Summary
OsNRAMP5 plays an important role in the translocation and distribution of Mn in rice plants in addition to its role in Mn uptake.
Key words: Cadmium, manganese, OsNRAMP5, rice, root-to-shoot translocation, transporter.
Abstract
Manganese (Mn) is an essential micronutrient for plants playing an important role in many physiological functions. OsNRAMP5 is a major transporter responsible for Mn and cadmium uptake in rice, but whether it is involved in the root-to-shoot translocation and distribution of these metals is unknown. In this work, OsNRAMP5 was found to be highly expressed in hulls. It was also expressed in leaves but the expression level decreased with leaf age. High-magnification observations revealed that OsNRAMP5 was enriched in the vascular bundles of roots and shoots especially in the parenchyma cells surrounding the xylem. The osnramp5 mutant accumulated significantly less Mn in shoots than the wild-type plants even at high levels of Mn supply. Furthermore, a high supply of Mn could compensate for the loss in the root uptake ability in the mutant, but not in the root-to-shoot translocation of Mn, suggesting that the absence of OsNRAMP5 reduces the transport of Mn from roots to shoots. The results suggest that OsNRAMP5 plays an important role in the translocation and distribution of Mn in rice plants in addition to its role in Mn uptake.
Introduction
Manganese (Mn), an essential micronutrient for most organisms, is involved in many biochemical pathways as an enzyme cofactor. Mn deficiency in plants can affect the biosynthesis of fructans and structural carbohydrates, causing leaves to become slack and soft (Marschner, 1995). In addition, Mn deficient plants are more susceptible to low-temperature stress and pathogen infection, resulting in lower crop yields (Hebbern et al., 2005; Marschner, 1995). However, when accumulated in excess, Mn can have various phytotoxic effects, including reduction of growth, photosynthesis and chlorophyll content, inhibition of enzyme activities, and damage to chloroplasts (Doncheva et al., 2005; Keen et al., 2000; Mora et al., 2009).
Recently, great progress has been made concerning the mechanism of Mn uptake in plants. A number of genes responsible for the uptake of Mn by roots have been identified (Cailliatte et al., 2010; Pedas et al., 2008). In barley (Hordeum vulgare), two separate Mn transport systems that mediate high-affinity and low-affinity Mn2+ influx at different concentrations were identified (Pedas et al., 2005). Further studies on Mn uptake in barley showed that HvIRT1, targeted to the plasma membrane, plays an important role in controlling Mn2+ uptake in roots. Heterologous expression of HvIRT1 in the Δsmf1 yeast mutant, which is deficient in Mn2+ uptake, restored its growth. In addition, a comparison of plants with differing ability to grow in soils with low Mn2+ showed that the expression of HvIRT1 was 40% greater in plants with an Mn-efficient genotype compared with plants with an Mn-inefficient genotype, and the higher expression of HvIRT1 was correlated with an increased Mn2+ uptake rate (Pedas et al., 2008). AtNRAMP1, a member of the gene family natural resistance associated macrophage protein (NRAMP) in Arabidopsis, was recently reported to be essential for Mn2+ uptake under Mn-deficient conditions (Cailliatte et al., 2010). AtNRAMP1 encodes a plasma membrane protein in the root cells and mediates a high-affinity transporter of Mn2+ with an apparent Km value of 28nM. Growth of the knockout mutant of AtNRAMP1 was inhibited under Mn-deficient conditions much more than the wild type.
Analysis of the tolerance mechanisms to Mn excess in plants is another focus of current research and has contributed substantially to the present understanding of intracellular Mn compartmentation (Pittman, 2005). Vacuoles play a very important role in Mn tolerance in plants. Expression of AtCAX2 in tobacco enables more Mn to accumulate in vacuoles and increases Mn tolerance compared with wild type (Hirschi et al., 2000). ShMTP1, a member of the cation diffusion facilitator (CDF) family from Stylosanthes hamata, conferred Mn2+ tolerance in yeast by internal sequestration rather than by efflux of Mn2+. When expressed in Arabidopsis, ShMTP1 is localized to the tonoplast and can also confer Mn tolerance by sequestering Mn2+ in this organelle (Delhaize, 2003). Another CDF member, AtMTP11, might be responsible for a secretory pathway-mediated mechanism in Mn detoxification in plants. AtMTP11 was targeted to a Golgi-like compartment, and AtMTP11-overexpressing plants were hyper-tolerant to Mn (Delhaize et al., 2007; Peiter et al., 2007).
A number of transporters have been found to be involved in Mn transport in rice (Oryza sativa L.). OsYSL2, a member of the yellow stripe 1-like (YSL) proteins, has a Mn2+ transport activity and contributes to the long-distance transport of Mn (Ishimaru et al., 2010; Koike et al., 2004). OsYSL6 is another member of the YSL family that has been shown to be a Mn-nicotianamine (NA) transporter required for the detoxification of high levels of Mn (Sasaki et al., 2011). Furthermore, OsNRAMP3 is able to transport Mn2+ and is specifically expressed in the vascular bundles, playing a role in the distribution of Mn between young and old tissues (Yamaji et al., 2013; Yang et al., 2013).
Recently, another member of the NRAMP family in rice, OsNRAMP5, was reported to be a Mn and Cd transporter involved in the root uptake of these metals from the medium (Ishikawa et al., 2012; Ishimaru et al., 2012a; Ishimaru et al., 2012b; Sasaki et al., 2012). Ishimaru et al. (2012b) found that OsNRAMP5 complemented the growth of yeast Δsmf1, whereas knockdown of OsNRAMP5 decreased Mn accumulation in both roots and shoots of rice plants. However, whether OsNRAMP5 participates in the distribution of Mn from roots and shoot and in the shoot tissues remains unknown. In the present study, the role of OsNRAMP5 in Mn distribution in rice plants was investigated.
Materials and methods
Plant materials
Wild-type, mutant and transgenic plants used in the present study were all based on the background of the cv. Zhonghua 11 (O. sativa L. ssp. japonica variety). The osnramp5 mutant is a T-DNA insertion mutant identified from the Rice Mutant Database (Wu et al., 2003; Zhang et al., 2006).
Hydroponic experiments
Hydroponic experiments were performed using a standard rice culture solution (1.44mM NH4NO3, 0.3mM NaH2PO4, 0.5mM K2SO4, 1.0mM CaCl2, 1.6mM MgSO4, 0.17mM Na2SiO3, 50 µM Fe-EDTA, 0.06 µM (NH4)6Mo7O24, 15 µM H3BO3, 8 µM MnCl2, 0.12 µM CuSO4, 0.12 µM ZnSO4, 29 µM FeCl3, 40.5 µM citric acid, pH 5.5) described by Yoshida et al. (1976). In the Mn concentration gradient experiments, a range of Mn concentrations (0.04, 0.08, 0.2, 0.4, 0.8, 1.6, 8, 80, and 800 µM) were imposed, with the 8 µM Mn treatment as the control. In some experiments 2nM of CdCl2 was added to the hydroponic solution to investigate Cd uptake.
Promoter fusion and RNA in situ hybridization
To create the OsNRAMP5-promoter:GUS construct, 2.06kb of the genomic sequence located upstream of the OsNRAMP5 initiation codon was amplified by PCR from Zhonghua 11 genomic DNA using primers Pnr5-F (5’-agggatcccgcaactcccacaactactg-3’) and Pnr5-R (5’-tcggatccgcttcctctcttagcttcttca-3’). The amplified promoter fragment was digested by BamH1 and introduced into the vector pDX2181 in the correct direction (Ye et al., 2012). Wild-type Zhonghua 11 calli was transformed with this construct. The transgenic plant tissues were incubated in a X-Gluc staining buffer at 37 °C for 4h (Jefferson et al., 1987).
