Within the human TCR-α/δ locus, ordered rearrangements requires RUNX1, which binds to the Dδ2-23RSS and interacts with RAG1 to enhance RAG1 deposition at this site. Absence of this RUNX1 binding site in the homologous murine Dδ1-23RSS offers an explanation for the lack of ordered TCR-δ gene assembly in mice.
Abstract
V(D)J recombination of TCR loci is regulated by chromatin accessibility to RAG1/2 proteins, rendering RAG1/2 targeting a potentially important regulator of lymphoid differentiation. We show that within the human TCR-α/δ locus, Dδ2-Dδ3 rearrangements occur at a very immature thymic, CD34+/CD1a−/CD7+dim stage, before Dδ2(Dδ3)-Jδ1 rearrangements. These strictly ordered rearrangements are regulated by mechanisms acting beyond chromatin accessibility. Importantly, direct Dδ2-Jδ1 rearrangements are prohibited by a B12/23 restriction and ordered human TCR-δ gene assembly requires RUNX1 protein, which binds to the Dδ2-23RSS, interacts with RAG1, and enhances RAG1 deposition at this site. This RUNX1-mediated V(D)J recombinase targeting imposes the use of two Dδ gene segments in human TCR-δ chains. Absence of this RUNX1 binding site in the homologous mouse Dδ1-23RSS provides a molecular explanation for the lack of ordered TCR-δ gene assembly in mice and may underlie differences in early lymphoid differentiation between these species.
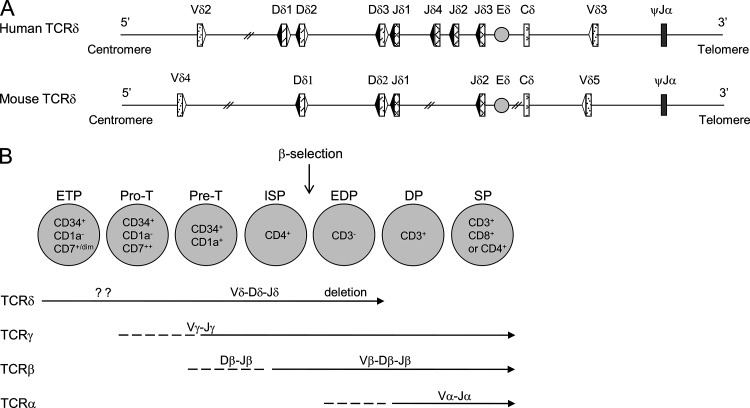
Human T lymphocyte ontogeny is a hierarchical thymic process in which the ordered somatic recombination of V, (D), and J gene segments at the TCR-δ, TCR-γ, TCR-β, or TCR-α loci determines the development into either γδ or αβ T cell lineages (Spits, 2002; Dik et al., 2005). TCR-δ rearrangement is the first to occur, at the CD34+, CD1a−, CD4/8 double-negative (DN) stage, followed by concurrent TCR-γ and TCR-β rearrangements coinciding with CD1a expression (Dik et al., 2005), and finally TCR-α rearrangements, which occur at a later CD4/8 double-positive (DP) stage, accompanied by TCR-δ locus deletion due to Vα-Jα recombination (Verschuren et al., 1997). All V, (D), and J gene segments are flanked with recombination signal sequences (RSSs) which are composed of conserved heptamer and nonamer motifs separated by a nonconserved spacer of either 12 or 23 base pairs. The V(D)J recombination process is initiated by the multimeric RAG1–RAG2 complex (RAG1/2) which binds to a 12RSS/23RSS pair (12/23 rule) and then introduces double-strand breaks (DSBs) simultaneously at the two coding segment–RSS junctions (Eastman et al., 1996; van Gent et al., 1996). The subsequent repair phase involves the non-homologous end joining complex as well as the TdT (terminal deoxynucleotidyl transferase) enzyme which increases antigen receptor diversity by adding N nucleotides at the coding segment junctions. The final products of V(D)J recombination are the CJ (coding joint) and the SJ (signal joint). In most cases, the latter is excised from the chromosome as a T cell receptor excision circle (TREC). Because TRECs are episomal and nonreplicative DNA, their quantity decreases during cell proliferation (Dik et al., 2005).
Regulation of V(D)J recombination is mediated by enhancer and promoter changes in chromatin structure which determine accessibility of the chromosomal RSS to RAG1/2 complexes (Yancopoulos and Alt, 1985; Hesslein and Schatz, 2001). In DN thymocytes, TCR-δ enhancer (Eδ) activation controls chromatin accessibility to support TCR-δ rearrangement (Bassing et al., 2003), whereas in DP thymocytes, activation of the TCR-α enhancer (Eα) is indispensable for the initiation of TCR-α rearrangement (Sleckman et al., 1997). Intrinsic RSS features can, however, be directly involved in the control of V(D)J recombination beyond chromatin accessibility (Krangel, 2003). This has been clearly demonstrated for TCR-β gene assembly. Indeed, direct Vβ-Jβ rearrangement is prohibited by a mechanism operating beyond the 12/23 rule and imposing Dβ segment usage (Bassing et al., 2000). This B12/23 restriction is independent of chromatin structure; it is based on RSS features and can be fully recapitulated with in vitro systems using chromatin-free DNA (Jung et al., 2003; Tillman et al., 2003).
The B12/23 restriction imposes a two-step process for TCR-β assembly but does not explain the ordering (D-J before V-DJ). It has recently been proposed that Dβ 23RSS binds a transcription factor (TF), c-Fos, which efficiently recruits RAG1 and enforces that Dβ-Jβ rearrangement occurs first (Wang et al., 2008). In both mice and humans, TCR-δ and TCR-β loci contain D gene segments (Lefranc, 2001) which harbor the same RSS distribution (Fig. 1). Despite this, it is generally considered that TCR-δ gene rearrangements are not ordered in mice (Chiei et al., 1987; Krangel et al., 2004), whereas it has been suggested that they could be ordered in human (Dik et al., 2005).
Figure 1.
TCR-δ configuration in humans and mice and the order of human thymic TCR rearrangements. (A) Schematic representation of human and mouse TCR-δ loci. 23RSS and 12RSS are represented, respectively, in white and black triangles. (B) Thymic maturation stages and their TCR rearrangements in humans.
To better characterize the control of V(D)J recombination at the human TCR-δ locus, we have assessed the kinetics of TCR-δ gene assembly during the early stages of thymopoiesis. We report here that human TCR-δ gene rearrangements are controlled by a B12/23 restriction and are ordered. Importantly the human but not mouse TCR-δ gene assembly ordering involves the TF RUNX1 which, through recruitment to the Dδ2-23RSS and interaction with RAG1, insures that Dδ2-Dδ3 rearrangement occurs before Dδ3-Jδ1 rearrangement. This specific RUNX1 Dδ2-23RSS interaction might provide molecular insight into the difference between murine and human early T cell ontogeny.
