Nfil3 is critical for normal development of innate lymphoid cell (ILC) progenitors. Nfil3-deficient mice have severely reduced lung and visceral adipose tissue ILC2s and gut-associated ILC3s, and compromised innate immunity to acute bacterial infection.
Abstract
The bZIP transcription factor Nfil3 (also known as E4BP4) is required for the development of natural killer (NK) cells and type 1 innate lymphoid cells (ILC1s). We find that Nfil3 plays a critical role in the development of other mucosal tissue-associated innate lymphocytes. Type 3 ILCs (ILC3s), including lymphoid tissue inducer (LTi)–like cells, are severely diminished in both numbers and function in Nfil3-deficient mice. Using mixed bone marrow chimeric mice, we demonstrate that Nfil3 is critical for normal development of gut-associated ILC3s in a cell-intrinsic manner. Furthermore, Nfil3 deficiency severely compromises intestinal innate immune defense against acute bacterial infection with Citrobacter rodentium and Clostridium difficile. Nfil3 deficiency resulted in a loss of the recently identified ILC precursor, yet conditional ablation of Nfil3 in the NKp46+ ILC3 subset did not perturb ILC3 numbers, suggesting that Nfil3 is required early during ILC3 development but not for lineage maintenance. Lastly, a marked defect in type 2 ILCs (ILC2s) was also observed in the lungs and visceral adipose tissue of Nfil3-deficient mice, revealing a general requirement for Nfil3 in the development of all ILC lineages.
The discovery and characterization of the innate lymphoid cell (ILC) family in recent years has greatly contributed to our understanding of antimicrobial, autoimmune, and tissue-protective immune responses at barrier surfaces (Spits and Di Santo, 2011; Spits et al., 2013). Although the common gamma chain cytokine receptor and the cytokine IL-7, but not RAG proteins, are required for ILC development (Spits and Cupedo, 2012), and the early inflammatory cues that control the wide spectrum of ILC responses (types 1, 2, and 3) are rapidly being elucidated (Sonnenberg et al., 2013; Spits et al., 2013; Walker et al., 2013), the transcriptional regulation of ILC development is less clear. The transcription factor inhibitor of DNA binding 2 (Id2) is required for the development of a common lymphoid precursor thought to represent an NK cell/ILC precursor (ILCP; Yokota et al., 1999; Moro et al., 2010; Satoh-Takayama et al., 2010), although an ILCP distinct from the NK cell precursor has been recently described (Klose et al., 2014). Additional factors such as RORγt, aryl hydrocarbon receptor (Ahr), and T-bet (Tbx21) specify ILC3 development (Eberl et al., 2004; Satoh-Takayama et al., 2008; Veldhoen et al., 2008; Luci et al., 2009; Sanos et al., 2009; Kiss et al., 2011; Lee et al., 2012; Qiu et al., 2012; Sciumé et al., 2012; Klose et al., 2013; Rankin et al., 2013), and factors such as Rora, Gata3, Tcf7, and Gfi control ILC2 development (Yang et al., 2011, 2013; Halim et al., 2012; Hoyler et al., 2012; Liang et al., 2012; Mjösberg et al., 2012; Wong et al., 2012; Furusawa et al., 2013; Klein Wolterink et al., 2013; Mielke et al., 2013; Spooner et al., 2013). In many of these studies, genetic ablation of individual transcription factors resulted in a defect in ILC subset numbers and/or function, resulting in susceptibility to pathogen challenge at mucosal surfaces.
Nfil3 (also known as E4BP4) is a basic leucine zipper transcription factor that has been shown to control an extensive range of cellular processes in lymphocyte subsets, including the transcription of IL-3 in T cells (Zhang et al., 1995), survival and class-switching in B cells (Ikushima et al., 1997; Kashiwada et al., 2010), development and response of macrophages and dendritic cell subsets (Kashiwada et al., 2011b; Kobayashi et al., 2011), modulation of TH2 responses (Kashiwada et al., 2011a; Motomura et al., 2011), and regulation of TH17 responses via circadian clock (Yu et al., 2013). However, arguably the most striking phenotype in Nfil3-deficient mice is the near complete loss of NK cells and ILC1s at steady-state (Gascoyne et al., 2009; Kamizono et al., 2009; Kashiwada et al., 2010; Firth et al., 2013; Fuchs et al., 2013). Thus, we investigated whether Nfil3 may regulate the development or homeostasis of additional innate lymphocyte populations. Here, we used Nfil3-deficient mice to demonstrate a critical role for the transcription factor Nfil3 in the development of group 1, 2, and 3 ILCs and resistance against intestinal pathogen challenge.
RESULTS AND DISCUSSION
Intestinal group 3 ILCs are severely reduced in Nfil3-deficient mice
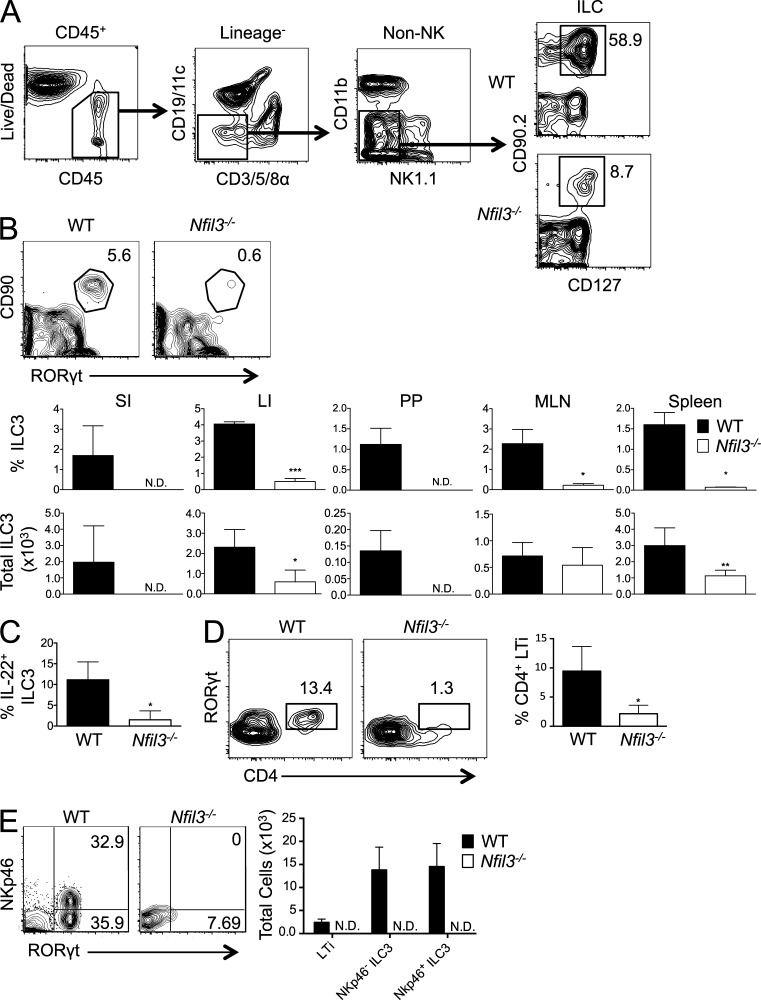
Consistent with previously reported findings (Gascoyne et al., 2009; Kamizono et al., 2009; Kashiwada et al., 2010; Firth et al., 2013; Fuchs et al., 2013), we found a dramatic deficiency in NK cells and group 1 ILCs (ILC1) in multiple tissues, including small intestine (SI), Peyer’s patches (PPs), lung, and spleen (unpublished data). Given that NK cells and ILC1s are found at extremely reduced frequency in Nfil3−/− mice at steady-state, we investigated whether Nfil3 was also required for development or homeostasis of other innate lymphocyte populations. Because ILCs (identified as lineage-negative cells that coexpress CD45, IL-7Rα [CD127], and Thy 1 [CD90]) are found in relatively high abundance at gut mucosal sites (Sonnenberg et al., 2013; Spits et al., 2013; Walker et al., 2013), we analyzed these innate lymphocytes (Fig. 1 A) in the lamina propria of SI and large intestine (LI), and in PPs of WT and Nfil3−/− mice. In contrast to WT mice, Nfil3−/− mice contained severely diminished ILC3 numbers in all intestinal sites examined (Fig. 1 B). The defect in ILC3 numbers in the gut was also observed in mesenteric LNs (MLNs) and spleen of Nfil3−/− mice (Fig. 1 B), suggesting that the defect was not due to an inability to properly home to mucosal sites. Furthermore, the few residual intestinal ILC3s identified phenotypically from Nfil3−/− mice were functionally impaired in their ability to produce IL-22 when stimulated ex vivo with IL-23 (Fig. 1 C). Within the RORγt+ ILC3 population, intestinal CD4-expressing lymphoid tissue inducer (LTi) cells from Nfil3−/− mice were also dramatically reduced compared with WT mice (Fig. 1 D), as were both NKp46− and NKp46+ ILC3s (Fig. 1 E), demonstrating the critical role of Nfil3 for the development of all type 3 ILCs.