Hybridization and immunological detection were performed as described in Xue et al. (2008). The probe of OsNRAMP5 was amplified from Zhonghua 11 by PCR using gene-specific primers in situ-F (5’-cgacgagcccttgccgta-3’) and in situ-R (5’-tctgcgagcgatctggacc-3’) and cloned into the pGEM-T vector (Promega). The sense and antisense probes were transcribed in vitro by T7 or SP6 transcriptase using a Digoxigenin RNA labelling kit (Roche).
RNA extraction and real-time PCR
Total RNA was extracted using Trizol reagent (Invitrogen). The first-strand cDNAs were synthesized with 3 µg of total RNA in 20 µl of reaction mixture using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer’s instructions. Real-time PCR was performed using the SYBR Premix Ex TaqTM (TaKaRa) with the following gene-specific primers: rqNR5-F (5’-cgctcccaaggtagagaagaagaa-3’) and rqNR-R (5’-atataaacgaacggctccgacgca-3’). The rice Ubiquitin5 gene was used as the internal control with following primers: qUbq-F (5’-aaccagctgaggcccaaga-3’) and qUbq-R (5’-acgattgatttaaccagtccatga-3’).
Elemental analysis
Shoots and roots were harvested separately and roots were washed with distilled water twice before sampling. All samples were dried at 80 °C for 3 d and then digested in 65% nitric acid in a MARS6 microwave (CEM) with a gradient of temperatures from 120 °C to 180 °C for 45min. After dilution in deionized water, the metal contents of the samples were determined by inductively coupled plasma mass spectrometry (ICP-MS, Agilent 7700 series, USA).
Results
The expression profile of OsNRAMP5 in different tissues of rice
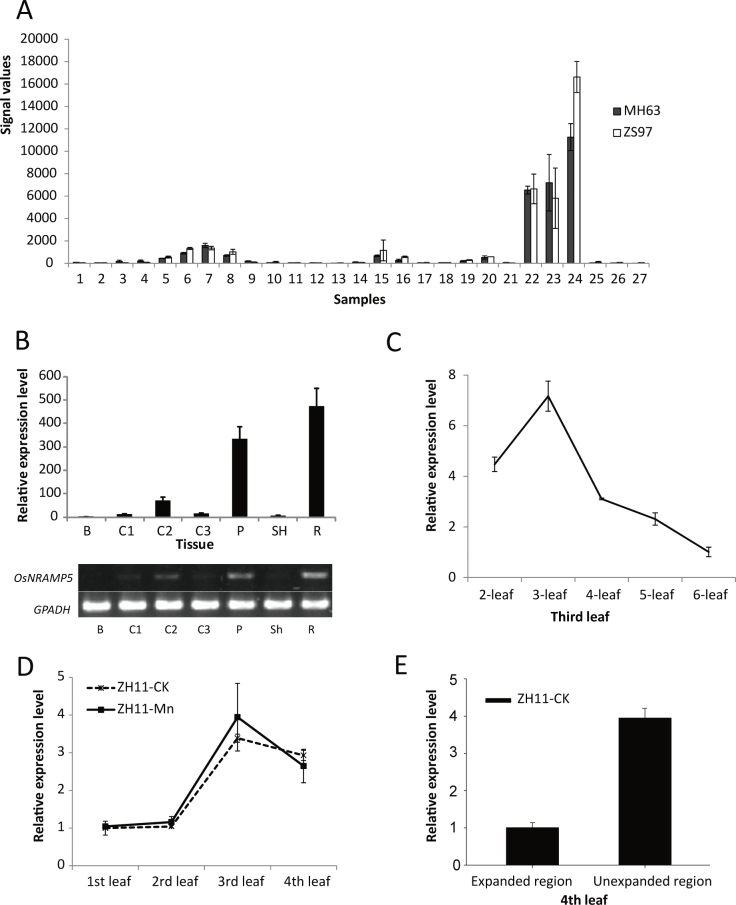
The expression pattern of OsNRAMP5 in different tissues of rice was first extracted from the CREP database (http://crep.ncpgr.cn; Wang et al., 2010); the microarray data have been verified by qPCR to reveal the expression profiles of various genes (Huang et al., 2009; Ma et al., 2009; Nuruzzaman et al., 2008). The results from the database for 27 tissues of cv. Minghui 63 (O. sativa L. ssp. indica) and cv. Zhenshan 97 (O. sativa L. ssp. indica), each with two replications, indicated that OsNRAMP5 was expressed in rice with a strong tissue-specific pattern (Fig. 1A). OsNRAMP5 exhibited the strongest expression signal in young reproductive tissues, e.g. panicle, spikelet, and hull. OsNRAMP5 also showed high expression levels in plumules and young shoots, leaves and roots, and a moderate level in stems at the heading stage. In contrast, little or no expression of OsNRAMP5 was found in the mature leaf, flag leaf, leaf sheath, or endosperm.
Fig. 1.
The transcript levels of OsNRAMP5 in different tissues of rice plants. (A) The transcript levels of OsNRAMP5 in 27 samples representing different organs or tissues at different developmental stages of the Minghui 63 (MH63) and Zhenshan 97 (ZS97) based on signal values from Wang et al. (2010). Numbers from 1 to 27 represent the following 27 samples: (1) calli, 15 days after subculture; (2) calli, screening stage; (3) calli, 5 days after regeneration; (4) seed, 72h after imbibition; (5) embryo and radicle after germination; (6) leaf and root at three-leaf stage; (7) root, seedling with 2 tillers; (8) shoot, seedling with 2 tillers; (9) sheath, young panicle at stage 3; (10) leaf, young panicle at stage 3; (11) leaf, 4–5cm young panicle; (12) flag leaf, 5 days before heading; (13) flag leaf, 14 days after heading; (14) sheath, 4–5cm young panicle; (15) stem, 5 days before heading; (16) stem, heading stage; (17) young panicle at stage 3; (18) young panicle at stage 4; (19) young panicle at stage 5; (20) panicle, 4–5cm young panicle; (21) stamen, one day before flowering; (22) panicle, heading stage; (23) spikelet, 3 days after pollination; (24) hull, one day before flowering; (25) endosperm, 7 days after pollination; (26) endosperm, 14 days after pollination; (27) endosperm, 21 days after pollination. (B) Quantitative RT-PCR analysis of the expression pattern of OsNRAMP5 in various tissues from Zhonghua 11 (ZH11). B, leaf blade; C1, culm I; C2, culm II; C3, culm III; P, panicle; Sh, sheath; R, root. (C) Quantitative RT-PCR analysis of the OsNRAMP5 transcriptional level in the third leaf of Zhonghua 11 seedling at the two-to-six-leaf stages. (D) The expression level of OsNRAMP5 in different leaves of rice plants at different Mn supplies; the 1st leaf is the oldest of the four leaves from the tiller and the 4th leaf is not fully expanded. (E) The expression pattern of OsNRAMP5 in different regions of the 4th leaf. Data are means ± SD of three biological replicates in real-time PCR.
To confirm the expression pattern, the specific accumulation of OsNRAMP5 mRNA was determined in seven different tissues of cv. Zhonghua 11 by RT-PCR and real-time PCR (Fig. 1B). OsNRAMP5 was highly expressed in roots and panicles, moderately in culms, but very little in leaf blades and sheaths. This pattern is highly consistent with the microarray data described above. To further understand the OsNRAMP5 expression pattern in leaves, the OsNRAMP5 transcript levels were monitored in the third leaf of wild-type seedlings during the growth stage from two to six leaf. At the two-leaf stage, the third leaf was wrapped inside the second leaf sheath and was not fully developed. At the three-leaf stage, the third leaf had fully expanded, and by the six-leaf stage, the third leaf had senesced slightly. Real-time RT-PCR showed relatively high transcript levels of OsNRAMP5 in the third leaf at the young stage, which then decreased gradually (Fig. 1C). The expression of OsNRAMP5 was also compared among different leaves of young wild-type plants. The expression of OsNRAMP5 was higher in younger than in older leaves under both Mn-sufficient and Mn-deficient conditions (Fig. 1D). Furthermore, the expression of OsNRAMP5 was clearly higher in the unexpanded than in the expanded part of the fourth leaf (Fig. 1E).