RESULTS
Human Dδ2-Dδ3 rearrangements occur before Dδ2-Jδ1 rearrangements at a specific CD34+ CD7+dim CD5+/− early thymic stage
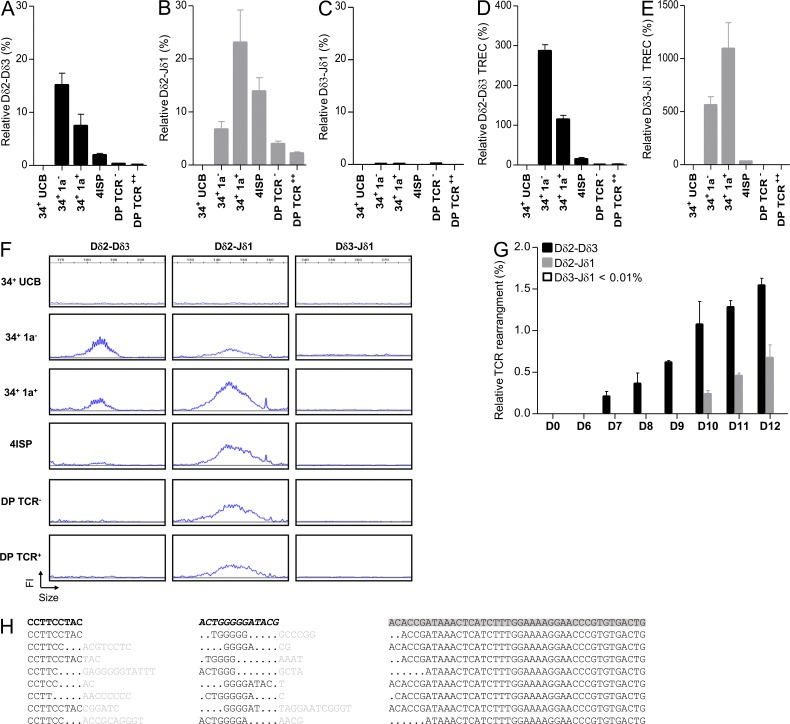
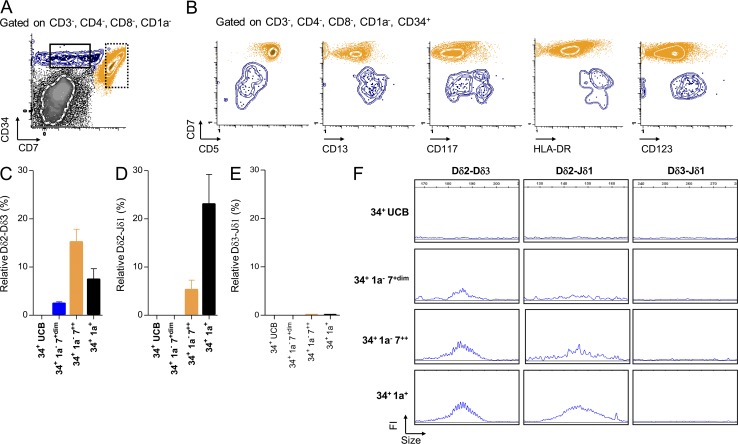
To determine whether human TCR-δ rearrangements are ordered, we first quantified early TCR-δ rearrangements (Dδ2-Dδ3, Dδ2-Jδ1, and Dδ3-Jδ1) in sorted (Fig. S1) human thymic subpopulations and CD34+ umbilical cord blood (UCB) cells. As previously reported (Dik et al., 2005), Dδ2-Dδ3 rearrangements were readily detected in CD34+/CD1a− thymocytes, whereas Dδ2-Jδ1 rearrangements were detected at a lower level. In CD34+/CD1a+ thymocytes, Dδ2-Jδ1 rearrangement reached maximum levels and Dδ2-Dδ3 rearrangement declined (Fig. 2, A and B). In contrast, virtually no Dδ3-Jδ1 rearrangement was detected in human thymic subpopulations (Fig. 2 C).These observations were confirmed by quantification of the related T cell excision circles (Fig. 2, D and E) and fluorescent PCR analysis (Fig. 2 F). We next cultured UCB CD34+ cells on OP9-DL1 and analyzed TCR-δ rearrangement during culture. Dδ2-Dδ3 rearrangements were first detected at day 7, followed by Dδ2-Jδ1 rearrangements 3 d later and again no Dδ3-Jδ1 rearrangement was identified (Fig. 2 G). To further confirm ordered TCR-δ rearrangement, we amplified, subcloned, and sequenced Dδ2-Jδ1 human thymic rearrangements. Dδ3 segments were detected in all Dδ2-Jδ1 rearrangements sequenced, suggesting that the assembly of Dδ2 and Jδ1 gene segments occurs in a two-step process which systematically includes the Dδ3 gene segment (representative data in Fig. 2 H). To identify an early thymic stage specific to Dδ2-Dδ3 rearrangements, we sorted human CD4/8 DN/CD34+ thymic precursors based on the level of CD7 expression, into CD34+/CD1a−/CD7+dim and CD34+/CD1a−/CD7++ subpopulations (Fig. 3 A). As expected, the CD34+/CD1a−/CD7+dim subset displayed an early T cell precursor (ETP) phenotype, based on weak CD5 expression and stem cell/myeloid antigen expression (Fig. 3 B). Interestingly ETP cells harbored only Dδ2-Dδ3 rearrangements, whereas CD34+/CD1a−/CD7++ thymocytes exhibited both Dδ2-Dδ3 and Dδ2-Jδ1 rearrangements (Fig. 3, C and D). Dδ3-Jδ1 rearrangements were not detected in these populations (Fig. 3 E). The monitoring of those rearrangements by multiplex fluorescent PCR analysis confirmed these data, and demonstrated that Dδ2-Dδ3 rearrangements start before Dδ2-Jδ1 rearrangements in a specific ETP subset (Fig. 3 F).
Figure 2.
Kinetics of TCR-δ rearrangements during T cell development. (A–C) Quantification of Dδ2-Dδ3 (A), Dδ2-Jδ1 (B), and Dδ3-Jδ1 (C) rearrangements by RQ-PCR from CD34+ UCB, DN (34+1a− and 34+1a+), immature SP (ISP), and DP thymocytes. Results (mean and SEM of triplicate reactions) are normalized to the Albumin gene throughout. (D and E) Quantification of Dδ2-Dδ3 (D) and Dδ3-Jδ1 (E) TREC by RQ-PCR. Presented data are from three experiments. Error bars represent SEM from three experiments. (F) Fluorescent PCR Genescan analysis (FI: fluorescence intensity) of Dδ2-Dδ3, Dδ2-Jδ1, and Dδ3-Jδ1 rearrangements present in sorted thymic populations. Data are representative of three experiments. (G) Quantification of Dδ2-Dδ3, Dδ2-Jδ1, and Dδ3-Jδ1 rearrangements at different days of CD34+ UCB culture on OP9-DL1. Presented data are from three independent experiments. (H) Sequence of Dδ2-Jδ1 thymic rearrangements. Data are representative of 52 sequences analyzed. Dδ2, Dδ3, and Jδ1 segments are, respectively, in bold black, italic bold black, and gray highlight. Points represent deleted nucleotides. Inserted nucleotides during recombination are in gray.
Figure 3.
Dδ2-Dδ3 rearrangements occur in the CD34+/CD1a−/CD7+dim stage. (A) FACS plots showing the gating strategy used for sorting populations (dots and SD curves). Black and dashed boxes represent CD34+/CD1a−/CD7+dim and CD34+/CD1a−/CD7++ subpopulations, respectively. (B) FACS plots representing phenotypic markers differentially expressed between CD34+/CD1a−/CD7+dim (blue) and CD34+/CD1a−/CD7++ (orange) populations. (C–E) Quantification by RQ-PCR normalized to albumin (mean and SEM) of Dδ2-Dδ3 (C), Dδ2-Jδ1 (D), and Dδ3-Jδ1 (E) rearrangements. Data are representative of three experiments. (F) Fluorescent PCR Genescan analysis (FI: fluorescence intensity) of Dδ2-Dδ3, Dδ2-Jδ1, and Dδ3-Jδ1 rearrangements. Data are representative of three experiments.
RUNX1 interacts with Dδ2-23RSS
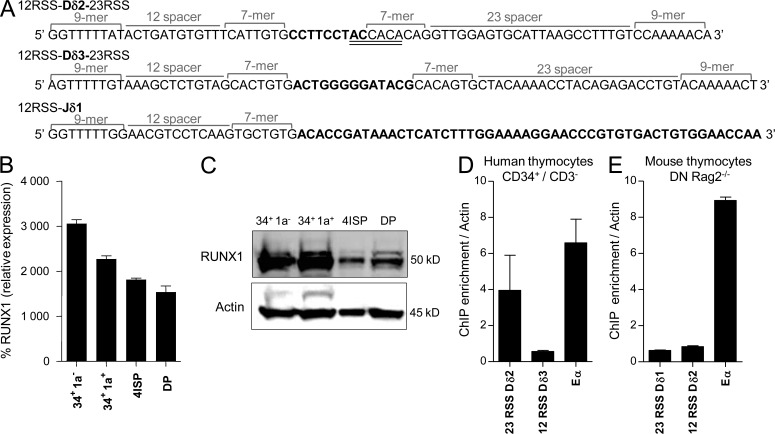
As Dδ2 to Dδ3 rearrangement occurs at a very immature stage of thymic maturation, harboring low levels of RAG transcripts (unpublished data), we hypothesized that early TCR-δ gene assembly requires a specific TF to allow efficient RAG1/2-loading onto RSS, as described for c-Fos and Dβ1 (Wang et al., 2008). To uncover putative TF binding sites within human Dδ2, Dδ3, and Jδ1 RSS, an in silico analysis was performed using the ConSite web-based tool. This identified a full consensus DNA binding site for RUNX1 (5′TG(T/C)GGT3′) spanning the human Dδ2-23RSS heptamer and Dδ2 coding sequence (Fig. 4 A). No RUNX1 binding site was found in the other Dδ and Jδ RSS/coding sequences. RUNX1 expression is high (at both transcript and protein levels) in human early thymic cells (Fig. 4, B and C) and as such coincides with early TCR-δ gene rearrangement. Chromatin immunoprecipitation (ChIP) assays performed with human CD34+ thymocytes and anti-RUNX1 antibody show that immunoprecipitated DNA is significantly enriched with Dδ2-23RSS DNA, compared with Dδ3-12RSS DNA (Fig. 4 D). Importantly, the RUNX1 binding site is not conserved in the mouse Dδ1 (homologous to human Dδ2) and, as expected, no Dδ1-23RSS enrichment was observed after RUNX1 ChIP in mouse Rag2−/− DN thymocytes (Fig. 4 E). These data demonstrate that RUNX1 can bind human Dδ2-23RSS at a stage when Dδ2 to Dδ3 rearrangement is taking place.
Figure 4.
