Abstract
Chronic lymphocytic leukemia (CLL) is characterized by the clonal expansion of CD5+CD23+ B cells in blood, marrow, and second lymphoid tissues. Gene-expression profiling and phenotypic studies suggest that CLL is probably derived from CD5+ B cells similar to those found in the blood of healthy adults. Next-generation sequencing has revealed recurrent genetic lesions that are implicated in CLL pathogenesis and/or disease progression. The biology of CLL is entwined with its microenvironment, in which accessory cells can promote leukemia cell growth and/or survival. Recently, much attention has been focused on the CLL B cell receptor (BCR) and on chemokine receptors that enable CLL cells to home to lymphoid tissues and to establish the leukemia microenvironment. Agents that can interfere with BCR signaling or chemokine– receptor signaling, or that target surface antigens selectively expressed on CLL cells, promise to have significant therapeutic benefit in patients with this disease.
Keywords: cell origin, genetic alterations, B cell signaling, microenvironment
INTRODUCTION
Chronic lymphocytic leukemia (CLL), the most common adult leukemia in the Western world (1, 2), is characterized by the clonal expansion of CD5+CD23+ B cells in blood, marrow, and secondary lymphoid tissues. Although CLL cells in the blood appear to be predominantly resting lymphocytes in G0 (3), focal aggregates of different-sized lymphocytes are scattered throughout the lymphoid tissue and are presumably where leukemia cells proliferate (2, 4–9). Heavy-water (deuterium) labeling studies have found that some patients have high rates of leukemia cell turnover; 0.1–1% of their entire leukemia cell population is produced each day (10). Because such growth kinetics can be observed in patients who have relatively stable disease, the high growth rate observed in such patients is presumably counterbalanced by a high rate of spontaneous apoptosis.
Some patients have progressive disease and require therapy relatively soon after diagnosis, whereas others have highly indolent disease that does not require treatment for many years (11, 12). This variability reflects intrinsic heterogeneity in the disease biology. For example, in approximately half of all cases, the leukemia cells express immunoglobulin (Ig) encoded by unmutated Ig heavy-chain variable-region genes (IGHVs), which are associated with more aggressive disease (13, 14). By contrast, patients with CLL cells that express Ig encoded by mutated IGHVs generally have more indolent disease. Our understanding of pathological mechanism(s) involved in CLL may have implications for the development of new treatments.
This review focuses on recent advances in our understanding of factors governing the pathogenesis and progression of CLL that have been made through analyses of leukemia cell gene expression and next-generation sequencing. Insights gained through these studies are guiding the development of targeted therapies that may be more effective and better tolerated than currently available treatments for patients with CLL.
ORIGIN OF CHRONIC LYMPHOCYTIC LEUKEMIA
CLL B cells express CD5, CD19, CD23, and low levels of surface Ig (2). This phenotypic profile differs from that of most normal B cell subsets. However, recent gene-expression profiling studies have confirmed that CLL is probably derived from CD5 B cells similar to those found in the blood of healthy adults.
CD5+ B Cells
The coexpression of CD5 and B cell surface antigens on leukemia cells prompted speculation that CLL is a malignancy of CD5+ B cells (15, 16); some investigators maintain that these cells constitute a distinct B cell lineage (17). However, similarities between CLL and splenic marginal zone (sMGZ) B cells led to a proposal that CLL arises from sMGZ B cells (18). On the basis of a study of specific IGHV rearrangements, other investigators proposed that CLL cells that use unmutated IGHVs are derived from conventional naïve B cells (19). However, gene-expression studies on CLL cells found that the leukemia cells that express either unmutated or mutated IGHVs share a common gene-expression profile, suggesting that they have a common origin (20, 21). Studies on CLL cells and normal human B cell subsets indicated that CLL cells have a gene-expression profile that is more similar to that of memory B cells than to that of CD5+ B cells isolated from neonatal cord blood (20). However, cord-blood CD5+ B cells appear to have a gene-expression profile that is distinct from that of CD5+ B cells found in the blood of healthy adults (22–24), and a more recent study of CLL cells and normal naïve B cells, sMGZ B cells, adult CD5+ B cells, class-switched B cells, and IgM+ memory B cells found that CLL cells have a gene-expression profile that is very similar to that of CD5+ B cells found in the peripheral blood of healthy adults (25). IGHV mutation analyses revealed that CD5+ blood B cells are clonally expanded and include a small number of post–germinal center B cells, a finding that is consistent with the existence of two classes of CLL defined by the presence or absence of IGHV hypermutation. On the basis of these studies, investigators now believe that CLL cells with unmutated IGHVs are derived from mature CD5+CD27− B cells with unmutated IGHVs and that CLL cells with mutated IGHVs are derived from a distinct, previously unrecognized, subset of CD5+CD27+ post– germinal center B cells with mutated IGHVs.
Monoclonal B Cell Lymphocytosis
The blood of a significant subset of healthy adults contains oligoclonal/monoclonal expansions of B lymphocytes with a surface-antigen phenotype similar to that of CLL cells. Such oligoclonal/monoclonal B cell expansions, known as monoclonal B cell lymphocytosis (MBL), have been identified in the blood mononuclear cells of approximately 13% of adult siblings of patients with CLL (26). MBL is observed at a lower rate (3.5%) of healthy adults older than 45 who have no family history of CLL (27, 28). Sometimes the B cells in MBL have chromosomal abnormalities similar to those observed in CLL (29), suggesting that MBL represents early CLL, or a precursor lesion that evolves into CLL, in some individuals. The risk of MBL developing into CLL may be influenced by polymorphisms in genes controlling key functions in B cell development. In any case, because adults with MBL develop CLL or other B cell lymphomas at a low rate (approximately 1% per year) (29), the B cells in MBL apparently must acquire additional genetic or epigenetic changes before becoming CLL B cells (Figure 1). The same may be true for B cells in MBL that subsequently evolves into other B cell lymphomas (30).
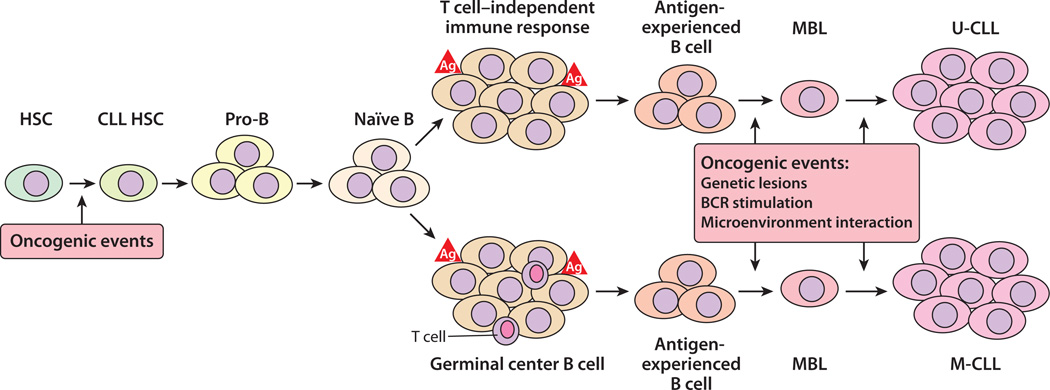
Figure 1.
A model for the development of chronic lymphocytic leukemia (CLL). Mutations that contribute to the development of CLL may occur at any stage of B cell development, including in hematopoietic stem cells (HSCs). HSCs bearing oncogenetic mutations may give rise to B cells with modest growth/survival advantages, which in the setting of T cell–independent or T cell–dependent immune stimulation may progress to monoclonal B cell lymphocytosis (MBL). CLL cells originating from B cells that have incurred immunoglobulin (Ig) somatic mutation express mutated Ig heavy-chain variable-region genes (IGHVs) and are defined as M-CLL, whereas CLL cells originating from B cells that have not undergone Ig somatic mutations express germ-line IGHVs and are defined as U-CLL. The expansion of a CLL (or MBL) clone is associated with de novo accumulation of additional genetic lesions, as well as continued interactions between the leukemic cells and accessory cells and antigen(s) in the leukemia microenvironment of lymphoid tissues (39, 58). Abbreviations: Ag, antigen; BCR, B cell receptor.
