Abstract
Botanical primary metabolites extensively exist in herbal medicine injections (HMIs), but often were ignored to control. With the limitation of bias towards hydrophilic substances, the primary metabolites with strong polarity, such as saccharides, amino acids and organic acids, are usually difficult to detect by the routinely applied reversed-phase chromatographic fingerprint technology. In this study, a proton nuclear magnetic resonance (1H NMR) profiling method was developed for efficient identification and quantification of small polar molecules, mostly primary metabolites in HMIs. A commonly used medicine, Danhong injection (DHI), was employed as a model. With the developed method, 23 primary metabolites together with 7 polyphenolic acids were simultaneously identified, of which 13 metabolites with fully separated proton signals were quantified and employed for further multivariate quality control assay. The quantitative 1H NMR method was validated with good linearity, precision, repeatability, stability and accuracy. Based on independence principal component analysis (IPCA), the contents of 13 metabolites were characterized and dimensionally reduced into the first two independence principal components (IPCs). IPC1 and IPC2 were then used to calculate the upper control limits (with 99% confidence ellipsoids) of χ2 and Hotelling T2 control charts. Through the constructed upper control limits, the proposed method was successfully applied to 36 batches of DHI to examine the out-of control sample with the perturbed levels of succinate, malonate, glucose, fructose, salvianic acid and protocatechuic aldehyde. The integrated strategy has provided a reliable approach to identify and quantify multiple polar metabolites of DHI in one fingerprinting spectrum, and it has also assisted in the establishment of IPCA models for the multivariate statistical evaluation of HMIs.
Introduction
Metabolic profiling is essential to ensure the quality, consistency, safety and efficacy of herbal medicine products; especially for the injection dosage form. Water extraction or decoction is the most favored method of preparation of herbal medicine injections (HMIs). Saccharides, amino acids, organic acids, and other primary metabolites are unavoidably extracted along with targeted secondary metabolites during the process of HMIs, such as Qingkailing injection [1], Danshen injection [2], Guanxinning injection [3], and Shuxuetong injection [4]. In general, amino acids are considered to provide tonic activities and act as key regulators of nutrient metabolism, and polysaccharides are believed to be one of the most important constitutes in some herbal materials for pharmacological activities. Some monosaccharides have the suppressive effect on cell-mediated immune reactions [5]. However, these primary metabolites in HMIs are often ignored to detect and set corresponding quality criteria in China Pharmacopeia and national standard. Due to the strong polarity and hydrophilicity, conventional reversed-phase performance liquid chromatography (HPLC) fingerprint [6], [7]and other analytical methodologies, including anion exchange chromatography [8], gas chromatography [9], [10], and capillary electrophoresis [11], [12], are limited to separate and detect primary metabolites unless with complicated pretreatment, derivatization reagents and laborious preparation procedures. To delineate various class metabolites in HMIs, different types of fingerprints are necessary for a holistic quality evaluation, which are difficult to realize during the practical industry processing. Therefore, a simple and fast approach is required to be capable of detecting saccharides, amino acids, and organic acids, together with mainly bioactive secondary metabolites in one fingerprinting spectrum simultaneously.
Proton nuclear magnetic resonance (1HNMR) spectroscopy provides access to detect all proton-bearing compounds, almost irrespective of the chemical compound class. Because of the signal intensity directly proportional to the number of nucleus contributing to a specific resonance [13], 1H NMR method achieved the identification and quantification of metabolites in a one-step acquisition. With simple sample preparation, 1H NMR facilitates high-throughput analysis for metabolic studies and quality control of various food [14]–[17] and herbal materials [18]–[20]. As a universal technique with the simplicity and rapidity of implementation, 1H NMR has expansive prospect of application in profiling of polar metabolites of HMIs.
Since the complexity of chemical composition, HMIs cannot be completely represented by a limited number of certain bioactive compounds [21]. To extract the feature variables, principal component analysis (PCA) [22] and independent component analysis (ICA) [23] are classical tools to reduce the dimension of multivariate. The components (PCs) in PCA method are mutually orthogonal, while ICA method contains the components to be statistically independent [24]. ICA has been found to be a successful alternative to PCA in eliminating the overlapping information between the components [25]. However, ICA faces some limitations due to some instability [26], the choice of number of components to extract and high dimensionality [27]. As a consequence, independent principal component analysis (IPCA) was proposed by Yao et al. in 2012 [28] to use PCA as a pre-processing step to reduce the dimension of the data, and then use ICA as a denoising process of PCA to separate relevant information. On simulation studies and real data sets, IPCA offered a better visualization of the data than ICA and with a smaller number of components than PCA. Owing to the benefit to generate denoised the loading vectors, we attempted to employ IPCA method to construct χ2 and Hotelling T2 control charts [29] for multivariate statistical analysis.
Danhong injection (DHI) is a patent injection made from the extracts of Radix Salviae Miltiorrhizae and Flos Carthami [30]. It has been widely used for the prevention and treatment of cardiovascular and cerebrovascular diseases in clinic [31]–[33]. In our previous work, ultra-performance liquid chromatography (UPLC) coupled with UV detection was adopted to identify 11polyphenolic acids in DHI [34]. However, the total weight of identified constituents accounted for only a low proportion (about 10%) of the solid content in DHI.
In this study, we describe a strategy to detect more hydrophilic primary metabolites in DHI based on quantitative 1H NMR spectroscopy. The absolute concentration of identified metabolites was calculated by using internal standard method. Linearity, precision, repeatability, stability and accuracy were carried out to validate the method. The contents of polar metabolites were further evaluated by the multivariate analysis tool IPCA to establish χ2 and Hotelling T2 control charts.
Materials and Methods
Materials and Chemicals
Thirty-six batches of DHI manufactured in 2011, 2012 and 2013 were provided by Heze Buchang Pharmaceutical Co. Ltd (Heze, China). The standards of valine, threonine, alanine, pyroglutamate, procatechuic aldehyde and asparagine were purchased from the National Institute for Food and Drug Control (Beijing, China). Salvianic acid and procatechuic acid were obtained from Zhongxin Innova Laboratories (Tianjin, RP China). Succinate and malonate were obtained from Dr. Ehrenstorfer GmbH (Augsburg, Germany). Fructose and glucose were purchased from Sigma (Aldrich, America). Rutinose was purchased from Hazard Communication (Tokyo, Japan).The purities of the compounds were all above 98%, using NMR analysis. Deuterium oxide (D2O, 99.9%) and sodium 3-trimethylsilyl [2,2,3,3-d 4] propionate (TSP) were purchased from Cambridge Isotope Laboratories (Miami, FL, USA). D2O was used as internal lock; TSP was as internal standard for chemical shift calibration and quantification.
Ethics
No specific permission was required for the described field studies. The field locations are neither privately owned nor protected, and neither endangered nor protected species were involved.
Sample preparation
All the injection samples were subjected to freeze-drying. The dried powders (18 mg) were accurately weighed and dissolved with 600 µL of D2O containing 0.58 mM TSP. Exactly 500 µL of sample solution was transferred into a standard 5 mm NMR tube (Vineland, NJ, USA). No buffer was used due to the stable pH of injection.
