Abstract
Immunological tests provide evidence of latent tuberculosis in one third of the global population, more than two billion individuals. Latent tuberculosis is defined by the absence of clinical symptoms but carries a risk of subsequent progression to clinical disease, particularly in the context of co-infection with HIV. Here we discuss the biology of latent tuberculosis as part of a broad spectrum of responses that occur following infection with Mycobacterium tuberculosis, resulting in formation of a range of physiologically distinct granulomatous lesions that provide environments with differential ability to support or suppress persistence of viable bacteria. We go on to show how this model can be used to inform a rational programme to discover drugs that will be effective in the eradication of M. tuberculosis infection.
Mycobacterium tuberculosis has evolved a highly efficient means of aerosol transmission that exploits immune-mediated damage to the lungs of individuals with active disease. Successful transmission can be detected by an antigen-specific T-cell response amongst exposed contacts, and its extent is reflected in the estimate that one third of the global population has developed such a response. Around one in ten of this infected population develop active tuberculosis (TB), most commonly within a few years after exposure but retaining a lifetime risk. This risk is significantly increased by immunosuppressive triggers including HIV infection, anti-TNF therapy for other diseases, and diabetes, and can be decreased by prolonged preventive therapy with isoniazid1. While current strategies for TB control focus on attempts to reduce transmission by prompt identification and treatment of infectious cases, it will be necessary to combine this with interventions to reduce the development of disease in the infected population if we are to approach the aim of TB elimination by 20502 (Box 1). In this review, we discuss strategies for the development and application of drugs to achieve this goal.
Box 1. The Global Plan to Stop TB.
Approximately 9 million people develop TB every year, resulting in almost 2 million deaths. Infection is spread by individuals with active disease, particularly “smear-positive” disease characterised by high numbers of bacteria in sputum. Infectiousness drops rapidly after initiation of drug treatment, and prompt diagnosis and effective therapy are the major goals of current TB control programmes. Epidemiological modelling suggests that detection of 70% of smear-positive cases, together with an 85% cure rate, will result in a self-limiting decline in incidence. The Global Plan to Stop TB aims to halve the prevalence and mortality of TB by 2015, in line with the UN Millennium Development Goals. A longer term aim of the Stop TB Partnership is to eliminate TB (defined as an incidence of less than 1 per million of the global population) by 2050. To achieve this goal, reduced transmission by detection and treatment will have to be combined with interventions that prevent progression to disease following exposure. Treatment of latent TB with isoniazid (INH) for nine months has been shown to reduce the risk of disease significantly; drugs that could achieve this goal by more feasible short-term therapy would provide a potent new weapon for TB control.
The tuberculosis spectrum
Latent TB is defined solely by evidence of immunological sensitisation by mycobacterial proteins in the absence of clinical signs and symptoms of active disease. Until recently sensitisation has been defined by reactivity in the tuberculin skin test (TST). A deficit of the TST is that the skin test reagent, purified protein derivative (PPD), contains cross-reactive antigens that are also present in non-pathogenic mycobacteria and so false positives can occur, for example, in those previously immunised with the Bacille Calmette Guérin (BCG) vaccine. A strength is that the prognostic value of recent conversion to positive in the TST is well-recognised3. A major disadvantage however is that such conversion is often absent in those at greatest risk of disseminated TB: HIV infected persons and children. Recently, more specific tests of sensitisation that rely on the in vitro release of interferon-gamma (IGRA) in response to M. tuberculosis antigens of greater specificity have been developed. There is consensus that these tests are less affected by prior exposure to BCG but their role in children and HIV infected persons remains debated4. Deficits common to both tests are that they do not distinguish latent infection from active disease, nor do they provide any direct evidence of the presence of viable bacilli: they determine simply that infection has at some time led to an acquired immune response that is detectable upon rechallenge with antigen.
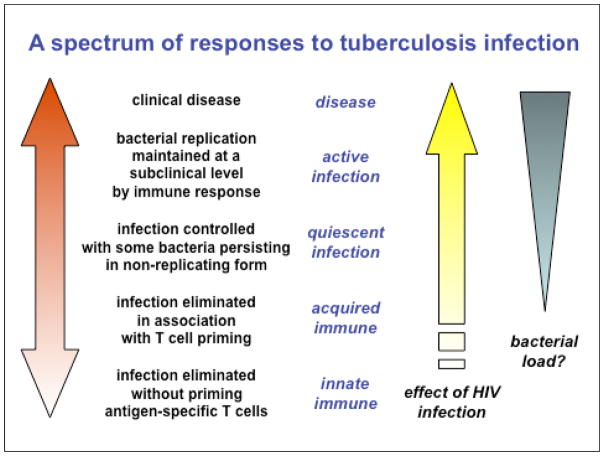
As a consequence of this purely clinical definition, TB is commonly conceptualised as a simple binary distribution between active disease and latent infection. While latent TB is generally loosely equated with bacterial containment in some inactive form, the current categorisation will include a diverse range of individuals from those who have completely cleared the infection to individuals who are incubating actively replicating bacteria in the absence of clinical symptoms5. Similarly, active TB in humans and non-human primates is characterised by diverse pathological presentations ranging from sterile tissue, through caseous hypoxic lesions containing variable numbers of bacteria, to liquefied cavities with a massive load of replicating organisms: it is attractive to propose that the clinical diversity reflects the relative numbers, type and anatomical distribution of lesions. M. tuberculosis infection may therefore be better viewed as a continuous spectrum extending from sterilising immunity to subclinical active disease through to fulminant active disease, with conventional designations of “latent infection” and “active disease” corresponding to partially overlapping regions of biological heterogeneity5 (Figure 1). HIV infection skews this distribution in favour of the bacillus. The greatly increased risk of disseminated disease in HIV infected persons is well-known but there is also an increasing recognition of very frequent subclinical active infection where this has been investigated by prevalence surveys6.
Figure 1. TB infection as a spectrum.
The outcome of infection by M. tuberculosis is generally represented as a bimodal distribution between active TB and latent TB on the basis of the presence or absence of clinical symptoms. We propose that latent TB is more usefully represented as part of a spectrum of responses to infection. One consequence of this model is that there may be a subpopulation within the group currently defined as having latent TB who should be preferentially targeted for preventive therapy. A second consequence is that efforts to develop drugs for effective treatment of latent TB would overlap the search for drugs that would shorten treatment times for active TB.
Two important consequences follow from this model. First, it can be proposed that treatment of latent infection will be most effective when directed towards the part of the spectrum corresponding to those at the highest risk of progression to disease, rather than to the entire two billion immunologically-positive people. Second, drug development strategies that target bacterial populations on the basis of physiological requirements for persistence in different lesion types may be equally applicable to preventive therapy of latent infection and to improved treatment of active disease.
Characterisation of TB lesions: new insights from imaging
Pathologists analysing post-mortem tissues from humans have studied latent TB lesions extensively. The latent lesions they have characterized represent a subset of the heterogeneous lesions seen in active disease, with wide variations reported in the recovery of viable bacteria7. Exciting new opportunities are provided by recent advances in modelling latency in cynomolgus macaque monkeys8, 9 particularly when combined with high-resolution computed tomography (CT) and positron emission tomography (PET). CT provides structural data on the lung and lymph nodes, including a spatial map of granulomas (Figure 2). PET probes mark areas based on the function or phenotype of cells and processes within the tissue of interest. For example, the most commonly used PET probe in human cancer diagnosis is 18F-fluorodeoxyglucose (FDG), which labels metabolically active cells. Integrating the CT and PET images provides a detailed structural and functional map of the tuberculous disease process in the lungs.
