A novel isoform of Sox5, Sox5t, and c-Maf activate RORγt to induce Th17 cells. Sox5−/− mice exhibit impaired Th17 differentiation and are thus resistant to EAE and delayed-type hypersensitivity.
Abstract
Stat3 signaling is essential for the induction of RORγt and subsequent Th17 cell differentiation. However, the downstream targets of Stat3 for RORγt expression remain largely unknown. We show here that a novel isoform of Sox5, named Sox5t, is induced in Th17 cells in a Stat3-dependent manner. In vivo, T cell–specific Sox5-deficient mice exhibit impaired Th17 cell differentiation and are resistant to experimental autoimmune encephalomyelitis and delayed-type hypersensitivity. Retrovirus-mediated induction of Sox5 together with c-Maf induces Th17 cell differentiation even in Stat3-deficient CD4+ T cells but not in RORγt-deficient CD4+ T cells, indicating that Sox5 and c-Maf induce Th17 cell differentiation as downstream effectors of Stat3 and as upstream inducers of RORγt. Moreover, Sox5 physically associates with c-Maf via the HMG domain of Sox5 and DNA-binding domain of c-Maf, and Sox5 together with c-Maf directly activates the promoter of RORγt in CD4+ T cells. Collectively, our results suggest that Sox5 and c-Maf cooperatively induce Th17 cell differentiation via the induction of RORγt as downstream targets of Stat3.
Th17 cells produce IL-17A and IL-17F and play a pathogenic role in a variety of autoimmune diseases (Dong, 2008; Korn et al., 2009; Littman and Rudensky, 2010). Activated CD4+ T cells need to be stimulated with IL-6/TGF-β (Bettelli et al., 2006; Mangan et al., 2006; Veldhoen et al., 2006) or IL-6/IL-1β/IL-23 (Ghoreschi et al., 2010) to develop into Th17 cells. Because the overexpression of RORγt (encoded by Rorc) induces Th17 cell differentiation, whereas RORγt-deficient mice lack Th17 cell differentiation (Ivanov et al., 2006), RORγt is assumed to be a lineage-specifying transcription factor of Th17 cells (Ciofani et al., 2012). Another ROR family member, RORα, is also involved in the generation of Th17 cells in conjunction with RORγt (Yang et al., 2008).
Regarding the mechanism underlying the induction of RORγt in developing Th17 cells, IL-6– and/or IL-21–mediated activation of Stat3 has been shown to play a central role (Laurence et al., 2007; Mathur et al., 2007; Yang et al., 2007; Zhou et al., 2007). Stat3 binds to intron 1 of the Rorc gene and allows permissive histone H3 lysine 4 trimethylation (H3K4me3) marks on Rorc, whereas Stat3 does not activate the promoter of Rorc (Durant et al., 2010; Lazarevic et al., 2011). With regard to the downstream pathways of Stat3, several genes including Nfkbiz, Rora, Batf, Irf4, Ahr, Maf, and HIF-1α have been demonstrated to be activated by Stat3 and implicated in Th17 cell differentiation (Brüstle et al., 2007; Veldhoen et al., 2008; Yang et al., 2008; Bauquet et al., 2009; Schraml et al., 2009; Durant et al., 2010; Dang et al., 2011). Among these transcription factors, HIF-1 has been shown to activate Rorc promoter (Dang et al., 2011). However, the downstream targets of Stat3 for RORγt induction have not been fully understood.
In this regard, we have previously performed DNA microarray analysis of IL-6–stimulated CD4+ T cells to identify transcription factors that are involved in Th17 cell differentiation as downstream targets of IL-6–Stat3 pathways (Hiramatsu et al., 2010). We have reported that c-Maf is highly expressed not only in IL-6–stimulated CD4+ T cells, but also in Th17 cells, and that c-Maf binds to the promoter and enhancer of IL-21 gene and induces IL-21 production in CD4+ T cells. In addition, Bauquet et al. (2009) have shown that c-Maf is required for the maintenance of Th17 cells by up-regulating IL-21 production. On the other hand, it has recently been shown that c-Maf negatively regulates several genes, including Il22 (Rutz et al., 2011), Batf, Rora, Runx1, Il1r1, Ccr6, and Tnf (Ciofani et al., 2012) in Th17 cells. However, interrelationship between c-Maf and RORγt during Th17 cell differentiation remains largely unknown.
Sox5 is a member of the SOX (SRY-related high-mobility-group [HMG]-box) family of transcription factors (Wegner, 2010). Sox5 belongs to SoxD group which is composed of Sox5, Sox6, and Sox13 (Lefebvre et al., 1998; Lefebvre, 2010). Sox5 has three functional domains, a HMG box DNA-binding domain and two coiled-coil domains, and the first coiled-coil domain mediates homo- and hetero-dimerization of SoxD proteins. SoxD proteins themselves do not have transactivation or transrepression domain and thus their activity is likely to be influenced by other molecules with which they interact. Sox5-deficient mice die after birth due to a cleft secondary palate and small thoracic cage (Smits et al., 2001; Dy et al., 2008), which is consistent with a finding that Sox5 is highly expressed in spermatids, neurons, oligodendrocytes, and chondrocytes. Regarding the relationship between Sox5 and autoimmune diseases, it has recently been shown that Sox5 is one of the most strikingly up-regulated transcription factors in whole blood in patients with multiple sclerosis (Riveros et al., 2010). In addition, our DNA microarray analysis of IL-6–stimulated CD4+ T cells has revealed that Sox5 is the most strongly induced transcription factor in CD4+ T cells upon IL-6 stimulation (unpublished data). However, the role of Sox5 in helper T cell differentiation is unknown.
In this study, we examined the role of Sox5 in the development of Th17 cells as well as in experimental autoimmune encephalomyelitis (EAE), which is a murine model of multiple sclerosis mainly caused by Th17 cell–mediated autoimmune responses. We show here that a novel isoform of Sox5 (named Sox5t) is expressed in Th17 cells and that T cell–specific Sox5-deficient mice are resistant to EAE. In addition, we show that Sox5t along with c-Maf induces RORγt expression and subsequent Th17 cell differentiation as downstream targets of IL-6–Stat3 pathways.
RESULTS
A novel isoform of Sox5 is expressed in CD4+ T cells in response to IL-6
To identify transcription factors downstream of IL-6–Stat3 pathways, we have performed DNA microarray analysis on IL-6–stimulated CD4+ T cells (Hiramatsu et al., 2010). We searched transcription factors that were up-regulated in CD4+ T cells in response to IL-6 and found that in addition to c-Maf, Sox5 was highly up-regulated in CD4+ T cells upon IL-6 stimulation (unpublished data). It has been reported that long form of murine Sox5 (L-Sox5: AJ010604) consists of 14 coding exons (Lefebvre et al., 1998; Dy et al., 2008; Fig. 1 A). To identify 5′ end of Sox5 expressed in IL-6–stimulated CD4+ T cells, we performed oligo-capping RACE and found that Sox5 contained a 2,148-bp open reading frame that differs from previously reported isoforms of Sox5 (Fig. 1 A). Hereafter, we designated this isoform as Sox5t. The transcription initiation site of Sox5t was 572-kb upstream from that of L-Sox5, suggesting that these two isoforms use different promoters. Sox5t lacked coding exon 1 of l-Sox5 that encodes N-terminal 13 aa but had another exon that encodes 49 amino acids between coding exon 8 and 9 of L-Sox5 (Fig. 1, B and C). We also found another isoform of Sox5t that lacked the insertion of the extra exon.
Figure 1.
A novel isoform of Sox5 is expressed in IL-6–stimulated CD4+ T cells. (A) Alignments between Sox5t and NCBI RNA reference sequences collection (RefSeq Genes) and mouse mRNAs in GenBank are shown. (B, top) Schema of the coding region of Sox5t and l-Sox5 (AJ010604.1). The locations of the primers used in qPCR analysis (Sox5t primer,l-Sox5 primer, and Sox5 HMG primer) are shown. (bottom) Sequence comparison of Sox5t and l-Sox5. Aligned are the first 360 nucleotides of Sox5t and 243 nucleotides of l-sox5 and the predicted amino acid sequence for Sox5t and l-Sox5. (C) Schematic comparison of Sox5t and l-Sox5. (D) The expression levels of Sox5t, l-Sox5, and Sox5 HMG in various tissues and CD4+ T cells. qPCR was performed by using Sox5t, l-Sox5, and Sox5 HMG primers indicated in B. Shown are means ± SD. Data are compiled from three independent experiments. (E) Naive CD4+ T cells from CD4cre Stat3fl/fl mice or Stat3fl/fl mice were stimulated under either neutral conditions, IL-6 conditions, or Th17 conditions for 4 d, and subjected to Western blot analysis for Sox5, c-Maf, and Hsp90 (as a control). Data are representative of three independent experiments.
To examine the expression of l-Sox5 and Sox5t in a various organs and cells, we designed primer pairs that are common to two isoforms (Sox5 HMG primers) and specific for l-Sox5 or Sox5t (L-Sox5 or Sox5t primers; Fig. 1 B) and examined their expression by quantitative PCR (qPCR). As shown in Fig. 1 D, Sox5t was highly expressed in IL-6–stimulated CD4+ T cells but not in naive CD4+ T cells. In contrast, l-Sox5 was expressed in the brain and liver but not in IL-6–stimulated CD4+ T cells (Fig. 1 D). qPCR for Sox5 HMG domain confirmed that not only the brain and liver but also IL-6–stimulated CD4+ T cells expressed high levels of Sox5 mRNA (Fig. 1 D). These results indicate that IL-6–stimulated CD4+ T cells express a novel isoform of Sox5, Sox5t.
Stat3 is required for the induction of Sox5 in Th17 cells
To determine whether Stat3 signaling is involved in Sox5 expression in CD4+ T cells, we examined the expression of Sox5 protein in CD4+ T cells in T cell–specific Stat3-deficient mice (CD4cre Stat3fl/fl mice) and littermate control mice (Stat3fl/fl mice). Naive CD4+ T cells were stimulated with anti-CD3 and -CD28 mAbs under neutral conditions (anti–IL-4 mAb and anti–IFN-γ mAb), IL-6 conditions (IL-6, anti–IL-4 mAb, and anti–IFN-γ mAb), or Th17 conditions (IL-6, TGF-β, anti–IL-4 mAb, and anti–IFN-γ mAb) for 4 d, and whole-cell lysates were subjected to Western blot analysis. Importantly, Sox5 was expressed in CD4+ T cells in IL-6 or Th17 conditions in Stat3fl/fl mice but not in CD4cre Stat3fl/fl mice (Fig. 1 E). As previously reported (Hiramatsu et al., 2010), c-Maf was modestly expressed in IL-6 conditions and strongly expressed in Th17 conditions in Stat3fl/fl mice but not in CD4cre Stat3fl/fl mice (Fig. 1 E). These results indicate that Stat3 signaling is required for the induction of Sox5, as well as c-Maf in IL-6–stimulated CD4+ T cells.
Severity of experimental autoimmune encephalomyelitis (EAE) and delayed-type hypersensitivity (DTH) is reduced in CD4cre Sox5fl/fl mice
To determine the roles of Sox5 expressed in T cells, we examined the phenotype of T cell–specific Sox5-deficient mice (CD4cre Sox5fl/fl mice) and found that the numbers of naive and memory CD4+ T cells, CD8+ T cells, γδ T cells, naturally occurring regulatory T cells, and lymphoid tissue inducer-like cells (LTi-like cells) were indistinguishable between CD4cre Sox5fl/fl mice and Sox5fl/fl mice (unpublished data).