ROR1+ B Cells
Recent studies have found that CLL cells also express receptor tyrosine kinase–like orphan receptor 1 (ROR1), which ordinarily is expressed only by cells during embryonic development (31–35). Further studies have found that ROR1 can also be expressed on a small subset of normal precursor B cells known as hematogones (36). These cells may coexpress CD5, CD10, and B cell differentiation antigens and typically are found in the marrow of pediatric patients recovering from myeloablative chemotherapy (37). These cells are present at very low levels in the blood of healthy adults (at frequencies of 0.01% to 0.1% of blood mononuclear cells). ROR1+ B cells isolated from the blood mononuclear cells of healthy donors have a heterogeneous phenotype; specifically, they may express CD19 and CD27, but not CD38, or they may express CD19 and CD38, but not CD27. In contrast to the subpopulation of ROR1+ B cells that express CD27, the subpopulation of ROR1+ B cells that express CD38 have relatively high levels of CD5 and CD23, both of which are considered markers of CLL B cells (38). These studies suggest that ROR1+CD38+ B cells in healthy adults may be the precursor of CLL. This hypothesis needs further testing, possibly through comparison between the transcriptome of ROR1+ normal B cells and that of ROR1+ CLL B cells and through functional studies on normal ROR1+ B cells and CLL B cells.
Stem Cells?
Xenogeneic transplantation studies have found that hematopoietic stem cells (HSCs) isolated from CLL patients are apparently primed to undergo differentiation into B cell progenitors that may subsequently develop into mono- or oligoclonal B cells with a CLL-like phenotype (39). The B cells that developed in such xenografts did not express the same IGHVs or have the same chromosomal abnormalities as those of the original CLL B cell clone, suggesting that these cells were generated from disparate clones of B cell progenitors that underwent de novo IGHV rearrangement. These data indicate that HSCs of patients with CLL could develop into B cells that have features of B cell CLL. Whether these cells constitute a type of stem cell for CLL is uncertain, but it does appear that other genetic or epigenetic changes are required for such B cells to become neoplastic (Figure 1). Moreover, studies on telomere length and telomerase expression (40) suggest that CLL cells (especially those that use unmutated IGHVs) have undergone extensive cell division prior to leukemic transformation.
IMMUNOGLOBULIN REPERTOIRE
B cells are crucial to adaptive immune responses. They are responsible for the production of antigen-specific antibodies that help eliminate exogenous, as well as autologous, threats to homeostasis (41). This fundamental function depends on a process that involves at least two major steps. The first requires generation of a diverse repertoire of antigen receptors. The second requires selection of B cells that express Ig that has appropriate binding activity for the selecting antigen. Indeed, the immune system relies on the formation of an extremely diverse population of B cells, each of which expresses one type of Ig on its surface that serves as the B cell receptor (BCR) for antigen in adaptive immune responses. The probability that two independent B cell clones carry exactly the same BCR is extremely low (e.g., less than 10−12). However, CLL cells isolated from different patients often express similar, if not identical, BCRs with common stereotypic features and/or structural similarities (42).
Certain IGHVs, such as IGHV1–69, IGHV4–34, and IGHV3–7, are used by CLL cells at higher frequencies than those observed in normal B cells (43–46). Furthermore, the incidence of somatic hypermutation is not uniform among IGHVs in CLL cells; for example, IGHV1–69 has few or no mutations, whereas IGHV3–7, IGHV3–23, and IGHV4–34 generally show substantial somatic mutations (46). In any case, one can identify shared (stereotypic) primary structures among the Ig expressed by CLL B cells that are not readily apparent in the highly diverse Ig repertoire of normal B cells. The marked restriction in the Ig gene repertoire of CLL cells highlights the role played by one or more common self-or environmental antigens in leukemic B cell selection.
ANTIGENS THAT MAY PLAY A ROLE IN LEUKEMIA B CELL SELECTION
Some of the Ig expressed in CLL can react with antigen expressed by cells undergoing apoptosis, including cytoskeletal proteins (47–50). Some Ig react with nonmuscle myosin heavy chain IIA, which is expressed on some apoptotic cells, namely myosin-exposed apoptotic cells (MEACs). Binding to MEACs is more commonly observed on CLL cells expressing unmutated IGHVs than on CLL cells expressing mutated IGHVs (51, 52). Ig with different stereotypic features have distinct patterns of antigen reactivity (47–51), suggesting that more than one antigen or antigenic epitope may be responsible for driving selection of the distinctive repertoire expressed in CLL.
In addition to self-antigen, several othermicrobial or virus-associated antigens may contribute to the selection of the Ig expressed in CLL. For example, CLL-associated Ig encoded by IGHV1–69 can react with various grampositive or gram-negative bacteria (53) or with highly conserved antigens of cytomegalovirus or other herpes viruses (54–56). Such antigens may also contribute to the selection of B cells in other pathological conditions (57).
GENETIC ALTERATIONS IN CHRONIC LYMPHOCYTIC LEUKEMIA
CLL cells commonly harbor deletions at 13q14, 11q22–q23, or 17p13 or may have an extra copy of chromosome 12 (trisomy 12); such genetic alterations are significantly associated with clinical outcome (1, 2, 59, 60). The advent of next-generation sequencing technologies, coupled with gene copy-number analyses, have identified additional genetic lesions in CLL, such as mutations in NOTCH1, SF3B1, and BIRC3 (61–64). Such mutations could be used as potential therapeutic targets or as biomarkers that can distinguish among patients who may have disparate clinical outcomes (61–67).
NOTCH1 encodes a ligand-activated transcription factor (NOTCH1) that regulates several downstream pathways that induce the differentiation of hematopoietic progenitors into immature T cells and of mature B cells into antibody-secreting cells (68, 69). Activating mutations in NOTCH1 occur in ∼60% of T-lineage acute lymphoblastic leukemias (70). In CLL, activating NOTCH1 mutations have been detected in ∼10% of newly diagnosed cases, but in 15% to 20% of progressive and/or relapsed CLL cases (61, 62, 66). NOTCH1 mutations are also more frequent in CLL cell populations that express unmutated IGHVs and that have trisomy 12 (61, 62, 66, 71, 72). Cases with NOTCH1 mutations appear to have a distinctive gene-expression profile (62, 72) and define a high-risk subgroup of patients with clinical outcomes comparable to those of cases with disruptions in TP53, independent of other risk factors (61, 62, 66). NOTCH1 mutations in CLL are restricted to the C-terminal PEST [proline (P), glutamate (E), serine (S), and threonine (T)] domain, which normally limits the intensity and duration of NOTCH1 signaling (61, 62, 66). Removal of the PEST domain impairs the degradation of NOTCH1, allowing for accumulation of the active form of NOTCH1 (70). One recurrent mutation (c.7544_7545delCT) accounts for ∼77% of all NOTCH1 mutations in CLL (45–47) and can be rapidly detected by a simple polymerase chain reaction–based strategy, providing a potential approach for a first-level screening of NOTCH1 alterations (66).
SF3B1 encodes the splicing factor 3B sub-unit 1 (SF3B1), which is a critical component of both major (U2-like) and minor (U12-like) spliceosomes that are required for the precise excision of introns from pre-mRNA (73). Mutations in SF3B1 were observed in ∼10% of newly diagnosed CLL cases and in ∼17% of cases with progressive, late-stage disease requiring therapy (64, 65). SF3B1 mutations are apparently acquired during clonal evolution, and the proportionate representation of sub-clones harboring SF3B1 mutations can increase over time, independently of cytoreductive therapy (74, 75). That such mutations play a role in leukemia pathogenesis and/or progression is supported by the clustering of these mutations in evolutionarily conserved hot spots localized within HEAT domains (64, 65). Because SF3B1 regulates the alternative splicing program of genes controlling cell-cycle progression and apoptosis, mutations in SF3B1 may enhance CLL cell proliferation and/or survival (64, 65).
