Summary
Excess red light induces a defence response by activating lipoxygenase at both the transcript and activity levels.
Key words: Arabidopsis, defence response, excess red light, lipoxygenase, MAPK, phytochrome B.
Abstract
Lipoxygenase (LOX), a non-haem-iron-containing dioxygenase, is activated under various biotic or abiotic stresses to trigger a series resistance response, but the molecular mechanism of LOX activation remains unclear. This work investigated the activation of LOX during the plant defence response induced by excess red light (RL). In conditions of RL-induced defence, Arabidopsis LOX activity and transcription levels of LOX2, LOX3, and LOX4 were both upregulated. Under RL, phytochrome B promoted the degradation of phytochrome-interacting factor 3 (PIF3), a factor that inhibited the expression levels of LOXs, and thus the transcription levels of LOX2, LOX3, and LOX4 were increased. Upon pathogen infection, the activity of mitogen-activated protein kinase 3 (MPK3) and MPK6 was increased in plants pre-treated with RL. Moreover, experiments with the inhibitor PD98059 and mutants mpk3 and mpk6-2 demonstrated that MPK3 and MPK6 were both responsible for LOX activation. Further results showed that, in response to RL, an increase in cytoplasmic calcium concentration and upregulation of calmodulin 3 (CaM3) transcript level occurred upstream of MPK3 and MPK6 activation. Collectively, these results suggested that activation of LOX both at the transcript level and in terms of activity modulates the defence response induced by RL, providing a new insight into the mechanistic study of LOX during plant defences.
Introduction
Plants, as sedentary organisms, have evolved a high flexibility in both metabolism and development to cope with the multiple environmental stimuli that they are exposed to (Genoud et al., 2002). Light signalling is fundamental to the growth and development of plants. Long-term exposure to either low light or excess white light (EL) has an adverse effect on plants. However, certain irradiation with moderate EL can enhance the plant defence response, and red light (RL) plays a major role (Szechyńska-Hebda et al., 2010). Phytochrome B (phyB), as the main receptor of RL, is essential for this process. Szechyńska-Hebda et al. (2010) found that photo-electrophysiological signalling is a component of signalling cascades that potentially regulates the defence response. However, the mechanisms mediating the defence response by phyB are still unclear. Plants monitor informational light signals from their surroundings using a range of sensory photoreceptors including phototrophin, crytochrome, and phytochrome (Leivar et al., 2012). RL and far-RL are sensed using the phytochrome family (phyA to phyE in Arabidopsis). Phytochromes perceive RL (660nm) and far-RL (720nm) of the solar spectrum, and monitor changes in light quality and quantity to control many aspects of growth and developmental responses such as germination, seedling de-etiolation, shade avoidance, and flowering time (Franklin and Quail, 2010; Strasser et al., 2010). Phytochromes photoconvert between two conformers reversibly: the inactive RL-absorbing Pr form and the biologically active far-RL-absorbing Pfr form. Photoconversion of Pr to Pfr takes place upon absorption of RL (Linschitz et al., 1966), and reversion of Pfr to Pr occurs in far-RL-enriched circumstances.
The Pr form of phytochromes is synthesized in the cytoplasm, and upon photoactivation to Pfr is translocated to the nucleus (Nagatani, 2004), where it associates with phytochrome-interacting factors (PIFs) (Soy et al., 2012). PIFs, a subset of basic helix–loop–helix transcription factors, preferentially bind with a G-box (CACGTG) DNA sequence element, which is a subclass of an E-box element (CANNTG) present in the promoters of some light-regulated genes. Interactions between the Pfr form of phyB and PIF3 bound to a G-box promoter motif are hypothesized to directly regulate transcript expression of light-responsive genes (Martínez-García et al., 2000; Quail, 2002).
Lipoxygenases (LOXs) catalyse peroxidation of many polyunsaturated fatty acids and some lipids to cause the production of oxylipins, a set of biologically active compounds (Yang et al., 2012). Oxylipins have many important physiological functions during signalling transduction in growth and development, senescence and death, and biotic or abiotic stress responses (Feussner and Wasternack, 2002; Porta and Rocha-Sosa, 2002; Duan et al., 2005; Liavonchanka and Feussner, 2006). The many different products of LOX could enhance the defence responses in plants, including direct inhibition of the pathogen and accumulation of phytoalexins (Alami et al, 1999; Lin and Ishii, 2009). There are six isoforms of LOX in Arabidopsis, and these can be classified as 9-LOXs or 13-LOXs according to the position at which oxygen is incorporated into substrates for LOX catalysis in plants (Feussner and Wasternack, 2002). LOX1 and LOX5 are 9-LOXs, while LOX2, LOX3, LOX4, and LOX6 are 13-LOXs (Bannenberg et al., 2009). As a key enzyme in the lipid peroxidation reaction, LOX plays an important role during the defence response. Its expression level dramatic rises in response to EL, indicating that it may play a role in this process (Rossel et al., 2007). However, whether LOX’s upregulation of transcript levels is induced by a specific spectrum or by EL in general is still unknown.
The LOX gene sequence may contain a G-box or a similar domain structure (Hou et al., 2010), which is assumed to combined with PIFs. This assumption provides a possible mechanism underlying the regulation of LOX gene expression by excess RL, i.e. RL promotes the degradation of PIFs, which suppress LOX transcription by combining with it, and the inhibited LOX is released, thus contributing to the increase in LOX transcript expression.
Protein kinases and phosphatases play a central role in signal transduction through the phosphorylation and dephosphorylation of proteins. The mitogen-activated protein kinase (MAPK) cascade, as the most conversed pathway, plays a crucial role in almost all eukaryotes by linking perception of external stimuli with changes in the cell (Taj et al., 2010). Each MAPK cascade consists of at least three kinases: MAPKKK, MAPKK, and MAPK. In the Arabidopsis genome, there are 20 MAPKs, 10 MAPKKs, and ~80 MAPKKKs (Colcombet and Hirt, 2008; Beckers et al., 2009). They play a pivotal role in the transduction of various extracellular stimuli, including many biotic and abiotic stresses, as well as a series of developmental responses (Taj et al., 2011b). Many studies in the literature have demonstrated that MAPKs (MPKs) take part in the regulation of innate immunity and adverse stress responses (Ichimura et al., 2002; Xing et al., 2008). Activation of MPK has been detected under different stimuli, for example, MPK3 and MPK6 are activated under some abiotic stresses, such as ethylene treatment, drought, or wounding; MPK6 is activated under heat shock; MPK4 and MPK6 are activated under conditions of cold, salt, or H2O2; and when under heavy metal stress, MPK2, MPK3, MPK4, and MPK6 are all activated. Infection with different kinds of pathogens can induce different pathways of MPKs. For example, fungal pathogens can induce MPK2, MPK3, MPK4, and MPK6 and bacterial pathogens can induce MPK2, MPK3, and MPK6 under normal circumstances (Taj et al., 2010). It is still unclear whether the most active MPK3 and MPK6 are activated during the enhancement of the defence response induced by RL. Ca2+ is a crucial second messenger (Hashimoto and Kudla, 2011), and previous studies have implicated it in the activation of MPK cascades during the response to various stimuli (Xing et al., 2008; Wang PC et al., 2010). The Ca2+ signatures are sensed, decoded, and transmitted to downstream signalling cascades by Ca2+ sensors. Calmodulin (CaM) acts as a prominent Ca2+ sensor protein in plant signal transduction. In Arabidopsis, CaM has several isoforms, and different isoforms interact with their particular targets upon different exogenous stimuli (Luan et al., 2002). The Ca2+ signature is believed to be necessary for the cellular signalling transduction in response to EL. However, analysis of the Ca2+–CaM response to RL, especially the signalling pathway leading to the defence reaction, is still lacking.
In this study, the possible molecular mechanisms underlying the process of RL-induced activation of LOX during enhancement of the defence response were investigated. Our results indicated that LOX was upregulated by RL at both the transcriptional level and activity level, and that it plays an important role in the RL-induced defence response.
Materials and methods
Plant materials and chemicals
Arabidopsis ecotype Columbia-0 (Col-0) and seeds of mutants phyB, mpk3, and mpk6-2 were obtained from the European Arabidopsis Stock Centre. phyB-ox-YFP (Wang FF et al., 2010), pif3 and pif3-ox-YFP (Soy et al., 2012) were sterilized and grown on solid Murashige and Skoog medium as described previously (Zhang and Xing, 2008). 4,′(2, 3-Dimethyltetramethylene)dipyrocatechol (NDGA) was obtained from Merk, linoleic acid, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetra-acetic acid (acetoxymethyl ester) (BAPTA-AM), and PD98059 were purchased from Sigma-Aldrich. Fluo-3-AM was obtained from Beyotime.
RL treatment
Arabidopsis rosettes were fully exposed to EL (1500 μmol photons m–2 s–1 for 1h) and excess RL (120 μmol photons m–2 s–1 for 4h, 650±20nm) supplied by light-emitting diode panels (Photon System Inst.). The above light conditions provided similar energy at the indicated spectral regions. Heat emission from the light source was insignificant.
