Abstract
UV irradiation is known to cause cyclobutane pyrimidine dimers (CPDs) and pyrimidine (6–4) pyrimidone photoproducts (6-4PPs), and plays a large role in the development of cancer. Tumor suppression, through DNA repair and proper cell cycle regulation, is an integral factor in maintaining healthy cells and preventing development of cancer. Transcriptional regulation of the genes involved in the various tumor suppression pathways is essential for them to be expressed when needed and to function properly. BRG1, an ATPase catalytic subunit of the SWI/SNF chromatin remodeling complex, has been identified as a tumor suppressor protein, as it has been shown to play a role in Nucleotide Excision Repair (NER) of CPDs, suppress apoptosis, and restore checkpoint deficiency, in response to UV exposure. Although BRG1 has been shown to regulate transcription of some genes that are instrumental in proper DNA damage repair and cell cycle maintenance in response to UV, its role in transcriptional regulation of the whole genome in response to UV has not yet been elucidated. With whole genome expression profiling in SW13 cells, we show that upon UV induction, BRG1 regulates transcriptional expression of many genes involved in cell stress response. Additionally, our results also highlight BRG1's general role as a master regulator of the genome, as it transcriptionally regulates approximately 4.8% of the human genome, including expression of genes involved in many pathways. RT-PCR and ChIP were used to validate our genome expression analysis. Importantly, our study identifies several novel transcriptional targets of BRG1, such as ATF3. Thus, BRG1 has a larger impact on human genome expression than previously thought, and our studies will provide inroads for future analysis of BRG1's role in gene regulation.
Introduction
SWI/SNF is part of a family of chromatin remodeling complexes which act as master regulators of gene expression in yeast and human cells [1], by modifying nucleosomes in an ATP-dependent fashion. Mammalian SWI/SNF complexes contain one of two ATP catalytic subunits, BRM (Brahma) or BRG1 (Brahma Related Gene), and also contain additional proteins called BAFs (BRM/BRG1 Associated Factors) consisting of core and accessory subunits [2], which function as a diverse array of biochemical and functional activities [3]–[6].
Several lines of evidence suggest that the SWI/SNF chromatin remodeling complexes are important for protection of genome integrity. Previous studies have shown SWI/SNF accumulation at DNA double strand breaks in yeast [7], [8]. In addition, SWI/SNF is involved in chromatin dynamics after exposure to ultraviolet (UV) radiation [9], [10] and appears to be associated with Rad4-Rad23, a heterodimer that recognizes UV lesions in yeast and facilitates nucleotide excision repair (NER) for such lesions [11]. Interestingly, it has been shown that the BAF subunits in SWI/SNF-like complexes play a protective role in genome integrity, since inactivation of the SWI/SNF-like BAF complexes renders human cells sensitive to DNA damaging agents, such as UV and ionizing radiation (IR) [12], [13]. BRG1 in particular, appears to be a crucial part of SWI/SNF-like BAF complexes, since expression of a dominant negative mutant of BRG1 inactivates of the BAF complexes and results in inefficient DNA double-strand break repair and sensitizes cells to ionizing radiation [14]. Cells lacking functional BRG1 are sensitive to various DNA damage agents, including ionizing radiation and doxorubicin [14], cisplatin [15], [16], and UV radiation [12]. BRG1 has also been shown to play a role in genome integrity by contributing to proper NER of CPDs induced by UV irradiation [17], [18], and has been shown to also respond to DNA damage by suppressing UV induced apoptosis, and restoring checkpoint deficiency [12].
Defects in DNA repair and improper regulation of cell cycle progression and apoptosis can lead to development of cancer, and thus much recent work has focused on the role of the BAF ATPase BRG1 in cancer [19]–[21]. BRG1 gene is frequently deleted or mutated in a variety of tumor cell lines, implicating BRG1 as a potential tumor suppressor gene [22], [23], and mouse models have confirmed the tumor suppressor activities of BRG1 [20], [24], [25]. Given that transcriptional response is a well-documented strategy for cells to survive exposure to various DNA damaging agents [26]–[28], thereby suppressing tumor formation, it becomes important to understand how BRG1 may play a role in regulating genes responsible for DNA repair and cell cycle progression while improving our efforts in combating cancer.