Binding of RUNX1 to the Dδ2-23RSS. (A) Sequences of RSS (12 and 23) and coding segments (bold) for Dδ2, Dδ3, and Jδ1. The RUNX1 putative binding site in Dδ2-23RSS is double underlined. (B) RQ-PCR for RUNX1 expression in human thymic subpopulations. Results (mean and SEM of triplicate reactions) are represented relative to the ABL1 housekeeping gene. (C) Western blot analysis of RUNX1 expression in human thymic subpopulations. Shown is a representative of three Western blots. (D) Analysis by ChIP-QPCR assays of RUNX1 binding in human CD34+/CD3− cells. Enrichment level was determined by comparison to a standard curve from input DNA. Dδ2-23RSS, Dδ3-12RSS, and Eα DNA were amplified by RQ-PCR and normalized to actin as negative control. IgG isotype control was performed to assess absence of nonspecific ChIP enrichment (not depicted). Errors bars represent SEM. (E) As in D, for mouse Rag2−/− DN thymocytes. ChIP experiments are representative of at least two independent experiments.
RUNX1 interacts with RAG1 protein and enhances RAG1 deposition to the Dδ2-23RSS
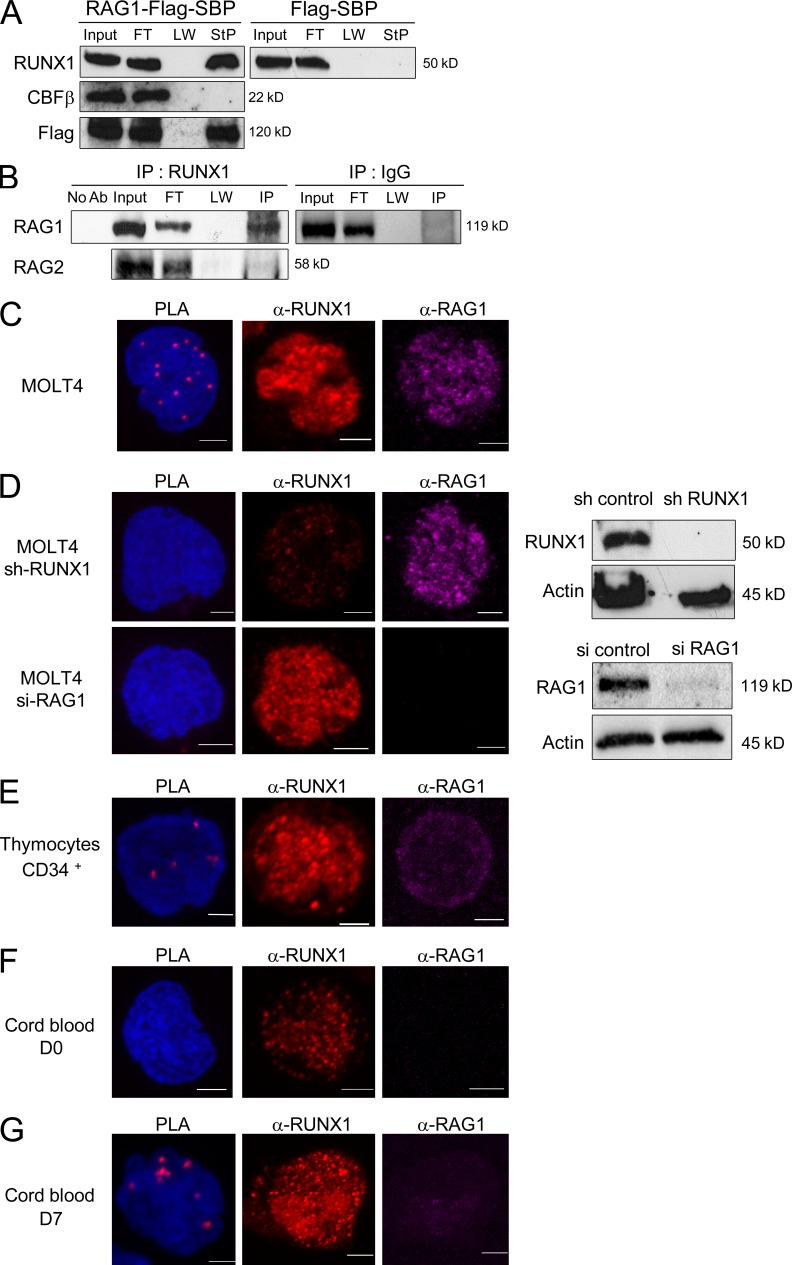
To explore further whether RUNX1 could interact with RAG proteins, streptavidin precipitation, coimmunoprecipitations (Co-IPs), and proximity ligation assays (PLAs) were performed. Streptavidin precipitation (StP) experiments demonstrated that RUNX1 protein, but not CBF-β, interacts with and immunoprecipitates RAG1 (Fig. 5 A). Furthermore, Co-IP assays using the MOLT-4 T lymphoblastic cell line, which coexpresses RUNX1 and RAG1, showed that IP performed with an anti-RUNX1 antibody results in the Co-IP of RAG1, but not RAG2 (Fig. 5 B). Finally, PLA confirmed the RUNX1–RAG1 interaction in MOLT-4 cells (Fig. 5 C) and showed that the interaction was lost after RUNX1 or RAG1 inactivation (Fig. 5 D). Importantly, PLA assays showed that RUNX1 interacts with RAG1 in CD34+ thymocytes (Fig. 5 E) and UCB cells co-cultured on OP9-DL1 (Fig. 5, F and G) for 7 d (concurring with initiation of Dδ2-Dδ3 rearrangement, as shown in Fig. 2 G).
Figure 5.
RUNX1–RAG1 interactions. (A) RAG1 interacts with RUNX1 but not CBF-β. Cell lysates from 293T cells cotransfected with vectors expressing RAG1-Flag-SBP, RUNX1, and CBF-β were precipitated with streptavidin (StP) beads and then immunoblotted (IB) with anti-RUNX1, anti–CBF-β, and anti-Flag antibodies. Input represents 1% of cell lysate used for StP. FT: flow through; LW: last wash. In the control experiment, 293T cells were transduced with Flag-SBP empty vector instead of RAG1-Flag-SBP. A representative of two independent experiments is shown. (B) MOLT-4 cells lysates were immunoprecipitated (IP) using anti-RUNX1 or control IgG antibody and then immunoblotted using an anti-RAG1 antibody. The input lanes correspond to 10% of cell extracts used in the Co-IP. No Ab: control IP experiment performed without anti-RUNX1 antibody. FT: flow-through; LW: last wash. A representative of two independent experiments is shown. (C–G) Duolink PLAs and confocal microscopy analysis of cells labeled with anti-RAG1 mAb (Alexa Fluor 555, purple) and anti-RUNX1 (Alexa Fluor 647, red). Shown is a representative of at least three independent experiments. Bars, 10 µm. (C and D) PLA using MOLT-4 cell line before (C) and after (D) inactivation of RUNX1 or RAG1. Western blots are shown on the right for RUNX1, RAG1, and actin expression from mock and knockdown cells. (E–G) PLA using CD34+ thymocytes (E) and CD34+ UCB after sort (D0; F) and after 7 d of culture (D7; G).
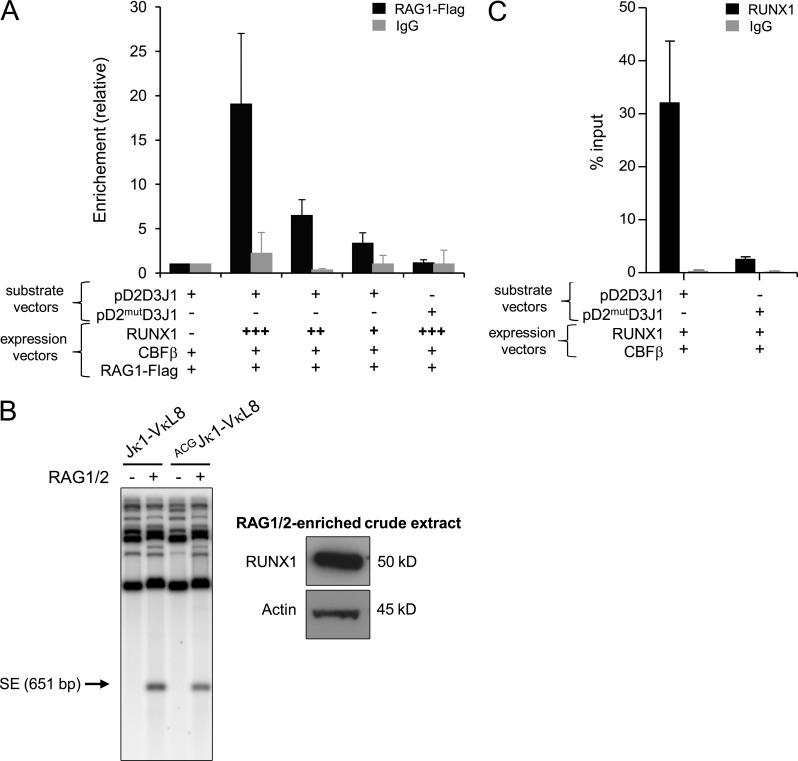
To next test the hypothesis that RUNX1 recruits RAG1 to Dδ2-23RSS, ChIP assays with anti–flag-RAG1 antibody were performed using 293T cells transfected with pD2D3J1 recombination substrate vector. We observed that RAG1 binding to Dδ2-23RSS is dependent on the presence of RUNX1 (Fig. 6 A). The presence of CBF-β, but not RAG2, was necessary for RAG1 recruitment (unpublished data). The ChIP assay was then performed with a pD2mutD3J1 vector harboring a mutated RUNX1 binding site. This mutation corresponds to the addition of a guanine located within the Dδ2 coding sequence and as such it keeps the Dδ2-23RSS intact. Coding flank composition can affect V(D)J recombination (Ezekiel et al., 1997); however, this mutation, which changes the first nucleotide of the coding flank, has no major impact on RAG cleavage efficiency (Fig. 6 B, left). This assay was performed with RAG1/2-enriched crude extract complemented with a thymic protein extract containing Runx1 protein (Fig. 6 B, right). In addition, this mutation results in significant decrease in RUNX binding to Dδ2-23RSS (Fig. 6 C). Importantly no enrichment of RAG1 binding to Dδ2-23RSS was observed, indicating that mutation of RUNX1 binding site abolished RAG1 recruitment onto the Dδ2-23RSS (Fig. 6 A). Collectively, these data strongly suggest that during early thymopoiesis, RUNX1 and RAG1 functionally interact to induce RAG1 loading onto the Dδ2-23RSS.