Pathogen growth and inoculation
The bacterial strain used in this study was Pseudomonas syringae pv. tomato DC3000 (Pst-DC3000, virulent), and it was grown at 28 °C in King’s B medium supplemented with appropriate antibiotics. Overnight log-phase cultures were collected by centrifugation, washed with 10mM MgCl2, and then diluted to a final optical density at 600nm (OD600) of 0.01 (for appearance determination) and 0.0001 (for pathogen growth assay). The procedures of pathogen inoculation and bacteria growth assays were as described previously (Mishina and Zeier, 2007).
Callose staining
The leaves of approximately 4-week-old plants of wild-type (WT), phyB, or phyB-ox-YFP were fixed in ethanol:acetic acid (3:1, v/v) and stained with 0.01% (w/v) aniline blue (Millet et al., 2010). The leaves were mounted on slides, and callose was observed with UV excitation (Sun et al., 2012).
RNA extraction and reverse transcription (RT)-PCR analysis
Total RNAs were extracted from detached Arabidopsis leaves using Trizol according to the supplier’s recommendations. The concentration of RNA was determined by measuring absorbance at 260nm. First-strand cDNA was synthesized with a SuperScript II First-Strand Synthesis System for qRT-PCR (Invitrogen). Quantitative RT-PCR was performed using a Roche LightCyclerTM 2.0 Real-time Detection System. The expression of target gene was normalized relative to the housekeeping gene ACTIN2 (Zhou et al., 2013). The primers used are listed in Supplementary Table S1 at JXB online.
LOX activity assays
The assay of LOX activity was performed as described previously (Skórzyńska-Polit et al., 2006) with a minor modification. The plant material was deep frozen in liquid nitrogen and ground in 0.2M boric acid buffer at pH 7.0. The homogenate was centrifuged at 15 000g for 20min, and the supernatant was used for determination of protein concentration and LOX activity. Protein concentration was determined according to Bradford (1976) using a Bio-Rad protein assay using BSA as a standard. The activity of LOX was measured spectrophotometrically at 234nm as described previously (Skórzyńska-Polit and Krupa, 2003). The reaction mixture contained 0.2M boric acid buffer (pH 8.0), 25 μl of plant extract, and 25 μl of linoleic acid as a substrate in a final volume of 1ml. The reaction was carried out at 30 °C for 4min. LOX activity was expressed as absorbance increase mg–1 of protein min–1.
Treatment with LOX inhibitor and MAPK cascade inhibitor
Before RL treatment and the later inoculation of Pst-DC3000, the Arabidopsis leaves were pre-sprayed with a solution containing NDGA (an inhibitor of LOX; dissolved in DMSO) or PD98059 (an inhibitor of the MAPK pathway; dissolved in DMSO) for 60min (Samuel et al., 2000; Matsumura et al., 2003; Miles et al., 2004). NDGA was used at a final concentration of 100 μM to inhibit the activity of LOX. A concentration gradient experiment was carried out for inhibitor PD98059 (Fig. S3). The results indicated that a concentration of 20 µM PD98059 could markedly inhibit the induction of LOX activity by inhibiting kinase activity, so this concentration was used in subsequent experiments.
Western blot and MAPK activity assay
Proteins were extracted from detached Arabidopsis leaves at the indicated time points after different treatments. Protein extracts were separated on a 10% SDS-PAGE mini-gel and then analysed by Western blotting. For detection of the phosphorylated proportion of MAPKs, as described by Li et al. (2012), blots were probed with anti-ACTIVE MAP kinase polyclonal Ab (pTEpY; Cell Signaling Technology, MA, USA), which recognizes activated MAPKs. Western blot experiments were carried out to show the MPK3/6 activity of the mpk6-2 and mpk3 mutant lines (Fig. S4). The results showed that the mpk6-2 mutant did not have MPK6 activity (47kDa) and mpk3 mutants did not have MPK3 (43kDa) activity compared with the WT. Together, these results demonstrated that the two bands were MPK3 and MPK6, so we used this antibody in our experiments. Subsequently, the blots were washed and incubated with an anti-rabbit horseradish peroxidase-conjugated secondary antibody.
Measurement of cytosolic calcium concentration ([Ca2+]cyt)
The method for [Ca2+]cyt detection was based on previous work (Zhou et al., 2013). Ca2+ was stained with Fluo-3-AM, which is hydrolysed to yield Fluo-3 capable of indicating changes in [Ca2+]cyt. The fluorescence intensity of Fluo-3 was measured by flow cytometry analysis.
Chromatin immunoprecipitation (ChIP)
For ChIP analysis, samples (1.5g) were cross-linked with 10ml of 1% formaldehyde under vacuum infiltration conditions. ChIP assays were performed as described previously (Shin et al, 2007) with a minor modifications of the three wash buffers: low wash buffer (150mM NaCl, 0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris/HCl, pH 8), high-salt wash buffer (500mM NaCl, 0.1% SDS, 1% Triton X-100, 2mM EDTA, 20mM Tris/HCl, pH 8), and LiCl wash buffer (0.25M LiCl, 1% v/v NP-40, 1% w/v sodium deoxycholate, 1mM EDTA, 10mM Tris/HCl, pH 8). The amount of each precipitated DNA fragment was determined by semi-quantitative PCR using LOX2, LOX3, and LOX4 primers.
Co-immunoprecipitation assay
A co-immunoprecipitation assay was performed as described previously (Liu et al., 2013) with some modifications. Total proteins were extracted from plants in extraction buffer [50mM Tris/HCl, pH 7.5–8.0, 100mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1mM EDTA, 1mM sodium orthovanadate, 50mM sodium fluoride, and 1mM phenylmethylsulfonyl fluoride, containing Protease Inhibitor Cocktail (Roche)]. Proteins extracts were inoculated with antibody for 4h, and protein A beads were then added. After incubation overnight at 4 °C, the beads were centrifuged and washed four times with PBS (pH 7.4). The immunoprecipitated proteins were detected by SDS-PAGE with an anti-ERK (extracellular signal-regulated kinase) antibody. Endogenous LOX protein was detected with a rabbit polyclonal anti-LOX antibody (Stenzel et al., 2003) after immunoprecipitation with anti-ERK antibody.
GenBank accession numbers
Sequence data from this article can be found in GenBank under the following accession numbers: PR1, AT2g14610; calmodulin 3 (CaM3), AT3g56800; Actin2, AT3g46520; LOX1, AT1g55020; LOX2, AT3g45140; LOX3, AT1g17420; LOX4, AT1g72520; LOX5, AT3g22400; LOX6, AT1g67560.
Results
EL protects Arabidopsis from pathogen infection, while RL is the main inducer
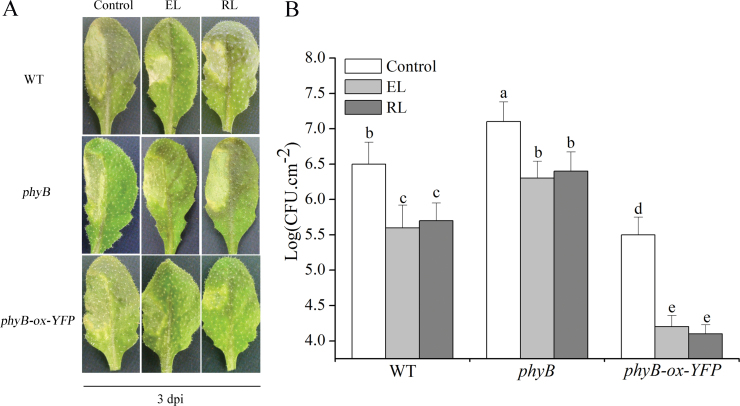
It has been demonstrated that EL regulates plant stress responses. The results of Szechyńska-Hebda et al. (2010) indicated that red but not blue EL induces a defence response. When Arabidopsis plants were inoculated with virulent Pst-DC3000, the WT plant leaves under normal light conditions turned yellow and finally wilted and died (Fig. 1A), whereas plants pre-irradiated with EL (1500 μmol m–2 s–1 for 1h) or excess RL (120 μmol m–2 s–1 for 4h; 660–680nm) showed minute yellow disease lesions at 3 d post-inoculation (dpi). The phyB null mutant plants showed more developed chlorotic lesions compared with WT plants, and RL-treated phyB plants showed no significant improvement on disease progression at 3 dpi. However, the chlorotic lesions were reduced in phyB-ox-YFP plants, which are transgenic plants overexpressing phyB::YFP (yellow fluorescent protein) fusion protein, and both EL and RL made this process more significant. In addition, the numbers of bacteria were significantly reduced in WT plants pre-irradiated with both EL and RL but not in phyB plants, compared with WT, and the reduction was more significant in phyB-ox-YFP plants (Fig. 1B). This finding was consistent with the disease symptoms shown in Fig. 1A. For the pathogen growth assays in Fig. 1B, the phyB Arabidopsis plants were more susceptible than WT plants, and either EL or RL had nearly the same impact on limitation of bacterial numbers. Together, these results illustrated that EL can induce an Arabidopsis defence response to pathogen and that excess RL is the main inducer, while phyB plays an important role during induction of the defence response.
Fig. 1.
