Summary
Cucumber fruit spine is multicellular and non-branched with no endoreduplication. Spines in the tbh mutant were tiny and branched. Meristem regulators and polarity genes regulate spine development in cucumber.
Key words: Cucumber, fruit spine, meristem regulator, polarity transcriptome, trichome.
Abstract
Trichomes are epidermal hair-like structures that function in plant defence against biotic and abiotic stresses. Extensive studies have been performed on foliar trichomes development in Arabidopsis and tomato, but the molecular mechanism of fruit trichome formation remains elusive. Cucumber fruit is covered with trichomes (spines) that directly affect the appearance and quality of cucumber products. Here, we characterized the fruit spine development in wild-type (WT) cucumber and a spontaneous mutant, tiny branched hair (tbh). Our data showed that the cucumber trichome was multicellular and non-glandular, with malformed organelles and no endoreduplication. Fruit spine development was generally homogenous and marked by a rapid base expansion stage. Trichomes in the tbh mutant were tiny and branched, with increased density and aberrant cell shape. Transcriptome profiling indicated that meristem-related genes were highly enriched in the upregulated genes in the tbh versus the WT, as well as in WT spines after versus before base expansion, and that polarity regulators were greatly induced during spine base expansion. Quantitative reverse transcription PCR and in situ hybridization confirmed the differential expression of CUP-SHAPED COTYLEDON3 (CUC3) and SHOOT MERISTEMLESS (STM) during spine development. Therefore, cucumber trichomes are morphologically different from those of Arabidopsis and tomato, and their development may be regulated by a distinct pathway involving meristem genes and polarity regulators.
Introduction
Trichomes are hair-like structures that are widely present on the surface of aerial organs of plant. As the first line of defence, trichomes function in protecting plants against insects, pathogens, and herbivores, reducing plant damage from UV irradiation, low temperature, and excessive transpiration (Hülskamp et al., 1999; Hülskamp, 2004; Schellmann and Hülskamp, 2005). In addition, trichomes may help plants attract pollinators and disperse seeds (Serna and Martin, 2006). Morphologically, trichomes exhibit a broad variation in shape, size, distribution, and density, ranging from flat plates to highly ramified outgrowths, comprising a single cell or multiple cells, and being secretary glandular or non-glandular (Hülskamp et al., 1999; Hülskamp, 2004; Schellmann and Hülskamp, 2005).
Extensive studies have been conducted on unicellular trichomes, especially the leaf trichomes of the model plant Arabidopsis thaliana (Hülskamp et al., 1999; Hülskamp, 2004; Schellmann and Hülskamp, 2005; Machado et al., 2009). Trichomes of Arabidopsis are single cells that originate from epidermis cells and are distributed on leaves, stems, and sepals in a regular pattern (Pesch and Hülskamp, 2004). During trichome morphogenesis, Arabidopsis leaf trichomes experience six distinct developmental stages comprising radial expansion of precursor, stalk emergence, branch formation, expansion of stalk and branches, pointed tip development, and mature trichome formation with a papillate surface (Szymanski et al., 1998). Mutant analyses have identified several regulators that function in distinct developmental processes including trichome initiation and/or formation, endoreduplication, branch formation, and growth directionality (Schwab et al., 2000; Szymanski et al., 2000). For example, wild-type (WT) trichomes are well spaced (around three cells in between trichomes) and almost never occur next to each other (Schellmann and Hülskamp, 2005), and such patterns are regulated by an activator/inhibitor system (Schellmann et al., 2002; Pesch and Hülskamp, 2004). The activators of trichome patterning consist of the WD-repeat protein TRANSPARENT TESTA GLABRA1 (TTG1) (Galway et al., 1994; Walker et al., 1999), the basic helix–loop–helix (bHLH) protein GLABRA3 (GL3) or ENHANCER OF GLABRA3 (EGL3) (Payne et al., 2000; Zhang et al., 2003), and two R2-R3 type-MYB transcription factors, GLABRA1 (GL1) and MYB23 (Oppenheimer et al., 1991), whereas the single-repeat MYB proteins TRIPTYCHON (TRY) (Schellmann S et al., 2002; Pesch and Hülskamp, 2011), CAPRICE (CPC) (Wada et al., 1997, 2002), ENHANCER OF TRY AND CPC1 (ETC1), ETC2, and CAPRICE-LIKE MYB3 (Esch et al., 2004; Kirik et al., 2004a, b ; Simon et al., 2007) act partially redundantly as negative regulators. After gaining the trichome cell fate, the incipient trichome cells switch from mitotic divisions to endoreduplication, reaching a DNA content of 32C (Hülskamp et al., 1994; Schellmann and Hülskamp, 2005), and this cell-cycle control is co-ordinately regulated by many factors including SIAMESE (SIM), a nuclear-localized plant-type cell-cycle regulator that represses endoreduplication cycles, and the plant cytohormone gibberellin signalling regulator SPINDLY (SPY), which promotes endoreduplication (Chien and Sussex, 1996; Jacobsen et al., 1996; Churchman et al., 2006). Furthermore, trichome DNA content resulting from endoreduplication cycles positively correlates with branch numbers. WT trichomes on rosette leaves usually have three to four branches, while mutants with increased DNA content such as try and spy have trichomes with up to eight branches (Hülskamp M et al., 1994; Perazza et al., 1999).
Compared with unicellular trichomes, the development and regulatory networks of multicellular trichomes in plants are much less understood, with most of the work being carried out in tomato. Multicellular trichomes in tomato are classified into I–VII types and, based on whether trichomes contain or secret a mixture of chemicals that confer resistance against insects or pathogens, they are classified into glandular (types I, IV, VI, and VII) or non-glandular (types II, III, and V) trichomes (Luckwill, 1943; Kang et al., 2010). The majority of trichome research in tomato has focused on the morphology, chemical composition, defence against herbivores, and developmental regulators of glandular trichomes, and it has been shown that there are both conserved and divergent regulatory pathways involved in different types of trichomes (Li et al., 2004; Kang et al., 2010; Yang et al., 2011; Deng et al., 2012; Tian et al., 2012). So far, little is known about the developmental process of multicellular non-glandular trichomes and the underlying regulatory molecular mechanisms.
Cucumber (Cucumis sativus L.) is one of the most important vegetable crops and has been cultivated worldwide for over 3000 years (Huang et al., 2009). Cucumber fruit are usually harvested at 1–2 weeks after anthesis and then used as fresh product or processed into pickles. The development of cucumber fruit follows a stereotypical pattern with visible external and internal morphological changes (Ando et al., 2012). During early fruit development, deep ridges along the length of the fruit cover the surface of the fruit, and densely spaced fruit spines (trichomes on cucumber fruits) are randomly scattered relative to the ridges (Ando et al., 2012). The fruit spine is of agricultural importance in affecting the appearance and flavour of cucumber products. To the best of our knowledge, there are no reports about the regulation of fruit spine development, and there is no detailed characterization of any cucumber mutant with disturbed trichome development. In this study, we compared and explored the developmental process and internal cell structures of cucumber trichomes in the WT and in a glabrous mutant, tiny branched hair (tbh). We further performed comparative transcriptome profiling analyses to identify genes and gene networks that may be involved in cucumber trichome development. We found that cucumber trichomes are multicellular and non-glandular, with no branches or endoreduplication. Development of fruit spines was generally homogenous and marked by a rapid spine base expansion stage. Furthermore, we found that meristem-related genes and polarity regulators participate in the development of fruit spine in cucumber.
Materials and methods
Plant materials
Cucumber inbred line R1407 (WT) and tbh mutant plants were grown at two generations every year in a greenhouse in the experimental field of the China Agricultural University in Beijing. Pest control and water control were carried out according to standard protocols.
Scanning electron microscopy (SEM)
SEM of cucumber trichomes was performed on young fruits with eight different lengths (0.5, 1.0, 1.6, 1.85, 2.3, 3.5, 4.3, and 6.5cm) and two stages of leaves (juvenile and mature). Samples were fixed with 2.5% glutaraldehyde at 4 °C for approximately 24h, washed with PBS (pH 7.2) three times and post-fixed in 1% (v/v) OsO4. The samples were then dehydrated through an ethanol series (30, 50, 70, 80, 90, and 100%, three times), critical-point dried using a desiccator (HCP-2; Hitachi), and coated with gold palladium (EIKO IB-3). Images were taken with a Hitachi S-4700 scanning electron microscope using a 2kV accelerating voltage.
Transmission electron microscopy (TEM)
Fruit spines were isolated from WT cucumber fruits of 1.6–1.8cm in length using fine tweezers under a dissecting microscope. Spines and leaves were fixed in 2.5% (w/v) glutaraldehyde and rinsed thoroughly with 0.1M phosphate buffer. Samples were post-fixed with 1% Hungry acid, washed in 0.1M phosphate buffer, dehydrated through an acetone series (30, 50, 70, 80, 90, and 100%), and then embedded in Spurr’s resin. Thin sections were cut with a LEICA UC6I microtome and examined with a JEM-123O scanning transmission electron microscope.
Flow cytometry analysis
Flow cytometry was performed as described previously (Galbraith et al., 2001) using a BD FACSCalibur analyser. Spines from 12 WT cucumber fruit of 1.6–1.8cm in length were isolated and pooled as a sample. To minimize contamination from trichome tissues, leaves from tbh mutant plants were used as a negative control. Nuclei from spines or leaves were prepared and stained with 4’,6-diamidino-2-phenylindole (DAPI) as described previously (Galbraith et al., 2001). Histograms of DAPI fluorescence indicated the relative DNA content in each sample.
Plant materials for digital gene expression (DGE) analysis
Two sets of transcriptome profiling experiments were carried out using the DGE approach. Samples were collected at the same time on the same day. The first set was to compare the transcriptome profiles of pericarps (epidermis plus trichome) from cucumber fruit in WT and the tbh mutant. Pericarps of around 0.2cm thick were peeled off from 1.6–1.8cm cucumber fruits. Pericarps of three fruits from different plants were pooled together as one biological sample for each genotype. The second set was used to compare spine-specific transcriptome profiles in two developmental stages. Fruit spines from WT cucumber fruit that were 1.6 or 0.5cm in length were isolated under a dissecting microscope, and spines from at least five fruits from different plants were pooled as one biological sample. Two biological replications were performed for each set of experiments.
