Summary
GSTF2/3 was identified to be a substrate of MSRB7 using our established CNBr digestion-based proteomic analysis, and the restoration of GSTF2/3 activity by MSRB7 was required for oxidative stress tolerance.
Key words: A. thaliana, glutathione transferase, LC-MS/MS, methyl viologen, oxidative stress, methionine sulfoxide reductase B (MSRB).
Abstract
Methionine sulfoxide reductases (MSRs) catalyse the reduction of oxidized methionine residues, thereby protecting proteins against oxidative stress. Accordingly, MSRs have been associated with stress responses, disease, and senescence in a taxonomically diverse array of organisms. However, the cytosolic substrates of MSRs in plants remain largely unknown. Here, we used a proteomic analysis strategy to identify MSRB7 substrates. We showed that two glutathione transferases (GSTs), GSTF2 and GSTF3, had fewer oxidized methionine (MetO) residues in MSRB7-overexpressing Arabidopsis thaliana plants than in wild-type plants. Conversely, GSTF2 and GSTF3 were highly oxidized and unstable in MSRB7-knockdown plants. MSRB7 was able to restore the MetO-GSTF2M100/104 and MetO-GSTF3M100 residues produced during oxidative stress. Furthermore, both GSTs were specifically induced by the oxidative stress inducer, methyl viologen. Our results indicate that specific GSTs are substrates of MSRs, which together provide a major line of defence against oxidative stress in A. thaliana.
Introduction
Reactive oxygen species (ROS), such as H2O2 and superoxide, are involved in signal transduction and defence mechanisms, although excess ROS can damage macromolecules, such as proteins, DNA, RNA, carbohydrates, and lipids (Moller and Sweetlove, 2010). This oxidative damage can lead to cell injury and even cell death. In the case of amino acids, both the free molecules and residues in polypeptides are targets of attack by ROS (Dat et al., 2000; Friguet, 2006), and in proteins, this oxidation can alter protein conformation and function. The methionine (Met) residues of proteins are particularly susceptible to ROS-mediated oxidation, which results in the formation of two diastereoisomeric forms of methionine sulfoxide, Met-S-sulfoxide (Met-S-O) and Met-R-sulfoxide (Met-R-O). These oxidized forms of Met (MetO) can alter the conformation of a protein and render it non-functional. Therefore, the sensitivity of a protein to oxidative stress is related to the number of constituent Met residues.
Methionine sulfoxide reductases A (MSRA) and MSRB, which are found in many organisms, can reduce Met-S-O and Met-R-O, respectively (Vogt, 1995; Sharov and Schoneich, 2000), thus restoring the functional states of non-functional oxidized proteins. MSRs are, therefore, integral parts of an important protein repair system that protects organisms against oxidative stress (Oien and Moskovitz, 2008). The phylogenetic relationships and subcellular locations of MSRA and MSRB enzymes in Arabidopsis thaliana have been reported (Rouhier et al., 2006; Tarrago et al., 2009a ), and genomic analyses have revealed the presence of nine A. thaliana MSRB genes. Proteins derived from two of the genes, MSRB1 and MSRB2, are predicted to be chloroplastic, whereas MSRB3 is predicted to be localized to the secretory pathway and is translocated to the endoplasmic reticulum; the six remaining MSRBs are likely to be cytosolic (Rouhier et al., 2006). The expressions of several A. thaliana MSR genes are modulated by abiotic stresses, including cold and high salinity, and by phytohormones such as abscisic acid (Oh et al., 2005). For example, MSRA4 is highly induced by high-light intensity or oxidative stress inducers such as methyl viologen (MV) and ozone, and its expression reduces the intracellular content of Met-S-O and confers protection against oxidative stress (Romero et al., 2004). In addition, A. thaliana msrb3 mutant accumulates more MetO and ROS than wild-type plants, independent of low temperature (Kwon et al., 2007), whereas the msrb1/msrb2 double mutant shows retarded growth and development under high-light and low-temperature conditions. The plastidial MSRBs are essential for maintaining plant growth through protection of the photosynthetic antennae (Laugier et al., 2010), and transgenic tomato (Solanum lycopersicum) constitutively expressing the pepper (Capsicum annuum) MSRB2 gene (CaMSRB2) was reported to have lower levels of ROS and enhanced resistance to pathogens (Oh et al., 2010). Finally, the MSR repair system has been reported to establish and preserve longevity in seeds (Chatelain et al., 2013).
Heat-shock protein 21 (HSP21) was the first specific substrate of plastidial MSRA identified in plants, and was shown to be crucial for plant resistance to oxidative stress. MSRA maintains the chaperone activity of HSP21 through the regeneration of the sulfoxidized N-terminal region, which contains a high proportion of Met residues (Gustavsson et al., 2002). Recently, 24 proteins that interact with the A. thaliana plastidial MSRB1 were isolated by affinity chromatography (Tarrago et al., 2012) and shown to be involved in photosynthesis, translation, and oxidative stress tolerance. Significantly, all of these interacting proteins have surface-exposed Met residues and higher-than-average Met contents, suggesting that they are more susceptible to oxidation by ROS and are dependent on plastidial MSRBs for repair (Tarrago et al., 2012). However, no substrate of the cytosolic MSRB family in plants has been identified to date.
Our previous study indicated that A. thaliana overexpressing MSRB7 (At4g21830) has a higher glutathione S-transferase (GST) activity and enhanced tolerance to oxidative stress (Li et al., 2012). GSTs have been shown to be important for maintaining redox homeostasis, reducing oxidative damage (Cummins et al., 1999), and protecting organisms against oxidative stress (Edwards and Dixon, 2005; Dixon et al., 2011; Chen et al., 2012). Accordingly, plant and animal GSTs are induced by various environmental stimuli, such as chilling, hypoxic stress, dehydration, wounding, pathogen attack, phytohormones and oxidative stress (Mauch and Dudler, 1993; Hayes et al., 2005; Sappl et al., 2009). In A. thaliana specifically, GSTs can be divided into seven classes: phi (F), tau (U), theta (T), zeta (Z), lambda (L), dehydroascorbate reductase and TCHQD (Dixon and Edwards, 2010), and can function as glutathione (GSH) transferases, GSH-dependent peroxidases, GSH-dependent isomerases, or GSH-dependent oxidoreductases (Edwards and Dixon, 2005). GSTs of stress-inducible plants may possess GSH-dependent peroxidase activities that act directly on H2O2, and at the same time are capable of utilizing GSH to reduce the organic hydroperoxides of fatty acids and nucleic acids (Dixon et al., 2002). In addition, A. thaliana lacking GSTF2 shows increased sensitivity to MV and HgCl2 treatments (Gong et al., 2005). These findings suggest an involvement of GSTFs in oxidative stress tolerance.
To investigate whether GSTs are the specific substrates of cytosolic MSRB7 under oxidative stress and to establish the mechanisms underlying the oxidative stress defence pathway, MSRB7 was subjected to functional analysis using proteomic, biochemical, and transgenic approaches.
Materials and methods
Plant materials and growth conditions
A. thaliana Heynh. ecotype Columbia plants were grown in controlled-environment chambers at 22 °C, 70% relative humidity, with a 16h photoperiod (approximately 120 μmol m–2 s–1). The floral dip transformation method (Clough and Bent, 1998) was used to generate transgenic A. thaliana lines. Ten-day-old seedlings were used for all experiments. For MV-tolerance experiments, plants were germinated and grown on Murashige and Skoog medium [4.3g Murashige and Skoog salt (Duchfa, Biochemie, Netherlands), Murashige and Skoog vitamins, 1% sucrose, 0.5g l–1 of MES, pH 5.7, 0.4% agar gel (Sigma-Aldrich, St Louis, MO, USA)] containing 10 μM MV. For protein stability experiments, plants were pre-treated with 10 μM MV for 8h, followed by 0.5mM cycloheximide (CHX) treatment for up to 36h.
RNA isolation and gene expression analysis
Total RNA was isolated from plant tissues using Trizol reagent according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA, USA). For reverse transcriptase (RT)-PCR, the cDNA was synthesized using a First-strand cDNA Synthesis kit (Promega, Madison, WI, USA). All gene-specific primers are listed in Supplementary Table S2 available at JXB online. Real-time PCR amplification was performed using SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) and monitored using an ABI 7500HT sequence detection system (Applied Biosystems). Data were analysed using ABI SDS 1.4 software (Applied Biosystems). Relative transcript levels were normalized to expression of the endogenous control genes Actin2 (At3g18780), EF1α (At5g60390), and 18S rRNA (AF206999) using the comparative cycle threshold (C t) method.
Plasmid constructions
The full-length MSRB7-encoding gene was isolated by RT-PCR (Supplementary Table S2 available at JXB online) from 2-week-old A. thaliana seedlings and subcloned into the binary vector pCAMBIA1390:35S (B7Ox) (Hsiao et al., 2007). The 5′ region including the 5′-untranslated region (5′-UTR) and partial coding sequence of MSRB7 was cloned into the pH7GWIWG vector (Invitrogen) to generate RNA interference knockdown plants (B7i) (Li et al., 2011). For β-glucuronidase (GUS) histochemical staining, the MSRB7 promoter region (2000bp upstream of the start codon) was cloned into the pHGWFS7 vector (Invitrogen) to regulate the expression of the GUS-encoding gene. These plasmids were transformed into Agrobacterium tumefaciens strain GV3101 (pMP90) by electroporation for use in A. thaliana transformation.
GUS histochemical staining and activity
MSRB7 promoter-driven GUS (B7pro-GUS), pCAMBIA1301 transgenic plants (CaMV35Spro-GUS; 1301), and wild-type plants were examined histochemically using GUS staining (0.1M sodium phosphate buffer, pH 7.0, 10mM EDTA, 0.5mM potassium ferrocyanide, 0.5mM potassium ferricyanide, 0.1% Triton X-100, 1mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid). GUS activity was determined as described previously (Lu et al., 1998).