Figure 6.
RAG1 deposition onto Dδ2-23RSS. (A) ChIP assays performed with anti-FLAG antibody (or control IgG antibody) and 293T cells transfected with the indicated recombination substrates and expression vectors. Enrichment at substrate vectors is shown relative to input substrate vectors DNA and normalized to GFP control. Error bars represent standard deviations from means. Presented data are from three independent experiments. (B) Jκ1-VκL8 recombination substrate containing RSS sequences of Jκ1 and VκL8 (Franchini et al., 2009) was mutated to harbor the ACG coding sequence found in the mutated version of Dδ2 gene segment. Left panel shows in vitro RAG cleavage assay of Jκ1-VκL8 and ACGJκ1-VκL8. The amounts of RAG1/2 crude extract used for each in vitro RAG cleavage assay (around 30 µg) were loaded on SDS-PAGE, and then Runx1 protein expression was analyzed by Western blot (right). Shown is a representative of two experiments. (C) ChIP-QPCR assays performed with anti-RUNX1 antibody (or control IgG antibody) and 293T cells transfected with the indicated recombination substrates and expression vectors. Enrichment at substrate vectors is shown relative to input substrate vectors DNA. Error bars represent standard deviations from means. IgG isotype control was performed to assess absence of nonspecific ChIP enrichment. Shown is a representative of two experiments.
RUNX1 loss of function leads to absence of human Dδ2-Dδ3 rearrangements
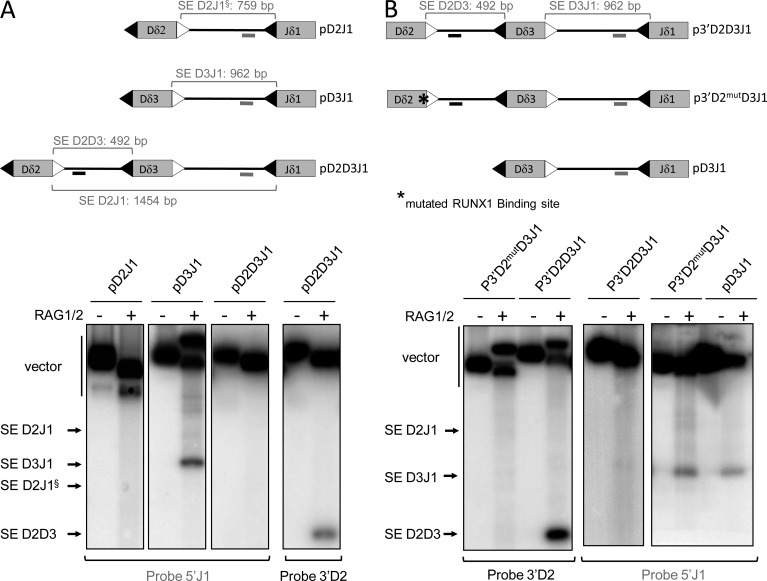
We showed that, in vivo, the assembly of Dδ2 and Jδ1 gene segments occurs in a two-step process systematically including the Dδ3 gene segment. We hypothesized that a B12/23 restriction prohibits direct Dδ2-Jδ1 rearrangement. To explore such a possibility, we performed in vitro RAG1/2-mediated DNA-coupled cleavage assays which recapitulate the first step of V(D)J recombination reaction (i.e., DSB formation) and hence result in the release of signal ends (SEs) and coding ends (CEs). Using pD3J1 or pD2J1 substrates, we observed the release of Dδ3-Jδ1 SE, but no Dδ2-Jδ1 SE, respectively (Fig. 7 A). With the pD2D3J1 substrate, we detected only the presence of Dδ2-Dδ3 SE, indicating that even if Dδ3 to Jδ1 rearrangement is possible, it does not proceed before Dδ2 to Dδ3 rearrangement (Fig. 7 A). Thus, the ordered assembly observed in vivo can be recapitulated in vitro with chromatin-free substrates.
Figure 7.
Inhibition of RUNX1 binding disrupts the order of TCR-δ rearrangement. (A and B) In vitro RAG1/2-mediated DNA-coupled cleavage assays. Top: schematic representation of plasmid substrates. 12 and 23 RSS are represented by black and white triangles, respectively. Positions of radiolabeled probes are indicated by gray (5′J1 probe) and black (3′D2 probe) lines. Sizes of SE fragments are indicated. Bottom: autoradiographs of Southern blot analyses of RAG1/2-mediated DNA coupled cleavage assays are representative of three independent experiments. Blots were hybridized with the probe specified below autoradiographs. Bands corresponding to SE products are indicated. § highlights SE D2J1 from pD2J1 vector. In B, RUNX1 binding site mutation is highlighted by an asterisk.
Next, to functionally test the role of the RUNX1 binding site in Dδ2-Dδ3 rearrangement, we used the mutated p3′D2mutD3J1 recombination substrate (Fig. 7 B). In vitro coupled cleavage assays performed with p3′D2mutD3J1, and as controls, p3′D2D3J1 and pD3J1, showed that disruption of the RUNX1 binding site prevents Dδ2-Dδ3 SE formation but makes possible the production of Dδ3-Jδ1 SE (Fig. 7 B). Therefore, the loss of RUNX1 binding onto Dδ2-23RSS disrupts the cleavage order observed with the pD2D3J1 substrate.
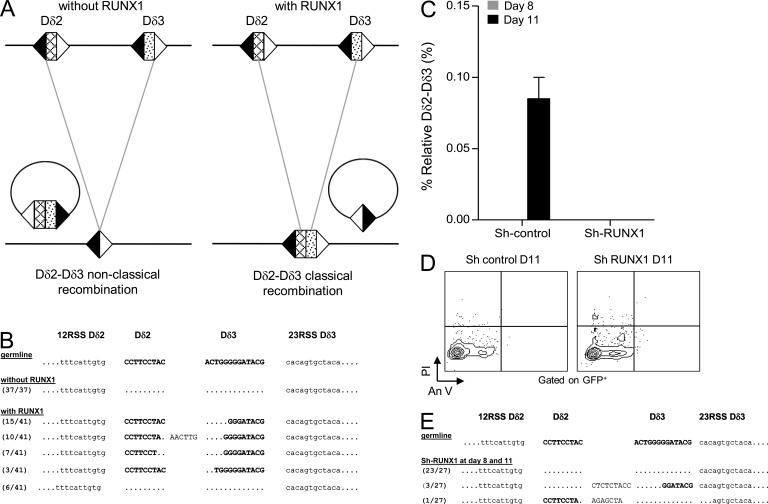
To establish the role of RUNX1 in endogenous TCR-δ locus rearrangements, we first took advantage of the BOSC23 cell line assay in which TCR-δ rearrangements can be induced (Langerak et al., 2001). As previously reported, when BOSC23 cells were transfected with RAG1/RAG2/E47-expressing vectors, we observed a nonclassical rearrangement between the upstream Dδ2-12RSS and the downstream Dδ3-23RSS, giving rise to Dδ2-Dδ3 SJ and elimination of the Dδ2-Dδ3 CJ as an episomal circle (Fig. 8, A [left] and B). However, when BOSC23 cells were cotransduced with the RUNX1-expressing vector, normal Dδ2-Dδ3 rearrangements occurred (Fig. 8, A [right] and B), indicating that RUNX1 is crucial for accurate TCR-δ locus rearrangement. To confirm this, RUNX1 was knocked down in UCB CD34+ cells which were then co-cultured on OP9-DL1 to promote T cell differentiation. Cells were collected at days 8 and 11 for quantification of Dδ2-Dδ3 rearrangements. By day 11, Dδ2-Dδ3 rearrangements were detected in sh-control-GFP conditions, whereas they were undetectable in RUNX1 knocked down cells (Fig. 8 C). Viability of sh-control-GFP and sh-RUNX1-GFP cells was monitored by Annexin-V/IP staining, which confirmed the absence of significant apoptosis in both conditions (Fig. 8 D). The absence of RUNX1 in the co-cultured cells led to formation of nonclassical rearrangements, where the coding segments Dδ2 and Dδ3 are deleted from the locus (Fig. 8 E). In ex vivo assays performed with nonlymphoid HEK293 cells (devoid of Runx1 protein), similar nonclassical rearrangements were observed; interestingly, in their discussion the authors anticipated our conclusion that a cofactor may dictate RAG binding to RSS (Olaru et al., 2005). Of note, the Dδ3-Jδ1 rearrangements observed above in chromatin-free substrate experiments were observed neither in RUNX1 knocked down CD34+ UBC co-cultured on OP9-DL1 cells nor in the BOSC23 cell line assay without RUNX1 (unpublished data). This difference between in vivo and in vitro experiments remains unexplained and requires further investigations.