Previous whole genome analyses have shown that BRG1 transcriptionally regulates genes involved in cellular proliferation and tumor suppression [29], [30]. Yet, only a limited number of genes regulated by BRG1 in response to UV radiation have been reported, and the impact of UV induced gene regulation by BRG1 on the human transcriptome as a whole is not well understood. In this study, we investigated the role of BRG1 in the transcriptional response to UV radiation with whole genome expression studies. Here we use a microarray approach to systematically compare UV-induced gene expression profiles in two isogenic cell lines derived from the adrenal cortical carcinoma cell line SW13, which lack BRG1 protein expression. Experiments with triplicates were designed to assess BRG1-mediated alterations in over 47,000 transcripts in response to UV radiation. Reverse transcriptase polymerase chain reaction (RT-PCR) was performed to confirm microarray data, and Chromatin Immuniprecipitation (ChIP) showed that BRG1 associated with promoter regions of these regulated genes. Here we show that BRG1 does indeed regulate transcription of genes important for UV damage repair and cell signaling upon UV induction, and identify several novel BRG1 transcriptional targets. These results will direct future experiments to fully understand BRG1's role as a tumor suppressor.
Materials and Methods
Cell lines and culture
Stable cell lines, SW13+pREP7 (vector), SW13+pREP7+BRG1 from previous study in which BRG1 isoform C expression has been confirmed [12], and SW13 and 293T cell lines (purchased from the ATCC), were cultured in Dulbecco's modified Eagle's medium (DMEM) (CELLGRO, Manassas, VA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and penicillin-streptomycin.
UV Treatment and 5-aza-2′-deoxycytidine treatment
As shown in Fig. 1, SW13+pREP7 and SW13+pREP7+BRG1 cells were irradiated with 10 J/m2 UV or mock treated. After treatment, warm media were added back and cells were incubated for 6 hours before being collected for RNA preparation. SW13 cells were treated with 50 µM demethylating agent 5-aza-2′-deoxycytidine (5-Aza) and incubated for 4 days before being collected for RNA extraction.
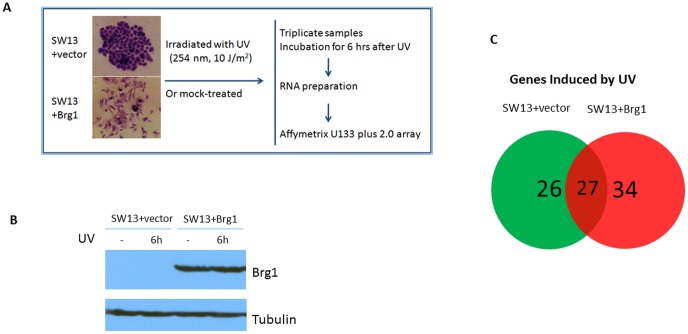
Figure 1. Microarray analysis of gene expression in response to UV irradiation in SW13 cells with or without BRG1.
(A) Morphological changes of SW13 cells after BRG1 re-expression and a schematic representation of sample preparation for Microarray analysis. SW13 cells with and without reintroduction of BRG1 were irradiated with UV or mock-treated. Three independent biological samples of these treatments were incubated for 6 hours in warm media to allow for recovery, and then collected for RNA extraction. RNA preparations were then used for Microarray analysis. Interestingly, this figure also shows the morphological changes associated with reintroduction of BRG1 in SW13 cells. (B) Western blotting confirming BRG1 protein expression. (C) Venn Diagram showing that BRG1 regulates the transcriptional response to UV irradiation in human cells. Shown here are the number of genes induced by UV in SW13 cells and SW13+BRG1 cells. 34 of these genes are BRG1-dependent genes, and are not expressed without BRG1 reintroduction. Out of the 87 total genes induced by UV, 61 of them are only induced in cells which include expression of BRG1. Thus, more than 70% of UV induced genes are regulated by BRG1.
Microarray analysis
Total RNA was prepared using TRIZOL reagent (Invitrogen, Carlsbad, CA), followed by the RNeasy kit (Qiagen, Valencia, CA), according to the manufacturer's instructions. Three independent biological replicates of SW13+pREP7 and SW13+pREP7+BRG1 were subjected to microarray analysis using Affymetrix U133 plus 2.0 gene chip. Target synthesis and GeneChip hybridization, washing, staining, and scanning were performed at the Molecular Biology Core at Washington State University. Microarray output was examined visually for excessive background noise and physical anomalies. The default MAS statistical values were used for all analyses. All probe sets on each array were scaled to a mean target signal intensity of 125, with the signal correlating to the amount of transcript in the sample. An absolute analysis using MAS was performed to assess the relative abundance of the 47,000 represented transcripts and variants, including 38,500 human genes, based on signal and detection (present, absent, or marginal). The resulting data from the absolute analysis were exported into Microsoft EXCEL and then imported into GeneSifter software (GeneSifter.net, Seattle, WA). Transcripts expressed differentially at a statistically significant level were determined using the Welch t-test (variances not assumed equal) with a P-value cutoff of 0.05.
