Loss of Nfil3 selectively reduces Peyer’s patch formation, impairing recruitment and distribution of lymphocytes and compromising immune responses to inflammatory and infectious agents.
Abstract
Innate lymphoid cell (ILC) populations protect against infection and are essential for lymphoid tissue formation and tissue remodeling after damage. Nfil3 is implicated in the function of adaptive immune lineages and NK cell development, but it is not yet known if Nfil3 regulates other innate lymphoid lineages. Here, we identify that Nfil3 is essential for the development of Peyer’s patches and ILC2 and ILC3 subsets. Loss of Nfil3 selectively reduced Peyer’s patch formation and was accompanied by impaired recruitment and distribution of lymphocytes within the patches. ILC subsets exhibited high Nfil3 expression and genetic deletion of Nfil3 severely compromised the development of all subsets. Subsequently, Nfil3−/− mice were highly susceptible to disease when challenged with inflammatory or infectious agents. Thus, we demonstrate that Nfil3 is a key regulator of the development of ILC subsets essential for immune protection in the lung and gut.
Protection of mucosal surfaces depends on the delicate balance between rapid induction of innate immune responses and priming of adaptive immune responses. This early immune response has largely been thought to be orchestrated by NK cells, but recent studies have uncovered that the innate lymphocyte family is more complex and diverse than previously appreciated. It comprises not only the prototypic innate lymphoid cell type 1 (ILC1), NK cells, and lymphoid tissue-inducer (LTi) cells but also nuocytes or natural helper cells and Rorγt+ ILCs. Recently, these lineages were classified into three ILC subsets: ILC1 (NK cell and IFN-γ–producing ILCs), ILC2 (nuocytes and natural helper cells), and ILC3 (Rorγt+LTi cells and natural cytotoxicity receptor [NCR] NKp46+ ILC3; Spits et al., 2013). ILCs exhibit many similarities to CD4+ T cell subsets in the array of cytokines they produce and their dependence on the same transcription factors for lineage commitment. A core set of transcription factors corresponding to those used in both CD4+ T cell and NK cell differentiation also significantly influence ILC subset differentiation. It is therefore likely that each cell type adopts a lineage-specific transcriptional program dictating the fate of ILC progenitors into the different subsets. As yet, however, the transcriptional networks that govern the development and effector function of the different ILC subsets are not well understood.
The transcription factor nuclear factor, IL-3 (Nfil3, also known as E4BP4) was originally described as necessary for the development of bone marrow-derived NK cells (Gascoyne et al., 2009) but is now known to also be involved in the development and the functions of other immune cells. Nfil3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development (Kashiwada et al., 2011) in B cells, IgE class switching (Kashiwada et al., 2010), and regulation of CD4+ T cell lineages (Motomura et al., 2011). Nfil3 has been proposed to regulate NK cell development by activating the transcription factor inhibitor of DNA binding 2 (Id2; Gascoyne et al., 2009); however, this is controversial as it is not required for hepatic NK cells, and Nfil3-deficient NK cells express normal levels of Id2 (Seillet et al., 2014). Id2 is essential for the development of all ILC subsets, including NK cells. Whether Nfil3 is involved in ILC subset differentiation is not yet known. In this report, we demonstrate that the absence of Nfil3 resulted in intrinsic defects in the development of all ILC subsets and altered Peyer’s patch formation, culminating in diminished protective immune responses during lung and intestinal inflammation. These results suggest that Nfil3, like Id2, plays a broad and essential role in regulating the gene network required for ILC development.
RESULTS AND DISCUSSION
Nfil3 has been shown to regulate multiple aspects of the development and function of adaptive immune cells and is likely to also impact the development of the innate arm of the immune system given its broad expression (Ikushima et al., 1997; Kashiwada et al., 2010; Kobayashi et al., 2011; Motomura et al., 2011; Seillet et al., 2013).
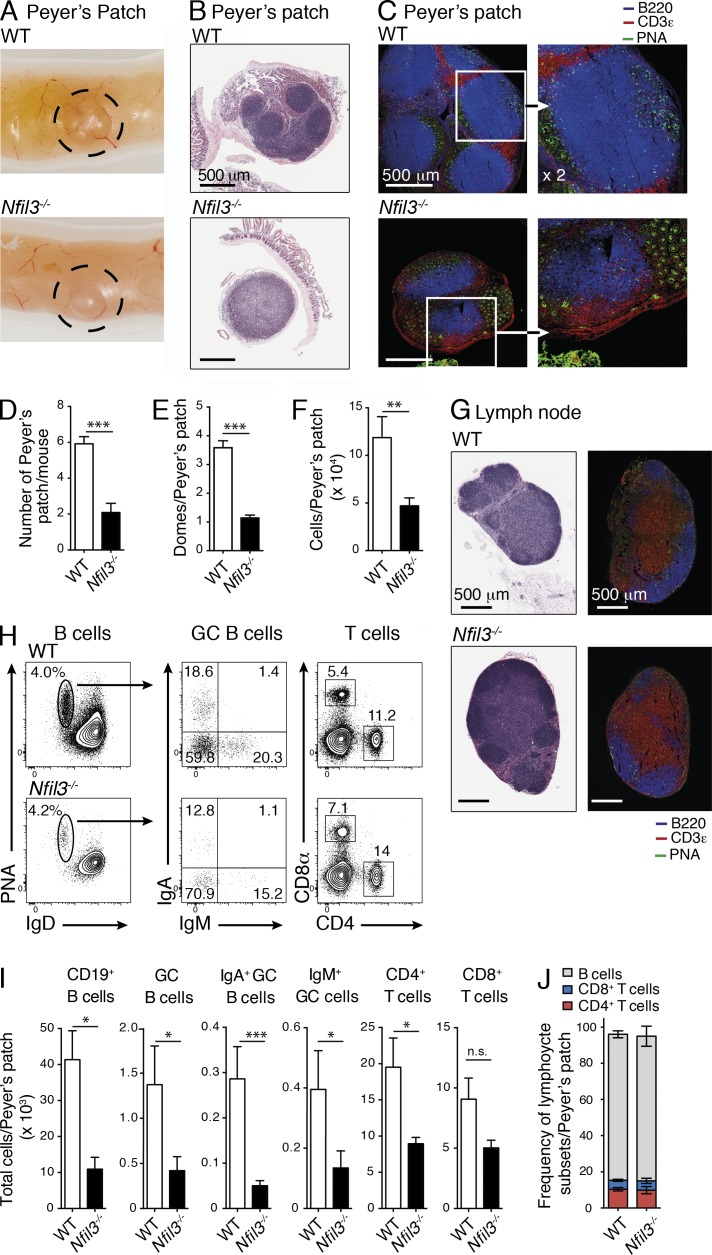
We first analyzed secondary lymphatic organs in Nfil3-deficient mice and detected clear defects in the formation of Peyer’s patches. These mice exhibited a significant reduction in the number, size, and cellularity of Peyer’s patches (∼3-fold reduction), although the frequency of T and B cell subsets and activation status was not altered (Fig. 1, A–J). Histological analyses also revealed that Nfil3-deficient mice failed to form multiple, clearly defined B cell follicles and associated lymphoepithelium that form domes over follicles in the Peyer’s patch. Instead, typically one or two B cell follicles were observed in each Peyer’s patch and T cell and B cell zones were less clearly defined in these dysregulated Peyer’s patches in the absence of Nfil3 (Fig. 1, B–E). In contrast, these alterations were not evident in Nfil3−/− peripheral LNs that appeared similar to WT LNs (Fig. 1 G). These data suggest that Nfil3 is a key regulator of Peyer’s patch formation and plays an important role in the organization of cells within the patches.