Figure 8.
RUNX1 is necessary for proper initiation of early T cell rearrangements. (A and B) Products of TCR-δ gene rearrangements in the BOSC23 non lymphoid human cell line. (A) Schematic representation of TCR-δ rearrangements when BOSC23 cells are transduced with RAG1-, RAG2-, and E47-expressing vectors (left) or with RAG1-, RAG2-, and RUNX1-expressing vectors (right). 12 and 23 RSS are represented by black and white triangles, respectively. (B) Sequences of detected Dδ2-Dδ3 rearrangements representative of two independent experiments. (C) RQ-PCR quantification of Dδ2-Dδ3 rearrangements in CD34+ UCB cells transduced with control sh-control-GFP and sh-RUNX1-GFP and cultured onto OP9-DL1 during 8 and 11 d (gray and black bars, respectively). Results (mean and SEM representative of three independent experiments) are normalized to the Albumin gene. (D) Flow cytometric analysis of CD34+ UCB cells after transduction with lentiviruses encoding the GFP protein and control or RUNX1-specific shRNAs. GFP+-transduced cells were gated for analysis of Annexin V/PI staining. Flow cytometry plots representative of three independent experiments. (E) Detection of the nonclassical Dδ2-Dδ3 rearrangements in the experiment of Runx1 knockdown in UCB CD34+ cells. Sequences are representative of three independent experiments.
Collectively, these results demonstrate that human TCR-δ rearrangements occur through an ordered two-step process which is controlled by beyond chromatin accessibility mechanisms. First, a B12/23 restriction impedes direct Dδ2 to Jδ1 rearrangements and thus insures Dδ3 gene segment utilization. Second, ordered rearrangements (Dδ2 to Dδ3 precedes Dδ3 to Jδ1) require the RUNX1 TF for RAG1 deposition on Dδ2-23RSS.
DISCUSSION
Among antigen receptor loci, only TCR-β, Ig heavy chain (IgH), and TCR-δ contain D segments. The TCR-δ and TCR-β loci share particularly similar structural features and harbor the same RSS distribution. The regulation of TCR-β and IgH, which both show allelic exclusion, has long been recognized to be ordered (Khor and Sleckman, 2002; Jung et al., 2006) both in mouse and humans. In contrast, the TCR-δ locus was considered to not be ordered. The vast majority of these studies, however, focused on mouse thymic maturation, and little data were available for early human T cell maturation (Chiei et al., 1987; Krangel et al., 2004). One study suggested that the earliest human TCR-δ rearrangements could be ordered, based on the identification of predominant Dδ2-Dδ3 rearrangements within a very immature (CD34+/CD1a−) thymic subset (Dik et al., 2005). We now demonstrate that Dδ2-Dδ3 rearrangements do indeed occur before Dδ2-Jδ1 rearrangements in a specific ETP subset of human thymocytes, highlighting differences in early mouse and human thymopoiesis (Blom et al., 1998). Importantly we show that this ordered TCR-δ rearrangement involves RUNX1, which binds to human Dδ2-23RSS (but not to the homologous mouse Dδ1-23RSS), recruits RAG1, and imposes Dδ2 to Dδ3 rearrangement before Dδ3 to Jδ1 rearrangement. A similar scenario was described for the ordering of TCR-β gene assembly by the c-Fos TF on Dβ 23RSS (Wang et al., 2008). Collectively, these data indicate that TF-mediated RAG1 deposition onto a given RSS represents a recurrent mechanism of control of the start of V(D)J recombination in thymopoiesis. It has been previously shown that TCR-δ rearrangement evolves in an age-dependent manner; in contrast to postnatal thymocytes, in fetal thymus TCR-δ chains contain only one Dδ segment (Dδ3) and display almost no N-nucleotide incorporation (Krangel et al., 1990). It is tempting to speculate that Runx1 is involved in this developmental shift from early fetal to postnatal pattern of TCR-δ rearrangement, and thus that TF-dependent recruitment of the RAG complex to RSS may represent mechanism of developmental control of V(D)J recombination.
The minor TCR-γδ–expressing T cell (γδT cell) population has been conserved throughout vertebrate evolution, indicating a nonredundant function of those cells compared with TCR-αβ T cells. γδT cells contribute to immune responses by combining innate and adaptive features (Vantourout and Hayday, 2013). TCR-γδ ligands are not yet fully characterized but they clearly differ from antigens recognized by αβT cells which consist of processed peptides presented by the major histocompatibility complex. The D region encodes the third complementary determining region (CDR3) and tandem use of two D gene segments is specific to TCR-δ chains. A consequence of our data are that, in human TCR-δ locus V(D)J recombination, RUNX1 imposes the use of two Dδ gene segments in all rearranged TCR-δ chains, suggesting an important functional role of the length (and diversity) of the TCR-δ CDR3. Importantly, the average mouse TCR-δ CDR3 is shorter than that observed in humans (Rock et al., 1994), suggesting an evolutionary advantage acquired (and imposed by the RUNX1 Dδ2-23RSS site) in humans compared with mice. These observations merit functional investigation to establish the role of this mechanism in human and mouse TCR-γδ immune responses.
RUNX1, RUNX2, and RUNX3 are members of the RUNX family of TF; they share a conserved Runt domain, which mediates DNA binding and heterodimerization with the CBF-β protein. RUNX1 is the predominantly expressed RUNX factor in the hematopoietic system, where it is essential for definitive hematopoiesis during embryogenesis. Runx1 gene inactivation in mice impedes the emergence of the first hematopoietic stem cells (HSCs) from the aorto-gonadal-mesonephros region. In adult mice, inactivation of Runx1 impairs lymphoid and megakaryocyte lineage maturation (Ichikawa et al., 2004; Growney et al., 2005) and leads to HSC exhaustion, although this is still under debate (Jacob et al., 2010; Cai et al., 2011). Deletion of Runx1 at either DN or DP stages of T cell differentiation using Lck-cre or CD4-cre transgenic mice, respectively, showed that Runx1 is required for DN3 to DN4 and DP to single positive (SP) CD4+ transitions (Taniuchi et al., 2002; Egawa et al., 2007). RUNX1 binding sites are present in human and murine TCR-α and TCR-β enhancers, which are essential for TCR-α and TCR-β expression (Sleckman et al., 1997; Tripathi et al., 2002). Herein, we identify a new function for RUNX1 during the early stages of human thymic development whereby it acts as a RAG1 cofactor for the start of TCR-δ rearrangement. A recent study described a role for Runx1 in early B lymphocyte development, whereby Runx1 activates the expression of the TF Ebf1 that is required for VH to DH-JH rearrangement (Seo et al., 2012). The mechanistic role of Runx1 in IgH locus assembly seems, however, to be quite distinct from the direct TCR-δ RSS-binding and RAG deposition described here.
This role for RUNX1 in “RAG1 deposition” on Dδ2-23RSS is likely to be particularly important at the early stages of T cell development, when RAG1 expression is low. Consistent with the “nonamer first” model (Schatz and Ji, 2011), we suggest that RAG1 interacts primarily with the nonamer, whereas RUNX1 binds to the coding/heptamer junction; the Dδ2-23RSS may, therefore, be able to accommodate both proteins, which could even cooperate in DNA binding, at least initially. Cooperative binding has been evidenced for RUNX1 and ETS1, whose interaction increases their affinity for their juxtaposed DNA binding sites, notably in TCR regulatory elements (Kim et al., 1999). Such cooperative interactions had up till now not been identified in the context of early TCR-δ rearrangements.
The probability that a coding/heptamer junction harbors a RUNX1 binding site, d(A1C2C3A4C5A6), is not negligible because most of the RSS heptamers start with d(C3A4C5A6) and all of them possess at least the d(C3A4C5) sequence, which is absolutely required for RAG cleavage. Despite this, we found only one RSS carrying a RUNX1 binding site among the human TCR-δ Dδ and Jδ segments. An in-depth analysis of RUNX1-RSS sequences in Ig/TCR V, D, and J segments will further clarify the potentially variable roles of RUNX1 in human and mouse lymphoid development. Our data are compatible with different roles for RUNX1 in the initiation of T lymphopoiesis in mice and men, which if confirmed, has profound impact for the extrapolation of mouse models to human T lymphopoiesis.