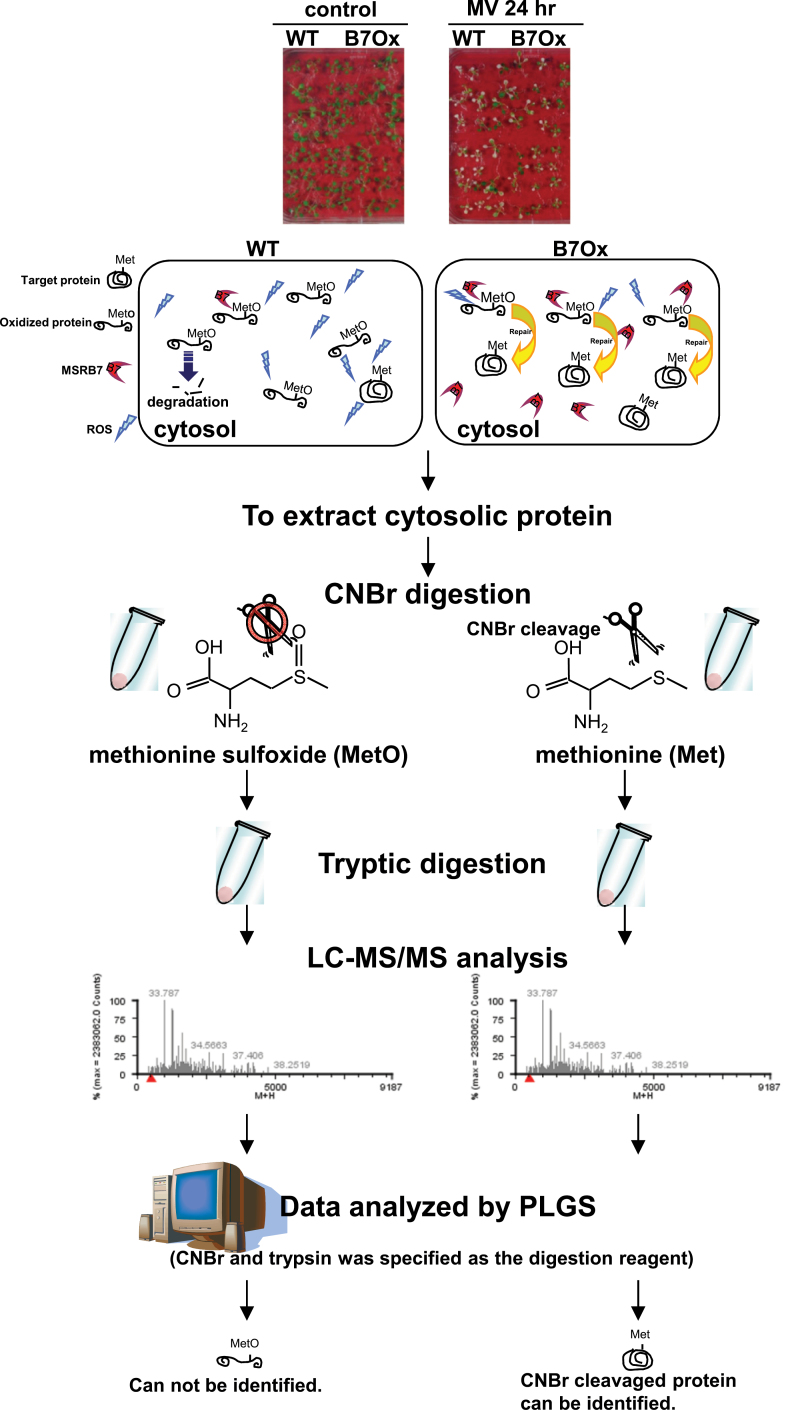
Comparative proteomic analysis using cyanogen bromide (CNBr) digestion
Cytosolic proteins were extracted from 10-d-old MV-treated seedlings using ice-cold buffer consisting of 50mM HEPES (pH 7.5), 300mM sucrose, 150mM NaCl, 10mM potassium acetate, 5mM EDTA, 1mM phenylmethylsulfonyl fluoride (PMSF), and 1× Protease Inhibitor Cocktail (Sigma-Aldrich). Proteins were digested overnight with 100mM CNBr in 50% trifluoroacetic acid in the dark at room temperature (Ogorzalek Loo et al., 1996), followed by trypsin digestion. Peptide mass fingerprinting was performed as described previously (Chen et al., 2010). Liquid chromatography tandem mass spectrometry (LC-MS/MS) was performed with a nanoflow LC system (nanoACQUITY UPLC; Waters, Millford, MA, USA) coupled to a hybrid Q-TOF mass spectrometer (Synapt HDMS G2; Waters, Manchester, UK). For the nanoflow LC system, mobile phase A contained water with 0.1% formic acid and phase B contained acetonitrile with 0.1% formic acid. The peptide samples were injected onto a trap column (Symmetry C18, 5 μm, 180 μm×20mm; Waters, Milford, MA, USA) and separated online with a reverse-phase column (BEH C18, 1.7 μm, 75 μm×250mm; Waters, Milford, MA, USA) at a flow rate of 300 nl min–1 using a 90min 15–90% acetonitrile/water gradient. The temperature of the separating column was maintained at 35 °C. For MS analysis, the LC column was online-coupled to the nanospray source of a hybrid Q-TOF mass spectrometer and 500fmol μl–1 of [Glu]fibrinopeptide B was continuously infused to the lockspray emitter at a flow rate of 250 nl min–1. The MS was switched to the lockspray source every 30 s and the [Glu]fibrinopeptide signal was used as a reference mass for calibration. The LC-MS data were collected in MSE mode: the low collision energy spectra were acquired at 4eV trapping energy and the high collision energy spectra were acquired by ramping the trapping energy from 10 to 30eV. The low and high collision energy scan range was from 50 to 1990 Th with a scan time of 1 s and a 0.02 s interscan time. The MSE data were processed using the ProteinLynx GlobalServer (PLGS, version 2.3; Waters, Manchester, UK). To process the chromatogram of each precursor and fragment ion, the minimal peak width was subjected to three scans and the expected peak was subjected to seven scans. The spectral noise was processed using the adaptive background subtraction provided by PLGS, and the maximum charge state for deisotoping was 6. For database searching, the IPI ARATH v.3.85 FASTA database (ftp://ftp.ebi.ac.uk/pub/databases/IPI) was used. CNBr and trypsin were used specifically as the digestion reagents, one missed cleavage was allowed, carbamidomethyl (C) was specified as the fixed modification, and oxidation (M) was considered as a variable modification. Peptides were considered identified if the identification confidence value was >95% in PLGS. Each peptide identified was further quantified by the ExpressionE tool in PLGS. All the quantified peptides were used to calculate the relative abundance of the proteins, and the protein abundance ratios were finally normalized by the ‘auto normalization’ function of PLGS.
Met residues in GSTs and their differential oxidation as analysed by MS
Cytosolic proteins were extracted by ice-cold cytosolic protein extraction buffer [PBS containing 5mM EDTA, 1mM PMSF, 1mM dithiothreitol (DTT), 1× Protease Inhibitor Cocktail (Sigma-Aldrich), 10% glycerol, and 0.01% Tween 20]. Protein samples were separated by SDS-PAGE. Proteins in the size range 25–30kDa were digested by trypsin and then analysed by LC-MS/MS. The MSE data were processed using PLGS. The amino acid sequences of GSTF2, GSTF3, and GSTF8 were referred for database searching. Coverage represents the number of times the peptide containing Met residues, both oxidized and reduced forms, was detected. The percentage of oxidization was calculated using the following formula: % oxidization=[number of MetO on GSTF2M100 / number of both oxidized and reduced forms of Met on GSTF2M100]×100.
Immunoblot analysis
Cytosolic proteins were extracted using cytosolic protein extraction buffer. Protein samples were separated by SDS-PAGE and electrotransferred to a polyvinylidene difluoride membrane. GSTF2, GSTF3, and MSRB7 were recognized with anti-GSTF2/3 antibody (Agrisera, Vännäs, Sweden) and in-house rabbit anti-MSRB7 antibody, respectively. Antibody-bound proteins were detected using a chemiluminescence system (Millipore Corporation, Billerica, MA, USA) following incubation with protein A-conjugated horseradish peroxidase (Invitrogen).
Production of recombinant proteins
The Met residues on GSTF2 and GSTF3 were mutated by PCR using the primers for site-directed mutagenesis listed in Supplementary Table S2 available at JXB online. The MSRB7 N terminus fused with a flag tag (flag–MSRB7), GSTF2, GSTF3, GSTF8, GSTF2 M100L (Met replaced by Leu), GSTF2 M104L, GSTF2 M100/104L, and GSTF3 M100L were cloned into the pET-53-DESTTM vector (Merck KGaA, Darmstadt, Germany). These vectors were transformed into the Escherichia coli Rosetta (DE3) strain. The recombinant His–flag–MSRB7, His–GSTF2, His–GSTF3, His–GSTF8, His–GSTF2M100L, His–GSTF2M104L, His–GSTF2M100/104L, and His–GSTF3M100L proteins were purified using Ni2+-affinity columns. Recombinant MSRB7 proteins were detected using anti-flag or anti-His antibodies (Sigma-Aldrich).
Bimolecular fluorescence complementation (BiFC) and protoplast transient assay
The full-length coding regions of MSRB7, GSTF2, GSTF3, and GSTF8 were cloned into BiFC vectors (Supplementary Fig. S1 available at JXB online) (Walter et al., 2004). Protoplasts were isolated using the tape–A. thaliana sandwich method and co-transformed with plasmid expressing nuclear-localizing marker [(bZIP63–CFP (cyan fluorescent protein)] (Walter et al., 2004) and BiFC plasmids using the polyethylene glycol method (Wu et al., 2009). After incubation at room temperature for 16h under light, the protoplasts were observed with a Zeiss LSM510 META laser scanning confocal microscope (Carl Zeiss, Jena, Germany).
Yeast two-hybrid assay
The ProQuest two-hybrid system (Invitrogen) was used in a yeast two-hybrid assay. MSRB7 was cloned into pDEST22 as bait, and GSTF2, GSTF3, and GSTF8 were cloned into pDEST32 as prey. The construct pairs were co-transformed into yeast strain MaV203 according to the manufacturer’s instructions (Invitrogen). Positive yeast transformants were selected on SD minimal (–Leu–Trp) and (–Leu–Trp–His) medium and experiments were performed with three biological repeats. Appropriate controls were included by co-transforming pEXP32/Krev1 with pEXP22/ RalGDs-WT (wild type with strong interaction), pEXP22/RalGDs-m1 (mutant with weak interaction), and pEXP22/RalGDs-m2 (mutant with no interaction).
GST activity
For in vitro GST activity assays, recombinant GST protein was oxidized by treatment with 0.5mM hypochlorous acid (HOCl) for 40min at room temperature and a final concentration of 5mM Met was added to terminate the oxidative reaction. Sixty micrograms of oxidant-treated GST was co-incubated with 20 μg of MSRB7 in PBS containing 10mM MgCl2, 30mM KCl, and 10mM DTT for 1h at room temperature. The activity of recombinant GST was measured using a 1-chloro-2,4-dinitrobenzene (CDNB) assay as described previously (Habig et al., 1974). For in vivo GST activity assay, 10-d-old B7Ox, B7i, and 1301 seedlings were treated with 10 μM MV for 8h and then treated with CHX for 0, 12, 24, and 36h. The CHX-treated A. thaliana proteins were extracted using cytosolic protein extraction buffer and the GST activity was then measured using a CDNB assay.