It was reported that the expression of the high-affinity Mn transporter AtNRAMP1 in Arabidopsis roots was induced by Mn deficiency (Cailliatte et al., 2010). Similarly, many genes involved in Fe uptake also show enhanced expression under Fe-limited conditions (Koike et al., 2004). Time-course experiments were conducted to test whether the expression of OsNRAMP5 was affected by Mn or Fe deficiency. Real-time RT-PCR showed that the transcript level of OsNRAMP5 in roots was not obviously affected by the absence of Fe or Mn in the hydroponic solution (Supplementary Fig. S1 available at JXB online), which is consistent with the result described by Ishimaru et al. (2012b). By contrast, the expression of OsNRAMP5 in shoots was induced by Fe deficiency after 9 days in the –Fe treatment and the induction was further increased on day 13. By contrast, the expression of OsNRAMP5 was not affected by the deficiency of Mn (Supplementary Fig. S1 available at JXB online).
The expression pattern of OsNRAMP5 at the cellular level
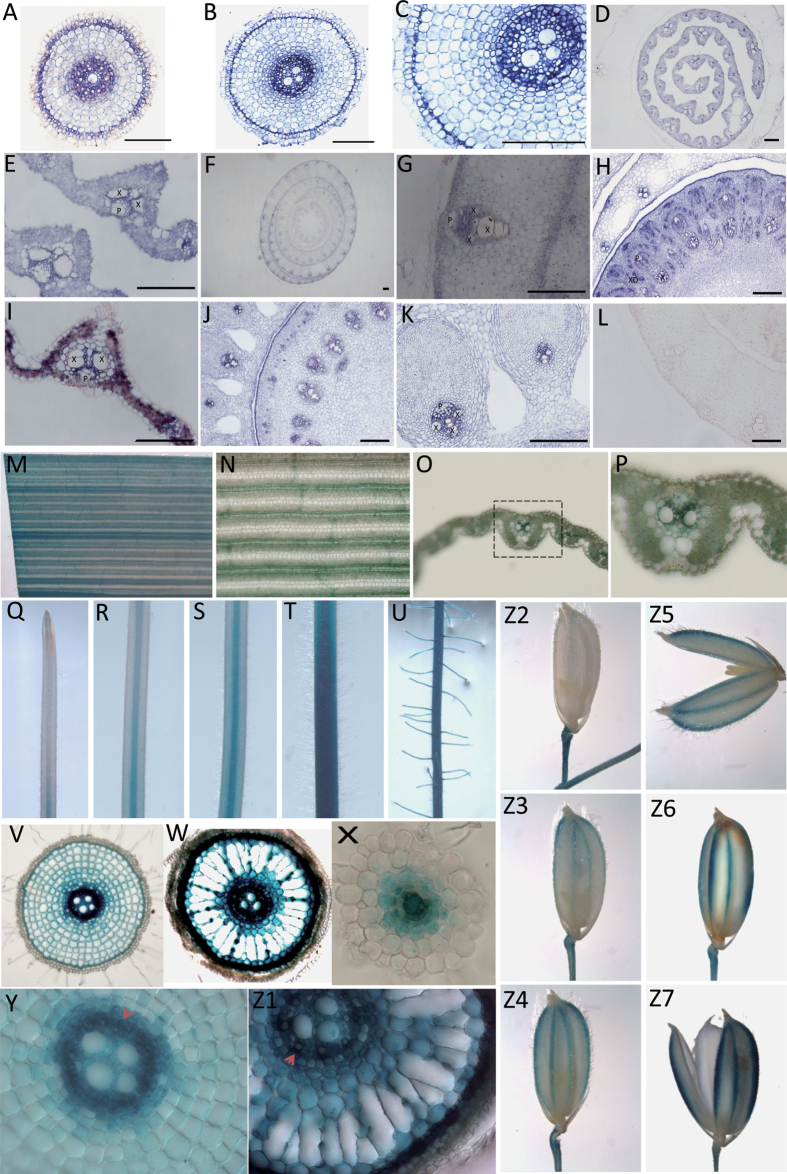
To study the cellular localization of OsNRAMP5 expression, RNA in situ hybridization was performed in roots, unexpanded leaves, young tillers, culms, expanded leaves, and the border region of nodes (Fig. 2A–L). The intensity of signals showed that OsNRAMP5 was highly expressed in the stele and sclerenchyma layer of roots, unexpanded leaves, culms, and the border region of nodes, but only moderately expressed in expanded leaves. There was little expression in the mature leaf sheaths. Furthermore, high-magnification observation showed an enrichment of OsNRAMP5 expression in the vascular bundles, especially in the parenchyma cells surrounding the xylem (Fig. 2E, G, I, K).
Fig. 2.
The expression pattern of OsNRAMP5. (A–L) In situ hybridization of OsNRAMP5. (A–C) Transverse section of roots. (A) The elongation zone of root. (B) The mature zone of root. (C) Magnified image of part of B. (D) Transverse section of unexpanded leaf. (E) Magnified image of part of D. (F) Transverse section of a young tiller. (G) Magnified image of part of F. (H) Transverse section of culm. (I) Transverse section of expanded leaf. (J) Transverse section of the border region of the node. (K) Magnified image of part of J. (L) A negative control detected by the sense probe. (M–Z7) Expression pattern of OsNRAMP5 detected in plants transformed with the construct OsNRAMP5-promoter:GUS by histochemical straining of GUS activity. (M–Z1) The localization of OsNRAMP5 in leaves and roots of a plant cultivated hydroponically for two weeks. (M) The expanded young leaf. (N) Magnified image of part of M. (O) Transverse section of leaf shown in M. (P) Enlarged view of a vascular bundle from O. (Q) The tip of root. (R) The elongation zone of root. (S, T) The basal (S) and the mid (T) mature zone of root. (U) The lateral roots. (V) The transverse section of R. (W) The transverse section of T. (X) The transverse section of a small lateral root described in U. (Y) Magnified image of part of V. (Z1) Magnified image of part of W. (Z2–Z7) The localization of OsNRAMP5 in hulls. (Z2) The hull, 5 days before heading. (Z3) The hull, 2 days before heading. (Z4, Z5) The hull at heading stage. (Z6, Z7) The hull and endosperm, 21 days after pollination. X, xylem region of regular vascular bundles; XD, xylem region of diffuse vascular bundles; P, phloem region of regular vascular bundles. Scale bars (A–L) =100 µm.
To complement the in situ hybridization results, a 2063-bp promoter region of OsNRAMP5 was used to direct beta-glucuronidase (GUS) expression in transgenic rice. GUS staining showed a strong expression of OsNRAMP5 in the leaf vascular bundles (Fig. 2M–P), consistent with the pattern revealed by in situ hybridization. In roots, the GUS activity was mainly detected in the elongation and maturation zones and in lateral roots, but not in root caps and the meristem zone (Fig. 2Q–U). Transverse sections of roots showed that the stele and sclerenchyma layer cells had the highest GUS activity (Fig. 2V–X), which confirmed the in situ hybridization results. Furthermore, high-magnification observation of transverse sections of roots identified the highest accumulation of OsNRAMP5 transcripts in the xylem of both the elongation and maturation zones (Fig. 2Y, Z1). In the maturation region of root, GUS activity was found in all cell types, including exodermis and endodermis. GUS activity was also detected in the vascular tissues of hulls from 2 d before heading to the stage of grain ripening (Fig. 2Z2–Z6). However, no GUS activity was detected in the endosperm (Fig. 2Z7).