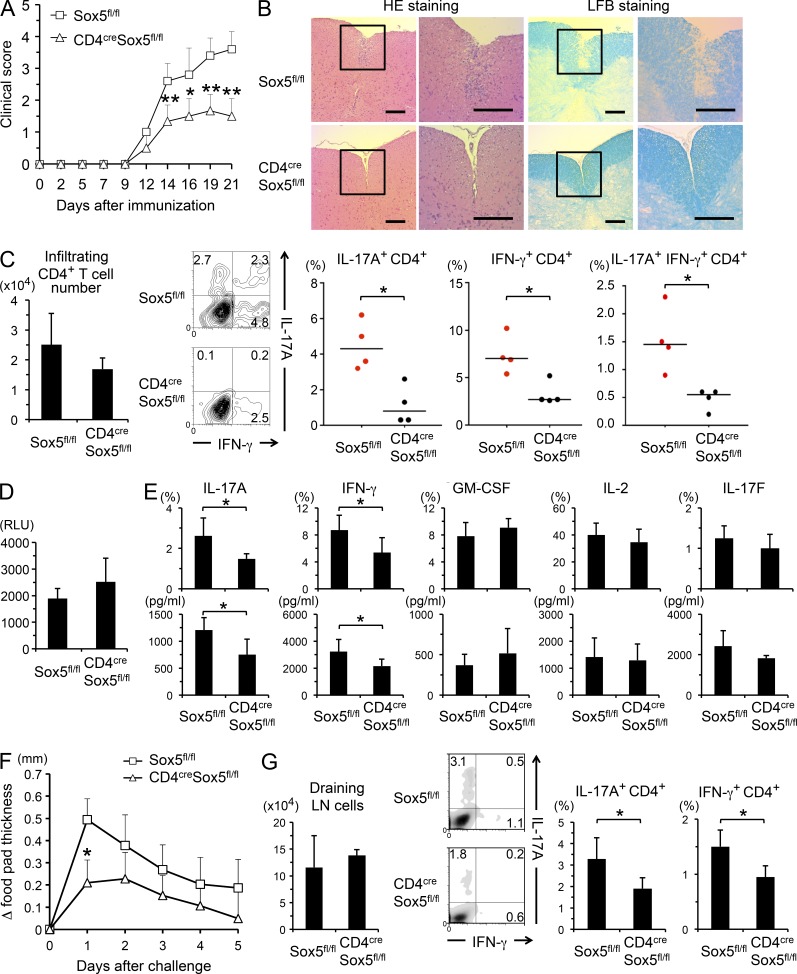
To determine the role of Sox5 in Th17 cell–mediated inflammatory diseases, EAE was induced in CD4cre Sox5fl/fl mice and Sox5fl/fl mice. As shown in Fig. 2 A, CD4cre Sox5fl/fl mice exhibited significantly reduced clinical scores of EAE as compared with Sox5fl/fl mice. Histopathological analyses of the spinal cord revealed that inflammatory cell infiltration and demyelination were reduced in CD4cre Sox5fl/fl mice as compared with those in Sox5fl/fl mice (Fig. 2 B). Importantly, although the number of infiltrating CD4+ T cells was not significantly decreased, the frequency of IL-17A–producing CD4+ T cells was significantly decreased in the brain and spinal cord of CD4cre Sox5fl/fl mice at 10 d after immunization (Fig. 2 C). In addition, the frequencies of IFN-γ–producing CD4+ T cells and IL-17A– and IFN-γ–double producing cells were decreased in CD4cre Sox5fl/fl mice (Fig. 2 C). Consistently, although MOG-induced proliferation of splenocytes was not significantly different between CD4cre Sox5fl/fl mice and Sox5fl/fl mice (Fig. 2 D), MOG-induced IL-17A and IFN-γ production from splenic CD4+ T cells was significantly decreased in CD4cre Sox5fl/fl mice (Fig. 2 E). Moreover, MOG-induced IL-17F production tended to be decreased in CD4cre Sox5fl/fl mice (Fig. 2 E), although the difference did not reach statistical significance.
Figure 2.
CD4cre Sox5fl/fl mice are resistant to EAE and DTH. (A) EAE disease course in Sox5fl/fl mice and CD4cre Sox5fl/fl mice. Disease severity was monitored and scored three times a week. Shown are means ± SD of clinical score. Data are compiled from six mice in each group from three independent experiments. *, P < 0.05; **, P < 0.01. (B) Shown are representative photomicrographs of hematoxylin and eosin (HE) and Luxol fast blue (LFB) staining of the spinal cord sections at 21 d after immunization. Bar, 200 µm. (C) Cells were harvested from the brain and spinal cord of EAE-induced mice at 10 d after immunization. (left) Means ± SD of the number of infiltrating CD4+ T cells. (middle and right) Harvested cells were stimulated with PMA/ionomycin for 4 h. Shown are representative FACS profiles of IFN-γ versus IL-17A staining (gated on CD4+ cells that are negative for fixable dead cell staining) and means ± SD of the percentages of the indicated cells in Sox5fl/fl mice and CD4cre Sox5fl/fl mice. Data are compiled from four mice in each group from two independent experiments. *, P < 0.05. Mann-Whitney U test. (D) Splenocytes from EAE-induced mice were stimulated with MOG peptide for 3 d and cell proliferation was evaluated. (E) CD4+ T cells isolated from the spleen of EAE-induced mice were stimulated with MOG peptide in the presence of irradiated splenocytes for 3 d. Shown are the percentages of CD4+ T cells that produce the indicated cytokines (top) and the levels of the indicated cytokines in the culture supernatants (bottom). Means ± SD. Data are compiled from six mice in each group from two independent experiments. *, P < 0.05. (F) Reduced DTH in CD4cre Sox5fl/fl mice. Δfootpad thickness of Sox5fl/fl mice and CD4cre Sox5fl/fl mice. Data are compiled from five mice in each group from two independent experiments. *, P < 0.05. (G) Cells were harvested from draining LNs of DTH-induced mice at 5 d after the challenge with TNP-KLH. (left) The number of draining LN cells. (middle and right) Representative FACS profiles of IFN-γ versus IL-17A on CD4+ cells and means ± SD of the percentages of IL-17A– and IFN-γ–producing CD4+ T cells in Sox5fl/fl mice and CD4cre Sox5fl/fl mice. Data are compiled from four mice in each group from two independent experiments. *, P < 0.05.
To further address the role of Sox5 in Th17 cell–mediated immune responses, we examined TNP-KLH–induced DTH, in which not only Th1 cells but also the IL-23–IL-17 cell axis plays a critical role (Ghilardi et al., 2004; McGeachy et al., 2009), in CD4cre Sox5fl/fl mice and Sox5fl/fl mice. As shown in Fig. 2 F, footpad swelling was significantly decreased in CD4cre Sox5fl/fl mice as compared with that in Sox5fl/fl mice. Although the total cell numbers harvested from draining LNs were not significantly different between CD4cre Sox5fl/fl mice and Sox5fl/fl mice, IL-17A–producing CD4+ T cells and IFN-γ–producing CD4+ T cells were significantly decreased in CD4cre Sox5fl/fl mice (Fig. 2 G). Collectively, these results suggest that Sox5 plays a role in the induction of Th17 cell–mediated in vivo immune responses.
Th17 cell differentiation is impaired in CD4cre Sox5fl/fl mice
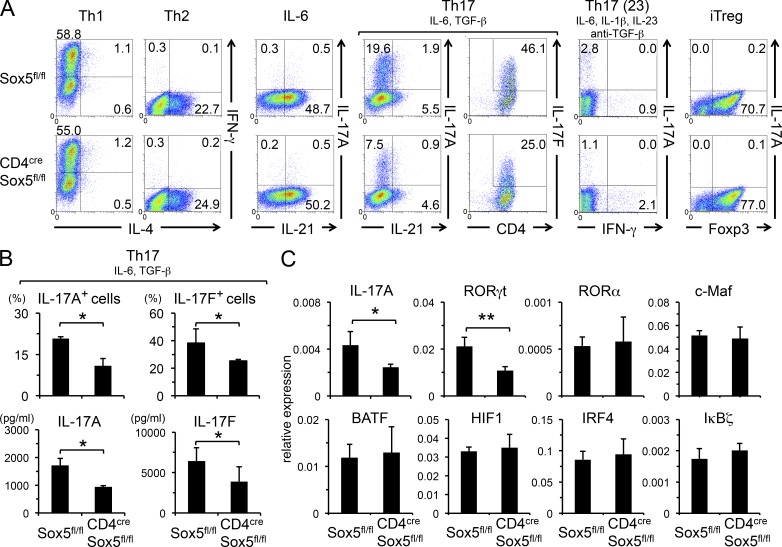
We next examined the role of Sox5 in helper T cell differentiation in vitro. Consistent with the reduced number of IL-17A–producing CD4+ T cells in vivo (Fig. 2), the numbers of IL-17A– and IL-17F–producing cells (Fig. 3, A and B), as well as the levels of IL-17A and IL-17F in the culture supernatants (Fig. 3 B) under Th17 conditions, were significantly decreased in CD4cre Sox5fl/fl mice as compared with those in Sox5fl/fl mice. Under another Th17 conditions (Th17 (23): IL-6, IL-1β, IL-23, and anti-TGF-β antibody; Ghoreschi et al., 2010), IL-17A–producing CD4+ T cells were also decreased in CD4cre Sox5fl/fl mice (Fig. 3 A). On the other hand, the differentiation of Th1 cells, Th2 cells, IL-21–producing T cells, and iTreg cells was indistinguishable between CD4cre Sox5fl/fl mice and Sox5fl/fl mice (Fig. 3 A). Among Th17 cell–related genes, mRNA levels for IL-17A and RORγt were significantly reduced in CD4+ T cells in CD4cre Sox5fl/fl mice, whereas the expression levels of RORα, c-Maf, BATF, HIF1, IRF4, and IκBζ mRNA in CD4+ T cells were not significantly different between CD4cre Sox5fl/fl mice and Sox5fl/fl mice (Fig. 3 C).
Figure 3.
Th17 cell differentiation is impaired in CD4cre Sox5fl/fl mice. (A–C) Naive CD4+ T cells from Sox5fl/fl mice and CD4cre Sox5fl/fl mice were stimulated under Th1, Th2, IL-6, Th17, Th17 (23), or iTreg conditions. (A) The expression of the indicated cytokines was evaluated by intracellular cytokine staining. Shown are representative FACS profiles of three independent experiments. (B) Shown are the percentages of IL-17A– and IL-17F–producing CD4+ T cells and the levels of IL-17A and IL-17F in the culture supernatants under Th17 conditions. Data are compiled from three mice in each group from two independent experiments. *, P < 0.05. (C) The expression levels of Th17 cell–related genes in CD4+ T cells under Th17 conditions were evaluated by qPCR. *, P < 0.05; n = 3 each.