Immunoprecipitation
His–flag–MSRB7, oxidized His–GSTF2, oxidized His–GSTF3, and oxidized His–GSTF8 recombinant proteins were incubated with rabbit anti-flag antibody in IP buffer (15mM HEPES, pH 7.4, 1mM EDTA, 1mM DTT, 10mM MgCl2, 30mM KCl, 1mM PMSF, and 1× Protease Inhibitor Cocktail) for 1h at room temperature. Protein G–Sepharose beads (Invitrogen) were added and the samples were incubated for 1h at room temperature for co-precipitation. The immunoprecipitate was immunoblotted and detected using mouse anti-His antibody.
H2O2 content
Ten-day-old A. thaliana plants treated with 10 μM MV for 24h were extracted by PBS. H2O2 content was measured using anAmplex Red Hydrogen Peroxide/Peroxidase assay kit (Invitrogen), according to the manufacturer’s instructions.
Statistical analyses
Data are presented as mean values±standard deviation (SD). Duncan’s test was performed to calculate the differences between distributions of data using SPSS v.12.0 software. P values of less than 0.05 were considered statistically significant.
Results
Identification of putative MSRB7 substrates by comparative proteomic analysis using CNBr digestion
To identify the substrates of MSRB7, cytosolic protein-enriched extracts were isolated from B7Ox and wild-type plants, digested with CNBr and trypsin, and analysed using LC-MS/MS. CNBr specifically hydrolyses the C terminus of Met but not MetO residues, and therefore proteins harbouring MetO residues are not hydrolysed by CNBr. A flowchart of the steps used for the comparative proteomic analysis using the CNBr digestion approach is presented in Fig. 1. The CNBr/trypsin-digested samples were first subjected to LC-MS/MS analysis, and the MSE data obtained were processed using the PLGS, with CNBr and trypsin specified as the digestion reagents for the database analysis search (Fig. 1). This strategy was designed to allow the identification of proteins/peptides that contained Met, as opposed to MetO, as a consequence of MSRB7 activity, and were therefore amenable to CNBr digestion, identification, and quantification (Fig. 1). This analysis led to the identification of a total of 188 such proteins (Supplementary Table S1 available at JXB online). To identify the possible interacting partners of MSRB7, putative targets that were present only in the proteomic data from the B7Ox sample, or were >1.5 fold higher in B7Ox than in the wild type, were selected for further analysis. Analysis of the Gene Ontology (GO) annotations in The Arabidopsis Information Resource (TAIR; (http://www.arabidopsis.org/tools/bulk/go/index.jsp) corresponding to these proteins indicated that 41 of the putative targets were related to stress responses, some of which, such as GST, peroxidase, and catalase, are known to be involved in ROS scavenging (Table 1). GSTF2 (At4g02520), GSTF3 (At2g02930), and GSTF8 (At2g47730) were more abundant in B7Ox than the wild type, suggesting that they may be interacting partners of MSRB7, and since GST activity was higher in the MSRB7-overexpressing plants than in the wild-type plants (Li et al., 2012), we hypothesized that GSTF2, GSTF3, and GSTF8 are direct substrates of MSRB7.
Fig. 1.
Flowchart of the steps used for comparative proteomic analysis using the CNBr digestion approach.
Table 1.
Potential substrates of MSRB7
Ten-day-old B7Ox and wild-type A. thaliana were treated with or without 10 μM MV for 24h. ND, not determined.
| Accession no. | Locus | Description | Score | B7Ox/WT MV 24h | B7Ox/WT MV 0 h | Unique |
|---|---|---|---|---|---|---|
| IPI00537995 | At1g35720 | Annexin D1 | 218.40 | B7Ox 24h | B7Ox 0h | B7Ox only |
| IPI00535149 | AT4g02520 | Glutathione S transferase F2 | 188.83 | B7Ox 24h | B7Ox 0h | B7Ox only |
| IPI00525727 | At4g37930 | Mitochondrial, serine hydroxymethyltransferase mitochondrial | 78.56 | B7Ox 24h | B7Ox 0h | B7Ox only |
| IPI00523477 | At5g38420 | Chloroplastic, ribulose bisphosphate carboxylase small chain 2B | 2906.54 | B7Ox 24h | 3.03 | |
| IPI00532772 | At1g66200 | Glutamine synthetase cytosolic isozyme 1 | 254.75 | B7Ox 24h | 1.47 | |
| IPI00532945 | At2g02930 | Glutathione S-transferase F3 | 170.04 | B7Ox 24h | ND | B7Ox 24h |
| IPI00520226 | At4g14960 | Tubulin α6 chain | 261.54 | B7Ox 24h | ND | B7Ox 24h |
| IPI00530621 | At1g19570 | Dehydroascorbate reductase 1 (DHAR1) | 155.99 | B7Ox 24h | 1.00 | |
| IPI00544626 | At3g01500 | Chloroplastic, isoform 1 of carbonic anhydrase | 756.83 | B7Ox 24h | 1.00 | |
| IPI00534087 | At5g56010 | Heat-shock protein 81 3 | 203.99 | 2.38 | B7Ox 0h | |
| IPI00533497 | At3g09260 | β-Glucosidase | 102.90 | 1.43 | B7Ox 0h | |
| IPI00544876 | At3g55800 | Chloroplastic, sedoheptulose 1,7 bisphosphatase | 111.61 | 1.41 | B7Ox 0h | |
| IPI00539020 | At1g67090 | Chloroplastic, ribulose bisphosphate carboxylase small chain 1A | 3213.8 | 1.63 | 1.59 | |
| IPI00521186 | At5g38430 | Chloroplastic, ribulose bisphosphate carboxylase small chain 1B | 2947.88 | 1.05 | 2.30 | |
| IPI00656928 | At4g35090 | Catalase 2 | 108.7 | 0.76 | 1.54 | |
| IPI00532582 | At4g21280 | Isoform 2 of oxygen evolving enhancer protein 3 | 296.93 | 0.75 | B7Ox 0h | |
| IPI00891841 | At5g38410 | Similar to ribulose bisphosphate carboxylase small chain 2B | 2291.48 | ND | B7Ox 0h | B7Ox 0h |
| IPI00532125 | At1g54040 | Epithiospecifier protein | 596.6 | ND | B7Ox 0h | B7Ox 0h |
| IPI00846574 | At5g14740 | β-Carbonic anhydrase 2 | 330.81 | ND | B7Ox 0h | B7Ox 0h |
| IPI00518163 | At2g39730 | Ribulose bisphosphate carboxylase oxygenase activase | 291.06 | ND | B7Ox 0h | B7Ox 0h |
| IPI00542532 | At1g24020 | MLP-like protein 423 | 245.59 | ND | B7Ox 0h | B7Ox 0h |
| IPI00516423 | At4g25050 | Acyl carrier protein 4 | 193.68 | ND | B7Ox 0h | B7Ox 0h |
| IPI00518090 | At1g13440 | Glyceraldehyde-3-phosphate dehydrogenase C2 (GAPC2) | 190.2 | ND | B7Ox 0h | B7Ox 0h |
| IPI00656779 | At2g21330 | Fructose bisphosphate aldolase | 160.8 | ND | B7Ox 0h | B7Ox 0h |
