Abstract
Owing to the demand for sustainable sex-control protocols in aquaculture, research in tilapia sex determination is gaining momentum. The mutual influence of environmental and genetic factors hampers disentangling the complex sex determination mechanism in Nile tilapia (Oreochromis niloticus). Previous linkage analyses have demonstrated quantitative trait loci for the phenotypic sex on linkage groups 1, 3, and 23. Quantitative trait loci for temperature-dependent sex reversal similarly reside on linkage group 23. The anti-Müllerian hormone gene (amh), located in this genomic region, is important for sexual fate in higher vertebrates, and shows sexually dimorphic expression in Nile tilapia. Therefore this study aimed at detecting allelic variants and marker-sex associations in the amh gene. Sequencing identified six allelic variants. A significant effect on the phenotypic sex for SNP ss831884014 (p<0.0017) was found by stepwise logistic regression. The remaining variants were not significantly associated. Functional annotation of SNP ss831884014 revealed a non-synonymous amino acid substitution in the amh protein. Consequently, a fluorescence resonance energy transfer (FRET) based genotyping assay was developed and validated with a representative sample of fish. A logistic linear model confirmed a highly significant effect of the treatment and genotype on the phenotypic sex, but not for the interaction term (treatment: p<0.0001; genotype: p<0.0025). An additive genetic model proved a linear allele substitution effect of 12% in individuals from controls and groups treated at high temperature, respectively. Moreover, the effect of the genotype on the male proportion was significantly higher in groups treated at high temperature, giving 31% more males on average of the three genotypes. In addition, the groups treated at high temperature showed a positive dominance deviation (+11.4% males). In summary, marker-assisted selection for amh variant ss831884014 seems to be highly beneficial to increase the male proportion in Nile tilapia, especially when applying temperature-induced sex reversal.
Introduction
Tilapias, famed for their rapid and efficient growth, their low position on the food chain and deemed the “aquatic chicken”, provide a possibility to nourish the poor and to conquer export markets. Even though intensive aquaculture farming resulted in rapid breeding and stunted populations, hybridisation or application of synthetic hormones (17α-methyl testosterone) allowed the production of mono-sex male broods [1]. The latter technique can effectively prevent fry production before harvest. Showcasing tilapia as the first aquaculture fish species certified by the Aquaculture Stewardship Council [2], this emphasizes the need for more sustainable sex control protocols. As such, a temperature treatment of the fry might be a future alternative [3], [4]. The complex sex determination (SD) in Nile tilapia is comprised of an interaction between genetic (major and minor factors) and environment-/temperature-dependent factors [5]. Due to its complex SD, Nile tilapia is a well-suited model species for the evolution of genetic or environmental sex-determining mechanisms. Although an increasing number of genomic tools has been developed for Nile tilapia, such as a genetic map with formerly 23 linkage groups (LG) [6], which has recently been resolved in 22 LGs in a high-density radiation hybrid map with 1358 genetic markers [7], BAC end sequences [8], and an assembled draft of its genome (http://www.broadinstitute.org/scientific-community/science/projects/mammals-models/vertebrates-invertebrates/tilapia-/tilapia-genom), a comprehensive understanding of the SD mechanism is still outstanding. On the one hand, linkage analysis demonstrated some success, with QTL found for phenotypic sex on LG 1, 3, and 23 in intraspecific and interspecific crosses of tilapia species [9]–[17]. On the other hand, neither assigning vertebrate SD candidate genes to tilapia linkage groups [18]–[20] nor the analysis of expressed sequence tags for ovary- and testis-specific libraries resulted in appreciable success so far [21]. Beside major QTL on LG1 and LG3 [22], Eshel et al. 2012 recently fine-mapped a SD region on LG23 [15]. The QTL mapped to 13–40 cM and more precisely peaked at 22 cM (F = 78.7; P<7.6×10−14). Linkage mapping located the SD QTL between markers GM597 and ARO124. The QTL region, located in scaffold 101 of the Nile tilapia genome assembly, harbours the anti-Müllerian hormone gene (amh), among others. A further study proposed that temperature-dependent sex reversal similarly seems to be influenced in at least some families by an even broader QTL region (between GM283 and UNH898) on LG23 [23].
The amh gene is involved in the development of the urogenital system during embryogenesis by suppressing the Müllerian ducts of mammals, birds and reptiles [24]–[26]. Both amh and its target in the TGF-ß pathway, the type II amh receptor (amhrII), are expressed in Sertoli cells of teleost [27]. Moreover, amh has been cloned in a number of fish species despite the fact that they lack Müllerian ducts [28]–[31]. The expression of the amh gene can be detected starting at 3 days post fertilisation (dpf) in XX and XY gonads [15]. The authors also demonstrated a significantly higher expression of amh in all-male vs. all-female batches at 3 dpf. Differences became more pronounced until 7 dpf [15]. Earlier investigations tended to show dimorphic expression in the gonads at later stages, i.e. after dmrt1 at 19 dpf [32]. Moreover, sustained up regulation of dmrt1 and amh during temperature sex-reversal from 13–15 dpf onwards and down regulation of foxl2 and/or cyp19a1a at 17–19 dpf were observed during sexual development of Nile tilapia [33]. Even in the brain, amh expression appeared to be clearly sexually dimorphic during the period from 10 to 15 dpf [34].
This study reports the detection of allelic variants in the coding and 5′ upstream region of amh. The development of a high-throughput genotyping assay for a single SNP and its association with the phenotypic sex let amh appear as a candidate for autosomal and temperature-induced sexual development in Nile tilapia.
Materials and Methods
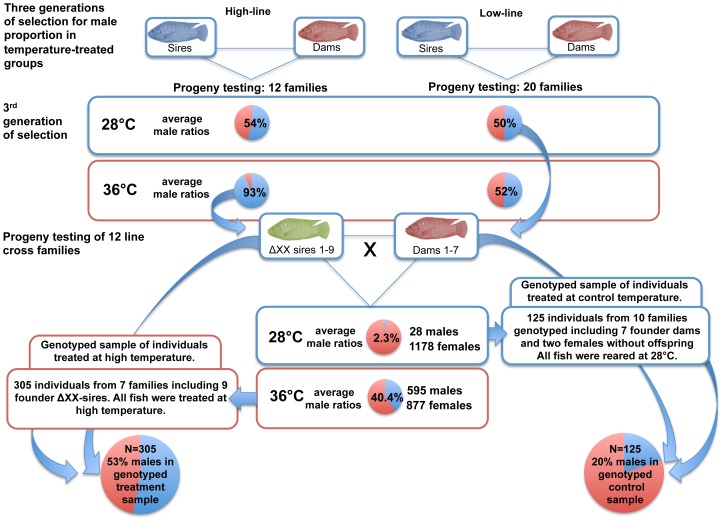
Stocks and Cross Design
The aim of the present study was to identify alleles in the amh gene with an influence on autosomal and temperature-dependent sex reversal. For this purpose genetically all-female (XX) populations were used, which had been tested for their chromosomal sex (XX vs. XY) via progeny testing (Figure 1). The genetically all-female population was derived from crosses between females from a selected line for low responsiveness to temperature (<60% males after 36°C treatment from 10–20 dpf) and temperature-sex reversed males from a line for high temperature-responsiveness (>90% males after 36°C treatment from 10–20 dpf) [3], [4]. The genetically female line was developed through mating of 9 sex-reversed (ΔXX) sires derived from 7 high-line families, which were treated at high temperature, to 7 females from 6 low-line families in order to produce a total of twelve families. One line cross family was derived from a within line mating (sire 105, dam 32, Table S1 in File S1). Briefly, progenies were obtained through artificial reproduction (Figure 1). Fertilised eggs were incubated for 10 days at 28°C. After yolk sac absorption, larvae from each family were randomly distributed into two groups (n∼110 larvae each). Temperature in the control groups was 28°C throughout the experiment, whereas treatment groups were kept at 36±0.5°C from 10–20 dpf according to [3]. From 20 dpf onwards, control and groups treated at high temperature were raised in separate tanks at 28°C for at least 2 months. From each family treated at high temperature 15 adult males and 15 adult females were phenotyped alive, assessing the urogenital papilla and the types of gametes each fish produced. All individuals that could not be confidently phenotyped, even after repeated inspection of the genital papilla, were excluded from further analysis. The remaining fish from the groups treated at high temperature and control groups were subjected to a lethal dose of phenoxyethanol (ethylene glycol monophenyl ether at 500 µL/L) and were immediately exanguated and dissected. The latter fish were phenotyped for their sex based on microscopical inspection of squashed gonads according to [35], classifying them into either testes or ovaries.