DNA-Chip Analyzer (dChip) analysis
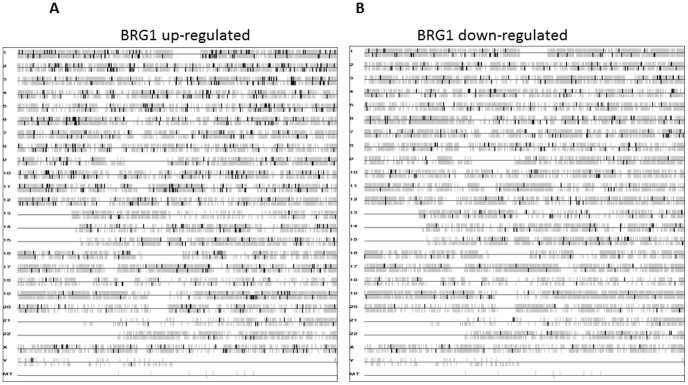
The bird's eye view of gene location in the genome, as seen in Fig. 2, was generated using dChip, a software package implementing model-based expression analysis of oligonucleotide arrays [31]. The transcription starting site is used for gene position. The vertical bar above the horizontal line means that the gene is on the forward strand, and vice versa.
Figure 2. Whole genome view of BRG1 regulatory gene targets (in the absence of UV irradiation) showing the locations of genes on each human chromosome.
The vertical bar above the horizontal line indicates genes on the forward strand, and vertical lines below the horizontal line indicate genes on the reverse strand. (A) Genes upregulated by BRG1. (B) Genes downregulated by BRG1.
Reverse transcription-PCR (RT-PCR) and quantitative real time PCR
Total RNA samples generated for the microarray experiment as described above, were also used for RT-PCR after DNase I digestion (Ambion, Austin, TX). Single-strand cDNA was synthesized using the Reverse Transcription System (Promega, Madison, WI) using oligo(dT)12–18 as primer. PCR was performed subsequently using RedTaq DNA polymerase (Sigma, St. Louis, MO) as described with primers listed in Table S1A. The PCR conditions were: 94°C, 30 sec; 52°C, 30 sec; and 72°C, 1 min, for 35 cycles. The cDNA was used as template in a quantitative real-time PCR using a MJ Mini Personal Thermal Cycler (Biorad). Reactions were done in a final volume of 20 µl in SsoFast EvaGreen Supermix (Biorad). Primer sequences are listed in the Table S1A. The mRNA expression levels for all samples was normalized to the levels of GAPDH.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation assays (ChIP) were performed using the protocol described on Upstate Cell Signaling website (http://www.millipore.com/techpublications/tech1/mcproto407) in 293T cell line. Immunoprecipitations were done using 4ug of the rabbit anti-BRG1 antibody (H-88) from Santa Cruz Biotechnology. The primers used to amplify E-cadherin, PML, RGC32/c13 orf15, EGFR, S100A2, TNIK, ARHGAP29/PARG1, and ATF3 from the promoter region are listed in Table S1B. The PCR conditions were similar to those described above.
Results and Discussion
DNA repair and transcriptional activation are two important cellular responses to deal with UV damage [32]. Since BRG1, an ATPase of the SWI/SNF-like BAF complexes, is a major co-regulator of transcription in mammalian cells [33], this study sought to examine its role in protecting human cells against UV radiation. To study the expression profile of genes in response to UV and mock treatment, we used isogenic cell lines, SW13+pREP7 (SW13+vector) and SW13+pREP7+BRG1 (Fig. 1A). BRG1 expression was confirmed by Western blot analysis and it is apparent that UV treatment did not affect BRG1 expression in SW13+Brg1 cells (Fig. 1B). By comparison with earlier studies, our investigation using oligonucleotide microarray technology (Affymetrix Human Genome U133 Plus 2.0 GeneChips) provides a more comprehensive profile of gene expression. We found that BRG1 regulates more than 70% of UV inducible genes, which are dispersed throughout the genome (Fig. 1C), some of which have been shown to be involved in UV damage repair and signaling. Surprisingly, by comparing gene expression in SW13+pREP7 and SW13+pREP7+BRG1 cells in the absence of UV treatment, this study shows that BRG1 regulates gene transcription in whole human genome, in a global fashion (Fig. 2). We show that altogether, about 4.8% of the human genome is transcriptionally regulated by BRG1, which is comparable with previous findings that the SWI/SNF complex regulates 6% of genes in yeast [34]. Thus, our study establishes that BRG1 availability in mammalian cells affects cellular transcriptional response to UV radiation. In addition, based on the genome expression data, our ChIP studies identified several novel transcriptional targets directly regulated by BRG1, warranting future experiments to fully understand BRG1's role as a tumor suppressor.