| IPI00518620 | At3g32980 | Peroxidase 32 | 147.69 | ND | B7Ox 0h | B7Ox 0h |
| IPI00539116 | At5g26000 | Myrosinase | 135.6 | ND | B7Ox 0h | B7Ox 0h |
| IPI00524641 | At2g21170 | Chloroplastic, triosephosphate isomerase | 127.79 | ND | B7Ox 0h | B7Ox 0h |
| IPI00523903 | At5g02490 | Heat-shock cognate 70kDa protein 2 | 121.8 | ND | B7Ox 0h | B7Ox 0h |
| IPI00536062 | At2g47730 | Glutathione S-transferase F8 | 117.32 | ND | B7Ox 0h | B7Ox 0h |
| IPI00526611 | At1g56410 | Early response to dehydrogenase 2 (HSP70) | 116.83 | ND | B7Ox 0h | B7Ox 0h |
| IPI00547926 | At3g18780 | Actin 2 | 115.01 | ND | B7Ox 0h | B7Ox 0h |
| IPI00538349 | At1g63940 | Monodehydroascorbate reductase 6 | 103.82 | ND | B7Ox 0h | B7Ox 0h |
| IPI00539389 | At1g16030 | Heat-shock protein 70B (HSP70B) | 96.9 | ND | B7Ox 0h | B7Ox 0h |
| IPI00539339 | At3g04790 | Ribose 5-phosphate isomerase related | 91.58 | ND | B7Ox 0h | B7Ox 0h |
| IPI00545934 | At5g12250 | Tubulin β6 chain | 84.79 | ND | B7Ox 0h | B7Ox 0h |
| IPI00530539 | At5g64290 | Dicarboxylate transport 2 | 84.74 | ND | B7Ox 0h | B7Ox 0h |
| IPI00525001 | At5g62690 | Tubulin β2β3 chain | 82.8 | ND | B7Ox 0h | B7Ox 0h |
| IPI00523675 | At4g23210 | Isoform 2 of cysteine rich receptor-like protein kinase 13 | 79.72 | ND | B7Ox 0h | B7Ox 0h |
| IPI00518916 | At5g24300 | Chloroplastic amyloplastic, soluble starch synthase | 78.84 | ND | B7Ox 0h | B7Ox 0h |
| IPI00517585 | At5g52250 | Transducin family protein | 76.69 | ND | B7Ox 0h | B7Ox 0h |
| IPI00530974 | At1g52770 | Phototropic responsive NPH3 family protein | 76.1 | ND | B7Ox 0h | B7Ox 0h |
MSRB7, GSTF2, and GSTF3 are induced by MV
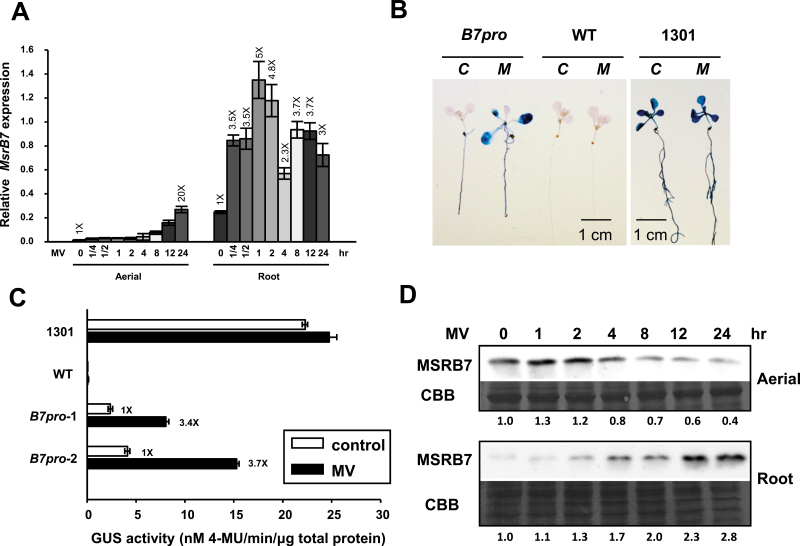
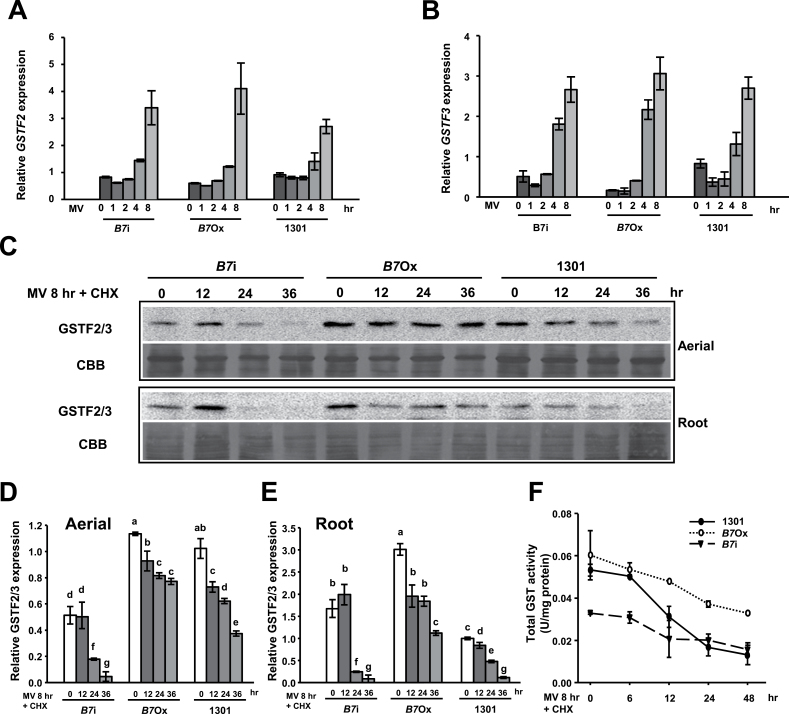
Real-time PCR analysis was performed to investigate whether the expression of MSRB7, GSTF2, and/or GSTF3 was affected by MV-induced oxidative stress. MSRB7 transcripts were highly expressed in roots and were strongly induced by 10 μM MV, with transcript levels increasing gradually during the first 2h of MV treatment and remaining high for 12–24h post-treatment (Fig. 2A). Conversely, MSRB7 transcripts were expressed at low levels in the aerial parts of plants, although prolonged MV treatment resulted in a gradual increase in transcript levels (Fig. 2A). We next examined the locations of MSRB7 expression using transgenic A. thaliana lines transformed with a GUS gene driven by the MSRB7 promoter (B7pro-GUS). Histochemical staining of 10-d-old transgenic seedlings indicated that MSRB7 was expressed in roots but not shoots under normal conditions (Fig. 2B), confirming the PCR results. However, upon treatment with MV for 8h, GUS expression was highly induced throughout the whole seedling (Fig. 2B, C), suggesting that MV induces MSRB7 expression. In addition, GUS staining of 6-week-old B7pro-GUS seedlings treated with MV revealed expression in the cauline and rosette leaves but not in the flowers or siliques (Supplementary Fig. S2 available at JXB online). Wild-type and transgenic plants transformed with the pCAMBIA1301 (CaMV35Spro-GUS; 1301) were used as negative and positive controls, respectively (Fig. 2B, C). Immunoblot analysis showed an increase in MSRB7 protein expression in the roots upon MV treatment (Fig. 2D), supporting the above observations (Fig. 2A, B). In the case of the aerial part, the MSRB7 protein abundance decreased upon MV treatment (Fig. 2D).
Fig. 2.
Induction of MSRB7, GSTF2, and GSTF3 by oxidative stress. (A) Expression patterns of MSRB7. Real-time PCR analysis of transcripts of 10-d-old A. thaliana plants treated with 10 μM MV for 15min to 24h. The data represent the means±SD (n=10) of three independent experiments. (B, C) Histochemical GUS staining and GUS activity. A. thaliana seedlings harbouring the MSRB7 promoter (B7pro)-driven GUS were untreated (C) or treated (M) with 10 μM MV for 8h and GUS expression (B) and activity (C) were determined. Wild-type (WT) and pCAMBIA1301 transgenic plants (CaMV35Spro-GUS; 1301) served as negative and positive controls, respectively. (D) Immunoblotting of MSRB7. Ten-d-old wild-type seedlings were treated with 10 μM MV for 0–24h and expression of the MSRB7 protein was detected with a specific anti-MSRB7 antibody. Protein stained with Coomassie Brilliant Blue (CBB) was used as a loading control.
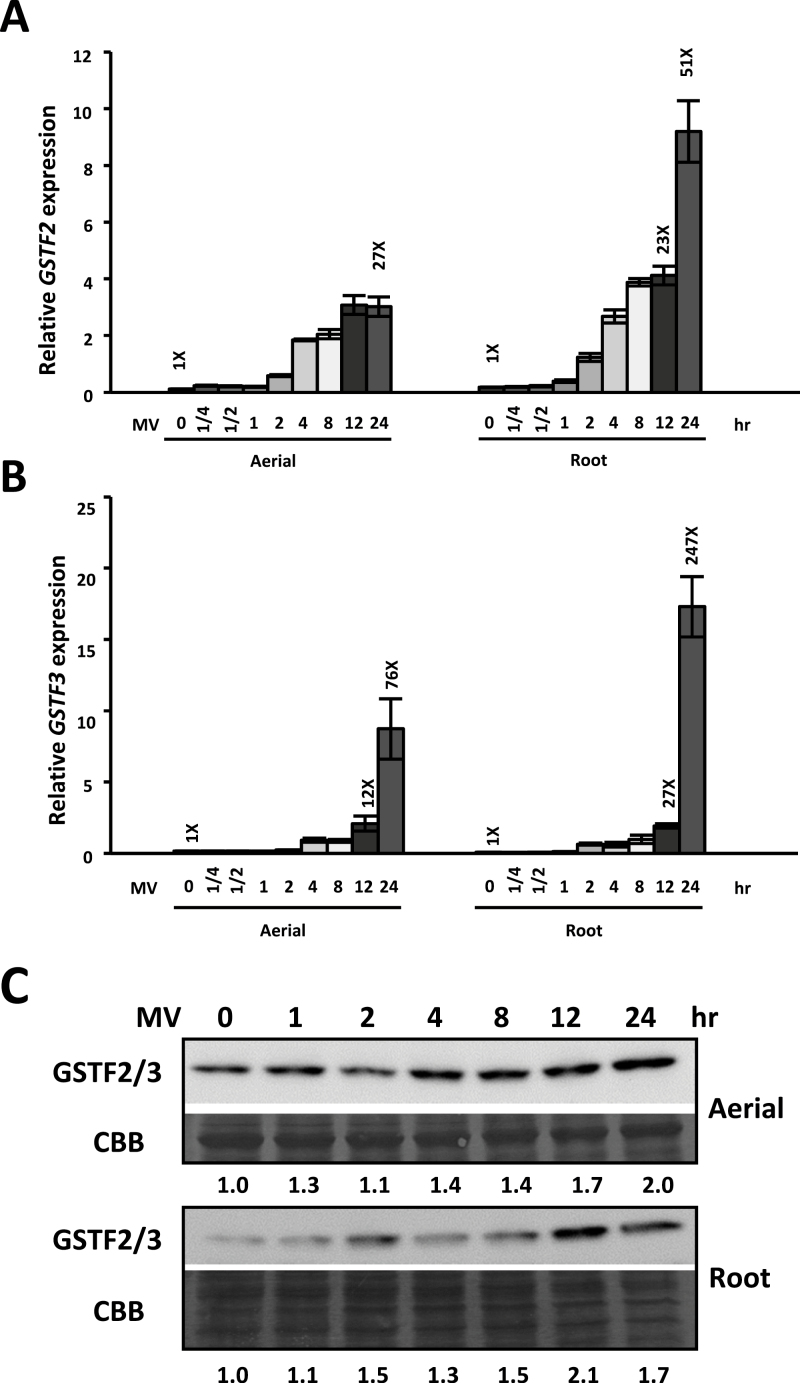
Based on sequence similarities/identities, GSTF2, GSTF3, and GSTF8 have been classified into the F class of the GST family (Dixon and Edwards, 2010). The polypeptides of GSTF2 and GSTF3, which are 92.5% identical with respect to their amino acid sequences, were both detected using the same GSTF2/3 antibody (Agrisera) (Supplementary Fig. S3 available at JXB online). MV treatment resulted in high expression levels and accumulation of GSTF2 and GSTF3 at both the transcript and protein levels, (Fig. 3); however, the expression pattern of GSTF8 showed no apparent changes following MV treatment (Supplementary Fig. S3B available at JXB online), indicating that the expression of GSTF2 and GSTF3, but not GSTF8, was MV inducible. The abundance of GSTF2/3 protein remained high in the aerial part of the plant, and gradually increased in the root part upon MV treatment, suggesting that these proteins, together with MSRB7, have a role in protecting plants against oxidative stress.
Fig. 3.
Induction of GSTF2 and GSTF3 by oxidative stress. (A, B) Expression patterns of GSTF2 and GSTF3. Real-time PCR analysis of transcripts in 10-d-old A. thaliana plants treated with 10 μM MV for 15min to 24h. The data represent the means±SD (n=10) of three independent experiments. (C) Immunoblotting of GSTF2/3. Ten-d-old wild-type seedlings were treated with 10 μM MV for 0–24h. GSTF2/3 expression was detected using an anti-GSTF2/3 antibody. Protein stained with Coomassie Brilliant Blue (CBB) was used as a loading control.