Figure 1. Experimental design to obtain a genetically female (XX) population in order to study temperature effects on the male proportion in a selected line of Nile tilapia.
Tissue samples from all individuals were collected from caudal fins and stored at −20°C until DNA extraction. The DNA was isolated from a sample of finclips by phenol-chloroform extraction [36]. The sample was comprised of 305 individuals from 7 out 12 line cross families including all 9 ΔXX founder sires of the 12 line cross families, which were all treated at high temperature. Furthermore DNA was extracted from 125 control individuals from 10 out of 12 line cross families including the 7 founder dams of the 12 line cross families and two additional females with no offspring. The numbers of fish genotyped from each family and corresponding control or treatment group are given in Table S1 and S5 in File S1. Water parameters measured during the experimental period were within the following range: oxygen >5.5 mg/L; pH 6.5–7.5; NH4+<0.5 mg/L; NO2−<0.25 mg/L. For first feeding, the fish were provided a diet rich in protein three times a day ad libitum (Tetra Werke, Germany; crude protein 48.5%). Generally, from day 20 until day 60, fish were fed ad libitum with a trout feed (Skretting F0 Aqua Brut, Norway; crude protein 48.5%) while from day 60 the fish were provided a carp feed (Skretting C2 Pro Aqua K18, Norway; crude protein 36%). All procedures were in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the German Animal Welfare Act [37]. This study was approved by the Institutional Animal Care and Use Committee of Goettingen University.
Sequencing of the amh gene
The genomic sequence of the amh gene was derived from Scaffold GL831234.1 of the Nile tilapia genome sequence deposited in the Ensembl database (http://www.ensembl.org; Orenil1.0 GCA_000188235.1; location of amh: Scaffold GL831234.1, 1.688.687–1.691.779). Gene-specific primers were designed to cover the coding sequence (cds), including intron and 5′UTR regions of the amh gene using the Primer3 software (see Table S2 in File S1). Each forward or reverse primer for sequencing of the amh gene was tailed at the 5′end with the M13 universal forward or reverse primer to enable direct bidirectional sequencing on an 3130xL Genetic Analyzer (Applied Biosystems, Germany) using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). DNA for sequencing of the amh gene was derived from 93 individuals comprised three families including the corresponding three dams and sires (Table S6 in File S1). PCR was carried out using 20 ng of genomic DNA, 1× PCR buffer containing MgCl2, 1× Q-solution, 10 pmol of each primer, 10 mM dNTPs and 2 U FastStart Taq DNA polymerase, in a final volume of 25 µl. All PCR components except the primers (MWG, Germany) and the 1× Q-solution (Qiagen, Germany) were purchased from Roche Diagnostics (Mannheim, Germany). PCR was performed using a Biometra T-3000 Thermocycler (Goettingen, Germany) with an initial denaturation at 95°C for 10 min, followed by 35 cycles of 92°C for 30 s; 60°C for 30 s and 72°C for 1 min with a final extension at 72°C for 5 min. The fragment identity was controlled via gel-electrophoresis on 1.5–2% agarose gels. PCR products were then purified with Exo-SAP-IT (USB, Germany). The obtained sequences were trimmed, contigs were built, and SNPs were manually identified using the program software suite DNASTAR Lasergene6 (DNASTAR, Inc., Germany).
Genotyping of SNP ss831884014 within amh
Comparative sequencing revealed six SNPs within the amh gene (Figure 2). To genotype SNP ss831884014, exhibiting the largest effect on the phenotypic sex, a fluorescence resonance energy transfer (FRET) was developed using a LightCycler 480 instrument (Roche Diagnostics, Mannheim, Germany). Primers Fret-for and Fret-rev were designed to flank SNP ss831884014 yielding a fragment of 122 bp length. A 5′ Rox labelled fluorescent anchor probe (acceptor) was designed with a phosphorylated 3′ tail. The fluorescent sensor probe (donor) was designed with a 3′ Fam modification (Table S3 in File S1). At the 3′-end of the sensor probe the donor fluorescent molecule is excited at a wavelength of 533 nm. The fluorescent acceptor molecule at the 5′-end of the anchor probe receives the emitted energy from the donor. The emission of the fluorescence signal from the acceptor molecule is measured by the LightCycler 480 instrument using a filter combination of 483–610 nm. The sensor probe was designed to match the G-allele of SNP ss831884014. In the case of a mismatch due to presence of the C-allele, a lower melting temperature is detected. The LightCycler 480 software was adjusted to automatically detect genotypes based on melting curve results. All genotypes were manually checked for genotyping errors, deciphered in triplicates via Sanger sequencing. Homozygous C/C individuals showed a fluorescence peak between 483 and 610 nm wavelength at 58°C, whereas homozygous G/G individuals showed one at 64°C, and heterozygous fish showed two peaks (Figure S1 in File S1). The melting curve analysis comprised of an initial denaturation step (95°C for 1 min), a step rapidly lowering the temperature to 40°C and holding for 30 sec, and a heating step slowly increasing the temperature up to 80°C under a continuous measurement of fluorescence (1 acquisition/°C). In total 337 individuals were investigated using the FRET assay, in order to obtain a total of 430 genotypes at SNP ss831884014 (including the before mentioned Sanger-sequenced individuals). Briefly, the PCR was run in a 25 µl reaction volume consisting of 1×PCR buffer containing MgCl2, 0.4 pmol of each primer and each FRET probe, 10 mM dNTPs, 2 U FastStart Taq DNA polymerase, H2O and 20 ng of DNA. All PCRs were carried out in 96 well plates in a LightCycler 480 with an initial denaturation at 95°C for 10 min, followed by 35 cycles of 92°C for 30 s, 56°C for 30 s and 72°C for 30 s.