Figure 2. PET/CT imaging.
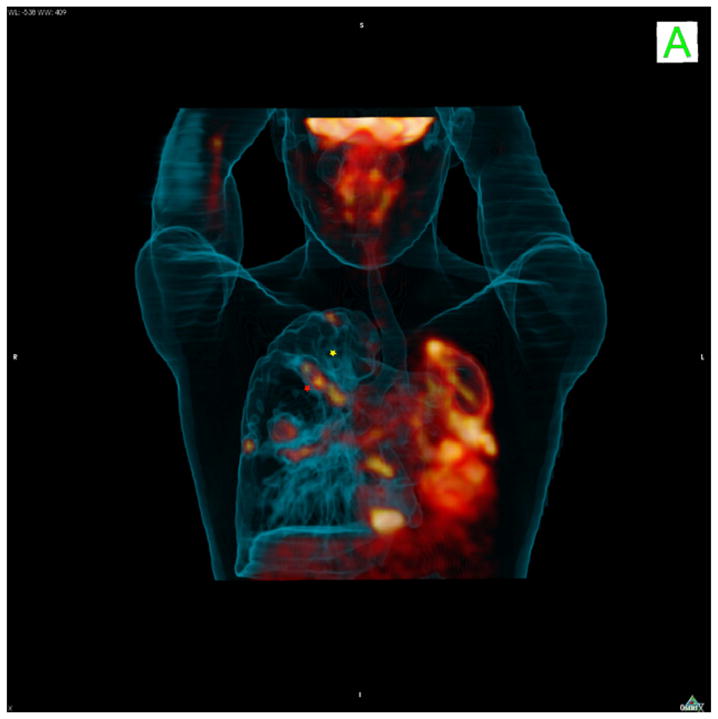
An [18F]-FDG-PET-CT scan of a human tuberculosis patient with extensive bilateral disease and a complete collapse of the left lung. The right lung also shows extensive disease throughout and illustrates the variability of FDG-PET uptake amongst lesions within even a single infected patient. The yellow star illustrates one lesion that fails to take up FDG that lies immediately adjacent to a string of three lesions that take up label avidly (red star). These different types of lesions respond with very different kinetics to chemotherapy suggesting they represent distinct bacterial subpopulations in different microenvironments.
The power of PET/CT imaging has rarely been applied to the study of infectious diseases, either in humans or in animal models. However, we can learn from studies in cancer and neurobiology. The very few reports of PET/CT imaging obtained from human latent TB patients are invariably concerned with the diagnostic dilemma of differentiating latent tuberculosis from malignancies10–12. A comparison of the CT findings from latent TB patients with those of active TB patients confirms the pathological results – latent TB includes a spectrum of CT findings representing a subset of those seen in active disease10. The lesions that are most often reported are solitary pulmonary nodules ranging from a few mm to 20mm in size, although linear branching and consolidations have also been found10. This probably represents selection bias because these lesions represent the biggest challenge to the diagnostic radiologist who must make a determination of TB or cancer. FDG-PET has been explored for its utility in improving the diagnostic accuracy of such exams and as a result some limited information is available regarding latent TB lesions. In such cases the conclusion has been that TB lesions are relatively difficult to distinguish from malignancy based on the uptake of FDG, which varies tremendously among lesions. In at least two patients with latent TB, FDG uptake was shown to be reduced following antituberculous chemotherapy13. Even based on this very limited sample the conclusion that latent infection represents a broad spectrum of conditions that overlaps with those seen in active disease may be made. In some patients this may represent a slowly progressing form of the disease, in others a chronic non-progressing “percolating” infection, and in others the remnant of a waning infection. It remains to be seen if any of these are truly the metastable “dormant” bacilli postulated to represent latency.
These studies represent only the beginning of what is possible using functional imaging. There are numerous other PET probes that target different types of cells and processes, including hypoxia, cellular proliferation, and angiogenesis. These probes can provide additional information about the lesions and the process of disease establishment, progression and treatment. PET/CT technology allows detailed analysis of changes in individual tuberculous lesions over time, and monitoring of the response of these lesions to treatment. Combining live imaging with analysis of resected and post-mortem tissues provides the opportunity to generate lesion-based profiles of TB-infected humans and animals and to map lesion categories defined by imaging to cellular and molecular parameters.
Important key factors in distinguishing among lesions include the response to treatment with existing and novel drugs, immune cells and responses, bacterial gene expression, the presence of hypoxic regions, and drug permeability and retention. Early studies dating from the ‘50s to ‘80s report the levels of rifampicin and isoniazid, the two most effective TB drugs, in different lesion types and lesion compartments, relative to plasma14–16. Though the readouts used at the time have intrinsic limitations in terms of sensitivity and accuracy, these studies clearly indicate that only a small fraction of the drug present in plasma is able to reach some of the lesion compartments, and the rate of penetration into diseased tissue and sequestered infection sites is both drug-specific and lesion-specific, in agreement with what has been reported for non-TB drugs in abscess fluid17, 18. There is a clear opportunity to refine and expand drug penetration studies in human and human-like TB lesions in animals, by taking advantage of modern, innovative and state of the art technologies for the quantification of small molecules. Modelling of the pharmacokinetics of 1st and 2nd line drugs in plasma, lung tissue and various lesion types will allow one to calculate coefficients of penetration for each drug in each lesion type, providing some insights with regard to the physico-chemical and other properties driving lesion penetration and to the location and type of lesions which are most resistant to drug dispersal.
A full analysis of tuberculous lesions will enhance interpretation of PET/CT images from infected persons, to determine efficacy of treatment or state of disease. The opportunity to visualize latent lesions in humans and in non-human primates, as well as possibly “watch” reactivation occur, will provide data that will greatly enhance our understanding of the latent infection, including the spectrum of latency, and reactivation.
Pre-clinical animal models
To determine what animal model(s) may be best suited for evaluating drug efficacy against latent TB, one needs to have a basic understanding of latent disease in humans. As discussed above, the pathology of TB in humans suggest that this form of the disease is characterized by a continuum of more or less quiescent and healed lesions (Figure 3)5. The few necropsies that were conducted on individuals with latent TB revealed consolidated fibrotic and necrotic lesions that had often become calcified19, 20. One commonly accepted paradigm is that these arrested granulomas contain the bacilli which are responsible for disease reactivation, and that these bacilli have become dormant in response to hypoxic conditions, though this remains to be formally demonstrated. Importantly, such structures could very well constitute effective barriers to the penetration of anti-TB drugs.
Figure 3. Tuberculous granulomas.
There are several granuloma types that can be found among humans and non-human primates, even within the same individual.
A. The classic tuberculous granuloma, found in active disease and latent infection, is the caseous granuloma, composed of epithelioid macrophages, neutrophils, a cuff of lymphocytes (CD4 and CD8 T cells, B cells) and sometimes surrounded by peripheral fibrosis. The center of this type of granuloma is caseous, a necrotic state that likely consists of dead macrophages and other cells. This area is hypoxic. Mycobacteria in this granuloma could be found in macrophages (either in contact with T cells or not) or in the hypoxic center, or possibly even in the fibrotic rim, leading to different microenvironments for the bacteria.