DGE library construction and sequencing
DGE library construction was performed as described previously (Eveland et al., 2010), with minor modifications. Briefly, total RNA was extracted using a Huayueyang RNA extraction kit (Huayueyang, China), and 6 µg of total RNA was used for constructing each library. Double-stranded cDNAs were synthesized using oligo(dT) beads (Invitrogen). The cDNAs were then digested with an anchoring restriction enzyme (NlaIII) and ligated to a 5’ adapter, which contains a recognition site for the type IIS restriction enzyme MmeI (New England Biolabs). Following MmeI digestion, a 3’ adapter was ligated. Tags flanked by both adapters were enriched by PCR for 15 cycles. The PCR products were run on a 6% polyacrylamide gel, and the ~105bp band was excised and purified. The DNA samples were sequenced on an Illumina HiSeq 2000 sequencer for 49 cycles.
Bioinformatics analysis of DGE data
Raw reads were trimmed for the adapter sequence and only 21 nt clean tags with a count of at least 2 in a library and without any ambiguous nucleotide (‘N’) were retained for further analysis. Clean tags were clustered into unique tags, which were mapped to the cucumber genome sequence (http://cucumber.genomics.org.cn, v.2i) (Huang et al., 2009) using the SOAP2 software (Li et al., 2009), allowing up to one mismatch. To recover tags that cover intron–exon junctions, unmapped tags were aligned to the annotated cucumber transcripts using SOAP2, allowing up to one mismatch. If a tag was mapped to multiple locations in the genome, the tag count was divided by the number of mapped locations. The expression value of a gene was calculated as the total count of all tags that were mapped to the sense strand of the gene region. Following the convention set as described (Eveland et al., 2010), gene boundaries were extended for 300bp on both 5’ and 3’ ends to maximize the capture of complete untranslated regions. EdgeR (Robinson et al., 2010) was used to identify genes that were differentially expressed in fruit spines of WT versus tbh mutant, and fruit spines from cucumber fruit of 0.5 versus 1.6cm long. The method of Benjamini and Hochberg (1995) was used for adjustment for multiple comparisons. A false discovery rate (FDR) of 0.05 was used as the significance cut-off.
Sequencing data were deposited in the Gene Expression Omnibus (GEO) database at the National Center for Biotechnology Information under accession number GSE49607.
Gene Ontology (GO) term enrichment analysis
Because the GO terms are not well annotated for cucumber genes, we used the best homologues in Arabidopsis for GO term enrichment analysis. We first used the cucumber annotated proteins to search the Arabidopsis proteins (TAIR10) using BLASTP with an e-value cut-off of 1e–5, and identified the best homologue (with the lowest e-value) in Arabidopsis for each cucumber gene. For the genes that were upregulated and downregulated, respectively, in each of the two pairwise transcriptome comparisons, we collected the corresponding Arabidopsis best homologues and used the GOEAST software (Zheng and Wang, 2008) to test for GO term enrichment. GOEAST was run with default parameters except for the use of algorithms to eliminate local dependencies between GO terms (Alexa et al., 2006).
Quantitative and semi-quantitative reverse transcription (RT)-PCR
To confirm the results of the DGE analyses, samples for RT-PCR were independently collected from pericarps or spines from the same batch of plants at the same time as in the DGE analyses. For Supplementary Table S4 (available at JXB online), RNA samples were exacted from the third true leaves or roots of 4-week-old cucumber seedlings. Total RNA was isolated with a Huayueyang RNA extraction kit and then reverse transcribed by Moloney murine leukemia virus reverse transcriptase using random primers. Quantitative RT-PCR (qRT-PCR)was performed on an Applied Biosystems 7500 real-time PCR system using SYBR Premix Ex Taq (TaKaRa). Both qRT-PCR and semi-quantitative PCR were repeated with three biological samples. The cucumber UBIQUITIN gene was used as reference control to normalize the expression data (Wan et al., 2010). The gene-specific primers are listed in Supplementary Table S5 available at JXB online.
In situ hybridization
Cucumber young fruits from WT and tbh mutant were fixed and hybridized as described previously (Zhang et al., 2013). In situ probes were generated through PCR amplification using gene-specific primers with T7 and SP6 RNA polymerase-binding sites. The gene-specific primers are listed in Supplementary Table S5 available at JXB online.
Results
Morphological characterization of the tbh mutant in cucumber
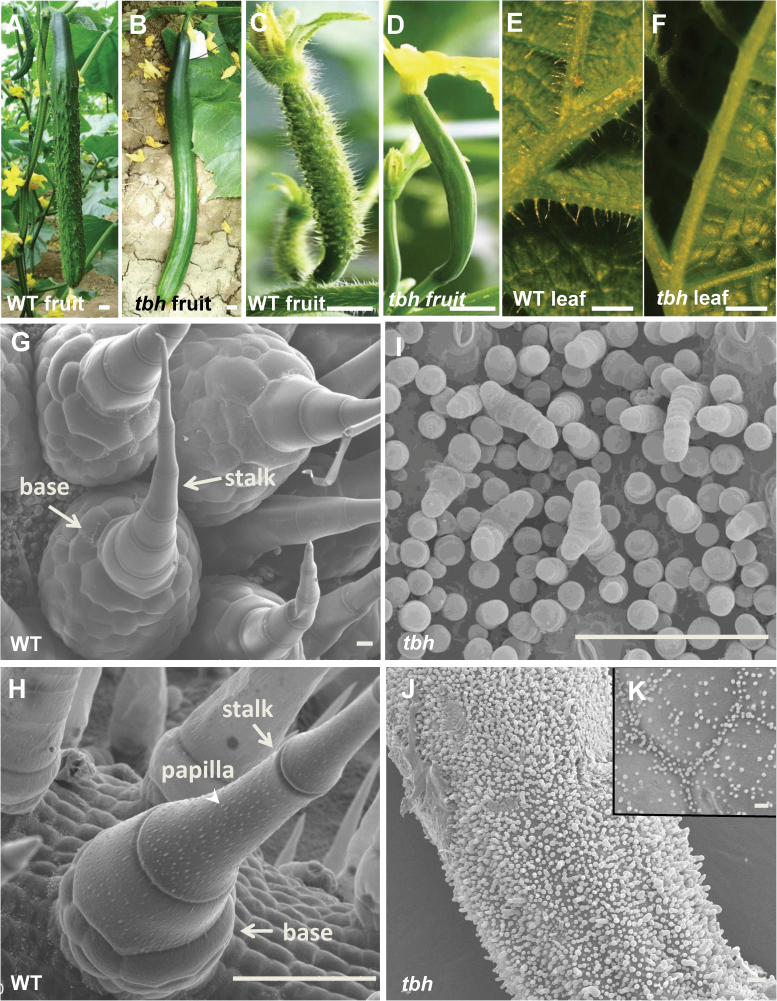
From the cucumber inbred line R1407, a spontaneous mutant, tbh, was identified by its ‘glabrous’ phenotype: hairless foliage and smooth fruit surface. We further stabilized the tbh mutation via six generations of selfing prior to this study. By the commercially mature stage, the WT fruit was dark green with spines and warts that were evenly distributed on the fruit surface (Fig. 1A and Supplementary Fig. S1A available at JXB online), whereas the tbh fruit appeared to be smooth and shining, with no noticeable spines or warts (Fig. 1B). During the stage of anthesis, WT young fruit was covered with densely spaced spines (Fig. 1C), while tbh fruit was glabrous (Fig. 1D). We next examined the cucumber trichomes with an optical microscope (Fig. 1E, F, Supplementary Fig. S1B–G available at JXB online). In the WT plants, leaves (Fig. 1E), tendrils (Supplementary Fig. S1B available at JXB online), stems (Supplementary Fig. S1C available at JXB online), and male flower buds (Supplementary Fig. S1D available at JXB online) were all covered with hairs, but in the tbh mutant plants, no evident trichomes were observed throughout the plant (Fig. 1F, Supplementary Fig. S1E–G available at JXB online). To characterize the morphology of cucumber trichomes in details, we observed fruit spines and leaf trichomes with SEM. Unlike the single-cell branched trichomes in Arabidopsis or multiple types of trichomes in tomato (Hülskamp et al., 1994; Kang et al., 2010; Tominaga-Wada et al., 2013), there were only two types of trichomes in cucumber, and both were multicellular (Supplementary Fig. S2A available at JXB online). Type I trichomes were tiny with a three-to-five-cell base topped with a four-to-eight-cell head (Supplementary Fig. S2B available at JXB online). Type I trichomes were shown to be involved in cuticle formation (Samuels et al., 1993). Type II trichomes were much bigger, with a conical shape, and were non-glandular and branchless (Fig. 1G, H). Type II trichomes were the dominant type that we observed in cucumber, and thus will be our focus hereafter. Each mature type II trichome was comprised of a base and a stalk. The base was made of hundreds of spherical-shaped cells whose sizes were smaller than those of the stalk cells. The stalk was consisted of three to seven cylindrical-shaped cells plus a pyramid-shaped apical cell (Fig. 1G, H). Therefore, the cucumber trichomes appeared sharp on the top. The leaf trichomes and fruit spines in cucumber had a similar structure and morphology, but there were three differences: (i) the size of the spine base was much bigger than that in the leaf trichome; (ii) white papilla (arrowhead in Fig. 1H) was observed on mature leaf trichomes but not on fruit spines (compare Fig. 1G and H); and (iii) trichome spacing was generally even on the fruit but uneven on cucumber leaves. There were more trichomes on the veins than other leaf areas (Fig. 1E and Supplementary Fig. S1A available at JXB online). In the tbh mutant, trichome morphology was strikingly defective. The most obvious characteristics of tbh trichomes were that they were ‘tiny’ with a greatly reduced number of cells, and that the cell shape and organization were so aberrant that it was difficult to separate the base and the stalk (Fig. 1I, K). Moreover, trichomes in the tbh mutant varied from no branches to up to four branches, and the apical cell was spherical (Fig. 1I). Hence, the top of the trichome in the tbh mutant was round, which was dramatically different from the ‘sharp tip’ in the WT. Furthermore, the density of trichome distribution in the tbh mutant was much higher compared with that in the WT, varying from 4-fold higher in the young leaves to around 118-fold more trichomes on the fruit of tbh during anthesis compared with those in WT, suggesting that TBH functions in both trichome outgrowth and trichome spacing. In addition, there was no noticeable difference between the leaf trichome and fruit spine in the tbh mutant with regard to trichome structure and morphology, despite the distribution pattern being different [distributed evenly on the fruit surface (Fig. 1J) but unevenly on the leaf surface (Fig. 1K)]. Since fruit spines directly affect the appearance and quality of the cucumber fruit, the rest of our study focused on the characterization of fruit spines.