Disruption of BIRC3 associates with unfavorable clinical outcome, independently of other risk factors (67). BIRC3 encodes the baculoviral inhibitor of apoptosis (IAP) repeat–containing 3 protein (BIRC3), which is a member of the IAP family of proteins that can inhibit apoptosis by binding to tumor necrosis factor (TNF) receptor–associated factors 1 and 2 (TRAF1 and TRAF2), possibly by interfering with activation of ICE-like proteases (caspases). BIRC3 can act as an E3 ubiquitin–protein ligase that regulates nuclear factor κB (NF-κB) signaling, acting to promote canonical NF-κB signaling while suppressing constitutive activation of noncanonical NF-κB signaling (76). Activation of canonical NF-κB signaling can promote the growth and survival of CLL cells in vitro and in vivo (77–79). BIRC3 mutations in CLL are predicted to disrupt the C-terminal RING domain, which is essential for proteasomal degradation of MAP3K14 (mitogen-activated protein kinase kinase kinase 14) by BIRC3 (67). Consistently, CLL cells harboring mutations in BIRC3 display constitutive NF-κB activation (67) and appear less responsive to conventional chemotherapy than CLL cells without such mutations. As such, inhibitors of NF-κB may have clinical activity in CLL, particularly in cases harboring alterations in BIRC3 (78–81).
Whole-exome sequencing studies performed on CLL cells have revealed mutations in several other genes, albeit at low frequencies (61–64). Mutations in MYD88 are observed in 3% to 10% of CLL cases at diagnosis (62, 64). MYD88 encodes a critical adaptor molecule of the Toll-like receptor (TLR) complex that is also mutated in other B cell malignancies, such as lymphoplasmacytic lymphoma (83), diffuse large B cell lymphoma (DLBCL), and marginal zone B cell lymphoma (83, 84). Mutations in other genes encoding proteins involved in the activation of TLR signaling and NF-κB have also been observed in DLBCL (85). In contrast to the mutations in NOTCH1 or SF3B1, mutations in MYD88 appear to be present in the vast majority (if not all) cells within the CLL clone; therefore, MYD88 appears to be an early driver mutation in at least a subset of CLL cases. Further evaluation of the frequency of MYD88 mutations at diagnosis and in late-stage disease may further clarify the role of TLR signaling in the pathogenesis or progression of CLL.
Mutations in genes encoding proteins involved in the Wnt signaling pathway have also been observed in CLL (20, 86). Although any one mutation might be observed in only a small proportion of cases, collectively the mutations in genes encoding any one of several proteins involved in Wnt signaling account for a fairly large proportion of the mutations observed in CLL (64). Prior studies identified activation of Wnt signaling in CLL cases, particularly those that express unmutated IGHV genes (87). The identified mutations may be responsible for the aberrant activation of Wnt pathway in CLL, so Wnt may be a target for the development of new therapies (87). Indeed, several inhibitors of Wnt signaling that have selective cytotoxicity for CLL cells have been identified (88, 89).
CLONAL EVOLUTION
Whole-genome sequencing (WGS) provides resolution of the subclonal architecture involved in CLL clonal evolution (90). Using WGS, Schuh et al. (91) examined three patients with CLL cells that expressed unmutated IGHVs. These patients provided leukemia samples at each of five time points for up to 7 years. Each patient received treatment with an alkylating agent, then fludarabine-based chemotherapy, and finally chemoimmunotherapy with anti-CD20 antibody. Across all time points, subclonal populations were observed; each sample harbored up to five distinct clonal subpopulations. Some mutations were detected in all the cells and at all time points and were considered to represent events in the founder subclone. Other mutations were considered secondary events because they were common to only a subset of leukemia cells in any one subpopulation and had varying allelic frequencies before and after treatment. Notably, although a several log reduction in white blood cell counts was observed in each patient after therapy, three very different temporal patterns of repopulation were observed. One patient had an apparently stable equilibrium of five subpopulations over the course of years, even in the setting of therapy. In contrast, in another patient one leukemia subclone was almost completely replaced by another following therapy. Finally, the third patient demonstrated a hybrid pattern, namely an apparently stable subclonal distribution that, over time, became distorted by the apparent expansion of one sub-clone, which became the dominant subclone prior to the patient’s death from CLL (91). These distinct subclonal patterns may account in part for the highly variable clinical course of CLL patients before and after therapy. This study also highlights the challenge in distinguishing early, initiating driver mutations from secondary events associated with subclonal diversification and disease progression.
B CELL RECEPTOR SIGNALING IN CHRONIC LYMPHOCYTIC LEUKEMIA
BCR signaling plays a crucial role in the pathogenesis of CLL (92). Engagement and signaling by the BCR trigger pathways that govern the fate of normal or leukemia B cells. Activation of BCR recruits kinases, such as spleen tyrosine kinase (SYK) and the SRC kinase LYN, that phosphorylate immunoreceptor tyrosine–based activation motifs (ITAMs) of the accessory proteins of the BCR complex, namely CD79a and CD79b. Phosphorylation of the ITAMs of CD79a and CD79b allows for recruitment of adaptor proteins and other kinases, such as Bruton’s tyrosine kinase or phosphatidylinositol 3-kinase (PI3K), to the BCR accessory molecule complex. Activation of these signaling molecules leads to downstream activation of AKT/mTOR (mammalian target of rapamycin), NF-κB, and/or ERK (extracellular signal–regulated kinase). Studies on CLL cells demonstrated that BCR signaling can be enhanced by the ζ-associated protein of 70 kD (ZAP-70) (Figure 2) (93, 94), which is expressed by approximately half of all CLL cases, particularly those that have a relatively aggressive clinical disease and use unmutated IGHV genes. ZAP-70 can enhance BCR signaling in CLL not by depending on its kinase activity, but rather by facilitating recruitment of other kinases, such as SYK, to the BCR complex. In any case, the enhanced signaling potential afforded by expression of ZAP-70 probably accounts for its association with aggressive disease (95).
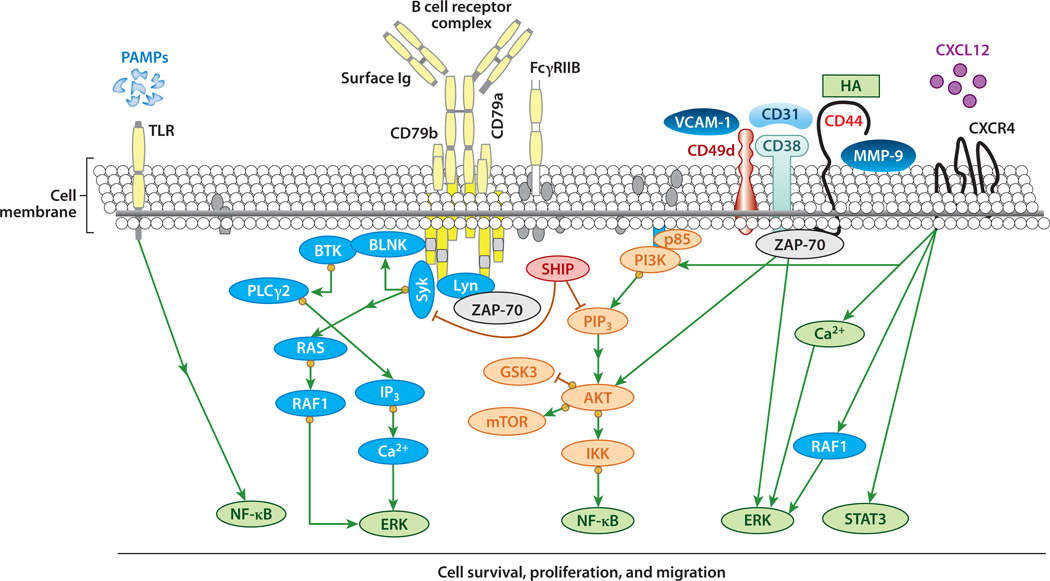
Figure 2.