Sox5t and c-Maf cooperatively induce IL-17A production
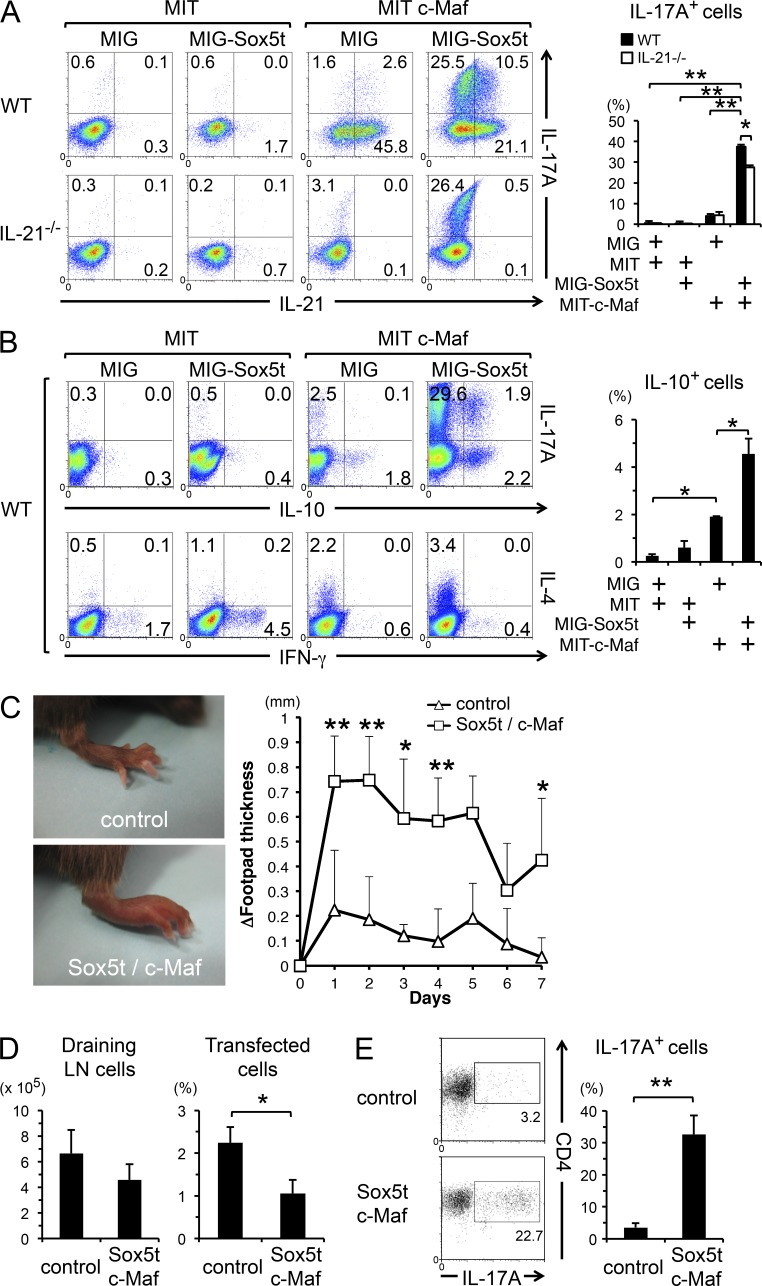
We next examined the effect of enforced expression of Sox5t on Th17 cell differentiation. Retrovirus-mediated Sox5t induction on WT CD4+ T cells did not induce IL-17A production under neutral conditions (Fig. 4 A), suggesting that the expression of Sox5t alone is insufficient for IL-17A production in CD4+ T cells. Because Th17 cells highly express c-Maf as well as Sox5t (Fig. 1 E), we examined the effect of co-induction of Sox5t and c-Maf on IL-17A production in WT CD4+ T cells under neutral conditions. Intriguingly, the enforced expression of Sox5t along with c-Maf significantly induced IL-17A production in CD4+ T cells, whereas the enforced expression of c-Maf alone only slightly induced IL-17A production (Fig. 4 A). We also found that the enforced expression of l-Sox5 along with c-Maf induced IL-17A production in CD4+ T cells, too (unpublished data), suggesting that the missing N-terminal region and the inserted region in Sox5t (Fig. 1 C) are not required for this function.
Figure 4.
Enforced expression of Sox5t and c-Maf induces Th17 cells. (A) Naive CD4+ T cells from IL-21−/− mice or WT mice were stimulated and co-infected with retroviruses of either MSCV-IRES-GFP (MIG) or MIG-Sox5t and with those of either MSCV-IRES-Thy1.1 (MIT) or MIT-c-Maf for 24 h. Cells were then cultured in neutral conditions for additional 3 d and the expression of IL-21 versus IL-17A in GFP+ Thy1.1+ cells was evaluated. Shown are representative FACS profiles and means ± SD of the percentages of IL-17A–producing CD4+ T cells. Data are compiled from five independent experiments. *, P < 0.05; **, P < 0.01. (B) Naive CD4 T cells from WT mice were stimulated and co-infected with retroviruses as described in Fig. 4 A. The expression of the indicated cytokines was evaluated by intracellular cytokine staining. Means ± SD of the percentages of IL-10–producing CD4+ T cells are shown (right). Data are compiled from three independent experiments. *, P < 0.05. (C–E) Naive CD4+ T cells from OT-II mice were stimulated and co-infected with retroviruses of either pMX-IRES-NGFR (pMX-IN) and MIT (control) or pMX-IN Sox5t and MIT c-Maf (Sox5t/c-Maf) for 48 h. After sorted NGFR- and Thy1.1 double-positive cells were adoptively transferred to C57BL/6 recipient mice, the mice were injected with OVA in IFA into the footpad. (C) Shown are representative photographs of footpads at 24 h after immunization and means ± SD of Δfootpad thickness. Data are compiled from five mice in each group from two independent experiments. *, P < 0.05; **, P < 0.01. (D) The numbers of cells harvested from the draining LNs and means ± SD of the percentages of CD4+ T cells that express both NGFR and Thy1.1 (transfected cells) in the draining LNs are shown. Data are compiled from three mice in each group. *, P < 0.05. (E) Cells harvested from the draining LNs were stimulated with PMA/ionomycin and IL-17A expression in CD4+ T cells was evaluated. Representative FACS profiles and means ± SD of the percentages of IL-17A–producing CD4+ T cells are shown. **, P < 0.01; n = 3 each.
Consistent with a previous study (Hiramatsu et al., 2010), enforced expression of c-Maf strongly induced IL-21 production (Fig. 4 A). Because c-Maf has been shown to participate in the development of Th17 cells by promoting IL-21 production (Bauquet et al., 2009), we examined whether Sox5t- and c-Maf–induced IL-17A production depends on IL-21. As shown in Fig. 4 A, IL-17A production in Sox5t- and c-Maf–expressing CD4+ T cells was modestly decreased but was still significantly observed in IL-21–deficient mice. Collectively, these results indicate that c-Maf and Sox5t could cooperatively induce IL-17A production independently of endogenously produced IL-21.
Because c-Maf has been shown to be involved in IL-4 and IL-10 production (Ho et al., 1996; Apetoh et al., 2010; Rutz et al., 2011), and because IFN-γ production is reduced in CD4cre Sox5fl/fl mice in vivo (Fig. 2), we next analyzed the production of IL-4, IL-10, and IFN-γ in Sox5t- and/or c-Maf–expressing CD4+ T cells. Consistent with previous studies, IL-4 and IL-10 production was induced by the enforced expression of c-Maf (Fig. 4 B). Interestingly, enforced expression of Sox5t and c-Maf synergistically induced IL-10 production in CD4+ T cells (Fig. 4 B). On the other hand, IFN-γ production was modestly induced by Sox5t, and Sox5t-mediated IFN-γ production was suppressed by c-Maf (Fig. 4 B).
Sox5t- and c-Maf–expressing CD4+ T cells show strong ability to induce delayed-type hypersensitivity
We next used an adoptive transfer model of DTH (Feuerer et al., 2006; McGeachy et al., 2009) to assess whether Sox5t- and c-Maf–expressing CD4+ T cells are pathogenic Th17 cells. As shown in Fig. 4 C, mice injected with Sox5t- and c-Maf–expressing OVA-specific CD4+ T cells exhibited strong OVA-mediated DTH as compared with mice injected with OVA-specific CD4+ T cells infected with control retroviruses. Whereas the cell recovery of transferred CD4+ T cells from the draining LNs was modestly decreased in the mice transferred with Sox5t- and c-Maf–expressing CD4+ T cells (Fig. 4 D), the frequency of IL-17A–producing cells was significantly increased in these mice (Fig. 4 E). These results suggest that Sox5t- and c-Maf–expressing CD4+ T cells develop into Th17 cells and play a pathogenic role in vivo.
Sox5t and c-Maf induce IL-17A production even in Stat3-deficient CD4+ T cells
Stat3 plays an essential role in the development of Th17 cells (Durant et al., 2010; Mathur et al., 2007; Yang et al., 2007; Zhou et al., 2007). Because both Sox5t and c-Maf were induced by IL-6–Stat3 signaling in Th17 cells (Fig. 1 E), we next examined whether enforced expression of Sox5t and c-Maf could bypass the requirement of Stat3 for IL-17A production in CD4+ T cells. As shown in Fig. 5 A, the enforced expression of Sox5t and c-Maf in CD4+ T cells significantly induced IL-17A production not only in control Stat3fl/fl mice but also in CD4cre Stat3fl/fl mice in neutral conditions. Similarly, as expected, enforced expression of RORγt induced IL-17A production in CD4+ T cells in both Stat3fl/fl mice and CD4cre Stat3fl/fl mice (Fig. 5 B). These results indicate that Sox5t along with c-Maf could compensate, at least in part, for the absence of Stat3 in IL-17A production, suggesting that Sox5t and c-Maf function as the downstream effector molecules of IL-6–Stat3 signaling for Th17 cell differentiation. On the other hand, c-Maf–induced IL-21 production was significantly diminished in CD4cre Stat3fl/fl mice (Fig. 5 A), consistent with previous studies showing the importance of endogenously produced IL-21 for the development of IL-21–producing cells (Korn et al., 2007; Suto et al., 2008).
Figure 5.
Enforced expression of Sox5t and c-Maf induces Th17 cell differentiation even in the absence of Stat3. (A and B) Naive CD4+ T cells from CD4cre Stat3fl/fl or Stat3fl/fl mice were co-infected with indicated retroviruses and the expression of IL-21 versus IL-17A was examined as described in Fig. 4 A. Means ± SD of the percentages of IL-17A–producing CD4+ T cells are shown on the right. Data are compiled from four (A) or three (B) independent experiments. **, P < 0.01. (C–E) Naive CD4+ T cells from CD4cre Sox5fl/fl mice and Sox5fl/fl mice (C) or WT mice and IL-21−/− mice (D and E) were transfected with siMaf or nonsilencing controls, and then stimulated in Th17 conditions for 24 h (C and D) or 84 h (E). The expression of IL-21 versus IL-17A was examined as described in Fig. 4 A. Means ± SD. Data are compiled from three independent experiments. *, P < 0.05.
Next, to assess the role of endogenously induced c-Maf and its relation to Sox5 in Th17 cell differentiation, we examined the effect of c-Maf knockdown on IL-17A and IL-21 production in CD4+ T cells in Sox5fl/fl mice and CD4cre Sox5fl/fl mice. c-Maf knockdown significantly suppressed IL-17A production in CD4+ T cells in Sox5fl/fl mice in the early phase (24 h) of Th17 differentiation (Fig. 5 C). Consistent with previous findings (Bauquet et al., 2009; Hiramatsu et al., 2010), c-Maf knockdown also decreased IL-21 production in CD4+ T cells in Sox5fl/fl mice (Fig. 5 C). Although c-Maf knockdown reproducibly suppressed IL-17A production in CD4cre Sox5fl/fl mice, the reduction rate tended to be less dramatic in CD4cre Sox5fl/fl mice as compared with that in Sox5fl/fl mice (Fig. 5 C), suggesting that c-Maf participates in IL-17A production partly by cooperating with Sox5. Importantly, c-Maf knockdown suppressed IL-17A production even in IL-21–deficient CD4+ T cells in the early phase (24 h) of Th17 cell differentiation (Fig. 5 D). In contrast, in the late phase (84 h) of Th17 cell differentiation, c-Maf knockdown did not suppress IL-17A production in IL-21–deficient CD4+ T cells (Fig. 5 E).
Sox5t- and c-Maf–mediated IL-17A production depends on RORγt
To elucidate the mechanisms underlying Sox5t- and c-Maf–mediated IL-17A production in CD4+ T cells, we examined the expression levels of Th17 cell–related genes in FACS-sorted Sox5t- and c-Maf–expressing CD4+ T cells. Sox5t- and c-Maf–expressing CD4+ T cells expressed significantly higher levels of RORγt as compared with CD4+ T cells expressing Sox5t or c-Maf, and the expression levels of RORγt in Sox5t- and c-Maf–expressing CD4+ T cells were similar to those in Th17 cells (Fig. 6 A). On the other hand, other Th17 cell–related genes such as RORα, BATF, HIF1, IRF4, and IκBζ were not significantly enhanced in Sox5t- and c-Maf–expressing CD4+ T cells (Fig. 6 B). Intracellular analysis revealed that the expression levels of RORγt proteins were enhanced in Sox5t- and c-Maf–expressing CD4+ T cells as compared with those in CD4+ T cells infected with control retroviruses (Fig. 6 C). To determine whether RORγt is indispensable for Th17 cell differentiation in Sox5t- and c-Maf–expressing CD4+ T cells, CD4+ T cells from RORγt-deficient mice or WT mice were infected with Sox5t and c-Maf retroviruses and the expression of IL-17A and IL-17F was examined. Importantly, RORγt-deficient CD4+ cells failed to express IL-17A even if they expressed Sox5t and c-Maf (Fig. 6 D). Collectively, these results suggest that Sox5 and c-Maf induce IL-17A production as downstream effectors of Stat3 and as upstream inducers of RORγt.