Figure 1.
Characterization of secondary lymphoid organs in WT and Nfil3-deficient mice. (A) Gross, (B) histological and (C) immunohistological analysis of Peyer’s patch in WT (C57BL/6) and Nfil3−/− mice. (B and G) Histological sections of Peyer’s patch (B) and LN (G) were stained with H&E, and (C and G) confocal images were stained with anti-B220 (blue), anti-CD3ε (red), and anti–peanut agglutinin (PNA, green). Bar, 500 µm. (C) Inset shown in higher magnification (×2) in right panel. (B, C, and G) H&E sections are representative of analyses performed on five mice/genotype; immunohistochemistry was performed on two mice/genotype. (D) Enumeration of the number of Peyer’s patch, (E) Peyer’s patch domes, and (F) total cells. (D–F) Data show mean ± SEM pooled from 2–4 independent experiments (n = 3–4 mice/genotype). Statistical differences were calculated using an unpaired Student’s t test. (H) Flow cytometric analyses of B and T cell subsets in Peyer’s patch of naive C57BL/6 and Nfil3−/− mice. (I) Enumeration of the total B (mean ± SEM of 3–9 mice/genotype pooled from 2 experiments) and T cell subsets (mean ± SEM of one of two similar experiments, n = 3–5 mice/genotype) within the Peyer’s patch. (J) Cumulative frequency of B and T cells from analyses in (I). Statistical differences were calculated with an unpaired Mann-Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
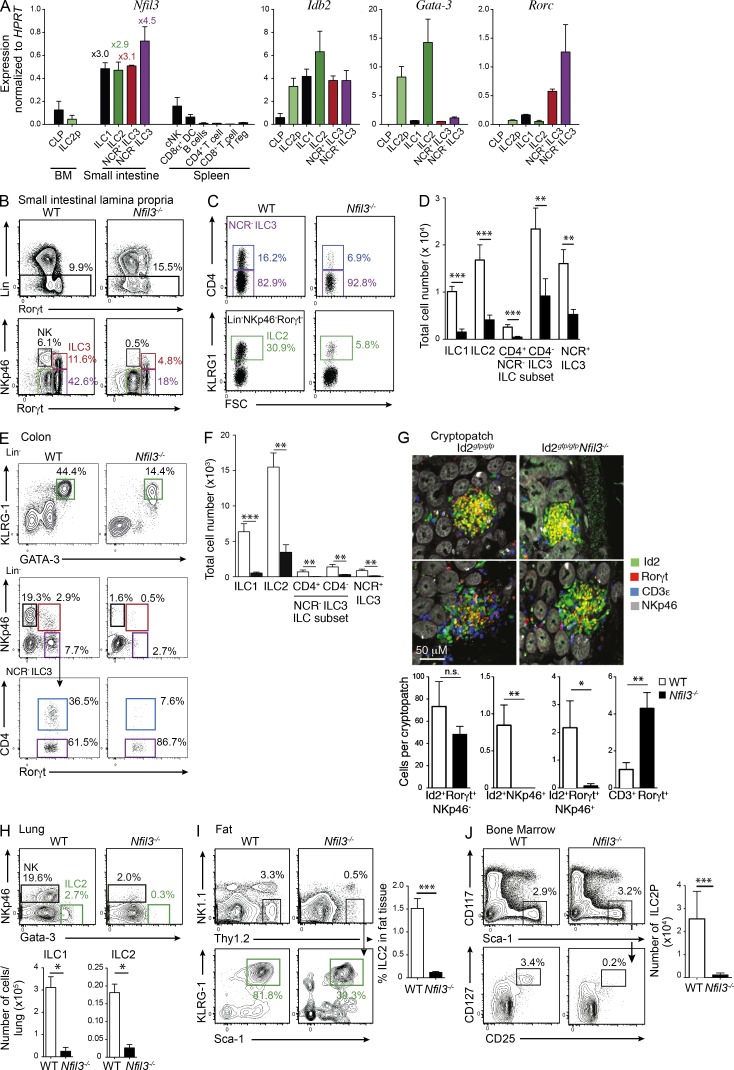
Due to the requirement of LTi cells, especially CD4+ LTi cells, for the proper generation of Peyer’s patches (Finke et al., 2002) and the high level of Nfil3 expression observed in all ILC populations (Fig. 2 A), including the ILC3-committed progenitor (Possot et al., 2011), we investigated whether Nfil3 was required for the development of the ILC subsets by measuring the prevalence of each subset in Nfil3−/− mice. We observed an overall reduction in the number of Rorγt+ ILCs in the small intestine (Fig. 2, B–D), colon (Fig. 2, E and F), and cryptopatches (Fig. 2 G) and in the absence of Nfil3 when compared with WT (C57BL/6, WT) mice although the number and frequency of Rorγt+CD4+ T cells was increased in the intestine of Nfi3-deficient mice. The size and number of lymphoid cells in the intestinal cryptopatches appeared unchanged between WT and Nfil3−/− mice, suggesting that Nfi3-deficient ILC have a similar capacity to form cryptopatches. Interestingly, within the ILC3 populations, the CD4+ subset was the most severely affected, exhibiting a >5–10-fold reduction compared with ∼2- and 3-fold difference in CD4− and NCR+ ILC subsets, respectively. This population is pivotal to Peyer’s patch development and its loss may significantly contribute to the altered development of the patches. In addition to the previously known defect in ILC1 development (Fuchs et al., 2013), the ILC2 population was the most dramatically affected by Nfil3 deficiency. We also observed a dramatic reduction in ILC2 (∼5-fold loss) in the small intestinal lamina propria (Fig. 2, C and D), as well in the lung (Fig. 2 H) and visceral adipose tissue (Fig. 2 I). In summary, our data indicate that Nfil3 is required for the development of all ILC populations.