B. The non-necrotizing granuloma is usually seen in active disease, and consists primarily of macrophages with some lymphocytes; this lesion can be seen in guinea pigs and mice, albeit with more lymphocytes. M. tuberculosis bacilli are within macrophages in this lesion.
C. Fibrotic lesions are seen mostly in latent tuberculosis but also in active disease, and are composed almost completely of fibroblasts, with a minimal number of macrophages. Although it is possible to culture bacilli from some fibrotic lesions, it is not clear where the bacilli reside, perhaps in macrophages or in the fibrotic area, or what the microenvironment is like.
While the mouse is by far the most practical and widely used model for drug discovery, TB in commonly used mouse strains does not recapitulate human pathology: the granulomas are poorly organized and exclusively cellular (Figure 3); they lack necrosis, fibrosis or hypoxia21; bacterial counts remain at high levels throughout the entire course of disease, and the mice ultimately die of progressive infection22. These traits are not reflective of the situation seen (or assumed) in human latent disease.
In the Cornell mouse model, a latency-like state is drug-induced, resulting in apparent eradication of cultivable bacilli with subsequent occasional spontaneous reactivation23. It is possible that some bacterial sub-populations in the Cornell model mimic those present in human latent TB, but in the absence of organized granulomas with caseous necrosis or mineralization, quantitative evaluation of drug efficacy against latent disease in this model is not reliable. Because the mouse remains such a practical species, efforts should focus on engineering the host and/or pathogen to provide more human-like immunopathology and environment-induced bacterial physiology24.
A rabbit model of latent TB has been proposed25, 26, characterized by persistent, host-contained, paucibacillary infection which can be reactivated upon immunosuppression. Though validation with standard TB drugs is required to assess its experimental utility, the model appears attractive to investigate drug efficacy against persistent mycobacteria.
Undoubtedly the most costly and resource intensive model, the non-human primate (NHP) is the only species which reproduces the clinical characteristics of human latent TB (to the extent that we understand them)8. When infected with low doses of M. tuberculosis, approximately half of cynomolgus macaques survive for up to three years, and anecdotal evidence suggests that this period is indefinite, without clinical or radiographic signs of disease. Like latently infected humans, they respond positively to the tuberculin skin test in the absence of clinical symptoms. Histologically, the lesions show thick fibrosis, mineralization and/or central caseation (Figure 3), which is also presumably the case in humans8. Hypoxia has been demonstrated in chronically infected macaques21, the model is currently being validated with standard anti-TB drugs, and gene expression studies to characterize the metabolic state of the bacilli are underway.
It thus appears that the NHP is the most reliable model for testing the ability of drug candidates to eradicate latent TB. But given the time, cost, and compound requirements, this can only be accomplished when a drug candidate is about to enter full clinical development. For evaluation during early drug discovery stages, there is a clear need for predictive and validated high-throughput in vitro assays. Combined with attractive pharmacokinetic and toxicological attributes, such assays must have the predictive value required to warrant efficacy studies in the NHP model.
Immunologic characterization of lesion types
The heterogeneity of lesion types among humans and non-human primates infected with M. tuberculosis is very similar. Macaques, when infected with a low dose of M. tuberculosis, develop active and latent disease in equal proportions. Both CD4 and CD8 T cells are found within the granuloma and these can be tested for effector function, specificity, and exhaustion. Differences amongst lesion types, as well as between latent and active disease monkeys may point to key features involved in the relative ability of granulomas to contain the infection. In addition, changes in immune functions during treatment may point to differences in drug mechanisms. We have observed increased lymphocytic infiltration of lesions during drug-induced healing, for example, supporting the hypothesis that killing of bacteria or release of antigens during therapy causes increased activation, proliferation or migration of lymphocytes to the lesions in the lungs.
The wealth of immunologic reagents for non-human primates provides the opportunity to test the necessity for various immune factors at different stages of disease, and to study the mechanism by which these factors affect control of infection. For example, tuberculosis can be seen in HIV-infected individuals prior to a substantial loss of CD4 T cells27, raising the possibility that CD4 T cell depletion alone is not responsible for the increased risk of tuberculosis in HIV patients; studies comparing CD4 T cell depletion with SIV infection (as a surrogate for HIV) in macaques will answer these questions. Similarly, the role of CD8 T cells can be studied using CD8-depleting antibodies in macaques. Although CD8 T cells have been implicated in control of tuberculosis in humans, no direct data support that these cells are necessary for containment of initial or latent infection. Finally, TNF neutralization, as a therapy for certain inflammatory diseases in humans, substantially increases the risk of reactivation TB28. Similar studies in macaques recapitulate the human data (Lin, Flynn, unpublished) and provide an opportunity to explore the mechanisms by which TNF is important in control of M. tuberculosis infection.
These studies, alongside investigation of the phenotypes and functions of immune cells in human granulomas from lung resection studies in combination with PET/CT imaging, provide a new look at the important host factors that work in concert with bacterial factors to determine the outcome of infection and maintenance of latent infection. It is likely that a large number of host factors contribute to initial containment of infection and prevention of reactivation. While the balance of factors that contribute to these states may be very different between different hosts, they may lead to the same outcome. Mathematical modelling can provide insights into the balance of host responses, as well as the relative importance of each response in acute and latent infection29–31.
Characterisation of subpopulations in latent TB
Much of the work on the transcriptional response of M. tuberculosis to various environmental conditions has focused on the hypoxic response, largely based on the pathologists’ intuitive deduction that some tuberculous lesions appeared hypoxic. Analyses of mRNA transcripts from human lesions have supported differential responses that are microenvironment-specific, showing that expression can vary even within a single lesion depending on the location of the bacilli relative to the outer lesion wall32, 33. Specific stresses or conditions can lead to major metabolic realignment that also translates into major changes in drug susceptibility34–37. These observations, coupled with the introduction of a simple in vitro system for hypoxic adaptation of M. tuberculosis, led to a hypothesis that latent TB represented a reservoir of organisms encased in caseous lesions under hypoxic conditions, while active TB was typified by bacilli replicating aerobically at the margin of liquefied cavities.
Although whole genome expression data are available from mice and macrophage infections38–41, evidence relating to the hypoxia hypothesis often relies on an interpretation of environmental conditions inferred from transcriptome analysis of just a handful of genes. This inference is complex for three reasons. Firstly, transcriptional responses to different environmental cues often involve overlapping gene sets. For example, the two-component regulatory system DosR-DosS/DosT that is central to hypoxic adaptation is also up-regulated by exposure to nitric oxide42; the siderophore-encoding mycobactin biosynthetic genes are up-regulated by hypoxia as well as by iron-limitation43; and genes of the methyl citrate cycle are regulated by hypoxia, acidic pH and iron restriction43, 44. As a result the “hypoxia regulon” is strongly induced even within apparently aerobic macrophage infections38. Secondly, it is now recognised that the hypoxia regulon comprises at least two subsets of genes, differentially up-regulated during initial adaptation and extended exposure to hypoxic environments38, 42, 45. The relevance of these adaptive responses have been validated by gene knockout studies in the mouse model46–50 though it should be noted that this model reproduces neither the pathology nor the complexity of human lesions21. In addition, expression data that are available from human samples only give bulk, averaged signals and don’t represent accurately the behaviour of sub-populations present even within a single lesion33.