The human genome is peppered with RUNX1 binding sites and RUNX1 is involved in the regulation of various genes during hematopoietic differentiation (Wong et al., 2011). The downside of RAG1–RUNX1 interaction is that RUNX1 might mis-target RAG1 and induce genomic instability by creating illegitimate DNA nicks or double-strand breaks at nonantigen receptor loci. Chromosomal rearrangements involving TCR loci are frequent in T-ALL and are not purely RAG mediated. We have recently shown that the vast majority of TCR-δ translocations occur during TCR-δ Dδ2-Dδ3, potentially suggesting a role for RUNX1 in their pathogenesis (Dadi et al., 2012: Le Noir et al., 2012). The RUNX1 gene is one of the most frequently mutated genes in human leukemia; RUNX1 loss-of-function or dominant-negative fusion proteins result in leukemia-prone cells which become fully leukemic upon acquisition of additional hits (Speck and Gilliland, 2002). More specifically, RUNX1 loss-of-function mutations occur in ∼25% of the most immature subset of acute myeloid leukemia (AML-M0; Preudhomme et al., 2000). Based on the data presented here, it is tempting to speculate that the maturation arrest in these rare AML may be, at least in part, related to a failure to initiate TCR-δ rearrangement.
MATERIALS AND METHODS
Purification of thymocyte fraction.
Thymii were obtained as surgical tissue discards from children, with informed consent from the parents and the ethical review board of Necker Enfants Malades Hospital at Paris Descartes. Thymocytes were prepurified by magnetic-activated cell-sorted beads before sorting (FACSAria III; BD). Purity after sort was >95%.
Plasmids.
The plasmids pD3J1, pD2D3J1, and p3′D2D3J1 were generated by PCR using placental DNA and cloned into p-GEMT-easy vector. The plasmid substrate p3′D2mutD3J1 was derived from p3′D2D3J1 using site-directed mutagenesis (Agilent Technologies). The plasmid pD2J1 was generated from pD2D3J1.
Vector pCMV5-RUNX1 was bought from Addgene (plasmid 12426). cDNA for CBF-β was cloned into pEGFPC1. RAG1-Flag-SBP and pHβAPneo-E47 were gifts from D. Payet-Bornet and A.W. Langerak, respectively.
Antibodies used.
For ChIP, immunoprecipitation, and WB, anti-RUNX1 (Abcam), RAG1 (D36D3; Cell Signaling Technology), anti-FLAG (Sigma-Aldrich), anti–CBF-β (Abcam), and normal rabbit IgG (Santa Cruz Biotechnology, Inc.) were used. For immunofluorescence, RAG1 (D36D3) and RUNX1 (Abcam) were used. For flow cytometry, CD1a-FITC (NA1/34), CD13-FITC (WM47; Dako); CD3–Alexa Fluor 700 (UCHT1), CD4-V450 (RPA-T4), CD5-PerCP-Cy5.5 (L17F12), CD7-PE (M-T701), CD8-APC (RPA-T8), CD34-PerCP-Cy5.5 (8G12), CD45-V500 (H130), HLA-DR-FITC (L243), CD34-APC (8G12; BD); CD117-PECy7 (104D2D1; Beckman Coulter); and CD123-APC (AC145; Miltenyi Biotec) were used.
TCR rearrangement and TREC quantification.
TCR-δ quantification (Dδ2-Dδ3, Dδ2-Jδ1, and Dδ3-Jδ1) was performed as previously described (Dik et al., 2005) with the listed sets of primers and probes. The following were used for Dδ2-Dδ3 rearrangements: Dδ2, 5′-CAAGGAAAGGGAAAAAGGAAGAA-3′; Dδ3, 5′-TTGCCCCTGCAGTTTTTGTAC-3′; and Dδ3 probe, 5′-ATACGCACAGTGCTACAAAACCTACAGAGACCT-3′. The following were used for Dδ2-Jδ1 rearrangements: Dδ2, 5′-AGCGGGTGGTGATGGCAAAGT-3′; Jδ1, 5′-TTAGATGGAGGATGCCTTAACCTTA-3′; and Jδ1 probe, 5′-CCCGTGTGACTGTGGAACCAAGTAAGTAACTC-3′. The following were used for Dδ3-Jδ1 rearrangements: Dδ3, 5′-GACTTGGAGAAAACATCTGGTTCTG-3′; and Jδ1 and the Jδ1 probe (listed above). TREC quantification was performed using an RQ-PCR mixture of 25 µl containing SYBR green (Applied Biosystems), 800 nM of each primer, 0.4 µg bovine serum albumin, and 100 ng genomic DNA. Normalization was performed with the Albumin gene for all targets.
The analysis of rearrangements by multiplex fluorescent PCR was performed by separation of fluorochrome-labeled single stand (denaturated) PCR products in a capillary sequencing polymer and detected via automated laser scanning. The results as a Gaussian distribution of multiple peaks represent many different PCR products in case of polyclonal rearrangements.
BOSC23 cells were transfected with appropriate plasmids using lipofectamine according to manufacturer’s conditions. 3 d after transfection, the cells were harvested and DNA was extracted. To verify the presence of rearrangements, PCR was performed as previously described (Langerak et al., 2001) with the following primers: Dδ25’ forward, 5′-AGCGGGTGGTGATGGCAAAGT-3′; and Dδ33’ reverse, 5′-TGGGACCCAGGGTGAGGATAT-3′. PCR product was cloned into p-GEMT-Easy vector and sequenced.
RAG1/2-mediated DNA cleavage in vitro assays.
RAG1/2-mediated coupled cleavage was performed as previously described (Franchini et al., 2009). Core RAG1 and RAG2 proteins were overexpressed in D10 cells (derived from B lymphoma M12 cell line; Leu and Schatz, 1995), and for the assay, RAG1/2-enriched crude extract was complemented with thymic protein extract. In brief, 1 µg plasmid was incubated with RAG1/2 for 2 h at 30°C in cleavage reaction buffer (50 mM HEPES-KOH, pH 7.5, 73 mM KCl, 2 mM NaCl, 10 mM MgCl2, and 1 mM DTT) supplemented with 1.5 mM rATP and 6 mg of a nuclear extract prepared from mouse WT thymocytes. Negative controls were performed using similar conditions without addition of the RAG1/2 extract. After extraction and precipitation, the DNA samples were analyzed by Southern blot using a Hybond N transfer membrane (GE Healthcare). Membranes were hybridized with TCR-δ–specific, radiolabeled probes 3′D2 (5′-TTGCTGGAGCTTGAC-3′) or 5′J1 (5′-GGGTAAGCAACAAGTGCC-3′).
ChIP.
ChIP assays were performed, with modifications, according to the manufacturer’s instructions (Agilent Technologies). In brief, thymocytes were cross-linked for 20 min with 1% formaldehyde and sonicated using an Ultrasonics sonicator (Vibra-cell VCX130; SONICS) to obtain a mean length for DNA fragments of ∼600 bp. After immunoprecipitation with anti-RUNX1 antibody, ChIP DNA was purified by phenol/chloroform extraction and a QiaQuick PCR Purification kit (QIAGEN). Samples were analyzed by RQ-PCR with the following human primers: Dδ2-23RSS forward, 5′-AGCGGGTGGTGATGGCAAAGT-3′; Dδ2-23RSS probe, 5′-AGAAGAGGGTTTTTATACTGATG-3′; Dδ2-23RSS reverse, 5′-AGACATACATAGCGGGTCAC-3′; Dδ3-12RSS forward, 5′-CTAACTGTCAGGACCCTTTGATCTT-3′; Dδ3-12RSS probe, 5′-ATACGCACAGTGCTACAAAACCTACAGAGACCT-3′; Dδ3-12RSS reverse, 5′-TTGCCCCTGCAGTTTTTGTAC-3′; Eα forward, 5′-TTCCATGACGTCATGGTTACC-3′; Eα reverse, 5′-GCGATGCTATCTCTAACTCAG-3′; Actin forward, 5′-CTCCCATTGTCTACCTCAGTTTC-3′; and Actin reverse, 5′-CTTATGTGCTGAGAAGGTGGTG-3′. Mouse thymocytes were purified from 4-wk-old Rag2−/− mice (Shinkai et al., 1992) bred on a C57BL/6J background. ChIP against Runx1 (Abcam) was performed as previously described (Koch et al., 2011). The ChIP sample was analyzed by RQ-PCR with the following mouse primers: negative control forward, 5′-CCCCTTTCTGAAGCACTCTG-3′; negative control reverse, 5′-TAAGGCGTCATTTCCCAAAG-3′; Eα forward, 5′-TGCTGACATGGGCAAACAGGTC-3′; Eα reverse, 5′-ACTCCTCTTTCCAGAGGATGTGGC-3′; RSS-Dδ1forward, 5′-TGGGTATGGCAGAGGGTGGT-3′; RSS-Dδ1 reverse, 5′-TGCCATCACAGTGAAACACAGCCG-3′; RSS-Dδ2 forward, 5′-TGTAGCACCGTGATCGGAGGGA-3′; and RSS-Dδ2 reverse, 5′-AGGCCTGGGAGACGGTTCTT-3′. For ChIP analysis of RAG1 binding, 293T cells were transfected with appropriate plasmids using lipofectamine (Invitrogen). 30 h after transfection, cells were cross-linked with 1% formaldehyde for 10 min then lysed and sonicated. Finally, immunoprecipitation of RAG1-Flag-SBP was performed using anti-FLAG antibody (Sigma-Aldrich). For analysis of ChIP samples by RQ-PCR, the forward primer complementary to T7 promoter of p-GEMT-easy vector T7 forward (5′-TAATACGACTCACTATAGG-3′) and the Dδ2 reverse primer (5′-AACACATCAGTATAAAAACCC-3′) were used.