Figure 2.
Nfil3 is essential for the development of innate lymphocytes. (A) Quantitative PCR analysis of Nfil3, Idb2 (Id2), Gata3, and Rorγt in the indicated populations purified from the indicated tissues of Rorγtgfp/+ mice. The fold change relative to conventional NK cells is shown for each ILC subset. Data show the mean ± SEM of samples analyzed in triplicate, pooled from two independent experiments. (B) Flow cytometric analysis of total hematopoietic cells (top) and Lin− (CD3−CD19−) CD45+ cells (bottom) isolated from the small intestine of WT or Nfil3-deficient mice and stained for intracellular expression of Rorγt. (C) Frequency of CD4+ (top) and KLRG1+ (bottom) cells within the NKp46−Rorγt+ ILC3 and NKp46−Rorγt− cells, respectively. (D) Enumeration of total ILC1 (CD45+Lin−Rorγt−NKp46+), ILC2 (CD45+Lin−Rorγt−NKp46−KLRG1+), CD4+ or CD4− ILC3 (CD45+Lin−Rorγt+NKp46−), and NCR+ ILC3 (CD45+Lin−Rorγt+NKp46+) subsets isolated from the small intestine of WT and Nfil3−/− mice. Data are pooled from three experiments (n = 3–15 mice/genotype). Statistical differences were tested using an unpaired student’s t test. (E) Representative flow cytometric profiles of ILC populations isolated from the lamina propria of the colon. (F) Total cell number and frequency of ILC populations in colon. (E and F) Data show the mean ± SEM 4 mice/genotype from one of two similar experiments.. (G) Immunohistological (top) and cellular composition (bottom) of cryptopatches in Id2gfp/gfpNfil3+/+ and Id2gfp/gfpNfil3−/− mice. Sections were stained with anti-Rorγt, CD3ε, and NKp46 and confocal images were acquired. Id2-GFP represents endogenous expression. Sections show 2 of 5 mice analyzed for each genotype. Bar, 50 µm. Enumeration of the ILC and lymphocytes in cryptopatches show mean ± SEM of 15 images/genotype. Statistical differences were determined using an unpaired Student’s t test. (H) Expression of NKp46 and Gata-3 in Lin−(CD3,CD19,Gr1) CD45+ cells isolated from the lungs of WT or Nfil3−/− mice (left) and total number of ILC1 (CD45+Lin− NKp46+) and ILC2 (CD45+Lin−NKp46− Gata-3+) recovered from lungs of WT or Nfil3−/− mice (right). Data are representative of 3 independent experiments (n = 4 mice/genotype). (I) Flow cytometric analysis of ILC2 (CD45+Lin−Thy1.2+KLRG1+Sca-1+) localized to visceral adipose tissue (left) and their frequency (right). Data are pooled from 2 independent experiments (n = 4 mice/genotype). (J) Frequency (left) and number (top right) of ILC2P (CD3−CD19−Gr1−CD11b−B220−Ter119−CD122−Sca-1+CD127+CD25+) isolated from bone marrow of WT and Nfil3−/− mice. Id2-GFP expression within the ILC2P populations (bottom left). Data are pooled from 3 experiments showing the mean ± SEM (n = 5 WT; n = 10 Nfil3−/− mice). Statistical differences were calculated using a two-tailed Mann-Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The fact that all ILC subsets are affected by the loss of Nfil3 expression suggested that the development of a common precursor might be impaired in Nfil3-deficient mice. To date, the proposed common innate lymphoid progenitor (CILP) has not been identified, thus precluding the definitive analysis of the role of Nfil3 in the development of this elusive ILC precursor. However, the precursor of the ILC2 (ILC2P) has recently been defined as a Lin−CD25+CD127+Sca1+Gata-3+Id2+ subset in the bone marrow (Hoyler et al., 2012). Analysis of lineage negative bone marrow of Id2gfp/gfpNfil3+/+ and Id2gfp/gfpNfil3−/− mice showed that ILC2Ps were severely reduced in the absence of Nfil3 (Fig. 2 J). We speculated that this early loss of the ILC2P would likely impair the capacity to generate the peripheral ILC2 pool, although a very small number of residual ILC2P were detected in Nfil3−/− mice, and these cells maintained their expression of Id2-GFP. Thus, loss of Nfil3 results in impaired development of multiple ILC populations including the early precursor population required for the development of all ILC2.