A significant recent accomplishment was the demonstration that a subset of lesions in higher vertebrate animal species (rabbits, guinea pigs and cynomolgus macaques) are, in fact, hypoxic and sensitive to Metronidazole – a drug selective for hypoxically adapted TB21. In the definitive experiment the oxygen tension was measured directly in rabbit lesions by surgically inserting a highly sensitive oxygen probe into lesions in live animals. This sort of analysis helps tremendously to define the bulk environment in a certain lesion. These studies confirm the hypoxia hypothesis for a subset of lesions across animal model systems (except mice), and are consistent with the view that hypoxia makes at least some contribution to human TB. Recent transcriptome data from active disease in humans adds further layers of complexity. Expression signatures of M. tuberculosis isolated directly from surgically-removed lesions from patients with TB suggests that the bacilli likely experience nutrient deprivation and hypoxia in addition to various antimicrobal defense systems, conditions that damage DNA, and conditions that encourage extensive remodelling of the cell wall51. A more recent study focused on M. tuberculosis isolated from patient sputum samples revealed an enrichment for lipid bodies and a surprising up-regulation of the DosR hypoxia regulon52. This throws substantial doubt on the simple hypothesis that sputum-borne organisms represent actively replicating bacilli from the margins of cavities open to a bronchus while latent infection represents non-replicating bacilli in closed caseous [GT], hypoxic lesions.
These studies have given rise to a competing hypothesis that latent bacilli reside in a spectrum of lesions with perhaps very different microenvironmental conditions driving them into a state of non-, or slow replication. A corollary of this hypothesis is that a subset of the lesions in active disease is identical to those found in latent infection. The next challenge is to translate information from lesion analysis and transcriptional profiling into a metabolic framework as a platform for drug discovery.
Detection of transcripts for genes within a metabolic pathway does not on its own necessarily implicate it as a drug target; the essential nature of the pathway during the relevant stage of host pathogenesis needs to be verified. Thus, although genes of the methylcitrate cycle appear to be upregulated in vivo, deletion of the corresponding genes does not affect virulence in the mouse model50. Similarly, the recycling pathway for NAD biosynthesis has been shown to be highly up-regulated at both the gene expression as well as the enzyme activity level during parasitism of host tissues, yet inactivation of this pathway did not affect virulence in mice53. Genome-wide mutagenesis studies have been instrumental in elucidating the essential nature of genes that are required for initial growth in the mouse model54, but results of such studies can only be interpreted in the light of the environment in which the transposon library was analyzed, and differences in the availability of nutrients between different host models and in different lesions must be taken into account when extrapolating these data to human infection.
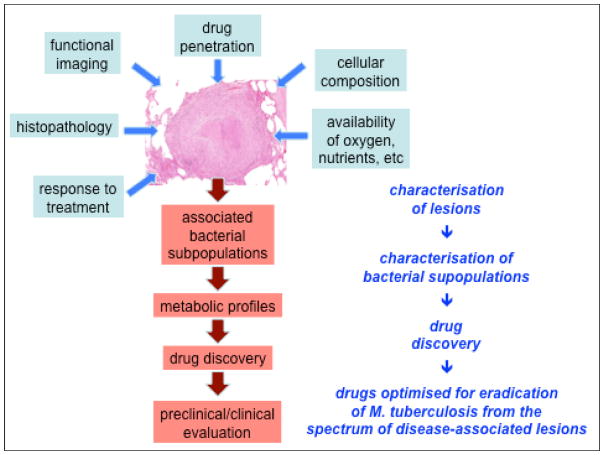
Most attempts at drug target validation have been based on studies that interrogated the role of individual enzymes. Understanding the full metabolic activity of an organism in a particular physiologic state would present the ultimate means of drug target discovery instead of piecemeal reconstruction of potential bottlenecks under these conditions (Figure 4). A genome-scale network model of M. tuberculosis metabolism was designed based on the principles of flux-balance analysis to predict growth rate, substrate consumption and biomass production in a pseudo-steady state55. The model was calibrated with experimental data generated from chemostat cultures of M. bovis BCG and its utility was further demonstrated by predicting gene essentiality, which correlated with genome wide mutagenesis data and correctly predicted the essentiality of several anti-tubercular drug targets including isoniazid (and thus ethionamide), pyrazinamide, ethambutol and cycloserine. Flux balance analysis has also been used to study potential drug targets in subsystems of the entire metabolic network. Raman et al56 developed a metabolic network based on the mycolic acid biosynthetic pathway to predict bottlenecks in this sub-system. Such metabolic models are of course only useful in the context of knowledge of the nutrient availability under a particular physiologic state, and metabolomic studies of lung lesions will provide the groundwork for developing genome-scale metabolic networks relevant for drug discovery. Such measurements in vivo would be exceptionally technically challenging. Ex vivo studies of M. tuberculosis recovered from lesions have indicated that metabolism immediately after recovery from the host to some extent recapitulates aspects of its in vivo metabolic activity53, suggesting that ex vivo metabolomic studies of M. tuberculosis may provide clues to the metabolic capacity of this pathogen.
Figure 4. A systems approach to the pathology of tuberculosis.
Progress in understanding the complex biology of human tuberculosis – and applying this knowledge to drug development – will depend on being able to integrate multiple sources of data into in silico models that allow us to prioritise experiments and predict effective interventions.
Discovery of drugs against latent TB
The most ambitious objectives for TB drug discovery are to identify new drugs that effectively eradicate latent TB infection and reduce chemotherapy of active disease to two weeks. To achieve these goals we need to identify and kill those subpopulations of M. tuberculosis that are not eliminated efficiently with current antibiotics, and thus are responsible for the prolonged treatments required for the various forms of TB (Figure 5). A key issue complicating the discovery of cidal drugs is that we only have a limited understanding of the death-causing events that are induced by the existing drugs57, 58. The physiological complexity underlying cidal activity is elegantly illustrated in a recent analysis of the mechanism of action of aminoglycosides59. At the heart of this difficulty is the fact that we lack a sensitive test to determine when a bacterium is ‘dead’, other than showing that it does not form a colony when placed on agar. Kohanski et al60 described a possible unifying death pathway in bacteria via formation of hydroxyl radicals by oxidation of iron, with DNA and protein damage as the direct mechanism of death. Whether this holds true in a general sense remains to be seen. However, it appears likely that cidal compounds need to “corrupt” or “derail” cellular processes in a way that the bacterium cannot control or counteract. Rather than simple inhibition of an enzyme activity to block a metabolic process, effective drugs might, for instance, have to trigger build-up of toxic intermediates, cause release of bactericidal metabolites (e.g nitric oxide61) or collapse central homeostatic systems62. Such activities are currently difficult to predict due to our limited understanding of metabolism. Novel cellular readouts, correlates of death, are urgently required to biologically annotate and evaluate hits.
Figure 5. A lesion-based framework for study of latent TB and drug development.
Latent and active TB in humans and non-human primates comprises a heterogeneous mixture of lesions that generate a range of physiological microenvironments associated with bacterial replication, persistence and killing. Characterisation of the different types of lesion using state-of-the-art imaging, cellular and molecular techniques will provide a framework for understanding the biology of TB and for the development of drugs targeted against relevant bacterial subpopulations.
