGCTATTAACGGAAGCGGAGCAACAAATTTCAGC-3′ and reverse 5′-TGGGCCGCCGGGTTATTAGTAGAAACAAGG-3′. The two PCR fragments were recombined into pDZ via Infusion HD cloning (Takara Bio Inc.). Clones were sequence verified and rescued via 293T transfection and amplification in embryonated chicken eggs as previously described (Heaton et al., 2013b). Rescued virus was dilution purified and titered on MDCK cells. For all experiments, viral titer was determined via standard plaque assay on MDCK cells.
Mice, virus infection, and diphtheria treatment.
Wild-type C57BL/6, B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J and C57BL/6-Gt(ROSA)26Sortm1(HBEGG)Awai/J mice were purchased from The Jackson Laboratory. Mice were anesthetized with ketamine/xylazine and infected i.n. with 500 PFU of IAV-Cre unless otherwise indicated. 5 d post-infection, mice were administered 100 ng diphtheria toxin (Sigma-Aldrich) i.p. and 10 ng i.n. Body weight was monitored over the course of infection, and 80% initial body weight was designated as the humane endpoint. All experiments involving animals were performed in accordance with the Icahn School of Medicine Animal Care and Use Committee.
Lung sectioning and histology.
Mice were euthanized via CO2 inhalation and lungs were inflated before removal. For H&E staining, lungs were inflated and fixed with 4% PFA in PBS. Lungs were paraffin embedded and 5-µm sections were cut. For pathological scoring, two lung sections 100 µm apart were analyzed. For frozen sectioning and tdTomato+ cell localization, lungs were inflated with 10% OCT in PBS. Lungs were embedded in OCT and frozen at −80C. 5 µm sections were dried on slides, fixed for 3 min in 1% PFA, and mounted in Prolong Gold with DAPI (Invitrogen).
Flow cytometry.
Lungs were digested with Type-IV collagenase (Worthington) for 30 min with agitation, and then processed into single-cell suspension and incubated with a rat anti–mouse CD45 antibody (30-F11; BD). Live/dead violet dye was used as per the manufacturer’s instructions (Life Technologies). All flow cytometry data were acquired on a LSR II (BD), sorting performed on a FACS Aria (BD), and analyzed using FlowJo software (Tree Star).
Next generation mRNA sequencing.
Mice were infected and cells FACS sorted as described above. 500 of the indicated cells were sorted into a 96-well plate, containing 20 µl of RNA reaction buffer. RNA was extracted and reverse transcribed via SMARTer Ultra Low RNA kit for Illumina Sequencing (Takara Bio Inc.) as per the manufacturer’s instructions. cDNA was sheared via Covaris sonication and prepared for sequencing using the TruSeq DNA sample Preparation kit (Illumina) as per the manufacturer’s instructions. Samples were sequenced via 100 nt single-end run on a HiSeq 2000. The raw data are available under accession nos: GSM1422381, GSM1422382, GSM1422383, GSM1422384, GSM1422385, GSM1422386, GSM1422387, GSM1422388, GSM1422389. The reads were mapped to the mouse transcriptome and the influenza genome using Bowtie. All software was parallelized and run on an internal high performance-computing cluster at Mount Sinai. Heat map visualization of the sequencing data were generated using the method described by Pavlidis and Noble (2003).
Microscopy.
All images of tdTomato+ cells in cell culture and lung sections were captured on an Olympus IX-70 camera. All H&E-stained sections were captured on an Axioplan 2 camera (Carl Zeiss). Images were captured with the same exposures and processed with ImageJ (National Institutes of Health). The same thresholds were applied to all images in a given experiment with the exception of the DAPI channel, which due to nonuniform staining was adjusted to be visible in all sections.
qRT-PCR.
RNA was extracted, reverse transcribed and assessed by qPCR as previously described (Langlois et al., 2012). In brief, cDNA was amplified using primers specific for tubulin, IRF-7, and ISG15 with KAPA SYBR FAST qPRC Master Mix (KAPA Biosystems) and analyzed on a realplex2 (Eppendorf). Delta delta cycle threshold (ΔΔCT) values were calculated with replicates over tubulin. Values represent the fold change over mock-infected samples.
Cytokine and chemokine quantification.
Protein levels of cytokines/chemokines in culture medium were evaluated using a multiplex bead array assay. All the antibodies and cytokine standards were purchased as antibody pairs from R&D Systems or PeproTech. Individual bead sets (Luminex) were coupled to cytokine-specific capture antibodies according to the manufacturer’s recommendations and as previously described (Biancotto et al., 2007). Conjugated beads were washed and kept at 4°C until use. Biotinylated polyclonal antibodies were used at twice the concentrations recommended for a classical ELISA according to the manufacturer. All assay procedures were performed in assay buffer containing PBS supplemented with 1% normal mouse serum (Invitrogen), 1% normal goat serum (Invitrogen), and 20 mM Tris-HCl (pH 7.4). The assays were run using 1,500 beads per set of each of cytokines measured per well in a total volume of 50 µl. The plates were read on a Luminex MAGPIX platform. For each bead set, >50 beads were collected. The median fluorescence intensity of these beads was recorded and used for analysis with the Milliplex software using a 5P regression algorithm.