Fig. 1.
Morphological characterization of the tbh mutant. (A, B) Commercially mature fruit of WT (A) and tbh mutant (B). (C–F) Light microscope images of young cucumber fruit around anthesis (C, D) and the back of leaves (E, F) in WT (C, E) and tbh mutant (D, F). (G–K) SEM images of fruit spines (G, I, J) and leaf trichomes (H, K) in WT (G, H) and tbh mutant (I–K). Bars, 1cm (A–F); 50 µm (G–K). (This figure is available in colour at JXB online.)
Internal cell structure of cucumber trichomes
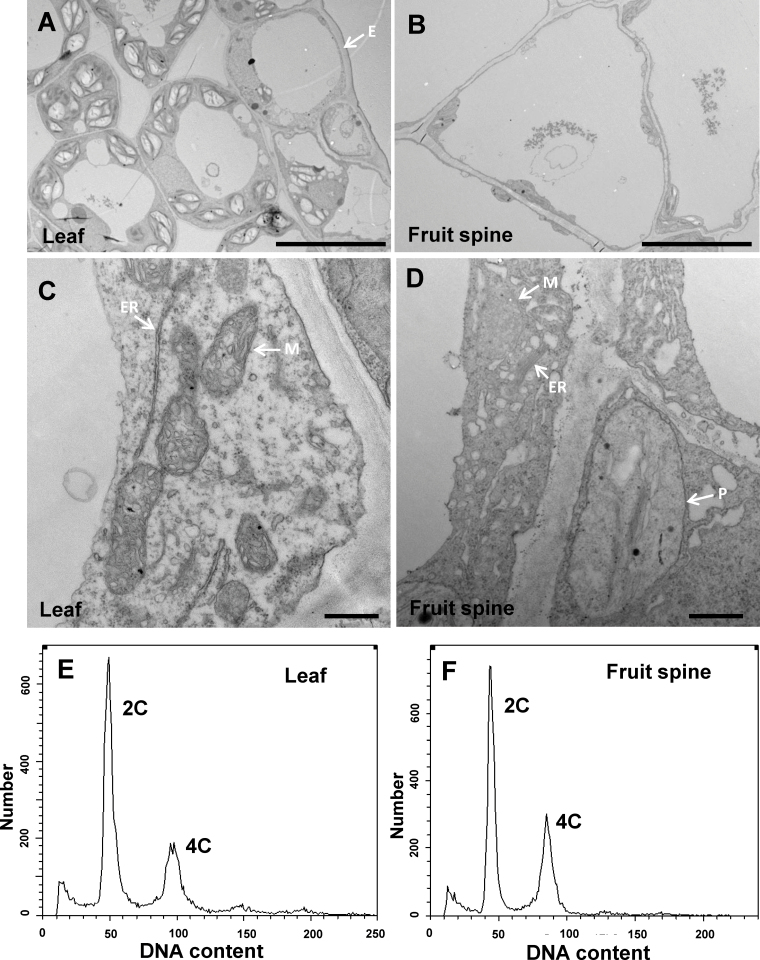
The unicellular Arabidopsis trichomes have been thought to originate from the epidermal cells (Marks, 1997; Hülskamp et al., 1999; Larkin et al., 2003), but comparison of the internal structure of trichomes and epidermal cells is lacking due to technical difficulties of fixing individual trichome cells. Taking advantage of the multicellular property of cucumber trichomes and the large base in fruit spines, we explored the cellular structure of cucumber trichomes using TEM (Fig. 2A–D). Compared with the abaxial leaf epidermal cells (Fig. 2A, C), the base cells of fruit spine were much larger, with an enormous central vacuole and the internal organelles being squeezed close to the cell wall (Fig. 2B). The endoplasmic reticulum and mitochondria appeared to be shorter and swollen in the cucumber trichome cells, probably due to the presence of a large central vacuole (Fig. 2D). Furthermore, misshaped plastids were often observed in the spine base cells but not in the leaf epidermal cells. The light green colour of the cucumber spine base supported the existence of coloured plastids or even chloroplasts in the spine cells (Supplementary Fig. S1A available at JXB online). The striking differences in morphology and cellular structure between cucumber and Arabidopsis trichomes suggest that cucumber trichomes may have a distinct origin from the epidermal-derived Arabidopsis trichomes and may have a divergent developmental mechanism.
Fig. 2.
Internal structure of cucumber trichome cells. (A–D) Transmission electron micrographs of the abaxial leaf cells (A, C) or the base cells of fruit spine (B, D) from WT cucumber. Bars, 10 µm (A, B); 500nm (C, D). (E, F) Flow cytometric analysis of the DNA content of nuclei in the tbh mutant leaf tissue (E) and WT fruit spines (F). The histograms show the DNA content of each nucleus. E, epidermal cell; P, plastid; ER, endoplasmic reticulum; M, mitochondria.
Previous studies have shown that the nuclear DNA content, an indicator of endoreduplication, is often positively correlated with cell size (Melaragno et al., 1993), and that Arabidopsis trichome cells undergo four rounds of endoreduplication resulting in a genomic DNA content of 32C (Hülskamp et al., 1994). To determine whether endoreduplication contributed to the enlarged cucumber trichome cells, we measured the DNA content of mature fruit spines using flow cytometry. Surprisingly, no difference in nuclear ploidy levels was observed between fruit spines and leaf tissue (2C and 4C) (Fig. 2E, F), suggesting that cucumber trichome cells did not undergo any endoreduplication.
Developmental stages of cucumber fruit trichomes
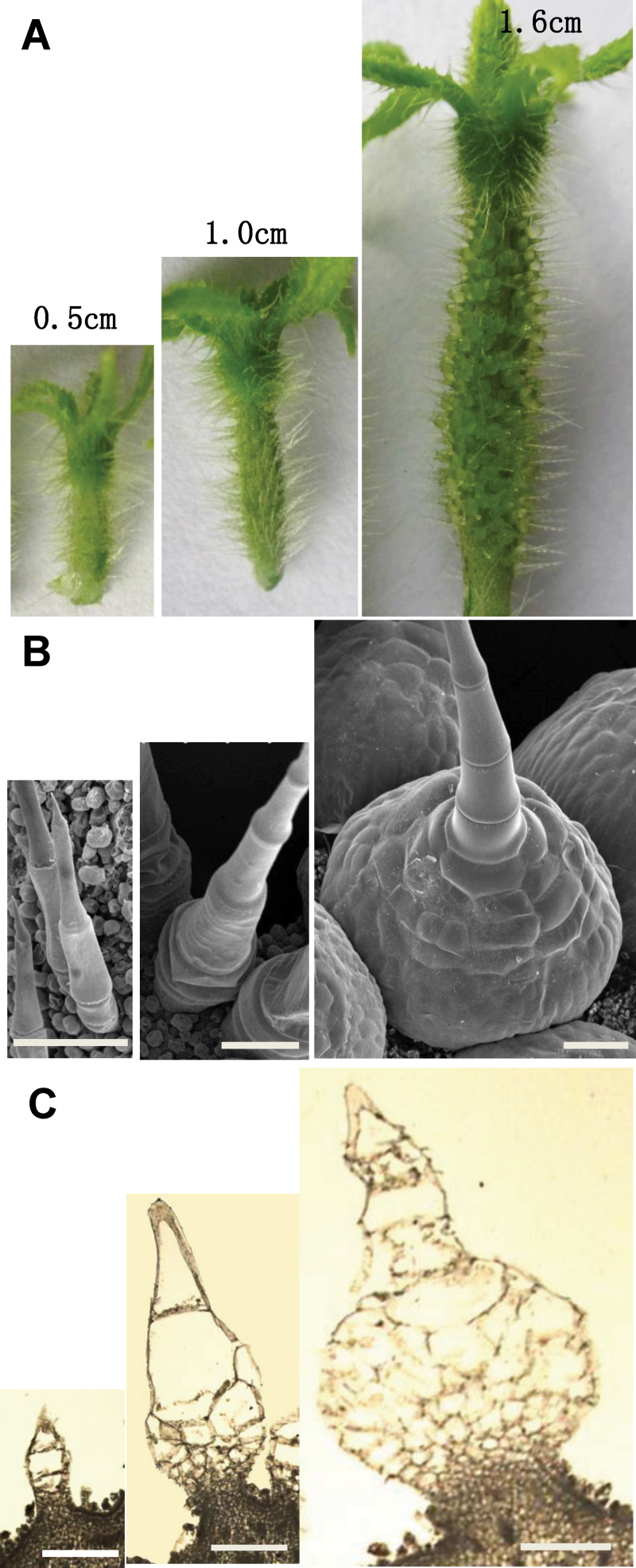
To characterize further the developmental process of cucumber trichomes in detail, we observed the spines of eight developmental stages of WT cucumber fruit (based on fruit size) using SEM (Supplementary Fig. S3 available at JXB online). Unlike leaf trichomes, which usually undergo distinct stages at the same time, even on the same leaf, cucumber fruit spines were generally homogenous (at the same developmental stage) on the same fruit (Supplementary Fig. S3 available at JXB online). The most dramatic change during spine development was the expansion of the spine base. When the fruit length was about 0.5cm long (approximately 7 d before anthesis), the spine consisted of a few cells that were organized in one tile, with a ‘sharp’ stalk on the top and a single-cell base (Fig. 3, left). The spine base experienced a rapid expansion through cell division when the fruit length was between 0.5 and 1.5cm (Fig. 3, middle). When the fruit length reached around 1.6cm (approximately 4 d before anthesis), the spine base became a dome-shaped body that contained hundreds of cells and could be easily observed by bare eyes (Fig. 3, right). The spine base continued to expand until the fruit reached around 4.3cm (during anthesis), after which its shape and size were maintained as relatively constant (Supplementary Fig. S3 available at JXB online). Based on these observations, we defined a fruit length of 0.5–1.6cm as the key stage for spine base expansion in our cucumber cultivar.
Fig. 3.