NMR measurements
1H NMR spectra were acquired at 298 K on a Bruker AV III 600 MHz NMR spectrometer (600.23 MHz for proton frequency) with a 5 mm broadband BBFO probehead. All pulse sequences were from Bruker pulse program library. A standard one dimensional composite pulse sequence (zgcppr) was employed to suppress the residual water signal. The 90° pulse width was adjusted to about 13 µm for each sample. Sixty-four scans were collected into 32k data points using a spectral width of 12335 Hz, a relaxation delay of 1.0 s and an acquisition time of 2.66 s. A 0.3 Hz line-broadening function was applied to all spectra for Fourier transformation (FT) followed by phasing and baseline correction.
For proton signals assignment purposes, a set of two dimensional (2D) spectra, including 1H-1H correlation spectroscopy (COSY), 1H-1H total correlation spectroscopy (TOCSY), 1H J-resolved (J-res), and 1H-13C heteronuclear single quantum coherence (HSQC), were acquired for selected samples and processed with similar parameters as described previously [35].
Spin-lattice relaxation time (T 1) values of the quantified protons of individual constituent and TSP were measured using a classical inversion recovery pulse sequence with 10 relaxation delays (τ) ranging from 0.01 to 20 s.
Quantification of the metabolites
Because the intensity of a given 1H NMR signal is directly proportional to its contributing number of protons, the amount of metabolites in DHI can be measured by the signal areas of given metabolites and an internal reference with known concentration. Thirteen metabolites in DHI were selected for quantification. The important parameters for data acquisition and processing of 1H NMR spectra must be set appropriately to obtain accurate and precise measurements [36]. Most of all, the relaxation delay τ should be long enough to ensure complete relaxation for all the signals of interest.1H NMR measurements are done in a longer acquisition time by choosing τ≥5× longest T 1. For shortening the acquisition time, 1H NMR spectra can be acquired in an incompletely relaxed condition, and the absolute concentrations should be calculated taking the T 1 values in consideration [37]. With the effective magnetization reading pulse of 90°, the quantification of chemical constituents in this study can be performed by using the following equation:
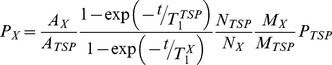 |
where PX and PTSP are the mass concentrations of metabolite and TSP, AX and ATSP are the integral areas for targeted signal of metabolite and for methyl groups of TSP, NX and NTSP are the proton numbers of metabolite and of methyl groups of TSP, MX and MTSP are the molar masses of metabolite and TSP, 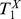 and
and 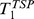 are the spin-lattice relaxation times for proton X and methyl protons of TSP, respectively; t is total relaxation time (relaxation delay plus acquisition time).
are the spin-lattice relaxation times for proton X and methyl protons of TSP, respectively; t is total relaxation time (relaxation delay plus acquisition time).
The quantitative 1H NMR method was checked for linearity, precision, repeatability, stability, and accuracy. Precision, repeatability and stability were calculated as the relative standard deviation (RSD). Recovery test was employed to determine the accuracy, and four typical compounds, alanine, glucose, salvianic acid and procatechuic aldehyde, were chosen to evaluate the average recovery.
Assay for multivariate quality control
36 batches of DHI samples were split into two phases, Phase I and Phase II. Phase I as training set included 25 batches of qualified products manufactured continuously in the years of 2012 and 2013. Phase II as testing set included 6 batches of qualified products in 2012 and 2013, and 5 batches of expired products manufactured in 2011.
The quantitative resulting data of 13 metabolites was imported to R 3.0.2 software loaded with packages of MVA, MSQC and mixOmics (www.r-project.org) for multivariate statistical analysis. For simplifying the multivariate problem, principal component analysis (PCA) and independence principal component analysis (IPCA) were performed to reduce the dimensionality of data. The scores of principal components characterized the whole data were then imported to χ2 and hotelling T2 control charts to calculate the upper control limits (with 99% confidence ellipsoids). Moreover, as one of the requisites in control chart is the independence of the data, the independence of selected components in PCA and IPCA models were validated by autocorrelation function (ACF). The out-of-control samples in Phase II were examined by the upper control limits achieved from Phase I.
Result and Discussion
Proton signal assignments and chemical identification
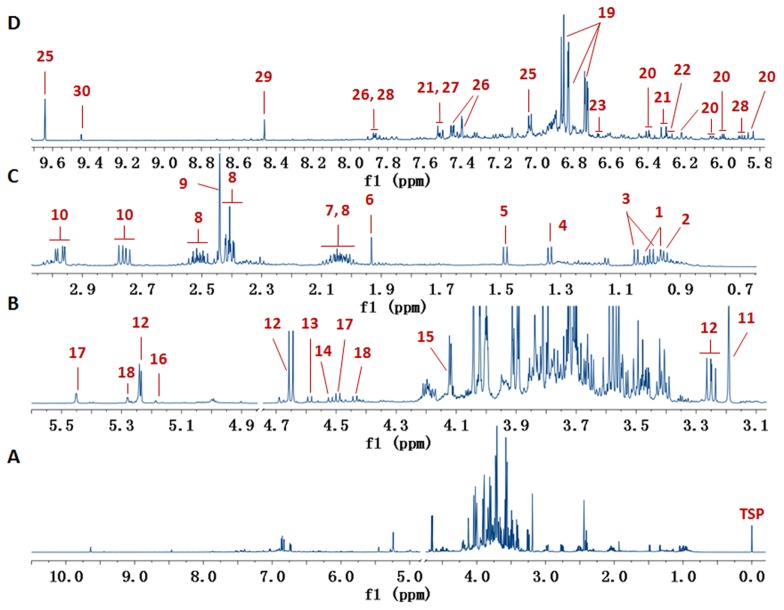
A representative 1H NMR spectrum of DHI was shown in Fig.1. The resonance signals were assigned to 30 metabolites based on the elucidation with extensive 2D NMR experiments (1H-1H COSY, 1H-1H TOCSY, 1H J-resolved, HSQC),the literature data in our former work [35], and in-house database. In the range of δ 3.2–5.8, the spectrum is dominated by 5monosaccharides and 2 disaccharides, including glucose, galactose, arabinose, fructose, rhamnose, rutinose, and rutinulose. In the high-field region (δ 0.5–3.2), 8 amino acids (isoleucine, leucine, valine, threonine, alanine, proline, pyroglutamate, asparagine) and 3 organic acids (acetate, succinate, malonate) were observed. In the low-field region (δ 5.8–10.0), 7 polyphenolic acids, including salvianic acid, salvianolic acid B, salvianolic acid A, rosmarinic acid, lithospermic acid, procatechuic acid and procatechuic aldehyde, together withuridine and 5-(hydroxymethyl)-2-furaldehyde (5-HMF), were identified. Moreover, 3 organic acids (4-hydroxybenzoic acid, 4-hydroxycinnamic acid, and formate) were observed as well in the low-field region. The chemical shifts of the identified 30 metabolites by1H NMR were listed in Table 1.To our knowledge, 7 saccharides and 6 organic acids were reported for the first time in DHI. Without the need of any sample pretreatment or pre-column derivatization, the established 1H NMR method provided an approach to determine 7 saccharides, 6 organic acids, 8 amino acids, 1nucleoside, 1 carbohydrate derivatives (5-HMF) and 7 polyphenolic acids in DHI simultaneously.