B cell signaling in chronic lymphocytic leukemia (CLL). Ligation of B cell receptor (BCR) by antigen recruits kinases such as spleen tyrosine kinase (SYK) and the SRC kinase LYN that phosphorylate immunoreceptor tyrosine–based activation motifs (ITAMs) on the cytoplasmic domains of the immunoglobulin (Ig) coreceptors CD79a and CD79b. Such phosphorylation recruits and activates Bruton’s tyrosine kinase (BTK) and phosphatidylinositol 3-kinase (PI3K), subsequently activating many downstream targets, including AKT/mTOR (mammalian target of rapamycin), nuclear factor κB (NF-κB), and extracellular signal–regulated kinase (ERK). This signaling can be enhanced by ζ-associated protein of 70 kD (ZAP-70). CD38, CD49d, CD44, and matrix metalloproteinase (MMP)-9 may form a supramolecular complex with ZAP-70. This complex can also recruit ZAP-70 to the plasma membrane, where it can enhance BCR signaling. Following binding to any or all of these receptors by ligands released by accessory cells in the leukemia microenvironment, AKT and ERK undergo enhanced activation. CXCR4 can also directly interact with CXCL12 to induce calcium mobilization, activation of PI3K/AKT, ERK, and serine phosphorylation of signal transducer and activator of transcription 3 (STAT3). Activation of Toll-like receptor (TLR) can also enhance or induce activation of NF-κB. Abbreviations: GSK, glycogen synthase kinase; HA, hyaluronic acid; IKK, inhibitor of κB kinase; IP3, inositol triphosphate; PAMP, pathogen-associated molecular pattern; PIP3, phosphatidylinositol (3,4,5)-triphosphate; PLC, phospholipase C; SHIP, SH2-containing inositol phosphatase; VCAM, vascular cell adhesion molecule.
Stimulation of CLL via activation of BCR signaling or by coculture with accessory cells, such as nurselike cells, can enhance CLL cell expression of important chemokines, namely CCL3 and CCL4 (also known as MIP-1α and −1β, respectively) (96). Release of these chemokines by activated CLL cells attracts to the leukemia microenvironment additional accessory cells, such as regulatory T cells (97, 98). Activated CLL cells isolated from lymph nodes appear to express high levels of CLL3 and CCL4 (99), suggesting that BCR also plays an important role in creating the microenvironment that in turn supports CLL cell growth and/or survival.
CD38 is a marker of unfavorable prognosis and an indicator of activation and proliferation of CLL cells (100–102). A ligand for CD38 is CD31, which is expressed by nurselike cells and other accessory cells in the leukemia microenvironment (83). Ligation of CD38 on CLL cells can activate ZAP-70 and ERK1/2 signaling pathways to enhance CLL cell proliferation and chemotaxis (5, 84); interaction between CD38 and ZAP-70 may enhance BCR signaling (93, 84). More recent research has identified a macromolecular complex of CD38, CD49d, CD44, and matrix metalloproteinase (MMP)-9 on the CLL cells that express ZAP-70 (103, 104); this complex may promote additional cross talk between BCR signaling and CD44 (Figure 2).
CD44 is a receptor for hyaluronic acid (HA), which is found on the basement membranes of high endothelial venules of lymphoid tissue in the leukemia microenvironment. The interactions between CD38 and CD44, MMP-9, and CD49d may influence CLL cell migration, invasion, and homing. Thus, the enhanced signaling leading to growth or survival of CLL cells that is afforded by complexes composed of CD38, CD44, MMP-9, CD49d, and ZAP-70 may help explain why the expression of ZAP-70 and/or CD38 is associated with more aggressive disease.
The CXCR4 chemokine receptor CD184 is expressed at high levels on CLL cells isolated from blood (105). However, proliferating Ki-67+ CLL cells found in bone marrow or in lymphatic tissues express substantially lower levels of CXCR4 and CXCR5, probably reflecting chemokine-induced surface-receptor downmodulation of these chemokine receptors (106). CXCR4 signaling, which can be inhibited by pertussis toxin, induces calcium mobilization, activation of PI3Ks (49, 105) and p44/42 MAPKs (107), and STAT3 (serine phosphorylation of signal transducer and activator of transcription 3) (108). CXCR4 signaling plays a major role in leukemia cell chemotaxis and migration beneath cells that express the ligand for CXCR4, namely CXCL12 (105). CXCL12 also induces prosurvival signaling in CLL via CXCR4 (107, 109). Labeling studies involving ingestion of heavy water revealed high uptake by CLL cells with low levels of CXCR4 and high expression levels of CD38 and CD5 (110). Because BCR signaling can also downmodulate expression of CXCR4 (111, 112), the increased uptake of heavy water by CLL cells that express low levels of CXCR4 is consistent with a model in which chemokines and antigen ligation of the CLL cell BCR help drive leukemia cell growth in the characteristic proliferation centers found in the lymphoid tissues of CLL patients (99).
TLRs are cell-surface receptors that form part of the innate immune system; they bind structurally conserved microbial antigens and, in turn, activate innate immune responses. Recent studies have found that various TLRs are expressed and functional in CLL; stimulation of these TLRs could induce surface expression of activation markers such as CD25 and CD80 (113).
Given the role played by BCR signaling and the microenvironment in sustaining the growth or survival of CLL cells, the signaling pathways activated by these receptors represent new targets for therapy (92). Drugs that can inhibit BCR signaling, such as ibrutinib (formerly known as PCI-32765) and idelalisib (formerly known as GS-1101 or CAL-101), appear to exert significant activity in clinical trials involving CLL patients (92). In addition to their capacity to inhibit stimulation leading to leukemia cell proliferation and survival, such drugs can affect the capacity of CLL cells to home to the tissue microenvironment. Patients treated with such agents generally experience rapid reductions in lymph node size and spleen mass that are often associated with increased lymphocytosis (114). With continued treatment, the lymphocytosis may decrease, possibly reflecting the spontaneous cell death of leukemia cells caused by their inability to access survival/growth signals provided by the CLL microenvironment.
THE CHRONIC LYMPHOCYTIC LEUKEMIA MICROENVIRONMENT
The tissue microenvironment plays a central role in the pathogenesis of CLL (5). As mentioned above, whereas the vast majority of circulating CLL cells in blood are nondividing and resting, tissue CLL cells proliferate at a relatively high rate. CLL cell proliferation occurs in microanatomical sites termed proliferation centers or pseudofollicles (5, 115). Proliferating Ki-67+ CLL cells are in intimate contact with accessory cells, such as T cells (97, 98), stromal cells of mesenchymal origin (116), and/or monocyte-derived nurselike cells (117). Gene-expression studies have found that CLL cells isolated from lymph nodes and, to a lesser extent, the marrow have higher expression of E2F and MYC than do CLL cells in the blood (99). In such tissues, particularly in the secondary lymphoid tissues, there appears to be enhanced leukemia cell BCR signaling and activation of receptors of the TNF family, such as the transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI), B cell maturation antigen (BCMA), and B cell–activating factor (BAFF) receptor 3 (99; reviewed in Reference 5).
Chemokine receptors and adhesion molecules expressed by CLL cells are critical for homing and retention of CLL cells within the tissue compartments (118). CLL cells receive prosurvival signals via contact with accessory stromal cells in the leukemia microenvironment (5, 107, 119, 120). For example, nurselike cells express the chemokines CXCL12 and CXCL13, whereas marrow stromal cells express predominantly CXCL12. Nurselike cells and marrow stromal cells attract CLL cells via the G protein–coupled chemokine receptors CXCR4 and CXCR5, which are expressed at high levels on CLL cells. Nurselike cells also express the TNF family member BAFF and a proliferation-inducing ligand, providing survival signals to CLL cells via the corresponding receptors (BCMA, TACI, BAFF receptor). Integrins, particularly very late antigen 4 integrins (CD49d), expressed on the surface of CLL cells cooperate with chemokine receptors in establishing cell– cell adhesion through respective ligands on the stromal cells (vascular cell adhesion molecule 1 and fibronectin). Marrow stromal cells also express high levels of Wnt5a, which can promote CLL cell survival by stimulating signaling through ROR1 expressed by CLL cells (33).
