Summary
Overexpression of SVP3 affects kiwifruit flower and fruit development. The reduced petal pigmentation results from interference with transcription of the kiwifruit flower tissue-specific R2R3 MYB regulator.
Key words: Actinidia, bud break, dormancy, flowering, kiwifruit, MYB, Nicotiana, petal colour, SVP.
Abstract
SVP-like MADS domain transcription factors have been shown to regulate flowering time and both inflorescence and flower development in annual plants, while having effects on growth cessation and terminal bud formation in perennial species. Previously, four SVP genes were described in woody perennial vine kiwifruit (Actinidia spp.), with possible distinct roles in bud dormancy and flowering. Kiwifruit SVP3 transcript was confined to vegetative tissues and acted as a repressor of flowering as it was able to rescue the Arabidopsis svp41 mutant. To characterize kiwifruit SVP3 further, ectopic expression in kiwifruit species was performed. Ectopic expression of SVP3 in A. deliciosa did not affect general plant growth or the duration of endodormancy. Ectopic expression of SVP3 in A. eriantha also resulted in plants with normal vegetative growth, bud break, and flowering time. However, significantly prolonged and abnormal flower, fruit, and seed development were observed, arising from SVP3 interactions with kiwifruit floral homeotic MADS-domain proteins. Petal pigmentation was reduced as a result of SVP3-mediated interference with transcription of the kiwifruit flower tissue-specific R2R3 MYB regulator, MYB110a, and the gene encoding the key anthocyanin biosynthetic step, F3GT1. Constitutive expression of SVP3 had a similar impact on reproductive development in transgenic tobacco. The flowering time was not affected in day-neutral and photoperiod-responsive Nicotiana tabacum cultivars, but anthesis and seed germination were significantly delayed. The accumulation of anthocyanin in petals was reduced and the same underlying mechanism of R2R3 MYB NtAN2 transcript reduction was demonstrated.
Introduction
Plant MADS-domain transcription factors act as key regulators of many developmental processes (Smaczniak et al., 2012). The STMADS subfamily of MADS-box genes are predominantly expressed in vegetative tissues and play roles in vegetative development and transition to flowering in diverse plant species (Carmona et al., 1998; Hartmann et al., 2000; Mao et al., 2000; Yu et al., 2002; Masiero et al., 2004; Aswath and Kim, 2005; Liu et al., 2007; Trevaskis et al., 2007; He et al., 2010; Cohen et al., 2012; Cooke et al., 2012).
Relatively small changes in sequence can have a major effect on STMADS gene function. In Arabidopsis thaliana, the two STMADS genes SVP and AGL24 act in an opposite manner despite their close homology. SVP plays a key role as a repressor of the transition to flowering; a single amino acid substitution causes the loss of function resulting in an early flowering phenotype (Hartmann et al., 2000; Méndez-Vigo et al., 2013). In contrast, AGL24 acts as a promoter of flowering because its inactivation results in late flowering (Yu et al., 2002). Both proteins have further roles during early stages of flower development to repress expression of class B and C floral homeotic genes and maintain floral meristem identity (Gregis et al., 2009). Protein interactions and formation of complexes with other MADS-domain proteins are essential for STMADS function (Liu et al., 2009; Gregis et al., 2013). Interaction with APETALA1 (AP1) in particular resulted in floral reversion phenotypes observed in transgenic Arabidopsis upon ectopic expression of SVP-like genes (Liu et al., 2007; Gregis et al., 2008; Fornara et al., 2008; Wu et al., 2012; Jaudal et al., 2013). For these and perhaps other reasons, the function of STMADS observed upon ectopic expression in Arabidopsis might be different from their function in the species of origin (Trevaskis et al., 2007; Jaudal et al., 2013).
In woody perennial plants, the STMADS subfamily genes have been reported to act as growth inhibitors, which correlated with maintenance of plant dormancy (Mazzitelli et al., 2007; Bielenberg et al., 2008; Diaz-Riquelme et al., 2009; Horvath et al., 2010; Li et al., 2010; Sasaki et al., 2011; Yamane et al., 2011). Deletion of the six SVP-like DORMANCY-ASSOCIATED MADS-BOX (DAM) genes in peach (Prunus persica) resulted in failure to enter dormancy under cold or short-day induction, and the expression of a subset of these genes was elevated during endodormancy (Bielenberg et al., 2008; Li et al., 2009; Yamane et al., 2011). Similarly, a negative correlation of expression with endodormancy release was observed for six tandemly arrayed DAM genes predicted to act as transcriptional repressors in Japanese apricot (Prunus mume) (Yamane et al., 2008; Sasaki et al., 2011). Ectopic expression of one of these genes in transgenic poplar resulted in premature growth cessation and terminal bud set, demonstrating the role in growth inhibition (Sasaki et al., 2011). However, neither of these genes was functionally characterized by ectopic expression in the plant species of origin.
In the kiwifruit species Actinidia chinensis and A. deliciosa, four SVP genes (SVP1–SVP4) have been identified, with potentially distinct roles in bud dormancy and flowering (Wu et al., 2012). Their expression was mostly confined to vegetative tissues, with SVP3 showing the highest relative expression. SVP1, SVP2, and SVP4 were elevated in buds over the winter dormancy period. In contrast, SVP3 accumulation in buds did not demonstrate seasonal changes, but delayed flowering in transgenic Arabidopsis and was able to rescue the Arabidopsis svp41 mutant. Ectopic expression resulted in floral reversion phenotypes, caused by the strong capacity for heterodimerization with Arabidopsis MADS box proteins responsible for flower development (Wu et al., 2012).
In the current study, functional analysis of SVP3 was performed using ectopic transgenic analysis in kiwifruit species A. deliciosa and A. eriantha, which have different chilling requirements and bloom time, and a model plant species tobacco (Nicotiana tabacum). Normal vegetative growth and floral transition, but abnormal flower development, reduced petal pigmentation, and abnormal fruit and seed development were observed. The underlying mechanism of reduced anthocyanin accumulation in petals was examined. Based on the results obtained, the biological function of SVP3 is discussed.