Figure 1.
Nfil3 is required for intestinal ILC3 and LTi cell development. (A) Gating strategy for analysis of the total ILC population (CD90.2+ CD127+ cells within the CD45+ Lineage− population) is shown. (B) Flow cytometric plots show the percentage of RORγt+ ILC3s within the CD45+ Lineage− population in the PPs. Graphs show percentage and absolute number of ILC3s within the CD45+ Lineage− population for SI, LI, PP, MLNs, and spleen from WT and Nfil3−/− mice. (C) Graph shows the percentage of IL-22–producing cells within the MLN ILC3 population of WT and Nfil3−/− mice after IL-23 stimulation. (D) Percentages of intestinal CD4+ RORγt+ LTi cells within the total ILC population of WT and Nfil3−/− mice are shown. (E) Plots show the percentage of SI NKp46− and NKp46+ ILC3s, and graph shows absolute numbers of LTi, NKp46− ILC3, and NKp46+ ILC3. All data are representative of n = 3–5 mice per group, with error bars showing standard deviation, repeated in 2 (panel E) or 4 (panels B-D) independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
Cell-intrinsic requirement for Nfil3 in ILC3 development
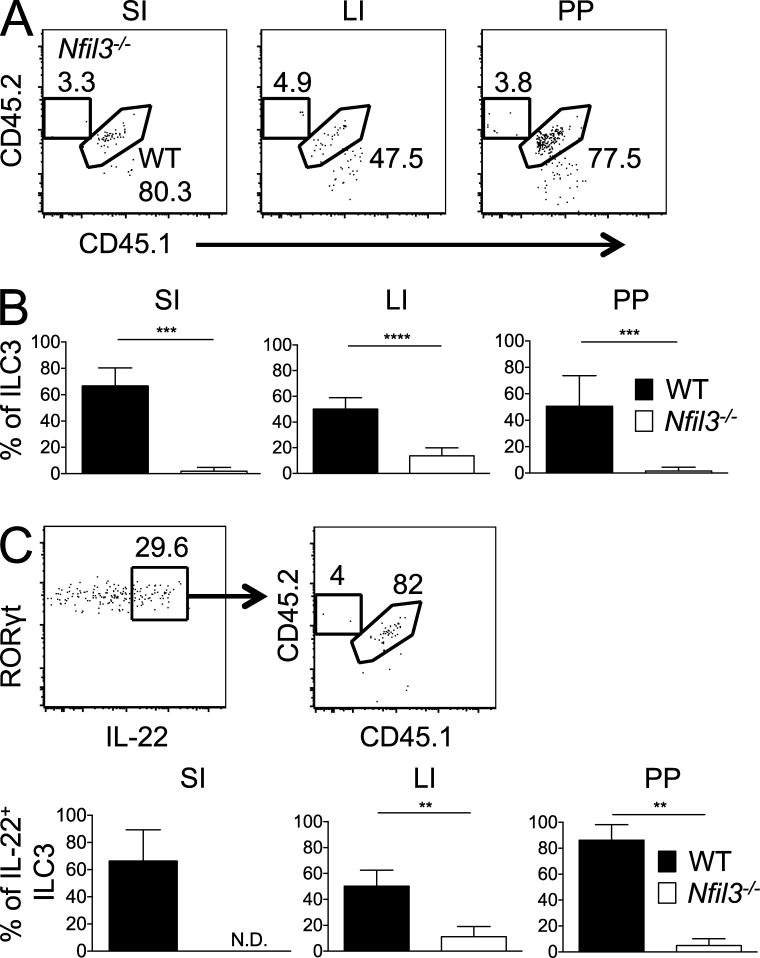
To rule out the possibility that ILC-extrinsic factors in Nfil3−/− mice may underlie the observed ILC3 defects, we generated mixed BM chimeric mice where lethally irradiated, congenically distinct recipient mice (CD45.1) received a 1:1 mixture of BM from WT (CD45.1 × 2) and Nfil3−/− (CD45.2) mice. We analyzed the mice 8–12 wk after BM transplantation (BMT), as we have previously observed development of donor ILC3s in recipient intestines at this time after BMT (Hanash et al., 2012). Although there were no substantial differences in myeloid, T, or B cell chimerism (not depicted), intestinal ILC3s from the WT donor population greatly outnumbered the ILC3s from the Nfil3−/− donor population (Fig. 2 A). In the chimeric mice, ILC3 development from Nfil3−/− donor marrow was impaired in multiple compartments, including SI, LI, and PP, compared with the WT donor population (Fig. 2 B). Furthermore, upon ex vivo stimulation of total ILC3s with IL-23, the IL-22–producing cells were overwhelmingly found within the WT population (Fig. 2 C). Because the mixed chimera setting possesses both WT stromal and hematopoietic elements, our findings imply that Nfil3 acts in a cell-intrinsic manner to drive ILC3 development and/or homeostasis.
Figure 2.