Knockout of OsNRAMP5 increased the sensitivity to Mn and Fe deficiency
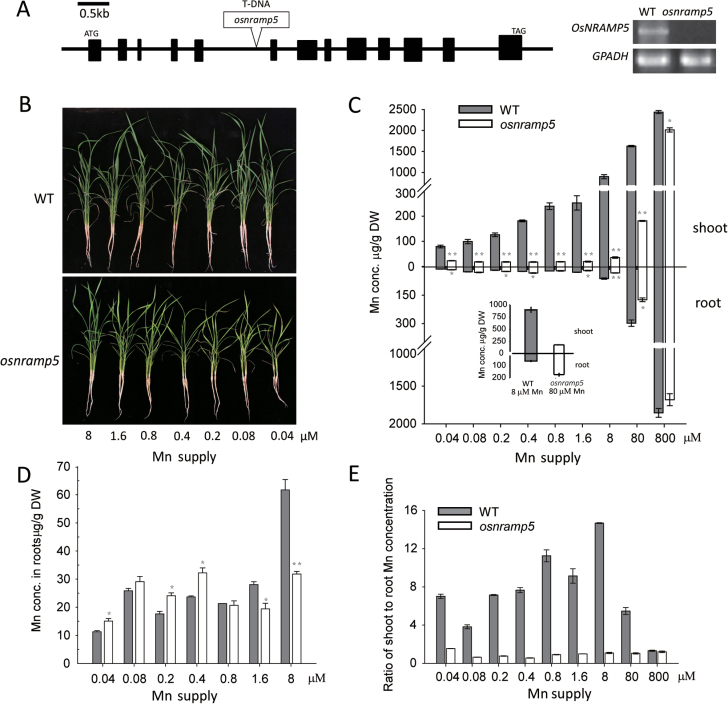
To further investigate the biological function of OsNRAMP5, a T-DNA insertion mutant, named osnramp5 from RMD (Rice Mutant Database: http://rmd.ncpgr.cn; Zhang et al., 2006), was identified. In this mutant, the T-DNA was inserted in the fifth intron of OsNRAMP5 (Fig. 3A), different from the allelic nramp5 mutant described by Sasaki et al. (2012). Transcripts of OsNRAMP5 were not detectable in the osnramp5 mutant by RT-PCR, indicating that the gene has been knocked out. When supplied with a sufficient level of Mn (8 µM), osnramp5 showed a similar phenotype to wild-type plants (Supplementary Fig. S2 available at JXB online). At lower levels of Mn supply (0.04–1.6 µM), the osnramp5 mutant plants showed reduced growth compared with the wild-type plants, suggesting that the mutant was more sensitive to Mn deficiency. The mutant was also slightly more sensitive to Fe deficiency, but not to the deficiencies of potassium (K), calcium (Ca), copper (Cu), magnesium (Mg), or zinc (Zn) (Supplementary Fig. S2 available at JXB online). These phenotypic differences were further supported by the concentrations of the metals in plants. Mn concentration was much lower in both roots and shoots of osnramp5 compared with wild-type plants, especially in the shoots in which Mn concentration was only about 4% of that in the wild-type plants (Supplementary Fig. S3 available at JXB online). By contrast, there were no significant differences in the concentrations of K, Ca, Mg, Cu, or Zn in either the control or the nutrient-deficient treatments (Supplementary Fig. S3 available at JXB online).
Fig. 3.
Identification of a knock-out line of OsNRAMP5. (A) The structure of OsNRAMP5. 13 exons (boxes) and 12 intron (lines between boxes) are contained in OsNRAMP5 and a T-DNA inserted in the 5th intron of the osnramp5 plant results in a complete loss of OsNRAMP5 transcriptional level as shown by RT-PCR. The growth phenotype (B), Mn concentrations (C), and the ratio of shoot to root Mn concentrations (E) were analysed at different Mn supplies. The Mn concentrations of wild type plants at 8 µM Mn and mutant plants at 80 µM Mn were extracted from C and are shown in C insert. (D) Enlarged view of root Mn concentrations from (C). Data are means ± SD of three biological replicates, and five plants were mixed in one replication for metal determination. One and two asterisks indicate values are significantly different from the wild type (WT) at the levels of P<0.05 and P<0.01, respectively (t-test).
To identify the role of OsNRAMP5 in Mn uptake and translocation in rice, a Mn concentration gradient experiment was performed. Plants were initially grown with the normal nutrient solution containing 8 µM Mn for 12 d and then transferred to treatments containing 0.04–800 µM Mn for another 18 d. Whereas wild-type plants grew normally in all treatments, growth of the osnramp5 plants decreased obviously when Mn concentration in the nutrient solution was lowered to 8 µM or less (Fig. 3B). In these treatments typical symptoms of Mn deficiency (stunted growth and leaf yellowing) were observed only in the osnramp5 plants. Moreover, shoot Mn concentrations of the osnramp5 plants were markedly lower than those of wild-type (3.5–25-fold difference; Fig. 3B, C), indicating that Mn deficiency was the cause of the growth reduction in the mutant. However, the response of root Mn concentration was different. In the low Mn treatments (0.04–0.8 µM), osnramp5 roots had either similar or significantly higher concentration of Mn than wild-type roots (Fig. 3D). Only in the sufficient-high Mn treatments (1.6–80 µM) did osnramp5 accumulate lower concentrations of Mn in the roots than wild-type (Fig. 3C). osnramp5 roots accumulated a much higher Mn concentration in the 80 µM Mn treatment than did wild-type plants grown under 8 µM Mn supply, but this did not rescue the shoot Mn concentration to the level of wild-type plants grown with 8 µM Mn (Fig. 3C insert). The ratios of shoot-to-root Mn concentrations were significantly higher, by 4.7–13.1 times, in wild-type than osnramp5 in all Mn treatments except the highest level (800 µM) (Fig. 3E). These results suggest that that OsNRAMP5 not only contributes to Mn uptake but also to the root-to-shoot translocation of Mn.
Uptake of Fe and Cd mediated by OsNRAMP5 was affected by Mn concentration
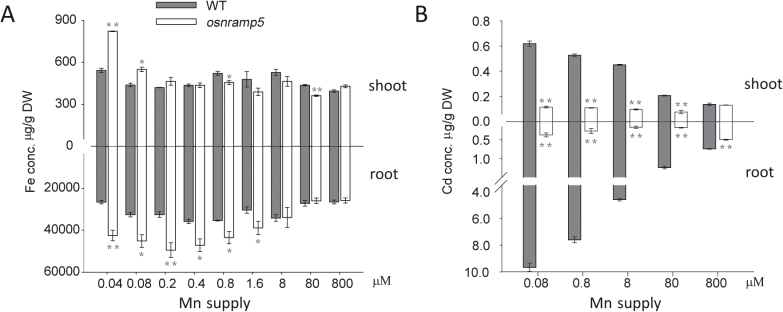
The NRAMP family transporters in Arabidopsis have been shown to transport Fe, Mn, and Cd (Lanquar et al., 2004; Lanquar et al., 2010; Thomine et al., 2003). To investigate the role of OsNRAMP5 in Fe and Cd transport, Fe and Cd concentrations of osnramp5 and wild-type plants were determined under different Mn supplies. The concentration of Fe in the roots of osnramp5 was significantly higher than that of wild type when plants were grown at low Mn concentrations (≤1.6 µM); at high Mn concentrations there were no significant differences (Fig. 4A). In shoots, a significantly higher Fe concentration was found in the mutant only in the two lowest Mn treatments. Compared with wild type, osnramp5 plants contained significantly lower Cd concentrations in both roots and shoots (Fig. 4B). Increasing the concentration of Mn in the nutrient solution markedly decreased the accumulation of Cd in both roots and shoots of wild type, but not in osnramp5 plants. As a result, the difference between wild-type and osnramp5 in root and shoot Cd concentrations decreased from 26 and 5.4 times, respectively, in the 0.04 µM Mn treatment to only 1.5 and 1.1 times in the 800 µM Mn treatment.