To add an additional level of complexity, it has recently become clear that bacteria also show intra-population diversity. Heterogeneity is observed at the single cell level within a single microenvironment. Phenotypic heterogeneity, defined as non-heritable and reversible variation in cellular parameters, is crucial for the persistence of bacterial populations under selective pressures, including antibiotic stress63. Stochastically-determined differences in growth rate or expression of a specific enzyme, for example, may result in a subpopulation of clonal organisms (referred to as “persisters”) even within a single microenvironment that might show differential antibiotic susceptibility. Differential behaviour of single bacilli is masked in standard batch culture readouts that derive mean population values by pooling large numbers of cells. Importantly, new technologies that permit the analysis of bacterial behaviour at single cell resolution have made this ‘persister’ problem more accessible64. Single cell analyses to determine the underlying population structure responsible for average cellular behaviour are an emerging field in TB63 and need to be implemented as an integral part in the drug discovery process for the evaluation and prioritization of leads.
Whole cell screens
Translating this individual lesion-based concept into something useful for drug discovery remains a critical challenge. While the target-led, genomics- and genetics-driven approach has provided the central strategy for drug discovery programmes in the pharmaceutical industry in recent decades, it has been markedly ineffective in the area of antibacterials, where potent enzyme inhibitors frequently fail to translate into agents that will kill, or even inhibit growth65. Furthermore, many successful antibacterials have multiple targets and several key TB drugs, such as isoniazid, require metabolic activation after uptake by the bacillus to exert their cidal effects. The concept of an isolated single target approach to TB drug discovery might therefore be questioned66. Whole cell screens allow for the simultaneous screening of all targets essential for a particular physiological state of the organism. Together, these considerations have encouraged a move towards high-throughput screens based on monitoring of activity against whole bacteria, with subsequent identification of the mechanism of action of active compounds employing whole genome sequencing of spontaneous resistant mutants67, 68, affinity purification69 and comparative transcriptional fingerprinting with drugs for which the mechanism of action is known70. The phenotypic, whole cell screening approach has the advantage that it eliminates the first hurdle encountered in target-based approaches, ‘translating’ activity against a target into whole cell activity: one starts with compounds that kill the pathogen. The follow-up target deconvolution can result in the identification of novel, compound-induced death mechanisms, not necessarily predictable by genetics71. An example of such a phenotypic screening project is work in progress within the Grand Challenges consortium (Box 2): a high-throughput screen was carried out to identify hits that trigger the collapse of ATP homeostasis, the Achilles heel of hypoxic non-growing bacilli62. Now corresponding targets are being identified employing genetics and affinity purification. In addition, a biochemical respiratory vesicle-based assay and a series of M. tuberculosis strains containing externally regulatable respiratory chain functions are employed to identify the subset of hits that target energy metabolism.
Box 2. Drugs for Latent TB: A Grand Challenge in Global Health.
A project to study fundamental biology and drug development strategies for latent TB was initiated in 2005 as part of a programme to tackle Grand Challenges in Global Health*. The project, funded jointly by the Bill & Melinda Gates Foundation and the Wellcome Trust, brought together eight academic groups from four continents with industrial expertise provided by the Novartis Institute for Tropical Diseases (NITD). The project focused in particular on exploring the role of hypoxia in latent TB, and was organised in the form of three interlinked components.
The aim of the first component was to characterise TB lesions in freshly resected lung tissue from humans and non-human primates with active and latent TB. The presence of hypoxic regions was demonstrated in lesions from non-human primates that matched the histopathology of a subset of lesions seen in humans with both active TB and latent infection. Efforts are continuing to determine whether particular patterns of bacterial transcription are associated with different lesion types.
The second component is directed towards drug discovery using several approaches. One is based on use of conditional expression systems to identify genes that are required for survival of M. tuberculosis under hypoxic conditions, and targeting associated enzymes in high-throughput screens. Another is based on high-throughput screens using whole cell cultures, to identify compounds that inhibit the ATP homeostasis essential for viability during non-replicating persistence. Finally, there is a program to optimize nitroimidazoles to release nitric oxide, which has been shown to bactericidal for non-replicating, hypoxic organisms.
The final component addresses strategies for evaluation of drugs with activity against hypoxic M. tuberculosis. This is achieved using PET/CT imaging to monitor the effect of drugs on individual lesions in a non-human primate model and in a clinical trial of the effect of adding metronidazole during salvage therapy for patients with multidrug-resistant TB. The potential use of immune biomarkers in field trials of preventive therapy is also being explored.
Many of the ideas and approaches outlined in this review are based on results and unique collaborative environment facilitated by the Grand Challenges project.
*http://www.grandchallenges.org/CureInfection/Challenges/Therapies/Pages/Tuberculosis.aspx
Target-based screens
Even though whole cell screens have been successful, they also have limitations. Bacterial growth can be inhibited by different mechanisms and secondary screens are required to identify the few useful compounds among the many that exhibit broad toxicity65. For those compounds that are not generally toxic, target identification often requires several of the experimental strategies mentioned above, all of which can and frequently do fail. In addition, lead optimization is greatly facilitated if the target of a compound is known and therefore more straightforward for hits from biochemical high-throughput screens than for hits from whole cell screens. In effect, the biology of target-selection becomes much simpler at the expensive of the chemistry of drug development. Target-based screens are also much more sensitive and compatible with novel strategies for hit generation, such as fragment-based screens72, which cannot be used in whole cell screens.
Why then have target-led approaches been so inefficient? In part, because cell permeability is not a factor in biochemical screens and is difficult to engineer into a compound without knowing the physicochemical rules that determine uptake or having simple uptake assays. In addition, even if they can enter the bacterial cell, good enzyme inhibitors do not always make good antibacterial agents. For example, the Grand Challenges consortium and NITD recently identified a large number of scaffolds that efficiently inhibited pantothenate kinase (PanK) and peptide deformylase (Def) in vitro. It has not been possible to isolate M. tuberculosis mutants with transposon insertions in the genes encoding PanK or Def73, which suggested that PanK and Def were essential and that chemical inactivation of either enzyme should prevent growth of M. tuberculosis. However, in contrast to a transposon insertion, which nearly always completely eliminates protein function, chemical inhibition of an enzyme within a cell is almost always incomplete, especially with inhibitors that have not yet been optimized for activity against their targets. In the case of PanK none of the inhibitors reduced growth of M. tuberculosis, despite their chemical diversity. Inefficient uptake might have contributed to the lack of activity of some PanK inhibitors, but this was not the case for the Def inhibitors74. These compounds were taken up by the bacilli and stopped their growth74, but their growth-inhibition kinetics were slow, the compounds were not cidal and resulted only in moderate efficacy in the TB mouse model (unpublished).
Therefore, we don’t know which genes encode functions that are not only essential for growth but are also vulnerable to chemical inhibition and, thus, represent good targets for biochemical screens. Gene silencing tools can help to solve this problem as they allow partial inhibition of a target in cellular assays and within animal models75, 76.
Our ability to transform compounds identified in target based screens into antibacterial agents will increase once we better understand how small molecules penetrate the bacterial cell envelope and which of the growth essential processes are susceptible to chemical inhibition. Information from transcriptional and metabolic profiling will help to predict vulnerable targets. Those targets should first be analyzed by genetic or, preferably, chemical knockdown to verify vulnerability to partial inhibition. It is important that these validation experiments are performed under several conditions representative of the proposed in vivo environment. Targets that are required for survival in diverse conditions, and are thus essential independent of the specific physiological state of the pathogen, obviously have particular attraction. For example, nicotinamide cofactors are essential for growth of M. tuberculosis and sudden NAD starvation causes cell death. Furthermore, inhibitors of NAD synthase are bactericidal in growing and nonreplicating cells53. NAD synthase should allow us to test whether biochemical screens against targets that are vulnerable to chemical inhibition and essential under different in vitro conditions more frequently identify enzyme inhibitors that also kill M. tuberculosis.