Effect of exposure to EL and RL on disease progression in leaves of WT, phyB, and phyB-ox-YFP plants. (A) After exposure to EL (1500 μmol photons m–2 s–1for 1h), and excess RL (120 μmol photons m–2 s–1 for 4h), WT and phyB, phyB-ox-YFP plants were inoculated with virulent Pst-DC3000 (OD600=0.01 in 10mM MgCl2). Leaves were infected on their left halves, and samples were collected at 3 d post-inoculation (dpi). (B) Bacterial growth quantification of Pst-DC3000-inoculated (OD600=0.0001) leaves after exposure to EL and RL. Samples were collected at 3 dpi for the assay. Each value is the mean±standard deviation (SD) of three replicates. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test: P<0.05). CFU, colony-forming units. (This figure is available in colour at JXB online.)
RL-induced defence responses are dependent on LOX activity
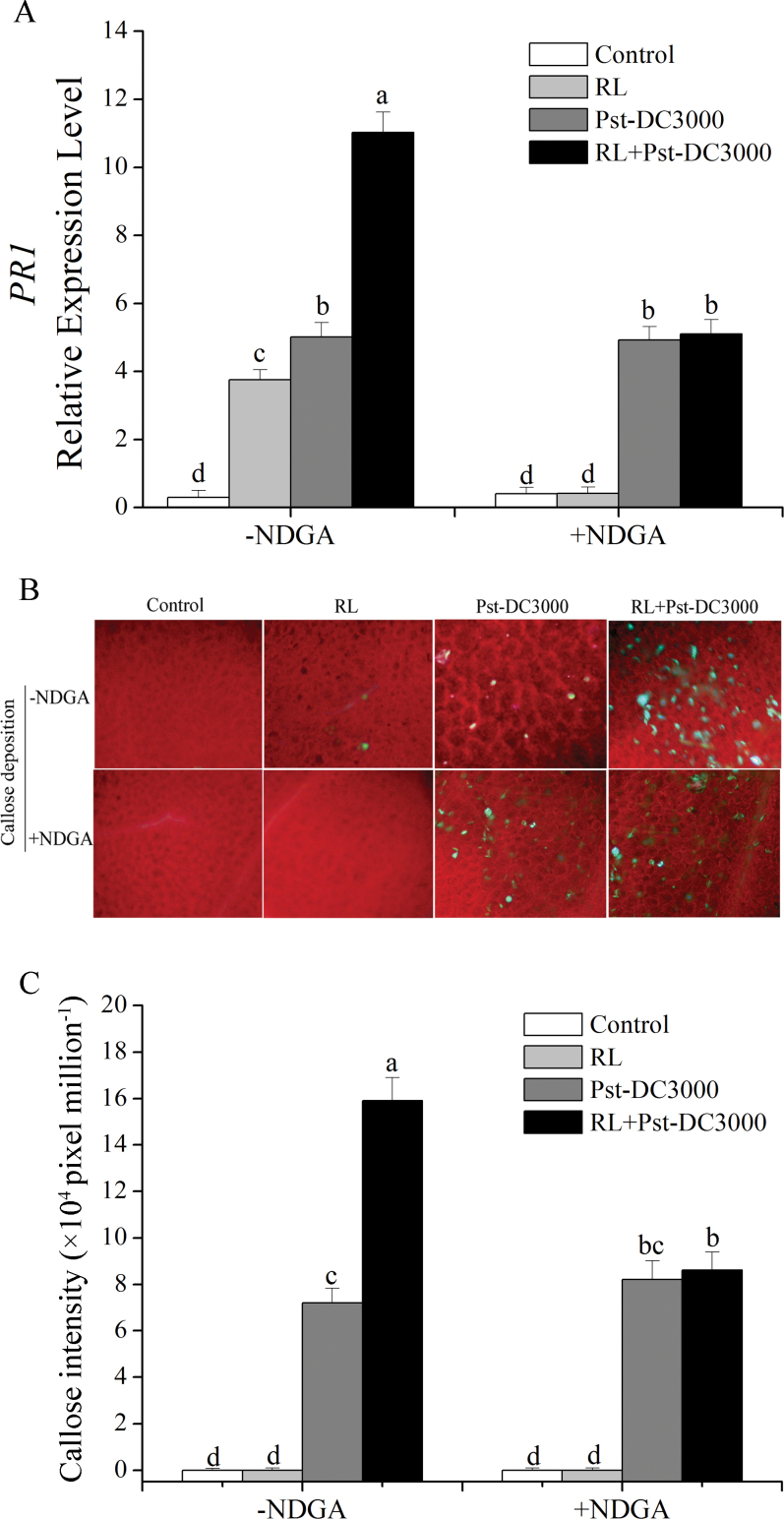
In our research, as the markers of enhancement of defence response, transcript expression of PR1 (pathogenesis-related protein 1) and deposition of callose were analysed. First, we detected the effect of different pre-treatment times of RL on plant resistance to pathogen infection, with transcript expression levels of PR1 as the indicator (Supplementary Fig. S1 at JXB online). We found that the expression level increased significantly with the extension of RL treatment time and the level gradually became stable, which indicated that 4h of RL could activate the plant defence response against the invading pathogen. Therefore, we chose RL (120 μmol m–2 s–1 for 4h; 660–680nm) as the inducer of the defence response in the following experiments. As shown in Fig. 2A, analysis of PR1 gene expression by RT-PCR revealed that, when plants were inoculated with Pst-DC3000, the PR1 transcript was significantly increased in plants with pre-irradiation of RL, whereas a lower level of expression was found in plants without RL. However, when plants were pre-treated with NDGA, a non-selective inhibitor of LOX (Xu et al., 2005; Keereetaweep et al., 2010; Gao et al., 2011; Wang et al., 2012), the induction of PR1 transcripts was insignificant. We also detected callose deposition. When plants were inoculated with Pst-DC3000, more callose deposition was observed in plants pre-treated with RL than in plants without exposure to RL. Pre-treatment with NDGA could erase the deposition of more callose induced by RL (Fig. 2B, C). The results in Fig. 2 indicated that NDGA application alone did not induce any evident responses in plants, whereas RL-induced defence responses were arrested by the inhibitor. Clearly, RL treatment primes Arabidopsis plants for augmented induction of defence responses when challenged by the pathogen, and activation of LOX was necessary during the process.
Fig. 2.
Effect of LOX inhibitor (NDGA) on the RL-induced defence response in Arabidopsis. (A) Quantitative RT-PCR data showing the influence of suppression of LOX activity on PR1 gene expression during the defence response in WT plants. Plants were pre-sprayed with or without NDGA (50 μM) and treated as follows: control, no treatment; RL, 120 μmol photons m–2 s–1 for 4h; Pst-DC3000 inoculation, OD600=0.01 in 10mM MgCl2; RL+Pst-DC3000, inoculation after RL. (B) Callose-staining imaging of leaves from plants under different treatments. (C) Callose deposition in the leaves in (B) was quantified by determining the number of pixels per million pixels in digital photographs. Data are means±SD of three experiments. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test: P<0.05). (This figure is available in colour at JXB online.)
Expression and activity of LOX are induced by RL
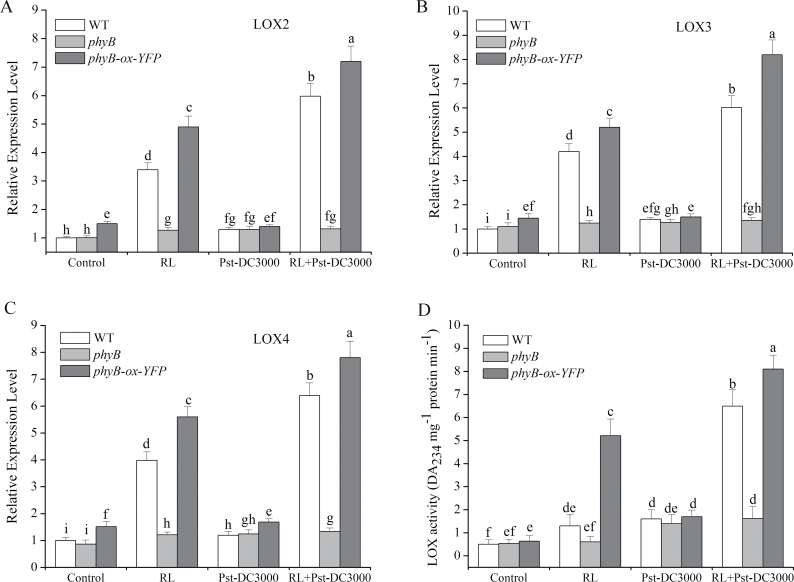
Application of the LOX inhibitor NDGA had a negative influence on the enhancement of defence induced by RL (Fig. 2). This was preliminary evidence that the induction of defence was dependent on LOX. To verify this further, we detected expression of six isoforms of LOX in Arabidopsis (Fig. 3A–C and Supplementary Fig. S2 at JXB online). The elevated expression levels of LOX2, LOX3, LOX4 were most evident in plants with RL treatment before Pst-DC3000 inoculation in comparison with either RL treatment only or Pst-DC3000 inoculated directly (Fig. 3A–C). Conversely, such a reinforcing effect was ruled out in phyB plants and amplified in phyB-ox-YFP plants. Transcripts of LOX1, LOX5, and LOX6 were almost unaffected by RL (Fig. S2). Examination of LOX activity was followed. The results showed that, when inoculated with Pst-DC3000, the activity of LOX was increased significantly in RL-treated plants compared with non-treated plants (Fig. 3D). The upregulation of LOX activity exhibited the same tendency as induction of LOX2, LOX3, and LOX4 transcripts. These data together demonstrated that LOX was upregulated both in transcription and activity during the defence response induced by RL.