Key developmental stages of spine base expansion in cucumber fruits. Digital camera images (A), SEM images (B) and paraffin sections (C) of cucumber spine before base expansion (left, fruit length around 0.5cm), during base expansion (middle, fruit length around 1.0cm), and after rapid base expansion (right, fruit length around 1.6cm). Bars, 200 µm (B, C).). (This figure is available in colour at JXB online.)
Transcriptome analyses of cucumber fruit spines
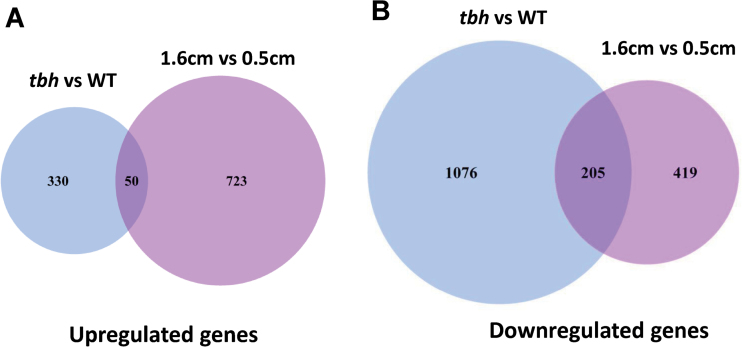
To identify genes and gene networks that were involved in the multicellular fruit spine development in cucumber, we performed two sets of genome-wide expression analyses using the DGE approach (Eveland et al., 2010). One set was to compare the transcriptome profiles of the pericarps (epidermis plus spines) of 1.6–1.8cm cucumber fruits between WT and the tbh mutant, and the other set was to examine WT spine-specific expression profiles before (fruit length around 0.5cm) versus after (fruit length around 1.6cm) spine base expansion. Two biological replicates were performed for each set, and thus eight DGE libraries were sequenced (two comparisons×two tissues×tworeplicates). We generated 6.95–7.49 million raw reads from each library. After adapter sequence and low-quality tags had been removed, we obtained 6.80–7.33 million clean tags that were 21 nt long and with counts of at least 2 in a library (Supplementary Table S1 available at JXB online). We clustered clean tags into unique tags, which were mapped to the cucumber genome and annotated transcripts. We summarized the total counts of tags that were mapped to the sense strand of each annotated cucumber gene region, which represented the expression levels in each sample. Using an FDR of 0.05 as the significance cut-off, we found that the number of downregulated genes (1281) was dramatically more than that of upregulated genes (380) in the tbh mutant compared with the WT (Fig. 4, Supplementary Table S2 available at JXB online), suggesting that the TBH locus may function as a major activator during fruit spine development. In the 2 (1.6 vs 0.5cm) of transcriptome comparison, we also found that 1397 genes were differentially expressed, in which 773 genes were upregulated and 624 genes were downregulated in the spines of 1.6cm long cucumber fruit compared with the spines of 0.5cm fruit (Fig. 4, Supplementary Table S3 available at JXB online).
Fig. 4.
Venn diagrams of differentially expressed genes that were significantly upregulated (A) or downregulated (B) (FDR <0.05) in the tbh mutant (blue) or in the spines of 1.6cm fruit (purple). (This figure is available in colour at JXB online.)
To validate the differentially expressed genes (DEGs) identified by DGE, we performed qRT-PCR assays using independently generated cucumber pericarp and fruit spine samples during the same developmental stages as those used in DGE. We randomly chose 20 DEGs for qRT-PCR analysis, in which eight were from set 1 (tbh vs WT) and 12 were from set 2 (1.6 vs 0.5cm). The qRT-PCR and DGE data showed close agreement (Pearson correlation coefficient 0.996, P=1.59E–21) (Table 1), indicating that the DGE results were highly reliable.
Table 1.
qRT-PCR confirmation of differentially expressed cucumber genes identified by DGE
| Gene ID | Putative annotation | DGE P value | DGE fold change | qRT-PCR fold change |
|---|---|---|---|---|
| tbh vs WT | ||||
| Csa1G064670 | ATML1 (MERISTEM LAYER 1) | 1.6E–05 | 3.2 | 3.2±0.41 |
| Csa6G514870 | PDF2 (PROTODERMAL FACTOR 2) | 1.5E–05 | 2.3 | 2.2±0.09 |
| Csa4G256430 | BOP2 (BLADE ON PETIOLE2) | 3.0E–03 | 2.7 | 4.1±0.57 |
| Csa6G355380 | CUC3 (CUP-SHAPED COTYLEDON3) | 6.4E–39 | –1638.5 | –481.4±1.59 |
| Csa2G167190 | BMY7 (BETA-AMYLASE 7) | 7.9E–13 | –15.8 | –5.4±0.43 |
| Csa1G628000 | LRP1 (LATERAL ROOT PRIMORDIUM 1) | 7.5E–20 | –6.5 | –7.5±0.37 |
| Csa6G501990 | HB-5 (HOMEOBOX PROTEIN 5) | 2.4E–51 | –142.9 | –5.8±1.78 |
| Csa3G824850 | MYB106 (MYB DOMAIN PROTEIN 106) | 3.3E–19 | –12.7 | –20.6±1.19 |
| 1.6cm vs 0.5 cm | ||||
| Csa7G041370 | STM (SHOOT MERISTEMLESS) | 3.4E–09 | 37.8 | 22.8±0.89 |
| Csa6G012240 | LBD21 (LOB DOMAIN-containing PROTEIN 21) | 5.2E–07 | 10.4 | 12.6±0.94 |
| Csa5G172270 | GA5(GA REQUIRING 5) | 1.3E–05 | 8.4 | 3.4±0.44 |
| Csa3G895630 | ATHB-8 (HOMEOBOX GENE 8) | 4.5E–07 | 6.3 | 2.1±0.75 |
| Csa6G496390 | ANT (AINTEGUMENTA) | 1.2E–04 | 4.0 | 3.7±0.71 |
| Csa6G426940 | YAB2 (YABBY2) | 6.7E–04 | 3.7 | 3.0±0.88 |
| Csa2G348930 | KNAT6 (KNOTTED1-LIKE HOMEOBOX GENE 6) | 2.0E–03 | 3.6 | 3.1±0.61 |
| Csa3G194380 | KAN1 (KANADI1) | 1.8E–03 | 3.0 | 4.5±0.65 |
| Csa3G143580 | SHY (SHORT HYPOCOTYL) | 1.4E–05 | –5.1 | –7.4±0.63 |
| Csa1G064670 | ATML1 (MERISTEM LAYER 1) | 2.2E–05 | –6.4 | –11.2±0.63 |
| Csa1G397130 | SHY2 (SHORT HYPOCOTYL 2) | 1.6E–06 | –7.1 | –3.7±0.79 |