Figure 1. Representative 1H NMR spectra of Danhong injection.
Peaks: 1, Isoleucine; 2, Leucine 3, Valine; 4, Threonine; 5, Alanine; 6, Acetate; 7, Proline; 8, Pyroglutamate; 9, Succinate; 10, Asparagine; 11, Malonate; 12, Glucose; 13, Galactose; 14, Arabinose; 15, Fructose 16, Rhamnose 17, Rutinose 18, Rutinulose; 19, Salvianic acid; 20, Salvianolic acid B; 21, Rosmarinic acid; 22, Lithospermic acid 23, Salvianolic acid A; 24, Procatechuic acid; 25, Procatechuic aldehyde; 26, 4-Hydroxybenzoic acid; 27, 4-Hydroxycinnamic acid; 28, Uridine; 29, Formate; 30, 5-(Hydroxymethyl)-2-furaldehyde.
Table 1. Chemical shifts for 30 metabolites identified in the 1H NMR spectra of Danhong injection.
| Peak | Metabolites | Assignment | δ 1H (multiplicitya) | Assigned with |
| 1 | Isoleucine | δ-CH3, γ′-CH3, γ-CH2, β-CH, α-CH | 0.96 (t, 6.1 Hz), 1.01 (d, 6.8 Hz), 1.26 (m), 1.47 (m), 1.96 (m), 3.66 (m) | COSY, TOCSY, J-res, HSQC |
| 2 | Leucine | δ, δ′-CH3, β-CH2,γ-CH | 0.95 (m), 1.67 (m), 3.72 (m) | COSY, TOCSY, J-res, HSQC |
| 3 | Valine | γ′-CH3, γ-CH3, β-CH, α-CH | 1.00 (d, 7.0 Hz), 1.05 (d, 7.0 Hz), 2.26 (m), 3.60 (d, 4.2 Hz) | COSY, TOCSY, J-res, HSQC |
| 4 | Threonine | γ-CH3, β-CH, α-CH | 1.36 (d, 6.5 Hz), 4.25 (m), 3.58 (d, 6.4 Hz) | COSY, TOCSY, J-res, HSQC |
| 5 | Alanine | β-CH3, α-CH | 1.48 (d, 7.5 Hz), 3.78 (m) | COSY, TOCSY, J-res, HSQC |
| 6 | Acetate | CH3 | 1.93 (s) | J-res, HSQC |
| 7 | Proline | γ-CH2, δ-CH2, β-CH2, α-CH | 2.00 (m), 3.34 (m), 3.42 (m), 2.35 (m),4.14 (m) | COSY, TOCSY, J-res, HSQC |
| 8 | Pyroglutamate | γ-CH2, β-CH2, α-CH | 2.04 (m), 2.51 (m), 2.41 (m), 4.18 (m) | COSY, TOCSY, J-res, HSQC |
| 9 | Succinate | CH2 | 2.44 (s) | J-res, HSQC |
| 10 | Asparagine | β-CH2, α-CH | 2.76 (dd, 14.0, 8.2 Hz), 2.97 (dd, 14.0, 4.3 Hz), 4.00 (m) | COSY, TOCSY, J-res, HSQC |
| 11 | Malonate | CH2 | 3.19 (s) | J-res, HSQC |
| 12 | Glucose | αC1H, αC2H, αC3H, αC4H, αC5H, βC1H, βC2H, βC3H | 5.24 (d, 3.6 Hz), 3.55 (m), 3.72 (m), 3.42 (m), 3.85 (m), 4.65 (d, 7.8 Hz), 3.25 (dd, 7.8, 9.2 Hz), 3.48 (m) | COSY, TOCSY, J-res, HSQC |
| 13 | Galactose | αC1H, αC2H, αC3H, βC1H, βC2H, βC3H, βC4H | 5.27 (d, 3.8 Hz), 3.83 (m), 4.02 (m), 4.59 (d, 7.8 Hz), 3.50 (m), 3.65 (m), 3.93 (m) | COSY, TOCSY, J-res, HSQC |
| 14 | Arabinose | αC1H, αC2H, αC3H, βC1H, βC2H, βC3H | 5.25 (d, 3.6 Hz), 3.82 (m), 4.02 (m), 4.52 (d, 7.8 Hz), 3.51 (m), 3.68 (m) | COSY, TOCSY, J-res, HSQC |
| 15 | Fructose | C1H, C3H, C4H, C5H, C6H | 3.58 (m), 3.69 (m), 3.82 (m), 4.11 (m), 3.90 (m), 4.11 (m), 3.82 (m), 4.00 (m), 3.69 (m), 3.82 (m), 4.02 (m) | COSY, TOCSY, J-res, HSQC |
| 16 | Rhamnose | αC1H, αC2H, αC3H, αC4H, αC5H, βC2H, βC3H, βC4H, βC5H | 5.18 (d, 1.7 Hz), 3.91 (m), 3.79 (m), 3.43 (m), 3.85 (m), 3.92 (m), 3.59 (m), 3.37 (m), 3.43 (m) | COSY, TOCSY, J-res, HSQC |
| 17 | Rutinose | Rha- C1H, Glc-C1H | 5.42 (d, 1.7 Hz), 4.50 (d, 7.4 Hz) | COSY, TOCSY, J-res, HSQC |
| 18 | Rutinulose | Rha- C1H, Fru-C1H | 5.25 (d, 1.7 Hz), 4.45 (d, 7.4 Hz) | COSY, TOCSY, J-res, HSQC |
| 19 | Salvianic acid | H-2, H-5, H-6, H2-7, H-8 | 6.81 (d, 1.5 Hz), 6.86 (d, 8.0 Hz), 6.74 (dd, 8.0, 1.5 Hz), 2.85 (m), 3.04 (m), 5.42 (m) | COSY, TOCSY, J-res, HSQC |
| 20 | Salvianolic acid B | H-2, H-3, H-5, H-6, H-β, H-α, H-5″, H-6″, H2-7″, H-8″, H-2″′, H-5″′, H-6″′, H-7″′, H-8″′ | 6.01 (d, 5.6 Hz), 4.33 (d, 5.6 Hz), 7.06, 6.92, 6.99 (d, 16.0 Hz), 5.85 (d, 16.0 Hz), 6.93, 6.76, 2.90 (m), 3.10 (m), 5.00 (m), 6.24 (d, 2.6), 6.42 (d, 8.0), 6.12 (dd, 8.0, 2.6 Hz), 2.98 (m), 2.56 (m), 4.90 (m) | COSY, TOCSY, J-res, HSQC |
| 21 | Rosmarinic acid | H-2, H-5, H-6, H-β, H-α, H-2′, H-5′, H-6′, H2-7′ | 6.98 (d, 1.5 Hz), 6.72 (d,8.0 Hz), 6.89 (dd, 8.0, 1.5 Hz), 7.49 (d, 16.0 Hz), 6.32 (d, 16.0 Hz), 6.90 (d, 1.5 Hz), 6.64 (d, 8.0 Hz), 6.55 (dd, 8.0, 1.5 Hz), 2.91 (m), 3.13 (m) | COSY, TOCSY, J-res, HSQC |
| 22 | Lithospermic acid | H-5, H-6, H-β, H-α, H-2′, H-5′, H-6′, H2-7′, H-8′, H-2″, H-5″, H-6″ | 6.52 (d, 8.5 Hz), 6.57 (d, 8.5 Hz), 7.22 (d, 16.0 Hz), 6.32 (d, 16.0 Hz), 6.74 (d, 1.5 Hz), 6.65 (d, 8.0 Hz), 6.52 (dd, 8.0, 1.5 Hz), 2.81 (m), 2.87 (m), 4.85 (m), 6.66 (d, 1.5 Hz), 6.55 (d, 8.0 Hz), 6.46 (dd, 8.0, 1.5 Hz) | COSY, TOCSY, J-res, HSQC |