Immunofluorescence.
Cells were fixed on slides using poly-L-lysine, and Duolink (Duolink II; Olink Biosciences) assay was performed according to the manufacturer’s instructions. In brief, cells were fixed with formaldehyde 2% for 15 min, permeabilized with Triton X-100 1% for 10 min, and incubated with RUNX1 (1/1,000) and RAG1 (1/25) antibodies for 1 h at room temperature. Images were collected on a confocal microscope (LSM 700; Carl Zeiss) with Zen 2011 software using 63× objectives at room temperature. Images were processed using ImageJ (National Institutes of Health).
Cell lines and T cell differentiation.
The MOLT-4 cell line was cultured in RPMI-1640 medium supplemented with 50 µg/ml streptomycin, 50 IU penicillin, and 20% FBS at 37°C in a humidified atmosphere of 5% CO2. BOSC23 and 293T cell lines were maintained in Dulbecco’s modified Eagle medium with 10% FBS and antibiotics.
T cell differentiation was performed as previously described (Six et al., 2011). In brief, CD34+ UCB cells were sorted and cultured on confluent OP9-DL1 in home made α-MEM medium (Invitrogen) supplemented with 20% FBS (Hyclone; Thermo Fisher Scientific) and cytokines (5 ng/ml rhFLT3-L, 10 ng/ml rhSCF, and 2 ng/ml rhIL7; Miltenyi Biotec). DNA was extracted at different days of culture. Cord blood samples, harvested with informed consent, were obtained from Saint-Louis Hospital Cord Blood Bank, which is authorized by the French Regulation Agency (reference TCG/12/R/004).
Immunoprecipitation.
Protein extracts from 2 × 108 MOLT-4 cells were prepared with RIPA buffer (50 mM Tris-HCl, pH 7.6, 1% NP 40, 150 mM NaCl, 0.1% SDS20, and 1× inhibitory protease cocktail [complete EDTA free; Roche]) and were incubated for 2 h at 4°C with 10 µg anti-RUNX1 covalently linked on protein G agarose beads (EMD Millipore). After 4 washes in 100 mM NaCl and 15 mM Tris-HCl, pH 7.8, the bound proteins were eluted and detected by Western blot analysis using anti-RAG1 antibody.
293T cells were cotransfected with expression vectors for RUNX1, CBF-β, RAG1-Flag-SBP, or as control the empty Flag-SBP vector. After 24 h of incubation, 4 × 107 293T cells were lysed with RIPA buffer and protein extracts were incubated for 2 h at 4°C with 50 µl magnetic beads (Dynabeads M-208 Streptavidin; Invitrogen). After streptavidin precipitation, bound proteins were detected by Western blot analysis using anti-FLAG, anti-RUNX1, and anti–CBF-β antibodies.
Lentiviral infection and nucleofection.
Mission shRNA-RUNX1 (TRCN0000013660) was purchased from Sigma-Aldrich. MISSION shRNA-pLKO.1-puro-GFP control transduction particles were used as a negative control. CD34+ UCB cells were infected, after 5 d culture in OP9-DL1, in cellgro medium (CellGenix) supplemented with 100 ng/ml FLT3-L and 100 ng/ml SCF cytokines. 48 h later, GFP+ cells were sorted and cultured on OP9-DL1 with cytokines.
MOLT4 cells were nucleofected with siRNA against RAG1 (SASI_Hs01_00024301; Sigma-Aldrich). Nucleofection was performed twice in an interval of 12 h with Amaxa Cell Line Nucleofector kit L (Lonza) according to manufacturer’s instructions.
Online supplemental material.
Fig. S1 shows gating strategy for cell sorting. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20132585/DC1.
Supplementary Material
Acknowledgments
We thank the Institute for Research at Necker-Enfants Malades (IRNEM) Cell Imaging platform.
S. Le Noir was supported by grants from the “ARC Association de Recherche contre le Cancer” and the “Fondation pour la recherche médicale (FRM)”. This work was supported in the Necker team by grants from the “Fondation de France/Comité Leucémie,” the “ARC Association de Recherche contre le Cancer,” the Association Laurette Fugain, the Institut Nationale de Cancer (INCa) translational research program (CARAMELE), and the Kay Kendal Leukemia Research Fund. Work in the B. Nadel laboratory is supported by institutional grants from INSERM and CNRS, and grants from INCa and the “Fondation de France.”
The authors declare no competing financial interests.
Author contributions: S. Le Noir, A. Cieslak, and D. Payet-Bornet performed cellular and molecular biology experiments, analyzed data, and wrote the manuscript; L. Lhermitte, A. Trinquand, J. Bond, and C. Reimann performed cytometry analysis. P. Villarese and A. Cieslak performed fluorescent PCR; E. Verhoeyen provided lentiviral shRNAs; E. Macintyre, B. Nadel, I. André-Schmutz, and E. Six analyzed data. All authors validated the manuscript. D. Payet-Bornet and V. Asnafi oversaw conceptual development of the project.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DN
- double negative
- DP
- double positive
- DSB
- double-strand break
- ETP
- early T cell precursor
- PLA
- proximity ligation assay
- RSS
- recombination signal sequence
- SE
- signal end
- SP
- single positive
- TF
- transcription factor
- TREC
- T cell receptor excision circle
- UCB
- umbilical cord blood
References
- Bassing C.H., Alt F.W., Hughes M.M., D’Auteuil M., Wehrly T.D., Woodman B.B., Gärtner F., White J.M., Davidson L., and Sleckman B.P.. 2000. Recombination signal sequences restrict chromosomal V(D)J recombination beyond the 12/23 rule. Nature. 405:583–586 10.1038/35014635 [DOI] [PubMed] [Google Scholar]
- Bassing C.H., Tillman R.E., Woodman B.B., Canty D., Monroe R.J., Sleckman B.P., and Alt F.W.. 2003. T cell receptor (TCR) α/Δ locus enhancer identity and position are critical for the assembly of TCR Δ and α variable region genes. Proc. Natl. Acad. Sci. USA. 100:2598–2603 10.1073/pnas.0437943100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blom B., Res P.C., and Spits H.. 1998. T cell precursors in man and mice. Crit. Rev. Immunol. 18:371–388 10.1615/CritRevImmunol.v18.i4.50 [DOI] [PubMed] [Google Scholar]
- Cai X., Gaudet J.J., Mangan J.K., Chen M.J., De Obaldia M.E., Oo Z., Ernst P., and Speck N.A.. 2011. Runx1 loss minimally impacts long-term hematopoietic stem cells. PLoS ONE. 6:e28430 10.1371/journal.pone.0028430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiei Y.H., Iwashima M., Wettstein D.A., Kaplan K.B., Elliott J.F., Born W., and Davis M.M.. 1987. T-cell receptor Δ gene rearrangements in early thymocytes. Nature. 330:722–727 10.1038/330722a0 [DOI] [PubMed] [Google Scholar]
- Dadi S., Le Noir S., Payet-Bornet D., Lhermitte L., Zacarias-Cabeza J., Bergeron J., Villarèse P., Vachez E., Dik W.A., Millien C., et al. . 2012. TLX homeodomain oncogenes mediate T cell maturation arrest in T-ALL via interaction with ETS1 and suppression of TCRα gene expression. Cancer Cell. 21:563–576 10.1016/j.ccr.2012.02.013 [DOI] [PubMed] [Google Scholar]
- Dik W.A., Pike-Overzet K., Weerkamp F., de Ridder D., de Haas E.F., Baert M.R., van der Spek P., Koster E.E., Reinders M.J., van Dongen J.J., et al. . 2005. New insights on human T cell development by quantitative T cell receptor gene rearrangement studies and gene expression profiling. J. Exp. Med. 201:1715–1723 10.1084/jem.20042524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q.M., Leu T.M., and Schatz D.G.. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 380:85–88 10.1038/380085a0 [DOI] [PubMed] [Google Scholar]
- Egawa T., Tillman R.E., Naoe Y., Taniuchi I., and Littman D.R.. 2007. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204:1945–1957 10.1084/jem.20070133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezekiel U.R., Sun T., Bozek G., and Storb U.. 1997. The composition of coding joints formed in V(D)J recombination is strongly affected by the nucleotide sequence of the coding ends and their relationship to the recombination signal sequences. Mol. Cell. Biol. 17:4191–4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchini D.M., Benoukraf T., Jaeger S., Ferrier P., and Payet-Bornet D.. 2009. Initiation of V(D)J recombination by Dβ-associated recombination signal sequences: a critical control point in TCRβ gene assembly. PLoS ONE. 4:e4575 10.1371/journal.pone.0004575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Growney J.D., Shigematsu H., Li Z., Lee B.H., Adelsperger J., Rowan R., Curley D.P., Kutok J.L., Akashi K., Williams I.R., et al. . 2005. Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood. 106:494–504 10.1182/blood-2004-08-3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesslein D.G., and Schatz D.G.. 2001. Factors and forces controlling V(D)J recombination. Adv. Immunol. 78:169–232 10.1016/S0065-2776(01)78004-2 [DOI] [PubMed] [Google Scholar]