BRG1 transcriptionally regulates stress response genes that are induced by UV radiation
Transcriptional response is one of several ways cells deal with stress associated with DNA damage. Other responses include activation of DNA repair pathways and cell cycle checkpoints to arrest cell cycle progression and apoptosis [32]. Previously, we have shown that BRG1 suppresses UV induced apoptosis in SW13 cells [35], but the underlying mechanisms remain undefined. Gene expression profiling showed that UV radiation significantly (>2 fold induction, p<0.05) induces expression of 53 genes in SW13+pREP7 cells, 61 genes in SW13+pREP7+BRG1 cells (Tables S2A and S2B, respectively). Interestingly, 34 genes induced by UV are BRG1-dependent (Fig. 1C and Table 1). Many of those UV-inducible genes are transcription factors, which directly enhance/repress the transcription of their target genes, thus regulating their ability to exert biological functions in cellular responses to protect cells again the toxic effect of UV radiation. Also among these BRG1-dependent UV inducible genes, ATF3, Gadd45a and p21 are known to play important roles in cellular resistance to UV radiation [36]–[38], and our microarray studies show that they exhibit higher levels of expression in SW13 cells when BRG1 is re-introduced. These data confirm our previous RT-PCR observations that BRG1 regulates expression of p21 and Gadd45a [35], and that BRG1 binds to Gadd45a and p21 promoter regions to regulate their expression [12], [39], [40].
Table 1. 34 Genes Induced by UV Radiation in a BRG1-Dependent Manner.
| Probe Set ID | Gene Symbol | Gene Title |
| 202284_s_at | CDKN1A | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) |
| 202672_s_at | ATF3 | Activating transcription factor 3 |
| 202708_s_at | HIS2H2BE | Histone cluster 2, H2be |
| 202859_x_at | IL8 | Interleukin 8 |
| 203725_at | GADD45a | Growth arrest and DNA inducible, alpha |
| 204621_s_at | NR4A2 | Nuclear receptor subfamily 4, group A, member 2 |
| 205193_at | MAFF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) |
| 207064_s_at | AOC2 | Amine oxidase, copper containing 2 (retina-specific) |
| 211506_s_at | IL8 | Interleukin 8 |
| 214169_at | UNC84A | Unc-84 homolog A (C. elegans) |
| 220533_at | Transcribed locus | |
| 36711_at | MAFF | v-maf musculoaponeurotic fibrosarcoma oncogene homolog F |
| 38037_at | HBEGF | Heparin-binding EGF-like growth factor |
| 223218_s_at | NFKBIZ | Nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| 228839_s_at | LOC642361 | Hypothetical gene supported by AF064843; AK025716 /// hypothetical LOC642361 |
| 230604_at | Transcribed locus | |
| 231233_at | Transcribed locus | |
| 231417_at | Transcribed locus | |
| 232097_at | TOX4 | TOX high mobility group box family member 4 |
| 237116_at | LOC646903 | Hypothetical LOC646903 |
| 238012_at | DPP7 | Dipeptidyl-peptidase 7 |
| 238633_at | EPC1 | Enhancer of polycomb homolog 1 (Drosophila) |
| 239814_at | Transcribed locus, strongly similar to XP_531062.1 PREDICTED: hypothetical protein [Pan troglodytes] | |
| 242255_at | LOC100130837 | Hypothetical protein LOC 100130837 |
| 242594_at | FAM44A | Family with sequence similarity 44, member 4 |
| 243404_at | Transcribed locus | |
| 243947_s_at | Transcribed locus | |
| 1552362_a_at | LEAP2 | Liver expressed antimicrobial peptide 2 |
| 1554020_at | BICD1 | Bicaudal D homolog 1 (Drosophila) |
| 1554980_a_at | ATF3 | Activating transcription factor 3 |
| 1556213_a_at | BTG3 | BTG family, member 3 |
| 1556216_s_at | cDNA clone IMAGE: 5261375 | |
| 1556346_at | Partial mRNA; ID YG39-1A | |
| 1556588_at | C15orf37 | Chromosome 15 open reading frame 37 |
| 1557104_at | ZNF397OS | Zinc finger protein 397 opposite strand |
| 1560129_at | mRNA; cDNA DKFZp313H0240 (from clone DKFZp313H0240) | |
| 1565759_at | RPL 13 | Ribosomal protein L 13 |
Although it has been shown that ATF3 acts cooperatively with BRG1 to activate promoters, and plays a role in recruitment of BRG1 to gene promoters [41], this study identifies for the first time that under UV induction, BRG1 controls ATF3 expression. ATF3, a transcription factor in the ATF/CREB family, plays a significant role in cells stress response. ATF3 protein is expressed at a low levels in normal and quiescent cells, but can be rapidly and highly induced in response to multiple and diverse extracellular signals [42] including UV irradiation [43]. Importantly, overexpression of ATF3 protein suppresses cell growth [44]. Interestingly, BRG1 binds to the ATF3 promoter both, with and without UV irradiation, but binds better under UV conditions and higher expression of ATF3 is seen under these conditions as well (Fig. 3). This seems to suggest that there are perhaps other transcription factors activated by UV that aid BRG1 in binding to the ATF3 promoter. Further studies are necessary to fully understand this data, but our data newly demonstrate that BRG1 plays a direct role in activating expression of ATF3 under UV conditions.