| 23 | Salvianolic acid A | H-5, H-6, H-β, H-α, H-2′, H-5′, H-6′, H2-7′, H-8′, H-2″, H-5″, H-6″, H-7″, H-8″ | 6.71 (d, 8.0 Hz), 7.05 (d, 8.0 Hz), 7.86 (d, 16.0 Hz), 6.20 (d, 16.0 Hz), 6.66 (d, 1.5 Hz), 6.59 (d, 8.0 Hz), 6.48 (dd, 8.0, 1.5 Hz), 2.90 (m), 3.01 (m), 5.11 (dd, 8.5, 3.5 Hz), 7.00 (d, 1.0 Hz), 6.69 (d, 8.0 Hz), 6.82 (dd, 8.0, 1.5 Hz), 6.60 (d, 16.0 Hz), 7.08 (d, 16.0 Hz) | COSY, TOCSY, J-res, HSQC |
| 24 | Procatechuic acid | H-2, H-5, H-6 | 7.41 (d, 2.0 Hz), 6.80 (d, 8.0 Hz), 7.45 (dd, 8.0, 2.0 Hz) | COSY, TOCSY, J-res, HSQC |
| 25 | Procatechuic aldehyde | H-2, H-5, H-6, CHO | 7.10 (d, 2.0 Hz), 6.80 (d, 8.0 Hz), 7.08 (dd, 8.0, 2.0 Hz), 9.63 (s) | COSY, TOCSY, J-res, HSQC |
| 26 | 4-Hydroxybenzoic acid | H-3, 5, H-2, 6 | 7.83 (d, 8.8 Hz), 6.91 (d, 8.8 Hz) | COSY, TOCSY, J-res, HSQC |
| 27 | 4-Hydroxycinnamic acid | H-3, 5, H-2, 6, H-β, H-α | 7.54 (d, 9.0 Hz), 6.89 (d, 9.0 Hz), 7.60 (d, 16.0 Hz), 6.37 (d, 16.0 Hz) | COSY, TOCSY, J-res, HSQC |
| 28 | Uridine | H-5, H-6, Xyl-C1H | 5.88 (d, 8.1 Hz), 7.88 (d, 8.1 Hz), 5.91 (d, 4.2 Hz) | COSY, TOCSY, J-res, HSQC |
| 29 | Formate | CHO | 8.46 (s) | J-res, HSQC |
| 30 | 5-(Hydroxymethyl)-2-furaldehyde | H-3, H-4, CHO | 7.23 (d, 3.5 Hz), 6.51 (d, 3.5 Hz), 9.45 (s) | COSY, TOCSY, J-res, HSQC |
Multiplicity: singlet (s), doublet (d), triplet (t), doublet of doublets (dd), quintet (q), multiplet (m).
Quantitative 1H NMR analysis and method validation
Due to the narrow chemical shift range of 1H NMR and frequent signal overlap, it is a challenge to quantify all the constituents in a mixture. In our study, 13 metabolites with fully separated signals were selected for quantification. In order to improve the efficiency of 1H NMR method, the spectra were acquired in an incompletely relaxed state. As a consequence, the spin-lattice relaxation time (T 1) value must be accurately measured and taken into account for quantitative analysis. The T 1 values were determined by the inversion-recovery experiments (Table 2). Accordingly, the absolute concentrations of the 13 metabolites were calculated from three parallel samples of each batch (Table S1 in File S1).
Table 2. Proton spin-lattice relaxation time (T 1) values for 13 selected metabolites and the method validation parameters for 1HNMR method.
| Metabolites | δ(ppm) | T 1 (s) (n = 3) | Regression equation (n = 3) | r 2 | Intra-day precision (n = 6) RSD (%) | Inter-day precision (n = 3) RSD (%) | Repeatability (n = 6) RSD (%) | Stability (n = 6) RSD (%) | Average recoverya (%) ± SD (n = 3) |
| Valine | 1.03 (3H) | 0.944±0.049 | y = 7.9917x - 0.1254 | 0.9996 | 0.79 | 0.84 | 1.83 | 1.67 | - |
| Threonine | 1.31 (3H) | 1.323±0.102 | y = 7.9895x - 0.1288 | 0.9995 | 0.49 | 0.98 | 1.94 | 0.99 | - |
| Alanine | 1.47 (3H) | 1.451±0.120 | y = 11.536x - 0.0821 | 0.9997 | 0.61 | 0.35 | 1.46 | 0.58 | 106.6±1.2 |
| Pyroglutamate | 2.41 (2H) | 2.249±0.044 | y = 3.9348x - 0.0387 | 0.9980 | 0.20 | 0.29 | 1.48 | 0.37 | - |
| Succinate | 2.42 (4H) | 1.844±0.268 | y = 9.7223x - 0.2087 | 0.9993 | 0.89 | 0.93 | 1.85 | 2.21 | - |
| Asparagine | 2.76 (1H) | 0.562±0.019 | y = 1.8370x - 0.1871 | 0.9979 | 0.60 | 1.49 | 2.75 | 1.78 | - |
| Malonate | 3.19 (2H) | 2.116±0.136 | y = 0.7533x - 0.0317 | 0.9972 | 0.20 | 0.56 | 1.66 | 1.43 | - |
| Fructose | 4.10 (3H) | 3.303±0.210 | y = 1.2045x - 0.2608 | 0.9998 | 0.45 | 0.34 | 1.69 | 2.06 | - |
| Glucose | 5.24 (1H) | 1.684±0.206 | y = 0.5486x - 0.0912 | 0.9997 | 0.32 | 0.68 | 2.59 | 1.32 | 106.6±2.5 |
| Rutinose | 5.44 (1H) | 1.447±0.164 | y = 2.4419x - 0.1850 | 0.9992 | 0.59 | 0.76 | 2.49 | 1.78 | - |
| Salvianic acid | 6.74 (1H) | 1.843±0.139 | y = 1.2845x - 0.1858 | 0.9994 | 0.23 | 0.34 | 2.01 | 1.23 | 98.3±2.1 |
| Procatechuic acid | 7.41 (1H) | 3.542±0.326 | y = 0.5172x - 0.0083 | 0.9983 | 0.67 | 0.93 | 1.93 | 1.99 | - |
| Procatechuic aldehyde | 9.63 (1H) | 2.932±0.436 | y = 1.2468x - 0.0013 | 0.9992 | 0.78 | 0.85 | 1.72 | 1.82 | 91.3±4.0 |
Recovery (%) = [(found−original)/spiked]×100.