Another important molecule involved in the interactions between leukemia cells and their microenvironment is CD44. CD44 is a receptor for HA that also interacts with other surface and cytosolic proteins expressed by CLL cells, such as CD38; CD49d; MMP-9; and, most recently, ZAP-70 (104). A humanized monoclonal antibody (mAb), RG7356, that is specific for CD44 has direct cytotoxic activity for CLL cells; however, RG7356 has little effect on normal B cells. The cytotoxic activity of this mAb was not mitigated when the CLL cells were cocultured with marrow stromal cells or stimulated with HA or BCR ligation (104). The selective toxicity in vitro and complete clearance of CLL cells by this mAb in vivo should encourage clinical evaluation of this mAb in the treatment of CLL patients.
CONCLUSION
The clinical heterogeneity of CLL patients appears to reflect the diversity of the molecular abnormalities that drive pathogenesis and progression of CLL. Through an improved understanding of the molecular genetics and cell biology of CLL, we now have new biomarkers with which to stratify patients, as well as new targets for the development of novel and more selective treatments for patients with this disease.
ACKNOWLEDGMENTS
The authors acknowledge Andrew Greaves for help in drawing figure images. Our work was supported by the National Cancer Institute grant P01CA081534 and the UC San Diego Foundation Blood Cancer Research Fund.
Glossary
- CLL
chronic lymphocytic leukemia
- Ig
immunoglobulin
- IGHV
Ig heavy-chain variable-region gene
- sMGZ
splenic marginal one
- MBL
monoclonal B cell lymphocytosis
- ROR1
receptor tyrosine kinase–like orphan receptor 1
- HSCs
hematopoietic stem cells
- BCR
B cell receptor
- MEACs
myosin-exposed apoptotic cells
- TNF
tumor necrosis factor
- TLR
Toll-like receptor
- WGS
whole-genome sequencing
- SYK
spleen tyrosine kinase
- ITAMs
immunoreceptor tyrosine–based activation motifs
- PI3K
phosphatidylinositol 3-kinase
- ZAP-70
ζ-associated protein of 70 kD
- mAb
monoclonal antibody
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N. Engl. J. Med. 2005;352:804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4th ed Vol. 2. Geneva, Switz: WHO; 2008. [Google Scholar]
- 3.Dighiero G, Travade P, Chevret S, Fenaux P, Chastang C, Binet JL. B-cell chronic lymphocytic leukemia: present status and future directions. French Cooperative Group on CLL. Blood. 1991;78:1901–1914. [PubMed] [Google Scholar]
- 4.Ponzoni M, Doglioni C, Caligaris-Cappio F. Chronic lymphocytic leukemia: the pathologist’s view of lymph node microenvironment. Semin. Diagn. Pathol. 2011;28:161–166. doi: 10.1053/j.semdp.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 5.Burger JA, Ghia P, Rosenwald A, Caligaris-Cappio F. The microenvironment in mature B cell malignancies: a target for new treatment strategies. Blood. 2009;114:3367–3375. doi: 10.1182/blood-2009-06-225326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ratech H, Sheibani K, Nathwani BN, Rappaport H. Immunoarchitecture of the “pseudofollicles” of well-differentiated (small) lymphocytic lymphoma: a comparison with true follicles. Hum. Pathol. 1988;19:89–94. doi: 10.1016/s0046-8177(88)80322-8. [DOI] [PubMed] [Google Scholar]
- 7.Schmid C, Isaacson PG. Proliferation centres in B cell malignant lymphoma, lymphocytic (B-CLL): an immunophenotypic study. Histopathology. 1994;24:445–451. doi: 10.1111/j.1365-2559.1994.tb00553.x. [DOI] [PubMed] [Google Scholar]
- 8.Soma LA, Craig FE, Swerdlow SH. The proliferation center microenvironment and prognostic markers in chronic lymphocytic leukemia/small lymphocytic lymphoma. Hum. Pathol. 2006;37:152–159. doi: 10.1016/j.humpath.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 9.Vandewoestyne ML, Pede VC, Lambein KY, Dhaenens MF, Offner FC, et al. Laser microdissection for the assessment of the clonal relationship between chronic lymphocytic leukemia/small lymphocytic lymphoma and proliferating B cells within lymph node pseudofollicles. Leukemia. 2011;25:883–888. doi: 10.1038/leu.2011.14. [DOI] [PubMed] [Google Scholar]
- 10.Messmer BT, Messmer D, Allen SL, Kolitz JE, Kudalkar P, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J. Clin. Investig. 2005;115:755–764. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zenz T, Mertens D, Kuppers R, Döhner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 12.Zenz T, Gribben JG, Hallek M, Döhner H, Keating MJ, Stilgenbauer S. Risk categories and refractory CLL in the era of chemoimmunotherapy. Blood. 2012;119:4101–4107. doi: 10.1182/blood-2011-11-312421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 14.Barrett-Connor E, Stuenkel C. Hormones and heart disease in women: Heart and Estrogen/Progestin Replacement Study in perspective. J. Clin. Endocrinol. Metab. 1999;84:1848–1853. doi: 10.1210/jcem.84.6.5667. [DOI] [PubMed] [Google Scholar]
- 15.Caligaris-Cappio F, Gobbi M, Bofill M, Janossy G. Infrequent normal B lymphocytes express features of B chronic lymphocytic leukemia. J. Exp. Med. 1982;155:623–628. doi: 10.1084/jem.155.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caligaris-Cappio F. B chronic lymphocytic leukemia: a malignancy of anti-self B cells. Blood. 1996;87:2615–2620. [PubMed] [Google Scholar]
- 17.Dono M, Burgio VL, Colombo M, Sciacchitano S, Reverberi D, et al. CD5+ B cells with the features of subepithelial B cells found in human tonsils. Eur. J. Immunol. 2007;37:2138–2147. doi: 10.1002/eji.200636887. [DOI] [PubMed] [Google Scholar]
- 18.Chiorazzi N, Ferrarini M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 2011;117:1781–1791. doi: 10.1182/blood-2010-07-155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forconi F, Potter KN, Wheatley I, Darzentas N, Sozzi E, et al. The normal IGHV1-69-derived B cell repertoire contains stereotypic patterns characteristic of unmutated CLL. Blood. 2010;115:71–77. doi: 10.1182/blood-2009-06-225813. [DOI] [PubMed] [Google Scholar]
- 20.Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, et al. Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells. J. Exp. Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J. Exp. Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–4398. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ha YJ, Mun YC, Seong CM, Lee JR. Characterization of phenotypically distinct B cell subsets and receptor-stimulated mitogen-activated protein kinase activation in human cord blood B cells. J. Leukoc. Biol. 2008;84:1557–1564. doi: 10.1189/jlb.0706457. [DOI] [PubMed] [Google Scholar]
- 24.Marie-Cardine A, Divay F, Dutot I, Green A, Perdrix A, et al. Transitional B cells in humans: characterization and insight from B lymphocyte reconstitution after hematopoietic stem cell transplantation. Clin. Immunol. 2008;127:14–25. doi: 10.1016/j.clim.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 25.Seifert M, Sellmann L, Bloehdorn J, Wein F, Stilgenbauer S, et al. Cellular origin and pathophysiology of chronic lymphocytic leukemia. J. Exp. Med. 2012;209:2183–2198. doi: 10.1084/jem.20120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rawstron AC, Yuille MR, Fuller J, Cullen M, Kennedy B, et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B lymphocyte expansion. Blood. 2002;100:2289–2290. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 27.Rawstron AC, Green MJ, Kuzmicki A, Kennedy B, Fenton JA, et al. Monoclonal B lymphocytes with the characteristics of “indolent” chronic lymphocytic leukemia are present in 3.5% of adults with normal blood counts. Blood. 2002;100:635–639. doi: 10.1182/blood.v100.2.635. [DOI] [PubMed] [Google Scholar]