Figure 3. BRG1 controls ATF3 expression under UV conditions.
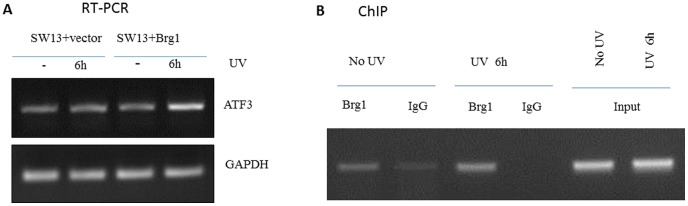
(A) RT-PCR analysis of ATF3 gene expression in SW13+vector and SW13+BRG1 cells. Both cells were collected 6 hours after UV treatment. RNA was purified and RT-PCR products were analyzed on agarose gel. (B) Binding of BRG1 to ATF3 gene promoter region. 293T cells were UV irradiated and incubated for 6 hours followed by Chromatin Immunoprecipitation (ChIP) assay with anti-BRG1 antibody.
Since SW13 cells have shown attenuated repair of CPDs, while BRG1 restoration has been shown to correct this deficiency [12], it might be possible to infer that BRG1 transcriptionally regulates expression of genes involved in NER. From our microarray data, we singled out the expression profile of NER genes. Although BRG1 expression leads to small changes in NER gene expression, none of the changes seem to be significant (>2 fold with a p value <0.05). Therefore, under our experimental conditions, we cannot conclude that BRG1 transcriptionally regulates expression of NER genes.
BRG1 expression changes global gene expression in the absence of UV radiation
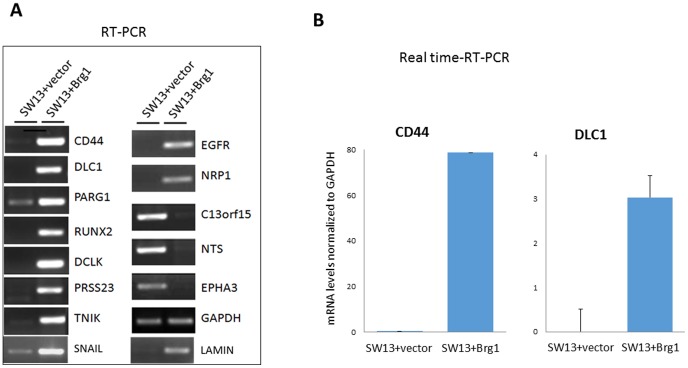
The design of this study also allowed us to identify novel transcriptional targets of BRG1 in the absence of UV treatment, by querying the global gene expression profile of the SW13+pREP7+BRG1 cells in comparison with SW13+pREP7 cells. Based on comparison of the SW13+vector vs. SW13+pREP7+BRG1 transcript abundance ratio, a value of >2.00 identified 1004 genes with reduced transcript levels and 599 genes with increased transcript levels in the SW13+pREP7+BRG1 (Tables 2 and S3A). Among these, CD44 [45], Gadd45 [30], [46] and p21 [39], [46] have previously been shown to be up-regulated by BRG1 in SW13 cells, validating our system for the study of gene regulation by BRG1. RT-PCR analysis of several highly up-regulated and down-regulated genes (CD44, DLC1, PARG1, RUNX2, DCLK, PRSS23, TNIK, EGFR, NRP1, C13orf15, NTS, EPHA3, SNAIL, LAMIN, and GAPDH), was performed to confirm the microarray data, and have shown to be consistent with the microarray data (Fig. 4A). Real time RT-PCR quantitation again confirms that BRG1 up-regulates expression of CD44 and DLC1 (Fig. 4B).