Cell-intrinsic Nfil3 is required for ILC development
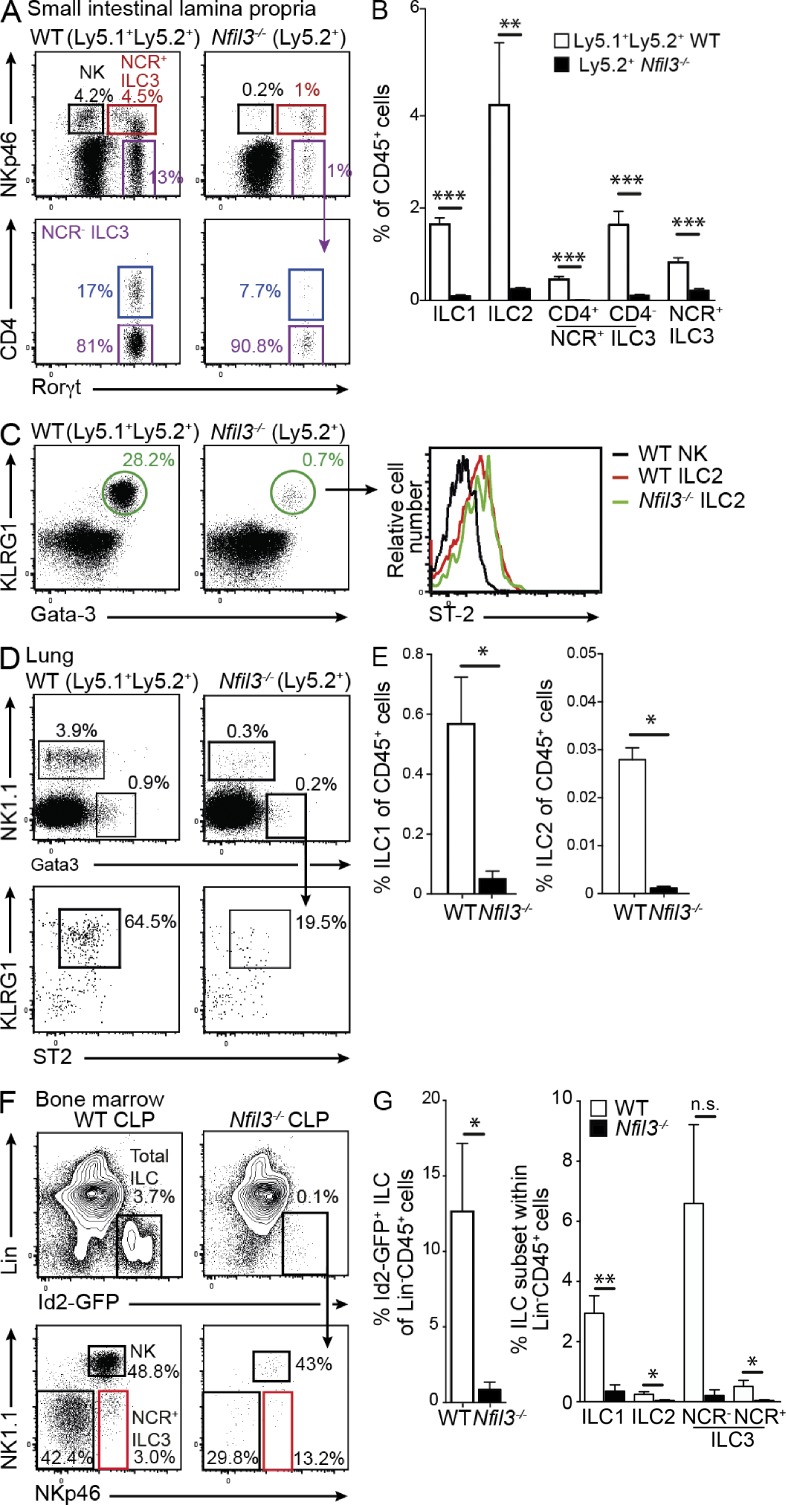
To determine whether the phenotype we observed in Nfil3−/− mice was a direct result of a cell-intrinsic role for Nfil3, we generated mixed bone marrow chimeras in which we transplanted lethally irradiated Ly5.1+/+ recipient mice with a 1:1 ratio of WT (Ly5.1+/Ly5.2+) and Nfil3−/− (Ly5.2+) bone marrow (Fig. 3, A and B). After 8 wk, WT cells effectively reconstituted the different ILC subsets in gut and lung, but Nfil3-deficient cells failed to do so and showed more than a 10-fold reduction in the competitive setting (Fig. 3, A–E). Other hematopoietic lineages such as mature splenic B cells that do not rely on Nfil3 for development, however, reconstituted at the expected ratio of 1:1 (unpublished data) indicating that the developmental defect was specific to ILCs.
Figure 3.
Nfil3 regulation of ILC development is cell intrinsic. Lethally irradiated WT (Ly5.1+) recipient mice were reconstituted with an equal mix of WT (Ly5.1+Ly5.2+) and Nfil3−/− (Ly5.2+) bone marrow cells. After 8 wk, the proportion of ILC1, ILC2, and ILC3 were determined in the small intestinal lamina propria (A–C) and the lung (D and E). (A and C) Dot plots show representative profiles of ILC3 (A) and ILC2 (C, left) in Nfil3-sufficient and -deficient compartments gated on Lin− (CD3−CD19−) CD45+ hematopoietic cells. (B) Frequency of ILC subsets within the WT (Ly5.1+Ly5.2+) and Nfil3−/− (Ly5.2+/+) populations in mixed chimeric mice. Data show the mean ± SEM (n = 3 mice/genotype) of one representative of two experiments. Statistical differences were tested using an unpaired student’s t test. (C) Expression of ST-2 within the KLRG1+Gata-3+ ILC2 subset (right). (D) Expression of NK1.1 and Gata-3 in Lin− (CD3−CD19−Gr1−) CD45+ cells isolated from the lungs and KLRG1 and ST2 expression on ILC2 (bottom). (E) Frequency of the different ILC1 and ILC2 in lung in each WT (Nfil3+/+) and Nfil3−/− compartment in mixed chimeras. (A–E) Analyses show representative profiles of two experiments with 6 mice/genotype. (F and G) Flow cytometrically purified CLP (Lin−CD127+Flt3/Flk2+Sca1intCD117int) isolated from bone marrow of Id2gfp/gfp mice were adoptively transferred into Rag2γc−/− mice. After 2 wk, the reconstitution of ILC subsets in the small intestinal lamina propria was analyzed. Representative flow cytometric plots show the proportion of total Id2-GFP+CD45+Lin− (CD3−CD19−CD11c−Gr1−) ILC (top) and ILC subsets within that gate (bottom) in WT and Nfil3−/− mice. (F) Data show representative flow cytometric profiles from (G) individuals pooled from two experiments and show mean ± SEM (n = 4 mice/genotype). Statistical differences were tested using an unpaired Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., not significant.
We recently showed that Nfil3 is already expressed in the common lymphoid progenitor (CLP) but that their development was not affected in Nfil3-deficient mice (Seillet et al., 2014). In addition to total bone marrow chimeras, CLP were purified from bone marrow of Id2gfp/gfpNfil3+/+ or Id2gfp/gfpNfil3−/− mice and adoptively transferred into Rag2−/−γc−/− recipient mice that lack T and B cells, as well as all ILC populations (Fig. 3, F and G). We observed that Nfil3-deficient Id2gfp/gfpNfil3−/− CLPs failed to efficiently give rise to ILC subsets after adoptive transfer. Thus, loss of ILC in mucosal tissues in the absence of Nfil3 arose from a failure in differentiation after the CLP stage and is not due to a defect in the hematopoietic stem cell and/or multipotent progenitor compartment.