Figure 6.
RORγt is essential for Sox5t- and c-Maf-mediated IL-17A production. (A–C) Naive CD4+ T cells from WT mice were co-infected with retroviruses of either pMX-IN or pMX-IN-Sox5t and with those of either MIT or MIT-c-Maf. (A and B) 2 d after the infection, NGFR+ Thy1.1+ cells were sorted and the expression levels of the indicated genes were assessed by qPCR. As controls, mRNAs from WT CD4+ T cells stimulated in neutral conditions or Th17 conditions were used. Means ± SD. Data are compiled from three independent experiments. (C) 2 d after the infection, the expression of RORγt in NGFR+ Thy1.1+ cells was determined by intracellular staining. (D) Splenic naive CD4+ T cells from RORγt-deficient or WT mice were co-infected with retroviruses of indicated vectors as described in Fig. 4 A. The expression of IL-17A versus IL-17F on Thy1.1+ GFP+ cells was evaluated. Data shown are representative of three independent experiments. (E–G) Naive CD4+ T cells from WT mice were co-infected with pMX-IN-HA-Sox5t and MIT-c-Maf in neutral conditions and subjected to ChIP-seq analysis. Shown are ChIP-seq binding tracks and MACS peak for HA-Sox5t and c-Maf at Rorc locus (E), Il10 locus (F), and Il17a-Il17f loci (G). VISTA plot of CNSs between human and mouse Rorc gene locus (pink plot) is shown (E, bottom). (H) Integration data of ChIP-seq and RNA-seq. Naive CD4+ T cells from WT mice were co-infected with pMX-IN/MIT or pMX-IN-HA-Sox5t/MIT-c-Maf for 24 h. Cells were then cultured in neutral conditions for additional 18 h and subjected to RNA-seq analysis. Transcripts were selected as described in the Materials and methods. Shown is a heat map of the levels of transcripts that were validated as direct targets of Sox5t and c-Maf by ChIP-seq analysis.
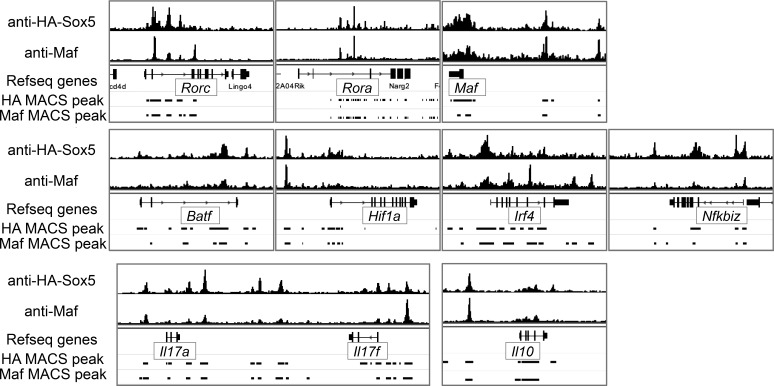
To elucidate the mechanisms underlying Sox5- and c-Maf–mediated RORγt expression, we searched for conserved noncoding sequence (CNS) between mouse and human Rorc gene loci and found 10 CNSs (Fig. 6 E). CNS1, CNS2, and CNS3 are located between Gm659 and Rorc gene locus and CNS3 is located in the promoter region of RORγ. CNS4 is a cluster of five short conserved sequences located in the promoter region of RORγt. CNS5, CNS6, CNS7, and CNS8 are located in intron 1, CNS9 is located in intron 3, and CNS10 is located in intron 9. We then performed RNA-seq and chromatin immunoprecipitation (ChIP)-seq analyses to examine how Sox5t and c-Maf induce RORγt expression in neutral conditions. Because anti-Sox5 antibody suitable to ChIP is not available, HA-tagged Sox5t, and c-Maf were retrovirally transduced into CD4+ T cells and the binding regions of Sox5t and those of c-Maf were identified by ChIP-seq analysis. As shown in Fig. 6 E, both Sox5t and c-Maf bound to CNS4, the promoter region of RORγt gene, and CNS6 of RORγt gene in neutral conditions. Sox5t and c-Maf also bound to Il10 locus (Fig. 6 F), which is consistent with the finding that IL-10 production is enhanced in Sox5t- and c-Maf–expressing CD4+ T cells (Fig. 4 B). In contrast, Sox5t and c-Maf did not bind to Il17a-Il17f loci in neutral conditions (Fig. 6 G).
By integrating the data of ChIP-seq and the gene expression profile of Sox5- and c-Maf–expressing CD4+ T cells by RNA-seq in neutral conditions, we found that 26 transcripts, including Rorc were directly up-regulated and 40 transcripts were directly repressed by Sox5t and c-Maf in CD4+ T cells (Fig. 6 H). These results suggest that RORγt is one of the direct targets of Sox5- and c-Maf in CD4+ T cells. On the other hand, in Th17 conditions, ChIP-seq analysis of c-Maf and HA-Sox5t in HA-Sox5t-expressing CD4+ T cells revealed that in addition to Rorc, several Th17 cell–related gene loci, including Il17a-Il17f, Rora, Maf, Batf, Hif1a, Irf4, and Nfkbiz, as well as IL-10 locus, were bound to by HA-Sox5 and c-Maf (Fig. 7).
Figure 7.
Both Sox5t and c-Maf bind to several Th17 cell–related gene loci in CD4+ T cells under Th17 conditions. Naive CD4+ T cells from WT mice were infected with pMX-IN-HA-Sox5t in Th17 conditions and subjected to ChIP-seq analysis. Shown are ChIP-seq binding tracks and MACS peak for HA-Sox5t and c-Maf at the indicated gene loci.
Sox5t and c-Maf synergistically activate RORγt promoter in CD4+ T cells
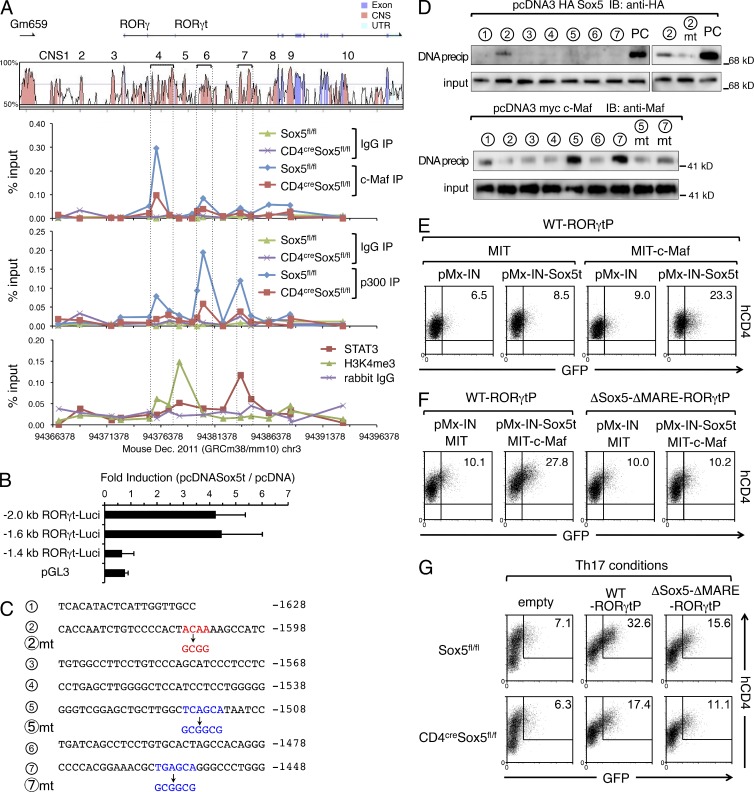
To determine the role of Sox5 in c-Maf binding to Rorc locus, we next performed ChIP-qPCR analysis of c-Maf in CD4+ T cells from CD4cre Sox5fl/fl mice and control Sox5fl/fl mice under Th17 conditions (Fig. 8 A). Consistent with the data of ChIP-seq, c-Maf strongly bound to CNS4 and weakly bound to CNS6 in control CD4+ T cells from Sox5fl/fl mice (Fig. 7 A). Importantly, the binding of c-Maf to CNS4 and CNS6 was reduced in CD4+ T cells from CD4cre Sox5fl/fl mice (Fig. 8 A). We also found that histone acetyltransferase p300, which is predictive of tissue-specific regulatory activity (Visel et al., 2009), bound to CNS4, CNS6, and CNS7 of Rorc locus under Th17 conditions (Fig. 8 A), and that the binding of p300 was reduced by the absence of Sox5 (Fig. 8 A), consistent with a previous finding that c-Maf recruits p300 to regulatory elements (Chen et al., 2002). In agreement with these findings, the elevation of trimethylation of histone H3 Lys4 (H3K4me3), a mark of active gene loci, was observed in CNS4-CNS5 of Rorc gene in CD4+ T cells under Th17 conditions (Fig. 8 A). On the other hand, Stat3 did not bind to RORγt promoter, but bound to CNS7 in CD4+ T cells under Th17 conditions (Fig. 8 A), consistent with previous studies (Durant et al., 2010; Lazarevic et al., 2011). Collectively, these results suggest that c-Maf binds to the promoter region of RORγt cooperatively with Sox5.
Figure 8.
Sox5 and c-Maf activate the promoter of RORγt gene. (A, top) VISTA plot of CNSs of Rorc gene locus. (middle panels) Naive CD4+ T cells from CD4cre Sox5fl/fl or Sox5fl/fl mice were stimulated under Th17 conditions for 8 h. ChIP-qPCR assay for CNS of Rorc gene locus was performed with anti–c-Maf antibody, anti-p300 antibody, or control rabbit IgG. (bottom) Naive CD4+ T cells from WT mice were stimulated under Th17 conditions for 8 h. ChIP-qPCR assay for CNS of Rorc gene locus was performed with anti-Stat3 antibody, anti-H3K4me3 antibody, or control rabbit IgG. Results are expressed as the percent input for each ChIP fraction. Data are representative of three independent experiments. (B) Luciferase assay of indicated RORγt-Luci in EL4 cells. Data are compiled from three independent experiments. (C) Sequence of RORγt promoter (from −1648 to −1448). Putative Sox5 binding site is indicated by red and putative MAREs are indicated by blue. Mutated sequences are shown in lower lines. (D, top) Representative data of DNA precipitation assay of HA-Sox5 with biotinylated double-stranded DNA probes containing a portion of RORγt promoter or its mutant at Sox5-binding site. As a positive control (PC), FXO+ probe, which harbors two Sox consensus sites, was used. (bottom) Representative data of DNA-precipitation assay of c-Maf with biotinylated double-stranded DNA probes containing a portion of RORγt promoter or its mutants at MARE sites. Shown are data representative of three independent experiments. (E) Naive CD4+ T cells were co-infected with retroviruses of either pMX-IN or pMX-IN-Sox5t and with those of either MIT or MIT-c-Maf for 24 h. Cells were reinfected with WT-RORγtP for 24 h, and GFP versus hCD4 in NGFR+ Thy1.1+ cells was evaluated by flow cytometry. (F) Naive CD4+ T cells were co-infected with retroviruses of indicated vectors and analyzed as described in Fig. 7 E. (G) Naive CD4+ T cells from Sox5fl/fl mice and CD4cre Sox5fl/fl mice were stimulated in neutral conditions for 48 h and infected with retroviruses of hCD4-pA-GFP (empty), WT-RORγtP, or ΔSox5-ΔMARE-RORγtP under Th17 conditions for additional 24 h. The expression levels of GFP were evaluated as described in Fig. 7 E. Data shown are representative of three independent experiments.