| Csa6G367090 | AP1 (APETALA1) | 1.2E–04 | –7.2 | –4.7±1.18 |
Meristem regulators and transcription factors implicated in cucumber fruit spine development
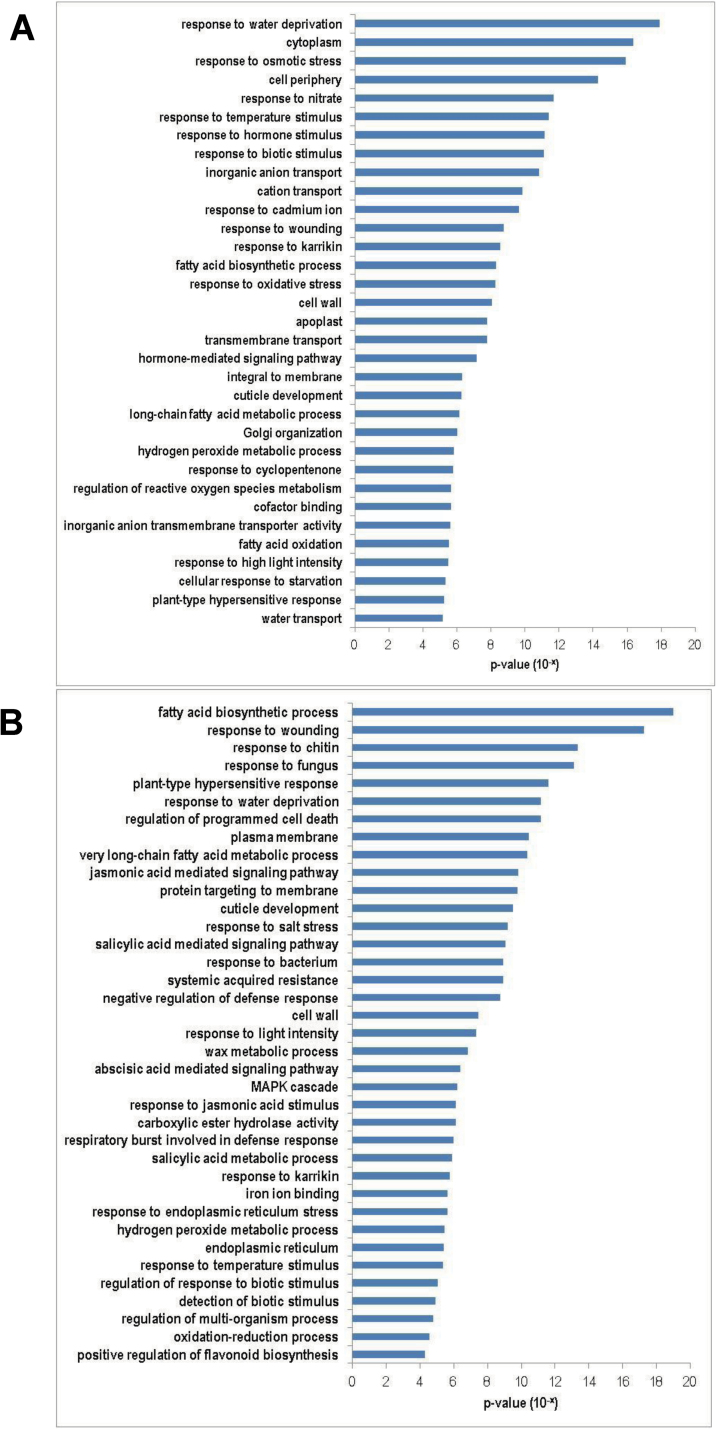
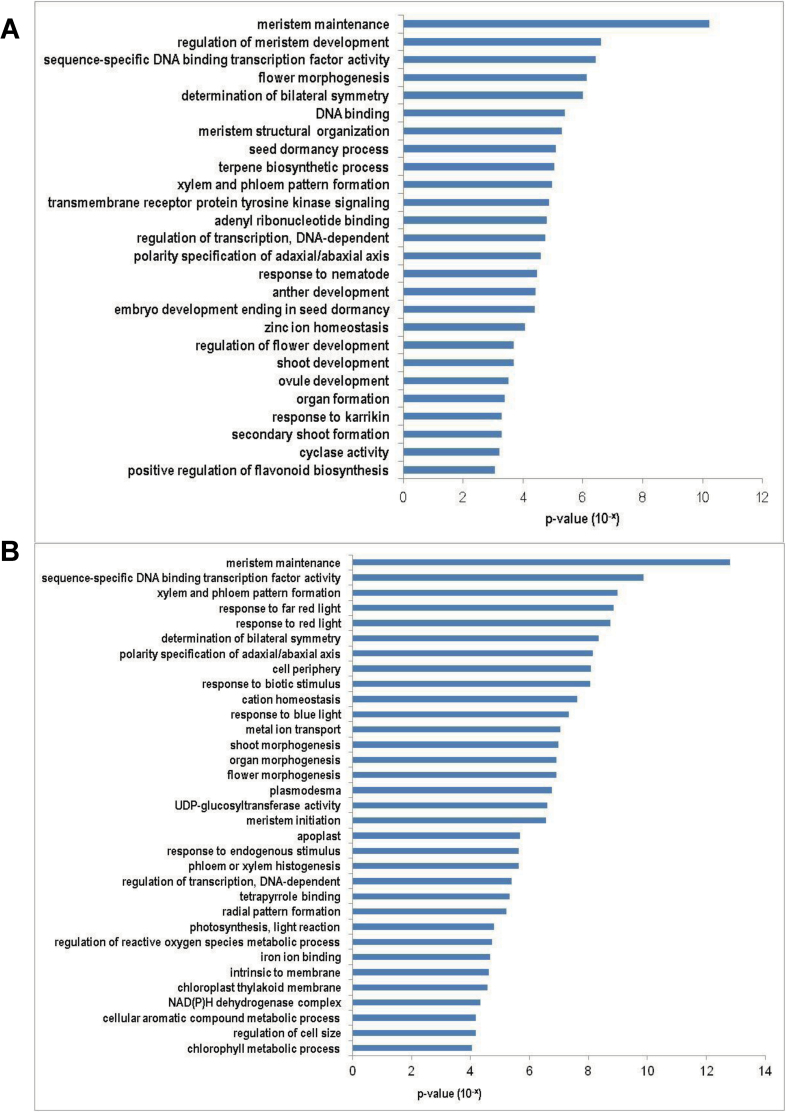
To analyse the functions of DEGs identified by DGE, GO term enrichment analyses were carried out for the up- and downregulated genes in both sets of data. Stress-related and metabolic pathway genes were significantly enriched in the DEGs that were downregulated in both tbh mutant (Fig. 5A) and spines of 1.6cm fruit (after base expansion) (Fig. 5B). For example, GO terms ‘response to water deprivation’ (P=1.2E–18), ‘cytoplasm’ (P=4.1E–17), and ‘response to osmatic stress’ (P=1.7E–16) were the top three significantly enriched groups in genes that were downregulated in the tbh mutant (Fig. 5A), whereas ‘fatty acid biosynthetic process’ (P=1.0E–19), ‘response to wounding’ (P=5.6E–18), and ‘response to chitin’ (P=4.4E–14) were the most significantly enriched GO terms in the downregulated genes in the spines of 1.6cm fruit. In contrast, meristem regulators and transcription factors were significantly enriched in the genes that were upregulated in both the tbh mutant (Fig. 6A) and the spines of 1.6cm fruit (after base expansion) (Fig. 6B, Tables 2 and 3). The most significantly enriched GO term was ‘meristem maintenance’ for both sets of data, with a P value of 6.12E–11 and 1.56E–13 in the tbh vs WT and the 1.6 versus 0.5cm comparisons, respectively. Accordingly, many well-known meristem regulators were significantly induced in the tbh mutant (Table 2) and in the spines of 1.6cm fruit (Table 3). For example, the expression levels of REPLUMLESS (RPL) and MERISTEM LAYER 1(ATML1) were 3.6- and 3.2-fold higher, respectively, in the tbh mutant compared with those in the WT (Table 2), and the expression of SHOOT MERISTEMLESS (STM) and ZWILLE (ZLL) increased 37.8- and 9.0-fold, respectively, in the spines of 1.6cm fruit compared with those in 0.5cm fruit (Table 3). The GO term ‘sequence-specific DNA-binding transcription factor activity’ was also significantly enriched in the genes that were upregulated in both tbh mutant (P=3.67E–07) and the spines of 1.6cm fruit (P=1.34E–10), suggesting that meristem genes and transcription factors mediate the fruit spine development in cucumber. Notably, despite many GO terms being commonly enriched in the upregulated genes in the tbh mutant and in spines of 1.6cm fruit (Fig. 6), only a few genes were shared in the two sets of upregulated genes (Fig. 4). Taken together with the fact that the tbh mutant has ‘tiny’ trichomes with a greatly reduced number of cells, and the spines of 1.6cm fruit have enormous base expansion with tremendously increased cell numbers compared with the spines of 0.5cm fruit, our data imply that distinct members of meristem regulators and transcription factors may regulate the TBH-mediated trichome development and spine base expansion in cucumber, and that some of them may function in opposite directions during cell division and trichome development.
Fig. 5.
GO terms that were significantly enriched (P<0.01) in the downregulated genes in the tbh mutant (A) or in the spines of 1.6cm long fruit (B). GO terms were sorted based on P values.
Fig. 6.
GO terms that were significantly enriched (P<0.01) in the upregulated genes in the tbh mutant (A) or in the spines of 1.6cm fruit (B). GO terms were sorted based on P values.
Table 2.
Examples of developmental regulators that were identified by DGE to be differentially expressed in the fruit spines of tbh mutant and WT
| Gene ID | Gene name | Fold change | P value |
|---|---|---|---|
| Meristem maintenance and regulation | |||
| Csa4G297540 | RPL (REPLUMLESS) | 3.6 | 5.0E–04 |
| Csa3G144740 | ZLL (ZWILLE) | 2.0 | 1.9E–03 |
| Csa1G064670 | ATML1 (MERISTEM LAYER 1) | 3.2 | 1.6E–05 |
| Csa6G497080 | BAM1 (BARELY ANY MERISTEM 1) | 1.8 | 3.3E–03 |
| Csa1G536820 | ARF6 (AUXIN RESPONSE FACTOR 6) | 1.9 | 1.9E–03 |
| Csa4G256430 | BOP2 (BLADE ON PETIOLE2) | 2.7 | 2.9E–03 |
| Csa3G736760 | AP2 (APETALA 2) | 2.6 | 1.1E–04 |
| Csa5G118190 | LPR1 (Low Phosphate Root1) | 2.5 | 2.5E–04 |
| Sequence-specific DNA-binding transcription factor activity | |||
| Csa4G645830 | KNAT1 (KNOTTED-LIKE FROM ARABIDOPSIS THALIANA) | 3.1 | 7.7E–05 |
| Csa4G638510 | MYB77 (myb domain protein 77) | 2.9 | 2.5E–07 |
| Csa3G812750 | MYB105 (myb domain protein 105) | 3.2 | 1.7E–04 |
| Csa3G354520 | SPCH (SPEECHLESS) | 2.6 | 1.2E–04 |
| Csa3G809420 | SPL9 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 9) | 2.2 | 4.7E–04 |
| Csa2G369820 | ATHB22 (HOMEOBOX PROTEIN 22) | 2.1 | 6.9E–04 |
| Csa2G021550 | bHLH family protein (bHLH096) | 2.1 | 8.5E–04 |
| Csa5G636510 | AtHB34 (ARABIDOPSIS THALIANA HOMEOB-OX PROTEIN 34) | 2.1 | 3.3E–04 |
Table 3.