Nfil3-regulated ILC populations play important roles at mucosal surfaces
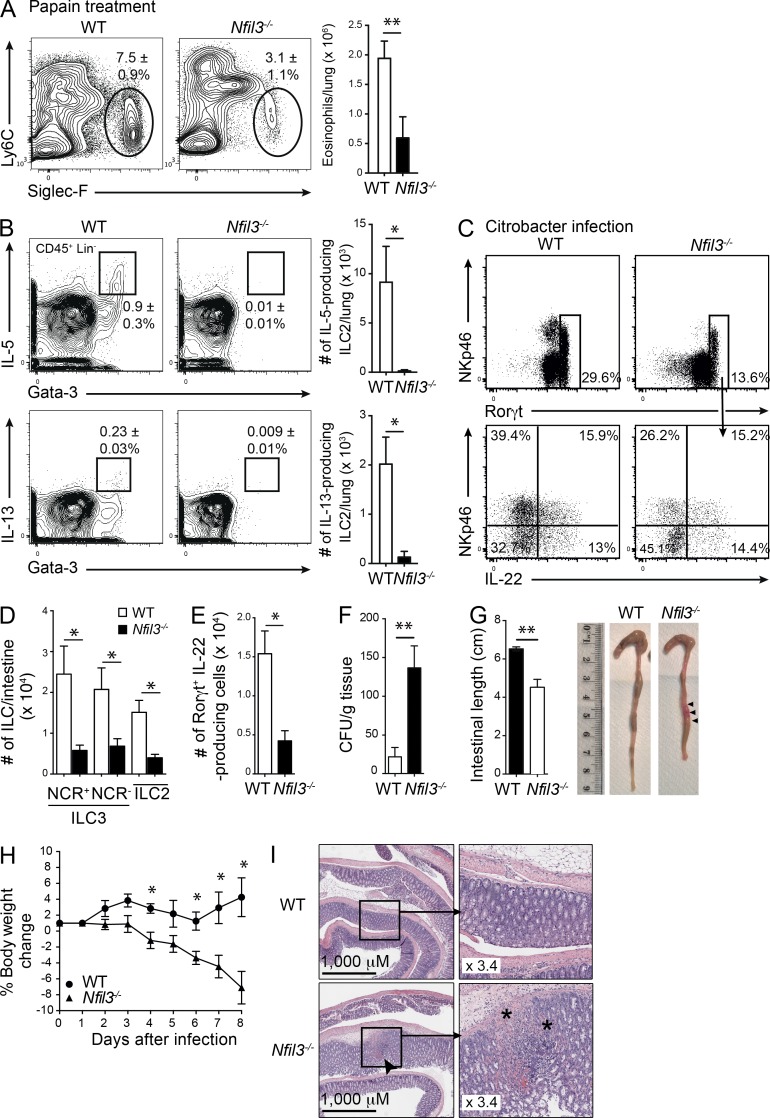
ILCs are localized in mucosal tissues and are thought to play important roles in immune protection. Therefore, we sought to determine whether loss of ILC subsets that accompanies Nfil3-deficiency influences the capacity to mount protection in the lung and gut. First, we examined whether the absence of Nfil3 altered the inflammation induced by intranasal challenge with papain. At steady state, the total number and frequency of eosinophils was similar in both Nfil3−/− and WT mice (Nfil3−/− 3.1 ± 0.3 × 105, 1.5 ± 0.3%; C57BL/6 3.3 ± 0.6 × 105,1.3 ± 0.1%) and the number of Nfil3−/− eosinophils was similar to WT eosinophils in papain-treated mixed bone marrow chimeras (unpublished data). We first measured eosinophil recruitment which was reduced by ∼2-fold in Nfil3−/− mice, indicating a defect in ILC2 activity (Fig. 4 A). In treated mice, ILC2 in the lungs of both WT and Nfil3-deficient mice were analyzed and, as anticipated, ILC2 were barely detectable in Nfil3-deficient mice (Fig. 4 B). This coincided with ablation of IL-5– and IL-13–producing ILC2 in the absence of Nfil3, explaining the reduced eosinophil recruitment (Fig. 4 B, right). Next, we investigated the mucosal response to C. rodentium infection in the gut of WT and Nfil3−/− mice (Fig. 4, C–G). In the absence of Nfil3, the total number of IL-22–producing ILC3 was significantly reduced (Fig. 4, D and E), indicating that although most ILC3 were lost in the absence of Nfil3, the remaining Rorγt+ ILC3 retained a capacity to produce IL-22 but this was not sufficient to prevent enhanced bacterial translocation (Fig. 4 F), shortening of the colon (Fig. 4 G), and weight loss (Fig. 4 H) in the Nfil3-deficient mice. These effects were accompanied by erosion of the intestines in Nfil3−/− mice but not in WT mice (Fig. 4 I). Collectively, these findings indicate that loss of Nfil3 results in the failure to mount a protective mucosal immune response due to significant reduction in ILC development.
Figure 4.
Nfil3−/− mice fail to mount effective immune responses in the absence of ILC2 and ILC3. (A) Representative flow cytometric profiles of eosinophils recovered in the lungs of Papain-treated WT and Nfil3−/− mice 1 d after the final treatment (left). Profiles are gated on CD45+ total live cells. Total number of eosinophils recovered from lungs showing mean ± SEM (right). (B) Expression of IL-5 and IL-13 among CD45+CD3−CD19− hematopoietic cells stained for intracellular Gata-3 (left). Total number of IL-5– and IL-13–expressing ILC2 in the lung (right). (A and B) Data show the mean ± SEM of data from 1 of 2 independent experiments (n = 4 mice/genotype). (C) IL-22 cytokine production by ILCs isolated from the small intestine of WT and Nfil3−/− mice gated on Lin− (CD3−CD19−) CD45+ cells 8 d after intragastric infection with C. rodentium. Cells were stimulated with IL-23 ex vivo in the presence of Brefeldin A. (D) Enumeration of the number of ILCs found in the intestine 8 d after infection of Nfil3−/− and WT mice with C. rodentium. (E) Enumeration of the number of IL-22–producing ILC3 cells recovered from the small intestine after infection. (F) Bacterial load in spleen, (G) colon length, and (H) weight loss in WT and Nfil3−/− mice after C. rodentium infection. (I) Representative H&E staining of the colon of C. rodentium–infected WT and Nfil3−/− mice on day 8. Bar, 1,000 µm; inset magnification 3.4×. Arrow indicates site of damage; * shows damage and lymphocyte infiltration. (C–I) Data are representative of two independent experiments (n = 3–4 mice/genotype). Error bars show the mean ± SEM. Statistical differences were tested using an unpaired Student’s t test. n.s. not significant. *, P < 0.05; **, P < 0.01.