Fig. 3.
Induction of transcription levels and activity of LOX by RL. (A–C) Transcript levels of LOX2, LOX3, and LOX4 in WT, phyB, and phyB-ox-YFP plants. (D) LOX activity analysis in WT, phyB, and phyB-ox-YFP plants. Total RNA and proteins were extracted from the leaves of full-grown Arabidopsis after different treatments as follows: control, no treatment; RL, 120 μmol photons m–2 s–1 for 4h; Pst-DC3000 inoculation, OD600=0.01 in 10mM MgCl2; RL+Pst-DC3000, inoculation after RL. Arabidopsis ACTIN2 was used as an internal control. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test: P<0.05). Values represent means±SD of three independent experiments.
PIF3 can bind to the sequence of LOX to inhibit its expression, while RL relieves the inhibition
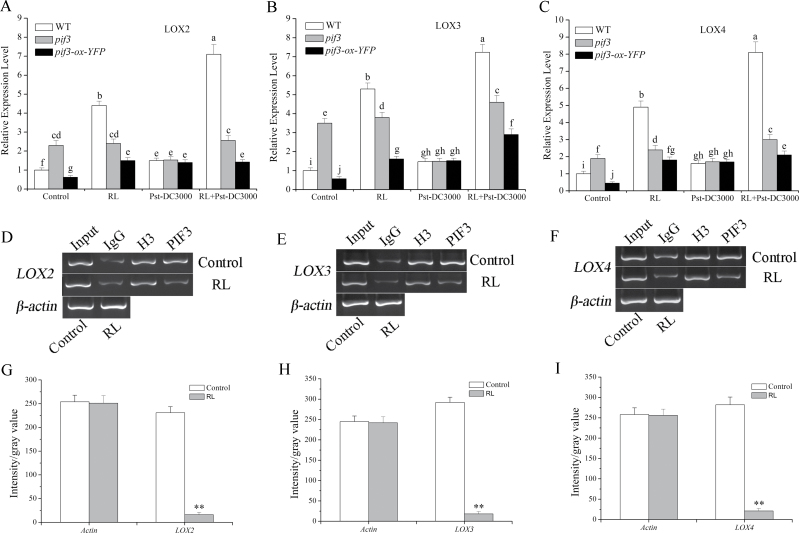
We showed that phyB-mediated LOX transcript expression was induced by RL (Fig. 3A–C). PIFs act as negative transcription factors and bind a G-box (CACGTG) DNA sequence element in many light-regulated genes. Hou et al., (2010) demonstrated that the sequence of the LOX gene may contain a G-box or similar domain structure, an interesting finding that led us to wonder whether PIFs could directly bind to special sequence in these genes to inhibit their expression. As the most important and widely studied PIF, PIF3 was analysed in our work. As shown in Fig. 3, the LOX2, LOX3, and LOX4 transcripts were upregulated under RL and there was a more significant increase following pathogen inoculation. First, we tested expression levels of LOX2, LOX3, and LOX4 in pif3 (T-DNA insertion mutant) and pif3-ox-YFP (overexpressing a YFP–PIF3 fusion) plants. The data in Fig. 4A–C, together with that in Fig. 3, illustrated that the function of phyB signalling in the induction of LOX2, LOX3, and LOX4 transcripts by RL was achieved through the negative regulation of PIF3. As shown in Fig. 3A–C, all three genes from plants pre-treated with RL showed a more conspicuous transcription level compared with non-pre-treated plants. A ChIP experiment was performed to test whether PIF3 could combine with the DNA sequence of LOX2, LOX3, and LOX4. We performed comparative PIF3 ChIP analyses on pif3-ox-YFP lines. ChIPs from the control proved to be highly enriched in LOX2, LOX3, and LOX4, supporting the conclusion that PIF3 was binding to LOX2, LOX3, and LOX4 sequences (Fig. 4D–F) and inhibited their expression (Fig. 4A–C). Thus, based on the results, PIF3 can bind to the promoters of LOX2, LOX3, and LOX4 to inhibit their expression, and RL relieved this inhibition and promoted their expressions through the phyB–PIF3 signalling pathway.
Fig. 4.
PIF3 inhibits LOX transcript expression by binding to its gene sequence. (A–C) PIF3 inhibited LOX2, LOX3, and LOX4 transcript levels. Expression levels were detected 4h after different treatments as indicated above using quantitative RT-PCR. Arabidopsis ACTIN2 was used as an internal control. Expression levels for each treatment were normalized to a RL-treated (0h) plant. (D–F) The binding of PIF3 and LOX gene sequence in normal light (control) or RL-treated (120 μmol photons m–2 s–1 for 4h) samples of the pif3-ox-YFP plant determined by ChIP. The co-immunoprecipitated DNA was detected by agarose gel electrophoresis. Input indicated samples before immunoprecipitation; IgG, H3, and PIF3 indicate samples immunoprecipitated with IgG antibody, H3 antibody and YFP antibody, respectively. (G–I) Quantitative analysis of the LOX2 (D), LOX3 (E) and LOX4 (F) genes of PIF3 samples are shown in (G), (H), and (I), respectively, with Image J software. Three gel photographs were taken for quantitative analysis, and values represent means±SD. Asterisks indicate significant differences between the control and RL treatment (Student’s paired t-test: *P<0.05, **P <0.01).
MPK3 and MPK6 are responsible for LOX activation during RL-induced defence
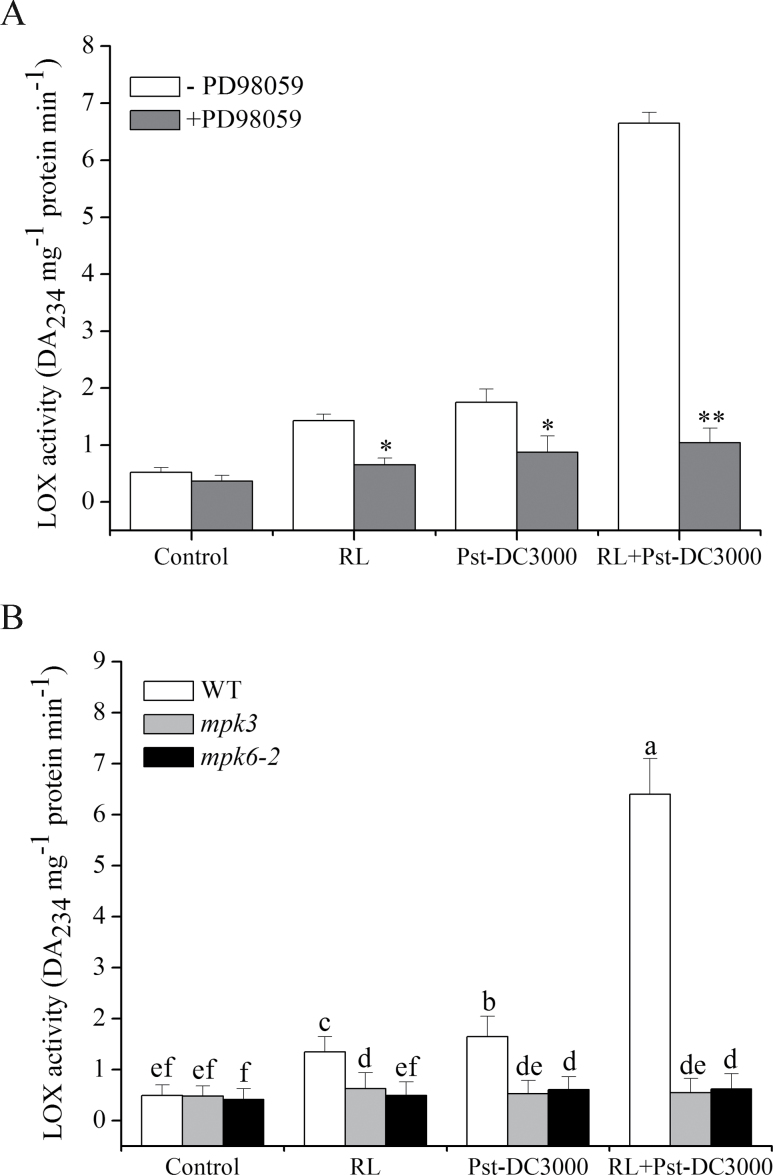
In order to investigate whether MAPK cascades were related to the RL-induced activation of Arabidopsis LOX, a common inhibitor of the MAPK cascade, PD98059, was used. PD98059 is a selective inhibitor of the MAPK-activating enzyme MEK and consequently of the MAPK cascade (Dudley et al., 1995), which inhibits the activation of MAPK and subsequent phosphorylation of MAPK substrates (Alessi et al., 1995; Kultz et al., 1998; Ye et al., 2013). First, the change in LOX activity was analysed. The results in Fig. 5A indicated that plants pre-irradiated with RL enhanced LOX activity when inoculated with Pst-DC3000, but the increase in LOX activity was effectively inhibited by PD98059. The data indicated that MAPK cascades were involved in LOX activation during the RL-induced defence response, but it remains to be established which specific MPK is involved in this process. MPK3 and MPK6 are reported to be activated in the Arabidopsis stress response (Li et al., 2012) and they are both critical in priming plants for full induction of the defence response during induced resistance (Beckers et al., 2009). The LOX activity in leaves of mpk6-2 (T-DNA insertion mutant) and mpk3 (MPK3-lacking mutant) plants under different treatments was measured. As shown in Fig. 5B, a substantial increase in LOX activity was observed in RL-treated leaves of WT when inoculated with Pst-DC3000, whereas no significant increase was observed in leaves of mpk3 and mpk6-2 plants, suggesting that both MPK3 and MPK6 were indispensable for RL-induced LOX activation when plants were challenged with pathogen.