Fig. 4.
The concentrations of Fe and Cd at different Mn supplies. The concentrations of Fe (A) and Cd (B) of wild-type and osnramp5 mutant plants were analysed at different Mn supplies. Data are means±SD of three biological replicates, and five plants were mixed in one replication for metal determination. One and two asterisks indicate values are significantly different from the WT at the levels of P<0.05 and P<0.01, respectively (t-test).
Because Fe and Cd accumulations were affected by different Mn supply, the expression of some other Fe and Cd transporters in rice root were investigated. The expression levels of OsIRT2, OsNRAMP1, OsYSL2, and OsYSL15 were suppressed by low Mn conditions, but the expression levels of OsIRT1, OsYSL16, and OsYSL18 was unaffected (Supplementary Fig. 4 available at JXB online). The results were consistent with those of rice plants grown under Mn deficiency (Supplementary Fig. 5 available at JXB online). By contrast, the expression levels of OsIRT2, OsNRAMP1, OsYSL2, and OsYSL15 were highly induced by Fe deficiency. Moreover, much higher levels of expression of these four genes were observed in osnramp5 mutant than wild type (Supplementary Fig. 6 available at JXB online).
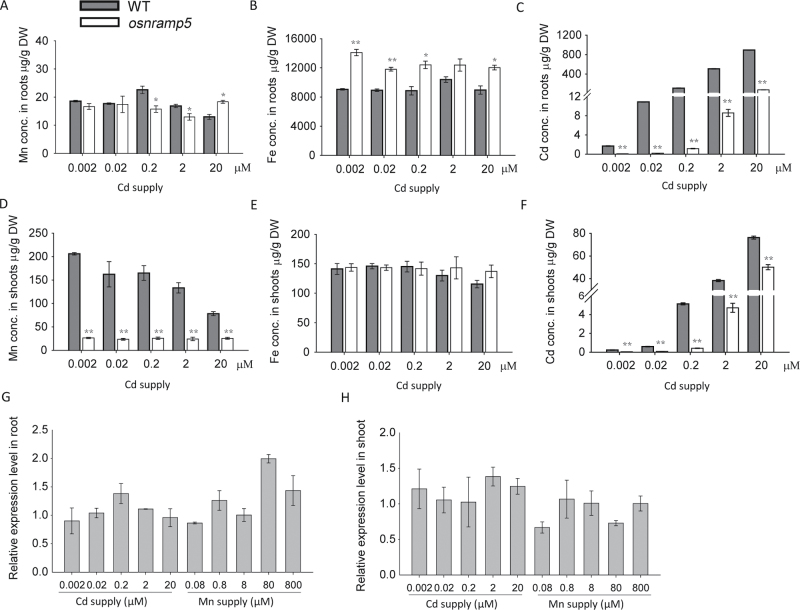
To investigate whether Mn accumulation was affected by different Cd supply, a Cd concentration gradient experiment was conducted (Fig. 5). Wild-type and osnramp5 plants were grown under normal conditions for two weeks and then transferred to normal nutrient solution with different Cd concentrations for another two weeks. As expected, the Cd concentrations of roots and shoots in both wild-type and mutant plants increased with Cd supply (Fig. 5C, F). However, osnramp5 plants accumulated much less Cd in both roots and shoots than wild-type plants, representing only 1.0–8.6% for root and 8.2–65.7% for shoot of the wild-type’s Cd concentrations. Cd treatment had no significant effect on Mn concentration in roots (Fig. 5A), but decreased Mn concentration in shoots of wild-type plants. This antagonistic effect was not seen in the mutant (Fig. 5D). No such suppression was observed for Fe concentrations (Fig. 5B, E).
Fig. 5.
The metal concentrations and expression levels at different Cd supplies. Plants were grown under normal condition for two weeks and then transferred to different Cd supplies for additional two weeks. Mn, Fe, and Cd concentrations were analysed in roots (A–C) or shoots (D–F) of rice plants at different Cd supplies. Expression levels of OsNRAMP5 of roots and shoots at different Mn or Cd supplies were analysed in (G) and (H) respectively. Data are means±SD of three biological replicates, and five plants were mixed in one replication for metal determination. One and two asterisks indicate values are significantly different from the WT at the levels of P<0.05 and P<0.01, respectively (t-test).
Fe deficiency affected Mn accumulation in both knockout and wild-type lines
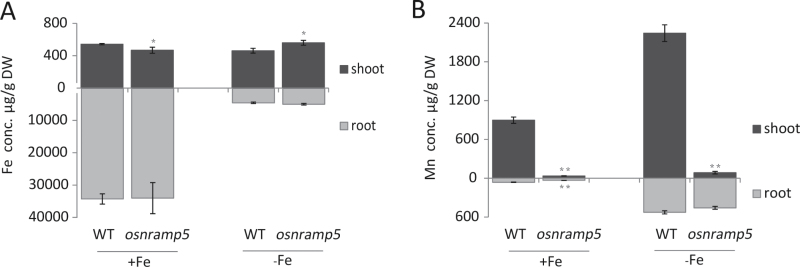
Osnramp5 and wild-type plants were cultivated for 12 d under normal conditions and then transferred to Fe-deficient (–Fe) or normal (+Fe) nutrient solution for an additional 18 d. Osnramp5 plants seemed more sensitive to Fe deficiency than wild type (Supplementary Fig. S2 available at JXB online). Fe concentrations in roots were markedly lower in the –Fe treatment but there was no significant difference between the mutant and wild-type plants (Fig. 6A). Compared with plants grown under normal conditions, both the mutant and wild-type plants grown in –Fe solution accumulated almost tenfold more Mn in roots (Fig. 6B). In addition, Fe-deficient conditions narrowed the difference in the root Mn concentration between the mutant and wild-type plants; the osnramp5 plants accumulated 87% of the Mn level of wild-type in the –Fe treatment but only 52% in the +Fe treatment. The shoots showed a similar tendency under Fe-deficient conditions, in which Mn concentration increased in both mutant and wild-type plants (Fig. 6B). Despite the large increase in Mn concentration in osnramp5 roots under Fe-deficient conditions, the effect on shoot Mn concentration was very small (Fig. 6B). In this treatment shoot Mn concentration of osnramp5 was only 4% of that in the wild type, suggesting that osnramp5 plants lost the ability to transport Mn from roots to shoots.
Fig. 6.
The concentrations of Mn and Fe in response to Fe deficiency. Plants were grown under normal condition for 12 d and then transferred to Fe-free (–Fe) medium or continued Fe-replete (+Fe) medium for additional 18 d. The Fe (A) and Mn (B) concentrations were determined in both roots and shoots. Data are means ± SD of three biological replicates, and five plants were mixed in one replication for metal determination. One and two asterisks indicate values are significantly different from the WT at the levels of P<0.05 and P<0.01, respectively (t-test).