Met residues in GSTF2 and GSTF3 are repaired by MSRB7 in vivo
To investigate the role of MSRB7 in the reducing of MetO residues in plant GSTF2 and GSTF3, MetO levels in the GSTs were quantified by MS. Ten-day-old B7Ox, B7i, and 1301 (vector-only control) plants were treated with MV for 24h, and protein samples from these and untreated plants were collected and subjected to tryptic digestion followed by LC-MS/MS analysis. The percentage of MetO was calculated from the identified peptides containing specific Met residues, and the coverage was defined as the number of times the peptide containing these Met residues was found (MetO and Met). In addition to the first Met residue, GSTF2, GSTF3, and GSTF8 contain two (GSTF2M100/104), one (GSTF3M100), and six (GSTF8M49/59/67/84/173/176) Met residues, respectively (Supplementary Fig. S4 available at JXB online). We observed that MV treatment resulted in an increase in the average percentage of MetO residues in all three proteins. In particular, GSTF2M104 and GSTF3M100 were susceptible to oxidization in B7i (34.5 and 60%, respectively) and 1301 (17.2 and 26.2%, respectively) plants under oxidative stress, and even in the absence of MV-induced oxidative stress, GSTF3M100 was highly oxidized (66.7%) in B7i plants. The percentages of MetO in GSTF2 and GSTF3 were significantly lower in B7Ox plants than in B7i and 1301 plants, while the MetO residues in GSTF8 were less frequently converted back to the reduced Met state in B7Ox plants (Table 2). These observations indicated that GSTF2 and GSTF3 may be direct substrates of MSRB7 in plants under oxidative stress.
Table 2.
Met residues in GSTs and their differential oxidation as revealed by MS
Ten-day-old B7Ox, B7i and 1301 plants were treated with or without 10 μM MV for 24h. Experiments were repeated three times. Results are shown as % oxidation. The values in brackets is the coverage, representing the number of times the peptide containing Met residues was found in both the oxidized and the reduced form, is shown in parentheses.
| Met position | 1301 | B7Ox | B7i | |||
|---|---|---|---|---|---|---|
| Control | MV treated | Control | MV treated | Control | MV treated | |
| GSTF2 | ||||||
| 100 | 0 (21) | 17.2 (29) | 0 (24) | 0 (44) | 0 (17) | 0 (81) |
| 104 | 0 (21) | 17.2 (29) | 0 (24) | 4.8 (44) | 0 (17) | 34.5(81) |
| Average | 0 (21) | 17.2 (29) | 0 (24) | 2.4 (44) | 0 (17) | 17.2 (81) |
| GSTF3 | ||||||
| 100 | 0 (12) | 26.2 (42) | 0 (24) | 16.1 (31) | 66.7 (24) | 60.0 (20) |
| Average | 0 (12) | 26.2 (42) | 0 (24) | 16.1 (31) | 66.7 (24) | 60.0 (20) |
| GSTF8 | ||||||
| 49 | 0 (11) | 0 (18) | 0 (13) | 0 (15) | 0 (20) | 16.7 (30) |
| 59 | 0 (23) | 24.1 (29) | 0 (12) | 22.6 (31) | 0 (18) | 33.3 (27) |
| 67 | 0 (21) | 0 (29) | 0 (8) | 22.6 (31) | 16.7 (30) | 28.6 (21) |
| 84 | 0 (28) | 36.1 (108) | 0 (14) | 33.9 (124) | 7.0 (43) | 46.2 (184) |
| 173 | 25.9 (27) | 44.7 (94) | 0 (7) | 40.4 (73) | 14.6 (41) | 36.3 (113) |
| 176 | 0 (38) | 44.7 (94) | 0 (12) | 17.6 (85) | 0 (49) | 27.0 (152) |
| Average | 4.3 (24.7) | 24.9 (62) | 0 (11) | 22.9 (59.8) | 6.4 (33.5) | 31.3 (87.8) |
GSTF2, GSTF3, and GSTF8 interact with MSRB7
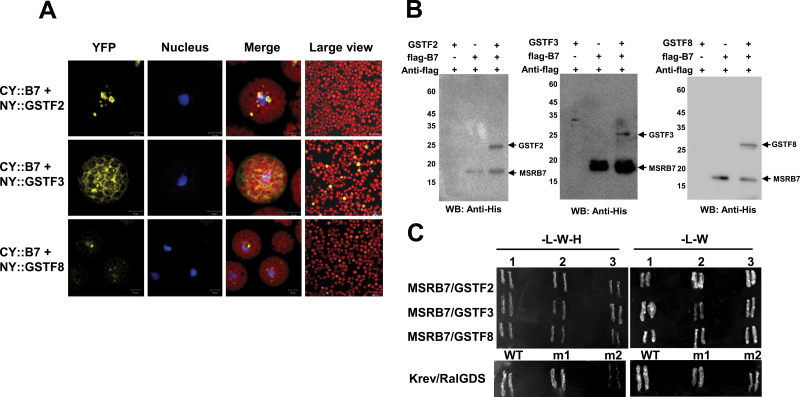
To verify whether the candidate proteins described above interacted with MSRB7, BiFC, co-immunoprecipitation, and yeast two-hybrid assays were performed. A protoplast transient assay revealed the presence of GSTF2, GSTF3, GSTF8, and MSRB7 in the cytosol (Supplementary Fig. S5 available at JXB online). GSTF8 has previously been reported to be localized in both the cytosol and plastids (Thatcher et al., 2007), and in our study where the nuclear marker (bZIP63–CFP)-expressing plasmid and BiFC plasmids were co-transformed into A. thaliana protoplasts, we observed that GSTF2 and GSTF3 interacted with MSRB7 in the cytosol, while GSTF8 interacted with MSRB7 in close proximity to the chloroplast (Fig. 4A and Supplementary Fig. S6 available at JXB online). In addition, we observed that the three GSTF proteins co-immunoprecipitated with MSRB7 and were detected using an anti-His antibody (Fig. 4B). Finally, when MSRB7 was used as the bait protein in the yeast two-hybrid assay, weak interactions with GSTF2, GSTF3, and GSTF8 were observed (Fig. 4C), supporting our observations from the BiFC and co-immunoprecipitation experiments. Together, these results indicated that GSTF2, GSTF3, and GSTF8 are likely to be substrates of MSRB7.
Fig. 4.
Interaction of MSRB7 with GSTs. (A) BiFC assay for interaction of MSRB7 with GSTs. Yellow indicates CY-B7 [MSRB7 fused with the C-terminal fragment of yellow fluorescence protein (YFP)] and NY-GSTF2/3/8 (GSTF2/3/8 fused with the N-terminal fragment of YFP) dimerization, as determined by BiFC. Red denotes chloroplast autofluorescence and blue denotes nuclear localizing marker (bZIP63–CFP). (B) Co-immunoprecipitation of MSRB7, GSTF2, GSTF3, and GSTF8. Recombinant GSTs proteins were co-precipitated with flag–MSRB7 using an anti-Flag antibody and detected with an anti-His antibody. WB, Western blot. (C) Yeast two-hybrid assay verifying the interactions of GSTF2, GSTF3, and GSTF8 with MSRB7. Controls were performed by co-transforming pEXP32/Krev1 with pEXP22/ RalGDs-WT (strong interaction), pEXP22/RalGDs-m1 (weak interaction), and pEXP22/RalGDs-m2 (no interaction). L, Leu; W, Trp; H, His.
MSRB7 can restore the activities of oxidized GSTF2 and GSTF3 in vitro
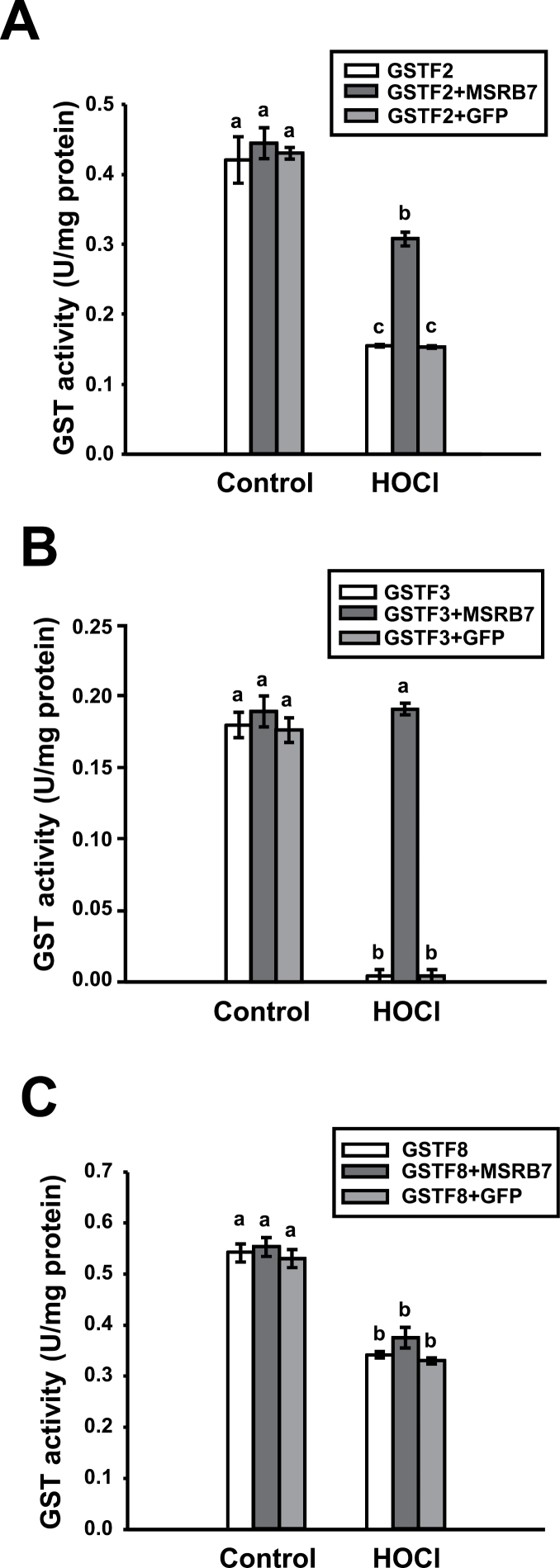
HOCl is an oxidant that preferentially targets Met residues in proteins, and it has been shown that proteins that are susceptible to oxidative damage can undergo HOCl-mediated oxidation of Met to different extents and lose their functions (Khor et al., 2004). Recombinant GSTF2, GSTF3, GSTF8, and MSRB7 proteins, as well as green fluorescent protein (GFP), were expressed in E. coli and purified by Ni2+-affinity chromatography. The GST proteins were pre-treated with HOCl to abolish their enzymatic activities, followed by treatment with 5mM Met to terminate the oxidative reaction. To determine whether MSRB7 can restore the enzymatic activities of the MetO–GSTF proteins, HOCl-oxidized or untreated GSTs were co-incubated with MSRB7 or GFP, and the enzymatic activities of the GSTFs were assayed. As expected, HOCl treatment compromised the GST activities (Fig. 5), suggesting that the oxidative damage resulted in loss of protein function. However, when the oxidized proteins were treated with MSRB7, the activities of GSTF2 and GSTF3 were restored to 71 and 100%, respectively (Fig. 5A, B), while no such affect occurred upon treatment with GFP protein, suggesting that the enzymes were repaired specifically by MSRB7 (Fig. 5A, B). Although GSTF8 interacts with MSRB7 (Fig. 4), MetO–GSTF8 activity was not restored upon treatment with MSRB7 (Fig. 5C).