Fig. 5.
MPK3 and MPK6 are indispensable for LOX activation in Arabidopsis. (A) Leaves were pre-incubated with or without PD98059 (20 μM) for 1h, and proteins were then extracted from the leaves at 2h post-treatment. (B) Changes in LOX activity in WT, mpk3, and mpk6-2 plants under different treatments. Plants were treated as follows: control, no treatment; RL, 120 μmol photons m–2 s–1 for 4h; Pst-DC3000 inoculation, OD600=0.01 in 10mM MgCl2; RL+Pst-DC3000, inoculation after RL. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test: P<0.05).
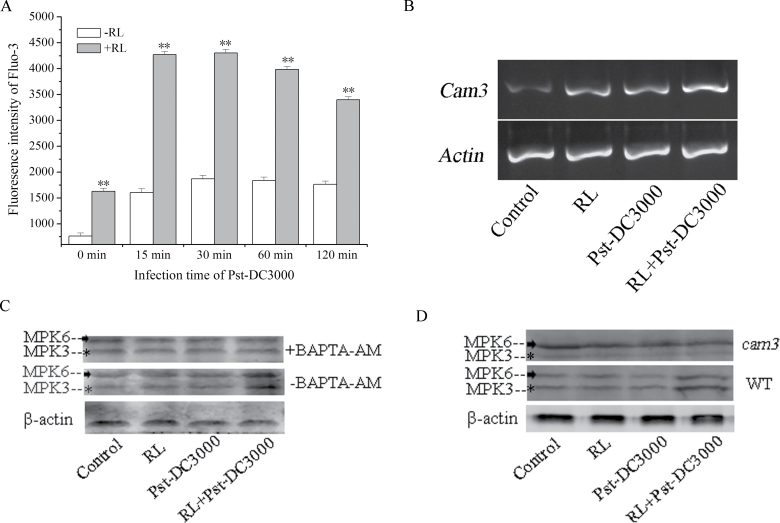
Activation of MPK3 and MPK6 is related to Ca2+–CaM3
The results given above demonstrated that MPK3 and MPK6 were both implicated in RL-induced activation of LOX. Plant MPKs have high homology to mammalian ERK1/2 MPKs, and ERK1/2 antisera that recognize the dually phosphorylated forms (pTEpY) of activated MPKs can be used to monitor plant MPK activity (Li et al., 2012). Hence, the endogenous kinase activity of MPK3 and MPK6 was determined using anti-ACTIVE MAP kinase polyclonal antibody (pTEpY). As shown in Fig. 6C, D, in response to RL, while a transient increase in both the MPK3 activity (43kDa band) and MPK6 activity (47kDa band) were observed in RL-treated WT followed by inoculation, compared with the control, no increase in kinase activity of MPK3 and MPK6 was observed in plants either treated with RL only or inoculated with Pst-DC3000 directly. This indicated that MPK3 and MPK6 were activated during the RL-induced defence response.
Fig. 6.
Ca2+–CaM3 is involved in activation of MPK3 and MPK6 during the RL-induced defence response. (A) Estimation of RL-induced changes in Ca2+ level by flow cytometry using Fluo-3. Protoplasts were incubated with Fluo-3-AM (at a final concentration of 5 μM) for 60min at room temperature and then subjected to flow cytometry analysis. Data represent means±SD of three independent experiments. Statistical analysis was performed with Student’s paired t-test. An asterisk indicates a significant difference from the control (**P<0.05). (B) Induction of CaM3 transcription by RL. Total RNAs were isolated from WT leaves under different treatments and semi-quantitative RT-PCR was performed. (C) MPK3 and MPK6 activity was measured in WT plants with (+BAPTA) or without (–BAPTA) 1mM BAPTA pre-treatment. (D) MPK3 and MPK6 activity was measured in WT plants and cam3 mutants. Proteins were extracted from leaves treated as follows: control, no treatment; RL, 120 μmol photons m–2 s–1 for 4h; Pst-DC3000 inoculation, OD600=0.01 in 10mM MgCl2 for 2h; RL+Pst-DC3000, 2h inoculation after RL. Each data point is the mean±SD of three independent replicates. Different letters indicate statistically significant differences between treatments (Duncan’s multiple range test: P<0.05).
We detected an increase in [Ca2+]cyt and upregulation of the CaM3 transcript level (Fig. 6A, B), which may function upstream of activation of MPK3 and MPK6 in response to RL, when plants were inoculated with Pst-DC3000. These observations compelled us to test the effect of application of the Ca2+ scavenger BAPTA-AM on activation of MPK3 and MPK6. BAPTA-AM is a lipophilic compound capable of crossing cell membranes; inside the cell, non-specific esterases cleave the AM moiety, thereby forming the ionized Ca2+-binding compound BAPTA (Tymianski et al., 1994; Kawano et al., 2000; Yue et al., 2012). The data in Fig. 6C showed that BAPTA-AM inhibited activation of MPK3 and MPK6 effectively, while the activation of MPK3 and MPK6 was impaired in cam3 (CaM3-lacking mutant) relative to WT (Fig. 6D). Together, MPK3 and MPK6 were apparently activated during RL-induced defence and the signalling pathway was related to Ca2+–CaM3.
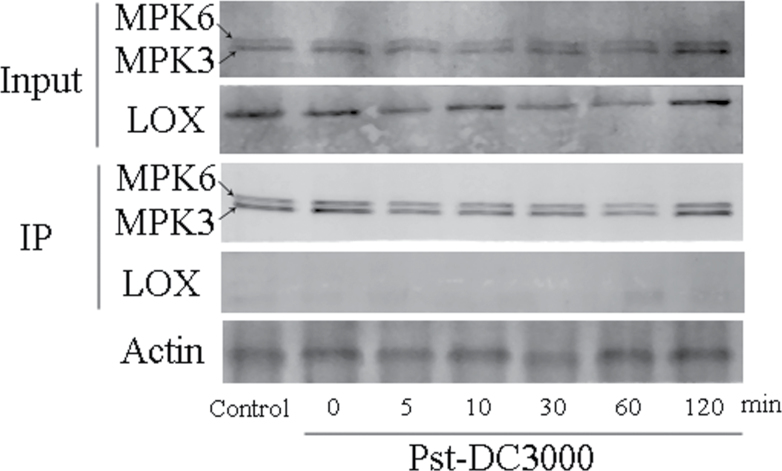
LOX is not activated through directly binding with MPK3/MPK6
Signalling transduction related to MPKs cascades is generally decoded by phosphorylation of downstream molecules. In our research, induction of LOX activity was concomitant with activation of MPK3 and MPK6, but whether LOX was regulated by MPK3 and MPK6 directly or indirectly was unknown. To gain more information about the regulation pattern, co-immunoprecipitation experiments were performed using WT lines. As shown in Fig. 7, we detected activation of MPK3, MPK6, and LOX before co-immunoprecipitation but no LOX in immunoprecipitates. Therefore, LOX could not be co-precipitated with MPK3 and MPK6, as a direct interaction was not observed in immunoprecipitates.
Fig. 7.
MPK3 and MPK6 are not directly combined with LOX. Co-immunoprecipitation experiments were performed using WT lines. Input indicates samples before immunoprecipitation with anti-ERK antibody, and IP indicates co-immunoprecipitated samples from different treatments. Proteins were extracted from RL-pre-treated leaves at the indicated times after inoculation with Pst-DC3000.
Discussion
Szechyńska-Hebda et al. (2010) demonstrated that plants possess a complex and dynamic light training and memory system that involves quantum redox, reactive oxygen species, and hormonal and photo-electrophysiological signalling to optimize light acclimation and defences. In their research, defence responses were induced by excess RL but not blue light. However, the molecular mechanisms underlying the induced process of defences have not been fully resolved. Our findings indicated that phyB plays an important role in the enhancement of defences (Fig. 1). PhyB acts in many aspects of plant growth and development. Recent studies have reported that phyB may participate in the defence response (Griebel and Zeier, 2008; Kazan and Manners, 2011). PhyB usually regulates the expressions of related genes by combining with PIFs. In our study, the data showed that phyB signalling was indispensable for RL-induced activation of LOX (Fig. 3) and PIF3 clearly functioned downstream of phyB (Fig. 4).