Figure 2. Gene structure, pairwise linkage disequilibrium (r2) between SNPs, and functional annotation of the Nile tilapia amh gene derived from the Ensembl database.
a) Gene structure of the amh gene derived from Scaffold GL831234.1. Exons 1–7 (E1–7) are represented by filled grey boxes. Blue and red dashed lines show positions of polymorphic SNPs in the amh gene. SNP ss831884015 (C>T) is located −225 bp upstream of the start codon ATG, SNP ss831884018 was located in the intron between exon 6 and 7. Variants ss831884014 and ss831884019 were found in exons 6 and 7. SNP ss831884014 (G>C) and ss831884019 (C>T) were missense mutations leading to amino acid changes from glutamic acid to glutamine (codons Gaa/Caa) and alanine to valine (codons gCg/gTg). Table 1 deals with the functional annotation of all six SNPs. b) Pairwise linkage disequilibrium heat map of four polymorphic allelic variants in the Nile tilapia amh gene. c) Partial DNA to protein translation of exon 6 in the Nile tilapia amh gene, depicting a non-synonymous amino acid substitution at position 376 of the putative amh-protein (indicated by black arrow) coded by an allelic variant ss831884014 of scaffold GL831234.1. d) Partial comparative alignment of the amh genomic exon 6 sequences for six teleost species derived from the Ensembl database. e) Partial comparative alignment of the amh protein sequence for six teleost species derived from the Ensembl database.
Statistical analysis
For the segregating SNPs (n = 4) the gene diversity, the allele- as well as genotype frequencies were analysed using the SAS/Genetics 9.3 software (SAS Inst., Inc., Cary, NC, USA). Phased parental haplotypes were reconstructed using the software package PLINK for 93 animals treated at high temperature [38]. The 93 individuals comprised three families including the corresponding three dams and sires. R2-values were calculated in SAS/Genetics 9.3 software as a measure of linkage disequilibrium between segregating SNPs [39], [40]. Associations of segregating SNPs with the phenotypic sex of the fish were investigated, fitting a generalized linear model (GLM) with binominal error distribution and logit function in SAS version 9.3. First, a stepwise logistic regression analysis was carried out to detect associations between any of the SNP genotypes coded as 0 (CC), 1 (GC), or 2 (GG) and sex coded as a binary trait (0 = male, 1 = female). Second, a gene substitution model was applied to detect sex specific effects and those of significant SNP driven from the stepwise regression. A stepwise logistic regression revealed a significant effect of SNP ss831884014 and ss831884018 on the probability of developing a male functional phenotype (stepwise logistic regression, binary logit, chi-square 9.81, p<0.0017). Both SNPs showed a significant effect of equal magnitude on the male proportion in progenies treated at high temperature as they were in full linkage disequilibrium (LD, r2 = 1, no recombination event occurred between the two SNPs). However, SNP ss831884018 was not further considered as the potentially causal variant due to its position in the non-coding region (intron 6; Table 1). The following model was applied:
| (1) |
where  is the probability of obtaining the male phenotype, φ is the overall mean effect,
is the probability of obtaining the male phenotype, φ is the overall mean effect, 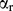 is the fixed effect of the treatment (levels: 28°C or 36°C);
is the fixed effect of the treatment (levels: 28°C or 36°C); 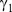 is the linear regression coefficient for association between the probability of obtaining the male phenotype and SNP ss831884014 genotype, Xrs is the effect of SNP ss831884014 genotype (levels: 0 (C/C), 1 (G/C), 2 (G/G));
is the linear regression coefficient for association between the probability of obtaining the male phenotype and SNP ss831884014 genotype, Xrs is the effect of SNP ss831884014 genotype (levels: 0 (C/C), 1 (G/C), 2 (G/G)); 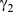 is the linear regression coefficient for the fixed interaction effect. For derivation of the gene substitution effect only the significant parameters were considered in the final model. Since the inverse link is nonlinear and does not give an equal substitution effect, in addition the model parameters were estimated by a general linear model (GLM) using a normally distributed response variable with an identity link function.
is the linear regression coefficient for the fixed interaction effect. For derivation of the gene substitution effect only the significant parameters were considered in the final model. Since the inverse link is nonlinear and does not give an equal substitution effect, in addition the model parameters were estimated by a general linear model (GLM) using a normally distributed response variable with an identity link function.
Table 1. Functional annotation of six allelic variants in the Nile tilapia amh gene.
| SNP ID | Allele1 | Consequence1 | Position | Position in cDNA | Position in CDS | Position in protein | Amino acid change1 | Codon change1 |
| ss831884015 | C/T | upstream gene variant | 5′UTR | - | - | - | - | - |
| ss831884016 | A/T | intron variant | Intron 4 | - | - | - | - | - |
| ss831884017 | C/G | synonymous variant | Exon 5 | 696 | 696 | 232 | G (no change) | ggC/ggG |
| ss831884014 | G/C | missense variant | Exon 6 | 1126 | 1126 | 376 | E/Q | Gaa/Caa |
| ss831884018 | G/A | intron variant | Intron 6 | - | - | - | - | - |
| ss831884019 | T/C | missense variant | Exon 7 | 1157 | 1157 | 386 | A/V | gCg/gTg |
(1The annotation of functional effects was carried out using SNPEff 2.0.5 (http://SNPeff.sourceforge.net/) using the Oreochromis niloticus genome sequence deposited in the Ensembl database (scaffold GL831234.1)).
Additive and dominance effects of alleles were calculated according to the following model:
| (2) |
where  is the probability of obtaining the male phenotype, φ is the overall mean effect,
is the probability of obtaining the male phenotype, φ is the overall mean effect, 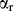 is the fixed effect of the treatment (levels: 28°C or 36°C from 10–20 dpf);
is the fixed effect of the treatment (levels: 28°C or 36°C from 10–20 dpf);  is the fixed effect of SNP ss831884014 genotype (levels: 0 (C/C), 1 (G/C), 2 (G/G));
is the fixed effect of SNP ss831884014 genotype (levels: 0 (C/C), 1 (G/C), 2 (G/G)); 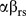 is the fixed effect of interaction between genotype and treatment. First, least square means were estimated on the logit scale and then back-transformed using the inverse link function
is the fixed effect of interaction between genotype and treatment. First, least square means were estimated on the logit scale and then back-transformed using the inverse link function  to the original scale (probability) applying the least square means (LSmeans) statement. Significant differences between least square means were tested using a t-test by inclusion of the PDIFF option in the LSmeans statement. Standard errors of least square means were calculated as described by [41]. Significant deviation of estimates of dominance effects from zero was tested using a t-test as described in [42].
to the original scale (probability) applying the least square means (LSmeans) statement. Significant differences between least square means were tested using a t-test by inclusion of the PDIFF option in the LSmeans statement. Standard errors of least square means were calculated as described by [41]. Significant deviation of estimates of dominance effects from zero was tested using a t-test as described in [42].