Assessment of drug activity against latent TB
The advent of a new compound potentially active against latent tuberculosis in humans would present considerable logistic challenges in evaluation. Historically clinical trials to assess the value of a candidate for prophylactic intervention in patients with latent tuberculosis have involved large numbers of patients and extensive follow-up studies. The two largest trials involved more than 50,000 participants and a ten year follow-up period77, 78. The last few years have witnessed a considerable expansion in the broad field of tuberculosis diagnostics and biomarkers, the main thrust being to better diagnose tuberculosis and to find a biological indicator of successful treatment response in active disease79. There is less research activity directed towards the discovery of markers that may change during treatment of latent infection and thus suggest successful treatment. Nevertheless there are interesting data that demonstrate that the IFN-γ response to MTB antigens approximately halves during drug treatment of latent TB80–83. This decrease may mark successful treatment: the chemotherapeutic reduction of bacillary numbers congruent with less necessity to mount an immune response. In our own study we also documented a transient initial doubling at one month, possibly due to an increase in antigen release attendant on bacillary death82. These findings predicated ongoing –omics based and larger clinical studies to determine whether a combination of markers monitored intensely can provide useful information.
The historical clinical trials of isoniazid77 collected relatively sparse information on subjects and evaluated efficacy based solely on relapse rates presuming that all participants were more or less equally latent at the start and equally adherent with medication. In some small studies monotherapy with isoniazid was found to have a dramatic impact (as large as a 100% reduction in the incidence of active disease in 261 participants at a Dutch marine training camp84) while in others the same regimen was found to have a very modest impact85. The reasons for these discrepant outcomes are not widely discussed other than to say that adherence or duration were likely confounders.
But what if conventional wisdom on this point is incorrect and rather than poor adherence or duration a confounder was the different points in the spectrum of latency within which the individual cohorts started? The implication would be that specific groups of patients are at different risks for development of active disease. We propose that it is not one-third of the global population that is at imminent risk for the development of active disease, the number is much smaller and the key issue is diagnosis of those patients who would benefit from intervention. The “spectrum of latency” concept allows one to envision a combination of imaging-enabled and perhaps biomarker tools to identify patients that have sub-clinical, but no less active, disease and targeting chemoprophylaxis to that specific population. Rather than requiring mass chemotherapy of one-third of the planet’s population to achieve eradication, we could target the much smaller one in ten that will otherwise perpetuate the epidemic. In no small way, imaging has allowed us to imagine a solution to the problem of developing new chemotherapy for latent tuberculosis.
Acknowledgments
We are grateful to colleagues in the Grand Challenges in Global Health project “Drugs for Treatment of Latent Tuberculosis” for stimulating discussion and to the Bill & Melinda Gates Foundation and the Wellcome Trust for financial support. We would also like to thank our Novartis colleagues in Singapore, Basel, Cambridge, and La Jolla for discussions and collaboration.
GLOSSARY TERMS
- caseation/caseous
necrotic degeneration of bodily tissue into a soft crumbly cheese-like mass where cellular outline is lost. In tuberculosis, this refers to the necrotic center of a granuloma, and is mainly driven by the immune response and typical of tuberculosis pathogenesis
- coefficient of penetration
the speed at which a compound moves through a tissue, generally due to diffusion but also to facilitated transport. Its units are cm/sec
- CT
Computed tomography; an imaging technique where X-ray scans of a subject are compiled to generate a three dimensional picture of various body organs and structures (i.e, brain, lungs, etc.)
- enduring hypoxic response
A set of 230 genes induced in M. tuberculosis during prolonged exposure to hypoxia
- granuloma
An organized structure comprised of lymphocytes, macrophages, neutrophils, and sometimes fibroblasts, often with a necrotic centre, that arises in response to continued antigenic stimulation in the presence of macrophages, for example in response to M. tuberculosis infection
- hypoxia
A localized environment containing low oxygen
- hypoxia regulon
A cluster of 48 genes controlled by the transcriptional regulator DosR that is upregulated in response to hypoxia but also during exposure to nitric oxide, carbon monoxide, SDS or low pH
- paucibacillary
containing just a few bacilli, as is the case of granulomas caused by a variety of mycobacterial diseases
- PET
Positron emission tomography; a nuclear medicine imaging technique using a positron-emitting probe that produces a three dimensional image of biological processes within the scanned subject
- PPD
Purified protein derivative; a precipitate of non-specific molecules from sterilized and filtered cultures of M. tuberculosis
- TST
Tuberculin skin test; a method of diagnosing M. tuberculosis infection by injecting tuberculosis antigens (PPD) intradermally; a delayed type hypersensitivity response, dependent on the presence of sensitized T cells, is seen in those infected with M. tuberculosis. This does not distinguish latent infection from active tuberculosis
References
- 1.Comstock GW, Baum C, Snider DE., Jr Isoniazid prophylaxis among Alaskan Eskimos: a final report of the bethel isoniazid studies. Am Rev Respir Dis. 1979;119:827–30. doi: 10.1164/arrd.1979.119.5.827. [DOI] [PubMed] [Google Scholar]
- 2.Dye C, Williams BG. Eliminating human tuberculosis in the twenty-first century. J R Soc Interface. 2008;5:653–62. doi: 10.1098/rsif.2007.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stead WW. Management of health care workers after inadvertent exposure to tuberculosis: a guide for the use of preventive therapy. Ann Intern Med. 1995;122:906–12. doi: 10.7326/0003-4819-122-12-199506150-00003. [DOI] [PubMed] [Google Scholar]
- 4.Pai M, Zwerling A, Menzies D. Systematic review: T-cell-based assays for the diagnosis of latent tuberculosis infection: an update. Ann Intern Med. 2008;149:177–84. doi: 10.7326/0003-4819-149-3-200808050-00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young DB, Gideon HP, Wilkinson RJ. Eliminating latent tuberculosis. Trends in Microbiology. 2009 doi: 10.1016/j.tim.2009.02.005. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Mtei L, et al. High rates of clinical and subclinical tuberculosis among HIV-infected ambulatory subjects in Tanzania. Clin Infect Dis. 2005;40:1500–7. doi: 10.1086/429825. [DOI] [PubMed] [Google Scholar]
- 7.Vandiviere HM, Loring WE, Melvin I, Willis S. The treated pulmonary lesion and its tubercle bacillus. II. The death and resurrection. Am J Med Sci. 1956;232:30–7. doi: 10.1097/00000441-195607000-00006. passim. [DOI] [PubMed] [Google Scholar]
- 8.Capuano SV, 3rd, et al. Experimental Mycobacterium tuberculosis infection of cynomolgus macaques closely resembles the various manifestations of human M. tuberculosis infection. Infect Immun. 2003;71:5831–44. doi: 10.1128/IAI.71.10.5831-5844.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin PL, et al. Early events in Mycobacterium tuberculosis infection in cynomolgus macaques. Infect Immun. 2006;74:3790–803. doi: 10.1128/IAI.00064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goo JM, et al. Pulmonary tuberculoma evaluated by means of FDG PET: findings in 10 cases. Radiology. 2000;216:117–21. doi: 10.1148/radiology.216.1.r00jl19117. [DOI] [PubMed] [Google Scholar]