Materials and methods
Gene isolation and vector construction
The A. chinensis SVP3 coding sequence was cloned under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (35S:SVP3) into the pART277 binary vector (Gleave, 1992) as previously described (Wu et al., 2012). The resulting plasmid was transformed into Agrobacterium tumefaciens strain EHA105 for transformation into A. deliciosa and A. eriantha, and into GV3101 for transformation into N. tabacum. A construct with a reporter gene uidA (GUS; β-gluronidase) under the control of the CaMV 35S promoter (35S:GUS) in appropriate Agrobacterium strains was used to transform control plants (Wang et al., 2006). Actinidia eriantha SVP3 (GenBank accession no. KJ123703) was amplified from the bud cDNA using gene-specific oligonucleotide primers, 5’-ATGGCGAGAGAGAAGATCAAGA-3’ (forward) and 5’-TGGTGTGACATTTCAAGTTCG-3’ (reverse). The predicted amino acid sequence alignment with A. deliciosa and A. chinensis SVP3 homologues was performed using Vector NTI version 9.0. (Invitrogen) Clustal W (opening 15, extension penalty 0.3).
Plant transformation and growth conditions
The transformation procedures for A. deliciosa and A. eriantha were as previously described (Wang et al., 2006, 2007). Once roots were established, transgenic plants were transferred to soil and grown in the containment glasshouse at the Plant & Food Research, Mt Albert, Auckland, New Zealand for 18 months. Flowering time and floral phenotype were assessed in the following spring season. Nicotiana tabacum ‘Maryland Mammoth’ and ‘Samsun’ transformations were carried out on young leaf discs excised from in vitro grown shoots (Horsch et al., 1985). Transgenic tobacco plants were grown in a glasshouse at 20 °C under long-day (16/8h light/dark; for ‘Samsun’) or short-day conditions (8/16h light/dark; for ‘Maryland Mammoth’). The seeds from these transgenic plants were collected and germinated on half-strength Murashige and Skoog (MS) agar medium (Murashige and Skoog, 1962) supplemented with 50 μg ml–1 kanamycin. Following the segregation tests, two homozygous lines of both cultivars at T2 generations were used for flowering time and floral phenotype assessment.
RNA extraction and expression studies
Actinidia eriantha root, stem, leaf, axillary bud, shoot apex, flower, and fruit tissue collection was carried out on vines growing in the glasshouse at the Plant & Food Research, Mt Albert, Auckland, New Zealand, during the spring and summer season of 2011–2012. Actinidia chinensis root, stem, leaf, flower, fruit, and axillary and apical bud collection was carried out on vines growing at the Plant & Food Research orchard near Kerikeri, New Zealand, during the spring and summer season of 2005–2006.
Total RNA was extracted from kiwifruit tissue as previously described (Chang et al., 1993). Total RNA was isolated from tobacco flowers using the Trizol reagent (Invitrogen). A 5 μg aliquot of total RNA was treated with DNase I (Ambion) and reverse-transcribed at 37 °C using the BluePrint® Reagent kit for reverse transcription–PCR (RT–PCR; TaKaRa) according to the manufacturer’s instructions. Amplification and quantification were carried out using the LightCycler® 480 System and LightCycler® 480 SYBR Green I Master Mix (Roche Diagnostics). A minimum of three technical replicate reactions were performed and a non-template control was included in each run. Thermal cycling conditions were 95 °C for 5min, followed by 50 cycles of 95 °C for 10 s, 60 °C for 10 s, and 72 °C for 20 s, and a melting temperature cycle, with constant fluorescence data acquisition from 65 °C to 95 °C. The data were analysed using the Target/Reference ratio calculated with the LightCycler® 480 software 1.5 (Roche Diagnostics). The expression was normalized to previously characterized reference genes, kiwifruit actin (Wu et al., 2012) and tobacco Ntα-Tub1 (Pattanaik et al., 2010).
Oligonucleotide primers for kiwifruit SVP3, actin, anthocyanin pathway genes, and regulatory genes MYB110 and MYB10 were as previously described (Montefiori et al., 2011; Wu et al., 2012; Fraser et al., 2013). Oligonucleotide primers for tobacco NtAn1a, NtAN1b, NtAn2, and Ntα-Tub1 were as described by Bai et al. (2011) and Pattanaik et al. (2010).
Biochemical analyses
For chlorophyll quantification, petals collected at visually similar developmental stages were frozen and ground in liquid nitrogen. Ground tissues were suspended in 80% acetone with 2.5M phosphate buffer pH 7.5 and incubated for 1h in the dark, followed by centrifugation at 16 000 g for 10min. The absorbance of 200 μl of the supernatant was measured spectrophotometrically at 663nm and 646nm (SpectraMax 384, Molecular Devices, Sunnyvale, CA, USA). The total chlorophyll (Ca+b) concentration was calculated using the equation Ca+b (μg ml–1) =7.15A 663+18.71A 646 (Lichtenthaler and Rinderle, 1988).
Anthocyanins and flavonoids were extracted from lyophilized samples and analysed by high-performance liquid chromatography (HPLC) and liquid chromatography–mass spectrometry (LC-MS) as described previously (McGhie et al., 2011; Dare et al., 2013). Quantification of cyanidin 3-O-galactoside was achieved by calibration at 520nm by HPLC analysis. The peaks were identified by a comparison of retention times with authentic standards of cyanidin 3-O-galactoside (Polyphenols Laboratories, Sandnes, Norway).
Microscopic analyses
Tobacco petal epidermis was obtained by careful peeling and observed under a light microscope (Olympus Vanox AHT3, Olympus Optical Co. Ltd, Tokyo, Japan). Photographs were taken using a CoolSNAP Colour camera system (Roper Scientific Ltd, Tucson, AZ, USA).