Mn and Cd showed different distributions in leaves and were affected by the absence of OsNRAMP5
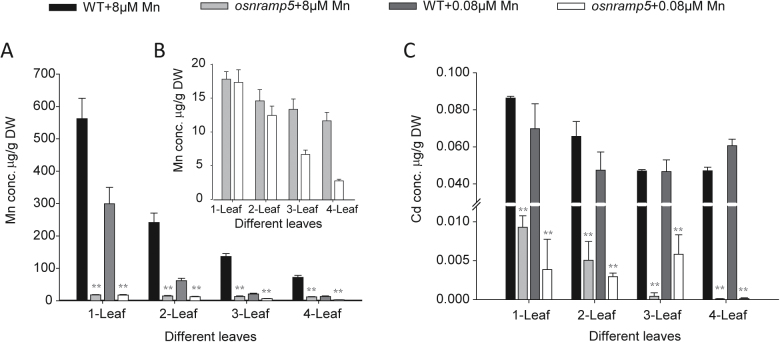
When grown without Mn, the young leaves of osnramp5 plants withered (data not shown). The chlorophyll concentrations of the third and fourth leaves of osnramp5 were significantly lower than wild type under Mn-deficient conditions (Supplementary Fig. S7 available at JXB online). Elemental concentrations of different leaves of wild type and osnramp5 plants grown with two levels of Mn were determined to investigate the possible role of OsNRAMP5 in Mn distribution in leaves. As expected, wild-type plants had markedly higher Mn concentrations in all leaves than the mutant (Fig. 7). Mn concentration also showed an increasing trend from young to old leaves. Unlike wild type, Mn concentrations in osnramp5 plants differed little among different leaves at 8 µM Mn supply, whereas the third and fourth leaves contained significantly lower Mn concentrations than other leaves at 0.08 µM Mn supply (Fig. 7B). As expected, Cd in leaves was significantly lower in osnramp5 plants compared with wild type (Fig. 7C).
Fig. 7.
The metal concentrations in different leaves. Plants were grown under normal condition for two weeks and then shifted to different Mn supplies for additional two weeks. (A) Mn concentrations; (B) part of A; (C) Cd concentrations. Data are means±SD of three biological replicates, and five plants were mixed in one replication for metal determination. One and two asterisks indicate values are significantly different from the WT at the levels of P<0.05 and P<0.01, respectively (t-test).
OsNRAMP5 contributed to Mn accumulation in hulls and brown rice
OsNRAMP5 showed strong expression in hulls (Fig. 1 and Fig. 2), suggesting that it may be involved in Mn accumulation in hulls. This hypothesis was tested by analysing the element concentrations of hulls and brown rice of the mutant and wild-type plants grown under field conditions. In wild-type plants, the concentrations of Mn, Fe, and Cd in hulls were far greater than those in brown rice (Supplementary Fig. S8 available at JXB online). There were significant differences in Mn concentrations between mutant and wild-type plants both in hulls and brown rice. Compared with wild-type plants, the knockout line showed an approximately 82 and 52% reduction in Mn concentrations in hulls and brown rice, respectively (Supplementary Fig. S8A available at JXB online). The Fe concentrations in brown rice was about 80% higher in osnramp5 than in wild-type plants, but no significant difference was found in hulls (Supplementary Fig. S8B available at JXB online). Significant differences were also found in Cd concentrations of hulls and brown rice between wild type and osnramp5 plants, with the mutant having 76% and 35% of the wild type Cd concentrations in hulls and brown rice, respectively (Supplementary Fig. S5C available at JXB online).
The distributions of Mn, Fe, and Cd in root were affected by the expression pattern of OsNRAMP5 in root
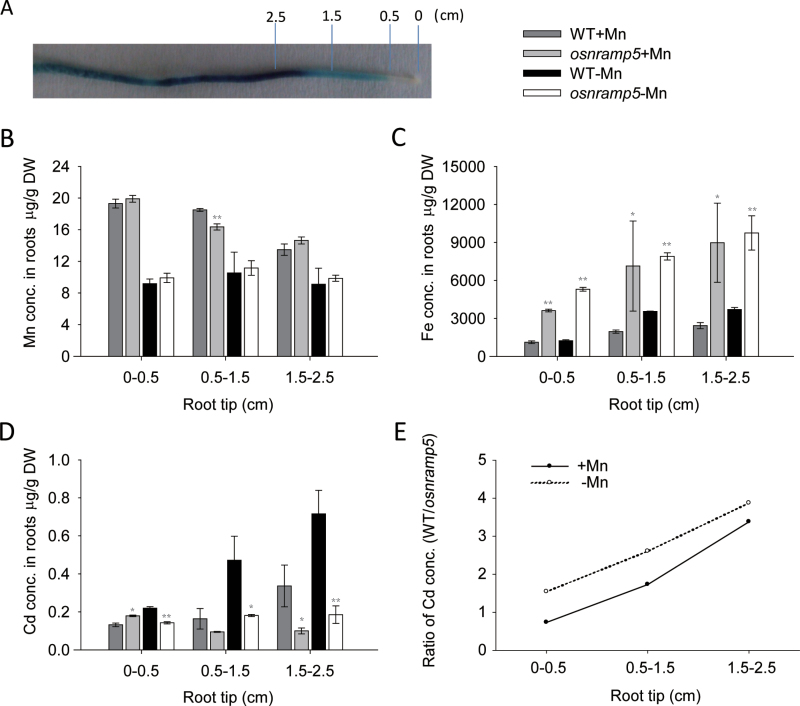
In roots, the GUS activity was mainly detected in the maturation zone and, to a lesser extent, also in the elongation zone, with no activity being observed in the root cap or the meristem zone (Fig. 2). Metal concentrations in the three regions of roots were determined, showing different distribution patterns for Mn, Fe, and Cd. When grown with 8 µM Mn supply, both mutant and wild-type plants had the highest Mn concentrations in the root tip (0–0.5cm), and the concentration decreased with the distance from the apex (Fig. 8B). Under low Mn supply (0.08 µM), the difference between different root zones disappeared. Moreover, root tips of the plants grown with high Mn supply showed a two-fold increase in Mn concentration from those grown under low Mn supply. There was little difference in the Mn concentration of root tips between wild type and mutant plants. By contrast, mutant plants showed much higher Fe concentrations than wild-type plants in all three root zones (Fig. 8C). As expected, Cd concentrations of wild type were significantly higher than those of mutant plants in the root zone where OsNRAMP5 was expressed, but not in the root tips where OsNRAMP5 was not expressed. Moreover, the Cd concentrations of wild type increased with the distance from the root apex (Fig. 8D), which was consistent with the expression level of OsNRAMP5 as described in Fig. 8A, but no similar pattern was observed in osnramp5 plants. Additionally, a higher Mn supply decreased Cd concentration in the three root zones of wild type. The ratio of root Cd concentrations between wild type and mutant plants increased from the root tip to the maturation zone in both Mn treatments (Fig. 8E).
Fig. 8.
The distribution of metals in different root zones. (A) Root expression pattern of OsNRAMP5. The expression pattern was detected in plants transformed with the construct OsNRAMP5-promoter:GUS by histochemical straining of GUS activity. 0, 0.5, 1.5, 2.5 represent the distance from root tip by centimetre. (B–D) Mn, Fe, and Cd concentrations in different root zones. (E) The ratio of Cd concentrations between wild type and knock-out line of OsNRAMP5. Data are means±SD of three biological replicates, each sample in one replication was harvested from 30 plants. (This figure is available in colour at JXB online.)
Discussion
The role of OsNRAMP5 in Mn uptake and translocation
OsNRAMP5 has been reported as a major transporter for root uptake of Mn from the external medium with an apparent K m of 1.08 µM (Sasaki et al., 2012). The present study confirmed the essential role of OsNRAMP5 for Mn uptake, but also showed that OsNRAMP5 was involved in the internal translocation of Mn.