Table 2. Comparison of BRG1 regulated gene expression with UV treatment and without.
| No UV | UV 6 hours | Overlapping genes | |
| Up-regulated genes | 599 | 806 | 62 |
| Down-regulated genes | 1004 | 1249 | 108 |
Figure 4. Validation of the Microarray data by RT-PCR.
(A) Florescence intensity of RT-PCR products as a measure of mRNA expression confirming microarray data. Upon reintroduction of BRG1 into SW13 cells, regulation of gene expression differs from those cells without BRG1. GAPDH was loaded as a control. (B) Real time RT-PCR quantitation of CD44 and DLC1 mRNA levels.
Identification of novel BRG1 transcriptional targets
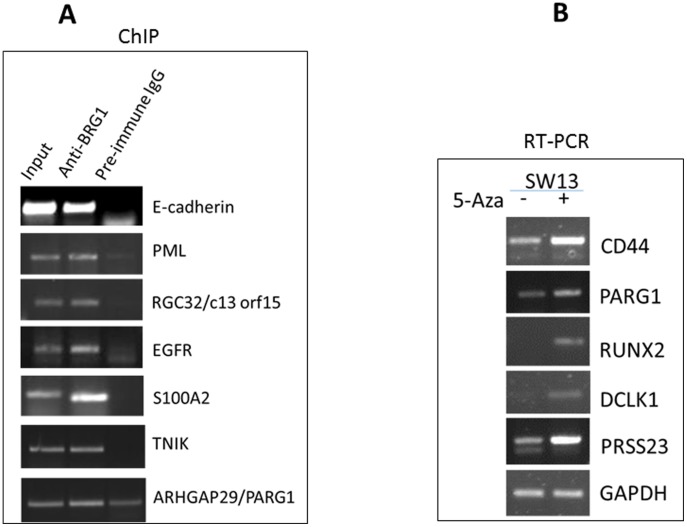
This study identifies many novel BRG1 transcriptional targets, including ATF3, TNIK, PARG, and S100A2, that may help explain the roles of BRG1 in DNA damage response. To determine if SWI/SNF complex containing BRG1 interacts directly with its target gene promoter, chromatin immunoprecipitation (ChIP) assays were performed and several of the genes found to be up-regulated in the global expression study were evaluated. Interestingly, eight of these genes exhibited binding of BRG1 to their promoters: E-cadherin, PML, RGC32/c13 orf15, EGFR, S1002A, TNIK, ARHGAP29/PARG1, and ATF3. As shown in Fig. 5A, the promoter sequence was significantly enriched in the immunoprecipitates with anti-BRG1 antibody in 293T cell lines. This study indicates that BRG1 has a direct role in transcriptional regulation of several of its target genes, and likely more of them, as well.
Figure 5. Validation of the Microarray data by RT-PCR and ChIP.
(A) Chromatin Immunoprecipitation studies show that BRG1 binds to the promoter of these genes, and regulates their expression. (B) RT-PCR showing that treatments with 5-aza-2′-deoxycytidine (5-Aza) induces expression of some of the identified BRG1 target genes in SW13 cells, which lack endogenous BRG1 expression.
It has been previously shown that the promoters of E-cadherin and CD44 are hypermethylated when BRG1 (or BRM) is absent from cells, and that BRG1 associates with the CpG islands in the promoter regions of the genes [47]. These authors proposed that the loss of SWI/SNF-mediated transcriptional activation increases DNA methylation in cancer cells. After confirming BRG1's association with promoters of several genes, (Fig. 5A), we performed demethylating studies with these genes, using 5-aza-2′deoxycytidine (as previously done, [47]) in SW13 cells that lack BRG1 expression. Similar to the findings in the previous studies, we also found induced expression of these genes upon demethylation treatment (Fig. 5B). This, in correlation with our microarray data and our RT-PCR data (Fig. 4) which show that BRG1 regulates expression of E-cadherin and CD44, led us to hypothesize that perhaps BRG1 regulates expression of these genes through the same mechanism proposed previously: by demethylating sequences in the promoter regions.