Linearity
1H NMR as method itself is linear and no calibration is necessary for the determination of molar ratios of mixtures. Thus, the 13 metabolites in five different molar ratios confirmed the linearity of NMR spectroscopy. Good linearity was achievable, as indicated by the equations and satisfactory correlation coefficients (r 2).
Precision
The intraday and interday precision was determined by analyzing six replicates on the same day and on three consecutive days respectively. The intraday precision for the contents of 13 metabolites ranged from 0.20% to 0.89%, and the interday precision ranged from 0.29% to 1.49%. The RSD values were adequate and indicated the suitability of the method.
Repeatability
Six samples prepared from the same batch showed RSD values ranging from 1.46% to 2.75%, indicating a high repeatability.
Stability
One sample was analyzed to determine stability on three consecutive days. The RSD values of the analytes were in the range of 0.37% to 2.21%.
Accuracy
Considering the limited volume of NMR tube and the cost of using deuterium reagents to dilute sample continuously, four metabolites of different types were employed, including alanine, glucose, salvianic acid and procatechuic aldehyde. The recovery was calculated as the ratio of the response of the selected four compounds in the spiked DHI samples against that of the standards at the same levels. The average recoveries were found to be 106.6% (±1.2), 106.6% (±2.5), 98.3% (±2.1) and 91.3% (±4.0) for alanine, glucose, salvianic acid and procatechuic aldehyde, respectively, indicating acceptable recovery.
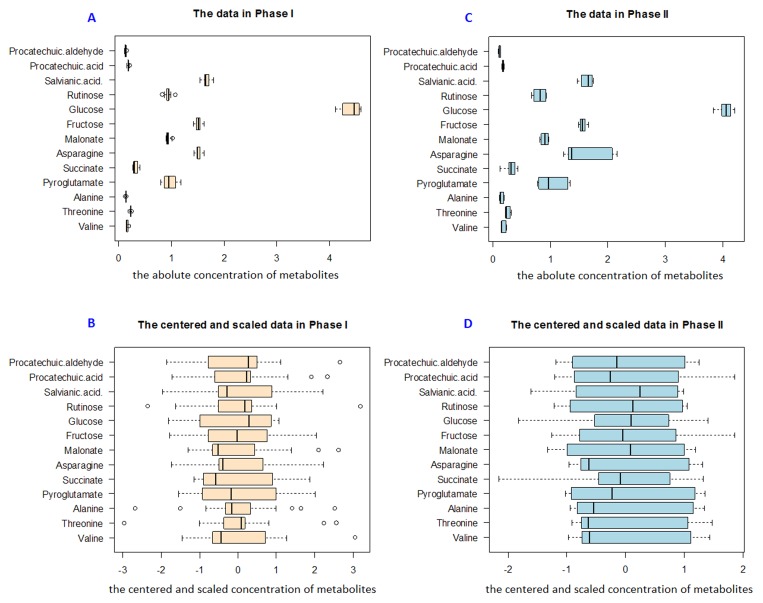
According to the results, the concentration of glucose was extremely high in DHI as shown in Fig. 2A and 2B, and the amount of saccharides, amino acids and organic acids represented about 60% of the total solid content of DHI (Table S1 in File S1).
Figure 2. Quantitative data of selected 13 metabolites.
Boxplots of the quantitative data before and after centering and scaling in Phase I (A, B) and Phase II (C, D).
Control Charts based on PCA and IPCA
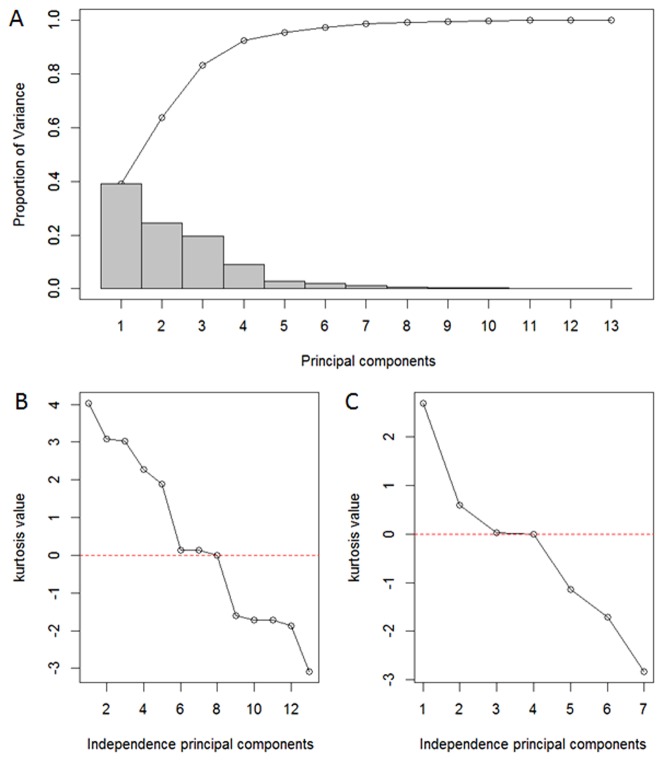
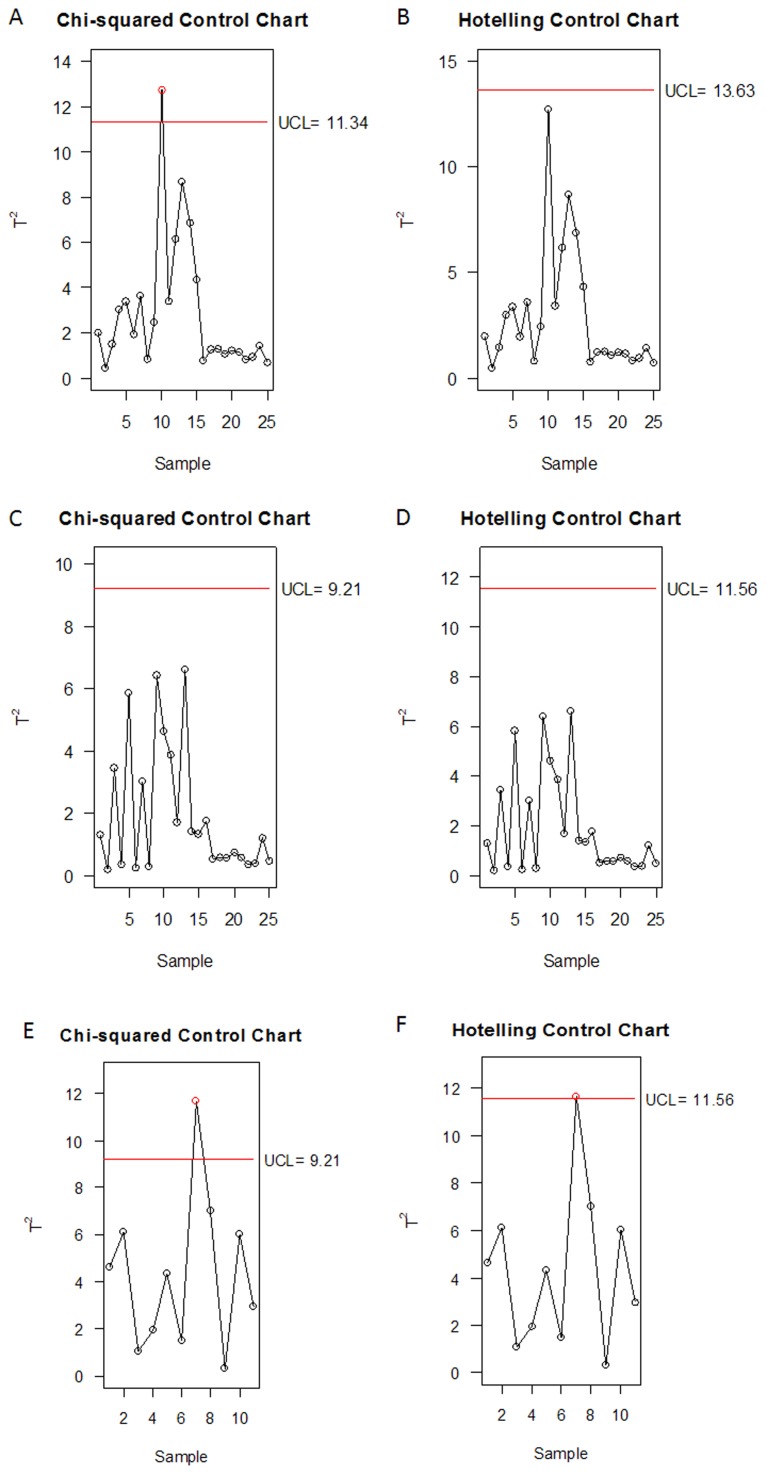
With the enhancement in quality control of DHI, the analysis should be performed through a multivariate approach, that is, the above 13 metabolites must be analyzed together, not independently. The concentrations of 13 metabolites were mean centered and unit-variance scaled before being analyzed by PCA and IPCA (Fig. 2C and 2D). To avoid the loss of significant information, the percent specified of the principal components (PCs) cumulative proportion of explain variance is normally fixed on 80% in PCA model [29]. Thus, the first three PCs in Phase I (Fig. 3A) were selected to construct χ2 (Fig. 4A) and Hotelling T2 (Fig. 4B) control charts. Since the independence of the data is one of the requisites in control chart, we assessed the marginal independence of each necessary PC to indicate the model validation [38]. Correlograms (Fig. 5 PC1-PC3) showed that PC1 fell outside of the confidence bands, which indicated that there was an evidence of autocorrelation or dependence of PC1, and PCA model using Phase I data was not achieved.
Figure 3. Component contributions of PCA and IPCA.
(A) Pareto chart of principal components (PCs) in PCA indicated the first three PCs in Phase I explained 80.99% variances. (B) Kurtosis measurements of all extracted independence principal components (IPCs) in IPCA with Phase I data. The kurtosis of IPC8 was close to zero, so the first 7 components of IPCA were used to choose exact numbers of IPCs. (C) Kurtosis measurements of the first 7 IPCs showed that the kurtosis of IPC3 was close to zero, and the first 2 components were sufficient with IPCA.
Figure 4. Multivariate quality control charts.
The χ2 and Hotelling control charts of the first three principle components (PCs) in Phase I (A and B), and the first two independence principle components (IPCs) in Phase I (C and D) and Phase II (E and F). Red solid lines represented the upper control limits (UCL) of control charts with 99% confidence region.
Figure 5. Autocorrelation function (ACF) validation of PCA and IPCA.
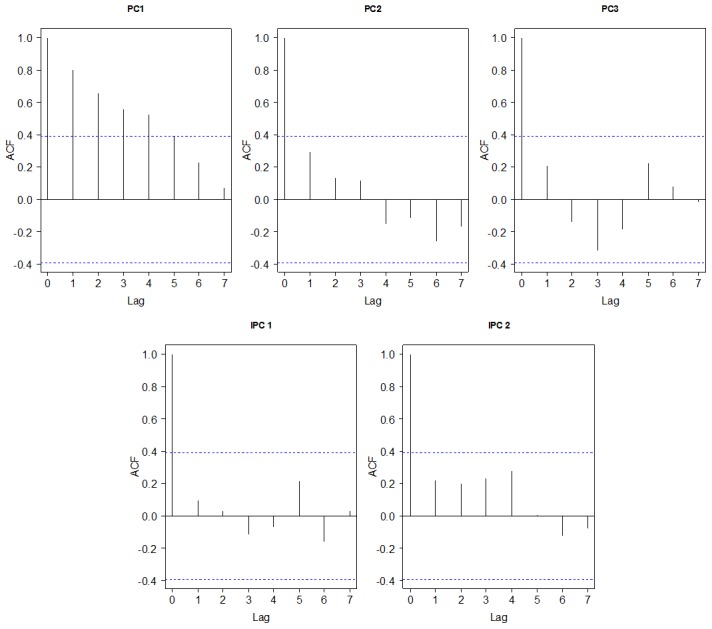
Correlograms for the first three principle components (PC1, PC2, PC3) and the first two independence principle components (IPC1, IPC2) in Phase I. Blue dashed lines represented the confidence bands of ACF. PC1 fell outside of the confidence bands with autocorrelation or dependence, it was said that PCA model was not successful.
In order to remove the autocorrelation effects of PCA, we employed IPCA to generate denoised and independent loading vectors [28]. The kurtosis measure of loading vectors was used to decide the number of independent principal components (IPCs). The kurtosis of all extracted IPCs was plotted in Fig. 3B, whereas the kurtosis of IPC7 was close to zero. By using the first 7 components of IPCA, the exactly choosing number of IPCs was obtained (Fig. 3C). Since the kurtosis of IPC3 was close to zero, the first 2 components were sufficient with IPCA. The presence of autocorrelation was assessed as shown in Fig. 5 (IPC1 and IPC2), which indicated that there was no evidence of relation between the adjacent observations. Therefore the original 13 dimension of our data had been reduced to a two-dimensional problem.