- 28.Ghia P, Prato G, Scielzo C, Stella S, Geuna M, et al. Monoclonal CD5+ and CD5− B lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 2004;103:2337–2342. doi: 10.1182/blood-2003-09-3277. [DOI] [PubMed] [Google Scholar]
- 29.Rawstron AC, Bennett FL, O’Connor SJ, Kwok M, Fenton JA, et al. Monoclonal B cell lymphocytosis and chronic lymphocytic leukemia. N. Engl. J. Med. 2008;359:575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 30.Roulland S, Navarro JM, Grenot P, Milili M, Agopian J, et al. Follicular lymphoma–like B cells in healthy individuals: a novel intermediate step in early lymphomagenesis. J. Exp. Med. 2006;203:2425–2431. doi: 10.1084/jem.20061292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baskar S, Kwong KY, Hofer T, Levy JM, Kennedy MG, et al. Unique cell surface expression of receptor tyrosine kinase ROR1 in human B cell chronic lymphocytic leukemia. Clin. Cancer Res. 2008;14:396–404. doi: 10.1158/1078-0432.CCR-07-1823. [DOI] [PubMed] [Google Scholar]
- 32.Daneshmanesh AH, Mikaelsson E, Jeddi-Tehrani M, Bayat AA, Ghods R, et al. ROR1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int. J. Cancer. 2008;123:1190–1195. doi: 10.1002/ijc.23587. [DOI] [PubMed] [Google Scholar]
- 33.Fukuda T, Chen L, Endo T, Tang L, Lu D, et al. Antisera induced by infusions of autologous Ad-CD154-leukemia B cells identify ROR1 as an oncofetal antigen and receptor for Wnt5a. Proc. Natl. Acad. Sci. USA. 2008;105:3047–3052. doi: 10.1073/pnas.0712148105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Chen L, Cui B, Chuang HY, Yu J, et al. ROR1 is expressed in human breast cancer and associated with enhanced tumor-cell growth. PLoS ONE. 2012;7:e31127. doi: 10.1371/journal.pone.0031127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang S, Chen L, Wang-Rodriguez J, Zhang L, Cui B, et al. The onco-embryonic antigen ROR1 is expressed by a variety of human cancers. Am. J. Pathol. 2012;181:1903–1910. doi: 10.1016/j.ajpath.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broome HE, Rassenti LZ, Wang HY, Meyer LM, Kipps TJ. ROR1 is expressed on hematogones (non-neoplastic human B lymphocyte precursors) and a minority of precursor-B acute lymphoblastic leukemia. Leuk. Res. 2011;35:1390–1394. doi: 10.1016/j.leukres.2011.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sevilla DW, Colovai AI, Emmons FN, Bhagat G, Alobeid B. Hematogones: a review and update. Leuk. Lymphoma. 2010;51:10–19. doi: 10.3109/10428190903370346. [DOI] [PubMed] [Google Scholar]
- 38.Milleck J, Kreuzer K-A, Med D, Assenmacher M, Schmitz J, Jaha PS. Characterization of peripheral B cells expressing CLL-associated receptor tyrosine kinase ROR1 in healthy donors; Presented at Meet. Am. Soc. Hematol.Atlanta: 2012. [Google Scholar]
- 39.Kikushige Y, Ishikawa F, Miyamoto T, Shima T, Urata S, et al. Self-renewing hematopoietic stem cell is the primary target in pathogenesis of human chronic lymphocytic leukemia. Cancer Cell. 2011;20:246–259. doi: 10.1016/j.ccr.2011.06.029. [DOI] [PubMed] [Google Scholar]
- 40.Damle RN, Batliwalla FM, Ghiotto F, Valetto A, Albesiano E, et al. Telomere length and telomerase activity delineate distinctive replicative features of the B-CLL subgroups defined by immunoglobulin V gene mutations. Blood. 2004;103:375–382. doi: 10.1182/blood-2003-04-1345. [DOI] [PubMed] [Google Scholar]
- 41.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Widhopf GF, 2nd, Goldberg CJ, Toy TL, Rassenti LZ, Wierda WG, et al. Nonstochastic pairing of immunoglobulin heavy and light chains expressed by chronic lymphocytic leukemia B cells is predicated on the heavy chain CDR3. Blood. 2008;111:3137–3144. doi: 10.1182/blood-2007-02-073130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kipps TJ. Immunoglobulin genes in chronic lymphocytic leukemia. [discussion 631-32];Blood Cells. 1993 19:615–25. [PubMed] [Google Scholar]
- 44.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu. Rev. Immunol. 2003;21:841–894. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 45.Stevenson FK, Caligaris-Cappio F. Chronic lymphocytic leukemia: revelations from the B cell receptor. Blood. 2004;103:4389–4395. doi: 10.1182/blood-2003-12-4312. [DOI] [PubMed] [Google Scholar]
- 46.Fais F, Ghiotto F, Hashimoto S, Sellars B, Valetto A, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J. Clin. Investig. 1998;102:1515–1525. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Myhrinder AL, Hellqvist E, Sidorova E, Söderberg A, Baxendale H, et al. A new perspective: Molecular motifs on oxidized LDL, apoptotic cells, and bacteria are targets for chronic lymphocytic leukemia antibodies. Blood. 2008;111:3838–3848. doi: 10.1182/blood-2007-11-125450. [DOI] [PubMed] [Google Scholar]
- 48.Catera R, Silverman GJ, Hatzi K, Seiler T, Didier S, et al. Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol. Med. 2008;14:665–674. doi: 10.2119/2008-00102.Catera. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu CC, Catera R, Hatzi K, Yan XJ, Zhang L, et al. Chronic lymphocytic leukemia antibodies with a common stereotypic rearrangement recognize nonmuscle myosin heavy chain IIA. Blood. 2008;112:5122–5129. doi: 10.1182/blood-2008-06-162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seiler T, Woelfle M, Yancopoulos S, Catera R, Li W, et al. Characterization of structurally defined epitopes recognized by monoclonal antibodies produced by chronic lymphocytic leukemia B cells. Blood. 2009;114:3615–3624. doi: 10.1182/blood-2009-01-197822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chu CC, Catera R, Zhang L, Didier S, Agagnina BM, et al. Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: implications for patient outcome and cell of origin. Blood. 2010;115:3907–3915. doi: 10.1182/blood-2009-09-244251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatopoulos K. Antigens in CLL: themes and variations. Blood. 2010;115:3855–3856. doi: 10.1182/blood-2010-02-270249. [DOI] [PubMed] [Google Scholar]
- 53.Landgren O, Rapkin JS, Caporaso NE, Mellemkjaer L, Gridley G, et al. Respiratory tract infections and subsequent risk of chronic lymphocytic leukemia. Blood. 2007;109:2198–2201. doi: 10.1182/blood-2006-08-044008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kostareli E, Hadzidimitriou A, Stavroyianni N, Darzentas N, Athanasiadou A, et al. Molecular evidence for EBV and CMV persistence in a subset of patients with chronic lymphocytic leukemia expressing stereotyped IGHV4-34 B cell receptors. Leukemia. 2009;23:919–924. doi: 10.1038/leu.2008.379. [DOI] [PubMed] [Google Scholar]
- 55.Steininger C, Rassenti LZ, Vanura K, Eigenberger K, Jager U, et al. Relative seroprevalence of human herpes viruses in patients with chronic lymphocytic leukaemia. Eur. J. Clin. Investig. 2009;39:497–506. doi: 10.1111/j.1365-2362.2009.02131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Steininger C, Widhopf GF, 2nd, Ghia EM, Morello CS, Vanura K, et al. Recombinant antibodies encoded by IGHV1-69 react with pUL32, a phosphoprotein of cytomegalovirus and B cell superantigen. Blood. 2012;119:2293–2301. doi: 10.1182/blood-2011-08-374058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kostareli E, Gounari M, Janus A, Murray F, Brochet X, et al. Antigen receptor stereotypy across B cell lymphoproliferations: the case of IGHV4-59/IGKV3-20 receptors with rheumatoid factor activity. Leukemia. 2012;26:1127–1131. doi: 10.1038/leu.2011.311. [DOI] [PubMed] [Google Scholar]