LOX is widespread in both animals and plants (Liavonchanka and Feussner, 2006) and catalyses the key step of lipid peroxidation (Gao et al., 2011). It catalyses the production of substances called oxylipins. Oxylipins, a series of versatile molecules, are engaged in many aspects during plant growth and development. The oxylipin synthetic pathway mediates plant defence responses to diverse biotic and abiotic stresses (Blée, 2002; Creelman and Mulpuri, 2002; Howe and Schilmiller, 2002). Oxylipins are believed to play pivotal roles in defences (Krumm et al., 1995; Bate and Rothstein, 1998) and they act as signal molecules and/or protective compounds, or as constituents of cutin (Blée, 2002). Hause et al., (2000) also showed that distinctive oxylipin profiles were produced by different external stimuli and by developmental cues. In this work, we found that RL upregulated LOX both at the transcription level and in terms of activity (Fig. 3), suggesting the possible involvement of phyB-mediated LOX activation in the RL-induced Arabidopsis defence response.
PIF3 acts as a negative factor and inhibits expression of some genes by binding to their promoters (Martínez-García et al., 2000; Quail, 2002). ChIP experiments (Fig. 4D–F) indicated that PIF3 binds to gene sequences of LOX2, LOX3, and LOX4, and expression of these three genes was prevented in control plants compared with RL-treated plants. The data above suggested that the gene sequences of LOX2, LOX3, and LOX4 may contain a G-box domain or a similar structure. A well-recognized mechanism of phyB signalling pathway is that activated phyB enters into nucleus and combines with PIF3 to target its degradation; this degradation is caused by phosphorylation and ubiquitination of PIF3 (Nicholson et al., 2011; Zhang et al., 2013). In our work, under normal circumstances, PIF3 combined with a region of the LOX2, LOX3, and LOX4 gene sequences to suppress their expressions. When plants were exposed to a certain intensity of RL, activated phyB entered into nucleus and caused degradation of PIF3; inhibition LOX was relieved, thus leading to an enhancement of expression of these LOX genes (Fig. 3A–C).
MPK cascades can be activated by various stimuli and play central roles in the process whereby extracellular stimuli are transduced into intracellular responses (Widmann et al., 1999; Asai et al., 2002; Nakagami et al., 2005). Our experiments demonstrated that MPK cascades also participate in the activation of LOX during the RL-induced defence response (Fig. 5A). Among the various MPK proteins, MPK3 and MPK6 are well-established signalling proteins in Arabidopsis, and can be activated by various stimuli. In RL-treated plants, the activation of both MPK3 and MPK6 was proved to be responsible for the upregulation of LOX activity and the subsequent execution of a defence response (Fig. 5B). A typical MPK signalling module consists of three kinases: an MAPKKK, an MAPKK, and an MAPK. MAPKs function at the bottom of the kinase cascade and are activated by MAPKKs through phosphorylation. The activation of MAPKKs is, in turn, regulated by MAPKKKs via phosphorylation. In our study, which MAPKK and MAPKKK are involved in the activation of MPK3 and MPK6 was unclear. MAPKK4/5 is possible engaged upstream of MPK3 and MPK6, because a large body of research has discovered that MAPKK4/5 activates MPK3 and MPK6 during plant pathogen signalling (Meng and Zhang, 2013; Vidhyasekaran, 2014). As an important signal messenger, Ca2+ can function upstream of the activation of the MPK cascade under different stimuli (Xing et al., 2008; Wang PC et al., 2010). Under RL, we found increased [Ca2+]cyt (Fig. 6A) and upregulation of CaM3 (Fig. 6B), which functioned in the upstream activation of MPK3 and MPK6 during the defence response. MPKs are proline-directed serine/threonine kinases phosphorylating serine or threonine in the dipeptide motif S/T-P (Bardwell, 2006), Arabidopsis LOX sequence show the presence of phosphorylation site for MPK (Taj et al., 2011b), an immunoprecipitation between MPK3/6 and LOX was done. We have not detected the binding of MPK3/6 and LOX at the time point we selected, which may due to the instantaneous interaction between them during defence response (Taj et al., 2011a), which means the time of interaction is too short to capture. Another possible explanation is that MPK3 and MPK6 do not facilitate LOX activation by binding with it directly but may function through downstream WRKY transcription factors. The WRKY transcription factors are also activated by MAPK-dependent phosphorylation and function downstream of MAPK during the defence response (Ishihama and Yoshioka, 2012). Whether WRKY is involved in the activation of LOX and the mechanisms in this regulation pathway needs further research.
Our experimental results showed that when plants pre-irradiated with RL were infected by the pathogen, the activity of LOX was significantly increased compared with that in plants directly inoculated with the pathogen. Whereas a comparative significant increase in LOX2, LOX3, and LOX4 was induced in RL-treated plants before pathogen infection, higher levels were induced after infection (Fig. 3). This means that when plants were exposed to RL, transcripts of several LOX genes increased significantly, while no obvious changes were detected in activity, indicating that plants are preparing for combat with the pathogen and have become very sensitive to pathogen invasion. Once the pathogen had infected the plant, the plants produced a stronger and more rapid response. As Beckers et al. (2009) proposed and demonstrated, accumulation of mRNA was primed for activation of the defence responses. In our study, accumulation of LOX2, LOX3, and LOX4 transcripts induced by RL was primed for a later defence response to the pathogen. A stronger LOX activity, subsequent PR1 expression, and callose deposition were triggered as later defence responses (Figs 2 and 3). Plants primed by treatments that induce resistance show a faster and/or stronger activation of defence responses when subsequently challenged by pathogens or abiotic stresses (Conrath et al., 2002, 2006). As a part of induced resistance responses, priming has been studied in many plants (Kuć, 1987; Zimmerli et al., 2000; Verhagen et al., 2004) for a number of years, but the molecular mechanism of priming has been presented recently (Beckers et al., 2009), and is associated with MPK3 and MPK6 during the development of chemically induced resistance in Arabidopsis. In our study, activation of MPK3 and MPK6 promoted LOX activity in the RL-induced defence response.
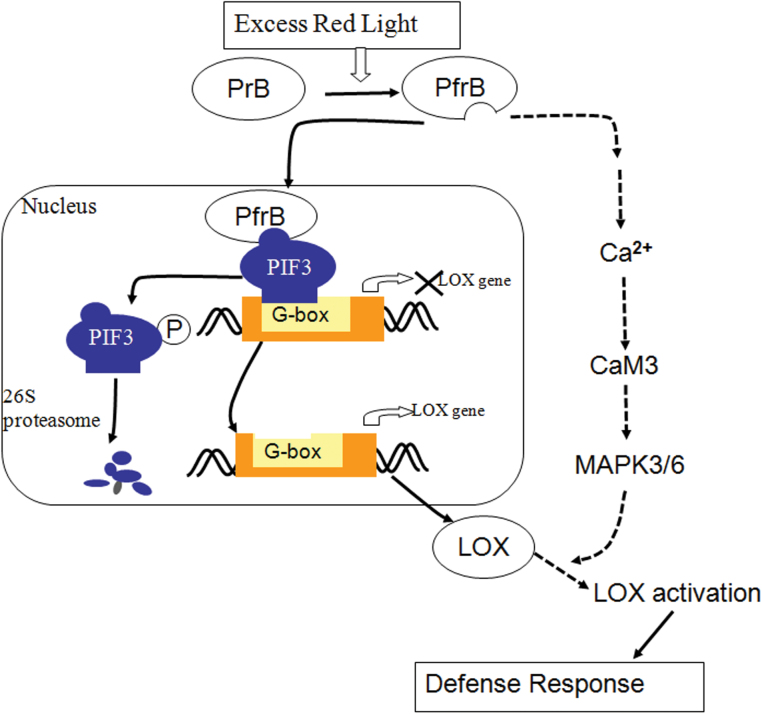
Our investigations provide evidence that LOX is responsible for defences induced by excess RL. According to the experimental results, a potential cascade of cellular events during enhancement of the defence response is suggested. As summarized in the model presented in Fig. 8, under the condition of RL, phyB is photo-activated and translocates to nucleus; it then binds with PIF3 and promotes its degradation. The degradation of PIF3 promotes expression of LOX2, LOX3, and LOX4, because these genes are inhibited by binding of PIF3 to the LOX DNA sequence. When plants are challenged with a pathogen, [Ca2+]cyt in cytoplasm is increased rapidly, which activates MPK3 and MPK6, thereby promoting the LOX enzyme capacity. As a result, plants can induce PR1 expression and callose deposition effectively, which means an enhancement of the defence response. Our results contribute to the corroboration of the signalling mechanism of induced defences by RL and highlight an important role of LOX in the process. Obviously, this method of enhancing the plans defence response is easy to implement and will have wide-ranging use in crop cultivation.
Fig. 8.