Fig. 5.
Restoration of oxidized-GST enzymatic activity by MSRB7 in vitro. The enzymatic activities of GSTF2 (A), GSTF3 (B), and GSTF8 (C). HOCl-treated recombinant GSTF2, GSTF3, and GSTF8 proteins were co-incubated with MSRB7 for 1h at 25 °C. Enzymatic activities were determined. Recombinant GFP protein was used as a negative control. Data are means±SD (n=3) of three independent experiments. Data were analysed statistically using Duncan’s test and different letters indicate significant differences at P<0.05.
MSRB7 maintains the stability of GSTF2 and GSTF3 in vivo
Since the transcript levels of GSTF2 and GSTF3 in wild-type A. thaliana are upregulated by oxidative stress (Fig. 3), we evaluated their expression in 1301 (vector-only control), B7Ox, and B7i plants upon MV treatment. GSTF2 and GSTF3 expression was induced by MV treatment (Fig. 6A, B); however, there were no significant differences in the transcript levels in the 1301, B7Ox, and B7i plants, suggesting that the expression of GSTF2 and GSTF3 was not affected by MSRB7. Given that GSTF2 and GSTF3 can interact with MSRB7 (Fig. 4), we examined whether MSRB7 affected the stability of these proteins. Ten-day-old B7Ox, B7i, and 1301 seedlings were first treated with or without 10 μM MV for 8h to induce GSTF2/3 expression followed by treatment with CHX, an inhibitor of de novo protein synthesis, for 0, 12, 24, and 36h. Immunoblot analysis revealed that the endogenous GSTF2/3 protein levels were steadily maintained throughout the 24h post-CHX treatment in 1301, B7Ox, and B7i plants under normal conditions (Supplementary Fig. S7 available at JXB online). However, when the plants were treated with MV, GSTF2/3 protein abundance decreased substantially in both the aerial parts and the roots of B7i plants and, importantly, GSTF2/3 protein levels were more stable in both the aerial parts and the roots of B7Ox plants than in control 1301 plants (Fig. 6C). Quantitative analysis further showed that GSTF2/3 protein levels were markedly more stable in B7Ox than in 1301 and B7i plants under oxidative stress (Fig. 6D, E), suggesting that MSRB7 plays a role in maintaining GSTF2 and/or GSTF3 protein stability in vivo upon oxidative stress.
Fig. 6.
Maintenance of GST stability by MSRB7 in vivo. (A, B) Expression patterns of GSTF2 and GSTF3. Real-time PCR analysis of GSTF2 and GSTF3 transcripts in 10-d-old B7Ox, B7i, and 1301 plants treated with 10 μM MV for 8h. (C) Immunoblotting of GSTF2/3 in aerial parts and roots. Ten-day-old 1301, B7Ox, and B7i seedlings were pre-treated with 10 μM MV for 8h, followed by treatment with 0.5mM CHX for 0–36h. GSTF2/3 was detected using an anti-GSTF2/3 antibody. Protein stained with Coomassie Brilliant Blue (CBB) was used as a protein loading control. (D, E) Relative expression of GSTF2/3. The relative amounts of GSTF2/3 in the aerial parts and roots were determined using immunoblot analysis, and quantified using G:Box iChemi XL (Syngene). Data were analysed statistically using Duncan’s test and different letters indicate significant differences at P<0.05. (F) Total GST activity of MSRB7 transgenic plants. Ten-day-old 1301, B7Ox, and B7i seedlings were pre-treated with 10 μM MV for 8h followed by treatment with 0.5mM CHX for 0–48h. GST activity was measured. Data represent the means±SD (n=10) of three independent experiments.
Prior to CHX treatment, GST activity was substantially lower in B7i plants than in 1301 plants under oxidative stress, but there was no significant difference in GST activity between 1301 and B7Ox plants (Fig. 6F). After CHX treatment, the B7Ox plants exhibited higher GST activity than the 1301 and B7i plants, while GST activity in the 1301 plants decreased rapidly. The activities remained lower in B7i than in 1301 and B7Ox plants (Fig. 6F). After CHX treatment for 24h, we observed no significant difference in GST activity between the 1301 and B7i plants (Fig. 6F). Together, these results suggest that GSTF2/3 activity in plants is maintained and stabilized by MSRB7 under oxidative stress.
Met residues of GSTF2 and GSTF3 are important for maintaining GST activity, and their oxidized states are reduced by MSRB7
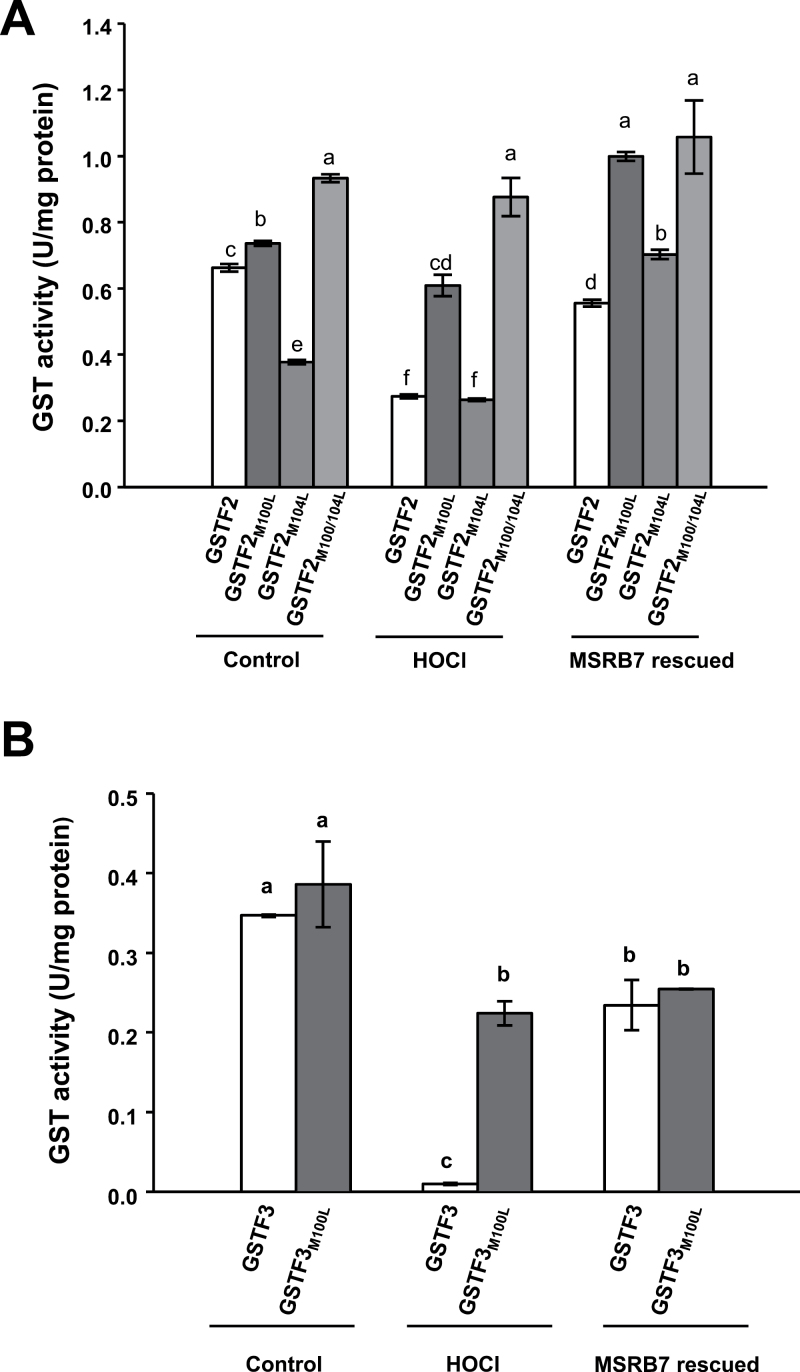
Since oxidation of GSTF2 and GSTF3 proteins affects their enzymatic activities, we examined the importance of different constituent Met residues under oxidative stress. To this end, recombinant proteins of wild-type GSTF2 and GSTF3, and mutants in which Met residues were replaced with another non-polar amino acid, Leu (GSTF2M100L, GSTF2M104L, GSTF2M100/104L, and GSTF3M100L), were generated and purified. Under normal control conditions, the enzymatic activities of all the mutant proteins, except for GSTF2M104L, were similar to those of the wild-type GSTF2/3 (Fig. 7). This may suggest that oxidation or reduction of GSTF2M104 affects the protein conformation, thereby influencing its enzymatic activity. Upon treatment with HOCl, the enzymatic activities of GSTF2 and GSTF3 were reduced to 42 and 3%, respectively, but were restored to 84 and 70%, respectively, after co-incubation with MSRB7 (Fig. 7). The activities of GSTF2M100L, GSTF2M104L, and GSTF2M100/104L proteins were slightly affected by the HOCl treatment, and co-incubation with MSRB7 resulted in slight increases in the activities of GSTF2M100L and GSTF2M104L (Fig. 7A). Since the enzymatic activities of these mutants lacking either one or both Met residues were only slightly compromised by the HOCl treatment, it can be concluded that these Met residues are important substrates for oxidative stress and that their oxidized state affects the enzymatic function of GSTF2.
Fig. 7.
Reduction of Met residues of GSTF2 and GSTF3 by MSRB7. The activities of wild-type and mutated GSTs are shown. HOCl-oxidized recombinant GSTF2, GSTF2M100L (Met replaced with Leu), GSTF2M104L, and GSTF2M100/104L (A), and GSTF3 and GSTF3M100L (B) proteins were co-incubated with MSRB7 for 1h at 25 °C and assayed for GST activity. Data denote means±SD (n=3) of three independent experiments. Data were analysed statistically using Duncan’s test and different letters indicate significant differences at P<0.05.