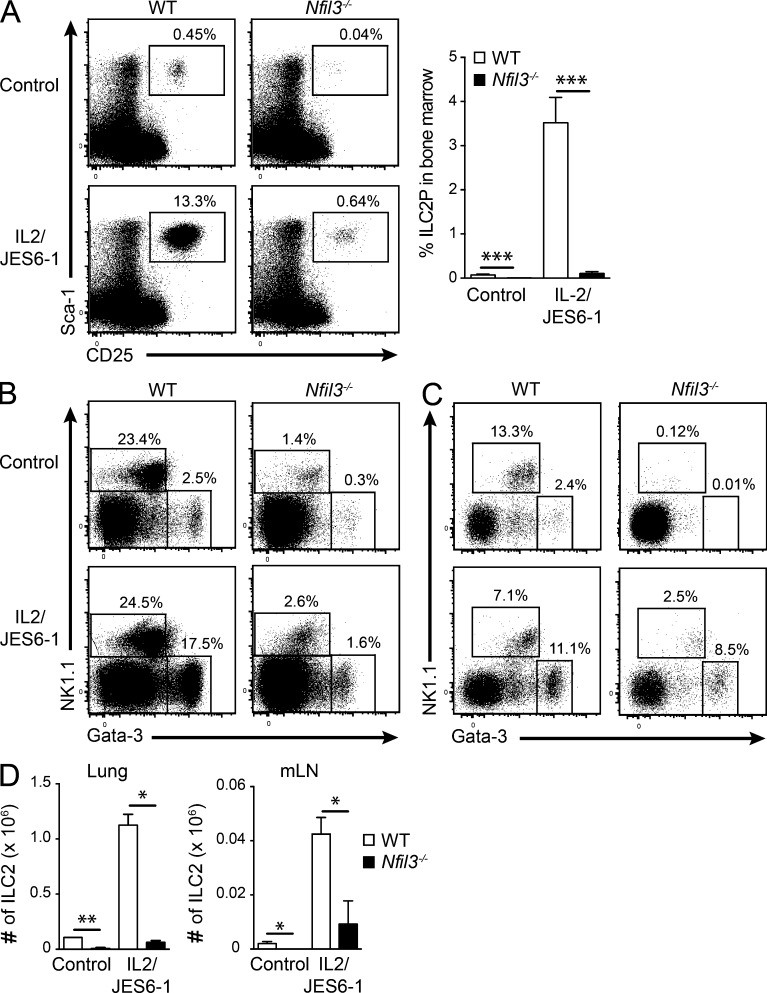
Interestingly, a very small but distinct population of ILCs remained in Nfil3−/− mice, suggesting that if ILCs can pass early checkpoints during development, they do not require Nfil3 for maintenance in the periphery, as has recently been showed in mature NK cells (Firth et al., 2013). Thus, we speculated that IL-2–IL-2mAb complex treatment, which can expand NK cells and ILC2 (Boyman et al., 2006; Nussbaum et al., 2013), would allow us to test their requirement for Nfil3 in mature ILC2. The frequency and the total number of ILC2 in the lung and mesenteric LNs (mLN) were analyzed (Fig. 5, A–D). Strikingly, ILC2 and ILC2P expanded ∼10-fold compared with untreated controls in both WT and Nfil3-deficient mice. IL-2/IL-2mAb treatment was not sufficient to return ILC2 cell numbers to the levels observed in WT mice, but a comparable fold expansion was observed in both Nfil3−/− and WT ILC2, indicating that these mature ILC2 had a similar ability to expand. Thus, residual Nfil3−/− ILC2 responded effectively to normal cytokine cues suggesting that they do not further require Nfil3 after activation or expansion in the periphery.
Figure 5.
Expansion of ILC populations after IL-2–IL-2mAb complex stimulation. WT and Nfil3−/− mice were injected i.p. for 2–3 wk with PBS or IL-2–JES6-1 to stimulate ILC expansion. (A) The proportion of ILC2P (CD3−CD19−Gr1−CD11b−B220−Ter119−CD122− cells) in the bone marrow, (B) ILC1 and ILC2 (CD45+CD3−CD19−Gr1− cells) in lung, and (C) mesenteric LNs (mLN) of cytokine complex-treated and control PBS-treated WT and Nfil3−/− mice. (D) Total number of ILC1 and ILC2 in the lung and mLNs of treated mice. (A–D) Data show representative flow cytometric profiles or mean ± SEM pooled from 2 independent experiments (n = 4 mice/genotype). Statistical differences were calculated using a two-tailed Mann-Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Several transcription factors have emerged as critical for the development of the ILC family, pointing to the complex regulation required for the generation of individual specialized subsets. Our findings identify Nfil3 as an essential factor for the development of innate immunity by impacting on all ILC subsets. Unlike loss of Id2 and Rorγt, in which secondary lymphoid tissues such as LNs and Peyer’s patch do not form, rudimentary Peyer’s patch were found in the absence of Nfil3, albeit smaller and less prevalent, than in WT mice. Interestingly, LNs appeared relatively unaffected by the loss of Nfil3, suggesting that Nfil3 acts specifically in Peyer’s patch organogenesis. Although the residual ILCs present at these sites remained functional in terms of cytokine production (IL-5, IL-13, and IL-22) in both naive (unpublished data) and challenged mice, their dramatic decrease in number reduced protective immune responses during lung and intestinal inflammation in the Nfil3−/− mice. Thus, as has recently been reported in NK cells (Firth et al., 2013; Seillet et al., 2014), it appears that Nfil3 is not absolutely required in mature ILCs at steady state, but most likely plays an essential role early in the development of ILC precursors, as we have demonstrated for the ILC2 lineage.
MATERIALS AND METHODS
Mice.
Id2gfp mice (Seillet et al., 2013), Rorc(γt)gfp mice (Eberl et al., 2004), and Nfil3−/− mice (Gascoyne et al., 2009) are on a C57BL/6 background and have been previously described. All mouse strains were bred and maintained in-house. Mice were used at 8–12 wk old. All procedures involving animals were approved by the Animal Ethics Committee of the Walter and Eliza Hall Institute of Medical Research.
Generation of bone marrow chimeras.
Mixed bone marrow chimeras were established by reconstituting lethally irradiated (2 × 0.55 Gy) Ly5.1+ recipient mice with a 1:1 mixture of Nfil3−/− (2.5 × 106) and WT (Ly5.1+Ly5.2+, 2.5 × 106) bone marrow cells and allowed to reconstitute for 8 wk.