Results
Phenotypes: Sex ratios in controls and groups treated at high temperature
Temperature had a significant effect on the sex ratio of the genetically female (XX) line of Nile tilapia consisting of 12 families investigated in the present study. The overall male percentage when reared at control temperature of 28°C, was 2.3%. Losses from 10 dpf until sexing in control groups were 8.6%. In the corresponding full sib groups treated at high temperature, the male percentage was 40.4%, while mortalities were 4.4%.
In the genotyped sample, which was derived from the before mentioned genetically female (XX) line of Nile tilapia consisting of 12 families, a male proportion of 20% in controls, and 53% in the group treated at high temperature was observed. Similarly, the temperature treatment significantly affected the proportion of phenotypic males (p<0.0001).
Functional annotation of amh variants
In total, sequencing of the amh gene revealed six SNPs (ss831884014 to ss831884019, Table 1, Figure 2). SNP ss831884015 (C>T) was detected in the 5′ upstream region, located −225 bp upstream of the start codon, and was classified as intergenic modifier. Two of the identified SNPs were located in introns (intron 4 SNP ss831884016, and intron 6 SNP ss831884018). Finally, three variants were assigned to exons 5 (ss831884017), 6 (ss831884014) and 7 (ss831884019). SNP ss831884017 was a synonymous mutation at position 696 of the coding sequence, corresponding to position 232 of the protein. Moreover, SNP ss831884014 (G>C) and ss831884019 (C>T) were missense mutations leading to amino acid changes from glutamine to glutamic acid (codons Gaa/Caa) and alanine to valine (codons gCg/gTg).
Frequency distribution of geno- and haplotypes
Four out of six variants (SNP ss831884015, ss831884014, ss831884018, and ss831884019) segregated in the investigated 93 individuals from the genetically female Nile tilapia population (Table 2). Moreover, four phased haplotypes from parent to offspring were reconstructed (C-C-A-C, C-G-G-C, C-G-G-T, T-G-G-T). Variants ss831884014 and ss831884018 were in full linkage disequilibrium (LD). SNPs ss831884015 and ss831884019 also exhibited a high degree of LD, showing an r2-value of 0.7. The LD between SNPs ss831884014 and ss831884019 as well as between ss831884018 and ss831884019 was intermediate, showing an r2-value of 0.54. In contrast, LD between SNPs ss831884015 and ss831884014, as well as ss831884015 and ss831884018 was lower, showing an r2-value 0.21 (Figure 2, Table S4 in File S1).
Table 2. Allele frequencies, polymorphism information content (PIC), heterozygosity and allelic diversity for four allelic variants in the amh gene detected in a genetically all-female (XX) Nile tilapia population.
| SNP locus | n individuals | Allele frequency | PIC | Heterozygosity | Allelic diversity |
| ss831884015 | 93 | 0.90 (C) | 0.1595 | 0.1935 | 0.1748 |
| ss831884014 | 931 | 0.71 (C) | 0.3245 | 0.4194 | 0.4075 |
| 4302 | 0.58 (C) | 0.3677 | 0.5186 | 0.4856 | |
| ss831884018 | 93 | 0.71 (A) | 0.3245 | 0.4194 | 0.4075 |
| ss831884019 | 93 | 0.84 (C) | 0.2340 | 0.3226 | 0.2706 |
(1 Initially 93 individuals were Sanger-sequenced for the amh gene. The genotyped sample was comprised of three families including the corresponding three dams and sires; 2 In total 337 individuals were investigated using the FRET-assay, in order to obtain a total of 430 genotypes at SNP ss831884014 (including the before mentioned Sanger-sequenced individuals)).
Association of amh variant ss831884014 with autosomal and temperature-dependent sex reversal
A fluorescence resonance energy transfer (FRET) based genotyping assay was developed for SNP ss831884014 and validated on a representative sample of fish, comprised of 125 individuals reared at control temperature (28°C) and 305 individuals reared at 36°C from 10–20 dpf. The statistical analysis revealed a highly significant effect of the treatment and genotype on the phenotypic sex (Table 3). In contrast, no significant genotype×treatment interaction, and thus no difference in association between fish reared at control or high temperature, was detected (interaction term: df = 424, F = 1.83, p = 0.1618, Table 3). Furthermore, using a reduced model including only the two main factors SNP and treatment as well as applying a Bonferroni correction resulted in significant differences between the group treated at high temperature and the control (p<0.0001), and SNP-genotypes (CC vs. GG; p<0.0007; GC vs. GG; p<0.0067), as no significant interaction between the SNP and treatment was observed (Table 3).
Table 3. Effect of SNP ss831884014 genotypes, temperature treatment (28°C vs. 36°C from 10–20 dpf) and their interaction on the phenotypic sex in Nile tilapia.
| Scale | Effect | Equation 1 | Equation 2 | ||||
| df Numerator | F-statistics | Pr>F | df Numerator | F-statistics | Pr>F | ||
| Logit link | treatment | 1 | 4.60 | <.0326 | 1 | 15.49 | <.0001 |
| function | SNP genotype | 1 | 12.66 | 0.0004 | 2 | 5.33 | 0.0025 |
| treat*SNP genotype | 1 | 2.63 | 0.1059 | 2 | 1.83 | 0.1618 | |
| Identity link | treatment | 1 | 8.18 | <.0044 | 1 | 24.02 | <.0001 |
| function | SNP genotype | 1 | 12.15 | 0.0005 | 2 | 6.87 | 0.0012 |
| treat*SNP genotype | 1 | 0.68 | 0.4102 | 2 | 1.35 | 0.2593 | |
(The effects were estimated using a GLM with binominal error distribution and logit link function and were validated using a GLM with identity link function; Equation 1: Linear regression model  ; Equation 2: Fixed effect model
; Equation 2: Fixed effect model 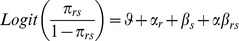 .
.
Generally, the genotype CC showed a more pronounced effect on the percentage of males. Thus genotypic values over both treatments, given as LSmeans, were 0.4224 for the CC- compared to a significantly lower value of 0.1699 for the GG-genotype (p<0.0007). The genotypic LSmean of the heterozygous GC-genotype was 0.3604, which differed significantly from the genotype GG (p<0.0067).
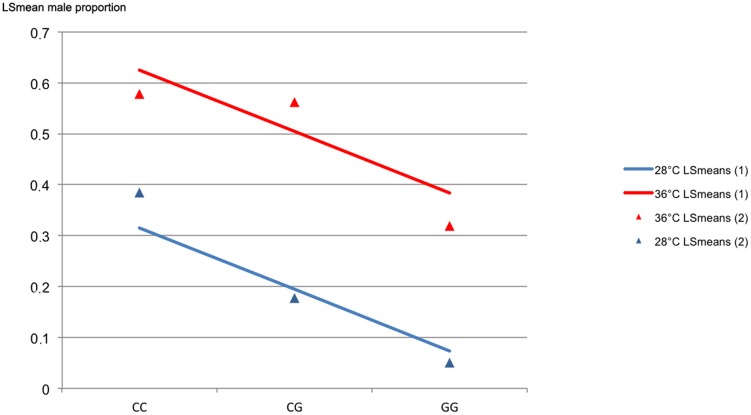
Although no significant interaction between treatments was detected, the genotypic LSmeans derived for both the group treated at high temperature and the control group are given separately in figure 3. The CC-genotype exhibited the highest and the GG-genotype the lowest genotypic LSmean male proportion, irrespective of the treatment. Thus, homozygous CC individuals showed an LSmean proportion of males (57.8%) when treated at high temperature. In comparison, homozygous CC specimens kept at 28°C exhibited LSmeans of 38.4%. The LSmean male proportions dropped to 31.9% and 5.0% for individuals possessing the homozygous genotype GG, in the group treated at high temperature and the control group, respectively.