Yeast two-hybrid assay
The full-length open reading frame of kiwifruit SVP genes and kiwifruit AGAMOUS (AG), FRUITFULL class (FUL and FUL-like), SEPALLATA class (SEP1, SEP3, and SEP4), APETALA3 (AP3), PISTILLATA (PI), and CENTRORADIALIS (CEN) were amplified and cloned as described previously (Varkonyi-Gasic et al., 2011, 2013; Wu et al., 2012). The bait vector and prey vector were mated and selected on synthetic complete (SC) plates lacking leucine and tryptophan. The affinity of two protein interactions was determined on selective SC medium lacking leucine, tryptophan, and histidine, and in the presence of 1, 3, and 5mM 3-aminotriazole (3AT) at 30 °C. All assayed proteins were tested for autoactivation, homodimerization, and heterodimerization capacity. The strength of interactions was compared with control plasmids of pEXP™32/Krev1 with pEXP™22/RalGDS-wt, pEXP™22/RalGDS-m1, and pEXP™22/RalGDS-m2 (Invitrogen).
Results
Overexpression of SVP3 has no significant effect on growth or dormancy in A. deliciosa
To investigate the role of kiwifruit SVP3, transgenic A. deliciosa was generated using the SVP3 cDNA driven by the CaMV 35S promoter (Wu et al., 2012). Ten independent transgenic lines with high SVP3 transgene expression levels were generated and compared with six control lines (Fig. 1A). Monitoring during callus formation, plantlet growth in tissue culture, and upon transfer to soil revealed no obvious difference between 35S:SVP3 lines and control lines. Eight 35S:SVP3 and three control lines were further monitored for spring bud break. The number of breaking buds or developing shoots was recorded weekly and is presented as a percentage of total buds. Large variation in the time of the first visible bud break, positions of breaking buds, number of developing branches, and rate of shoot outgrowth was observed for all lines; however, the first visible bud break time of the 35S:SVP3 lines was in the range recorded for control lines and was followed by normal growth of multiple branches (Fig. 1B, C). Therefore, it was concluded that SVP3 does not have a significant effect on growth and dormancy in A. deliciosa under glasshouse conditions.
Fig. 1.
Overexpression of SVP3 in Actinidia deliciosa. (A) Relative expression of SVP3 in 35S:SVP3 and control plants. The expression was normalized against kiwifruit actin. Error bars represent the standard error (SE) of four replicate reactions. (B) Bud break time measured as a percentage of total buds with visible shoot outgrowth. (C) A subset of 35S:SVP3 and control A. deliciosa plants, photographed on the same day in spring. Arrowheads indicate positions of visible bud break and shoot outgrowth. Scale bar=10cm.
SVP3 affects reproductive development in transgenic A. eriantha
Actinidia deliciosa has a high chilling requirement (Wall et al., 2008) and late bud break, and fails to flower under glasshouse conditions. Therefore, to evaluate further the role of SVP3, the non-cultivated kiwifruit species A. eriantha was chosen for transformation, as it provided advantages including low chilling requirement and early bud break, fast maturity, and prolific flowering in glasshouse conditions (Wang et al., 2006). Actinidia eriantha SVP3 shares almost identical sequence and expression pattern with the previously studied cultivated kiwifruit species A. deliciosa and A. chinensis (Supplementary Fig. S1A, B available at JXB online; Wu et al., 2012). Six transgenic lines with moderate to high levels of SVP3 transgene expression and three control lines were generated (Fig. 2A). None of the lines showed any obvious difference during callus formation, growth in tissue culture, and establishment in the soil; similar growth rate and architecture were observed (Supplementary Fig. S1 available at JXB online), and in all lines the time of bud break and flowering time (appearance of visible floral buds) were comparable (Table 1). The number of flowers was highly variable and not correlated with transgene expression levels, but the duration of flower development and appearance of flowers were altered significantly in transgenic lines, particularly in the highest expressing Line 1 (Table 1).
Fig. 2.
Overexpression of SVP3 in Actinidia eriantha. (A) Relative expression of SVP3 in 35S:SVP3 and control plants. The expression was normalized against kiwifruit actin. Error bars represent the standard error (SE) of four replicate reactions. (B–D) Morphology of flower buds (B, C) and fully open flowers (D). (E, F) Fruit morphology. (G–I) Seed morphology (G, H) and weight (I). Error bars represent the standard error (SE), n=3. Scale bars=5mm (B, C), 10mm (D–F), and 0.5mm (G, H). (J) Protein interactions detected by yeast two-hybrid assays. ++, very strong interaction; +, strong interaction; –, no interaction.
Table 1.
Phenotypic analysis of six lines of 35S:SVP3 transgenic A. eriantha compared with three control linesBud break time represents the number of days from 100% leaf drop to the first visible bud break. Flowering time represents the number of days from the first visible bud break to the first visible flower bud. Anthesis time represents the number of days from visible flower buds to fully open flowers.
| A. eriantha transgenic lines | Bud break time (d) | Flowering time (d) | No. of flowers | Anthesis time (d) | Petal colour |
|---|---|---|---|---|---|
| Line 1 | 38 | 25 | 12 | 55.1±10.3 | White/green |
| Line 2 | 32 | 23 | 2 | 49.7±4.1 | Light pink |
| Line 3 | 38 | 24 | 1 | 43 | Light pink |
| Line 4 | 38 | 26 | 2 | 47.6±6.8 | Light pink |
| Line 5 | 32 | 21 | 10 | 46.2±7.6 | Light pink |
| Line 6 | 35 | 21 | 2 | 45.1±9.3 | Light pink |
| Control 1 | 35 | 28 | 12 | 27.4±4.1 | Pink |
| Control 2 | 40 | 22 | 35 | 21.4±5.0 | Pink |
| Control 3 | 32 | 24 | 5 | 27.8±6.2 | Pink |
Data are expressed as the mean± SE.
For further detailed analysis, Line 1 and Line 2 were chosen as representatives of high and intermediate SVP3 transgene expression, respectively. Flower buds in Line 1 and to a lesser extent in Line 2 were not fully enclosed with sepals (Fig. 2B); the number and order of developing floral organs appeared normal (Fig. 2C), but flowers in Line 1 remained smaller (Fig. 2D). Fruit in Line 1 was misshapen and smaller compared with control or Line 2 fruit (Fig. 2E, F) and the transgenic seed was smaller, lighter (Fig. 2G–I), and failed to germinate.