The knockout line of OsNRAMP5 was highly sensitive to Mn deficiency (Fig. 3B). In the low range of Mn supply (0.04–1.6 µM), the Mn concentrations in shoots of wild-type plants increased markedly with increasing Mn supply, but no significant increase was found in osnramp5 shoots. In addition, a high Mn supply could compensate for the lost capacity of Mn uptake in roots of the mutant but not the root-to-shoot translocation (Fig. 3C insert). A similar phenomenon occurred in the Fe-deficient osnramp5 plants, which accumulated plentiful Mn in roots but did not rescue the shoot Mn concentration (Fig. 6B). The results obtained here suggest that OsNRAMP5 was involved not only in Mn uptake from the external solution but also in Mn translocation from roots to shoots. This hypothesis was also supported by the expression pattern of OsNRAMP5 in roots: OsNRAMP5 was expressed at high levels in stele cells especially in the xylem region, implying that OsNRAMP5 possibly participated in the xylem-mediated root-to-shoot transport (Fig. 2). In addition, Ishimaru et al. (2012b) found that Mn concentrations in the xylem sap from OsNRAMP5i plants were markedly lower than those of wild-type plants. This report also presented the expression pattern of OsNRAMP5 in roots by the GUS reporter gene driven by the OsNRAMP5 promoter, showing GUS activity around the xylem in the stele region. By contrast, Sasaki et al. (2012) showed that OsNRAMP5 was specifically expressed in the exodermis and endodermis as detected by immunostaining. In the present study, OsNRAMP5 expression was also observed in the exodermis and endodermis in the maturation zone of roots, although the level of expression was lower than that in the stele. This difference between the present study and that of Sasaki et al. (2012) may be caused by the different detection levels on expression pattern: in situ hybridization and OsNRAMP5 promoter:β-GUS transgenic experiments in the present study were both at transcript level but the other study used detection at the protein level.
Although Mn is an essential microelement for plants, over-accumulation of Mn in leaves can cause severe toxicity (Pittman, 2005). Rice plants have evolved strategies to tolerate high Mn in leaves. For example, OsYSL6 is implicated in the detoxification of excess Mn (Sasaki et al., 2011). However, the mechanism of Mn distribution in leaves under normal conditions of Mn supply is still poorly understood. OsYSL6 was found to be expressed in leaves with the expression level increasing with leaf age. By contrast, the present study showed that the expression of OsNRAMP5 decreased with leaf age (Fig. 1). The higher expression of OsNRAMP5 in the developing leaves suggests that OsNRAMP5 might play an important role in preferentially transporting Mn to young leaves. This assumption is supported by RNA in situ hybridization showing that OsNRAMP5 was enriched in the vascular bundles of leaves, especially in the parenchyma cells surrounding the xylem (Fig. 2). OsNRAMP5 may therefore participate in the process of Mn unloading from the xylem vessels or in the uptake of Mn from the apoplast to the parenchyma cells. However, more direct evidence is needed. In addition, leaves of wild-type rice plants showed a pattern of Mn concentration increasing with leaf age, which was not observed in the osnramp5 mutant (Fig. 7). This difference implies that OsNRAMP5 contributed to Mn accumulation and distribution in different leaves of rice plants.
Rice hull is important for the development of grain, and its photosynthesis contributes to grain filling (Pengelly et al., 2011). It has been reported that knocking down OsYSL2 decreased the concentrations of Mn and Fe in polished rice (Ishimaru et al., 2010). The present study showed a high expression level of OsNRAMP5 in the panicle, almost as high as that in roots, suggesting that OsNRAMP5 may also play an important role in distributing Mn in this tissue. Both the hulls and brown rice of the OsNRAMP5 knockout line contained much lower concentrations of Mn than wild type (Supplementary Fig. S8A available at JXB online), although this could be a cumulative effect of decreased root uptake, decreased root-to-shoot translocation and impaired distribution of Mn in the panicle.
Role of OsNRAMP5 in Fe homeostasis
There is strong evidence that Mn and Fe share some of the membrane transporters for the entrance into the root cells (Lanquar et al., 2010). Indeed, Fe deficiency resulted in increased accumulation of Mn in rice roots of both the wild type and the osnramp5 mutant, which can be explained by the induction of other Fe/Mn transporters, such as OsYSL2 (Supplementary Fig. S5 available at JXB online). OsNRAMP5 has been shown to possess transport activities for both Fe and Mn when expressed in yeast (Ishimaru et al., 2012b). However, the total Fe concentrations in roots and shoots were hardly affected by the loss of function of OsNRAMP5. In fact, when supplied with a low level of Mn, Fe concentration of the mutant roots was increased more than that in wild type (Fig. 4A), an observation that is consistent with the results of Sasaki et al. (2012). This can be explained by the induction of some Fe transporters, such as OsIRT2, OsYSL2, OsYSL15, and OsNRAMP1 (Supplementary Fig. S6 available at JXB online), in osnramp5 plants especially under Mn-deficient conditions. That hypothesis was also supported by our data showing that a much higher Fe concentration in osnramp5 root tips than wild type, where OsNRAMP5 was not expressed (Fig. 8). Taken together, the results show a strong interaction between Fe and Mn and that the loss of function of OsNRAMP5 affects Fe homeostasis by influencing the expression of other Fe transporters.
Role of OsNRAMP5 in Cd uptake
Cd is harmful for most living organisms, including rice plants (Clemens et al., 2013). OsNRAMP5 has been reported to be a major transporter for Cd uptake in rice (Ishikawa et al., 2012; Sasaki et al., 2012). This is supported by the results from the present study showing a marked decrease in Cd concentrations in both roots and shoots of the osnramp5 mutant (Fig. 5). However, Ishimaru et al. (2012a) reported decreased Cd concentration in roots but increased Cd concentration in shoots in a knockdown line of this gene (Ishimaru et al., 2012b). This inconsistency may be explained by the difference between OsNRAMP5 knockdown and knockout lines. In the knockdown lines, Cd is taken up from soil by OsNRAMP5, and may be transported to the shoot by Fe deficiency-induced transporters.
A significant antagonism was observed between Cd and Mn in both roots and shoots in wild type, but this antagonism was not observed in the OsNRAMP5 knockout line. In wild type, increasing Mn supply resulted in decreased Cd concentration, whereas Mn uptake was also suppressed by a high Cd supply (Fig. 4B and 5D). Moreover, the expression of OsNRAMP5 was not affected by different Mn or Cd supply (Fig. 4G, 5G, H). The lack of Mn/Cd antagonism in the knockout mutant suggests that the antagonism observed in wild type occurs via the competition for transport by OsNRAMP5. Another possible mechanism is that high Cd and Mn were both toxic for rice plants and caused inactivation of OsNRAMP5 at the protein level to limit excessive Cd or Mn absorption.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. The expression pattern of OsNRAMP5 under Mn-deficient or Fe-deficient conditions.
Figure S2. The tolerance test of wild-type and osnramp5 to various metal deficiencies.
Figure S3. Metal concentrations in plants under different metal deficiencies.
Figure S4. The expression levels of some Fe transporters in rice roots at different Mn supplies.
Figure S5. The transcript levels of Fe transporters in rice roots under Mn and Fe deficiencies.
Figure S6. The transcript levels of Fe transporters in roots of osnramp5 mutant and wild type plants.
Figure S7. Chlorophyll contents in different leaves of wild type and osnramp5 mutant.
Figure S8. Metal concentrations of rice plants grown under field conditions.
Table S1. The primers of Fe transporters used in this study.
Acknowledgements
This work was supported by grants from the National High Technology Research and Development Program of China (2012AA10A303), Special Fund for Agro-scientific Research in the Public Interest (201403015), the National Natural Science Foundation of China (31172017) and transgenic project 2013ZX08001005.