Figure 3. Relationship between male proportion and genotype at amh variant ss831884014 in Nile tilapia, reared at 28°C or 36°C from 10 to 20 days post fertilisation.
Linear least-square regression of the genotypic values (LSmean male proportion) for genotypes C/C, G/C, and G/G at locus ss831884014 of the amh gene (Scaffold GL831234.1). The genotypic values were derived from the regression models by either excluding the interaction between SNP and treatment (LSmeans 1: red and blue line) or by including the interaction between SNP and treatment (LSmeans 2: red and blue triangles) as effect class. Numbers of individuals per genotype were 26/79/20 and 114/144/47 for genotypes C/C, G/C, and G/G, for the control group (28°C) and the group reared at high temperature (36°C), respectively.
Gene substitution effect, homozygous additive allele effect, and dominance effect of amh variant ss831884014 for autosomal and temperature-dependent sex reversal
An analysis of covariance was carried out to estimate the gene substitution effect, where the effect of the SNP-genotype was considered as the regression term (equation 2). The estimated gene substitution effect was 12% for both treatments. Genotypic values derived from the corresponding regression model were 63%, 50%, and 38% for genotypes CC, GC, and GG in fish treated at high temperature. In contrast to this, lower genotypic values of 32%, 19%, and 7% were observed in fish kept at the control temperature of 28°C for genotypes CC, GC, and GG, respectively. Moreover, the genotype effect of SNP ss831884014 on the male proportion was significantly higher in groups treated at high temperature, yielding 31% higher male ratios (LSmeans) on average of the three genotypes (Figure 3). Furthermore, the homozygous additive effect of the C-allele was estimated, which deals with the increase in male percentage contributed by each copy of the C-allele. This effect was calculated as the mean between the two homozygous genotypes (a = μCC−μGG)/2). In controls the homozygous additive allele effect showed a value of 16.7% (S.E. = 6.9), whereas a value of 12.9% (S.E. = 4.04) was observed in fish treated at high temperature (Table 4). In addition to the additive effects of alleles, dominance is of theoretical and practical importance (i.e. in breeding). A significant dominance deviation of 11.4% (S.E. = 5.6) was estimated for the fish treated at high temperature. In contrast, no significant dominance deviation was detected in individuals reared at control temperature (−4.01%, S.E. = 8.6).
Table 4. Dominance and additive effects of the variant ss831884014 on the male proportion in Nile tilapia reared at 28°C or 36°C from 10–20 dpf.
| Genetic parameter | Treatment | Estimate1 | Back transformed Lsmeans2 | Estimate3 | t-value1 | p-Value1 | |
| Dominance | = μCG−(μCC+μGG)/2 | 28°C | 0.1719 (0.6250) | −0.0400 | −0.0400 (0.0868) | 0.28 | 0.7834 |
| = μCG−(μCC+μGG)/2 | 36°C | 0.4709 (0.2484) | 0.1135 | 0.1135 (0.0560) | 1.90 | 0.0586 | |
| Homozygous additive allele effect | = (μCC−μGG)/2 | 28°C | 1.2372 (0.5512) | 0.1673 | 0.1673 (0.0693) | 2.24 | 0.0253 |
| = (μCC−μGG)/2 | 36°C | 0.5381 (0.1830) | 0.1299 | 0.1299 (0.0404) | 2.94 | 0.0035 | |
(Values given brackets are standard errors of the estimates; 1) Parameters were derived from GLM with binominal error distribution and logit link function; 2) Dominance and homozygous additive allele effects were derived from back transformed LSmeans; 3) Parameters were derived from GLM with identity link function).
Discussion
Tilapias have the potential to become the most important aquaculture species group world-wide [43]. They serve as a means for poverty alleviation and concurrently constitute a world-wide commodity, being sold as frozen fillets to export markets in the US, Europe, and Asia [44]. More than 50% of seafood consumed comes from aquaculture; tilapia is a major mainstay of this production.
Tilapia differ from other aquaculture species insofar as they can thrive under poor conditions and on diets that contain low shares of fish meal [2], but a major sustainability constraint - i.e. hormonal sex-reversal - is yet to be overcome. The present study reports novel allelic variants in the Nile tilapia amh gene. One of the variants is associated with the sex and might enable marker-assisted selection. A strategy is proposed to increase the male proportion by the development of a straightforward genotyping assay combined with temperature treatment of fry.
Association of amh variant ss831884014 with autosomal and temperature-dependent sex reversal
The largest effect on the phenotypic sex of Nile tilapia was observed in homozygous carriers of the C-allele at SNP ss831884014, a missense variant that leads to an amino acid change from glutamine to glutamic acid at position 376 of the putative amh protein. Moreover, a ten-day temperature treatment of the sexually undifferentiated fry carrying the homozygous CC genotype significantly increased the probability of developing the male phenotype. Although the homozygous effect of the C-allele was higher in the control groups (16.7% compared to 12.9% males), the average genotypic values were 31% higher (LSmean male ratios) in groups treated at high temperature.
Two factors were identified which suggest that the C-allele (in homozygous and heterozygous allelic state) in our population has the potential to significantly increase the proportion of males: 1) Genotypic values of 63% and 32% males for the CC-genotype and 38% and 7% for the alternative GG-genotype in the group treated at high temperature and control group, respectively; 2) A significant dominance deviation (+11.4% of males) of the heterozygous (CG) genotype in the fish treated at high temperature.
Comparative genomics and functional role of the amh variant ss831884014
A comparative analysis of the amh protein revealed that the G-allele might be the ancestral allele at locus ss831884014 (Figure 2, panel e). Species such as tilapia, stickleback, and medaka have the amino acid glutamic acid (E: coded by Gaa, Gag) in their protein sequence, whereas puffer fish and platy fish exhibit the acidic amino acid aspartate (D: coded by Gac, Gat). Hence, this might indicate that glutamine (Q), which is coded by the nucleotide codon Caa, is more recent in the Nile tilapia line investigated here and might have taken over a critical role in autosomal and temperature-dependent sex reversal.
The amh gene was identified as a potentially sex-modifying cue in a number of earlier studies [15], [16], [18], [22], [32]–[34]. On the one hand, expression analysis confirmed amh as a functional marker of maleness at a precocious age of 3 dpf and later during sex differentiation [15], [32]–[34]. On the other hand, evidence for sex-specific differences in the amh gene at the genomic level was lacking so far. An earlier identified variant (AM232733), similarly located in exon 6 of the gene, proved not to be a trigger for sexual fate in Nile tilapia [18]. Moreover, the Nile tilapia population investigated here (Lake Manzala population), showed no segregation of variant AM232733. However, among 10 genes putatively involved in sex determination or differentiation, amh is located in the centre of an SD QTL on LG23 [15].