- Ichikawa M., Asai T., Saito T., Seo S., Yamazaki I., Yamagata T., Mitani K., Chiba S., Ogawa S., Kurokawa M., and Hirai H.. 2004. AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat. Med. 10:299–304 10.1038/nm997 [DOI] [PubMed] [Google Scholar]
- Jacob B., Osato M., Yamashita N., Wang C.Q., Taniuchi I., Littman D.R., Asou N., and Ito Y.. 2010. Stem cell exhaustion due to Runx1 deficiency is prevented by Evi5 activation in leukemogenesis. Blood. 115:1610–1620 10.1182/blood-2009-07-232249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., Bassing C.H., Fugmann S.D., Cheng H.L., Schatz D.G., and Alt F.W.. 2003. Extrachromosomal recombination substrates recapitulate beyond 12/23 restricted VDJ recombination in nonlymphoid cells. Immunity. 18:65–74 10.1016/S1074-7613(02)00507-1 [DOI] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., and Alt F.W.. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570 10.1146/annurev.immunol.23.021704.115830 [DOI] [PubMed] [Google Scholar]
- Khor B., and Sleckman B.P.. 2002. Allelic exclusion at the TCRβ locus. Curr. Opin. Immunol. 14:230–234 10.1016/S0952-7915(02)00326-6 [DOI] [PubMed] [Google Scholar]
- Kim W.Y., Sieweke M., Ogawa E., Wee H.J., Englmeier U., Graf T., and Ito Y.. 1999. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 18:1609–1620 10.1093/emboj/18.6.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F., Fenouil R., Gut M., Cauchy P., Albert T.K., Zacarias-Cabeza J., Spicuglia S., de la Chapelle A.L., Heidemann M., Hintermair C., et al. . 2011. Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat. Struct. Mol. Biol. 18:956–963 10.1038/nsmb.2085 [DOI] [PubMed] [Google Scholar]
- Krangel M.S.2003. Gene segment selection in V(D)J recombination: accessibility and beyond. Nat. Immunol. 4:624–630 10.1038/ni0703-624 [DOI] [PubMed] [Google Scholar]
- Krangel M.S., Yssel H., Brocklehurst C., and Spits H.. 1990. A distinct wave of human T cell receptor γ/Δ lymphocytes in the early fetal thymus: evidence for controlled gene rearrangement and cytokine production. J. Exp. Med. 172:847–859 10.1084/jem.172.3.847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krangel M.S., Carabana J., Abbarategui I., Schlimgen R., and Hawwari A.. 2004. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor α/Δ locus. Immunol. Rev. 200:224–232 10.1111/j.0105-2896.2004.00155.x [DOI] [PubMed] [Google Scholar]
- Langerak A.W., Wolvers-Tettero I.L., van Gastel-Mol E.J., Oud M.E., and van Dongen J.J.. 2001. Basic helix-loop-helix proteins E2A and HEB induce immature T-cell receptor rearrangements in nonlymphoid cells. Blood. 98:2456–2465 10.1182/blood.V98.8.2456 [DOI] [PubMed] [Google Scholar]
- Le Noir S., Ben Abdelali R., Lelorch M., Bergeron J., Sungalee S., Payet-Bornet D., Villarèse P., Petit A., Callens C., Lhermitte L., et al. . 2012. Extensive molecular mapping of TCRα/δ- and TCRβ-involved chromosomal translocations reveals distinct mechanisms of oncogene activation in T-ALL. Blood. 120:3298–3309 10.1182/blood-2012-04-425488 [DOI] [PubMed] [Google Scholar]
- Lefranc M.P.2001. Nomenclature of the human T cell receptor genes. Curr. Protoc. Immunol. 1(Appendix):1O. [DOI] [PubMed] [Google Scholar]
- Leu T.M., and Schatz D.G.. 1995. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol. Cell. Biol. 15:5657–5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaru A., Petrie H.T., and Livák F.. 2005. Beyond the 12/23 rule of VDJ recombination independent of the Rag proteins. J. Immunol. 174:6220–6226 10.4049/jimmunol.174.10.6220 [DOI] [PubMed] [Google Scholar]
- Preudhomme C., Warot-Loze D., Roumier C., Grardel-Duflos N., Garand R., Lai J.L., Dastugue N., Macintyre E., Denis C., Bauters F., et al. . 2000. High incidence of biallelic point mutations in the Runt domain of the AML1/PEBP2 α B gene in Mo acute myeloid leukemia and in myeloid malignancies with acquired trisomy 21. Blood. 96:2862–2869. [PubMed] [Google Scholar]
- Rock E.P., Sibbald P.R., Davis M.M., and Chien Y.H.. 1994. CDR3 length in antigen-specific immune receptors. J. Exp. Med. 179:323–328 10.1084/jem.179.1.323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz D.G., and Ji Y.. 2011. Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11:251–263 10.1038/nri2941 [DOI] [PubMed] [Google Scholar]
- Seo W., Ikawa T., Kawamoto H., and Taniuchi I.. 2012. Runx1-Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. J. Exp. Med. 209:1255–1262 10.1084/jem.20112745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai Y., Rathbun G., Lam K.P., Oltz E.M., Stewart V., Mendelsohn M., Charron J., Datta M., Young F., Stall A.M., et al. . 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 68:855–867 10.1016/0092-8674(92)90029-C [DOI] [PubMed] [Google Scholar]
- Six E.M., Benjelloun F., Garrigue A., Bonhomme D., Morillon E., Rouiller J., Cacavelli L., Blondeau J., Beldjord K., Hacein-Bey-Abina S., et al. . 2011. Cytokines and culture medium have a major impact on human in vitro T-cell differentiation. Blood Cells Mol. Dis. 47:72–78 10.1016/j.bcmd.2011.04.001 [DOI] [PubMed] [Google Scholar]
- Sleckman B.P., Bardon C.G., Ferrini R., Davidson L., and Alt F.W.. 1997. Function of the TCR α enhancer in αβ and γΔ T cells. Immunity. 7:505–515 10.1016/S1074-7613(00)80372-6 [DOI] [PubMed] [Google Scholar]
- Speck N.A., and Gilliland D.G.. 2002. Core-binding factors in haematopoiesis and leukaemia. Nat. Rev. Cancer. 2:502–513 10.1038/nrc840 [DOI] [PubMed] [Google Scholar]
- Spits H.2002. Development of αβ T cells in the human thymus. Nat. Rev. Immunol. 2:760–772 10.1038/nri913 [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., and Littman D.R.. 2002. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633 10.1016/S0092-8674(02)01111-X [DOI] [PubMed] [Google Scholar]
- Tillman R.E., Wooley A.L., Khor B., Wehrly T.D., Little C.A., and Sleckman B.P.. 2003. Cutting edge: targeting of Vβ to Dβ rearrangement by RSSs can be mediated by the V(D)J recombinase in the absence of additional lymphoid-specific factors. J. Immunol. 170:5–9 10.4049/jimmunol.170.1.5 [DOI] [PubMed] [Google Scholar]
- Tripathi R., Jackson A., and Krangel M.S.. 2002. A change in the structure of Vβ chromatin associated with TCR β allelic exclusion. J. Immunol. 168:2316–2324 10.4049/jimmunol.168.5.2316 [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Ramsden D.A., and Gellert M.. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 85:107–113. [DOI] [PubMed] [Google Scholar]
- Vantourout P., and Hayday A.. 2013. Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol. 13:88–100 10.1038/nri3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verschuren M.C., Wolvers-Tettero I.L., Breit T.M., Noordzij J., van Wering E.R., and van Dongen J.J.. 1997. Preferential rearrangements of the T cell receptor-Δ-deleting elements in human T cells. J. Immunol. 158:1208–1216. [PubMed] [Google Scholar]
- Wang X., Xiao G., Zhang Y., Wen X., Gao X., Okada S., and Liu X.. 2008. Regulation of Tcrb recombination ordering by c-Fos-dependent RAG deposition. Nat. Immunol. 9:794–801 10.1038/ni.1614 [DOI] [PubMed] [Google Scholar]
- Wong W.F., Kohu K., Chiba T., Sato T., and Satake M.. 2011. Interplay of transcription factors in T-cell differentiation and function: the role of Runx. Immunology. 132:157–164 10.1111/j.1365-2567.2010.03381.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancopoulos G.D., and Alt F.W.. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 40:271–281 10.1016/0092-8674(85)90141-2 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.