Examples of developmental regulators that were identified by DGE to be differentially expressed in the spines of 1.6 and 0.5cm cucumber fruit
| Gene ID | Gene name | Fold change (1.6/0.5cm) | P value |
|---|---|---|---|
| Meristem maintenance | |||
| Csa7G041370 | STM (SHOOT MERISTEMLESS) | 37.8 | 3.4E–09 |
| Csa1G408710 | ZLL (ZWILLE) | 9.0 | 1.8E–03 |
| Csa1G025070 | PIN1 (PIN-FORMED 1) | 19.9 | 1.7E–08 |
| Csa6G139130 | LBD4 (LOB DOMAIN-CONTAINING PROTEIN 4) | 6.3 | 4.6E–03 |
| Csa4G103350 | DWF1 (DWARF 1) | 3.7 | 1.2E–03 |
| Csa5G608050 | NIK1 (NSP-INTERACTING KINASE 1) | 9.2 | 4.6E–06 |
| Csa6G109640 | SCL6 (scarecrow-like transcription factor 6) | 3.5 | 1.8E–03 |
| Csa5G642730 | NIK1 (NSP-INTERACTING KINASE 1) | 6.6 | 1.9E–06 |
| Csa5G092940 | BRL2 (BRI1-LIKE 2) | 149.6 | 1.7E–06 |
| Sequence-specific DNA-binding transcription factor activity | |||
| Csa4G046650 | MNP (MONOPOLE) | 119.1 | 9.9E–06 |
| Csa4G652000 | WRKY35 (WRKY DNA-binding protein 40) | 53.5 | 4.3E–10 |
| Csa1G505950 | Dof-type zinc finger domain-containing protein | 19.1 | 8.6E–09 |
| Csa2G356610 | WOX4 (WUSCHEL RELATED HOMEOBOX 4) | 157.2 | 1.0E–06 |
| Csa3G895630 | ATHB-8 (HOMEOBOX GENE 8) | 6.3 | 4.5E–07 |
| Csa6G081510 | SHR (SHORT ROOT) | 214.4 | 1.5E–07 |
| Csa6G135460 | APL (ALTERED PHLOEM DEVELOPMENT) | 214.4 | 4.6E–08 |
| Csa1G423190 | AIL5 (AINTEGUMENTA-LIKE 5) | 157.2 | 1.2E–06 |
| Csa2G035350 | MYB50 (MYB DOMAIN PROTEIN 50) | 286.7 | 6.2E–09 |
| Csa6G526230 | WRKY35 (WRKY DNA-binding protein 35) | 256.3 | 9.7E–08 |
| Csa6G426940 | YAB2 (YABBY2) | 3.7 | 6.7E–04 |
| Vascular patterning and polarity specification | |||
| Csa6G141360 | REV (REVOLUTA) | 4.7 | 1.4E–05 |
| Csa6G525430 | PHB (PHABULOSA) | 3.4 | 9.2E–04 |
| Csa3G194380 | KAN1 (KANADI1) | 3.0 | 1.8E–03 |
| Csa4G644740 | ANT (AINTEGUMENTA) | 8.2 | 4.5E–06 |
| Csa6G496390 | ANT (AINTEGUMENTA) | 4.0 | 1.2E–04 |
| Csa3G009500 | TRN2 (TORNADO 2) | 17.7 | 3.2E–06 |
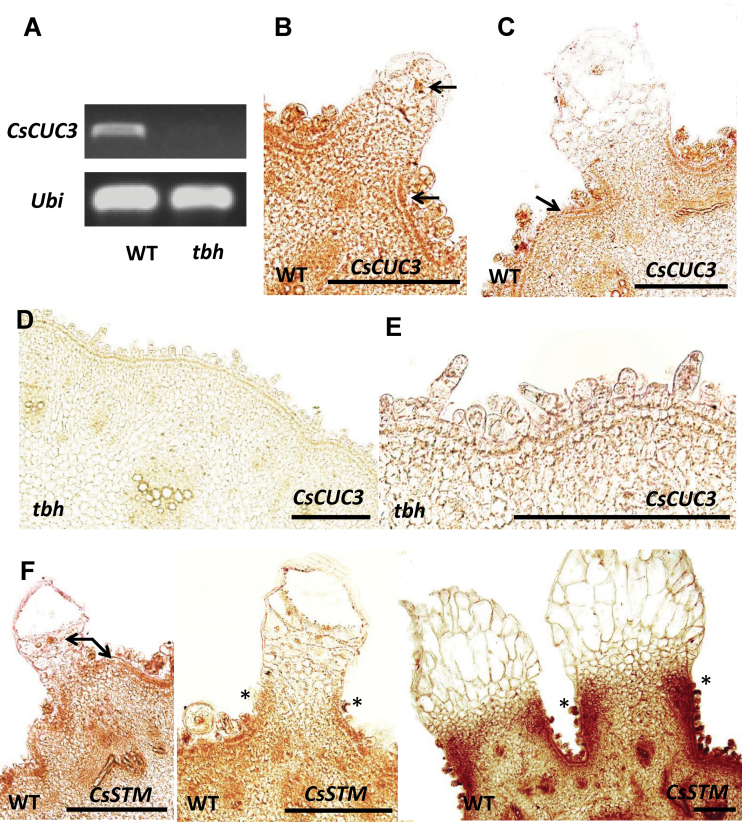
To explore further the role of meristem genes in trichome development, we examined the expression patterns of two meristem regulators, CUP-SHAPED COTYLEDON3 (CUC3) and STM by in situ hybridization (Fig. 7). CUC3 encodes a member of the NAC domain transcription factor family that is expressed in the meristem–organ boundaries and has been shown to regulate meristem organization, organ separation, and branching in Arabidopsis (Vroemen et al., 2003; Hibara et al., 2006). STM, which is expressed throughout the meristem but is downregulated in the organ primordia, encodes a member of the class I KNOX homeodomain transcription factors, which functions as a key regulator for meristem formation and maintenance (Clark et al., 1996; Long et al., 1996). In our DGE data, the expression of cucumber CUC3 (CsCUC3) was over 1638-fold decreased in the tbh mutant, and we confirmed this downregulation via qRT-PCR (481-fold reduction) and semi-quantitative RT-PCR (Table 1, Fig. 7A). In situ hybridization showed that CsCUC3 was strongly expressed in the epidermis of WT cucumber fruit, as well as in the nucleus of spine cells (arrows in Fig. 7B, C) but was almost abolished in the tbh mutant (Fig. 7D, E). This was consistent with the dramatic reduction in CsCUC3 expression in the tbh mutant revealed by DGE and RT-PCR data, suggesting that the activation of CsCUC3 by the TBH-related pathway may be important for fruit spine development in cucumber. Another meristem regulator, STM, was shown to be induced by 37.8-fold as detected by DGE and 22.8-fold as detected by qRT-PCR in the spines of 1.6cm fruit versus 0.5cm fruit (Table 1). Consistently, in situ hybridization showed that, in young spines before base expansion (on 0.5cm fruit), cucumber STM (CsSTM) was present in the epidermis and nucleus of spine cells (arrows in Fig. 7F, left). During spine development, CsSTM was strongly expressed in the junction between the spine base and fruit wart (denoted by asterisks in Fig. 7F, middle and right). This pattern is similar to the high accumulation of STM at the leaflet initiation sites in complex leaves (Bharathan et al., 2002; Hake et al., 2004), implying that STM may promote fruit base expansion in cucumber.
Fig. 7.
Expression analyses of CsCUC3 and CsSTM during fruit spine development in cucumber. (A) Semi-quantitative RT-PCR of CsCUC3 in pericarps of WT and tbh mutant. (B–F) Gene expression patterns as detected by in situ hybridization. CsCUC3 was expressed in the epidermis and nucleus of spine cells in developing fruit spines of WT (arrows in B and C), but was undetectable in the tbh mutant (D, E). (F) Transcript accumulation of CsSTM was localized in the epidermis and nucleus of spine cells (arrows), and was strongly induced at the junction of the fruit base and wart during fruit base expansion (asteroids) in WT cucumber. Bars, 200 µm. (This figure is available in colour at JXB online.)
Vascular patterning and polarity genes were induced during base expansion of cucumber fruit spines
Among the genes that were upregulated in the spines of 1.6cm fruit compared with those of 0.5cm fruit, genes that belong to GO terms ‘xylem and phloem pattern formation’ (P=1.0E–09) and ‘polarity specification of adaxial/abaxial axis’ (P =7.0E–09) were significantly enriched (Fig. 6B). Consequently, many well-known regulators for polarity specification and vascular patterning were significantly upregulated during fruit base expansion (Table 3). For example, the expression of cucumber homologues of the adaxial/xylem development marker REVOLUTA (REV) and the abaxial identity gene KANADI1 (KAN1) was 4.7- and 3.0-fold higher, respectively, in the spines of 1.6cm fruit (Table 3) (Emery et al., 2003; Juarez et al., 2004; Nakata and Okada, 2012). In situ hybridization showed that CsKAN was expressed predominately in the nucleus of spine cells as well as in the epidermis (arrows in Supplementary Fig. S4 available at JXB online). Due to the presence of huge central vacuoles and squeezed internal organelles in the mature fruit spine, we were unable to locate the nucleus in the sections and were thus unable to detect a polarity pattern of CsKAN expression using in situ hybridization technology. However, we did qRT-PCR analyses and confirmed that CsKAN was indeed upregulated (4.5-fold) during fruit base expansion (Table 1), suggesting that polarity regulators may be involved in the fruit spine development in cucumber.
Discussion
Distinct morphology and developmental process of cucumber trichomes
Trichomes are the epidermal appendages that are mainly distributed on leaves, stems, and sepals (Hülskamp et al., 1999; Hülskamp, 2004). Here, we found that cucumber trichomes not only cover the above-mentioned aerial organs but are also scattered on the surface of fruit (Fig. 1 and Supplementary Fig. S1 available at JXB online). Whether spines are present on cucumber fruit affects consumer preference and is thus of great economic importance. For example, in some European regions, people prefer glabrous cucumber without spines, whereas Asia customers desire cucumber with spines and warts. Cucumber trichomes belong to the multicellular non-glandular type of trichome. Each trichome consists of a stalk with four to eight cells lining up in a row and a dome-shaped base (Fig. 1G, H). Non-glandular trichomes have been shown to function in defence against insect herbivores, in protection against UV light and low temperature, and in facilitating seed dispersal in other plants (Hülskamp et al., 1999; Hülskamp, 2004; Serna and Martin, 2006), and whether the non-glandular cucumber trichomes have the same functions warrants further studies.
Taking advantage of the large trichome base on cucumber fruit, we were able to explore the internal structure of trichomes using TEM. Fruit trichome cells display an enlarged cell size with a huge central vacuole and malformed internal organelles (Fig. 2A–D). Notably, abnormal-coloured plastids are often observed in the fruit spine cells (Fig. 2D and Supplementary Fig. S1A available at JXB online) but are very rare in epidermal cells, suggesting that cucumber trichomes may have a distinct origin from the epidermal-derived Arabidopsis trichomes (Marks, 1997; Hülskamp et al., 1999; Larkin et al., 2003). In addition, unlike the four rounds of endoreduplications in Arabidopsis trichome cells (32C) (Hülskamp et al., 1994), cucumber trichome cells did not appear to undergo any endoreduplication (Fig. 2E, F). Consistently, cell-cycle-related genes were not differentially expressed in either of the DGE comparisons (tbh mutant vs WT, and 1.6 vs 0.5cm long fruit), supporting the suggestion that uni- and multicellular trichomes may be under different regulatory controls.
Compared with the complex developmental process of Arabidopsis trichomes (Szymanski et al., 1998), formation of cucumber trichomes is relatively simple and can be generally divided into two steps: (i) initiation of the stalk and the base; and (ii) expansion of the base (Fig. 3). More importantly, the developmental phase of cucumber spines on each fruit surface is generally homogeneous, which makes it possible to isolate trichome populations in each specific developmental stage. Because cucumber is a horticultural crop of worldwide importance, and fruit spines directly affect the appearance and quality of cucumber fruit, detailed characterization of cucumber fruit trichomes will not only help understand the underlying molecular mechanisms of the development of multicellular non-glandular trichomes but will also pave the way for creating new cucumber varieties with desired trichome growth and density through breeding and genetic engineering. Therefore, cucumber fruit trichomes may serve as a model system for studying the development of multicellular non-glandular trichomes.