Cell-intrinsic role for Nfil3 in development of ILC3s. (A) Percentages of intestinal WT (CD45.1 × 2) and Nfil3−/− (CD45.2) ILC3 populations in mixed BM chimeric mice are shown. The CD45.1+ population in each plot represents WT host ILC3s. (B) Graphs show percentages of WT and Nfil3−/− ILC3 derived from donor BM in the SI, LI, and PP of chimeras. (C) Representative plots and graphs show the percentage of IL-22–producing cells within the intestinal WT and Nfil3−/− ILC3 populations after IL-23 stimulation. All data are representative of n = 3–5 mice per group, with error bars showing standard deviation, repeated in 2 (C) or 3 (A and B) independent experiments. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Nfil3 is essential for resistance against intestinal pathogens
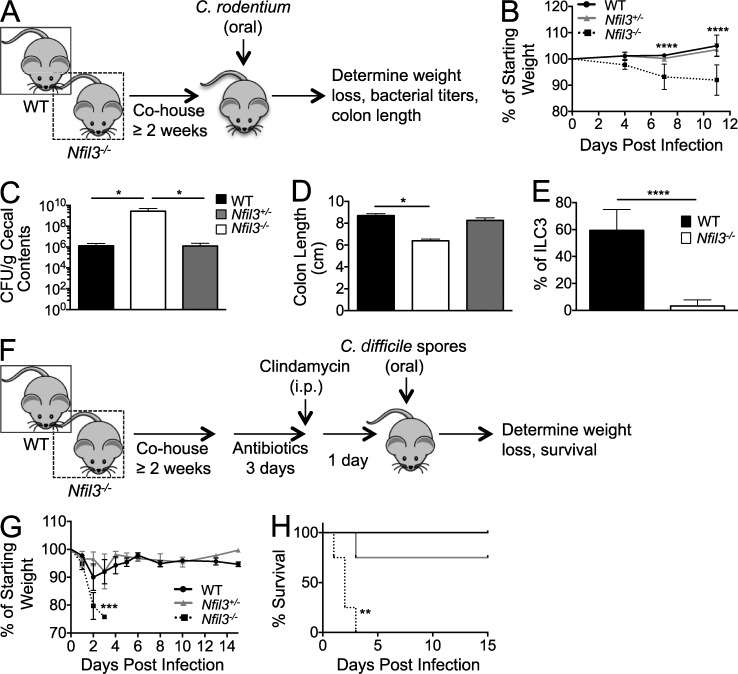
ILC3s have been shown to be critical for host protection against the murine enteric pathogen Citrobacter rodentium, as mice lacking ILC3s or depleted of ILCs become susceptible to bacterial dissemination and mortality (Satoh-Takayama et al., 2008; Cella et al., 2009; Sonnenberg et al., 2011; Qiu et al., 2012; Sonnenberg et al., 2012). Given the defective ILC3 numbers in Nfil3−/− mice compared with WT mice, we next investigated whether Nfil3−/− mice were more susceptible to oral challenge with C. rodentium. In our studies, WT and Nfil3−/− mice, along with Nfil3+/− heterozygous control mice containing intact ILC3 development (unpublished data), were cohoused for a minimum of 2–3 wk before infection to ensure normalization of mouse commensal microbial communities (Elinav et al., 2011; Ubeda et al., 2012). After oral C. rodentium infection, all three experimental cohorts were assessed for disease status and bacterial titers (Fig. 3 A). Within 4 d post infection (PI), Nfil3−/− mice began to lose body weight at a greater rate than WT mice or Nfil3+/− littermates (Fig. 3 B) despite comparable C. rodentium titers in all experimental groups early after infection (not depicted). The Nfil3−/− mice showed significantly greater weight loss at days 7 and 11 PI, whereas WT and Nfil3+/− mice maintained body weight (Fig. 3 B). All groups were sacrificed at day 11 PI and Nfil3−/− mice had higher bacterial titers within cecal contents (Fig. 3 C), with some showing bacterial dissemination to the liver (not depicted), compared with control groups. Consistent with C. rodentium–induced colitis, infected Nfil3−/− mice had shorter colons relative to WT and Nfil3+/− mice (Fig. 3 D), even though we have not observed shorter colons in uninfected Nfil3−/− mice (not depicted). Finally, WT but not Nfil3−/− ILC3s dominated the total intestinal ILC3 population in chimeric mice infected with C. rodentium (Fig. 3 E), suggesting that inflammation generated during infection is unable to expand or recruit gut ILC3s lacking Nfil3. The inability of ILC3s to undergo prolific expansion was confirmed by the lack of BrdU incorporation in mice infected with either C. rodentium or MCMV (unpublished data), the latter of which was previously shown to drive Ly49H+ NK cells to expand in Nfil3−/− mice (Firth et al., 2013).
Figure 3.
Nfil3−/− mice are susceptible to intestinal pathogens. (A) Schematic of C. rodentium experiment. (B) Body weight of mice from WT, Nfil3+/−, and Nfil3−/− groups was assessed during the course of C. rodentium infection. (C and D) Infected WT, Nfil3+/−, and Nfil3−/− mice were sacrificed on day 11 PI, and C. rodentium colony forming units (CFU) in cecal content was determined (C), and colon length measured (D). (E) Mixed WT:Nfil3−/− chimeric mice were infected with C. rodentium, and percentages of WT and Nfil3−/− cells within the total colonic ILC3 population on day 2 PI are shown. (F) Schematic of C. difficile experiment. (G and H) Body weight (G) and survival (H) of mice from WT, Nfil3+/−, and Nfil3−/− groups was assessed during the course of C. difficile infection. All data are representative of n = 3–5 mice per group, with error bars showing standard deviation (B–E) and SEM (G), repeated in 2 independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Next, we tested susceptibility of Nfil3−/− mice against pathogenic bacteria using a clinically relevant model of intestinal Clostridium difficile infection. C. difficile is an opportunistic gram-positive bacterium that can cause severe colitis and diarrhea when the normal microbiota is disrupted after antibiotic treatment (Rupnik et al., 2009), and the incidence of infection in hospital settings is increasing, especially among BMT patients (Kelly and LaMont, 2008). As with the C. rodentium model, experimental mice were first cohoused for 2–3 wk; mice were then treated with an antibiotic regimen (diagrammed in Fig. 3 F) previously shown to disrupt the intestinal microbiota and induce susceptibility to C. difficile spores and colitis (Buffie et al., 2012). Antibiotic-treated Nfil3−/− mice orally challenged with a pathogenic strain of C. difficile demonstrated extreme weight loss within 48–72 h PI, in contrast to WT and Nfil3+/− heterozygous mice (Fig. 3 G). Within 3 d PI, all of the Nfil3−/− mice succumbed to C. difficile infection, whereas control groups recovered from initial weight loss (Fig. 3 H). Together with the C. rodentium studies, infection with C. difficile demonstrates that the transcription factor Nfil3 contributes to host protection against multiple intestinal bacterial pathogens.