BRG1 is involved with regulation of many pathways
Our study shows that approximately 4.8% of the human genes are transcriptionally regulated by BRG1. Interestingly, analysis of our microarray data shows that BRG1 does not regulate genes within localized ‘hot-spots’, but rather plays a regulatory role of transcription throughout the whole genome, as seen in Fig. 2. Indeed, functional classification of the BRG1 responsive genes into different pathways (Table 3) indicates that BRG1 regulates various cellular pathways such as, MAPK signaling, cell cycle, regulation of actin cytoskeleton, cell adhesion, apoptosis, GTPase-mediated signal transduction, and transcriptional regulation. BRG1 does not regulate an exclusive signaling pathway; instead, multiple cellular pathways are affected upon re-expression of BRG1 in SW13 cells. Thus, this study highlights the notion that BRG1/BAF complexes serves as a fundamental component of various pathways.
Table 3. BRG1 regulates multiple cellular pathways.
| KEGG Pathway | Total Genes Regulated by BRG1 | Up-regulated | Down-regulated |
| MAPK Signaling Pathway | 46 | 29 | 17 |
| Focal Adhesion | 44 | 35 | 9 |
| Regulation of actin cytoskeleton | 37 | 25 | 12 |
| Cytokine-cytokine receptor interaction | 33 | 26 | 7 |
| Axon guidance | 29 | 15 | 14 |
| Wnt signaling pathway | 29 | 18 | 11 |
| Calcium signaling pathway | 28 | 15 | 13 |
| Neuroactive ligand-receptor interaction | 28 | 18 | 10 |
| Cell communication | 26 | 17 | 9 |
| Cell cycle | 26 | 5 | 21 |
| Pancreatic cancer | 18 | 14 | 4 |
| Colorectal cancer | 16 | 11 | 5 |
| Basal cell carcinoma | 15 | 10 | 5 |
| Melanoma | 15 | 11 | 4 |
| Glioma | 14 | 11 | 3 |
| Renal cell carcinoma | 14 | 12 | 2 |
| Small cell lung cancer | 14 | 11 | 3 |
| Non-small cell lung cancer | 11 | 10 | 1 |
| Prostate cancer | 11 | 11 | 0 |
Our microarray data indicate that BRG1 plays a role in regulating expression of many genes in the MAPK signaling pathway (Table 3). Deregulation of the MAPK pathways has been widely associated with tumorigenesis and has found to be deregulated in approximately 1/3 of cancers [48]. JNK has been shown to bind to and phosphorylate p53 in response to stresses such as UVB radiation and DNA damage [49]. This interaction with p53 is thought to increase its transcriptional expression and stability [50]. Thus, proper BRG1 expression is important for proper maintenance of cell signaling and p53 activity, through the MAPK pathway.
Activation of cell cycle checkpoint to arrest cell cycle progression and apoptosis are other ways that cells deal with stress associated with DNA damage [32]. Our cDNA microarray data (Table 1) not only agree with previous observations that BRG1 reintroduction into SW13 cells induces gene expression of p21, p15 and GADD45a [30], but also reveal more BRG1-regulated genes that are involved in cell cycle control, including p16, SMAD3, CCNG1(Cyclin G1), CCNG2 (Cyclin G2) and PML (Promyelocytic leukemia) genes.
Consistent with previous reports [30], our microarray and RT-PCR experiments demonstrate that BRG1 induces expression of several genes involved in cell adhesion and differentiation. Given that reintroduction of BRG1 induces changes in overall cell shape and morphology [30], [51], [52], and the cells become flatter with altered cytoskeletal organization (Fig. 1), BRG1's regulation of genes involved in cell morphology and development is crucial in understanding how cell function is differentially regulated. As reported in earlier studies [30], [47], our data show upregulation of CD44 and E-cadherin by BRG1 (Table S3B), and that BRG1 binds to the promoters of the genes (Fig. 5A). CD44 associates with moesin, which links the plasma membrane with the actin cytoskeleton. Cadherin-based adherens junction found between polarized epithelial cells, are also intimately associated with the actin cytoskeleton.