- 11.Hara T, Kosaka N, Suzuki T, Kudo K, Niino H. Uptake rates of 18F-fluorodeoxyglucose and 11C-choline in lung cancer and pulmonary tuberculosis: a positron emission tomography study. Chest. 2003;124:893–901. doi: 10.1378/chest.124.3.893. [DOI] [PubMed] [Google Scholar]
- 12.Yang CM, Hsu CH, Lee CM, Wang FC. Intense uptake of [F-18]-fluoro-2 deoxy-D-glucose in active pulmonary tuberculosis. Ann Nucl Med. 2003;17:407–10. doi: 10.1007/BF03006610. [DOI] [PubMed] [Google Scholar]
- 13.Park IN, Ryu JS, Shim TS. Evaluation of therapeutic response of tuberculoma using F-18 FDG positron emission tomography. Clin Nucl Med. 2008;33:1–3. doi: 10.1097/RLU.0b013e31815c5128. [DOI] [PubMed] [Google Scholar]
- 14.Canetti G, Parrot R, Porven G, Le Lirzin M. Rifamycin levels in the lung and tuberculous lesions in man. Acta Tuberc Pneumol Belg. 1969;60:315–22. [PubMed] [Google Scholar]
- 15.Kislitsyna NA. Comparative evaluation of rifampicin and isoniazid penetration into the pathological foci of the lungs in tuberculosis patients. Probl Tuberk. 1985:55–7. [PubMed] [Google Scholar]
- 16.Kislitsyna NA, Kotova NI. Rifampicin and isoniazid concentration in the blood and resected lungs in tuberculosis with combined use of the preparations. Probl Tuberk. 1980:63–5. [PubMed] [Google Scholar]
- 17.Sauermann R, et al. Antibiotic abscess penetration: fosfomycin levels measured in pus and simulated concentration-time profiles. Antimicrob Agents Chemother. 2005;49:4448–54. doi: 10.1128/AAC.49.11.4448-4454.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagner C, Sauermann R, Joukhadar C. Principles of antibiotic penetration into abscess fluid. Pharmacology. 2006;78:1–10. doi: 10.1159/000094668. [DOI] [PubMed] [Google Scholar]
- 19.Cotran RS, Kumar V, Robbins SL. Pathologic Basis of Disease. Company, W.B.S; Philadelphia: 1989. [Google Scholar]
- 20.Dannenberg AM., Jr . Pathogenesis of Human Pulmonary Tuberculosis. ASM Press; Washington D. C: 2006. pp. 36–64. A comprehensive review of five decades of literature on the pathogenesis of tuberculosis in humans and in the rabbit model, including comparisons with other animal models. [Google Scholar]
- 21.Via LE, et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect Immun. 2008;76:2333–40. doi: 10.1128/IAI.01515-07. A conclusive demonstration that hypoxia is a relevant phenotype in several non-murine animal models of TB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhoades ER, Frank AA, Orme IM. Progression of chronic pulmonary tuberculosis in mice aerogenically infected with virulent Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78:57–66. doi: 10.1016/s0962-8479(97)90016-2. [DOI] [PubMed] [Google Scholar]
- 23.Radaeva TV, Nikonenko BV, Mischenko VV, Averbakh MM, Jr, Apt AS. Direct comparison of low-dose and Cornell-like models of chronic and reactivation tuberculosis in genetically susceptible I/St and resistant B6 mice. Tuberculosis (Edinb) 2005;85:65–72. doi: 10.1016/j.tube.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 24.Sissons J, et al. Multigenic control of tuberculosis resistance: analysis of a QTL on mouse chromosome 7 and its synergism with sst1. Genes Immun. 2009;10:37–46. doi: 10.1038/gene.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manabe YC, et al. The aerosol rabbit model of TB latency, reactivation and immune reconstitution inflammatory syndrome. Tuberculosis (Edinb) 2008;88:187–96. doi: 10.1016/j.tube.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kesavan AK, Brooks M, Tufariello J, Chan J, Manabe YC. Tuberculosis genes expressed during persistence and reactivation in the resistant rabbit model. Tuberculosis (Edinb) 2009;89:17–21. doi: 10.1016/j.tube.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenberg P, et al. How soon after infection with HIV does the risk of tuberculosis start to increase? A retrospective cohort study in South African gold miners. J Infect Dis. 2005;191:150–8. doi: 10.1086/426827. [DOI] [PubMed] [Google Scholar]
- 28.Keane J, et al. Tuberculosis associated with infliximab, a tumor necrosis factor alpha-neutralizing agent. N Engl J Med. 2001;345:1098–104. doi: 10.1056/NEJMoa011110. The first demonstration that suppressing TNF-a levels was correlated with reactivation of “latent” TB, the implication being that there is a delicate balance between immune function and the development of disease. [DOI] [PubMed] [Google Scholar]
- 29.Marino S, et al. Differences in reactivation of tuberculosis induced from anti-TNF treatments are based on bioavailability in granulomatous tissue. PLoS Comput Biol. 2007;3:1909–24. doi: 10.1371/journal.pcbi.0030194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sud D, Bigbee C, Flynn JL, Kirschner DE. Contribution of CD8+ T cells to control of Mycobacterium tuberculosis infection. J Immunol. 2006;176:4296–314. doi: 10.4049/jimmunol.176.7.4296. [DOI] [PubMed] [Google Scholar]
- 31.Ray JC, Flynn JL, Kirschner DE. Synergy between individual TNF-dependent functions determines granuloma performance for controlling Mycobacterium tuberculosis infection. J Immunol. 2009;182:3706–17. doi: 10.4049/jimmunol.0802297. Illustrates the use of modelling to understand the complex dynamics of tuberculous granulomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timm J, et al. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci U S A. 2003;100:14321–6. doi: 10.1073/pnas.2436197100. While far from exhaustive this was the first study to attempt to relate the transcriptional responses in humans to conditions relevant to disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fenhalls G, et al. In situ detection of Mycobacterium tuberculosis transcripts in human lung granulomas reveals differential gene expression in necrotic lesions. Infect Immun. 2002;70:6330–8. doi: 10.1128/IAI.70.11.6330-6338.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie Z, Siddiqi N, Rubin EJ. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2005;49:4778–80. doi: 10.1128/AAC.49.11.4778-4780.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paramasivan CN, Sulochana S, Kubendiran G, Venkatesan P, Mitchison DA. Bactericidal action of gatifloxacin, rifampin, and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49:627–31. doi: 10.1128/AAC.49.2.627-631.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Herbert D, et al. Bactericidal action of ofloxacin, sulbactam-ampicillin, rifampin, and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1996;40:2296–9. doi: 10.1128/aac.40.10.2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wayne LG, Sramek HA. Metronidazole is bactericidal to dormant cells of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1994;38:2054–8. doi: 10.1128/aac.38.9.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schnappinger D, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohde KH, Abramovitch RB, Russell DG. Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe. 2007;2:352–64. doi: 10.1016/j.chom.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Tailleux L, et al. Probing host pathogen cross-talk by transcriptional profiling of both Mycobacterium tuberculosis and infected human dendritic cells and macrophages. PLoS ONE. 2008;3:e1403. doi: 10.1371/journal.pone.0001403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Talaat AM, et al. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J Bacteriol. 2007;189:4265–74. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voskuil MI, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–13. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, An essential gene in Mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect Immun. 2002;70:3371–81. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisher MA, Plikaytis BB, Shinnick TM. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184:4025–32. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rustad TR, Harrell MI, Liao R, Sherman DR. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE. 2008;3:e1502. doi: 10.1371/journal.pone.0001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Converse PJ, et al. Role of the dosR-dosS two-component regulatory system in Mycobacterium tuberculosis virulence in three animal models. Infect Immun. 2009;77:1230–7. doi: 10.1128/IAI.01117-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malhotra V, et al. Disruption of response regulator gene, devR, leads to attenuation in virulence of Mycobacterium tuberculosis. FEMS Microbiol Lett. 2004;231:237–45. doi: 10.1016/S0378-1097(04)00002-3. [DOI] [PubMed] [Google Scholar]