Proposed working model for LOX activation of both transcript levels and activity during the RL-induced defence response. (This figure is available in colour at JXB online.)
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Impact of different times of RL on PR1 expression.
Supplementary Fig. S2. Induction of transcription levels of LOX1, LOX5, and LOX6 by RL.
Supplementary Fig. S3. Effect of different concentrations of PD98059 on activation of LOX.
Supplementary Fig. S4. Activation of MPK3 and MPK6 during the defence response induced by RL in WT, mpk6-2, and mpk3 plants.
Supplementary Table S1. Primers for several genes.
Acknowledgements
We thank Professor Ivo Feussner for kindly providing anti-LOX antibody, Professor Peter H. Quail for providing the seeds of Arabidopsis pif3, and pif3-ox-YFP, Professor Hongquan Yang for providing Arabidopsis phyB-ox-YFP seeds. This research is supported by the Program for Changjiang Scholars and Innovative Research Team in University (IRT0829), the Key Program of NSFC-Guangdong Joint Funds of China (U0931005), and the National High Technology Research and Development Program of China (863 Program) (2007AA10Z204).
Glossary
Abbreviations:
- [Ca2+]cyt
cytosolic calcium concentration
- CaM
calmodulin
- ChIP
chromatin immunoprecipitation
- dpi
days post-inoculation
- EL
excess light
- ERK
extracellular signal-regulated kinase
- LOX
lipoxygenase
- MAPK/MPK
mitogen-activated protein kinase
- NDGA
4,′(2, 3-dimethyltetramethylene)dipyrocatechol
- phyB
phytochrome B
- PIF
phytochrome-interacting factor 3
- Pst
Pseudomonas syringae pv. tomato
- RL
red light
- RT-PCR
reverse transcription-PCR
- SD
standard deviation
- WT
wild type.
References
- Alami I, Jouy N, Clerivet A. 1999. The lipoxygenase pathway is involved in elicitor-induced phytoalexin accumulation in plane tree (Platanus acerifolia) cell-suspension cultures. Journal of Phytopathology 147, 515–519 [Google Scholar]
- Alessi DR, Cuenda A, Cohen P, Dudley DT, Saltiel AR. 1995. PD098059 is a specific inhibitor of the activation of mitogen-activated protein kinase kinase in vitro and in vivo. Journal of Biological Chemistry 270, 27489–27494 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signaling cascade in Arabidopsis innate immunity. Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- Bannenberg G, Martínez M, Hamberg M, Castresana C. 2009. Diversity of the enzymatic activity in the lipoxygenase gene family of Arabidopsis thaliana . Lipids 44, 85–95 [DOI] [PubMed] [Google Scholar]
- Bardwell L. 2006. Mechanisms of MAPK signaling specificity. Biochemical Society Transactions 34, 837–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate NJ, Rothstein SJ. 1998. C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. The Plant Journal 16, 561–569 [DOI] [PubMed] [Google Scholar]
- Beckers GJM, Jaskiewicz M, Liu YD, Underwood WR, He SY, Zhang SQ, Conrath U. 2009. Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana . The Plant Cell 21, 944–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blée E. 2002. Impact of phyto-oxylipins in plant defense. Trends in Plant Science 7, 315–322 [DOI] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254 [DOI] [PubMed] [Google Scholar]
- Colcombet J, Hirt H. 2008. Arabidopsis MAPKs: a complex signaling network involved in multiple biological processes. Biochemical Journal 413, 217–226 [DOI] [PubMed] [Google Scholar]
- Conrath U, Beckers GJM, Flors V, et al. 2006. Priming: getting ready for battle. Molecular Plant–Microbe Interactions 19, 1062–1071 [DOI] [PubMed] [Google Scholar]
- Conrath U, Pieterse CMJ, Mauch-Mani B. 2002. Priming in plant pathogen interactions. Trends in Plant Science 7, 210–216 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mulpuri R. 2002. The oxylipin pathway in Arabidopsis . American Society of Plant Biologists 1, e0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan H, Huang MY, Palacio K, Schuler MA. 2005. Variations in CYP74B2 (HYDROPEROXIDE LYASE) gene expression differentially affect hexenal signaling in the Columbia and Landsbergerecta Ecotypes of Arabidopsis . Plant Physiology 139, 1529–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. 1995. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proceedings of the National Academy of Sciences, USA 92, 7686–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feussner I, Wasternack C. 2002. The lipoxygenase pathway. Annual Review of Plant Biology 53, 275–297 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. 2010. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany 61, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao GL, Zhang SC, Wang CF, Yang X, Wang YQ, Su XJ, Du JJ, Yang CW. 2011. Arabidopsis CPR5 independently regulates seed germination and postgermination arrest of development through LOX pathway and ABA signaling. PLoS ONE 6, e19406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genoud T, Buchala AJ, Chua NH, Métraux JP. 2002. Phytochrome signalling modulates the SA-perceptive pathway in Arabidopsis . The Plant Journal 31, 87–95 [DOI] [PubMed] [Google Scholar]
- Griebel T, Zeier J. 2008. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiology 147, 790–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K, Kudla J. 2011. Calcium decoding mechanisms in plants. Biochimie 93, 2054 –2059 [DOI] [PubMed] [Google Scholar]
- Hause B, Stenzel I, Miersch O, Maucher H, Kramell R, Ziegler J, Wasternack C. 2000. Tissue-specific oxylipin signature of tomato flowers: allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. The Plant Journal 24, 113–126 [DOI] [PubMed] [Google Scholar]
- Hou XL, Lee LYC, Xia KF, Yan YY, Yu H. 2010. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental Cell 19, 884–894 [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL. 2002. Oxylipin metabolism in response to stress. Current Opinion in Plant Biology 5, 230–236 [DOI] [PubMed] [Google Scholar]
- Ichimura K, Shinozaki K, Tena G, et al. 2002. Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends in Plant Science 7, 301–308 [DOI] [PubMed] [Google Scholar]
- Ishihama N, Yoshioka H. 2012. Post-translational regulation of WRKY transcription factors in plant immunity. Current Opinion in Plant Biology 15, 431–437 [DOI] [PubMed] [Google Scholar]
- Kawano T, Pinontoan R, Uozumi N, Miyake C, Asada K, Kolattukudy PE, Muto S. 2000. Aromatic monoamine-induced immediate oxidative burst leading to an increase in cytosolic Ca2+ concentration in tobacco suspension culture. Plant Cell Physiol 41, 1251–1258 [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2011. The interplay between light and jasmonate signalling during defence and development. Journal of Experimental Botany 62, 4087–4100 [DOI] [PubMed] [Google Scholar]
- Keereetaweep J, Kilaru A, Feussner I, Venables BJ, Chapman KD. 2010. Lauroylethanolamide is a potent competitive inhibitor of lipoxygenase activity. FEBS Letters 584, 3215–3222 [DOI] [PubMed] [Google Scholar]
- Krumm T, Bandemer K, Boland W. 1995. Induction of volatile biosynthesis in the Lima bean (Phaseolus lunatus) by leucine-and isoleucine conjugates of 1-oxo- and 1-hydroxyindan-4-carboxylic acid: evidence for amino acid conjugates of jasmonic acid as intermediates in the octadecaboid signaling pathway. FEBS Letters 377, 523–529 [DOI] [PubMed] [Google Scholar]
- Kuć J. 1987. Translocated signals for plant immunization. Annals of the New York Academy of Sciences 494, 221–223 [Google Scholar]
- Kultz D, Madhany S, Burg MB. 1998. Hyperosmolality causes growth arrest of murine kidney cells. Induction of GADD45 and GADD153 by osmosensing via stress-activated protein kinase 2. Journal of Biological Chemistry 273, 13645–13651 [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E, Cohn MM, Quail PH. 2012. Phytochrome signaling in green Arabidopsis seedings: impact assessment of a mutually negative phyB–PIF feedback loop. Molecular Plant 5, 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yue HY, Xing D. 2012. MAP Kinase 6-mediated activation of vacuolar processing enzyme modulates heat shock-induced programmed cell death in Arabidopsis . New Phytologist 195, 85–96 [DOI] [PubMed] [Google Scholar]
- Liavonchanka A, Feussner I. 2006. Lipoxygenases: occurrence, functions and catalysis. Journal of Plant Physiology 163, 348–357 [DOI] [PubMed] [Google Scholar]
- Lin TC, Ishii H. 2009. Accumulation of H2O2 in xylem fluids of cucumber stems during ASM-induced systemic acquired resistance (SAR) involves increased LOX activity and transient accumulation of shikimic acid. European Journal of Plant Pathology 125, 119–130 [Google Scholar]
- Linschitz H, Kasche V, Butler WL, Siegelman HW. 1966. The kinetics of phytochrome conversion. Journal of Biological Chemistry 241, 3395–3403 [PubMed] [Google Scholar]