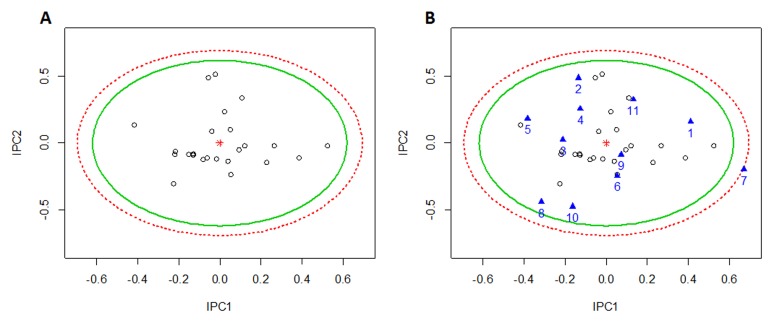
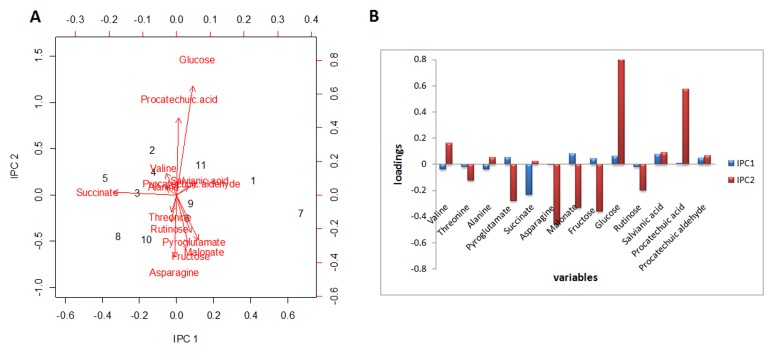
Then the first two IPCs were taken to establish the in-control state (Phase I). According to χ2 (Fig. 4C) and Hotelling T2 (Fig. 4D) control charts with the 99% confidence region, the upper control limits (UCL) were determined as 9.21 and 11.56, respectively. The first two IPCs were consequently controlled through 2D ellipsoids (Fig. 6).The χ2 control ellipse (UCL = 9.21) could be used as process region, and the T2 control ellipse with less restrictive (UCL = 11.56) was used as tolerance region. In both cases, all the points in Phase I fell inside the confidence ellipsoids (Fig. 6A). The samples in Phase II (Table S2 in File S1) were monitored by employing the UCLs of both χ2 and T2 charts obtained from Phase I (Fig. 4E and 4F), and the points of Phase II were added into 2D ellipsoids (Fig. 6B). The seventh sample fell outside the 99th confidence ellipsoids of both the process and tolerance regions, indicating the presence of out-of-control sample (batch 110402). The decomposition of T2 value showed that the out-of-control variability was associated to the IPC1 since p-value was equal to 0.0032 (Table 3). The same result was also obtained through IPCA loading plots as shown in Fig. 7. Although the loading plot could not exactly determine which metabolites were responsible for the variation, it still showed the contents of succinate, malonate, glucose, fructose, salvianic acid and protocatechuic aldehyde made more contributions to the independent loading vectors of IPC1.
Figure 6. The confidence ellipses for quality control.
Scatterplots for the first two IPC scores with the confidence ellipses in Phase I (A) and Phase II (B). The χ2 control ellipse used as process region was shown in green solid line, while the Hotelling T2 control ellipse used as tolerance region was in red dashed line. The numbers represented the samples in Phase II listing in Table S2 in File S1, and the seventh sample fell outside the tolerance region.
Table 3. Decomposition of the seventh sample in Phase II.
Figure 7. The loading plots and values of IPCA.
(A) The IPCA loading plots for the first two IPCs in Phase II. The numbers represented the samples in Phase II listing in Table S2 in File S1. The influence of metabolites was showing in red arrow lines. (B) Bar diagram showed the loading values of metabolites in first two independence principle components.
Conclusion
Based on quantitative 1H NMR analysis, a reliable approach for simultaneous determination of amino acids, organic acids, saccharides, and botanic secondary metabolites of HMIs in one fingerprinting spectrum has been developed and validated by using Danhong injection as a model. The method had takenT 1 values into account when calculated the contents of feature metabolites, which allowed the assay with good linearity, precision, repeatability, stability and accuracy. Unlike HPLC fingerprinting methods, the 1H NMR approach has the significant advantages of less analysis time (about 5 min) without chromatographic separation, and no requirement of standard materials used to quantitative analysis. In combination with IPCA, two kinds of multivariate control charts (χ2 and Hotelling T2) were also successfully carried out for detecting off-test HMI samples by employing the independence principle components. The decomposition of T2 value and IPCA loading vectors can reflect the significance variations of overall metabolite profiling, although it cannot decide the mutative metabolites exactly. The established multivariate models have the prospects for extracting sufficient characterization to monitor more feature metabolites in HMIs.
Supporting Information
Quantification of 13 chemical markers selected in the 1H NMR spectra of DHI samples in Phase I (Table S1) and Phase II (Table S2).
(DOC)
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All data are available within the Supporting Information files.
Funding Statement
The authors gratefully acknowledge financial support provided by the National Natural Science Foundation of China (Grant No. 81303181), the National Key Technology R&D Program in the 12th Five Year Plan of China (Grant No. 2013ZX09201020, 2014ZX09201-023). The National Natural Science Foundation website address is: http://www.nsfc.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Zhang L, Wang X, Su J, Liu H, Zhang Z, et al. (2014) One single amino Acid for estimation the content of total free amino acids in qingkailing injection using high-performance liquid chromatography-diode array detection. J Anal Methods Chem 2014: 951075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zeng S, Wang L, Chen T, Wang Y, Mo H, Qu H (2012) Direct analysis in real time mass spectrometry and multivariate data analysis: a novel approach to rapid identification of analytical markers for quality control of traditional Chinese medicine preparation. Anal Chim Acta 733: 38–47. [DOI] [PubMed] [Google Scholar]
- 3. Chen X, Lou Z, Zhang H, Tan G, Liu Z, et al. (2011) Identification of multiple components in Guanxinning injection using hydrophilic interaction liquid chromatography/time-of-flight mass spectrometry and reversed-phase liquid chromatography/time-of-flight mass spectrometry. Rapid Commun Mass Spectrom25: 1661–1674. [DOI] [PubMed] [Google Scholar]
- 4. Jin Y, Zhang J, Jia Z, Chen Y, Wu C (2013) Determination of saccharide in Shuxuetong injection, hirudo and pheretima by HPLC with pre-column derivatization method. Chin New Drugs J22: 111–115. [Google Scholar]
- 5. Baba T, Yoshida T, Yoshida T, Cohen S (1979) Suppression of cell-mediated immune reactions by monosaccharides. J Immunol 122: 838–841. [PubMed] [Google Scholar]
- 6. Inglis AS, Bartone NA, Finlayson JR (1988) Amino acids analysis using phenylisothiocyanate prederivatization: Elimination of the drying steps. J Biochem Biophys Methods 15: 249–254. [DOI] [PubMed] [Google Scholar]
- 7. Mopper K, Johnson L (1983) Reversed-phase liquid chromatographic analysis of dns-sugars : Optimization of derivatization and chromatographic procedures and applications to natural samples. J Chromatogr A 256: 27–38. [Google Scholar]
- 8. Lee YC (1990) High-performance anion-exchange chromatography for carbohydrate analysis. Anal Biochem 189: 151–162. [DOI] [PubMed] [Google Scholar]