Floral reversion phenotypes observed upon expression of STMADS genes in Arabidopsis result from interaction with Arabidopsis floral homeotic MADS box proteins, particularly AP1 (Fornara et al., 2008; Gregis et al., 2009; Wu et al., 2012). To test if phenotypes observed upon ectopic expression of SVP3 in A. eriantha arose from similar MADS box protein interactions, yeast two-hybrid analysis was performed using previously characterized kiwifruit floral MADS box genes (Varkonyi-Gasic et al., 2011). SVP3 interacted with kiwifruit AG and SEP proteins SEP1, SEP3, and SEP4 with similar strong interaction intensities; a weaker interaction was identified with FUL and FUL-like, but no interaction was detected with AP3, PI, and the negative control CEN (Fig. 2J).
SVP3 affects petal colour in transgenic A. eriantha by transcriptional repression of key genes in the anthocyanin pathway
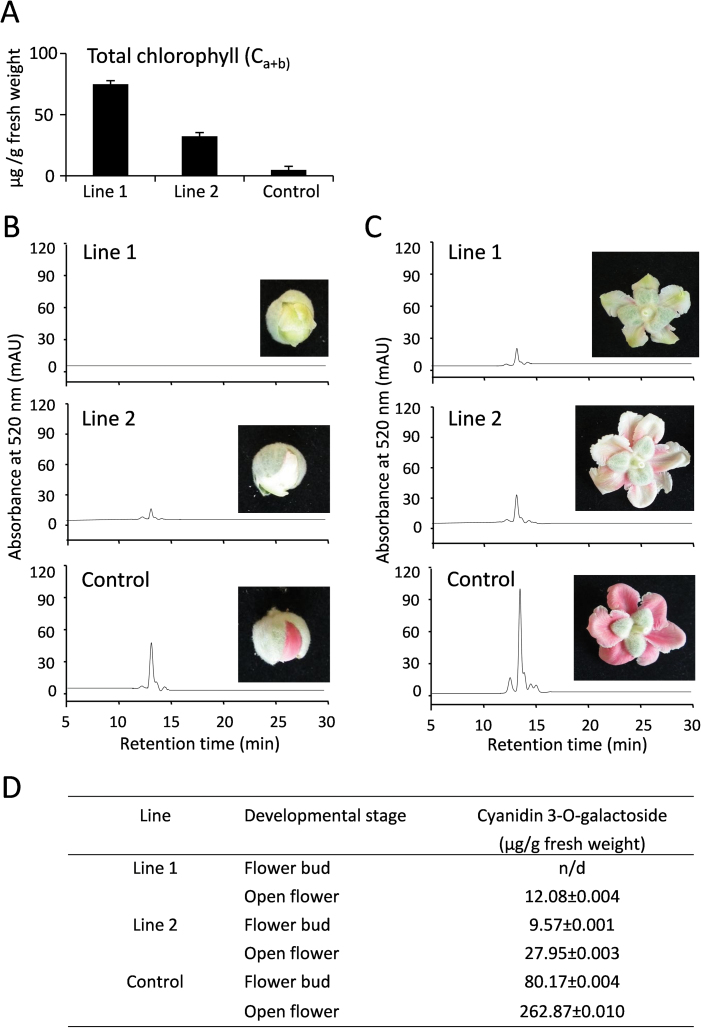
In addition to the described phenotypes, the colour of the petals of 35S:SVP3 lines was dramatically reduced (Fig. 2D, Table 1) and the chlorophyll content was higher (Fig. 3A).
Fig. 3.
Biochemical analysis of petal pigmentation. (A) Total chlorophyll content in petals of fully opened flowers. Error bars represent the SE of three independent measurements. (B, C) HPLC assay for cyanidin 3-O-galactoside correlated with visual phenotypes of flower buds (B) and open flowers (C). (D) Quantification of cyanidin 3-O-galactoside using LC-MS. Data represent the means ±SE (n=3).
The presence and intensity of the peak corresponding to the major kiwifruit anthocyanin, cyanidin 3-O-galactoside, correlated with the visual petal phenotype in flower buds and open flowers (Fig. 3B, C), and further quantification confirmed differential accumulation of cyanidin 3-O-galactoside (Fig. 3D) and several minor flavonoid compounds (Supplementary Fig. S2 available at JXB online).
Analysis of the relative expression of the genes encoding enzymes of the anthocyanin biosynthetic pathway (Montefiori et al., 2011) revealed similar expression profiles in representative 35S:SVP3 and control lines, with some variation between flower bud and open flower samples. The exception was the transcript of F3GT1, which was absent or barely detectable in flower buds of 35S:SVP3 lines, in contrast to relatively high accumulation observed in the control flower bud (Fig. 4A). Analysis of regulatory genes of the anthocyanin biosynthetic pathway (Fraser et al., 2013) revealed high relative expression of MYB110a and low relative expression of MYB10 in flower buds of the control line. In 35S:SVP3 lines these transcripts were absent or barely detectable, and in the control line both transcripts showed a decrease in open flowers (Fig. 4B).
Fig. 4.
Relative expression of genes responsible for anthocyanin biosynthesis in kiwifruit. (A) Relative expression of kiwifruit flavonoid biosynthetic pathways genes. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavonoid 3-hydroxylase; F3’H, flavonoid 3’-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; F3GT1 and F3GGT1; flavonoid 3-O glycosyltransferases. (B) Relative expression of regulatory MYB genes. Relative expression of SVP3 is presented below. The expression was normalized against kiwifruit actin. Error bars represent the SE of three replicate reactions.