TBH may function as a major activator during fruit spine development
Mutant analyses have identified both positive and negative regulators of Arabidopsis trichome development (Schwab et al., 2000; Szymanski et al., 2000). Although different cucumber varieties display enormous divergence in fruit trichome formation, no trichome mutant with developmental defects has been characterized yet. Here, we obtained and purified a spontaneous mutation tbh, and found that the morphology and development of trichomes was dramatically changed in the tbh mutant (Fig. 1). Disruption of the TBH locus led to tiny trichomes with a greatly reduced number of cells, aberrant cell shapes and organization, and branched trichomes (Fig. 1), suggesting that TBH may be required for cell division and directional growth of cucumber trichomes. In addition, transcriptome analyses by DGE showed that the downregulated genes were greatly outnumbered by the upregulated genes in the tbh mutant (Fig. 4, Supplementary Table S2 available at JXB online), implying that TBH may act as a primary activator during fruit spine development in cucumber. Considering that trichomes initiate but arrest their further development in the tbh mutant (Fig. 1), and that there are more trichomes in the tbh mutant than in WT, it is possible that TBH regulates both the initiation and outgrowth of trichomes in cucumber. Based on the activator–inhibitor model, initiated trichome cells may activate factors that inhibit trichome development in neighbouring cells (Hülskamp and Schnittger, 1998; Hülskamp, 2004). Therefore, TBH may act immediately after trichome initiation and its function is required for later development of trichomes as well as the activation of some inhibiting factor(s). Disruption of TBH function leads to arrested trichome growth and reduced inhibition for surrounding cells, which causes increased trichome density.
New roles of meristem genes during fruit trichome development
The shoot apical meristem (SAM) is the ultimate origin for all above-ground parts of the plant body. It produces lateral organ primordia from the peripheral zone (PZ) while maintaining a pool of stem cells in the central zone (CZ) (Fletcher, 2002). The meristem regulator STM promotes the indeterminate meristematic cell fate in the SAM by limiting expression of ASYMMETRIC LEAVES 1 (AS1) and AS2 in primordia. Both AS1 and AS2 inhibit the expression of KNOX family genes, which include STM, in the initiating primordia so as to stimulate cell differentiation and patterning (Byrne et al., 2000, 2002; Ori et al., 2000). Overexpression of STM leads to switching the cell fate from determinate to indeterminate meristematic in leaf tissues (Smith et al., 1992; Sinha et al., 1993; Long et al., 1996). Here, we found that ‘meristem maintenance’ genes were significantly induced in the tbh pericarps (epidermis plus spines) compared with in the WT (Fig. 6, Table 2). Therefore, the tiny trichomes with arrested development in the tbh mutant may be caused by the upregulated expression of meristem genes, which enables the trichomes to recapitulate a shoot meristem programme. Similarly, meristem maintenance genes were induced in the 1.6cm fruit compared with the 0.5cm fruit (Fig. 6, Table 3). However, only very few genes were commonly induced in both sets of comparisons (Fig. 4). Given that the CZ of SAM displays a greatly reduced number of cell divisions whereas the PZ cells divide much more frequently (Fletcher, 2002; Reddy and Meyerowitz, 2005), together with the fact that the tbh mutant had a reduced number of trichome cells with inert cell division, and that spines of 1.6cm fruit undergo rapid base expansion with active cell division, it is plausible that distinct members of meristem genes were upregulated in the tbh mutant and during spine base expansion. While negative regulators of cell division were induced in the tbh mutant, similar to those functioning in the CZ, positive regulators for cell division were promoted in the spines of 1.6cm fruit, similar to those acting in the PZ. In addition, it has been shown that the meristem–organ boundary gene CUC3 promotes adventitious shoot formation and cell division in Arabidopsis (Daimon et al., 2003). Here we found that transcripts of CsCUC3 were abolished in the tbh mutant (Table 1, Fig. 7). However, neither the genomic sequence nor the 2.6kb promoter region of CsCUC3 showed any difference between WT and the thb mutant (data not shown), implying that TBH regulates the expression of CsCUC3 epigenetically or is an upstream regulator that is essential for CsCUC3 expression and therefore cell division. During the expansion of the spine base, the meristem marker CsSTM was induced over 22-fold as detected by DGE and qRT-PCR (Table 1), and in situ hybridization showed that CsSTM was highly accumulated in the junction between the spine base and fruit wart (Fig. 7F). Previous studies have indicated that overexpression of STM leads to ectopic cell divisions (knots) in leaf tissue (Smith et al., 1992; Sinha et al., 1993), and that STM was induced at the leaflet initiation sites in complex leaves (Bharathan et al., 2002; Hake et al., 2004). These data suggested that, similar to the leaflet initiation, spine base expansion is mediated by CsSTM activation in cucumber. Interestingly, we did not detect any differential expression of the homologues of well-characterized trichome regulators such as TTG1 (Galway et al., 1994; Walker et al., 1999), GL1, GL2, GL3, and EGL3 (Payne et al., 2000; Zhang et al., 2003), TRY (Schellmann et al., 2002; Pesch and Hülskamp, 2011), and CPC (Wada et al., 1997, 2002) in either set of DGE data. One possibility is that most of these genes act in the trichome initiation or shortly after initiation, and our samples for the DGE analyses were collected at a time point that was too late to detect any changes in their expression. Another possibility is that the unicellular Arabidopsis trichomes and multicellular cucumber spines may be under the control of distinct transcription networks. Consistent with this notion, unlike the common regulatory mechanism for trichome and root hair development in Arabidopsis (Hülskamp, 2004; Pesch and Hülskamp, 2004), neither meristem genes nor known trichome regulators showed significantly differences in the roots of tbh mutant and WT (Supplementary Table S4 available at JXB online). Future studies using reverse genetics strategies will help uncover the regulatory roles of meristem genes and known trichome regulators during cucumber fruit spine development.
Conserved requirement of polarity regulators for organ expansion
Polarity specification of the adaxial–abaxial axis has been well characterized in leaf patterning (Yamaguchi et al., 2012). Generally, the vascular bundles are also polarized, with the xylem being adaxial and the phloem being abaxial. Previous studies have identified adaxial-specific genes such as PHABULOSA, PHAVOLUTA, and REVOLUTA (REV) (Eshed et al., 2001; McConnell et al., 2001; Otsuga et al., 2001), and abaxial-specific regulators such as FILAMENTOUS FLOWER and KANADI (Sawa et al., 1999; Emery et al., 2003; Juarez et al., 2004; Nakata and Okada, 2012). A model has been proposed in which juxtaposition of adaxial and abaxial developmental fields is required for laminar expansion and margin development in leaves (Eshed et al., 2001; Emery et al., 2003; Nakata and Okada, 2012). Disruption of either adaxial- or abaxial-specific genes causes a partial or completely filamentous leaf phenotype that is defective in laminar expansion (Sawa et al., 1999; McConnell et al., 2001; Otsuga et al., 2001; Emery et al., 2003). In this study, we found that GO terms ‘xylem and phloem pattern formation’ and ‘polarity specification of adaxial/abaxial axis’ were significantly enriched among the upregulated genes in the spines of 1.6cm fruit (Fig. 5B, Table 3), and we verified that both adaxial and abaxial identity genes were promoted during spine base expansion (Tables 1 and 3, Fig. 6). These data suggested that, similar to the laminar expansion in leaf development, both adaxial and abaxial regulators are required for cucumber spine base expansion. It would be interesting to explore whether auxin mediates the adaxial and abaxial juxtaposition in cucumber trichome development as in the leaves (Wang et al., 2011), and what are the expression patterns of the above-mentioned polarity markers in the mature dome-shaped spine base of cucumber fruit in future studies.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Trichome distribution in WT cucumber and the tbh mutant.
Supplementary Fig. S2. Two types of trichomes in cucumber as observed by scanning electron microscopy.
Supplementary Fig. S3. Developmental stages of cucumber fruit spines.
Supplementary Fig. S4. Gene ontology (GO) terms that were significantly enriched (P<0.01) in the downregulated genes in the tbh mutant or in the spines of 1.6cm long fruit.
Supplementary Table S1. Summary of digital gene expression sequencing data.
Supplementary Table S2. List of genes that are differentially expressed in fruit spines of the WT and tbh mutant.
Supplementary Table S3. List of genes that are differentially expressed in fruit spines of two different developmental stages.
Supplementary Table S4. qRT-PCR analysis of meristem genes and trichome regulators in the leaf and root of tbh mutant versus those in WT cucumber.
Supplementary Table S5. List of primers used in this study.
Acknowledgements
The cucumber genome and annotation data were kindly provided by The Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences (IVF-CAAS). The authors thank members of the Zhang laboratory for technical assistance and stimulating discussions, and Drs Adrienne Roeder and Xuexian Li for critical reading and comments on the manuscript. This study was supported by the National Basic Research of China 973 programme (012CB113900), National Natural Science Foundation of China (31171399), Chinese Universities Scientific Fund (2014JD047) and (2013RC030) to XZ; by the University of California Agricultural Experiment Station fund to RL; and by National Basic Research Program of China (2009CB119000) to HR.
Glossary
Abbreviations:
- bHLH
basic helix–loop–helix
- CZ
central zone
- DAPI
4’,6-diamidino-2-phenylindole
- DGE
digital gene expression
- FDR
false discovery rate
- GO
gene ontology
- PZ
peripheral zone
- qRT-PCR
quantitative reverse transcription-PCR
- SAM
shoot apical meristem
- SEM
scanning electron microscopy
- TEM
transmission electron micrograph
- WT
wild type.