Reference
- Cailliatte R, Schikora A, Briat JF, Mari S, Curie C. 2010. High-affinity manganese uptake by the metal transporter NRAMP1 is essential for Arabidopsis growth in low manganese conditions. Plant Cell 22, 904–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Aarts MG, Thomine S, Verbruggen N. 2013. Plant science: the key to preventing slow cadmium poisoning. Trends in Plant Science 18, 92–99 [DOI] [PubMed] [Google Scholar]
- Delhaize E. 2003. Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. The Plant Cell Online 15, 1131–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Gruber BD, Pittman JK, White RG, Leung H, Miao Y, Jiang L, Ryan PR, Richardson AE. 2007. A role for the AtMTP11 gene of Arabidopsis in manganese transport and tolerance. The Plant Journal 51, 198–210 [DOI] [PubMed] [Google Scholar]
- Doncheva S, Georgieva K, Vassileva V, Stoyanova Z, Popov N, Ignatov G. 2005. Effects of succinate on manganese toxicity in pea plants. Journal of Plant Nutrition 28, 47–62 [Google Scholar]
- Hebbern CA, Pedas P, Schjoerring JK, Knudsen L, Husted S. 2005. Genotypic differences in manganese efficiency: field experiments with winter barley (Hordeum vulgare L.). Plant and Soil 272, 233–244 [Google Scholar]
- Hirschi KD, Korenkov VD, Wilganowski NL, Wagner GJ. 2000. Expression of Arabidopsis CAX2 in tobacco. Altered metal accumulation and increased manganese tolerance. Plant Physiology 124, 125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Zhao X, Yu H, Ouyang Y, Wang L, Zhang Q. 2009. The ankyrin repeat gene family in rice: genome-wide identification, classification and expression profiling. Plant Molecular Biology 71, 207–226 [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Ishimaru Y, Igura M, Kuramata M, Abe T, Senoura T, Hase Y, Arao T, Nishizawa NK, Nakanishi H. 2012. Ion-beam irradiation, gene identification, and marker-assisted breeding in the development of low-cadmium rice. Proceedings of the National Academy of Sciences, USA 109, 19166–19171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Bashir K, Nakanishi H, Nishizawa NK. 2012a. OsNRAMP5, a major player for constitutive iron and manganese uptake in rice. Plant Signaling & Behavior 7, 763–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimaru Y, Masuda H, Bashir K, Inoue H, Tsukamoto T, Takahashi M, Nakanishi H, Aoki N, Hirose T, Ohsugi R, Nishizawa NK. 2010. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. The Plant Journal 62, 379–390 [DOI] [PubMed] [Google Scholar]
- Ishimaru Y, Takahashi R, Bashir K, Shimo H, Senoura T, Sugimoto K, Ono K, Yano M, Ishikawa S, Arao T, Nakanishi H, Nishizawa NK. 2012b. Characterizing the role of rice NRAMP5 in manganese, iron and cadmium transport. Scientific Reports 2, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keen CL, Ensunsa JL, Clegg MS. 2000. Manganese metabolism in animals and humans including the toxicity of manganese. Metal Ions in Biological Systems 37, 89–121 [PubMed] [Google Scholar]
- Koike S, Inoue H, Mizuno D, Takahashi M, Nakanishi H, Mori S, Nishizawa NK. 2004. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. The Plant Journal 39, 415–424 [DOI] [PubMed] [Google Scholar]
- Lanquar V, Lelievre F, Barbier-Brygoo H, Thomine S. 2004. Regulation and function of AtNRAMP4 metal transporter protein. Soil Science and Plant Nutrition 50, 1141–1150 [Google Scholar]
- Lanquar V, Ramos MS, Lelievre F, Barbier-Brygoo H, Krieger-Liszkay A, Kramer U, Thomine S. 2010. Export of vacuolar manganese by AtNRAMP3 and AtNRAMP4 is required for optimal photosynthesis and growth under manganese deficiency. Plant Physiology 152, 1986–1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma K, Xiao J, Li X, Zhang Q, Lian X. 2009. Sequence and expression analysis of the C3HC4-type RING finger gene family in rice. Gene 444, 33–45 [DOI] [PubMed] [Google Scholar]
- Marschner H. 1995. Mineral Nutrition of Higher Plants. London: Academic Press 324–332 [Google Scholar]
- Mora MD, Rosas A, Ribera A, Rengel Z. 2009. Differential tolerance to Mn toxicity in perennial ryegrass genotypes: involvement of antioxidative enzymes and root exudation of carboxylates. Plant and Soil 320, 79–89 [Google Scholar]
- Nuruzzaman M, Gupta M, Zhang CJ, Wang L, Xie WB, Xiong LH, Zhang QF, Lian XM. 2008. Sequence and expression analysis of the thioredoxin protein gene family in rice. Molecular Genetics and Genomics 280, 139–151 [DOI] [PubMed] [Google Scholar]
- Pedas P, Hebbern CA, Schjoerring JK, Holm PE, Husted S. 2005. Differential capacity for high-affinity manganese uptake contributes to differences between barley genotypes in tolerance to low manganese availability. Plant Physiology 139, 1411–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedas P, Ytting CK, Fuglsang AT, Jahn TP, Schjoerring JK, Husted S. 2008. Manganese efficiency in barley: identification and characterization of the metal ion transporter HvIRT1. Plant Physiology 148, 455–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiter E, Montanini B, Gobert A, Pedas P, Husted S, Maathuis FJ, Blaudez D, Chalot M, Sanders D. 2007. A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proceedings of the National Academy of Sciences, USA 104, 8532–8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly JJ, Kwasny S, Bala S, Evans JR, Voznesenskaya EV, Koteyeva NK, Edwards GE, Furbank RT, von Caemmerer S. 2011. Functional analysis of corn husk photosynthesis. Plant Physiology 156, 503–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman JK. 2005. Managing the manganese: molecular mechanisms of manganese transport and homeostasis. New Phytologist 167, 733–742 [DOI] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Xia J, Ma JF. 2011. OsYSL6 is involved in the detoxification of excess manganese in rice. Plant Physiology 157, 1832–1840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Yamaji N, Yokosho K, Ma JF. 2012. Nramp5 is a major transporter responsible for manganese and cadmium uptake in rice. Plant Cell 24, 2155–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomine S, Lelievre F, Debarbieux E, Schroeder JI, Barbier-Brygoo H. 2003. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. The Plant Journal 34, 685–695 [DOI] [PubMed] [Google Scholar]
- Wang L, Xie WB, Chen Y, Tang WJ, Yang JY, Ye RJ, Liu L, Lin YJ, Xu CG, Xiao JH, Zhang QF. 2010. A dynamic gene expression atlas covering the entire life cycle of rice. The Plant Journal 61, 752–766 [DOI] [PubMed] [Google Scholar]
- Wu CY, Li XJ, Yuan WY, et al. 2003. Development of enhancer trap lines for functional analysis of the rice genome. The Plant Journal 35, 418–427 [DOI] [PubMed] [Google Scholar]
- Xue WY, Xing YZ, Weng XY, et al. 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nature Genetics 40, 761–767 [DOI] [PubMed] [Google Scholar]
- Yamaji N, Sasaki A, Xia JX, Yokosho K, Ma JF. 2013. A node-based switch for preferential distribution of manganese in rice. Nature Communications 4, 2442. [DOI] [PubMed] [Google Scholar]
- Yang M, Zhang W, Dong H, Zhang Y, Lv K, Wang D, Lian X. 2013. OsNRAMP3 is a vascular bundles-specific manganese transporter that is responsible for manganese distribution in rice. PLoS One 8, e83990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye RJ, Zhou F, Lin YJ. 2012. Two novel positive cis-regulatory elements involved in green tissue-specific promoter activity in rice (Oryza sativa L ssp.). Plant Cell Reports 31, 1159–1172 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Forno DA, Cock JH, Gomez KA. 1976. Laboratory Manual for Physiological Studies of Rice , 3 rd ed Manila: International Rice Research Institute [Google Scholar]
- Zhang JW, Li CS, Wu CY, Xiong LZ, Chen GX, Zhang QF, Wang SP. 2006. RMD: a rice mutant database for functional analysis of the rice genome. Nucleic Acids Research 34, D745–D748 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.