SVP3 does not alter flowering time but affects reproductive development and petal colour in transgenic tobacco
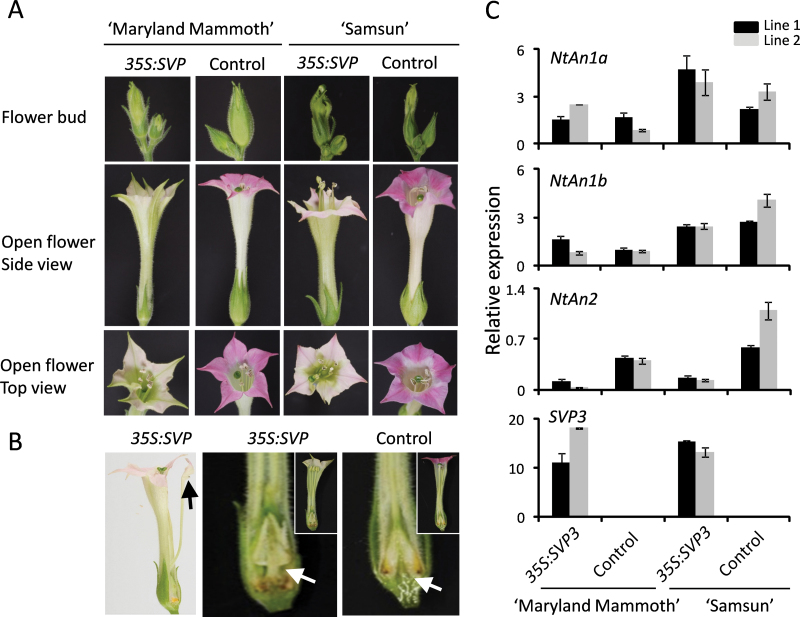
To investigate further whether kiwifruit SVP3 affects reproductive development and anthocyanin accumulation in a conserved manner, transgenic tobacco lines were generated using the 35S:SVP3 construct. A day-neutral tobacco ‘Samsun’ and a short-day variety ‘Maryland Mammoth’ were used. This approach provided the opportunity to establish if SVP3 affects flowering time in an annual representative of asterids, the clade of eudicots to which the perennial kiwifruit belongs. Eight independent transgenic lines of tobacco ‘Samsun’ and three of ‘Maryland Mammoth’ were generated. Two homozygous T2 lines of each cultivar with high transgene expression and two corresponding wild-type control lines were chosen for detailed analysis. No significant differences were observed in vegetative growth and flowering time, but the duration of flower development was extended, flower fertility was diminished, and seed germination was delayed (Table 2). Spike-shaped sepals in transgenic plants failed to enclose flower buds fully, the corolla remained green, and the distal part of the petal was altered in both colour and shape (Fig. 5A). Additional phenotypes were identified, including occasional homeotic conversion of stamen to petal and change in the ovary shape and its position relative to attachment of other floral parts (Fig. 5B). Changes in total chlorophyll, anthocyanin, and flavonoid content and presence of stomata in green petal segments were also noted (Supplementary Fig. S3 available at JXB online). To establish if reduced pigment accumulation resulted from transcriptional repression of key regulators of anthocyanin biosynthesis, quantitative PCR (qPCR) analysis of basic helix–loop–helix (bHLH) genes, NtAn1a and NtAn1b, and the R2R3 MYB gene, NtAn2 (Pattanaik et al., 2010; Bai et al., 2011), was performed. NtAn2 was significantly and consistently down-regulated in transgenic lines, while NtAn1a and NtAn1b expression showed only minor variations (Fig. 5C).
Table 2.
Phenotypic analysis of transgenic 35S:SVP tobacco ‘Maryland Mammoth’ and ‘Samsun’Flowering time was measured as the number of leaves when visible flower buds were observed. Anthesis time represents days from flower buds to fully open flowers. Total number of flowers includes sterile flowers. Seed germination was recorded as number of days to visible germination on half-strength MS medium.
| Cultivar | Genotype | Flowering time (d) | Anthesis time (d) | Total flowers | Sterile flowers | Seed germination |
|---|---|---|---|---|---|---|
| Maryland Mammoth | 35:SVP Line 1 | 19.5±0.19 | 7±1.4 | 27.5±0.5 | 20.5±2.2 | 16±0.3 |
| 35:SVP Line 2 | 20.8±1.0 | 8±0.7 | 40.7±4.4 | 28.3±9.2 | 18±0.2 | |
| Wild-type Line 1 | 20.9±0.3 | 5±1.5 | 27.4±1.6 | 3.7±0.7 | 8±0.0 | |
| Wild-type Line 2 | 22.0±0.6 | 4±1.2 | 31.7±3.4 | 2.5±0.6 | 8±0.0 | |
| Samsun | 35:SVP Line 1 | 33.0±1.1 | 6±1.0 | 23.7±1.5 | 13.2±7.2 | 9±0.2 |
| 35:SVP Line 2 | 42.0±0.8 | 7±1.4 | 26.7±3.0 | 12.1±6.3 | 11±0.4 | |
| Wild-type Line 1 | 34.0±0.1 | 4±0.6 | 21.7±3.7 | 2.8±0.4 | 6±0.0 | |
| Wild-type Line 2 | 36.0±1.5 | 4±1.6 | 24.7±3.8 | 3.0±0.2 | 6±0.0 |
Data represent the mean ±SE of six individuals for each genotype.
Fig. 5.
Expression of kiwifruit SVP3 in transgenic tobacco. (A) Phenotypic analysis of flower buds and open flowers. (B) Floral organ abnormalities: petaloid stamen (black arrow) and ovary position (white arrows). The cross-sections of whole flowers demonstrating colour change are presented in insets. (C) Relative expression of NtAn1a, NtAn1b, NtAN2, and SVP3 transgene in petals normalized against Ntα-Tub1. Error bars represent the SE for three replicate reactions.