To determine the binding site of Sox5 in RORγt promoter, we performed reporter assay and found that Sox5t activated −2 kb and −1.6 kb RORγt promoter but not −1.4 kb RORγt promoter (Fig. 8 B), suggesting that −1.6–−1.4-kb region of RORγt promoter is crucial for Sox5t binding. We thus performed DNA precipitation assay which covers the −1.6–−1.4-kb region of RORγt promoter (Fig. 8 D) to find the region bound by Sox5t. We found that Sox5t bound to probe #2 but not its mutant (#2mt) in which target sequence of Sox5 was destroyed (Fig. 8 D), suggesting that Sox5t directly binds to and activates the −1.6–−1.4-kb region of RORγt promoter. We also found three putative Maf-binding sequences (MARE) in −1.6–−1.4-kb region of RORγt promoter (Fig. 8 C). DNA precipitation assay revealed that c-Maf bound probe #5 and #7 but not their mutants (#5mt and #7mt; Fig. 8 D) in which MARE was destroyed (Fig. 8 C).
We next examined whether Sox5t and c-Maf activate RORγt promoter in primary CD4+ T cells. We used a reverse-strand retroviral reporter (hCD4-pA-GFP vector) that utilizes a truncated form of human CD4 (hCD4) to detect infected cells and GFP to detect promoter activity (Zhu et al., 2001). CD4+ T cells were infected with retroviruses of Sox5t (pMX-IN Sox5t) and/or c-Maf (MIT-c-Maf) and, 24 h later, the cells were reinfected with retrovirus of WT-RORγtP (hCD4-pA-GFP containing −2-kb fragment of RORγt promoter). As shown in Fig. 8 E, Sox5t and c-Maf synergistically enhanced the reporter activity of WT-RORγtP. Moreover, Sox5t- and c-Maf–mediated induction of the reporter activity was attenuated in ΔSox5-ΔMARE-RORγtP, a mutant of WT-RORγtP in which Sox5 and Maf binding sequences were destroyed (Fig. 8 F). Furthermore, under Th17 conditions, the reporter activity of ΔSox5-ΔMARE-RORγtP was significantly decreased as compared with that of WT-RORγtP in Sox5-sufficient CD4+ T cells and was modestly decreased in Sox5-deficient CD4+ T cells (Fig. 8 G). These results suggest that endogenously expressed Sox5 and c-Maf in CD4+ T cells are involved in the activation of RORγt promoter under Th17 conditions.
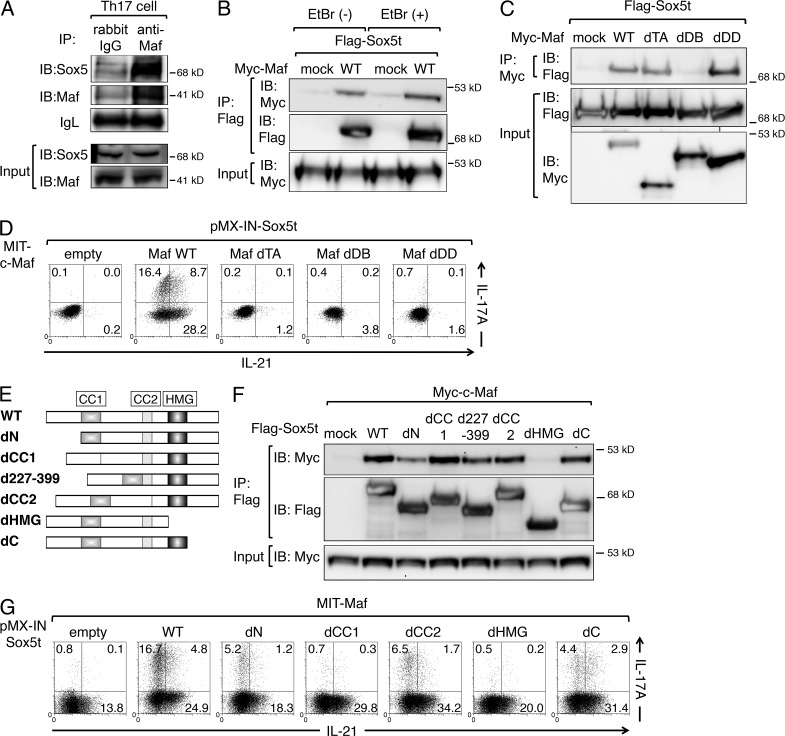
Sox5 physically associates with c-Maf
Because the binding sites of Sox5 and c-Maf are close to each other (Figs. 6 E, 7, and 8), we finally assessed the physical interaction of Sox5 and c-Maf. As shown in Fig. 9 A, Sox5 was co-immunoprecipitated with anti–c-Maf antibody in Th17 cells. When Flag-tagged Sox5t was co-expressed with Myc-tagged c-Maf in 293T cells, Myc-tagged c-Maf was co-immunoprecipitated with anti-Flag antibody even in the presence of ethidium bromide (Fig. 9 B), suggesting that the interaction between Sox5 and c-Maf does not depend on the presence of DNA. Co-immunoprecipitation assay using various deletion mutants of c-Maf (Rutz et al., 2011) revealed that its association with Sox5t was significantly attenuated in a mutant lacking the DNA-binding domain (dDB) but not in mutants lacking transactivation domain (dTA) or dimerization domain (dDD; Fig. 9 C). We also examined the ability of the c-Maf mutants to induce IL-17A production in the presence of Sox5t and found that WT c-Maf (Maf WT) but not Maf dTA, Maf dDB, or Maf dDD induced IL-17A production (Fig. 9 D). These results indicate that although the DNA-binding domain of c-Maf is crucial for binding to Sox5, all of transactivation domain, DNA-binding domain, and dimerization domain are indispensable for IL-17A production. Furthermore, IL-21 production was also severely impaired in cells expressing Maf dTA, Maf dDB, or Maf dDD, indicating that all of these domains are also indispensable for IL-21 production (Fig. 9 D).
Figure 9.
Sox5 physically associates with c-Maf. (A) Whole-cell lysates from Th17 cells were subjected to IP with anti-Maf antibody or control rabbit IgG and IB with anti-Sox5 or anti-Maf antibody. Input proteins (input) were also IB with anti-Sox5 or anti-Maf antibody. IgL, additional loading controls. (B) 293T cells were transfected with pNTAP (mock) or pNTAP-Myc-c-Maf (WT), together with p3xFlag-Sox5t. Whole cell extracts were IP with anti-Myc antibody in the presence or absence of ethidium bromide (EtBr), and IB with anti-Flag antibody. (C) 293T cells were transfected with pNTAP (mock) or indicated myc-tagged c-Maf mutants together with p3xFlag-Sox5t. Whole-cell extracts were immunoprecipitated with anti-Myc antibody and IB with anti-Flag antibody. Input proteins were also IB with anti-Flag antibody or anti–Myc-antibody. (D) Naive CD4+ T cells were co-infected with retroviruses of pMX-IN-Sox5t and indicated c-Maf mutants as described in Fig. 4 A. The expression of IL-17A versus IL-21 on NGFR+ Thy1.1+ cells was assessed by flow cytometry. (E) Schematic comparison of wild-type Sox5t (WT) and its deletion mutants. (F) 293T cells were transfected with p3xFlag (mock) or indicated Flag-tagged Sox5t mutants together with pNTAP-Myc-c-Maf. Whole-cell extracts were immunoprecipitated with anti-Flag antibody and IB with anti-Myc antibody or anti-Flag antibody. Input proteins were also IB with anti–Myc-antibody. (G) Naive CD4+ T cells were co-infected with retroviruses of MIT-c-Maf and indicated Sox5t mutants, and the expression of IL-17A versus IL-21 was examined as described in Fig. 4 A. Data shown are representative of five independent experiments.
Previous studies have shown that the first coiled–coil domain (CC1) but not the second coiled-coil domain (CC2) is required for SoxD dimerization (Lefebvre et al., 1998), whereas HMG domain is required for both DNA binding and association with other proteins (Wilson and Koopman, 2002). We found that a Sox5t mutant lacking HMG domain (dHMG; Fig. 9 E) failed to associate with c-Maf, whereas Sox5t mutants lacking N-terminal region (dN), CC1 (dCC1), amino acids 227–339 (d227-339), CC2 (dCC2), or C-terminal region (dC) could associate with c-Maf (Fig. 9 F). We also examined the ability of these Sox5t mutants to induce IL-17A production in the presence of c-Maf and found that Sox5t dCC1and Sox5t dHMG failed to induce IL-17A production (Fig. 9 G). Collectively, these results indicate that HMG domain of Sox5t and DNA-binding domain of c-Maf is indispensable for the association of Sox5t and c-Maf.
DISCUSSION
In this study, we show that Sox5 plays a critical role in the development of Th17 cell–mediated in vivo immune responses and that Sox5 along with c-Maf plays a crucial role in the differentiation of Th17 cells by inducing RORγt expression. It is well established that Stat3 is essential for RORγt expression during Th17 differentiation, although Stat3 itself does not activate RORγt promoter (Durant et al., 2010; Lazarevic et al., 2011). We found that Stat3 was required for the induction of Sox5 and c-Maf (Fig. 1 E). We also found that the enforced expression of Sox5 and c-Maf induced Th17 cell differentiation in neutral conditions not only in WT CD4+ T cells but also in Stat3-deficient CD4+ T cells although the induction in Stat3-deficient CD4+ T cells was somewhat less efficient than that in WT CD4+ T cells (Fig. 5 A). In contrast, the enforced expression of Sox5t and c-Maf could not induce Th17 cell differentiation in RORγt-deficient CD4+ T cells (Fig. 6 D). Importantly, the enforced expression of Sox5t and c-Maf induced the expression of RORγt in CD4+ T cells (Fig. 6 A and 6C) presumably by binding its regulatory elements (Fig. 6 E). Moreover, although Sox5t and c-Maf bound to the gene loci of other Th17 cell–related molecules such as RORα, BATF, IκBζ, and IRF4 in CD4+ T cells under Th17 conditions (Fig. 7), the expression levels of these molecules were not significantly increased by the enforced expression of Sox5t and c-Maf (Fig. 6 B). These results suggest that Sox5 and c-Maf are involved in IL-17A production as one of the downstream effectors of Stat3 signaling and the upstream activators of RORγt expression during Th17 cell differentiation independently of other Th17 cell–related molecules.
Regarding the roles of c-Maf in the differentiation of Th17 cells, a previous study using naive CD4+ T cells from bone marrow chimeric c-Maf–deficient mice has shown that c-Maf is not required for the differentiation of Th17 cells but is required for the maintenance of Th17 cells via the production of IL-21 (Bauquet et al., 2009). Consistently, we found that c-Maf knockdown suppressed IL-17A production from CD4+ T cells in WT mice but not in IL-21–deficient mice in the late phase of Th17 cell differentiation (Fig. 5 E). On the other hand, we found that in the early phase of Th17 differentiation, c-Maf knockdown modestly but reproducibly suppressed anti-CD3/CD28–mediated IL-17A production even in IL-21–deficient mice (Fig. 5 D). We also found that the enforced expression of c-Maf along with Sox5t induced IL-17A production even in the absence of IL-21 under neutral conditions (Fig. 4 A). These results suggest that the requirement of IL-21 for c-Maf–mediated Th17 cell differentiation may differ depending on the phase of Th17 cell differentiation. Alternatively, it is possible that the difference in the residual c-Maf levels between complete absence in knockout mice and partial reduction by RNAi knockdown may influence the IL-21–independent role of c-Maf in Th17 cell differentiation because c-Maf has been shown to function not only as an activator of IL-21 (Bauquet et al., 2009; Hiramatsu et al., 2010) but also as a negative regulator of Th17 cell–related genes, such as Rora, Runx1, Il1r1, Ccr6, and Tnf during Th17 cell differentiation (Ciofani et al., 2012).
We also found that c-Maf knockdown significantly decreased IL-17A in CD4cre Sox5fl/fl mice, but the reduction rate tended to be less dramatic in CD4cre Sox5fl/fl mice as compared with that in Sox5fl/fl mice (Fig. 5 C). These results suggest that c-Maf participates in IL-17A production in cooperation with Sox5 during Th17 cell differentiation. On the other hand, we found that IL-17A production was decreased in CD4cre Sox5f/f CD4+ T cells under Th17 (23) conditions (Fig. 3 A). Because it has been shown that c-Maf is not significantly induced in CD4+ T cells under Th17 (23) conditions (Ghoreschi et al., 2010), Sox5 may have another binding partner in this situation. Further studies that compare T cell–specific c-Maf–deficient mice, Sox5-deficient mice, and c-Maf– and Sox5-double deficient mice are required to elucidate the precise role of c-Maf expressed in CD4+ T cells in Sox5-mediated Th17 cell induction.