Development of the ILCP depends on Nfil3
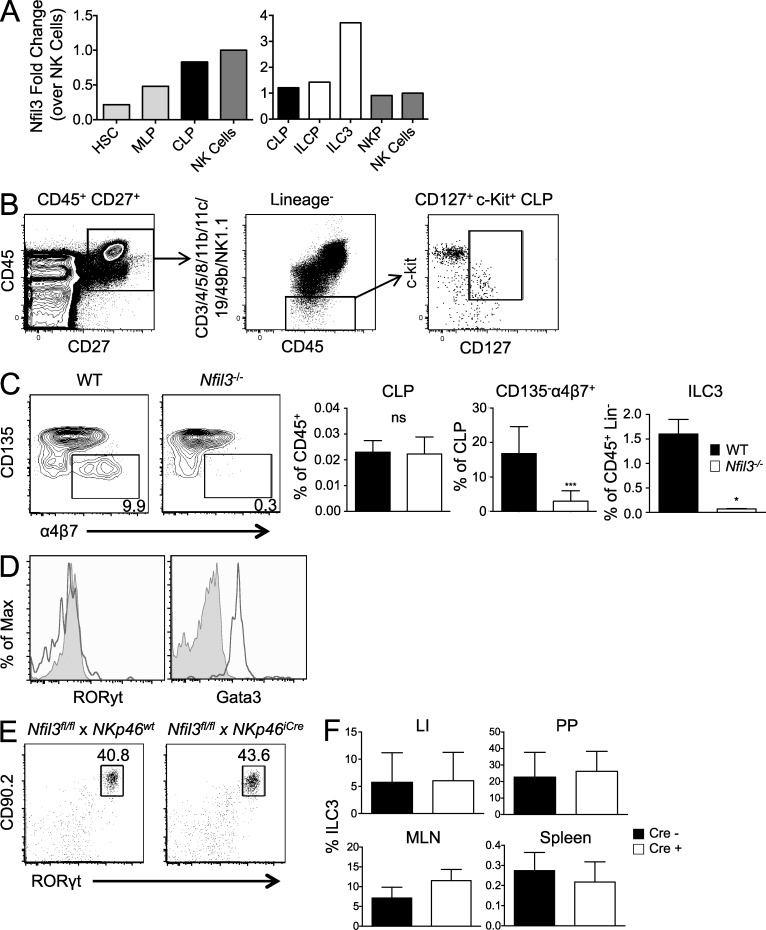
To better understand at which developmental stage Nfil3 is required for generation of mature ILC3s, we analyzed the expression level of Nfil3 mRNA in the earliest progenitor cells by microarray, and in ILCPs and mature ILC3s by qRT-PCR. We found that Nfil3 expression increases as the hematopoietic stem cell differentiates into the multilineage progenitor and then the Id2-expressing common lymphoid progenitor (CLP; Fig. 4 A). Indeed, these data are consistent with recent findings demonstrating Nfil3 expression as early as the CLP stage (Male et al., 2014). From the CLP to ILCP to mature ILC3 stage, Nfil3 expression continues to increase, with highest levels of Nfil3 in gut ILC3 (Fig. 4 A). Thus, we analyzed CLP (Lin− CD45+ CD27+ CD127+ c-Kit+) and ILCP (Lin− CD45+ CD27+ CD127+ c-Kit+ CD135− α4β7+) populations in the BM of WT and Nfil3−/− mice (Fig. 4 B), using surface markers previously described to delineate these precursors (Sawa et al., 2010; Fathman et al., 2011; Possot et al., 2011; Cherrier et al., 2012; Walker et al., 2013; Serafini et al., 2014). Whereas CLP numbers in the BM were comparable between WT and Nfil3−/− mice, ILCP numbers were strikingly reduced in Nfil3−/− mice (Fig. 4 C), suggesting that Nfil3 is required for the transition from the CLP stage to the ILCP stage. Thus, it is possible that Nfil3 is expressed earlier than and may regulate the expression of RORγt and Gata3, neither of which is expressed in CLP, although ILCP expressed Gata3 (Fig. 4 D), consistent with recent findings (Serafini et al., 2014).
Figure 4.
Nfil3 is critical during CLP to ILCP transition but not for maintenance of mature ILC3s. (A) Relative Nfil3 expression was determined by Immgen microarray dataset (left graph) and qRT-PCR (right graph) on indicated cell populations. Data are shown as fold change relative to Nfil3 expression in NK cells. (B) Gating strategy shown for analysis of CLP (CD127+ c-Kit+ cells within the CD45+ CD27+ Lineage− population) in BM. (C) Plots show percentage of BM ILCP (CD135− α4β7+) within the CLP population. Graphs show percentages of CLP and ILCP in BM and ILC3 in the spleen of WT and Nfil3−/− mice. (D) Histograms show expression of RORγt and Gata3 in CLP (tinted) and ILCP (black line). (E) Percentages of intestinal ILC3s in Nfil3fl/fl × Nkp46iCre mice and littermate control (without Cre) are shown. (F) Graphs show percentage of ILC3s in the indicated tissues from Nfil3fl/fl × Nkp46iCre mice and littermate controls. Data are representative of n = 3–5 mice per group, with error bars showing standard deviation, repeated in 2 (C–F) or 3 (A) independent experiments. *, P ≤ 0.05; ***, P ≤ 0.001.
Nfil3-independent maintenance of mature NKp46+ ILC3s
Expression of Nfil3 in mature ILC3s is greater than in conventional NK cells (Fig. 4 A; Klose et al., 2014), even though maintenance of mature NK cells is Nfil3-independent (Firth et al., 2013). Our data and previous studies have found that a large fraction of intestinal ILC3s express the activating receptor NKp46 (Fig. 1 E; Satoh-Takayama et al., 2008; Cella et al., 2009; Sanos et al., 2009; Sawa et al., 2010). To investigate whether Nfil3 is required for maintenance of a mature ILC3 population beyond the developmental stage when NKp46 is first expressed, we crossed Nkp46iCre mice (Narni-Mancinelli et al., 2011), which express Cre-recombinase under control of the NKp46 gene, to Nfil3fl/fl mice (Motomura et al., 2011). Nfil3fl/fl × Nkp46iCre mice contained a normal number of mature intestinal ILC3s compared with littermate control mice lacking Cre expression (Fig. 4 E). Similar ILC3 frequencies were also found in the spleen and MLNs of Nfil3fl/fl × Nkp46iCre mice and littermate controls (Fig. 4 F). Together, these data suggest that Nfil3 is required for ILC3s at a developmental stage preceding the acquisition of NKp46 expression, and that the maintenance of NKp46+ ILC3s is independent of Nfil3.