Discussion
Overexpression of SVP3 affects reproductive development but not floral transition in transgenic kiwifruit and tobacco
Ectopic expression of the STMADS gene subfamily often results in altered flowering time, severe floral abnormalities, delayed flower anthesis, and senescence (Hartmann et al., 2000; Yu et al., 2002; Lee et al., 2007; Fornara et al., 2008; He et al., 2010; Li et al., 2010), and affects duration of endodormancy in woody perennial species (Sasaki et al., 2011). Overexpression of SVP3 in A. deliciosa showed no major effect on vegetative growth and the timing of first visible bud break in glasshouse conditions, although insufficient winter chilling and large variation in the position and number of synchronously breaking buds might have affected the rate of visible shoot outgrowth. Overexpression of SVP3 in A. eriantha and tobacco showed no significant effect on plant growth, bud break, and flowering time, supporting the findings in A. deliciosa. However, flower, fruit, and seed development were severely affected. The size and shape of sepals were abnormal and they failed to enclose flower buds fully, consistent with the phenotype observed in Arabidopsis (Wu et al., 2012) and similar to other reports (Masiero et al., 2004; Liu et al., 2007; Trevaskis et al., 2007; Fornara et al., 2008). The petals contained vegetative features including increased chlorophyll content and stomata, similar to phenotypes observed upon overexpression of potato StMADS16 and sweet potato IbMADS3-1 in tobacco (Garcia-Maroto et al., 2000; Shin et al., 2011). Flower development was slower, resulting in significantly delayed anthesis and an increased proportion of sterile flowers, which failed to produce fruit (Fornara et al., 2008; Li et al., 2010). Additional phenotypes included underdeveloped and misshapen fruit, abnormal seed development, reduced seed germination rate, and extended seed germination, suggesting that SVP3 affected all stages of reproductive development. The severity of the observed phenotypes during reproductive development, combined with normal vegetative growth, wild-type bud break and flowering time, and relatively high levels of expression in vegetative tissues (Wu et al., 2012), indicate that SVP3 has a role in the regulatory network of vegetative organogenesis and early stages of floral transition as the key repressor of floral organ development. It is hypothesized that SVP3 acts to retain vegetative features during plant growth and floral transition, and has to be removed to enable subsequent stages of reproductive development. Ectopic expression phenotypes, combined with constitutive vegetative expression, suggest that there is no involvement of SVP3 in maintenance of winter dormancy and general growth inhibition. However, SVP3 may have a role in these processes as an interacting partner of other, rate-limiting proteins, and a loss-of-function phenotype would be required to understand SVP3 function fully.
SVP3 interferes with anthocyanin biosynthesis in petals via repression of an anthocyanin-related MYB transcription factor
Overexpression of MADS-box genes is often associated with floral abnormalities, including changes in petal pigmentation. Light green sepaloid petals were observed upon overexpression of Arabidopsis AGAMOUS-LIKE 20 (AGL20 or SOC-1) in Arabidopsis (Liu et al., 2007) and of the mustard AGL20 orthologue MADSA in short-day tobacco ‘Maryland Mammoth’ (Borner et al., 2008). Similarly, expression of STMADS genes such as potato StMADS16 and sweet potato IbMADS3-1 in tobacco resulted in development of chlorophyll-enriched petals (Garcia-Maroto et al., 2000; Shin et al., 2011). However, these reports did not investigate anthocyanin biosynthesis and accumulation in the petals of transgenic plants, and very little is known about potential MADS-box protein interference with the regulatory genes of the anthocyanin pathway. This study demonstrated that SVP3 reduced anthocyanin accumulation in petals of transgenic A. eriantha in a quantitative manner. The underlying mechanism was dramatically reduced expression of the key structural gene F3GT1 (Montefiori et al., 2011) and transcriptional repression of regulatory MYB transcription factors, in particular MYB110a, the key gene required for kiwifruit petal pigmentation (Fraser et al., 2013). A similar mechanism was observed in transgenic tobacco, where SVP3 interfered with transcription of the regulatory MYB NtAn2.
At this stage, it is unclear if SVP3 acts by direct or indirect repression of the regulatory MYB transcription. Currently, there is little knowledge about upstream regulators of anthocyanin-related R2R3 MYB expression. Some MADS box genes from the AP1/SQUA class have been implicated in regulation of anthocyanin accumulation. IbMADS10 expression correlated with red pigmentation in sweet potato, and ectopic expression resulted in anthocyanin accumulation in transgenic sweet potato callus and transgenic Arabidopsis (Lalusin et al., 2006, 2011). VmTDR4 expression was linked with colour development and anthocyanin-related gene expression in bilberry, while silencing of this gene reduced anthocyanin levels and altered expression of the regulatory R2R3 MYB transcription factor (Jaakola et al., 2010). STMADS proteins have a capacity to interact with other MADS box proteins, and interaction with AP1 affected flowering time and flower development in transgenic Arabidopsis (Fornara et al., 2008; Gregis et al., 2008; Wu et al., 2012). Therefore, a similar mechanism might be responsible for repression of MYB110a and NtAn2 transcription and reduced anthocyanin accumulation in transgenic kiwifruit and tobacco flowers. The yeast two-hybrid assays showed that SVP3 could interact with kiwifruit AP1/SQUA proteins FUL and FUL-like, and this interaction may interfere with activation of R2R3 MYB transcription. Even stronger interactions were observed with AG and SEP MADS box proteins required for normal flower and fruit development (Varkonyi-Gasic et al., 2011). Therefore, interference with establishment of homeotic MADS box protein complexes, as previously shown in Arabidopsis (Fornara et al., 2008; Gregis et al., 2008; Wu et al., 2012), is the likely mechanism by which SVP3 interferes with various aspects of normal reproductive development.
Further functional studies using transgenic kiwifruit and tobacco generated in the course of this study will aid in establishing direct targets and protein partners of SVP3, providing understanding of the pathways and plant processes regulated by SVP3. These plants also provide an exciting opportunity for deeper understanding of the regulation of anthocyanin-related MYB expression in flowers.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. SVP3 in Actinidia eriantha.
Figure S2. LC-MS analysis of flavonoid compounds in petals of 35:SVP3 and control A. eriantha.
Figure S3. Analysis of transgenic 35S:SVP3 tobacco.
Acknowledgements
The authors wish to thank Andrew Dare for help with HPLC analysis, Ian Hallett and Paul Sutherland for microscopy, Tim Holmes and Darren Snaith for photography, Sakuntala Karunairetnam and Andrew Gleave for cloning support, and Monica Dragulescu and Wade Wadasinghe for maintenance of plants in the glasshouse. Thanks to Mirco Montefiori and Richard Espley for providing valuable advice and critically reading this manuscript. This work was funded by the New Zealand Foundation for Research, Science and Technology, contract C10X0816 MeriNET.