Our findings indicate that Sox5 associates with c-Maf (Fig. 9) and directly activates the promoter of RORγt (Fig. 8). Regarding the downstream targets of Stat3 for Th17 cell differentiation, several signaling molecules including IκBζ, RORα, BATF, IRF4, Ahr, Maf, and HIF-1 have been listed as candidates (Brüstle et al., 2007; Veldhoen et al., 2008; Yang et al., 2008; Bauquet et al., 2009; Schraml et al., 2009; Durant et al., 2010; Dang et al., 2011). Among these transcription factors, HIF-1 has been shown to activate Rorc promoter. However, the HIF-1 binding site is ∼140 bp upstream from the transcription start site of RORγt and is far from the binding sites for Sox5t and c-Maf, which are located at 1,400–1,600 bp upstream from the start site (Fig. 8), suggesting that HIF-1 and Sox5t/c-Maf may independently activate RORγt promoter.
We also found that the first coiled–coil domain of Sox5 is required for the induction of IL-17A production in CD4+ T cells (Fig. 9 G). The coiled-coil domain is shown to mediate homodimerization as well as heterodimerization of the SoxD proteins (Lefebvre, 2010). Regarding the binding partner of Sox5 in CD4+ T cells, we found that other SoxD family members Sox13 and Sox6 were not expressed in CD4+ T cells at mRNA levels (data not shown), consistent with a previous study showing that αβ T cells do not express Sox13 (Melichar et al., 2007). Therefore, Sox5 is the only SoxD family protein expressed in CD4+ T cells and the homodimerization of Sox5 seems involved in the induction of RORγt expression during Th17 cell differentiation.
It has been shown that SoxD proteins have no known transactivation domain, but they do participate in the transcriptional activation or repression of the corresponding genes (Lefebvre, 2010). In this study, we show that Sox5 associates with c-Maf via the HMG domain of Sox5 and the DNA-binding domain of c-Maf (Fig. 9, C and F), consistent with previous studies showing the association of Maf and Sox family proteins. Rajaram and Kerppola (2004) have shown that Maf interacts with Sox2 and Sox3 and induces the expression of crystalline genes during lens development. In addition, it has been shown that long form of c-Maf (Lc-Maf), which has an extra 10 aa at the C terminus, interacts with Sox9 via the HMG domain of Sox9 and the basic region-leucine zipper (bZIP) of Lc-Maf and activates type II collagen gene (Huang et al., 2002; Yang and Cvekl, 2007). Because DNA-binding domain of Maf family proteins is highly conserved (Kurokawa et al., 2009), the interaction of Sox family proteins and Maf family proteins may be widely observed.
Although many studies have indicated the roles of Sox family proteins in the developmental processes (Lefebvre, 2010; Wegner, 2010), there are only a few studies that demonstrate the roles of Sox family proteins in the development and differentiation of T cells. Schilham et al. (1996, 1997) have shown that Sox4 is highly expressed in lymphocytes and regulates T cell differentiation in the thymus. Melichar et al. (2007) have shown that Sox13 and Sox5 are expressed in γδ T cells and that the development of γδ T cells is impaired in Sox13-deficient mice. In addition, Malhotra et al. (2013) have recently shown that Sox4 and Sox13 regulate Tγδ17 differentiation through the induction of RORγt and BLK, respectively. Moreover, Kuwahara et al. (2012) have reported that Sox4 negatively regulates the function of GATA3 and inhibits Th2 cell–mediated inflammation, whereas the differentiation of Th17 cells, Th9 cells, and Foxp3+ regulatory T cells is not affected by the deficiency of Sox4. Here, we have shown that Sox5 is involved in the differentiation of Th17 cells. Therefore, although further studies are needed, each Sox family protein may play a distinct role in the development and/or differentiation of effector CD4+ T cells.
In conclusion, we have shown that Sox5 is expressed in Th17 cells and acts together with c-Maf to induce RORγt expression during Th17 cell differentiation. Our results should add a new insight into the regulatory mechanism of Th17 cell differentiation and the pathogenesis of Th17 cell–mediated autoimmune diseases.
MATERIALS AND METHODS
Mice.
BALB/c mice and C57BL/6 mice were purchased from Charles River Laboratories. IL-21–deficient (IL-21−/−) mice were obtained from Mutant Mouse Regional Resource Centers, and CD4cre mice and Stat3fl/fl mice were purchased from The Jackson Laboratory. Sox5fl/fl mice (Dy et al., 2008), gifts from V. Lefebvre (Cleveland Clinic, Cleveland, OH), were backcrossed onto C57BL/6 mice for eight generations and then mated with CD4cre mice. Female CD4cre Sox5fl/fl mice and Sox5fl/fl mice were used in this study. RORγt-deficient mice and OT-II mice were gifts from Y. Iwakura (Tokyo University of Science, Tokyo, Japan) and M. Kubo (Tokyo University of Science), respectively. All mice were housed in microisolator cages under specific pathogen–free conditions. Chiba University Animal Care and Use Committee approved protocols of animal experiments.
Cell culture.
Naive CD4+ T cells were stimulated with plate-bound anti-CD3ε mAb (1 µg/ml) in the presence of anti-CD28 mAb (1 µg/ml) at 0.5–106 cells/ml in a 48-well plate under neutral conditions (10 µg/ml anti–IL-4 mAb and 10 µg/mlanti–IFN-γ mAb), IL-6 conditions (100 ng/ml IL-6, anti–IL-4 mAb, and anti–IFN-γ mAb), Th17 conditions (1 ng/ml IL-6, TGF-β, anti–IL-4 mAb, and anti–IFN-γ mAb), Th17 (23) conditions (20 ng/ml IL-6, 20 ng/ml IL-1β, 50 ng/ml IL-23, 5 ng/ml anti–TGF-β mAb, anti–IL-4 mAb, and anti–IFN-γ mAb), or iTreg conditions (3 ng/mlTGF-β), 10 ng/ml IL-2, anti–IL-4 mAb, and anti–IFN-γ mAb).
Reagents.
Antibodies to CD3ε (145-2C11), CD28 (37.51), CD11c (HL3), CD44 (IM7), CD62L (MEL-14), human NGFR (C40-1457), IL-4 (11B11), IL-4 (BDV4-1D11), and IFN-γ (XMG1.2) were purchased from BD. Antibodies to CD8α (53–6.7), B220 (RA3-6B2), Thy1.1 (OX-7), NKp46 (29A1.4), and IL-17A (TC11-18H10.1) were purchased from BioLegend. Antibodies to Foxp3 (FJK-16s), IL-17F (eBio18F10), and γδ TCR (UC7-13D5) were purchased from eBioscience. Murine IL-1β, IL-4, and IL-6 were purchased from PeproTech. Human TGF-β, murine IL-21, IL-23, anti–TGF-β mAb (1D11), and IL-21R-Fc chimera were purchased from R&D Systems.
Plasmids.
MSCV-IRES-GFP (MIG), MSCV-IRES-Thy1.1 (MIT), and pMX-IRES-NGFR (pMX-IN) are retrovirus vectors containing GFP, Thy1.1, and human NGFR, respectively, under the regulation of an internal ribosome entry site (IRES). Sox5t cDNA from pCR4Blunt-TOPO-Sox5t was subcloned into MIG and pMX-IN to create MIG-Sox5t and pMX-IN-Sox5t, respectively. HA-tag was fused to Sox5t by PCR amplification and subcloned into pMX-IN to create pMX-IN-HA-Sox5t. HA-Sox5t was also subcloned into pcDNA3 (Invitrogen). 3xFlag-tag was fused to Sox5t by subcloning it into p3xFlag CMV vector (Sigma-Aldrich). MIT-c-Maf and pNTAP Myc-c-Maf was described previously (Hiramatsu et al., 2010). MIG-RORγt was a gift from D. Littman (Skirball Institute, New York, NY). hCD4-pA-GFP-RV was a gift from K. Murphy (Washington University, St. Louis, MO; Zhu et al., 2001). The Rorc promoter construct (Rorc-luci) was a gift from V. Lazarevic (National Cancer Institute, National Institutes of Health, Bethesda, MD; Lazarevic et al., 2011). A 2-kb fragment of Rorc promoter from Rorc-luci was subcloned into hCD4-pA-GFP-RV to create hCD4-pA-GFP-RORγtP (WT-RORγtP).
Cell isolation.
CD62L+ CD25− TCR γδ− CD4+ T cells (naive CD4+ T cells) were isolated from lymph nodes or spleen of mice using a CD4+ CD62L+ T cell isolation kit II (Miltenyi Biotec) according to the manufacturer’s instruction. The resultant cells were > 98% pure CD4+ CD62L+ T cells by FACS analysis.
Intracellular cytokine analysis.
Intracellular cytokine staining for IL-21 was performed as described previously (Suto et al., 2008; Hiramatsu et al., 2010). In brief, cultured cells were washed and stimulated with PMA plus ionomycin for 5 h. Cells were stained with anti-CD4 PerCP or anti-Thy1.1 PerCP (BioLegend) for 30 min at 4°C. Cells were then fixed, permeabilized with Perm/Wash buffer (BD), and incubated with IL-21R/Fc chimera (R&D Systems) and anti–IL-4 PE, anti–IFN-γ PE, or anti–IL-17A PE for 30 min at 4°C. Cells were then washed with Perm/Wash buffer and stained with allophycocyanin-conjugated affinity-purified F(ab′)2 fragment of donkey anti–human IgG (H+L; Jackson ImmunoResearch Laboratories) for 30 min at 4°C. Cytokine profiles of CD4+ cells were analyzed on a FACSCalibur with FlowJo software (Tree Star).
Retrovirus-mediated gene expression.
Retrovirus-mediated gene induction for naive CD4+ T cells was performed by a RetroNectin-bound virus infection method (Takara Bio) as described previously (Suto et al., 2008). In brief, 48-well plates were coated with RetroNectin (25 µg/ml) and anti-CD3 mAb (1 µg/ml) overnight at 4°C. Medium containing retrovirus was added to the RetroNectin-coated plate and the plate was centrifuged for 2 h at 2,000 g at 32°C. After washing with PBS, naive CD4+ T cells that had been stimulated with plate-bound anti-CD3 mAb/anti-CD28 mAb under neutral conditions for 24 h were added to the retrovirus-bound RetroNectin/anti-CD3 mAb-coated plates and were cultured for 24 h at 37°C. Cells were then stimulated with anti-CD3 mAb/anti-CD28 mAb in the presence of indicated cytokines or neutralizing antibodies for an additional 3 d.
c-Maf knockdown analysis.
Two sets of siRNA for c-Maf and negative controls were used. siMaf_287-311 (stealth select RNAi Maf-MSS275601) and stealth RNAi negative control duplexes were purchased from Invitrogen. siMaf_110-132 (Rutz et al., 2011) and control siRNA were purchased from Operon Biotechnology. Either 100 pmolsiMaf_110-132 and siMaf_287-311 or 100 pmol negative controls were transfected to naive CD4+ T cells (5 × 105 cells) by using Neon transfection system (Invitrogen). After 4-h incubation under resting conditions, cells were stimulated with plate-bound anti-CD3ε mAb in the presence of anti-CD28 mAb under Th17 conditions for 24 h.
Induction of EAE.