References
- Alexa A, Rahnenfuhrer J, Lengauer T. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22, 1600–1607 [DOI] [PubMed] [Google Scholar]
- Ando K, Carr KM, Grumet R. 2012. Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genomics 518, 1471–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B 57, 289–300 [Google Scholar]
- Bharathan G, Goliber TE, Moore C, Kessler S, Pham T, Sinha NR. 2002. Homologies in leaf form inferred from KNOXI gene expression during development. Science 296, 1858–1860 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Barley R, Curtis M, Arroyo JM, Dunham M, Hudson A, Martienssen RA. 2000. Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971 [DOI] [PubMed] [Google Scholar]
- Byrne ME, Simorowski J, Martienssen RA. 2002. ASYMMETRIC LEAVES1 reveals knox gene redundancy in Arabidopsis. Development 129, 1957–1965 [DOI] [PubMed] [Google Scholar]
- Chien JC, Sussex IM. 1996. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiology 111, 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, et al. 2006. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana . Plant Cell 18, 3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. 1996. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 122, 1567–1575 [DOI] [PubMed] [Google Scholar]
- Daimon Y, Takabe K, Tasaka M. 2003. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant and Cell Physiology 44, 113–121 [DOI] [PubMed] [Google Scholar]
- Deng W, Yang Y, Ren Z, Audran-Delalande C, Mila I, Wang X, Song H, Hu Y, Bouzayen M, Li Z. 2012. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytologist 194, 379–390 [DOI] [PubMed] [Google Scholar]
- Emery JF, Floyd SK, Alvarez J, Eshed Y, Hawker NP, Izhaki A, Baum SF, Bowman JL. 2003. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Current Biology 13, 1768–1774 [DOI] [PubMed] [Google Scholar]
- Esch JJ, Chen MA, Hillestad M, Marks MD. 2004. Comparison of TRY and the closely related At1g01380 gene in controlling Arabidopsis trichome patterning. The Plant Journal 40, 860–869 [DOI] [PubMed] [Google Scholar]
- Eshed Y, Baum SF, Perea JV, Bowman JL. 2001. Establishment of polarity in lateral organs of plants. Current Biology 11, 1251–1260 [DOI] [PubMed] [Google Scholar]
- Eveland AL, Satoh-Nagasawa N, Goldshmidt A, Meyer S, Beatty M, Sakai H, Ware D, Jackson D. 2010. Digital gene expression signatures for maize development. Plant Physiology 154, 1024–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher JC. 2002. Shoot and floral meristem maintenance in arabidopsis. Annual Review of Plant Biology 53, 45–66 [DOI] [PubMed] [Google Scholar]
- Galbraith DW, Lambert GM, Macas J, Dolezel J. 2001. Analysis of nuclear DNA content and ploidy in higher plants. Curr Protoc Cytom Chapter 7, Unit 7.6. [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, S JW. 1994. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Developmental Biology 166, 740–754 [DOI] [PubMed] [Google Scholar]
- Hake S, Smith HM, Holtan H, Magnani E, Mele G, Ramirez J. 2004. The role of knox genes in plant development. Annual Review of Cell and Developmental Biology 20, 125–151 [DOI] [PubMed] [Google Scholar]
- Hibara K, Karim MR, Takada S, Taoka K, Furutani M, Aida M, Tasaka M. 2006. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 18, 2946–2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, et al. 2009. The genome of the cucumber, Cucumis sativus L. Nature Genetics 41, 1275–1281 [DOI] [PubMed] [Google Scholar]
- Hülskamp M. 2004. Plant trichomes: a model for cell differentiation. Nature Reviews Molecular Cell Biology 5, 471–480 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Miséra S, Jürgens G. 1994. Genetic dissection of trichome cell development in Arabidopsis. Cell 76, 555–566 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A, Folkers U. 1999. Pattern formation and cell differentiation: trichomes in Arabidopsis as a genetic model system. Int Rev Cytol 186, 147–178 [DOI] [PubMed] [Google Scholar]
- Hülskamp M, Schnittger A. 1998. Spatial regulation of trichome formation in Arabidopsis thaliana . Seminars in Cell and Developmental Biology 9, 213–220 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proceedings of the National Academy of Sciences, USA 93, 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. 2004. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428, 84–88 [DOI] [PubMed] [Google Scholar]
- Kang JH, Shi F, Jones AD, Marks MD, Howe GA. 2010. Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. Journal of Experimental Botany 61, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J. 2004a. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Developmental Biology 268, 506–513 [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Wester K, Schiefelbein J, Hülskamp M. 2004b. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Molecular Biology 55, 389–398 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Brown ML, Schiefelbein J. 2003. How do cells know what they want to be when they grow up? Lessons from epidermal patterning in Arabidopsis. Annual Review of Plant Biology 54, 403–430 [DOI] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. 2004. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J. 2009. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967 [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. 1996. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature 379, 66–69 [DOI] [PubMed] [Google Scholar]
- Luckwill LC. 1943. The genus Lycopersicon: a historical, biological and taxonomic survey of the wild and cultivated tomato. Aberd Univ Stud 120, 1–44 [Google Scholar]
- Machado A, Wu Y, Yang Y, Llewellyn DJ, Dennis ES. 2009. The MYB transcription factor GhMYB25 regulates early fibre and trichome development. The Plant Journal 59, 52–62 [DOI] [PubMed] [Google Scholar]
- Marks MD. 1997. Molecular genetic analysis of trichome development in Arabidopsis. Annual Review of Plant Physiology and Plant Molecular Biology 48, 137–163 [DOI] [PubMed] [Google Scholar]
- McConnell JR, Emery J, Eshed Y, Bao N, Bowman J, Barton MK. 2001. Role of PHABULOSA and PHAVOLUTA in determining radial patterning in shoots. Nature 411, 709–713 [DOI] [PubMed] [Google Scholar]
- Melaragno JE, Mehrotra B, Coleman AW. 1993. Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Okada K. 2012. The three-domain model: a new model for the early development of leaves in Arabidopsis thaliana . Plant Signaling & Behavior 7, 1423–1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer DG, Herman PL, Sivakumaran S, Esch J, Marks MD. 1991. A myb gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67, 483–493 [DOI] [PubMed] [Google Scholar]
- Ori N, Eshed Y, Chuck G, Bowman JL, Hake S. 2000. Mechanisms that control knox gene expression in the Arabidopsis shoot. Development 127, 5523–5532 [DOI] [PubMed] [Google Scholar]
- Otsuga D, DeGuzman B, Prigge MJ, Drews GN, Clark SE. 2001. REVOLUTA regulates meristem initiation at lateral positions. The Plant Journal 25, 223–236 [DOI] [PubMed] [Google Scholar]
- Payne CT, Zhang F, Lloyd AM. 2000. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1 and TTG1. Genetics 156, 1349–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perazza D, Herzog M, Hülskamp M, Brown S, Dorne A-M, Bonneville J-M. 1999. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics 152, 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M. 2004. Creating a two-dimensional pattern de novo during Arabidopsis trichome and root hair initiation. Current Opinion in Genetics & Development 14, 422–427 [DOI] [PubMed] [Google Scholar]
- Pesch M, Hülskamp M. 2011. Role of TRIPTYCHON in trichome patterning in Arabidopsis. BMC Plant Biology 11, 1471–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy GV, Meyerowitz EM. 2005. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science 310, 663–667 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels AL, Glass ADM, Ehret DL, Menzies JG. 1993. The effects of silicon supplementation on cucumber fruit: changes in surface characteristics. Annals of Botany 72, 433–440 [Google Scholar]
- Sawa S, Watanabe K, Goto K, Liu YG, Shibata D, Kanaya E, Morita EH, Okada K. 1999. FILAMENTOUS FLOWER, a meristem and organ identity gene of Arabidopsis, encodes a protein with a zinc finger and HMG-related domains. Genes & Development 13, 1079–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellmann S, Hülskamp M. 2005. Epidermal differentiation: trichomes in Arabidopsis as a model system. Int J Developmental Biology 49, 579–584 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. 2002. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO Journal 19, 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab B, Folkers U, Ilgenfritz H, Hülskamp M. 2000. Trichome morphogenesis in Arabidopsis. Philosophical Transactions of the Royal Society B: Biological Sciences 355, 879–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna L, Martin C. 2006. Trichomes: different regulatory networks lead to convergent structures. Trends in Plant Science 11, 274–280 [DOI] [PubMed] [Google Scholar]
- Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. 2007. Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Developmental Biology 311, 566–578 [DOI] [PubMed] [Google Scholar]
- Sinha NR, Williams RE, Hake S. 1993. Overexpression of the maize homeo box gene, KNOTTED-1, causes a switch from determinate to indeterminate cell fates. Genes & Development 7, 787–795 [DOI] [PubMed] [Google Scholar]
- Smith LG, Greene B, Veit B, Hake S. 1992. A dominant mutation in the maize homeobox gene, Knotted-1, causes its ectopic expression in leaf cells with altered fates. Development 116, 21–30 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Jilk RA, Pollock SM, Marks MD. 1998. Control of GL2 expression in Arabidopsis leaves and trichomes. Development 125, 1161–1171 [DOI] [PubMed] [Google Scholar]
- Szymanski DB, Lloyd AM, Marks MD. 2000. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends in Plant Science 5, 214–219 [DOI] [PubMed] [Google Scholar]
- Tian D, Tooker J, Peiffer M, Chung SH, Felton GW. 2012. Role of trichomes in defense against herbivores: comparison of herbivore response to woolly and hairless trichome mutants in tomato (Solanum lycopersicum). Planta 236, 1053–1066 [DOI] [PubMed] [Google Scholar]
- Tominaga-Wada R, Nukumizu Y, Sato S, Wada T. 2013. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One 8, e54019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vroemen CW, Mordhorst AP, Albrecht C, Kwaaitaal MA, de Vries SC. 2003. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 15, 1563–1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. 2002. Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129, 5409–5419 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277, 1113–1116 [DOI] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Graya JC. 1999. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11, 1337–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. 2010. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Analytical Biochemistry 399, 257–261 [DOI] [PubMed] [Google Scholar]
- Wang W, Xu B, Wang H, Li J, Huang H, Xu L. 2011. YUCCA genes are expressed in response to leaf adaxial–abaxial juxtaposition and are required for leaf margin development. Plant Physiology 157, 1805–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T, Nukazuka A, Tsukaya H. 2012. Leaf adaxial–abaxial polarity specification and lamina outgrowth: evolution and development. Plant and Cell Physiology 53, 1180–1194 [DOI] [PubMed] [Google Scholar]
- Yang C, Li H, Zhang J, et al. 2011. A regulatory gene induces trichome formation and embryo lethality in tomato. Proceedings of the National Academy of Sciences, USA 108, 11836–11841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. 2003. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130, 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhang X, Zhou Y, Ding L, Wu Z, Liu R, Meyerowitz EM. 2013. Transcription repressor HANABA TARANU controls flower development by integrating the actions of multiple hormones, floral organ specification genes, and GATA3 family genes in Arabidopsis. Plant Cell 25, 83–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q, Wang XJ. 2008. GOEAST: a web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Research 36, W358–W363 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.