References
- Aswath CR, Kim SH. 2005. Another story of MADS-box genes—their potential in plant biotechnology. Plant Growth Regulation 46, 177–188 [Google Scholar]
- Bai Y, Pattanaik S, Patra B, Werkman JR, Xie CH, Yuan L. 2011. Flavonoid-related basic helix–loop–helix regulators, NtAn1a and NtAn1b, of tobacco have originated from two ancestors and are functionally active. Planta 234, 363–375 [DOI] [PubMed] [Google Scholar]
- Bielenberg DG, Wang Y, Li ZG, Zhebentyayeva T, Fan SH, Reighard GL, Scorza R, Abbott AG. 2008. Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genetics and Genomes 4, 495–507 [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleißner R, Wisman E, Apel K, Melzer S. 2008. A MADS domain gene involved in the transition to flowering in Arabidopsis . The Plant Journal 24, 591–599 [DOI] [PubMed] [Google Scholar]
- Carmona MJ, Ortega N, Garcia-Maroto F. 1998. Isolation and molecular characterization of a new vegetative MADS-box gene from Solanum tuberosum L. Planta 207, 181–188 [DOI] [PubMed] [Google Scholar]
- Chang SJ, Puryear J, Cairney J. 1993. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter 11, 113–116 [Google Scholar]
- Cohen O, Borovsky Y, David-Schwartz R, Paran I. 2012. CaJOINTLESS is a MADS-box gene involved in suppression of vegetative growth in all shoot meristems in pepper. Journal of Experimental Botany 63, 4947–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JEK, Eriksson ME, Junttila O. 2012. The dynamic nature of bud dormancy in trees: environmental control and molecular mechanisms. Plant, Cell and Environment 35, 1707–1728 [DOI] [PubMed] [Google Scholar]
- Dare AP, Tomes S, Jones M, McGhie TK, Stevenson DE, Johnson RA, Greenwood DR, Hellens RP. 2013. Phenotypic changes associated with RNA interference silencing of chalcone synthase in apple (Malus×domestica). The Plant Journal 74, 398–410 [DOI] [PubMed] [Google Scholar]
- Diaz-Riquelme J, Lijavetzky D, Martinez-Zapater JM, Carmona MJ. 2009. Genome-wide analysis of MIKCC-type MADS box genes in grapevine. Plant Physiology 149, 354–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, Gregis V, Pelucchi N, Colombo L, Kater M. 2008. The rice StMADS11-like genes OsMADS22 and OsMADS47 cause floral reversions in Arabidopsis without complementing the svp and agl24 mutants. Journal of Experimental Botany 59, 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser LG, Seal AG, Montefiori M, et al. 2013. An R2R3 MYB transcription factor determines red petal colour in an Actinidia (kiwifruit) hybrid population. BMC Genomics 14, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Maroto F, Ortega N, Lozano R, Carmona MJ. 2000. Characterization of the potato MADS-box gene STMADS16 and expression analysis in tobacco transgenic plants. Plant Molecular Biology 42, 499–513 [DOI] [PubMed] [Google Scholar]
- Gleave AP. 1992. A versatile binary vector system with a T-DNA organizational-structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology 20, 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gregis V, Andrés F, Sessa A, et al. 2013. Identification of pathways directly regulated by SHORT VEGETATIVE PHASE during vegetative and reproductive development in Arabidopsis . Genome Biology 14, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM. 2008. AGAMOUS-LIKE24 and SHORT VEGETATIVE PHASE determine floral meristem identity in Arabidopsis . The Plant Journal 56, 891–902 [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Dorca-Fornell C, Kater MM. 2009. The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. The Plant Journal 60, 626–637 [DOI] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. 2000. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis . The Plant Journal 21, 351–360 [DOI] [PubMed] [Google Scholar]
- He C, Tian Y, Saedler R, Efremova N, Riss S, Khan MR, Yephremov A, Saedler H. 2010. The MADS-domain protein MPF1 of Physalis floridana controls plant architecture, seed development and flowering time. Planta 231, 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsch R, Fry J, Hoffmann N, Eichholtz D, Rogers SG, Fraley R. 1985. A simple and general method for transferring genes into plants. Science 227, 1229–1231 [DOI] [PubMed] [Google Scholar]
- Horvath DP, Sung S, Kim D, Chao W, Anderson J. 2010. Characterization, expression and function of DORMANCY ASSOCIATED MADS-BOX genes from leafy spurge. Plant Molecular Biology 73, 169–179 [DOI] [PubMed] [Google Scholar]
- Jaakola L, Poole M, Jones MO, et al. 2010. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiology 153, 1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaudal M, Monash J, Zhang L, Wen J, Mysore KS, Macknight R, Putterill J. 2013. Overexpression of Medicago SVP genes causes floral defects and delayed flowering in Arabidopsis but only affects floral development in Medicago . Journal of Experimental Botany 65, 429–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalusin A, Nishita K, Kim S-H, Ohta M, Fujimura T. 2006. A new MADS-box gene (IbMADS10) from sweet potato (Ipomoea batatas (L.) Lam) is involved in the accumulation of anthocyanin. Molecular Genetics and Genomics 275, 44–54 [DOI] [PubMed] [Google Scholar]
- Lalusin A, Ocampo E, Fujimura T. 2011. Arabidopsis thaliana plants over-expressing the IbMADS10 gene from sweet potato accumulate high level of anthocyanin. Philippine Journal of Crop Science 36, 30–36 [Google Scholar]