The role of the amh variant ss831884014 in the continuum of major genetic, minor genetic, and temperature-dependent factors
The present study does confirm - for the first time - that an allelic variant in the amh gene might be a major QTL for autosomal and temperature-dependent sex reversal in Nile tilapia. In the Patagonian pejerrey a functional duplication of the amh gene suggests that amhy may be the master sex-determining gene [45], but our results for tilapia point in another direction: multiple interacting loci each partially contributing to the formation of the sexual phenotype. Although the allelic variant reported here exerts a large effect on the formation of the sexual phenotype under both rearing environments, not all sex phenotypes can be explained using a simple one-locus model. Recently, restriction site associated sequencing revealed the main sex-determining locus flanked by two SNPs (Oni23063 and Oni28137) and located on linkage group 1 [46]. The authors showed that in one family, sex-reversed males which were treated at high temperature carried the female genotype at SNP Oni23063 and Oni28137, but did not show an association with the temperature-dependent sex [46]. The underlying gene of the main sex-determining locus on LG1 is unknown so far. Further studies are needed to verify if SNP ss831884014 on LG23 acts as autosomal sex modifying cue in this Nile tilapia line, which in addition or alternatively to the major genetic factor on LG1, described in [46], increases the probability of developing the male phenotype during temperature-dependent sex reversal. The missing significant interaction between the SNP-genotype and treatment effect detected in the present study (Table 3) suggests that the same genetic network (i.e. the TGF-ß pathway) might be at least partially active during autosomal and temperature-dependent sex reversal in this Nile tilapia line, yet with a drastically pronounced effect under elevated rearing temperatures from 10–20 dpf.
Further research should now focus on the functional role of the identified allelic amh variant during autosomal and temperature-dependent sex reversal. The underlying gene networks, such as the TGF-ß pathway, need to be investigated in greater depth, and their transcriptional, translational, and phenotypic consequences have to be evaluated to fully characterize the effect on the phenotypic sex described here. Targeted techniques such as RNA-seq [47], or morpholino-mediated knockdown of known target genes might reveal the functional role of genes and variants [48]. Another avenue of enquiry focuses upon the specific biochemical properties of the amh hormone. The combined use of temperature treatment and marker-assisted selection to introgress the C-allele at SNP ss831884014 into other strains of cultivated tilapia might help to increase the male proportion and therefore the overall productivity. Ultimately, increasing the yield through higher male proportions without counteracting reproductive traits would greatly contribute to increase the efficiency of Nile tilapia culture and protein availability in predominant small-scale aquaculture, while also minimizing negative environmental effects due to hormonal sex-reversal from expanding intensive aquaculture systems.
Conclusions
This study shows that an allelic variant in the amh gene is a major QTL for autosomal and temperature-dependent sex reversal in a selected line of Nile tilapia. If the applicability of this marker in other populations proofs successful, a combined strategy of marker-assisted selection and temperature treatment might be beneficial to increase the male proportion in aquaculture stocks of O.niloticus.
Supporting Information
Contains supporting Figures and Tables. Figure S1, Melt curve analysis output from the Lightcycler 480 system for genotypes at amh variant ss831884014. Different genotypes are illustrated using different colours: C/C = green curve, C/G and G/C = blue curve, and G/G = red curve. Homozygeous C/C individuals show a fluorescence peak between 483 and 610 nm wave length at 58°C, whereas homozygeous G/G individuals showed one at 64°C, and heterozygous fish showed two peaks. Table S1, Pedigree and sex ratios of the genetically female population reared at control (28°C) and elevated temperature (36°C) from 10 to 20 dpf. Table S2, Forward and reverse primers tailed with a universal M13 forward or reverse primer for bidirectional sequencing the amh gene in Nile tilapia. Table S3, Fret-primer for allelic variant 1690582 in the Nile tilapia amh gene, anchor and sensor probe sequences and positions on scaffold GL831234.1. Table S4, R2-measure of linkage disequilibrium between four segregating allelic variants in the amh gene of Nile Tilapia. The estimates were derived from a sample 93 temperature-treated Nile tilapia individuals. Table S5, Raw data for the genetically female study population reared at control (28°C) and elevated temperature (36°C) from 10 to 20 dpf. Table S6, Genotypes of four segregating SNPs in the amh gene of 93 individuals derived from three Nile tilapia families.
(ZIP)
Acknowledgments
We thank Birgit Reinelt for her technical assistance in the recirculation system and help during sampling of fish. We are grateful to E. Schütz for designing the FRET-probes.
Funding Statement
This work was funded by the German Research Foundation DFG (DFG WE4434/2-1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Cressey D (2009) Aquaculture: Future fish. Nature 458: 398–400 Available: http://www.nature.com/news/2009/090325/full/458398a.html. Accessed 2013 July 11. [DOI] [PubMed] [Google Scholar]
- 2.Cressey D (n.d.) Tilapia standard showcases continuing growth of aquaculture: Nature News Blog. Available: http://blogs.nature.com/news/2012/08/tilapia-standard-showcases-continuing-growth-of-aquaculture.html. Accessed 2013 July 10.