ILC2 populations are severely diminished in Nfil3-deficient mice
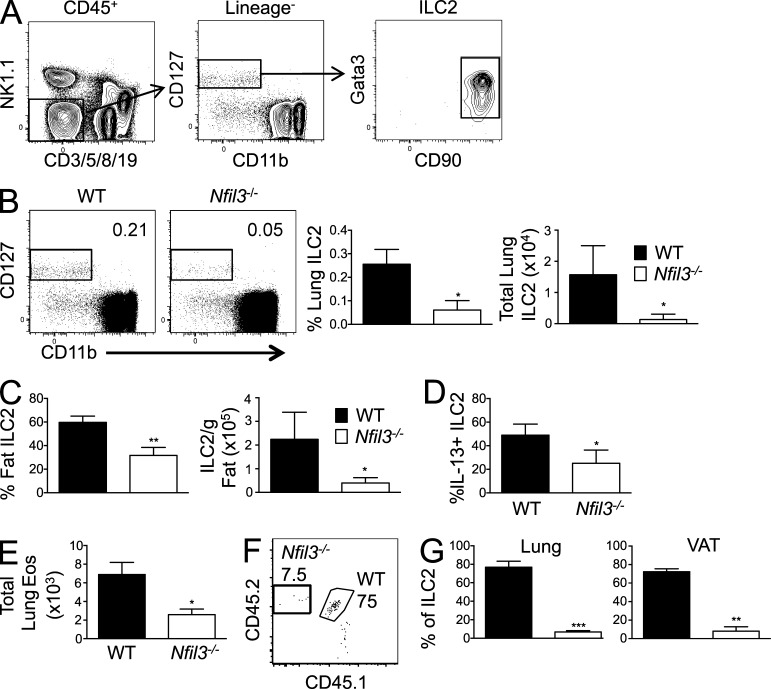
Given the dependence of ILC3s on Nfil3, as well as the previously reported dependence of type 1 ILCs on Nfil3 (Gascoyne et al., 2009; Kamizono et al., 2009; Kashiwada et al., 2010; Firth et al., 2013; Fuchs et al., 2013), we investigated whether type 2 ILCs are also diminished in Nfil3-deficient mice. ILC2s have been characterized as the predominant subset of ILC in healthy lungs, and can mediate lung inflammatory responses and pulmonary immunity against pathogens (Spits and Cupedo, 2012; Sonnenberg et al., 2013; Walker et al., 2013). We discovered that Nfil3−/− mice contain markedly reduced numbers of ILC2s (identified as Lineage-negative cells that coexpress CD45, IL-7Rα [CD127], Thy 1 [CD90], and Gata3; Fig. 5 A) in lung tissue relative to WT mice (Fig. 5 B). Because ILC2s have also been described to constitute a major source of Th2 cytokines in visceral adipose tissue (VAT; Moro et al., 2010; Molofsky et al., 2013), we investigated whether ILC2s were defective in the VAT of Nfil3−/− mice. Indeed, compared with WT mice, both ILC2 numbers and function (as measured by IL-13 secretion) were drastically diminished within VAT of Nfil3−/− mice (Fig. 5, C and D), demonstrating that the ILC2 defect in the absence of Nfil3 is not restricted to the lungs. A recent study showed that a consequence of ILC2 presence in the lungs is the regulation of basal eosinophil homeostasis (Nussbaum et al., 2013). When we assessed eosinophils in the lungs of Nfil3−/− mice, we found diminished numbers compared with WT mice (Fig. 5 E), suggesting that Nfil3 control of ILC2 development may contribute to regulating tissue eosinophil accumulation at steady-state. Using 1:1 WT:Nfil3−/− mixed chimeric mice, as described earlier (Fig. 2), we found that ILC2 in lung and VAT consisted of cells derived from WT marrow in significantly greater frequency than from Nfil3−/− marrow (Fig. 5, F and G), suggesting that like ILC3s, development of ILC2s requires Nfil3 activity via a cell-intrinsic mechanism. Altogether, these findings demonstrate that Nfil3 deficiency results in the disrupted development of ILC1, ILC2, and ILC3 subsets.
Figure 5.
Nfil3−/− mice are deficient in lung and fat ILC2s and eosinophils. (A) Gating strategy shown for analysis of lung ILC2 (CD90.2+ Gata3+ cells within the CD45+ Lineage− population). (B) Percentage (of CD45+) and absolute number of ILC2s in lungs of WT and Nfil3−/− mice are shown in plots and graph. (C and D) Graphs show total ILC2s (C) and IL-13–secreting ILC2s (after stimulation with PMA + Ionomycin; D) in VAT from WT and Nfil3−/− mice. (E) Absolute number of eosinophils (Lin− CD45+ CD90− NK1.1− CD11b+ SiglecF+) in lungs of WT and Nfil3−/− mice is shown. (F) Percentages of WT (CD45.1 × 2) and Nfil3−/− (CD45.2) lung ILC2 populations from mixed BM chimeric mice are shown. The CD45.1+ population in each plot represents WT host ILC2s. (G) Graph shows percentages of WT and Nfil3−/− lung and VAT ILC2s derived from donor BM in chimeric mice. All data are representative of n = 3–5 mice per group, with error bars showing standard deviation, repeated in 2 (E), 3 (F and G), or 4 (B–D) independent experiments. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
In summary, our study demonstrates a critical role for the bZIP transcription factor Nfil3 in the development of all innate lymphocyte subsets. The loss of the ILC3 subset in Nfil3−/− mice may account for the loss of intestinal integrity at steady-state and the development of spontaneous colitis which was recently reported (Kobayashi et al., 2014). Indeed, Nfil3 represents a susceptibility gene in Crohn’s disease and ulcerative colitis patients (Kobayashi et al., 2011). Although the precise mechanisms of Nfil3-mediated host protection from inflammatory bowel disease (IBD) remain to be elucidated, our current findings suggest that absence of the Nfil3-dependent ILCs may contribute toward greater risk of intestinal injury and colitis, and morbidity during pathogenic bacteria exposure.
Given the broad role for Nfil3 in regulating a diverse range of immune cells, our findings importantly demonstrate that the activity of Nfil3 in the general development of ILCs is cell-intrinsic and may not be required beyond the early ILC developmental stages, reminiscent of the Nfil3-independent lineage maintenance recently reported for NK cells (Firth et al., 2013). Thus, the transcription factor Nfil3 may play the role of a master promoter of ILC development, acting in an early ILCP similarly to Id2 (Yokota et al., 1999; Moro et al., 2010; Satoh-Takayama et al., 2010), or, as recently described, Gata3 (Serafini et al., 2014) and Plzf (Constantinides et al., 2014). Further elucidation of the regulation and targets of Nfil3 in ILC development will be valuable for determining the lineage relationships between ILC subsets. Our findings may be useful for understanding pathophysiology of inflammatory processes at mucosal surfaces and for developing therapeutic interventions for multiple causes of infectious and noninfectious intestinal injury, including IBD and graft versus host disease.
MATERIALS AND METHODS
Mice.
Wild-type C57BL/6 (B6), congenic (CD45.1 and CD45.1xCD45.2), Nfil3−/− (Kashiwada et al., 2010), Nfil3fl/fl (Motomura et al., 2011), and Nkp46iCre (Narni-Mancinelli et al., 2011) mice were bred and maintained at Memorial Sloan-Kettering Cancer Center (MSKCC). Mice were housed and maintained according to MSKCC guidelines, and all experiments were performed in accordance with MSKCC Institutional Animal Care and Use Committee approval and institutional guidelines. Mixed BM chimeric mice were generated, as previously described (Sun et al., 2009). In oral infection studies, mice were cohoused for a period of 2–3 wk before bacteria challenge to normalize bacteria flora between experimental groups. Wild-type controls were age- and sex-matched C57BL/6 mice in all experiments.
Bacterial infections and titers.