Discussion
MSRB7 participates in tolerance to chemically induced ROS
Abiotic stress can induce the accumulation of excess ROS, which are known to damage biomolecules such as Met residues in proteins (Moller and Sweetlove, 2010; Toda et al., 2010). Recent studies have shown that MSRB proteins in A. thaliana, rice, and pepper plants have functions related to oxidative stress or defence responses (Vieira Dos Santos et al., 2005; Kwon et al., 2007; Laugier et al., 2010; Oh et al., 2010), and the presence of large numbers of MSRB genes suggests that they may have multiple biological functions and protect plants against different types of stress conditions. Overexpression of MSRB genes can enhance tolerance to MV and act as non-antibiotic selectable marker genes (Li et al., 2013). This correlates well with the finding that, in A. thaliana and tomato plants overexpressing MSRB7, there is a decrease in H2O2 accumulation (Supplementary Fig. S8 available at JXB online) accompanied by an increase in GST activity and tolerance to oxidative stress (Li et al., 2012). However, the mechanism by which MSRB7 protects plants against chemically induced oxidative stress remains unclear. In this study, we focused on investigating how oxidized GSTs generated during oxidative stress are reduced by the cytosolic MSRB7 protein and examined the implications of such a mechanism in oxidative stress tolerance.
Based on our proteomic analysis, B7ox and B7i plants have higher and lower ratios of MetO residues, respectively, in their GSTF2 and GSTF3 proteins (Table 2). This suggests that MSRB7 can protect these proteins against Met oxidation, or that it can convert MetO back to Met following oxidative stress. During the reduction of MetO–GSTs, MSRB7 is itself oxidized, and oxidized proteins are usually unstable and likely to be degraded unless they are reduced by reductases (Kurepa and Smalle, 2008). However, since it has been shown that thioredoxins or glutaredoxin can reduce oxidized MSR proteins via a MSR redox cycle (Tarrago et al., 2009b ), it is possible that the oxidized MSRB7 is restored to its functional state through this cycle. We observed that MSRB7 protein abundance in the aerial parts was gradually decreased after 4h of MV treatment (Fig. 2D). We propose that this decrease in MSRB7 protein abundance is due to: (i) an inability of the MSR redox cycle to maintain the reduced state of the large amount of oxidized MSRB7 protein, causing it to be degraded; or (ii) adaptation of the plants to oxidative stress such that MSRB7 expression is no longer required. The gradual increase in MSRB7 mRNA expression in the aerial part in response to MV treatment (Fig. 2A) may be a compensatory response to supplement the degraded MSRB7 protein. Since the level of GSTF2 and GSTF3 expression in the aerial parts of the plant was slightly increased at 4h after MV treatment and remained high throughout (Fig. 3C), we propose that the plant was under constant oxidative stress and did not adapt to such stress.
Comparative proteomic analysis using CNBr digestion as an efficient strategy for the identification of MSRB7 substrates
To date, no substrates of plant cytosolic MSRBs have been identified. Our data suggest that MSRB7 may protect its target protein(s) against Met oxidation, which would explain the low and high percentages of MetO-containing proteins observed in B7ox and B7i plants, respectively, during oxidative stress (Table 2). Since CNBr does not cleave proteins/peptides containing MetO, these proteins are not identified by LC-MS/MS analysis. In addition, B7i plants contain a higher percentage of MetO, and are therefore not suitable for CNBr-digested comparative proteomic analysis. Using the CNBr approach, 41 stress-related proteins were identified as possible substrates of MSRB7 (Table 1) and GO annotation identified some of these putative substrates as ROS-scavenging proteins, which might account for the enhanced tolerance of B7ox plants to oxidative stress.
The total GST activity in B7Ox plants is significantly higher than in the wild-type and B7i plants (Li et al., 2012), and since GSTF2, GSTF3, and GSTF8 interact with MSRB7 (Fig. 4), they may all be important in protecting plants against oxidative stress. CNBr-cleavable GSTF2 and GSTF3 proteins were detected in B7Ox plants during oxidative stress (Table 1). This suggests that the MetO–GSTF2 and MetO–GSTF3 proteins generated during oxidative stress are reduced by the high levels of MSRB7 expressed in B7Ox plants. Despite the apparent interaction between GSTF8 and MSRB7 (Fig. 4), CNBr-cleavable GSTF8 was not detected in the MV-treated B7Ox plants (Table 1), while high levels, comparable to the other plants, of MetO–GSTF8 were observed (Table 2). The inability of MSRB7 to target repair MetO–GSTF8 protein efficiently may account for the failure to restore the GSTF8 enzymatic activity (Fig. 5C). The structure of GSTF8 has not yet been solved, but one possible explanation for our results is that the surface-exposed Met residue in GSTF8 is not involved in enzymatic activity. However, we cannot rule out the possibility that oxidative stress may have caused the oxidation of other amino acid residues that are not affected by MSRB7 action, hence reducing the enzymatic activity of GSTF8. We propose that the CNBr digestion approach is an efficient tool for comparative proteomics allowing for identification of physiological targets of MSRB7. A similar approach may be used to study the remaining 41 potential substrates of MSRB7. In addition, the functions of MSRB proteins present in other species may be similarly elucidated using this approach.
MSRB7 maintains the activity and stability of the substrates GSTF2 and GSTF3
Although some studies have reported that the substrates of MSRs are Met-rich proteins (Gustavsson et al., 2002; Sundby et al., 2005; Tarrago et al., 2012), it is interesting that the Met contents of GSTF2 (1.4%) and GSTF3 (0.9%) are lower than average for all proteins (1.7%) (Alamuri and Maier, 2006). The three-dimensional structures of GSTF2 and GSTF3 retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/Structure/index.shtml) revealed that GSTF2M100/104 and GSTF3M100 are surface-exposed residues (Supplementary Fig. S4). We therefore propose that the Met residues of GSTF2 and GSTF3 are located in the functional domain (the α-helical domain in the F class of GSTs), which is essential for secondary structure folding and protein function (Fig. 7 and Supplementary Fig. S4 available at JXB online). Despite the lack of one Met residue, MSRB7 was able to restore the enzymatic activities of GSTF2M100L and GSTF2M104L proteins to a level higher than those of the control group. One explanation for this is that these recombinant proteins generated from E. coli underwent some degree of oxidation during protein preparation, so the enzymatic activities of the control proteins were not maximal. In contrast, recombinant GSTF2M100/104L protein lacks two Met residues and is unlikely to contain any MetO, explaining the high enzymatic activities observed in all three groups (Fig. 7A). The GSTF3M100L protein lacking the Met residue is susceptible to oxidative damage despite the absence of MetO, and this damage is not affected by MSRB7 action (Fig. 7B), suggesting that the oxidative stress results in the oxidation of other amino acids that are not modified by MSRB7.
The fifty-four homologue (Ffh) protein, a signal recognition particle protein of E. coli, is reported to be a substrate of MSRs (Ezraty et al., 2004). This protein is remarkably unstable in an E. coli mutant lacking msra and msrb (Ezraty et al., 2004), suggesting that MSRs have roles in maintaining the stability of their substrates. It has been reported previously that GSTs themselves can become oxidized, causing them to either lose their antioxidant function, or become partially degraded (Dixon and Edwards, 2010). The Met residues of GSTs could be a critical requirement for enzymatic activity and plant survival under oxidative stress, so their oxidation is possibly reversed by other MSRs. The experiments shown in Fig. 5 show consistent >80% recovery for GST2/3 activities in response to MSRB7 treatment. We therefore postulate that HOCl may possess a preference for Met R-oxidation, and the enzymatic activities of these oxidized GSTF2 and GSTF3 proteins are, therefore, readily recovered by MSRB7. Our observations suggest that the Met residues of GSTF2 and GSTF3 are important for maintaining enzymatic activities, and their non-functional oxidized states are probably important targets for protein repair by MSRB7 (Figs 6 and 7). Both B7i and 1301 plants have a tendency to form MetO at GSTF2M104 and GSTF3M100 during oxidative stress in vivo (Table 2). We conclude that GSTF2, GSTF3, and MSRB7 are components of an oxidative stress tolerance mechanism that depends on the maintenance of GSTF2 and GSTF3 activity by MSRB7.
This study identified GSTF2 and GSTF3 as substrates of plant cytosolic MSRB during oxidative stress tolerance. However, the mechanism underlying the maintenance of substrate stability remains to be elucidated. Although MSRB8 shares 95% amino acid identity with MSRB7 and is MV inducible (Li et al., 2012), we believe that MSRB8 may act on substrates other than GSTF2 and GSTF3 as it is incapable of compensating for the loss of MSRB7 activity observed in B7i plants. Therefore, in the future, it will be also interesting to identify the substrates of MSRB8, and compare and contrast the functional roles of these two highly homologous proteins.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Constructs used in the BiFC assay.
Supplementary Fig. S2. Induction of the MSRB7 promoter by oxidative stress in cauline and rosette leaves but not in flowers and siliques.
Supplementary Fig. S3. Determination of the binding specificities of GSTF2/3-specific antibody against rGSTF2 and rGSTF3 recombinant proteins.
Supplementary Fig. S4. Amino acid sequences of GSTs and the three-dimensional structure of GSTF2.
Supplementary Fig. S5. Cytosolic locations of MSRB7, GSTF2, GSTF3, and GSTF8.
Supplementary Fig. S6. Controls of BiFC assays.
Supplementary Fig. S7. Amounts of GSTF2/3 are not significantly different following CHX treatment.
Supplementary Fig. S8. H2O2 content.
Supplementary Table S1. LC-MS/MS proteomic analysis of WT and B7Ox plants.
Supplementary Table S2. Primers used for PCR and real-time PCR.
Acknowledgements
We are very grateful to Fu-Hui Wu, Chen-Tai Lo, and Shu-Chen Shen for cloning the BiFC vector and providing excellent technical assistance with protoplast transformation and confocal microscopy. We thank Dr Hsou-min Li (Institute of Molecular Biology, Academia Sinica, Taiwan) and Ms Miranda Loney for discussion and manuscript scientific editing. We also thank PlantScribe (http://www.plantscribe.com) for manuscript editing. This work was supported by grant 98-2324-B-001-003-CC1 from the Development Program of Industrialization for Agricultural Biotechnology, Academia Sinica, Republic of China, and grant 99AS-1.1.1-FD-Z1 from the Agriculture and Food Agency, Council of Agriculture, Executive Yuan, Republic of China.
Glossary
Abbreviations:
- B7i
MSRB7 knockdown plant
- B7Ox
MSRB7 overexpressing plant
- BiFC
Bimolecular Fluorescence Complementation
- CFP
cyan fluorescent protein
- CHX
cycloheximide
- CNBr
cyanogen bromide
- CDNB
1-chloro-2,4-dinitrobenzene
- DTT
dithiothreitol
- GFP
green fluorescent protein
- GO
Gene Ontology
- GST
glutathione transferase
- GUS
β-glucuronidase
- HOCl
hypochlorous acid
- HSP
heat-shock protein
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MetO
methionine sulfoxide
- Met-R-O
Met-R-sulfoxide
- Met-S-O
Met-S-sulfoxide
- MSR
methionine sulfoxide reductase
- MV
methyl viologen
- PLGS
ProteinLynx GlobalServer
- PMSF
phenylmethylsulfonyl fluoride
- ROS
reactive oxygen species
- RT-PCR
reverse transcription-PCR
- SD
standard deviation.