- 48.Dahl JL, et al. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol. 2005;187:2439–47. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McKinney JD, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–8. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 50.Munoz-Elias EJ, Upton AM, Cherian J, McKinney JD. Role of the methylcitrate cycle in Mycobacterium tuberculosis metabolism, intracellular growth, and virulence. Mol Microbiol. 2006;60:1109–22. doi: 10.1111/j.1365-2958.2006.05155.x. [DOI] [PubMed] [Google Scholar]
- 51.Rachman H, et al. Unique transcriptome signature of Mycobacterium tuberculosis in pulmonary tuberculosis. Infect Immun. 2006;74:1233–42. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garton NJ, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. This work raises new questions about the metabolic consequences of activation of the dormancy regulon in M. tuberculosis and the presence of lipid bodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boshoff HI, et al. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J Biol Chem. 2008;283:19329–41. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Beste DJ, et al. GSMN-TB: a web-based genome-scale network model of Mycobacterium tuberculosis metabolism. Genome Biol. 2007;8:R89. doi: 10.1186/gb-2007-8-5-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raman K, Rajagopalan P, Chandra N. Flux balance analysis of mycolic acid pathway: targets for anti-tubercular drugs. PLoS Comput Biol. 2005;1:e46. doi: 10.1371/journal.pcbi.0010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lange RP, Locher HH, Wyss PC, Then RL. The targets of currently used antibacterial agents: lessons for drug discovery. Curr Pharm Des. 2007;13:3140–54. doi: 10.2174/138161207782110408. [DOI] [PubMed] [Google Scholar]
- 58.Walsh C. Antibiotics: Actions, Origins, Resistance. ASM Press; Washington, D.C: 2003. pp. 11–88. [Google Scholar]
- 59.Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–90. doi: 10.1016/j.cell.2008.09.038. A must-read illustration of the complexities of the mechanism by which antibiotics cause bacterial cells to die. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 61.Singh R, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–5. doi: 10.1126/science.1164571. Identification of novel anti-mycobactericidal mechanism under anaerobic coinditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao SP, Alonso S, Rand L, Dick T, Pethe K. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2008;105:11945–50. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dhar N, McKinney JD. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr Opin Microbiol. 2007;10:30–8. doi: 10.1016/j.mib.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 64.Brehm-Stecher BF, Johnson EA. Single-cell microbiology: tools, technologies, and applications. Microbiol Mol Biol Rev. 2004;68:538–59. doi: 10.1128/MMBR.68.3.538-559.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6:29–40. doi: 10.1038/nrd2201. A review that describes many of the challenges complicating antibacterial drug development. [DOI] [PubMed] [Google Scholar]
- 66.Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Discov. 2007;6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 67.Manjunatha UH, et al. Identification of a nitroimidazo-oxazine-specific protein involved in PA-824 resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2006;103:431–6. doi: 10.1073/pnas.0508392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Andries K, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–7. doi: 10.1126/science.1106753. First report of a promising new anti-mycobacterial agent with activity against replicating and non-replicating cultures. [DOI] [PubMed] [Google Scholar]
- 69.Terstappen GC, Schlupen C, Raggiaschi R, Gaviraghi G. Target deconvolution strategies in drug discovery. Nat Rev Drug Discov. 2007;6:891–903. doi: 10.1038/nrd2410. [DOI] [PubMed] [Google Scholar]
- 70.Boshoff HI, et al. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem. 2004;279:40174–84. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 71.Dartois V, Leong FJ, Dick T. In: Drug Discovery in Infectious Diseases. Seltzer P, editor. Wiley-VCH; Weinheim: 2009. In press. [Google Scholar]
- 72.Ciulli A, Abell C. Fragment-based approaches to enzyme inhibition. Curr Opin Biotechnol. 2007;18:489–96. doi: 10.1016/j.copbio.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 74.Pichota A, et al. Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results. Bioorg Med Chem Lett. 2008;18:6568–72. doi: 10.1016/j.bmcl.2008.10.040. [DOI] [PubMed] [Google Scholar]
- 75.Klotzsche M, Ehrt S, Schnappinger D. Improved tetracycline repressors for gene silencing in mycobacteria. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gandotra S, Schnappinger D, Monteleone M, Hillen W, Ehrt S. In vivo gene silencing identifies the Mycobacterium tuberculosis proteasome as essential for the bacteria to persist in mice. Nat Med. 2007;13:1515–20. doi: 10.1038/nm1683. Demonstration of the use of conditional gene expression systems to study mycobacterial pathogenesis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ferebee SH, Mount FW. Tuberculosis morbidity in a controlled trial of the prophylactic use of isoniazid among household contacts. Am Rev Respir Dis. 1962;85:490–510. doi: 10.1164/arrd.1962.85.4.490. [DOI] [PubMed] [Google Scholar]
- 78.Ferebee SH, Mount FW, Murray FJ, Livesay VT. A controlled trial of isoniazid prophylaxis in mental institutions. Am Rev Respir Dis. 1963;88:161–75. doi: 10.1164/arrd.1963.88.2.161. [DOI] [PubMed] [Google Scholar]
- 79.Wallis RS, et al. Biomarkers for tuberculosis disease activity, cure, and relapse. Lancet Infect Dis. 2009;9:162–72. doi: 10.1016/S1473-3099(09)70042-8. [DOI] [PubMed] [Google Scholar]
- 80.Ewer K, et al. Dynamic antigen-specific T-cell responses after point-source exposure to Mycobacterium tuberculosis. Am J Respir Crit Care Med. 2006;174:831–9. doi: 10.1164/rccm.200511-1783OC. [DOI] [PubMed] [Google Scholar]
- 81.Goletti D, et al. Isoniazid prophylaxis differently modulates T-cell responses to RD1-epitopes in contacts recently exposed to Mycobacterium tuberculosis: a pilot study. Respir Res. 2007;8:5. doi: 10.1186/1465-9921-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilkinson KA, et al. Effect of treatment of latent tuberculosis infection on the T cell response to Mycobacterium tuberculosis antigens. J Infect Dis. 2006;193:354–9. doi: 10.1086/499311. [DOI] [PubMed] [Google Scholar]
- 83.Higuchi K, Harada N, Mori T. Interferon-gamma responses after isoniazid chemotherapy for latent tuberculosis. Respirology. 2008;13:468–72. doi: 10.1111/j.1440-1843.2008.01244.x. [DOI] [PubMed] [Google Scholar]
- 84.Veening GJ. Long term isoniazid prophylaxis. Controlled trial on INH prophylaxis after recent tuberculin conversion in young adults. Bull Int Union Tuberc. 1968;41:169–71. [PubMed] [Google Scholar]
- 85.Comstock GW, Woolpert SF. In: The Mycobacteria: A Sourcebook. Kubica GP, Wayne LG, editors. Marcel Dekker; New York: 1984. pp. 1071–1082. [Google Scholar]