- 58.Gaidano G, Foa R, Dalla-Favera R. Molecular pathogenesis of chronic lymphocytic leukemia. J. Clin. Investig. 2012;122:3432–3438. doi: 10.1172/JCI64101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Döhner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 60.Caporaso N, Goldin L, Plass C, Calin G, Marti G, et al. Chronic lymphocytic leukaemia genetics overview. Br. J. Haematol. 2007;139:630–634. doi: 10.1111/j.1365-2141.2007.06846.x. [DOI] [PubMed] [Google Scholar]
- 61.Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J. Exp. Med. 2011;208:1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Puente XS, Pinyol M, Quesada V, Conde L, Ordóñez GR, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quesada V, Conde L, Villamor N, Ordóñez GR, Jares P, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2012;44:47–52. doi: 10.1038/ng.1032. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, Lawrence MS, Wan Y, Stojanov P, Sougnez C, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011;365:2497–2506. doi: 10.1056/NEJMoa1109016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossi D, Bruscaggin A, Spina V, Rasi S, Khiabanian H, et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: association with progression and fludarabine refractoriness. Blood. 2011;118:6904–6908. doi: 10.1182/blood-2011-08-373159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rossi D, Rasi S, Fabbri G, Spina V, Fangazio M, et al. Mutations of NOTCH1 are an independent predictor of survival in chronic lymphocytic leukemia. Blood. 2012;119:521–529. doi: 10.1182/blood-2011-09-379966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rossi D, Fangazio M, Rasi S, Vaisitti T, Monti S, et al. Disruption of BIRC3 associates with fludarabine chemorefractoriness in TP53 wild-type chronic lymphocytic leukemia. Blood. 2012;119:2854–2862. doi: 10.1182/blood-2011-12-395673. [DOI] [PubMed] [Google Scholar]
- 68.Rothenberg EV. T cell lineage commitment: identity and renunciation. J. Immunol. 2011;186:6649–6655. doi: 10.4049/jimmunol.1003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Santos MA, Sarmento LM, Rebelo M, Doce AA, Maillard I, et al. Notch1 engagement by Delta-like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proc. Natl. Acad. Sci. USA. 2007;104:15454–15459. doi: 10.1073/pnas.0702891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ferrando AA. The role of NOTCH1 signaling in T-ALL. Hematol. Am. Soc. Hematol. Educ. Program. 2009:353–361. doi: 10.1182/asheducation-2009.1.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Balatti V, Bottoni A, Palamarchuk A, Alder H, Rassenti LZ, et al. NOTCH1 mutations in CLL associated with trisomy 12. Blood. 2012;119:329–331. doi: 10.1182/blood-2011-10-386144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Del Giudice I, Rossi D, Chiaretti S, Marinelli M, Tavolaro S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2012;97:437–441. doi: 10.3324/haematol.2011.060129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 74.Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schwaederlé M, Ghia E, Rassenti LZ, Obara M, Dell’Aquila ML, et al. Subclonal evolution involving SF3B1 mutations in chronic lymphocytic leukemia. Leukemia. 2013;27:1214–1217. doi: 10.1038/leu.2013.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vallabhapurapu S, Karin M. Regulation and function of NF-κB transcription factors in the immune system. Annu. Rev. Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 77.Endo T, Nishio M, Enzler T, Cottam HB, Fukuda T, et al. BAFF and APRIL support chronic lymphocytic leukemia B cell survival through activation of the canonical NF-κB pathway. Blood. 2007;109:703–710. doi: 10.1182/blood-2007-04-081786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Enzler T, Kater AP, Zhang W, Widhopf GF, 2nd, Chuang HY, et al. Chronic lymphocytic leukemia of Emu-TCL1 transgenic mice undergoes rapid cell turnover that can be offset by extrinsic CD257 to accelerate disease progression. Blood. 2009;114:4469–4476. doi: 10.1182/blood-2009-06-230169. [DOI] [PubMed] [Google Scholar]
- 79.Zhang W, Kater AP, Widhopf GF, 2nd, Chuang HY, Enzler T, et al. B cell activating factor and v-Myc myelocytomatosis viral oncogene homolog (c-Myc) influence progression of chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2010;107:18956–18960. doi: 10.1073/pnas.1013420107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hewamana S, Lin TT, Jenkins C, Burnett AK, Jordan CT, et al. The novel nuclear factor κB inhibitor LC-1 is equipotent in poor prognostic subsets of chronic lymphocytic leukemia and shows strong synergy with fludarabine. Clin. Cancer Res. 2008;14:8102–8111. doi: 10.1158/1078-0432.CCR-08-1673. [DOI] [PubMed] [Google Scholar]
- 81.Hertlein E, Wagner AJ, Jones J, Lin TS, Maddocks KJ, et al. 17-DMAG targets the nuclear factor κB family of proteins to induce apoptosis in chronic lymphocytic leukemia: clinical implications of HSP90 inhibition. Blood. 2010;116:45–53. doi: 10.1182/blood-2010-01-263756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Treon SP, Xu L, Yang G, Zhou Y, Liu X, et al. MYD88 L265P somatic mutation in Waldenström’s macroglobulinemia. N. Engl. J. Med. 2012;367:826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 83.Deaglio S, Vaisitti T, Bergui L, Bonello L, Horenstein AL, et al. CD38 and CD100 lead a network of surface receptors relaying positive signals for B-CLL growth and survival. Blood. 2005;105:3042–3050. doi: 10.1182/blood-2004-10-3873. [DOI] [PubMed] [Google Scholar]
- 84.Malavasi F, Deaglio S, Damle R, Cutrona G, Ferrarini M, Chiorazzi N. CD38 and chronic lymphocytic leukemia: a decade later. Blood. 2011;118:3470–3478. doi: 10.1182/blood-2011-06-275610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470:115–119. doi: 10.1038/nature09671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gutierrez A, Jr, Tschumper RC, Wu X, Shanafelt TD, Eckel-Passow J, et al. LEF-1 is a prosurvival factor in chronic lymphocytic leukemia and is expressed in the preleukemic state of monoclonal B cell lymphocytosis. Blood. 2010;116:2975–2983. doi: 10.1182/blood-2010-02-269878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lu D, Zhao Y, Tawatao R, Cottam HB, Sen M, et al. Activation of the Wnt signaling pathway in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 2004;101:3118–3123. doi: 10.1073/pnas.0308648100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu D, Liu JX, Endo T, Zhou H, Yao S, et al. Ethacrynic acid exhibits selective toxicity to chronic lymphocytic leukemia cells by inhibition of the Wnt/β-catenin pathway. PLoS ONE. 2009;4:e8294. doi: 10.1371/journal.pone.0008294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lu D, Choi MY, Yu J, Castro JE, Kipps TJ, Carson DA. Salinomycin inhibits Wnt signaling and selectively induces apoptosis in chronic lymphocytic leukemia cells. Proc. Natl. Acad. Sci. USA. 2011;108:13253–13257. doi: 10.1073/pnas.1110431108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu CJ. CLL clonal heterogeneity: an ecology of competing subpopulations. Blood. 2012;120:4117–4118. doi: 10.1182/blood-2012-09-452805. [DOI] [PubMed] [Google Scholar]
- 91.Schuh A, Becq J, Humphray S, Alexa A, Burns A, et al. Monitoring chronic lymphocytic leukemia progression by whole genome sequencing reveals heterogeneous clonal evolution patterns. Blood. 2012;120:4191–4196. doi: 10.1182/blood-2012-05-433540. [DOI] [PubMed] [Google Scholar]
- 92.Woyach JA, Johnson AJ, Byrd JC. The B cell receptor signaling pathway as a therapeutic target in CLL. Blood. 2012;120:1175–1184. doi: 10.1182/blood-2012-02-362624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chen L, Apgar J, Huynh L, Dicker F, Giago-McGahan T, et al. ZAP-70 directly enhances IgM signaling in chronic lymphocytic leukemia. Blood. 2005;105:2036–2041. doi: 10.1182/blood-2004-05-1715. [DOI] [PubMed] [Google Scholar]