In C. rodentium studies, mice were inoculated by oral gavage with 108 CFU (in 200 µl) of an overnight LB culture of C. rodentium (strain DBS100). Infected mice were assessed for body weight, signs of morbidity, and bacterial titers. To determine C. rodentium titers, fecal or cecal contents were mechanically homogenized in PBS and 10-fold serial dilutions cultured overnight on MacConkey’s agar, as previously described (Sonnenberg et al., 2011). In C. difficile studies, mice were treated with antibiotic water (0.25 g/liter metronidazole, 0.25 g/liter neomycin, and 0.25 g/liter vancomycin) from day −6 to −3 and received 200 µg clindamycin i.p. on day −1 before infection with 200 CFU C. difficile spores (strain VPI 10463) by oral gavage. Infected mice were assessed for body weight and signs of morbidity (Buffie et al., 2012).
Isolation of ILC subsets and ex vivo stimulation.
Spleens, MLNs, and Peyer’s patches were mechanically crushed into single cell suspensions. Lungs, intestines, and fat were digested in collagenase type 4 (Worthington), collagenase D (Roche), and collagenase type 2 (Worthington), respectively. To assess production of cytokines, ILC2 and ILC3 cells were stimulated for 3 h at 37°C in complete RPMI + 10% FBS with 1:1,000 Brefeldin A (BD), 1:1,000 2-mercaptoethanol (Sigma-Aldrich), and 40 ng/ml IL-23 (for ILC3 stimulation) or 0.1 µg/ml PMA + 1 µg/ml ionomycin (for ILC2 stimulation), followed by intracellular staining. Unstimulated controls (media only) were used to determine gating strategy for flow cytometric plots in figures.
Flow cytometry.
Single cell suspensions were generated from indicated organs and incubated with the anti-Fc receptor antibody 2.4G2 before staining with indicated monoclonal antibodies (BioLegend, eBioscience, and BD) for 20 min on ice. In certain experiments, staining was performed on transcription factors and intracellular cytokines using the FoxP3 staining kit (eBioscience) according to manufacturer protocols. Lineage-negative cells are defined as lacking surface CD3, CD4, CD5, CD8, CD11b, CD11c, CD19, CD49b, Gr-1, and NK1.1. Samples were acquired using an LSRII flow cytometer with FACSDiva software (BD), and analysis was performed with FlowJo 9.6 software (Tree Star).
Quantitative real-time PCR.
BM CLP (lin− c-kit+ sca1+ flt3+ IL-7Ra+), ILCP (lin− c-kit+ sca1+ flt3− IL-7Ra+ α4β7+), and gut ILC3 (lin− Rorc+ IL-7Ra+) were sorted to ∼99% purity on an Aria II cytometer (BD). Cell lysis was subsequently performed using Tri-Reagent (Ambion), RNA was purified using the RNeasy kit (with on-column DNase I treatment; QIAGEN), and MuLV reverse transcription and oligo(dT)16 primers (Applied Biosystems) were used for cDNA synthesis. iQ Sybr Green SuperMix (Bio-Rad Laboratories) was used for qRT-PCR. Data were normalized to expression of β-actin and expressed as relative target abundance via the ΔΔCt method, where Ct (threshold cycle) is the cycle number at which the amplification curve intersects the threshold value. The primer sets used for qRT-PCR are the following: Nfil3 forward, 5′-AATTCATTCCGGACGAGAAG-3′; Nfil3 reverse, 5′-CGATCAGCTTGTTCTCCAAA-3′; β-actin forward, 5′-TGCGTGACATCAAAGAGAAG-3′; and β-actin reverse, 5′-CGGATGTCAACGTCACACTT-3′.
Statistical analysis.
Results are expressed as mean, with error bars showing ±SD unless otherwise indicated. Data were analyzed using a two-tailed unpaired Student’s t test with Welch’s correction or one-way ANOVA (with multiple comparisons where applicable). All analyses were performed using Prism 5.0b (GraphPad Software), and differences were considered significant when P ≤ 0.05.
Acknowledgments
We thank members of the Sun and Hanash labs for technical support and experimental assistance, and members of the MSKCC NK club for insightful comments and helpful discussions. Paul Rothman, Masato Kubo, Eric Vivier, and David Artis provided mice and bacteria critical to this study. We thank the ImmGen consortium for providing the microarray data used in this study.
G. Gasteiger is supported by an Irvington Fellowship of the Cancer Research Institute. M.A. Firth is supported by a fellowship from the Lucille Castori Center for Microbes, Inflammation, and Cancer. A.M. Hanash is supported by an American Society of Hematology Scholar Award. J.C. Sun is supported by the Searle Scholars Program and the Cancer Research Institute. National Institutes of Health grants R01-095706 (E.G. Pamer and M. Abt); R01-HL069929, R01-AI100288, R01-AI080455, and R01-AI101406 (M.R. van den Brink); K08-KHL115355 (A.M. Hanash); and R01-AI100874 (J.C. Sun and T.L. Geiger) supported this work.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BMT
- BM transplantation
- CLP
- common lymphoid progenitor
- ILC
- innate lymphoid cell
- ILCP
- ILC precursor
- LI
- large intestine
- LTi
- lymphoid tissue inducer
- MLN
- mesenteric LN
- PI
- post infection
- PP
- Peyer’s patch
- SI
- small intestine
- VAT
- visceral adipose tissue
References
- Buffie, C.G., Jarchum I., Equinda M., Lipuma L., Gobourne A., Viale A., Ubeda C., Xavier J., and Pamer E.G.. 2012. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 80:62–73 10.1128/IAI.05496-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cella, M., Fuchs A., Vermi W., Facchetti F., Otero K., Lennerz J.K., Doherty J.M., Mills J.C., and Colonna M.. 2009. A human natural killer cell subset provides an innate source of IL-22 for mucosal immunity. Nature. 457:722–725 10.1038/nature07537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrier, M., Sawa S., and Eberl G.. 2012. Notch, Id2, and RORγt sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J. Exp. Med. 209:729–740 10.1084/jem.20111594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinides, M.G., McDonald B.D., Verhoef P.A., and Bendelac A.. 2014. A committed precursor to innate lymphoid cells. Nature. 508:397–401 10.1038/nature13047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl, G., Marmon S., Sunshine M.J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORγt in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Elinav, E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathman, J.W., Bhattacharya D., Inlay M.A., Seita J., Karsunky H., and Weissman I.L.. 2011. Identification of the earliest natural killer cell-committed progenitor in murine bone marrow. Blood. 118:5439–5447 10.1182/blood-2011-04-348912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firth, M.A., Madera S., Beaulieu A.M., Gasteiger G., Castillo E.F., Schluns K.S., Kubo M., Rothman P.B., Vivier E., and Sun J.C.. 2013. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med. 210:2981–2990 10.1084/jem.20130417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, A., Vermi W., Lee J.S., Lonardi S., Gilfillan S., Newberry R.D., Cella M., and Colonna M.. 2013. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 38:769–781 10.1016/j.immuni.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa, J., Moro K., Motomura Y., Okamoto K., Zhu J., Takayanagi H., Kubo M., and Koyasu S.. 2013. Critical role of p38 and GATA3 in natural helper cell function. J. Immunol. 191:1818–1826 10.4049/jimmunol.1300379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne, D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., and Brady H.J.. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Halim, T.Y., MacLaren A., Romanish M.T., Gold M.J., McNagny K.M., and Takei F.. 2012. Retinoic-acid-receptor-related orphan nuclear receptor α is required for natural helper cell development and allergic inflammation. Immunity. 37:463–474 10.1016/j.immuni.2012.06.012 [DOI] [PubMed] [Google Scholar]