- Liu XC, Chen CY, Wang KC, et al. 2013. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA 15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 25, 1258–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan S, Kudla J, Rodriguez-Concepcion M, Yalovsky S, Gruissem W. 2002. Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14, 389–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-García JF, Huq E, Quail PH. 2000. Direct targeting of light signals to a promoter element-bound transcription factor. Science 288, 859–863 [DOI] [PubMed] [Google Scholar]
- Matsumura H, Nirasawa S, Kiba A, Urasaki N, Saitoh H, Ito M, Kawai-Yamada M, Uchimiya H, Terauchi R. 2003. Overexpression of Bax inhibitor suppresses the fungal elicitor-induced cell death in rice (Oryza sativa L.) cells. The Plant Journal 33, 425–434 [DOI] [PubMed] [Google Scholar]
- Meng XZ, Zhang SQ. 2013. MAPK cascades in plant disease resistance signaling. Annual Review of Phytopathology 51, 245–266 [DOI] [PubMed] [Google Scholar]
- Miles GP, Samuel MA, Jones AM, Ellis BE. 2004. Mastoparan rapidly activates plant MAP kinase signaling independent of heterotrimeric G proteins. Plant Physiology 134, 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet YA, Danna C H, Clay N K, Songnuan W, Simon MD, Werck -Reichhart D, Ausubel FM. 2010. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. The Plant Cell 22, 973–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina TE, Zeier J. 2007. Pathogen-associated molecular pattern recognition rather than development of tissue necrosis contributes to bacterial induction of systemic acquired resistance in Arabidopsis . Plant Journal 50, 500–513 [DOI] [PubMed] [Google Scholar]
- Nagatani A. 2004. Light-regulated nuclear localization of phytochromes. Current Opinion in Plant Biology 7, 708–711 [DOI] [PubMed] [Google Scholar]
- Nakagami H, Pitzschke A, Hirt H. 2005. Emerging MAP kinase pathways in plant stress signaling. Trends in Plant Science 10, 339–346 [DOI] [PubMed] [Google Scholar]
- Nicholson SJ, Hoecker U, Srivastava V. 2011. A novel phytochrome B allele in Arabidopsis thaliana exhibits partial mutant phenotype: a short deletion in N-terminal extension reduces phytochrome B activity. Plant Growth Regulation 65, 207–212 [Google Scholar]
- Porta H, Rocha-Sosa M. 2002. Plant lipoxygenases. Physiological and molecular features. Plant Physiology 130, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. 2002. Phytochrome photosensory signaling networks. Nature Reviews Molecular Cell Biology 3, 85–93 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Wilson PB, Hussain D, Woo NS, Gordon MJ, Mewett OP, Howell KA, Whelan J, Kazan K, Pogson BJ. 2007. Systemic and intracellular responses to photooxidative stress in Arabidopsis . Plant Cell 19, 4091–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel MA, Miles GP, Ellis BE. 2000. Ozone treatment rapidly activates MAP kinase signalling in plants. The Plant Journal 22, 367–376 [DOI] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G. 2007. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis . Plant Journal 49, 981–994 [DOI] [PubMed] [Google Scholar]
- Skórzyńska-Polit E, Krupa Z. 2003. Activity of lipoxygenase in Arabidopsis thaliana—a preliminary study. Cellular & Molecular Biology Letters 8, 279–284 [PubMed] [Google Scholar]
- Skórzyńska-Polit E, Pawlikowska-Pawlęga B, Szczuka E, Drążkiewicz M, Krupa Z. 2006. The activity and localization of lipoxygenases in Arabidopsis thaliana under cadmium and copper stresses. Plant Growth Regulation 48, 29–39 [Google Scholar]
- Soy J, Leivar P, González-Schain N, Sentandreu M, Prat S, Quail PH, Monte E. 2012. Phytochrome-imposed oscillations in PIF3 protein abundance regulate hypocotyl growth under diurnal light/dark conditions in Arabidopsis . The Plant Journal 71, 390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Hause B, Miersch O, Kurz T, Maucher H, Weichert H, Ziegler J, Feueener I, Wasternack C. 2003. Jasmonate biosynthesis and the allene oxide cyclase family of Arabidopsis thaliana. Plant Molecular Biology 51, 895–911 [DOI] [PubMed] [Google Scholar]
- Strasser B, Sanchez-Lamas M, Yanovsky MJ, Casal JJ, Cerdan PD. 2010. Arabidopsis thaliana life without phytochromes. Proceedings of the National Academy of Sciences, USA 107, 4776–4781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun AZ, Nie SJ, Xing D.2012. Nitric oxide-mediated maintenance of redox homeostasis contributes to NPR1-dependent plant innate immunity triggered by lipopolysaccharides. Plant Physiology 160, 1081–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szechyńska-Hebda M, Kruk J, Górecka M, Karpińska B, Karpiński S.2010. Evidence for light wavelength-specific photoelectrophysiological signaling and memory of excess light episodes in Arabidopsis . Plant Cell 22, 2201–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj G, Agarwal P, Grant M, Kumar A. 2010. MAPK machinery in plants: recognition and response to different stresses through multiple signal transduction pathways. Plant Signaling & Behavior 5, 1379–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taj G, Agarwal P, Grant M, Kumar A. 2011a. Co-expression and in-silico interaction studies for inter-linking the activation of MAPK3 and LOX genes during pathogenesis of Alternaria brassicae in Brassica juncea. Journal of Oilseed Brassica 2, 13–20 [Google Scholar]
- Taj G, Sharma S, Gaur VS, Kumar A. 2011b. Prediction of downstream interaction of transcription factors with MAPK3 in Arabidopsis thaliana using protein sequence information. International Journal of Bioinformatics Research 3, 167–177 [Google Scholar]
- Tymianski M, Spigelman I, Zhang L, Carlen PL, Tator CH, Charlton MP, Wallace MC. 1994. Mechanism of action and persistence of neuroprotection by cell-permeant Ca2 + chelators. Journal of Cerebral Blood Flow and Metabolism 14, 911–923 [DOI] [PubMed] [Google Scholar]
- Verhagen BWM, Glazebrook J, Zhu T, Chang HS, van Loon LC, Pieterse CMJ. 2004. The transcriptome of rhizobecteria-induced systemic resistance in Arabidopsis . Molecular Plant–Microbe Interactions 17, 895–908 [DOI] [PubMed] [Google Scholar]
- Vidhyasekaran P. 2014. Mitogen-activated protein kinase cascades in plant innate immunity. PAMP Signals in Plant Innate Immunity 21, 331–374 [Google Scholar]
- Wang FF, Lian HL, Kang CY, Yang HQ. 2010. Phytochrome B is involved in mediating red light-induced stomatal opening in Arabidopsis thaliana . Molecular Plant 3, 246–259 [DOI] [PubMed] [Google Scholar]
- Wang PC, Du YY, Li Y, Ren DT, Song CP. 2010. Hydrogen peroxide-mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis . Plant Cell 22, 2981–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XY, Wang N, Rui Q, Zhang P, Xu LL. 2012. Jasmonates modulate the promotion effects induced by SNP on root development of wheat under osmotic stress through lipoxygenase activation. Journal of Plant Biochemistry and Biotechnology 22, 295–303 [Google Scholar]
- Widmann C, Gibson S, Jarpe MB, Johnson GL. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiological Reviews 79, 143–180 [DOI] [PubMed] [Google Scholar]
- Xing Y, Jia WS, Zhang JH. 2008. AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis . The Plant Journal 54, 440–451 [DOI] [PubMed] [Google Scholar]
- Xu MJ, Dong JF, Zhu MY. 2005. Nitric oxide mediates the fungal elicitor-induced hypericin production of Hypericum perforatum cell suspension cultures through a jasmonic-acid-dependent signal pathway. Plant Physiology 139, 991–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XY, Jiang WJ, Yu HJ. 2012. The expression profiling of the lipoxygenase (LOX) family genes during fruit development, abiotic stress and hormonal treatments in cucumber (Cucumis sativus L.). International Journal of Molecular Sciences 13, 2481–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Li Z, Xing D. 2013. Nitric oxide promotes MPK6-mediated caspase-3-like activation in cadmium-induced Arabidopsis thaliana programmed cell death. Plant, Cell & Environment 36, 1–15 [DOI] [PubMed] [Google Scholar]
- Yue HY, Nie SJ, Xing D. 2012. Overexpression of Arabidopsis Bax inhibitor-1 delays methyl jasmonate-induced leaf senescence by suppressing the activation of MAP Kinase 6. Journal of Experimental Botany 63, 4463–4474 [DOI] [PubMed] [Google Scholar]
- Zhang LR, Xing D. 2008. Methyl jasmonate induces production of reactive oxygen species and alterations in mitochondrial dynamics that precede photosynthetic dysfunction and subsequent cell death. Plant and Cell Physiology 49, 1092–1111 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH. 2013. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis . PLoS Genetics 9, e1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Sun AZ, Xing D. 2013. Modulation of cellular redox status by thiamine-activated NADPH oxidase confers Arabidopsis resistance to Sclerotinia scerotiorum . Journal of Experimental Botany 64, 3261–3272 [DOI] [PubMed] [Google Scholar]
- Zimmerli L, Jakab G, Métraux JP, Mauch-Mani B. 2000. Potentiation of pathogen-specific defense mechanisms in Arabidopsis is β-aminobutyric acid. Proceedings of the National Academy of Sciences, USA 97, 12920–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.