References
- Alamuri P, Maier RJ. 2006. Methionine sulfoxide reductase in Helicobacter pylori: interaction with methionine-rich proteins and stress-induced expression. Journal of Bacteriology 188, 5839–5850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatelain E, Satour P, Laugier E, Ly Vu B, Payet N, Rey P, Montrichard F. 2013. Evidence for participation of the methionine sulfoxide reductase repair system in plant seed longevity. Proceedings of the National Academy of Sciences, USA 110, 3633–3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CJ, Tseng MC, Lin HJ, Lin TW, Chen YR. 2010. Visual indicator for surfactant abundance in MS-based membrane and general proteomics applications. Analytical Chemistry 82, 8283–8290 [DOI] [PubMed] [Google Scholar]
- Chen JH, Jiang HW, Hsieh EJ, Chen HY, Chien CT, Hsieh HL, Lin TP. 2012. Drought and salt stress tolerance of an Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiology 158, 340–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of A. thaliana thaliana. The Plant Journal 16, 735–743 [DOI] [PubMed] [Google Scholar]
- Cummins I, Cole DJ, Edwards R. 1999. A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. The Plant Journal 18, 285–292 [DOI] [PubMed] [Google Scholar]
- Dat J, Vandenabeele S, Vranova E, Van Montagu M, Inze D, Van Breusegem F. 2000. Dual action of the active oxygen species during plant stress responses. Cellular and Molecular Life Sciences 57, 779–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Edwards R. 2010. Glutathione transferases. The Arabidopsis Book , e0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Lapthorn A, Edwards R. 2002. Plant glutathione transferases. Genome Biology 3, REVIEWS3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon DP, Steel PG, Edwards R. 2011. Roles for glutathione transferases in antioxidant recycling. Plant Signaling & Behavior 6, 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Dixon DP. 2005. Plant glutathione transferases. Methods in Enzymology 401, 169–186 [DOI] [PubMed] [Google Scholar]
- Ezraty B, Grimaud R, El Hassouni M, Moinier D, Barras F. 2004. Methionine sulfoxide reductases protect Ffh from oxidative damages in Escherichia coli . EMBO Journal 23, 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friguet B. 2006. Oxidized protein degradation and repair in ageing and oxidative stress. FEBS Letters 580, 2910–2916 [DOI] [PubMed] [Google Scholar]
- Gong H, Jiao Y, Hu WW, Pua EC. 2005. Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro. Plant Molecular Biology 57, 53–66 [DOI] [PubMed] [Google Scholar]
- Gustavsson N, Kokke BP, Harndahl U, Silow M, Bechtold U, Poghosyan Z, Murphy D, Boelens WC, Sundby C. 2002. A peptide methionine sulfoxide reductase highly expressed in photosynthetic tissue in Arabidopsis thaliana can protect the chaperone-like activity of a chloroplast-localized small heat shock protein. The Plant Journal 29, 545–553 [DOI] [PubMed] [Google Scholar]
- Habig WH, Pabst MJ, Jakoby WB. 1974. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. Journal of Biological Chemistry 249, 7130–7139 [PubMed] [Google Scholar]
- Hayes JD, Flanagan JU, Jowsey IR. 2005. Glutathione transferases. Annual Review of Pharmacology and Toxicology 45, 51–88 [DOI] [PubMed] [Google Scholar]
- Hsiao P, Sanjaya, Su RC, Teixeira da Silva JA, Chan MT. 2007. Plant native tryptophan synthase β1 gene is a non-antibiotic selection marker for plant transformation. Planta 225, 897–906 [DOI] [PubMed] [Google Scholar]
- Khor HK, Fisher MT, Schoneich C. 2004. Potential role of methionine sulfoxide in the inactivation of the chaperone GroEL by hypochlorous acid (HOCl) and peroxynitrite (ONOO–). Journal of Biological Chemistry 279, 19486–19493 [DOI] [PubMed] [Google Scholar]
- Kurepa J, Smalle JA. 2008. To misfold or to lose structure?: Detection and degradation of oxidized proteins by the 20S proteasome. Plant Signaling & Behavior 3, 386–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SJ, Kwon SI, Bae MS, Cho EJ, Park OK. 2007. Role of the methionine sulfoxide reductase MsrB3 in cold acclimation in Arabidopsis . Plant and Cell Physiology 48, 1713–1723 [DOI] [PubMed] [Google Scholar]
- Laugier E, Tarrago L, Vieira Dos Santos C, Eymery F, Havaux M, Rey P. 2010. Arabidopsis thaliana plastidial methionine sulfoxide reductases B, MSRBs, account for most leaf peptide MSR activity and are essential for growth under environmental constraints through a role in the preservation of photosystem antennae. The Plant Journal 61, 271–282 [DOI] [PubMed] [Google Scholar]
- Li C-W, Lee S-H, Chan M-T. 2013. Utilization of the plant methionine sulfoxide reductase B genes as selectable markers in Arabidopsis and tomato transformation. Plant Cell, Tissue and Organ Culture 113, 555–563 [Google Scholar]
- Li CW, Lee SH, Chieh PS, Lin CS, Wang YC, Chan MT. 2012. Arabidopsis root-abundant cytosolic methionine sulfoxide reductase B genes MsrB7 and MsrB8 are involved in tolerance to oxidative stress. Plant and Cell Physiology 53, 1707–1719 [DOI] [PubMed] [Google Scholar]
- Li CW, Su RC, Cheng CP, Sanjaya, You SJ, Hsieh TH, Chao TC, Chan MT. 2011. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiology 156, 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lim EK, Yu SM. 1998. Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. Journal of Biological Chemistry 273, 10120–10131 [DOI] [PubMed] [Google Scholar]
- Mauch F, Dudler R. 1993. Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiology 102, 1193–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller IM, Sweetlove LJ. 2010. ROS signalling—specificity is required. Trends in Plant Science 15, 370–374 [DOI] [PubMed] [Google Scholar]
- Ogorzalek Loo RR, Stevenson TI, Mitchell C, Loo JA, Andrews PC. 1996. Mass spectrometry of proteins directly from polyacrylamide gels. Analytical Chemistry 68, 1910–1917 [DOI] [PubMed] [Google Scholar]
- Oh J-E, Hong S-W, Lee Y, et al. 2005. Modulation of gene expressions and enzyme activities of methionine sulfoxide reductases by cold, ABA or high salt treatments in Arabidopsis . Plant Science 169, 1030–1036 [Google Scholar]
- Oh SK, Baek KH, Seong ES, Joung YH, Choi GJ, Park JM, Cho HS, Kim EA, Lee S, Choi D. 2010. CaMsrB2, pepper methionine sulfoxide reductase B2, is a novel defense regulator against oxidative stress and pathogen attack. Plant Physiology 154, 245–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oien DB, Moskovitz J. 2008. Substrates of the methionine sulfoxide reductase system and their physiological relevance. Current Topics in Developmental Biology 80, 93–133 [DOI] [PubMed] [Google Scholar]
- Romero HM, Berlett BS, Jensen PJ, Pell EJ, Tien M. 2004. Investigations into the role of the plastidial peptide methionine sulfoxide reductase in response to oxidative stress in Arabidopsis . Plant Physiology 136, 3784–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Vieira Dos Santos C, Tarrago L, Rey P. 2006. Plant methionine sulfoxide reductase A and B multigenic families. Photosynthesis Research 89, 247–262 [DOI] [PubMed] [Google Scholar]
- Sappl PG, Carroll AJ, Clifton R, Lister R, Whelan J, Harvey Millar A, Singh KB. 2009. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. The Plant Journal 58, 53–68 [DOI] [PubMed] [Google Scholar]
- Sharov VS, Schoneich C. 2000. Diastereoselective protein methionine oxidation by reactive oxygen species and diastereoselective repair by methionine sulfoxide reductase. Free Radical Biology & Medicine 29, 986–994 [DOI] [PubMed] [Google Scholar]
- Sundby C, Harndahl U, Gustavsson N, Ahrman E, Murphy DJ. 2005. Conserved methionines in chloroplasts. Biochimica et Biophysica Acta 1703, 191–202 [DOI] [PubMed] [Google Scholar]
- Tarrago L, Kieffer-Jaquinod S, Lamant T, Marcellin MN, Garin JR, Rouhier N, Rey P. 2012. Affinity chromatography: a valuable strategy to isolate substrates of methionine sulfoxide reductases? Antioxidants & Redox Signaling 16, 79–84 [DOI] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Rey P. 2009. a Protein-repairing methionine sulfoxide reductases in photosynthetic organisms: gene organization, reduction mechanisms, and physiological roles. Molecular Plant 2, 202–217 [DOI] [PubMed] [Google Scholar]
- Tarrago L, Laugier E, Zaffagnini M, Marchand C, Le Marechal P, Rouhier N, Lemaire SD, Rey P. 2009. b Regeneration mechanisms of Arabidopsis thaliana methionine sulfoxide reductases B by glutaredoxins and thioredoxins. Journal of Biological Chemistry 284, 18963–18971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Carrie C, Andersson CR, Sivasithamparam K, Whelan J, Singh KB. 2007. Differential gene expression and subcellular targeting of Arabidopsis glutathione S-transferase F8 is achieved through alternative transcription start sites. Journal of Biological Chemistry 282, 28915–28928 [DOI] [PubMed] [Google Scholar]
- Toda T, Nakamura M, Morisawa H, Hirota M, Nishigaki R, Yoshimi Y. 2010. Proteomic approaches to oxidative protein modifications implicated in the mechanism of aging. Geriatrics & Gerontology International 10 (Suppl. 1 ), S25–S31 [DOI] [PubMed] [Google Scholar]
- Vieira Dos Santos C, Cuine S, Rouhier N, Rey P. 2005. The Arabidopsis plastidic methionine sulfoxide reductase B proteins. Sequence and activity characteristics, comparison of the expression with plastidic methionine sulfoxide reductase A, and induction by photooxidative stress. Plant Physiology 138, 909–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt W. 1995. Oxidation of methionyl residues in proteins: tools, targets, and reversal. Free Radical Biology & Medicine 18, 93–105 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438 [DOI] [PubMed] [Google Scholar]
- Wu FH, Shen SC, Lee LY, Lee SH, Chan MT, Lin CS. 2009. Tape–Arabidopsis sandwich—a simpler Arabidopsis protoplast isolation method. Plant Methods 5, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.