Female CD4cre Sox5fl/fl mice and littermate Sox5fl/fl mice (6–8-wk-old) were injected s.c. in the tail base with MOG35-55 peptide (100 µg/mouse, MEVGWYRSPFSRVVHLYRNGK) in complete Freund’s adjuvant (Chondrex, Inc.). 5 min and 48 h after the injection of MOG35-55 peptide, the mice were injected i.p. with pertussis toxin (200 ng/mouse; Sigma-Aldrich). Status of the mice was monitored and disease severity was scored three times a week as follows: 0 = no clinical signs, 1 = limp tail (tail paralysis), 2 = complete loss of tail tonicity or abnormal gait, 3 = partial hind limb paralysis, 4 = complete hind limb paralysis, 5 = forelimb paralysis or moribund, 6 = death (Okamoto et al., 2010). Cells were isolated from whole brain and spinal cord by Percoll gradient centrifugation and subjected to flow cytometric analysis.
MOG recall Assay.
Ten days after the induction of EAE, single cell suspensions of splenocytes (5 × 105) were stimulated with the MOG peptide (2.5 µg/ml) and cell proliferation was evaluated at 72 h after stimulation by CellTiter-Glo (Promega) according to the manufacturer’s instruction. For cytokine analyses, CD4+ T cells were purified from splenocytes and stimulated with the MOG peptide (2.5 µg/ml) in the presence of irradiated splenocytes for 72 h.
DTH.
Mice (8-wk-old) were immunized subcutaneously with 100 µg of TNP-KLH (Biosearch Technologies) emulsified in CFA at the tail base. 6 d later, mice were challenged with 50 µg of TNP-KLH in the footpad of hind leg and PBS in the footpad of the contralateral hind leg as controls. ΔFootpad thickness was determined by measuring the footpad thickness before and after the induction of DTH using an electronic micrometer (Niigata Seiki) in a blinded fashion.
Adoptive transfer model of DTH.
Naive CD4+ T cells from OVA-specific OT-II mice were stimulated with anti-CD3 mAb and anti-CD28 mAb under neutral conditions for 24 h and infected with retrovirus for 48 h. Retrovirus-infected cells (5 × 106 cells/mouse) were injected i.v. into C57BL/6 mice. 24 h later, the mice were injected s.c. with OVA (25 µg) in incomplete Freund’s Adjuvant into the footpad as descried previously (Feuerer et al., 2006).
Real-time PCR analysis.
Extraction of total cellular RNA, reverse transcription, and quantitative PCR analysis were performed as described previously (Suto et al., 2008). Sequences of qPCR primers for l-Sox5, Sox5t, and Sox5 HMG domain are listed in Table S1. Quantitative PCR was performed with an ABI PRISM 7300 sequence detection system (Applied Biosystems) or StepOnePlus real-time PCR system (Applied Biosystems). The levels of target genes were normalized to the levels of β-actin.
Analysis of CNS in Rorc gene.
We searched for CNS in Rorc gene loci by using the on-line-based mVISTA global alignment program (http://genome.lbl.gov/vista/index.shtml). We defined regions containing fragments longer than 100 bp with at least 75% sequence identity between paired sequences of mouse and human as CNS.
ChIP-qPCR analysis.
ChIP was performed using a ChIP assay kit (Millipore) and Dynabeads Protein G (Invitrogen) as described previously (Hiramatsu et al., 2010). The following antibodies were used for ChIP: anti-c-Maf antibody (M153; Santa Cruz Biotechnology, Inc.), anti-HA antibody (ab9110; Abcam), anti-p300 antibody (C20; Santa Cruz Biotechnology, Inc.), and anti-H3K4me3 antibody (17–614; Millipore). Sequences of qPCR primers for ChIP-qPCR analysis are listed in Table S2. DNA contents were measured by qPCR with a SYBR green reagent. Data were expressed as the percent input for each ChIP fraction.
ChIP-seq analysis.
For ChIP-seq analysis of HA-Sox5t and c-Maf in CD4+ T cells, naive CD4+ T cells were co-infected with pMX-IN-HA-Sox5t and MIT-c-Maf for 24 h and then cultured under neutral conditions for 7 h. For ChIP-seq analysis of HA-Sox5t and c-Maf in Th17 cells, naive CD4+ T cells were infected with pMX-IN-HA-Sox5t under Th17 conditions for 24 h, cultured under Th17 conditions for additional 2 d, and restimulated with plate-bound anti-CD3 mAb and anti-CD28 mAb for 90 min. ChIP was performed as described above. DNAs were sonicated to reduce the length to 100–500 bp with BioRuptor (Cosmo Bio Co.). ChIP-seq libraries were prepared by using a NEBNext ChIP-Seq library prep master mix set, multiples oligos for Illumina (New England BioLabs), and AMpure XP beads (Beckman Coulter) according to the manufacturer’s instructions. The ChIP-seq libraries were subjected to sequence analysis with an Illumina HiSeq1500 in a 50-base single-end mode. The obtained reads were mapped on the mouse reference genome sequence (mm10) using Bowtie. Read depths at each base were calculated and normalized to total depths for anti-HA or anti-c-Maf precipitated and total input DNA. Intervals from -5 kb relative to the transcriptional start site to +5 kb relative to the transcriptional end site of each annotated gene in the RefSeq database were analyzed. HA-Sox5 and c-Maf target genes were determined using MACS software (Zhang et al., 2008; p-value for peak calling set at 0.00001) and, in some cases, visual inspection. For visualization of HA-Sox5 and c-Maf binding sites, data were converted to a wiggle file format or BedGraph format at 25-base resolution and uploaded to the UCSC genome browser or Integrative Genome Viewer (IGV 2.3; Robinson et al., 2011).
RNA-seq analysis.
Naive CD4+ T cells from WT mice were co-infected with retroviruses of pMX-IN/MIT or pMX-IN-HA-Sox5t/MIT-c-Maf for 24 h, and then cultured in neutral conditions for additional 18 h. CD4+ T cells that were co-infected with pMX-IN/MIT or pMX-IN-Sox5t/MIT-c-Maf were isolated and total RNA was prepared from these cells using the PureLink RNA mini kit (Invitrogen). RNA-seq libraries were prepared using a SureSelect Strand Specific RNA Library Preparation kit (Agilent). Sequencing was performed on an Illumina HiSeq1500 using a TruSeq Rapid SBS kit (Illumina) in a 50-base single-end mode. mRNA profiles were calculated with a Cufflinks software and expressed as FPKM (fragments per kilobase of exon model per million mapped fragments).
Integration of ChIP-seq data and RNA-seq data.
Target genes of Sox5 and c-Maf were selected by integrating ChIP-seq data and RNA-seq data as follows: (1) transcripts whose expression levels were <1 FPKM were excluded; (2) transcripts whose difference in FPKM values between CD4+ T cells co-infected with pMX-IN/MIT and those co-infected with pMX-IN-HA-Sox5t/MIT-c-Maf is <4-fold were excluded; and (3) transcripts that were validated as direct targets of Sox5t and c-Maf by ChIP-seq analysis were selected. Log-transformed FPKM values were normalized and used for clustering analysis with GeneSpring software (ver.12.6; Agilent technologies).
Luciferase reporter assay.
EL4 cells (5 × 105 cells) were resuspended with the indicated plasmids (1 µg each for pcDNA3 vectors [pcDNA3-Sox5t or empty pcDNA3] and pGL3 vectors [−2.0 kb, −1.6 kb, or −1.4 kb RORγt-Luci] and 20 ng for pRL-TK vector) in resuspension buffer R and electroporated by using Neon transfection system (Invitrogen). After cells were cultured at 37°C for 48 h, cells were lysed and relative light units (RLU) were assessed with a dual luciferase assay system (Promega). Firefly luciferase activity of reporter constructs was normalized by Renilla luciferase activity of pRL-TK and data were shown as fold induction relative to pcDNA3-transfected cells. All values were obtained from experiments performed in triplicate and repeated at least three times.
Western blot and co-immunoprecipitation (Co-IP) analysis.
Western blot and Co-IP analysis were performed as described previously (Hiramatsu et al., 2010). To examine the interaction between c-Maf and Sox5, the following vectors were used: pNTAP-Myc-c-Maf (WT) or its mutants lacking transactivation domain (dTA), DNA-binding domain (dDB), or dimerization domain (dDD); and p3xFlag-Sox5t (WT) or its mutants lacking either N-terminal region (dN), the first coiled-coil domain (dCC1), aa 227–339 (d227-399), the second coiled–coil domain (dCC2), HMG domain (dHMG), or C-terminal region (dC). The following antibodies were used for immunoblotting and Co-IP: anti-Sox5 antibody (H-90; Santa Cruz Biotechnology Inc.), anti–c-Maf antibody (M153; Santa Cruz Biotechnology Inc.), anti–Flag-M2 antibody (Sigma-Aldrich), anti-Myc antibody (9E10; Santa Cruz Biotechnology Inc.), anti-HSP90 antibody (Santa Cruz Biotechnology Inc.), and HRP-conjugated goat anti–rabbit IgG (H+L; Zymed).
DNA precipitation assay.
293T cells were transfected with pcDNA3-HA-Sox5 or pcDNA3-Myc-c-Maf. Nuclear protein was incubated for 1 h at 4°C with a biotinylated double-stranded DNA probe of promoter regions of RORγt gene or its mutant on putative binding site of Sox5 or Maf recognition elements (MARE) and conjugated with streptavidin-agarose beads (Invitrogen) in binding buffer (100 mM NaCl, 10 mM Tris-HCl, pH 7.6, 0.1 mM EDTA, 1 mM dithiothreitol, 5% (vol/vol) glycerol, 1 mg/ml of BSA, and protease inhibitors). The beads were washed with binding buffer, and bound proteins were analyzed by immunoblotting with anti-HA antibody or anti–Maf-antibody.
5′-Rapid amplification of cDNA ends (5′-RACE).
5′-RACE of Sox5 mRNA was performed by Gene Racer kit (Invitrogen) according to the manufacturer’s instruction. In brief, total RNA from IL-6–stimulated CD4+ T cells was subjected to oligo-capping and RNA ligase-mediated RACE methods. Ligated mRNA was reverse transcribed with random primers. To obtain 5′ ends, the first-strand cDNA was amplified using Sox5R2430 primer (5′-GGCTCACCACAGTCTGTTGGCCCTTA-3′) and the Gene Racer 5′ primer. Nested PCR was performed using Sox5R2410 nested primer (5′-CCCTTATCAGTTGGCTTGTCCCGCAATG-3′) and Gene Racer 5′ nested primer. PCR products were subcloned into pCR4Blunt-TOPO vector for sequencing.
Data analysis.
Data are summarized as means ± SD. The statistical analysis of the results was performed by the unpaired Student’s t test and Mann-Whitney U test. P < 0.05 was considered significant.
Online supplemental material.
Table S1 lists sequences of qPCR primers for L-Sox5, Sox5t, and Sox5 HMG domain. Table S2 lists sequences of qPCR primers for ChIP-qPCR assay. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20130791/DC1.
Supplementary Material
Acknowledgments
We thank Dr. V. Lefebvre for Sox5fl/fl mice, Dr. Y. Iwakura for RORγt-deficient mice, Dr. M. Kubo for OT-II mice, Dr. M. Kuwahara for Sox5 deletion mutants, and Ms. M. Kobayashi, H. Furuta, and J. Iwata for technical help.