- 3. Wessels S, Hoerstgen-Schwark G (2007) Selection experiments to increase the proportion of males in Nile tilapia (Oreochromis niloticus) by means of temperature treatment. Aquaculture 272: S80–S87 Available: http://linkinghub.elsevier.com/retrieve/pii/S0044848607008216. Accessed 2011 July 18. [Google Scholar]
- 4. Wessels S, Hoerstgen-Schwark G (2011) Temperature dependent sex ratios in selected lines and crosses with a YY-male in Nile tilapia (Oreochromis niloticus). Aquaculture 318: 79–84 Available: http://linkinghub.elsevier.com/retrieve/pii/S0044848611003668. [Google Scholar]
- 5. Baroiller JF, D'Cotta H, Bezault E, Wessels S, Hoerstgen-Schwark G (2009) Tilapia sex determination: Where temperature and genetics meet. Comp Biochem Physiol A Mol Integr Physiol 153: 30–38 Available: http://www.ncbi.nlm.nih.gov/pubmed/19101647. Accessed 2011 August 16. [DOI] [PubMed] [Google Scholar]
- 6. Lee B-Y, Lee W-J, Streelman JT, Carleton KL, Howe AE, et al. (2005) A second-generation genetic linkage map of tilapia (Oreochromis spp.). Genetics 170: 237–244 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1449707&tool=pmcentrez&rendertype=abstract. Accessed 2011 August 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guyon R, Rakotomanga M, Azzouzi N, Coutanceau J-P, Bonillo C, et al. (2012) A high-resolution map of the Nile tilapia genome: a resource for studying cichlids and other percomorphs. BMC Genomics 13: 222 Available: http://www.ncbi.nlm.nih.gov/pubmed/22672252. Accessed 2012 August 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soler L, Conte MA, Katagiri T, Howe AE, Lee B-Y, et al. (2010) Comparative physical maps derived from BAC end sequences of tilapia (Oreochromis niloticus). BMC Genomics 11: 636 Available: http://www.biomedcentral.com/1471-2164/11/636. Accessed 2010 December 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shirak A, Palti Y, Cnaani A, Korol A, Hulata G, et al. (2002) Alleles and Distorted Sex Ratios in an Inbred Line of Tilapia (Oreochromis aureus). J Hered 97: 270–276. [DOI] [PubMed] [Google Scholar]
- 10. Lee B-Y, Penman DJ, Kocher TD (2003) Identification of a sex-determining region in Nile tilapia (Oreochromis niloticus) using bulked segregant analysis. Anim Genet 34: 379–383 Available: http://www.ncbi.nlm.nih.gov/pubmed/14510676. [DOI] [PubMed] [Google Scholar]
- 11. Lee B-Y, Hulata G, Kocher TD (2004) Two unlinked loci controlling the sex of blue tilapia (Oreochromis aureus). Heredity (Edinb) 92: 543–549 Available: http://www.ncbi.nlm.nih.gov/pubmed/15100706. Accessed 2012 December 18. [DOI] [PubMed] [Google Scholar]
- 12. Cnaani A, Zilberman N, Tinman S, Hulata G, Ron M (2004) Genome-scan analysis for quantitative trait loci in an F2 tilapia hybrid. Mol Genet Genomics 272: 162–172 Available: http://www.ncbi.nlm.nih.gov/pubmed/15449174. Accessed 2011 November 1. [DOI] [PubMed] [Google Scholar]
- 13. Karayücel İ, Ezaz T, Karayücel S, McAndrew BJ, Penman DJ (2004) Evidence for two unlinked “sex reversal” loci in the Nile tilapia, Oreochromis niloticus, and for linkage of one of these to the red body colour gene. Aquaculture 234: 51–63. [Google Scholar]
- 14. Cnaani A, Kocher TD (2008) Sex-linked markers and microsatellite locus duplication in the cichlid species Oreochromis tanganicae . Biol Lett 4: 700–703 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2614142&tool=pmcentrez&rendertype=abstract. Accessed 2012 January 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eshel O, Shirak A, Weller JI, Hulata G, Ron M, et al. (2012) Linkage and Physical Mapping of Sex Region on LG23 of Nile Tilapia (Oreochromis niloticus). G3 Genes|Genomes|Genetics 2: 35–42 Available: http://g3journal.org/cgi/doi/10.1534/g3.111.001545 Accessed 2012 January 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eshel O, Shirak A, Weller JI, Slossman T, Hulata G, et al. (2010) Fine-mapping of a locus on linkage group 23 for sex determination in Nile tilapia (Oreochromis niloticus). Anim Genet 1: 222–224 Available: http://doi.wiley.com/10.1111/j.1365-2052.2010.02128.x Accessed 2010 November 24. [DOI] [PubMed] [Google Scholar]
- 17. Liu F, Sun F, Li J, Xia JH, Lin G, et al. (2013) A microsatellite-based linkage map of salt tolerant tilapia (Oreochromis mossambicus×Oreochromis spp.) and mapping of sex-determining loci. BMC Genomics 14: 58 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3565888&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shirak A, Seroussi E, Cnaani A, Howe AE, Domokhovsky R, et al. (2006) Amh and Dmrta2 genes map to tilapia (Oreochromis spp.) linkage group 23 within quantitative trait locus regions for sex determination. Genetics 174: 1573–1581 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1667067&tool=pmcentrez&rendertype=abstract. Accessed 2012 October 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cnaani A, Lee B-Y, Ozouf-Costaz C, Bonillo C, Baroiller JF, et al. (2007) Mapping of sox2 and sox14 in tilapia (Oreochromis spp.). Sex Dev 1: 207–210 Available: http://www.ncbi.nlm.nih.gov/pubmed/18391531. Accessed 2013 January 3. [DOI] [PubMed] [Google Scholar]
- 20. Lee BY, Kocher TD (2007) Exclusion of Wilms tumour (WT1b) and ovarian cytochrome P450 aromatase (CYP19A1) as candidates for sex determination genes in Nile tilapia (Oreochromis niloticus). Anim Genet 38: 85–86 Available: http://www.ncbi.nlm.nih.gov/pubmed/17257199. Accessed 2013 January 3. [DOI] [PubMed] [Google Scholar]
- 21. Lee B-Y, Howe AE, Conte Ma, D'Cotta H, Pepey E, et al. (2010) An EST resource for tilapia based on 17 normalized libraries and assembly of 116,899 sequence tags. BMC Genomics 11: 278 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2874815&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cnaani A, Lee B-Y, Zilberman N, Ozouf-Costaz C, Hulata G, et al. (2008) Genetics of sex determination in tilapiine species. Sex Dev 2: 43–54 Available: http://www.ncbi.nlm.nih.gov/pubmed/18418034. Accessed 2011 August 4. [DOI] [PubMed] [Google Scholar]
- 23. Luehmann LM, Knorr C, Hoerstgen-Schwark G, Wessels S (2012) First evidence for family-specific QTL for temperature-dependent sex reversal in Nile tilapia (Oreochromis niloticus). Sex Dev 6: 247–256 Available: http://www.ncbi.nlm.nih.gov/pubmed/22797471. Accessed 2013 July 11. [DOI] [PubMed] [Google Scholar]
- 24. Josso N, Belville C, di Clemente N, Picard J-Y (2005) AMH and AMH receptor defects in persistent Müllerian duct syndrome. Hum Reprod Update 11: 351–356 Available: http://www.ncbi.nlm.nih.gov/pubmed/15878900. Accessed 2012 November 19. [DOI] [PubMed] [Google Scholar]
- 25. Smith Ca, Sinclair AH (2004) Sex determination: insights from the chicken. Bioessays 26: 120–132 Available: http://www.ncbi.nlm.nih.gov/pubmed/14745830. Accessed 2013 May 27. [DOI] [PubMed] [Google Scholar]
- 26. Shoemaker-Daly CM, Jackson K, Yatsu R, Matsumoto Y, Crews D (2010) Genetic network underlying temperature-dependent sex determination is endogenously regulated by temperature in isolated cultured Trachemys scripta gonads. Dev Dyn 239: 1061–1075 Available: http://www.ncbi.nlm.nih.gov/pubmed/20235200. Accessed 2011 August 8. [DOI] [PubMed] [Google Scholar]