For experiments in which the CLP were adoptively transferred into recipient mice, Lin−IL-7Rα+Flt3/Flk2+Sca1intCD117int bone marrow cells were purified by flow cytometric sorting from Id2gfp/gfpNfil3+/+ or Id2gfp/gfpNfil3−/− mice after lineage depletion, as previously described (Carotta et al., 2011). 3 × 104 CLPs were injected in Rag2γc−/− mice and ILC populations were analyzed 2 wk after transfer.
Tissue preparation.
Mononuclear cells from intestinal lamina propria were isolated as previously described (Mielke et al., 2013; Rankin et al., 2013). Lungs were cut into small fragments and digested for 30 min at 37°C with Collagenase III (1 mg/ml; Worthington) and DNase I (200 µg/ml; Roche). Cells were filtered by using a cell strainer after red blood cell lysis. Perigonadal adipose tissue was used as representative visceral adipose tissue. Adipose tissue was finely dissected with a scalpel blade and digested with 3 ml of PBS containing 0.2 mg/ml Collagenase IV and 4% BSA at 37°C for 45 min with gentle agitation. Digests were filtered through 70-µm cell strainers and centrifuged at 800 g for 15 min to enrich for immune cells in stromal vascular fractions. Single-cell suspensions were blocked with PBS containing 5 µg/ml anti-CD16/CD32 (2.4G2) and stained for 30 min on ice with fluorophore-conjugated antibodies. The following antibodies were used for the identification and purification of ILC2 cells: CD19 (ID3), B220 (RA3-6B2), CD3 (145-2C110), CD4 (GK1.4), CD11b (M1/70), Gr-1 (RB6-8C5), TCRβ (H57-597), NKp46 (29A1.4), NK1.1 (PK136), CD45.1 (A20), CD45.2 (104), CD117 (2B8), CD127 (A7R34), Sca1 (E13-161.7), KLRG1 (2F1), Thy1.2 (30H12), and ST2 (DJ8 or RMST2-2). Intracellular staining was performed using the Transcription Factor Staining Buffer Set (eBioscience) and antibodies to Gata-3 (TWAJ), Rorγt (AFKJS-9), IL-13 (eBio13A), and IL-5 (TRFK5). Intracellular cytokine staining for IL-13 and IL-5 was performed after stimulation for 4 h with PMA (50 ng/ml) and ionomycin (100 ng/ml), in the presence of Brefeldin A (1 mg). Cells were analyzed using a FACSCanto II (BD), and FlowJo software (Tree Star) was used for analysis. Flow cytometric sorting was performed with a FACSAria (BD).
Infection with Citrobacter rodentium.
Mice were inoculated with 2 × 109 colony forming units of C. rodentium by oral gavage. Mice were analyzed 8 d after infection and bacterial dissemination was determined by culture of livers and spleens from infected mice on agar plates containing nalidixic acid as previously described (Rankin et al., 2013).
Papain-induced lung inflammation.
Mice were treated intranasally with PBS or 20 µl of papain (5 mg/ml diluted in PBS) on three consecutive days. 24 h after the final treatment, lung cells were isolated and stained for analysis by flow cytometry with antibodies for CD45, CD11c, and Siglec F to identify infiltrating eosinophils.
In vivo treatment with IL-2/–anti–IL-2 complexes.
For expansion of ILC populations, mice were treated with IL-2–JES6-1 complexes as previously described (Boyman et al., 2006). For each injection, IL-2 (1 µg; PeproTech) was complexed with 5 µg JES-1 mAb (WEHI) at a molar ratio of 2:1 and incubated for 30 min at 37°C. Aliquots were stored at -70°C and resuspended in PBS immediately before use.
Quantitative RT-PCR.
Total RNA was prepared from purified ILC populations using the RNeasy mini kit (QIAGEN), and then cDNA was synthesized with oligo(dT) and Thermoscript reverse transcription (Invitrogen). Real-time PCR was done using SensiMix SYBR no-Rox kit (Bioline) and the following primer pairs: Nfil3 forward, 5′-GAACTCTGCCTTAGCTGAGGT-3′, Nfil3 reverse 5′-ATTCCCGTTTTCTCCGACACG-3′; Id2 forward, 5′-ACCAGAGACCTGGACAGAAC-3′, Id2 reverse, 5′-AAGCTCAGAAGGGAATTCAG-3′; Gata-3 forward, 5′-CTACCGGGTTCGGATGTAAG-3′, Gata-3 reverse, 5′-TGCTAGACATCTTCCGGTTTC-3′; Hprt forward, 5′-GGGGGCTATAAGTTCTTTGC-3′, Hprt reverse, 5′-TCCAACACTTCGAGAGGTCC-3′.
Analyses were done in triplicate and normalized mean expression was calculated using the Q-Gene application with Hprt as the reference gene (Simon, 2003).
Statistical analysis.
GraphPad Prism software was used for statistical analysis. The two-tailed Mann-Whitney U-test or unpaired Student’s t test were used for comparisons. All results are expressed as mean ± SEM.
Acknowledgments
We thank Ajith Vasanthakumar for technical advice and are indebted to the facilities of our institute, particularly those responsible for animal husbandry and flow cytometry.
L.C. Rankin was supported by a National Health and Medical Research Council (NHMRC; Australia) Dora Lush Training Scholarship; J.R. Groom was supported by an NHMRC Overseas Postgraduate Training Fellowship; L.A. Mielke was supported by a Cancer Australia grant (APP1050241); G.T. Belz was supported by an Australian Research Council Future Fellowship; and S. Carotta was supported by an NHMRC Career Development Fellowship (APP1011808). This work was supported by NHMRC project grants (APP1027472 and APP1047903) of the NHMRC of Australia, Victorian State Government Operational Infrastructure Support, and Australian Government NHMRC IRIIS.
The authors declare no competing financial interests.
Author contributions: C. Seillet, L.C. Rankin, J.R. Groom, L.A. Mielke, M. Chopin, J. Tellier, N.D. Huntington, G.T. Belz and S. Carotta designed, performed, interpreted the experiments and contributed helpful suggestions for the manuscript. G.T. Belz and S. Carotta wrote the paper.