- Hanash, A.M., Dudakov J.A., Hua G., O’Connor M.H., Young L.F., Singer N.V., West M.L., Jenq R.R., Holland A.M., Kappel L.W., et al. 2012. Interleukin-22 protects intestinal stem cells from immune-mediated tissue damage and regulates sensitivity to graft versus host disease. Immunity. 37:339–350 10.1016/j.immuni.2012.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyler, T., Klose C.S., Souabni A., Turqueti-Neves A., Pfeifer D., Rawlins E.L., Voehringer D., Busslinger M., and Diefenbach A.. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 37:634–648 10.1016/j.immuni.2012.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima, S., Inukai T., Inaba T., Nimer S.D., Cleveland J.L., and Look A.T.. 1997. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA. 94:2609–2614 10.1073/pnas.94.6.2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono, S., Duncan G.S., Seidel M.G., Morimoto A., Hamada K., Grosveld G., Akashi K., Lind E.F., Haight J.P., Ohashi P.S., et al. 2009. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J. Exp. Med. 206:2977–2986 10.1084/jem.20092176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada, M., Levy D.M., McKeag L., Murray K., Schröder A.J., Canfield S.M., Traver G., and Rothman P.B.. 2010. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. USA. 107:821–826 10.1073/pnas.0909235107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada, M., Cassel S.L., Colgan J.D., and Rothman P.B.. 2011a. NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. EMBO J. 30:2071–2082 10.1038/emboj.2011.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada, M., Pham N.L., Pewe L.L., Harty J.T., and Rothman P.B.. 2011b. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 117:6193–6197 10.1182/blood-2010-07-295873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, C.P., and LaMont J.T.. 2008. Clostridium difficile—more difficult than ever. N. Engl. J. Med. 359:1932–1940 10.1056/NEJMra0707500 [DOI] [PubMed] [Google Scholar]
- Kiss, E.A., Vonarbourg C., Kopfmann S., Hobeika E., Finke D., Esser C., and Diefenbach A.. 2011. Natural aryl hydrocarbon receptor ligands control organogenesis of intestinal lymphoid follicles. Science. 334:1561–1565 10.1126/science.1214914 [DOI] [PubMed] [Google Scholar]
- Klein Wolterink, R.G., Serafini N., van Nimwegen M., Vosshenrich C.A., de Bruijn M.J., Fonseca Pereira D., Veiga Fernandes H., Hendriks R.W., and Di Santo J.P.. 2013. Essential, dose-dependent role for the transcription factor Gata3 in the development of IL-5+ and IL-13+ type 2 innate lymphoid cells. Proc. Natl. Acad. Sci. USA. 110:10240–10245 10.1073/pnas.1217158110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose, C.S., Kiss E.A., Schwierzeck V., Ebert K., Hoyler T., d’Hargues Y., Göppert N., Croxford A.L., Waisman A., Tanriver Y., and Diefenbach A.. 2013. A T-bet gradient controls the fate and function of CCR6−RORγt+ innate lymphoid cells. Nature. 494:261–265 10.1038/nature11813 [DOI] [PubMed] [Google Scholar]
- Klose, C.S., Flach M., Möhle L., Rogell L., Hoyler T., Ebert K., Fabiunke C., Pfeifer D., Sexl V., Fonseca-Pereira D., et al. 2014. Differentiation of type 1 ILCs from a common progenitor to all helper-like innate lymphoid cell lineages. Cell. 157:340–356 10.1016/j.cell.2014.03.030 [DOI] [PubMed] [Google Scholar]
- Kobayashi, T., Matsuoka K., Sheikh S.Z., Elloumi H.Z., Kamada N., Hisamatsu T., Hansen J.J., Doty K.R., Pope S.D., Smale S.T., et al. 2011. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J. Immunol. 186:4649–4655 10.4049/jimmunol.1003888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Steinbach E.C., Russo S.M., Matsuoka K., Nochi T., Maharshak N., Borst L.B., Hostager B., Garcia-Martinez J.V., Rothman P.B., et al. 2014. NFIL3-deficient mice develop microbiota-dependent, IL-12/23-driven spontaneous colitis. J. Immunol. 192:1918–1927 10.4049/jimmunol.1301819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.S., Cella M., McDonald K.G., Garlanda C., Kennedy G.D., Nukaya M., Mantovani A., Kopan R., Bradfield C.A., Newberry R.D., and Colonna M.. 2012. AHR drives the development of gut ILC22 cells and postnatal lymphoid tissues via pathways dependent on and independent of Notch. Nat. Immunol. 13:144–151 10.1038/ni.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, H.E., Reinhardt R.L., Bando J.K., Sullivan B.M., Ho I.C., and Locksley R.M.. 2012. Divergent expression patterns of IL-4 and IL-13 define unique functions in allergic immunity. Nat. Immunol. 13:58–66 10.1038/ni.2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luci, C., Reynders A., Ivanov I.I., Cognet C., Chiche L., Chasson L., Hardwigsen J., Anguiano E., Banchereau J., Chaussabel D., et al. 2009. Influence of the transcription factor RORγt on the development of NKp46+ cell populations in gut and skin. Nat. Immunol. 10:75–82 10.1038/ni.1681 [DOI] [PubMed] [Google Scholar]
- Male, V., Nisoli I., Kostrzewski T., Allan D.S., Carlyle J.R., Lord G.M., Wack A., and Brady H.J.. 2014. The transcription factor E4bp4/Nfil3 controls commitment to the NK lineage and directly regulates Eomes and Id2 expression. J. Exp. Med. 211:635–642 10.1084/jem.20132398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke, L.A., Groom J.R., Rankin L.C., Seillet C., Masson F., Putoczki T., and Belz G.T.. 2013. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 191:4383–4391 10.4049/jimmunol.1301228 [DOI] [PubMed] [Google Scholar]
- Mjösberg, J., Bernink J., Golebski K., Karrich J.J., Peters C.P., Blom B., te Velde A.A., Fokkens W.J., van Drunen C.M., and Spits H.. 2012. The transcription factor GATA3 is essential for the function of human type 2 innate lymphoid cells. Immunity. 37:649–659 10.1016/j.immuni.2012.08.015 [DOI] [PubMed] [Google Scholar]
- Molofsky, A.B., Nussbaum J.C., Liang H.E., Van Dyken S.J., Cheng L.E., Mohapatra A., Chawla A., and Locksley R.M.. 2013. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. J. Exp. Med. 210:535–549 10.1084/jem.20121964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moro, K., Yamada T., Tanabe M., Takeuchi T., Ikawa T., Kawamoto H., Furusawa J., Ohtani M., Fujii H., and Koyasu S.. 2010. Innate production of TH2 cytokines by adipose tissue-associated c-Kit+Sca-1+ lymphoid cells. Nature. 463:540–544 10.1038/nature08636 [DOI] [PubMed] [Google Scholar]