- Lee JH, Yoo SJ, Park SH, Hwang I, Lee JS, Ahn JH. 2007. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis . Genes and Development 21, 397–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZG, Reighard GL, Abbott AG, Bielenberg DG. 2009. Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. Journal of Experimental Botany 60, 3521–3530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZM, Zhang JZ, Mei L, Deng XX, Hu CG, Yao JL. 2010. PtSVP, an SVP homolog from trifoliate orange (Poncirus trifoliata L. Raf.), shows seasonal periodicity of meristem determination and affects flower development in transgenic Arabidopsis and tobacco plants. Plant Molecular Biology 74, 129–142 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler HK, Rinderle U. 1988. The role of chlorophyll fluorescence in the detection of stress conditions in plants. CRC Critical Reviews in Analytical Chemistry 19, 29–85 [Google Scholar]
- Liu C, Xi WY, Shen LS, Tan CP, Yu H. 2009. Regulation of floral patterning by flowering time genes. Developmental Cell 16, 711–722 [DOI] [PubMed] [Google Scholar]
- Liu C, Zhou J, Bracha-Drori K, Yalovsky S, Ito T, Yu H. 2007. Specification of Arabidopsis floral meristem identity by repression of flowering time genes. Development 134, 1901–1910 [DOI] [PubMed] [Google Scholar]
- Mao L, Begum D, Chuang H, Budiman MA, Szymkowiak EJ, Irish EE, Wing RA. 2000. JOINTLESS is a MADS-box gene controlling tomato flower abscission zone development. Nature 406, 910–912 [DOI] [PubMed] [Google Scholar]
- Masiero S, Li MA, Will I, Hartmann U, Saedler H, Huijser P, Schwarz-Sommer Z, Sommer H. 2004. INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum . Development 131, 5981–5990 [DOI] [PubMed] [Google Scholar]
- Mazzitelli L, Hancock RD, Haupt S, et al. 2007. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. Journal of Experimental Botany 58, 1035–1045 [DOI] [PubMed] [Google Scholar]
- McGhie TK, Hudault SB, Lunken RC, Christeller JT. 2011. Apple peels, from seven cultivars, have lipase-inhibitory activity and contain numerous ursenoic acids as identified by LC-ESI-QTOF-HRMS. Journal of Agricultural and Food Chemistry 60, 482–491 [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B, Martínez-Zapater JM, Alonso-Blanco C. 2013. The flowering repressor SVP underlies a novel Arabidopsis thaliana QTL interacting with the genetic background. PLoS Genetics 9, e1003289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori M, Espley RV, Stevenson D, Cooney J, Datson PM, Saiz A, Atkinson RG, Hellens RP, Allan AC. 2011. Identification and characterisation of F3GT1 and F3GGT1, two glycosyltransferases responsible for anthocyanin biosynthesis in red-fleshed kiwifruit (Actinidia chinensis). The Plant Journal 65, 106–118 [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497 [Google Scholar]
- Pattanaik S, Kong Q, Zaitlin D, Werkman JR, Xie CH, Patra B, Yuan L. 2010. Isolation and functional characterization of a floral tissue-specific R2R3 MYB regulator from tobacco. Planta 231, 1061–1076 [DOI] [PubMed] [Google Scholar]
- Sasaki R, Yamane H, Ooka T, Jotatsu H, Kitamura Y, Akagi T, Tao R. 2011. Functional and expressional analyses of PmDAM genes associated with endodormancy in Japanese apricot (Prunus mume). Plant Physiology 157, 485–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin MR, Seo SG, Kim JS, Joen SB, Kang SW, Lee GP, Kwon SY, Kim SH. 2011. Alteration of floral organ identity by over-expression of IbMADS3-1 in tobacco. Transgenic Research 20, 365–376 [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K. 2012. Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139, 3081–3098 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Tadege M, Hemming MN, Peacock WJ, Dennis ES, Sheldon C. 2007. Short Vegetative Phase-like MADS-box genes inhibit floral meristem identity in barley. Plant Physiology 143, 225–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Moss SM, Voogd C, Wu R, Lough RH, Wang Y-Y, Hellens RP. 2011. Identification and characterization of flowering genes in kiwifruit: sequence conservation and role in kiwifruit flower development. BMC Plant Biology 11, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varkonyi-Gasic E, Moss S, Voogd C, Wang T, Putterill J, Hellens RP. 2013. Homologs of FT, CEN and FD respond to developmental and environmental signals affecting growth and flowering in the perennial vine kiwifruit. New Phytologist 198, 732–746 [DOI] [PubMed] [Google Scholar]
- Wall C, Dozier W, Ebel RC, Wilkins B, Woods F, Foshee W. 2008. Vegetative and floral chilling requirements of four new kiwi cultivars of Actinidia chinensis and A. deliciosa . Hortscience 43, 644–647 [Google Scholar]
- Wang T, Atkinson R, Janssen B. 2007. Choice of Agrobacterium strain for transformation of kiwifruit. Acta Horticulturae 753, 227–232 [Google Scholar]
- Wang T, Ran Y, Atkinson RG, Gleave AP, Cohen D. 2006. Transformation of Actinidia eriantha: a potential species for functional genomics studies in Actinidia . Plant Cell Reports 25, 425–431 [DOI] [PubMed] [Google Scholar]
- Wu RM, Walton EF, Richardson AC, Wood M, Hellens RP, Varkonyi-Gasic E. 2012. Conservation and divergence of four kiwifruit SVP-like MADS-box genes suggest distinct roles in kiwifruit bud dormancy and flowering. Journal of Experimental Botany 63, 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane H, Kashiwa Y, Ooka T, Tao R, Yonemori K. 2008. Suppression subtractive hybridization and differential screening reveals endodormancy-associated expression of an SVP/AGL24-type MADS-box gene in lateral vegetative buds of japanese apricot. Journal of the American Society for Horticultural Science 133, 708–716 [Google Scholar]
- Yamane H, Ooka T, Jotatsu H, Hosaka Y, Sasaki R, Tao R. 2011. Expressional regulation of PpDAM5 and PpDAM6, peach (Prunus persica) dormancy-associated MADS-box genes, by low temperature and dormancy-breaking reagent treatment. Journal of Experimental Botany 62, 3481–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Xu YF, Tan EL, Kumar PP. 2002. AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proceedings of the National Academy of Sciences, USA 99, 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.