Footnotes
Abbreviations used:
- CLP
- common lymphoid progenitor
- Id2
- inhibitor of DNA binding 2
- ILC
- innate lymphoid cell
- LTi
- lymphoid tissue-inducer
- mLN
- mesenteric LN
- NCR
- natural cytotoxicity receptor
- Rorγt
- retinoid-related orphan γt
References
- Boyman, O., Kovar M., Rubinstein M.P., Surh C.D., and Sprent J.. 2006. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 311:1924–1927 10.1126/science.1122927 [DOI] [PubMed] [Google Scholar]
- Carotta, S., Pang S.H., Nutt S.L., and Belz G.T.. 2011. Identification of the earliest NK-cell precursor in the mouse BM. Blood. 117:5449–5452 10.1182/blood-2010-11-318956 [DOI] [PubMed] [Google Scholar]
- Eberl, G., Marmon S., Sunshine M.-J., Rennert P.D., Choi Y., and Littman D.R.. 2004. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat. Immunol. 5:64–73 10.1038/ni1022 [DOI] [PubMed] [Google Scholar]
- Finke, D., Acha-Orbea H., Mattis A., Lipp M., and Kraehenbuhl J.. 2002. CD4+CD3− cells induce Peyer’s patch development: role of alpha4beta1 integrin activation by CXCR5. Immunity. 17:363–373 10.1016/S1074-7613(02)00395-3 [DOI] [PubMed] [Google Scholar]
- Firth, M.A., Madera S., Beaulieu A.M., Gasteiger G., Castillo E.F., Schluns K.S., Kubo M., Rothman P.B., Vivier E., and Sun J.C.. 2013. Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med. 210:2981–2990 10.1084/jem.20130417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, A., Vermi W., Lee J.S., Lonardi S., Gilfillan S., Newberry R.D., Cella M., and Colonna M.. 2013. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-γ-producing cells. Immunity. 38:769–781 10.1016/j.immuni.2013.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoyne, D.M., Long E., Veiga-Fernandes H., de Boer J., Williams O., Seddon B., Coles M., Kioussis D., and Brady H.J.. 2009. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat. Immunol. 10:1118–1124 10.1038/ni.1787 [DOI] [PubMed] [Google Scholar]
- Hoyler, T., Klose C.S., Souabni A., Turqueti-Neves A., Pfeifer D., Rawlins E.L., Voehringer D., Busslinger M., and Diefenbach A.. 2012. The transcription factor GATA-3 controls cell fate and maintenance of type 2 innate lymphoid cells. Immunity. 37:634–648 10.1016/j.immuni.2012.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikushima, S., Inukai T., Inaba T., Nimer S.D., Cleveland J.L., and Look A.T.. 1997. Pivotal role for the NFIL3/E4BP4 transcription factor in interleukin 3-mediated survival of pro-B lymphocytes. Proc. Natl. Acad. Sci. USA. 94:2609–2614 10.1073/pnas.94.6.2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada, M., Levy D.M., McKeag L., Murray K., Schröder A.J., Canfield S.M., Traver G., and Rothman P.B.. 2010. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc. Natl. Acad. Sci. USA. 107:821–826 10.1073/pnas.0909235107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada, M., Pham N.-L.L., Pewe L.L., Harty J.T., and Rothman P.B.. 2011. NFIL3/E4BP4 is a key transcription factor for CD8α+ dendritic cell development. Blood. 117:6193–6197 10.1182/blood-2010-07-295873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T., Matsuoka K., Sheikh S.Z., Elloumi H.Z., Kamada N., Hisamatsu T., Hansen J.J., Doty K.R., Pope S.D., Smale S.T., et al. 2011. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J. Immunol. 186:4649–4655 10.4049/jimmunol.1003888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielke, L.A., Groom J.R., Rankin L.C., Seillet C., Masson F., Putoczki T., and Belz G.T.. 2013. TCF-1 controls ILC2 and NKp46+RORγt+ innate lymphocyte differentiation and protection in intestinal inflammation. J. Immunol. 191:4383–4391 10.4049/jimmunol.1301228 [DOI] [PubMed] [Google Scholar]
- Motomura, Y., Kitamura H., Hijikata A., Matsunaga Y., Matsumoto K., Inoue H., Atarashi K., Hori S., Watarai H., Zhu J., et al. 2011. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat. Immunol. 12:450–459 10.1038/ni.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussbaum, J.C., Van Dyken S.J., von Moltke J., Cheng L.E., Mohapatra A., Molofsky A.B., Thornton E.E., Krummel M.F., Chawla A., Liang H.E., and Locksley R.M.. 2013. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 502:245–248 10.1038/nature12526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possot, C., Schmutz S., Chea S., Boucontet L., Louise A., Cumano A., and Golub R.. 2011. Notch signaling is necessary for adult, but not fetal, development of RORγt(+) innate lymphoid cells. Nat. Immunol. 12:949–958 10.1038/ni.2105 [DOI] [PubMed] [Google Scholar]
- Rankin, L.C., Groom J.R., Chopin M., Herold M.J., Walker J.A., Mielke L.A., McKenzie A.N., Carotta S., Nutt S.L., and Belz G.T.. 2013. The transcription factor T-bet is essential for the development of NKp46+ innate lymphocytes via the Notch pathway. Nat. Immunol. 14:389–395 10.1038/ni.2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seillet, C., Jackson J.T., Markey K.A., Brady H.J., Hill G.R., Macdonald K.P., Nutt S.L., and Belz G.T.. 2013. CD8α+ DCs can be induced in the absence of transcription factors Id2, Nfil3, and Batf3. Blood. 121:1574–1583 10.1182/blood-2012-07-445650 [DOI] [PubMed] [Google Scholar]
- Seillet, C., Huntington N.D., Gangatirkar P., Axelsson E., Minnich M., Brady H.J., Busslinger M., Smyth M.J., Belz G.T., and Carotta S.. 2014. Differential requirement for Nfil3 during NK cell development. J. Immunol. 192:2667–2676 10.4049/jimmunol.1302605 [DOI] [PubMed] [Google Scholar]
- Simon, P.2003. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics. 19:1439–1440 10.1093/bioinformatics/btg157 [DOI] [PubMed] [Google Scholar]
- Spits, H., Artis D., Colonna M., Diefenbach A., Di Santo J.P., Eberl G., Koyasu S., Locksley R.M., McKenzie A.N., Mebius R.E., et al. 2013. Innate lymphoid cells—a proposal for uniform nomenclature. Nat. Rev. Immunol. 13:145–149 10.1038/nri3365 [DOI] [PubMed] [Google Scholar]