- 94.Chen L, Huynh L, Apgar J, Tang L, Rassenti L, et al. ZAP-70 enhances IgM signaling independent of its kinase activity in chronic lymphocytic leukemia. Blood. 2008;111:2685–2692. doi: 10.1182/blood-2006-12-062265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dühren-von Minden M, Übelhart R, Schneider D, Wossning T, Bach MP, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489:309–312. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
- 96.Burger JA, Quiroga MP, Hartmann E, Burkle A, Wierda WG, et al. High-level expression of the T cell chemokines CCL3 and CCL4 by chronic lymphocytic leukemia B cells in nurselike cell cocultures and after BCR stimulation. Blood. 2009;113:3050–3058. doi: 10.1182/blood-2008-07-170415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Patten PE, Buggins AG, Richards J, Wotherspoon A, Salisbury J, et al. CD38 expression in chronic lymphocytic leukemia is regulated by the tumor microenvironment. Blood. 2008;111:5173–5181. doi: 10.1182/blood-2007-08-108605. [DOI] [PubMed] [Google Scholar]
- 98.Ghia P, Strola G, Granziero L, Geuna M, Guida G, et al. Chronic lymphocytic leukemia B cells are endowed with the capacity to attract CD4+, CD40L+ T cells by producing CCL22. Eur. J. Immunol. 2002;32:1403–1413. doi: 10.1002/1521-4141(200205)32:5<1403::AID-IMMU1403>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 99.Herishanu Y, Pérez-Galán P, Liu D, Biancotto A, Pittaluga S, et al. The lymph node microen-vironment promotes B cell receptor signaling, NF-κB activation, and tumor proliferation in chronic lymphocytic leukemia. Blood. 2011;117:563–574. doi: 10.1182/blood-2010-05-284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ibrahim S, Keating M, Do KA, O’Brien S, Huh YO, et al. CD38 expression as an important prognostic factor in B cell chronic lymphocytic leukemia. Blood. 2001;98:181–186. doi: 10.1182/blood.v98.1.181. [DOI] [PubMed] [Google Scholar]
- 101.Damle RN, Temburni S, Calissano C, Yancopoulos S, Banapour T, et al. CD38 expression labels an activated subset within chronic lymphocytic leukemia clones enriched in proliferating B cells. Blood. 2007;110:3352–3359. doi: 10.1182/blood-2007-04-083832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Deaglio S, Aydin S, Grand MM, Vaisitti T, Bergui L, et al. CD38/CD31 interactions activate genetic pathways leading to proliferation and migration in chronic lymphocytic leukemia cells. Mol. Med. 2010;16:87–91. doi: 10.2119/molmed.2009.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Buggins AG, Levi A, Gohil S, Fishlock K, Patten PE, et al. Evidence for a macromolecular complex in poor prognosis CLL that contains CD38, CD49d, CD44 and MMP-9. Br. J. Haematol. 2011;154:216–222. doi: 10.1111/j.1365-2141.2011.08725.x. [DOI] [PubMed] [Google Scholar]
- 104.Zhang S, Wu CCN, Fecteau J-F, Cui B, Chen L, et al. Targeting chronic lymphocytic leukemia cells with a humanized monoclonal antibody specific for CD44. Proc. Natl. Acad. Sci. USA. 2013;110:6127–6132. doi: 10.1073/pnas.1221841110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Burger JA, Burger M, Kipps TJ. Chronic lymphocytic leukemia B cells express functional CXCR4 chemokine receptors that mediate spontaneous migration beneath bone marrow stromal cells. Blood. 1999;94:3658–3667. [PubMed] [Google Scholar]
- 106.Bennett F, Rawstron A, Plummer M, de Tute R, Moreton P, et al. B cell chronic lymphocytic leukaemia cells show specific changes in membrane protein expression during different stages of cell cycle. Br. J. Haematol. 2007;139:600–604. doi: 10.1111/j.1365-2141.2007.06790.x. [DOI] [PubMed] [Google Scholar]
- 107.Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell’Aquila M, Kipps TJ. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell–derived factor 1. Blood. 2000;96:2655–2663. [PubMed] [Google Scholar]
- 108.Burger M, Hartmann T, Krome M, Rawluk J, Tamamura H, et al. Small peptide inhibitors of the CXCR4 chemokine receptor (CD184) antagonize the activation, migration, and antiapoptotic responses of CXCL12 in chronic lymphocytic leukemia B cells. Blood. 2005;106:1824–1830. doi: 10.1182/blood-2004-12-4918. [DOI] [PubMed] [Google Scholar]
- 109.Nishio M, Endo T, Tsukada N, Ohata J, Kitada S, et al. Nurselike cells express BAFF and APRIL, which can promote survival of chronic lymphocytic leukemia cells via a paracrine pathway distinct from that of SDF-1α. Blood. 2005;106:1012–1020. doi: 10.1182/blood-2004-03-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Calissano C, Damle RN, Hayes G, Murphy EJ, Hellerstein MK, et al. In vivo intraclonal and interclonal kinetic heterogeneity in B cell chronic lymphocytic leukemia. Blood. 2009;114:4832–4842. doi: 10.1182/blood-2009-05-219634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Quiroga MP, Balakrishnan K, Kurtova AV, Sivina M, Keating MJ, et al. B cell antigen receptor signaling enhances chronic lymphocytic leukemia cell migration and survival: specific targeting with a novel spleen tyrosine kinase inhibitor, R406. Blood. 2009;114:1029–1037. doi: 10.1182/blood-2009-03-212837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vlad A, Deglesne PA, Letestu R, Saint-Georges S, Chevallier N, et al. Down-regulation of CXCR4 and CD62L in chronic lymphocytic leukemia cells is triggered by B cell receptor ligation and associated with progressive disease. Cancer Res. 2009;69:6387–6395. doi: 10.1158/0008-5472.CAN-08-4750. [DOI] [PubMed] [Google Scholar]
- 113.Muzio M, Scielzo C, Bertilaccio MT, Frenquelli M, Ghia P, Caligaris-Cappio F. Expression and function of Toll-like receptors in chronic lymphocytic leukaemia cells. Br. J. Haematol. 2009;144:507–516. doi: 10.1111/j.1365-2141.2008.07475.x. [DOI] [PubMed] [Google Scholar]
- 114.Burger JA, Montserrat E. Coming full circle: 70 years of chronic lymphocytic leukemia cell redistribution, from glucocorticoids to inhibitors of B cell receptor signaling. Blood. 2012;121:1501–1509. doi: 10.1182/blood-2012-08-452607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stein H, Bonk A, Tolksdorf G, Lennert K, Rodt H, Gerdes J. Immunohistologic analysis of the organization of normal lymphoid tissue and non-Hodgkin’s lymphomas. J. Histochem. Cytochem. 1980;28:746–760. doi: 10.1177/28.8.7003001. [DOI] [PubMed] [Google Scholar]
- 116.Ruan J, Hyjek E, Kermani P, Christos PJ, Hooper AT, et al. Magnitude of stromal hemangiogenesis correlates with histologic subtype of non-Hodgkin’s lymphoma. Clin. Cancer Res. 2006;12:5622–5631. doi: 10.1158/1078-0432.CCR-06-1204. [DOI] [PubMed] [Google Scholar]
- 117.Tsukada N, Burger JA, Zvaifler NJ, Kipps TJ. Distinctive features of “nurselike” cells that differentiate in the context of chronic lymphocytic leukemia. Blood. 2002;99:1030–1037. doi: 10.1182/blood.v99.3.1030. [DOI] [PubMed] [Google Scholar]
- 118.Burger JA. Chemokines and chemokine receptors in chronic lymphocytic leukemia (CLL): from understanding the basics towards therapeutic targeting. Semin. Cancer Biol. 2010;20:424–430. doi: 10.1016/j.semcancer.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 119.Panayiotidis P, Jones D, Ganeshaguru K, Foroni L, Hoffbrand AV. Human bone marrow stromal cells prevent apoptosis and support the survival of chronic lymphocytic leukaemia cells in vitro. Br. J. Haematol. 1996;92:97–103. doi: 10.1046/j.1365-2141.1996.00305.x. [DOI] [PubMed] [Google Scholar]
- 120.Lagneaux L, Delforge A, Bron D, De Bruyn C, Stryckmans P. Chronic lymphocytic leukemic B cells but not normal B cells are rescued from apoptosis by contact with normal bone marrow stromal cells. Blood. 1998;91:2387–2396. [PubMed] [Google Scholar]