Statistical analysis.
Statistical analysis between datasets was performed using a one- or two-tailed Student’s t test where appropriate. Differences were considered to be statistically significant at P values at or below 0.05.
Online supplemental material.
Table S1 shows normalized RNA transcript numbers detected in FACS purified tdTomato− and tdTomato+ cell populations over a 21-d time course. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20140488/DC1.
Supplementary Material
Acknowledgments
We would like to thank Virginia Gillespie for performing the lung pathological scoring, Omar Jabado and the Genomics core facility, Carlos A. Rodriguez, and the Mount Sinai Microscopy Shared Resource Facility for technical assistance. We also thank Adeeb Rahman and the Flow Cytometry Shared Resource Facility for assistance in FACS experiments.
N.S. Heaton is a Merck fellow of the Life Sciences Research Foundation. R.A. Langlois is supported by the Research Training Program in Molecular and Cellular Hematology (T32-HL094283). This work was partially supported by a Centers for Excellence for Influenza Research and Surveillance grant (HHSN26620070010C to P. Palese), NIH program project grant (1P01AI097092 to P. Palese), PATH (P. Palese), and by the NIH (grant number A1093571 to B.R. tenOever).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- IAV
- influenza A virus
- ISG
- interferon-stimulated gene
References
- Biancotto, A., Grivel J.C., Iglehart S.J., Vanpouille C., Lisco A., Sieg S.F., Debernardo R., Garate K., Rodriguez B., Margolis L.B., and Lederman M.M.. 2007. Abnormal activation and cytokine spectra in lymph nodes of people chronically infected with HIV-1. Blood. 109:4272–4279 10.1182/blood-2006-11-055764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie, A.G., and Unterholzner L.. 2008. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 8:911–922 10.1038/nri2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower, M., Carney D.N., Oie H.K., Gazdar A.F., and Minna J.D.. 1986. Growth of cell lines and clinical specimens of human non-small cell lung cancer in a serum-free defined medium. Cancer Res. 46:798–806 [PubMed] [Google Scholar]
- Buch, T., Heppner F.L., Tertilt C., Heinen T.J., Kremer M., Wunderlich F.T., Jung S., and Waisman A.. 2005. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat. Methods. 2:419–426 10.1038/nmeth762 [DOI] [PubMed] [Google Scholar]
- Carrat, F., Vergu E., Ferguson N.M., Lemaitre M., Cauchemez S., Leach S., and Valleron A.J.. 2008. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am. J. Epidemiol. 167:775–785 10.1093/aje/kwm375 [DOI] [PubMed] [Google Scholar]
- Daly, H.E., Baecher-Allan C.M., Barth R.K., D’Angio C.T., and Finkelstein J.N.. 1997. Bleomycin induces strain-dependent alterations in the pattern of epithelial cell-specific marker expression in mouse lung. Toxicol. Appl. Pharmacol. 142:303–310 10.1006/taap.1996.8056 [DOI] [PubMed] [Google Scholar]
- Damjanovic, D., Small C.L., Jeyanathan M., McCormick S., and Xing Z.. 2012. Immunopathology in influenza virus infection: uncoupling the friend from foe. Clin. Immunol. 144:57–69 10.1016/j.clim.2012.05.005 [DOI] [PubMed] [Google Scholar]
- de Jong, M.D., Simmons C.P., Thanh T.T., Hien V.M., Smith G.J.D., Chau T.N.B., Hoang D.M., Chau N.V.V., Khanh T.H., Dong V.C., et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12:1203–1207 10.1038/nm1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberger, M., Allan W., Zijlstra M., Jaenisch R., and Doherty P.C.. 1991. Clearance of influenza virus respiratory infection in mice lacking class I major histocompatibility complex-restricted CD8+ T cells. J. Exp. Med. 174:875–880 10.1084/jem.174.4.875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, R., Bhatnagar J., Blau D.M., Greer P., Rollin D.C., Denison A.M., Deleon-Carnes M., Shieh W.J., Sambhara S., Tumpey T.M., et al. 2013. Cytokine and chemokine profiles in lung tissues from fatal cases of 2009 pandemic influenza A (H1N1): role of the host immune response in pathogenesis. Am. J. Pathol. 183:1258–1268 10.1016/j.ajpath.2013.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser, S.W., Korfhagen T.R., Bruno M.D., Dey C., and Whitsett J.A.. 1990. Structure and expression of the pulmonary surfactant protein SP-C gene in the mouse. J. Biol. Chem. 265:21986–21991 [PubMed] [Google Scholar]
- Hackett, B.P., Shimizu N., and Gitlin J.D.. 1992. Clara cell secretory protein gene expression in bronchiolar epithelium. Am. J. Physiol. 262:L399–L404 [DOI] [PubMed] [Google Scholar]