- 27. Kluever N, Pfennig F, Pala I, Storch K, Schlieder M, et al. (2007) Differential expression of anti-Müllerian hormone (amh) and anti-Müllerian hormone receptor type II (amhrII) in the teleost medaka. Dev Dyn an Off Publ Am Assoc Anat 236: 271–281 Available: http://www.ncbi.nlm.nih.gov/pubmed/17075875. [DOI] [PubMed] [Google Scholar]
- 28. Miura T, Miura C, Konda Y, Yamauchi K (2002) Spermatogenesis-preventing substance in Japanese eel. Development 129: 2689–2697 Available: http://www.ncbi.nlm.nih.gov/pubmed/12015296. [DOI] [PubMed] [Google Scholar]
- 29. Yoshinaga N, Shiraishi E, Yamamoto T, Iguchi T, Abe S, et al. (2004) Sexually dimorphic expression of a teleost homologue of Müllerian inhibiting substance during gonadal sex differentiation in Japanese flounder, Paralichthys olivaceus . Biochem Biophys Res Commun 322: 508–513 Available: 10.1016/j.bbrc.2004.07.162 Accessed 2013 July 11. [DOI] [PubMed] [Google Scholar]
- 30. Rodríguez-Marí A, Yan Y-L, Bremiller RA, Wilson C, Cañestro C, et al. (2005) Characterization and expression pattern of zebrafish Anti-Müllerian hormone (Amh) relative to sox9a, sox9b, and cyp19a1a, during gonad development. Gene Expr patterns GEP 5: 655–667 Available: http://www.ncbi.nlm.nih.gov/pubmed/15939378. [DOI] [PubMed] [Google Scholar]
- 31. Halm S, Rocha A, Miura T, Prat F, Zanuy S (2007) Anti-Müllerian hormone (AMH/AMH) in the European sea bass: its gene structure, regulatory elements, and the expression of alternatively-spliced isoforms. Gene 388: 36–41 Available: http://www.ncbi.nlm.nih.gov/pubmed/17157448. [DOI] [PubMed] [Google Scholar]
- 32. Ijiri S, Kaneko H, Kobayashi T, Wang D-S, Sakai F, et al. (2008) Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus . Biol Reprod 78: 333–341 Available: http://www.ncbi.nlm.nih.gov/pubmed/17942796. Accessed 2012 November 4. [DOI] [PubMed] [Google Scholar]
- 33. Poonlaphdecha S, Pepey E, Canonne M, Verdal HDe, Baroiller J, et al. (2013) Temperature induced-masculinization in the Nile tilapia causes rapid up-regulation of both dmrt1 and amh expressions. Gen Comp Endocrinol Available: http://www.ncbi.nlm.nih.gov/pubmed/23800559. [DOI] [PubMed] [Google Scholar]
- 34. Poonlaphdecha S, Pepey E, Huang S-H, Canonne M, Soler L, et al. (2011) Elevated amh gene expression in the brain of male tilapia (Oreochromis niloticus) during testis differentiation. Sex Dev 5: 33–47 Available: http://www.ncbi.nlm.nih.gov/pubmed/21178329. Accessed 2013 January 3. [DOI] [PubMed] [Google Scholar]
- 35. Guerrero RD, Shelton WL (1974) An Aceto-Carmine Squash Method for Sexing Juvenile Fishes. Progress Fish-Culturist 36: 56–56 Available: 10.1577/1548-8659(1974)36[56:AASMFS]2.0.CO;2 Accessed 2013 July 11. [DOI] [Google Scholar]
- 36. Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 Available: 10.1016/0003-2697(87)90021-2 Accessed 2013 July 11. [DOI] [PubMed] [Google Scholar]
- 37.“Tierschutzgesetz in der Fassung der Bekanntmachung vom 18. Mai 2006 (BGBl. I S. 1206, 1313), das zuletzt durch Artikel 4 Absatz 90 des Gesetzes vom 7. August 2013 (BGBl. I S. 3154) geändert worden ist”
- 38. Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lewontin RC (1964) The Interaction of Selection and Linkage. I. General Considerations; Heterotic Models. Genetics 49: 49–67 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1210557&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hill WG, Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38: 226–231 10.1007/BF01245622 [DOI] [PubMed] [Google Scholar]
- 41.Littell RC, Milliken GA, Stroup WW, Wolfinger RD, Schabenberger O. (2006) SAS for Mixed Models, Second Edition, Cary, NC: SAS Institute Inc. [Google Scholar]
- 42. Gandolfi G, Cinar MU, Ponsuksili S, Wimmers K, Tesfaye D, et al. (2011) Association of PPARGC1A and CAPNS1 gene polymorphisms and expression with meat quality traits in pigs. Meat Sci 89: 478–485 Available: http://www.deepdyve.com/lp/elsevier/association-of-ppargc1a-and-capns1-gene-polymorphisms-and-expression-0gLB0aUQS3/fulltext. Accessed 2013 July 11. [DOI] [PubMed] [Google Scholar]
- 43.Liping L, Fitzsimmons K (2011) Better science, better fish, better life. Proceedings of the ninth international Symposium on tilapia in aquaculture. Liping L, Fitzsimmons K, editors Shanghai: AquaFish Collaborative Research Support Program AquaFish. [Google Scholar]
- 44. Gjedrem T (2012) Genetic improvement for the development of efficient global aquaculture: A personal opinion review. Aquaculture 344–349: 12–22 Available: http://linkinghub.elsevier.com/retrieve/pii/S0044848612001548. Accessed 2013 February 20. [Google Scholar]
- 45. Hattori RS, Murai Y, Oura M, Masuda S, Majhi SK, et al. (2012) A Y-linked anti-Müllerian hormone duplication takes over a critical role in sex determination. Proc Natl Acad Sci U S A 109: 2955–2959 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3286941&tool=pmcentrez&rendertype=abstract. Accessed 2012 October 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palaiokostas C, Bekaert M, Khan MGQ, Taggart JB, Gharbi K, et al. (2013) Mapping and Validation of the Major Sex-Determining Region in Nile Tilapia (Oreochromis niloticus L.) Using RAD Sequencing. PLoS One 8: e68389 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3708939&tool=pmcentrez&rendertype=abstract. Accessed 2013 August 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2949280&tool=pmcentrez&rendertype=abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Summerton J, Weller D (1997) Morpholino antisense oligomers: Design, preparation, and properties. Antisense Nucleic Acid Drug Dev 195: 187–195 Available: http://www.ncbi.nlm.nih.gov/pubmed/?term=Morpholino+Antisense+Oligomers%3A+Design%2C+Preparation%2C+and+Properties. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Contains supporting Figures and Tables. Figure S1, Melt curve analysis output from the Lightcycler 480 system for genotypes at amh variant ss831884014. Different genotypes are illustrated using different colours: C/C = green curve, C/G and G/C = blue curve, and G/G = red curve. Homozygeous C/C individuals show a fluorescence peak between 483 and 610 nm wave length at 58°C, whereas homozygeous G/G individuals showed one at 64°C, and heterozygous fish showed two peaks. Table S1, Pedigree and sex ratios of the genetically female population reared at control (28°C) and elevated temperature (36°C) from 10 to 20 dpf. Table S2, Forward and reverse primers tailed with a universal M13 forward or reverse primer for bidirectional sequencing the amh gene in Nile tilapia. Table S3, Fret-primer for allelic variant 1690582 in the Nile tilapia amh gene, anchor and sensor probe sequences and positions on scaffold GL831234.1. Table S4, R2-measure of linkage disequilibrium between four segregating allelic variants in the amh gene of Nile Tilapia. The estimates were derived from a sample 93 temperature-treated Nile tilapia individuals. Table S5, Raw data for the genetically female study population reared at control (28°C) and elevated temperature (36°C) from 10 to 20 dpf. Table S6, Genotypes of four segregating SNPs in the amh gene of 93 individuals derived from three Nile tilapia families.
(ZIP)