This work was supported in part by Grants-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, the Japanese Government, Global COE Program (Global Center for Education and Research in Immune System Regulation and Treatment), and LGS (Leading Graduate School at Chiba University) Program, MEXT, Japan.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- Chromatin immunoprecipitation
- CNS
- conserved noncoding sequence
- DTH
- delayed-type hypersensitivity
- EAE
- experimental autoimmune encephalomyelitis
- H3K4me3
- histone H3 lysine 4 trimethylation
- IB
- immunoblotted
- LTi-like
- lymphoid tissue inducer-like
- SOX
- SRY-related high-mobility-group-box
References
- Apetoh, L., Quintana F.J., Pot C., Joller N., Xiao S., Kumar D., Burns E.J., Sherr D.H., Weiner H.L., and Kuchroo V.K.. 2010. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 11:854–861 10.1038/ni.1912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauquet, A.T., Jin H., Paterson A.M., Mitsdoerffer M., Ho I.C., Sharpe A.H., and Kuchroo V.K.. 2009. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH-17 cells. Nat. Immunol. 10:167–175 10.1038/ni.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli, E., Carrier Y., Gao W., Korn T., Strom T.B., Oukka M., Weiner H.L., and Kuchroo V.K.. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238 10.1038/nature04753 [DOI] [PubMed] [Google Scholar]
- Brüstle, A., Heink S., Huber M., Rosenplänter C., Stadelmann C., Yu P., Arpaia E., Mak T.W., Kamradt T., and Lohoff M.. 2007. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol. 8:958–966 10.1038/ni1500 [DOI] [PubMed] [Google Scholar]
- Chen, Q., Dowhan D.H., Liang D., Moore D.D., and Overbeek P.A.. 2002. CREB-binding protein/p300 co-activation of crystallin gene expression. J. Biol. Chem. 277:24081–24089 10.1074/jbc.M201821200 [DOI] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang, E.V., Barbi J., Yang H.Y., Jinasena D., Yu H., Zheng Y., Bordman Z., Fu J., Kim Y., Yen H.R., et al. 2011. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 146:772–784 10.1016/j.cell.2011.07.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C.2008. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat. Rev. Immunol. 8:337–348 10.1038/nri2295 [DOI] [PubMed] [Google Scholar]
- Durant, L., Watford W.T., Ramos H.L., Laurence A., Vahedi G., Wei L., Takahashi H., Sun H.W., Kanno Y., Powrie F., and O’Shea J.J.. 2010. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 32:605–615 10.1016/j.immuni.2010.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dy, P., Han Y., and Lefebvre V.. 2008. Generation of mice harboring a Sox5 conditional null allele. Genesis. 46:294–299 10.1002/dvg.20392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer, M., Eulenburg K., Loddenkemper C., Hamann A., and Huehn J.. 2006. Self-limitation of Th1-mediated inflammation by IFN-gamma. J. Immunol. 176:2857–2863 10.4049/jimmunol.176.5.2857 [DOI] [PubMed] [Google Scholar]
- Ghilardi, N., Kljavin N., Chen Q., Lucas S., Gurney A.L., and De Sauvage F.J.. 2004. Compromised humoral and delayed-type hypersensitivity responses in IL-23-deficient mice. J. Immunol. 172:2827–2833 10.4049/jimmunol.172.5.2827 [DOI] [PubMed] [Google Scholar]
- Ghoreschi, K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiramatsu, Y., Suto A., Kashiwakuma D., Kanari H., Kagami S., Ikeda K., Hirose K., Watanabe N., Grusby M.J., Iwamoto I., and Nakajima H.. 2010. c-Maf activates the promoter and enhancer of the IL-21 gene, and TGF-beta inhibits c-Maf-induced IL-21 production in CD4+ T cells. J. Leukoc. Biol. 87:703–712 [DOI] [PubMed] [Google Scholar]
- Ho, I.C., Hodge M.R., Rooney J.W., and Glimcher L.H.. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell. 85:973–983 10.1016/S0092-8674(00)81299-4 [DOI] [PubMed] [Google Scholar]
- Huang, W., Lu N., Eberspaecher H., and De Crombrugghe B.. 2002. A new long form of c-Maf cooperates with Sox9 to activate the type II collagen gene. J. Biol. Chem. 277:50668–50675 10.1074/jbc.M206544200 [DOI] [PubMed] [Google Scholar]
- Ivanov, I.I., McKenzie B.S., Zhou L., Tadokoro C.E., Lepelley A., Lafaille J.J., Cua D.J., and Littman D.R.. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133 10.1016/j.cell.2006.07.035 [DOI] [PubMed] [Google Scholar]
- Korn, T., Bettelli E., Gao W., Awasthi A., Jäger A., Strom T.B., Oukka M., and Kuchroo V.K.. 2007. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 448:484–487 10.1038/nature05970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn, T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Kurokawa, H., Motohashi H., Sueno S., Kimura M., Takagawa H., Kanno Y., Yamamoto M., and Tanaka T.. 2009. Structural basis of alternative DNA recognition by Maf transcription factors. Mol. Cell. Biol. 29:6232–6244 10.1128/MCB.00708-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara, M., Yamashita M., Shinoda K., Tofukuji S., Onodera A., Shinnakasu R., Motohashi S., Hosokawa H., Tumes D., Iwamura C., et al. 2012. The transcription factor Sox4 is a downstream target of signaling by the cytokine TGF-β and suppresses T(H)2 differentiation. Nat. Immunol. 13:778–786 10.1038/ni.2362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence, A., Tato C.M., Davidson T.S., Kanno Y., Chen Z., Yao Z., Blank R.B., Meylan F., Siegel R., Hennighausen L., et al. 2007. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 26:371–381 10.1016/j.immuni.2007.02.009 [DOI] [PubMed] [Google Scholar]
- Lazarevic, V., Chen X., Shim J.H., Hwang E.S., Jang E., Bolm A.N., Oukka M., Kuchroo V.K., and Glimcher L.H.. 2011. T-bet represses T(H)17 differentiation by preventing Runx1-mediated activation of the gene encoding RORγt. Nat. Immunol. 12:96–104 10.1038/ni.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, V.2010. The SoxD transcription factors—Sox5, Sox6, and Sox13—are key cell fate modulators. Int. J. Biochem. Cell Biol. 42:429–432 10.1016/j.biocel.2009.07.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre, V., Li P., and de Crombrugghe B.. 1998. A new long form of Sox5 (L-Sox5), Sox6 and Sox9 are coexpressed in chondrogenesis and cooperatively activate the type II collagen gene. EMBO J. 17:5718–5733 10.1093/emboj/17.19.5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littman, D.R., and Rudensky A.Y.. 2010. Th17 and regulatory T cells in mediating and restraining inflammation. Cell. 140:845–858 10.1016/j.cell.2010.02.021 [DOI] [PubMed] [Google Scholar]
- Malhotra, N., Narayan K., Cho O.H., Sylvia K.E., Yin C., Melichar H., Rashighi M., Lefebvre V., Harris J.E., Berg L.J., and Kang J.. Immunological Genome Project Consortium. 2013. A network of high-mobility group box transcription factors programs innate interleukin-17 production. Immunity. 38:681–693 10.1016/j.immuni.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan, P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Mathur, A.N., Chang H.C., Zisoulis D.G., Stritesky G.L., Yu Q., O’Malley J.T., Kapur R., Levy D.E., Kansas G.S., and Kaplan M.H.. 2007. Stat3 and Stat4 direct development of IL-17-secreting Th cells. J. Immunol. 178:4901–4907 10.4049/jimmunol.178.8.4901 [DOI] [PubMed] [Google Scholar]
- McGeachy, M.J., Chen Y., Tato C.M., Laurence A., Joyce-Shaikh B., Blumenschein W.M., McClanahan T.K., O’Shea J.J., and Cua D.J.. 2009. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat. Immunol. 10:314–324 10.1038/ni.1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melichar, H.J., Narayan K., Der S.D., Hiraoka Y., Gardiol N., Jeannet G., Held W., Chambers C.A., and Kang J.. 2007. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 315:230–233 10.1126/science.1135344 [DOI] [PubMed] [Google Scholar]
- Okamoto, K., Iwai Y., Oh-Hora M., Yamamoto M., Morio T., Aoki K., Ohya K., Jetten A.M., Akira S., Muta T., and Takayanagi H.. 2010. IkappaBzeta regulates T(H)17 development by cooperating with ROR nuclear receptors. Nature. 464:1381–1385 10.1038/nature08922 [DOI] [PubMed] [Google Scholar]
- Rajaram, N., and Kerppola T.K.. 2004. Synergistic transcription activation by Maf and Sox and their subnuclear localization are disrupted by a mutation in Maf that causes cataract. Mol. Cell. Biol. 24:5694–5709 10.1128/MCB.24.13.5694-5709.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveros, C., Mellor D., Gandhi K.S., McKay F.C., Cox M.B., Berretta R., Vaezpour S.Y., Inostroza-Ponta M., Broadley S.A., Heard R.N., et al. ANZgene Multiple Sclerosis Genetics Consortium. 2010. A transcription factor map as revealed by a genome-wide gene expression analysis of whole-blood mRNA transcriptome in multiple sclerosis. PLoS ONE. 5:e14176 10.1371/journal.pone.0014176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutz, S., Noubade R., Eidenschenk C., Ota N., Zeng W., Zheng Y., Hackney J., Ding J., Singh H., and Ouyang W.. 2011. Transcription factor c-Maf mediates the TGF-β-dependent suppression of IL-22 production in T(H)17 cells. Nat. Immunol. 12:1238–1245 10.1038/ni.2134 [DOI] [PubMed] [Google Scholar]
- Schilham, M.W., Oosterwegel M.A., Moerer P., Ya J., de Boer P.A., van de Wetering M., Verbeek S., Lamers W.H., Kruisbeek A.M., Cumano A., and Clevers H.. 1996. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 380:711–714 10.1038/380711a0 [DOI] [PubMed] [Google Scholar]
- Schilham, M.W., Moerer P., Cumano A., and Clevers H.C.. 1997. Sox-4 facilitates thymocyte differentiation. Eur. J. Immunol. 27:1292–1295 10.1002/eji.1830270534 [DOI] [PubMed] [Google Scholar]
- Schraml, B.U., Hildner K., Ise W., Lee W.L., Smith W.A., Solomon B., Sahota G., Sim J., Mukasa R., Cemerski S., et al. 2009. The AP-1 transcription factor Batf controls T(H)17 differentiation. Nature. 460:405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits, P., Li P., Mandel J., Zhang Z., Deng J.M., Behringer R.R., de Crombrugghe B., and Lefebvre V.. 2001. The transcription factors l-Sox5 and Sox6 are essential for cartilage formation. Dev. Cell. 1:277–290 10.1016/S1534-5807(01)00003-X [DOI] [PubMed] [Google Scholar]
- Suto, A., Kashiwakuma D., Kagami S., Hirose K., Watanabe N., Yokote K., Saito Y., Nakayama T., Grusby M.J., Iwamoto I., and Nakajima H.. 2008. Development and characterization of IL-21–producing CD4+ T cells. J. Exp. Med. 205:1369–1379 10.1084/jem.20072057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen, M., Hocking R.J., Atkins C.J., Locksley R.M., and Stockinger B.. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Veldhoen, M., Hirota K., Westendorf A.M., Buer J., Dumoutier L., Renauld J.C., and Stockinger B.. 2008. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 453:106–109 10.1038/nature06881 [DOI] [PubMed] [Google Scholar]
- Visel, A., Blow M.J., Li Z., Zhang T., Akiyama J.A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 457:854–858 10.1038/nature07730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner, M.2010. All purpose Sox: The many roles of Sox proteins in gene expression. Int. J. Biochem. Cell Biol. 42:381–390 10.1016/j.biocel.2009.07.006 [DOI] [PubMed] [Google Scholar]
- Wilson, M., and Koopman P.. 2002. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr. Opin. Genet. Dev. 12:441–446 10.1016/S0959-437X(02)00323-4 [DOI] [PubMed] [Google Scholar]
- Yang, Y., and Cvekl A.. 2007. Large Maf transcription factors: cousins of AP-1 proteins and important regulators of cellular differentiation. Einstein J. Biol. Med. 23:2–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., and Dong C.. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363 10.1074/jbc.C600321200 [DOI] [PubMed] [Google Scholar]
- Yang, X.O., Pappu B.P., Nurieva R., Akimzhanov A., Kang H.S., Chung Y., Ma L., Shah B., Panopoulos A.D., Schluns K.S., et al. 2008. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR alpha and ROR gamma. Immunity. 28:29–39 10.1016/j.immuni.2007.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, L., Ivanov I.I., Spolski R., Min R., Shenderov K., Egawa T., Levy D.E., Leonard W.J., and Littman D.R.. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8:967–974 10.1038/ni1488 [DOI] [PubMed] [Google Scholar]
- Zhu, H., Yang J., Murphy T.L., Ouyang W., Wagner F., Saparov A., Weaver C.T., and Murphy K.M.. 2001. Unexpected characteristics of the IFN-gamma reporters in nontransformed T cells. J. Immunol. 167:855–865 10.4049/jimmunol.167.2.855 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.