- Motomura, Y., Kitamura H., Hijikata A., Matsunaga Y., Matsumoto K., Inoue H., Atarashi K., Hori S., Watarai H., Zhu J., et al. 2011. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat. Immunol. 12:450–459 10.1038/ni.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narni-Mancinelli, E., Chaix J., Fenis A., Kerdiles Y.M., Yessaad N., Reynders A., Gregoire C., Luche H., Ugolini S., Tomasello E., et al. 2011. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. USA. 108:18324–18329 10.1073/pnas.1112064108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum, J.C., Van Dyken S.J., von Moltke J., Cheng L.E., Mohapatra A., Molofsky A.B., Thornton E.E., Krummel M.F., Chawla A., Liang H.E., and Locksley R.M.. 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 502:245–248 10.1038/nature12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot, C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A., and Golub R.. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt+ innate lymphoid cells. Nat. Immunol. 12:949–958 10.1038/ni.2105 [DOI] [PubMed] [Google Scholar]
- Qiu, J., Heller J.J., Guo X., Chen Z.M., Fish K., Fu Y.X., and Zhou L.. 2012. The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity. 36:92–104 10.1016/j.immuni.2011.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin, L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A., McKenzie A.N., Carotta S., Nutt S.L., and Belz G.T.. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14:389–395 10.1038/ni.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupnik, M., Wilcox M.H., and Gerding D.N.. 2009. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat. Rev. Microbiol. 7:526–536 10.1038/nrmicro2164 [DOI] [PubMed] [Google Scholar]
- Sanos, S.L., Bui V.L., Mortha A., Oberle K., Heners C., Johner C., and Diefenbach A.. 2009. RORγt and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat. Immunol. 10:83–91 10.1038/ni.1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Takayama, N., Vosshenrich C.A., Lesjean-Pottier S., Sawa S., Lochner M., Rattis F., Mention J.J., Thiam K., Cerf-Bensussan N., Mandelboim O., et al. 2008. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 29:958–970 10.1016/j.immuni.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Satoh-Takayama, N., Lesjean-Pottier S., Vieira P., Sawa S., Eberl G., Vosshenrich C.A., and Di Santo J.P.. 2010. IL-7 and IL-15 independently program the differentiation of intestinal CD3−NKp46+ cell subsets from Id2-dependent precursors. J. Exp. Med. 207:273–280 10.1084/jem.20092029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa, S., Cherrier M., Lochner M., Satoh-Takayama N., Fehling H.J., Langa F., Di Santo J.P., and Eberl G.. 2010. Lineage relationship analysis of RORγt+ innate lymphoid cells. Science. 330:665–669 10.1126/science.1194597 [DOI] [PubMed] [Google Scholar]
- Sciumé, G., Hirahara K., Takahashi H., Laurence A., Villarino A.V., Singleton K.L., Spencer S.P., Wilhelm C., Poholek A.C., Vahedi G., et al. 2012. Distinct requirements for T-bet in gut innate lymphoid cells. J. Exp. Med. 209:2331–2338 10.1084/jem.20122097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serafini, N., Klein Wolterink R.G., Satoh-Takayama N., Xu W., Vosshenrich C.A., Hendriks R.W., and Di Santo J.P.. 2014. Gata3 drives development of RORγt+ group 3 innate lymphoid cells. J. Exp. Med. 211:199–208 10.1084/jem.20131038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg, G.F., Monticelli L.A., Elloso M.M., Fouser L.A., and Artis D.. 2011. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 34:122–134 10.1016/j.immuni.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg, G.F., Monticelli L.A., Alenghat T., Fung T.C., Hutnick N.A., Kunisawa J., Shibata N., Grunberg S., Sinha R., Zahm A.M., et al. 2012. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 336:1321–1325 10.1126/science.1222551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg, G.F., Mjösberg J., Spits H., and Artis D.. 2013. SnapShot: innate lymphoid cells. Immunity. 39:622–622: e1 10.1016/j.immuni.2013.08.021 [DOI] [PubMed] [Google Scholar]
- Spits, H., and Cupedo T.. 2012. Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu. Rev. Immunol. 30:647–675 10.1146/annurev-immunol-020711-075053 [DOI] [PubMed] [Google Scholar]
- Spits, H., and Di Santo J.P.. 2011. The expanding family of innate lymphoid cells: regulators and effectors of immunity and tissue remodeling. Nat. Immunol. 12:21–27 10.1038/ni.1962 [DOI] [PubMed] [Google Scholar]
- Spits, H., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N., Mebius R.E., et al. 2013. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13:145–149 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]
- Spooner, C.J., Lesch J., Yan D., Khan A.A., Abbas A., Ramirez-Carrozzi V., Zhou M., Soriano R., Eastham-Anderson J., Diehl L., et al. 2013. Specification of type 2 innate lymphocytes by the transcriptional determinant Gfi1. Nat. Immunol. 14:1229–1236 10.1038/ni.2743 [DOI] [PubMed] [Google Scholar]
- Sun, J.C., Beilke J.N., and Lanier L.L.. 2009. Adaptive immune features of natural killer cells. Nature. 457:557–561 10.1038/nature07665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda, C., Lipuma L., Gobourne A., Viale A., Leiner I., Equinda M., Khanin R., and Pamer E.G.. 2012. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J. Exp. Med. 209:1445–1456 10.1084/jem.20120504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen, M., Hirota K., Westendorf A.M., Buer J., Dumoutier L., Renauld J.C., and Stockinger B.. 2008. The aryl hydrocarbon receptor links TH17-cell-mediated autoimmunity to environmental toxins. Nature. 453:106–109 10.1038/nature06881 [DOI] [PubMed] [Google Scholar]
- Walker, J.A., Barlow J.L., and McKenzie A.N.. 2013. Innate lymphoid cells—how did we miss them? Nat. Rev. Immunol. 13:75–87 10.1038/nri3349 [DOI] [PubMed] [Google Scholar]
- Wong, S.H., Walker J.A., Jolin H.E., Drynan L.F., Hams E., Camelo A., Barlow J.L., Neill D.R., Panova V., Koch U., et al. 2012. Transcription factor RORα is critical for nuocyte development. Nat. Immunol. 13:229–236 10.1038/ni.2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Saenz S.A., Zlotoff D.A., Artis D., and Bhandoola A.. 2011. Cutting edge: Natural helper cells derive from lymphoid progenitors. J. Immunol. 187:5505–5509 10.4049/jimmunol.1102039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q., Monticelli L.A., Saenz S.A., Chi A.W., Sonnenberg G.F., Tang J., De Obaldia M.E., Bailis W., Bryson J.L., Toscano K., et al. 2013. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity. 38:694–704 10.1016/j.immuni.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota, Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., and Gruss P.. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 397:702–706 10.1038/17812 [DOI] [PubMed] [Google Scholar]
- Yu, X., Rollins D., Ruhn K.A., Stubblefield J.J., Green C.B., Kashiwada M., Rothman P.B., Takahashi J.S., and Hooper L.V.. 2013. TH17 cell differentiation is regulated by the circadian clock. Science. 342:727–730 10.1126/science.1243884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., Zhang J., Kornuc M., Kwan K., Frank R., and Nimer S.D.. 1995. Molecular cloning and characterization of NF-IL3A, a transcriptional activator of the human interleukin-3 promoter. Mol. Cell. Biol. 15:6055–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]