- Heaton, N.S., Leyva-Grado V.H., Tan G.S., Eggink D., Hai R., and Palese P.. 2013a. In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J. Virol. 87:8272–8281 10.1128/JVI.00969-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton, N.S., Sachs D., Chen C.J., Hai R., and Palese P.. 2013b. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. USA. 110:20248–20253 10.1073/pnas.1320524110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T.. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Julkunen, I., Sareneva T., Pirhonen J., Ronni T., Melén K., and Matikainen S.. 2001. Molecular pathogenesis of influenza A virus infection and virus-induced regulation of cytokine gene expression. Cytokine Growth Factor Rev. 12:171–180 10.1016/S1359-6101(00)00026-5 [DOI] [PubMed] [Google Scholar]
- Kalina, M., Mason R.J., and Shannon J.M.. 1992. Surfactant protein C is expressed in alveolar type II cells but not in Clara cells of rat lung. Am. J. Respir. Cell Mol. Biol. 6:594–600 10.1165/ajrcmb/6.6.594 [DOI] [PubMed] [Google Scholar]
- Kim, J.H., Lee S.R., Li L.H., Park H.J., Park J.H., Lee K.Y., Kim M.K., Shin B.A., and Choi S.Y.. 2011. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE. 6:e18556 10.1371/journal.pone.0018556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois, R.A., Varble A., Chua M.A., García-Sastre A., and tenOever B.R.. 2012. Hematopoietic-specific targeting of influenza A virus reveals replication requirements for induction of antiviral immune responses. Proc. Natl. Acad. Sci. USA. 109:12117–12122 10.1073/pnas.1206039109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois, R.A., Albrecht R.A., Kimble B., Sutton T., Shapiro J.S., Finch C., Angel M., Chua M.A., Gonzalez-Reiche A.S., Xu K., et al. 2013. MicroRNA-based strategy to mitigate the risk of gain-of-function influenza studies. Nat. Biotechnol. 31:844–847 10.1038/nbt.2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen, L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R., et al. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13:133–140 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magdaleno, S.M., Wang G., Jackson K.J., Ray M.K., Welty S., Costa R.H., and DeMayo F.J.. 1997. Interferon-gamma regulation of Clara cell gene expression: in vivo and in vitro. Am. J. Physiol. 272:L1142–L1151 [DOI] [PubMed] [Google Scholar]
- Manicassamy, B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., and García-Sastre A.. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. USA. 107:11531–11536 10.1073/pnas.0914994107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich, M.N., Matrosovich T.Y., Gray T., Roberts N.A., and Klenk H.D.. 2004. Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc. Natl. Acad. Sci. USA. 101:4620–4624 10.1073/pnas.0308001101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, A.2000. Cre recombinase: the universal reagent for genome tailoring. Genesis. 26:99–109 [DOI] [PubMed] [Google Scholar]
- Pavlidis, P., and Noble W.S.. 2003. Matrix2png: a utility for visualizing matrix data. Bioinformatics. 19:295–296 10.1093/bioinformatics/19.2.295 [DOI] [PubMed] [Google Scholar]
- Sanders, C.J., Doherty P.C., and Thomas P.G.. 2011. Respiratory epithelial cells in innate immunity to influenza virus infection. Cell Tissue Res. 343:13–21 10.1007/s00441-010-1043-z [DOI] [PubMed] [Google Scholar]
- Shieh, W.J., Blau D.M., Denison A.M., Deleon-Carnes M., Adem P., Bhatnagar J., Sumner J., Liu L., Patel M., Batten B., et al. 2010. 2009 pandemic influenza A (H1N1): pathology and pathogenesis of 100 fatal cases in the United States. Am. J. Pathol. 177:166–175 10.2353/ajpath.2010.100115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smed-Sörensen, A., Chalouni C., Chatterjee B., Cohn L., Blattmann P., Nakamura N., Delamarre L., and Mellman I.. 2012. Influenza A virus infection of human primary dendritic cells impairs their ability to cross-present antigen to CD8 T cells. PLoS Pathog. 8:e1002572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, O., and Akira S.. 2009. Innate immunity to virus infection. Immunol. Rev. 227:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikenheiser, K.A., Vorbroker D.K., Rice W.R., Clark J.C., Bachurski C.J., Oie H.K., and Whitsett J.A.. 1993. Production of immortalized distal respiratory epithelial cell lines from surfactant protein C/simian virus 40 large tumor antigen transgenic mice. Proc. Natl. Acad. Sci. USA. 90:11029–11033 10.1073/pnas.90.23.11029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann, A., and Noack T.. 2010. The Clara cell: a “Third Reich eponym”? Eur. Respir. J. 36:722–727 10.1183/09031936.00146609 [DOI] [PubMed] [Google Scholar]
- Yatim, N., and Albert M.L.. 2011. Dying to replicate: the orchestration of the viral life cycle, cell death pathways, and immunity. Immunity. 35:478–490 10.1016/j.immuni.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Zinkernagel, R.M., and Doherty P.C.. 1979. MHC-restricted cytotoxic T cells: studies on the biological role of polymorphic major transplantation antigens determining T-cell restriction-specificity, function, and responsiveness. Adv. Immunol. 27:51–177 10.1016/S0065-2776(08)60262-X [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.