- 9. Ruiz-Matute AI, Hernandez-Hernandez O, Rodriguez-Sanchez S, Sanz ML, Martinez-Castro I (2011) Derivatization of carbohydrates for GC and GC-MS analyses. J Chromatogr B Analyt Technol Biomed Life Sci 879: 1226–1240. [DOI] [PubMed] [Google Scholar]
- 10. Hušek P (1991) Rapid derivatization and gas chromatographic determination of amino acids. J Chromatogr A 552: 289–299. [Google Scholar]
- 11. Soga T, Imaizumi M (2001) Capillary electrophoresis method for the analysis of inorganic anions, organic acids, amino acids, nucleotides, carbohydrates and other anionic compounds. Electrophoresis 22: 3418–3425. [DOI] [PubMed] [Google Scholar]
- 12. Harvey DJ (2011) Derivatization of carbohydrates for analysis by chromatography; electrophoresis and mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 879: 1196–1225. [DOI] [PubMed] [Google Scholar]
- 13. Simmler C, Napolitano JG, McAlpine JB, Chen S-N, Pauli GF (2014) Universal quantitative NMR analysis of complex natural samples. Curr Opin Biotech 25: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marcone MF, Wang S, Albabish W, Nie S, Somnarain D, et al. (2013) Diverse food-based applications of nuclear magnetic resonance (NMR) technology. Food Res Int 51: 729–747. [Google Scholar]
- 15. Bouteille R, Gaudet M, Lecanu B, This H (2013) Monitoring lactic acid production during milk fermentation by in situ quantitative proton nuclear magnetic resonance spectroscopy. J Dairy Sci 96: 2071–2080. [DOI] [PubMed] [Google Scholar]
- 16. Lopez-Rituerto E, Cabredo S, Lopez M, Avenoza A, Busto JH, et al. (2009) A thorough study on the use of quantitative 1H NMR in Rioja red wine fermentation processes. J Agric Food Chem 57: 2112–2118. [DOI] [PubMed] [Google Scholar]
- 17. Ohtsuki T, Sato K, Sugimoto N, Akiyama H, Kawamura Y (2012) Absolute quantification for benzoic acid in processed foods using quantitative proton nuclear magnetic resonance spectroscopy. Talanta 99: 342–348. [DOI] [PubMed] [Google Scholar]
- 18. Fan G, Zhang MY, Zhou XD, Lai XR, Yue QH, et al. (2012) Quality evaluation and species differentiation of Rhizoma coptidis by using proton nuclear magnetic resonance spectroscopy. Anal Chim Acta 747: 76–83. [DOI] [PubMed] [Google Scholar]
- 19. Bohni N, Cordero-Maldonado ML, Maes J, Siverio-Mota D, Marcourt L, et al. (2013) Integration of Microfractionation, qNMR and zebrafish screening for the in vivo bioassay-guided isolation and quantitative bioactivity analysis of natural products. PLoS ONE 8: e64006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chauthe SK, Sharma RJ, Aqil F, Gupta RC, Singh IP (2012) Quantitative NMR: an applicable method for quantitative analysis of medicinal plant extracts and herbal products. Phytochem Anal 23: 689–696. [DOI] [PubMed] [Google Scholar]
- 21. Jiang Y, David B, Tu P, Barbin Y (2010) Recent analytical approaches in quality control of traditional Chinese medicines-a review. Anal Chim Acta 657: 9–18. [DOI] [PubMed] [Google Scholar]
- 22.Jolliffe I (2002) Principal Component Analysis. second edition. Springer, NewYork.
- 23. Comon P (1994) Independent component analysis, a new concept? SignalProcess36: 287–314. [Google Scholar]
- 24. Kong W, Vanderburg C, Gunshin H, Rogers J, Huang X (2008) A review ofindependent component analysis application to microarray geneexpression data. Biotechniques 45: 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hyvärinen A, Karhunen J, Oja E (2001) Independent Component Analysis. first edition.John Wiley& Sons. NewYork.
- 26. Engreitz J, Daigle B Jr, Marshall J, Altman R (2010) Independent componentanalysis: Mining microarray data for fundamental human geneexpression modules. J Biomed Inform 43: 932–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee S, Batzoglou S (2003) Application of independent component analysis tomicroarrays. Genome Biol 4: R76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yao F, Coquery J, Le Cao KA (2012) Independent Principal Component Analysis for biologically meaningful dimension reduction of large biological data sets. BMC Bioinformatics 13: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santos-Fernández E (2013) Multivariate Statistical Quality Control Using R: Springer, New York.
- 30. Sun M, Zhang JJ, Shan JZ, Zhang H, Jin CY, et al. (2009) Clinical observation of Danhong Injection (herbal TCM product from Radix Salviae miltiorrhizae and Flos Carthami tinctorii) in the treatment of traumatic intracranial hematoma. Phytomedicine 16: 683–689. [DOI] [PubMed] [Google Scholar]
- 31. Chen Q, Yi D, Xie Y, Yang W, Yang W, et al. (2011) Analysis of clinical use of Danhong injection based on hospital information system. Chin J Chin Mater Med36: 2817–2820. [PubMed] [Google Scholar]
- 32. Li X, Tang J, Meng F, Li C, Xie Y (2011) Study on 10 409 cases of post-marketing safety Danhong injection centralized monitoring of hospital. Chin J Chin Mater Med 36: 2783–2785. [PubMed] [Google Scholar]
- 33. Zhao ZW, Cheng CS, Ma WL, Shan HM, Cheng ZZ, et al. (2013) Danhong injection, ligustrazine injection, combined adsorbable biomembranes prevented adhesion of tendons after the repair operation: a clinical research. Chin J Integr Med 33: 1212–1215. [PubMed] [Google Scholar]
- 34. Liu HT, Wang YF, Olaleye O, Zhu Y, Gao XM, et al. (2013) Characterization of in vivo antioxidant constituents and dual-standard quality assessment of Danhong injection. Biomed Chromatogr 27: 655–663. [DOI] [PubMed] [Google Scholar]
- 35. Jiang M, Wang C, Zhang Y, Feng Y, Wang Y, et al. (2014) Sparse partial-least-squares discriminant analysis for different geographical origins of Salvia miltiorrhiza by1H-NMR-based metabolomics. Phytochem Anal 25: 50–58. [DOI] [PubMed] [Google Scholar]
- 36. Pauli GF, Godecke T, Jaki BU, Lankin DC (2012) Quantitative 1H NMR. Development and potential of an analytical method: an update. J Nat Prod 75: 834–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dai H, Xiao C, Liu H, Tang H (2010) Combined NMR and LC-MS analysis reveals the metabonomic changes in Salvia miltiorrhiza Bunge induced by water depletion. J Proteome Res 9: 1460–1475. [DOI] [PubMed] [Google Scholar]
- 38.(2001) Multivariate Statistical Process Control with Industrial Application: Society for Industrial and Applied Mathematics, Philadelphia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantification of 13 chemical markers selected in the 1H NMR spectra of DHI samples in Phase I (Table S1) and Phase II (Table S2).
(DOC)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All data are available within the Supporting Information files.