This study also shows BRG1 upregulated expression of TNIK and PARG1 (Table S3B and Fig. 4A). TNIK is known to be an essential activator of the Wnt signaling pathway and an integral part of canonical and non-canonical Wnt pathways [53], [54] and PARG1 is a Rho GTPase activating protein. The upregulation of these two genes suggests that BRG1 could control one or both of them for intricate control of actin cytoskeleton. Our ChIP data confirms BRG1 upregulation of most of these genes, as seen by BRG1 binding to their promoters in Fig. 5A. Our microarray and RT-PCR studies (Table S3B and Fig. 4A) also show that BRG1 upregulates SNAIL and LAMIN genes. SNAIL is a transcription factor that is known to bind to E-cadherin promoters and repress its transcription, thus inducing Epithelial-mesenchymal transition (EMT) [55], which is a crucial event in tumor progression. LAMIN is fundamental component of the nuclear lamina, which is involved in cross-talk with several cancer-regulating pathways, and its altered expression has been associated with a wide variety of cancers [56]. High amounts of LAMIN A have been shown to interfere with cell migration [57], a crucial process that occurs in EMT. Thus, proper levels of SNAIL and LAMIN are important for EMT to remain in a healthy balance, and BRG1 may regulate EMT and cancer metastasis through transcriptional regulation of these genes. Indeed, our pathway analysis showed that BRG1 regulates many genes involved in several cancer types (Table 3), and BRG1 is frequently deleted or mutated in a variety of tumor cell lines, including carcinomas of pancreas, lungs, prostate, breast [22]. BRG1 functions as a tumor suppressor, and regulation of gene expression through chromatin remodeling is critical for tumor progression. The functional balance of BRG1-regulated genes might dictate the likelihood of a given cell becoming cancerous or not.
BRG1 regulates a significant number of p53 targets
SWI/SNF-like BAF complexes have been shown to physically interact with p53 [58], [59], suggesting that p53 may recruit BAF complexes to its targeted promoters, and that the SWI/SNF complex may function as a tumor suppressor via interactions with p53. In fact, in vivo studies have shown BRG1 recruitment to p53-dependent promoters [59]. Therefore, we examined if BRG1 affects transcriptional expression of p53 target genes. A comparison of BRG1 transcriptional targets and reported p53 targets (Table 4) reveals that a significant number of p53-regulated genes are also regulated by BRG1, such as ATF3, PML, p21, Gadd45a. ATF3 has been shown to bind to p53 and prevent its ubiquitination upon DNA damage, thereby stabilizing p53 and enhancing its transcriptional activity and tumor suppressor function [60]. Recently, ATF3 has also been implicated as a crucial co-transcription factor for p53 upon DNA damage [36]. PML is well-known to form complexes with p53 upon UV radiation, thus activating and stabilizing p53 for further tumor suppressor action [61]. Another overlapping gene target, p21, is well-known to be recruited by p53 to arrest cell growth in the event of DNA damage [37]. In addition, Gadd45a, a ubiquitously expressed protein involved in growth arrest and apoptosis induced by UV damage, is highly intertwined with the p53 pathway, helping to activate it, which at the same time being regulated by it [38]. These overlapping gene targets suggest that p53 may require BRG1 to regulate its downstream targets. Thus, our findings support the notion that the tumor suppressor role of BRG1 is mediated in part through the p53 pathway [62].
Table 4. List of BRG1 regulated p53 targets.
| Gene Name | GenBank ID | Affimetrix | Gene Description | References |
| CAV1 | NM_001753 | 203065_s_at | Caveolin 1, caveolae protein, 22kDa | [65] |
| MET | BG170541 | 203510_at | Met proto-oncogene (hepatocyte growth factor receptor) | [66] |
| IFI16 | NM_005531 | 206332_s_at | Interferon, gamma-inducible protein 16 | [67] |
| IGFBP3 | M31159 | 210095_s_at | ATP-binding cassette, sub-family C (CTFR/MRP), member 3 | [68] |
| S100A2 | NM_005978 | 204268_at | S100 calcium binding protein A2 | [69] |
| PMAIP1 | AI857639 | 204285_s_at | Phorbol-12-myristate-13-acetate-induced protein 1 | [70] |
| TGFA | M31172 | 205015_s_at | Transforming growth factor, alpha | [71] |
| MVP | NM_017458 | 202180_s_at | Major vault protein | [72], [73] |
| IER3 | NM_003897 | 201631_s_at | Immediate early response 3 | [74] |
| GADD45a | NM_001924 | 203725_at | Growth arrest and DNA damage-inducible, alpha | [75] |
| EGFR | NM_005228 | 201984_s_at | Epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | [76] |
| LIF | NM_002309 | 205266_at | Leukemia inhibitory factor (cholinergic differentiation factor) | [77] |
| PML | NM_002675 | 206503_x_at | Promyelocytic leukemia | [78] |
| BNIP3L | ALI32665 | 221478_at | BCL2/adenovirus E1B 19kDa interaction protein 3-like | [79] |
| IL15 | Y09908 | 217371_s_at | H. sapiens mRNA for interleukin 15/PROD = interleukin 15 | [80] |
| STEAP3 | NM_018234 | 218424_s_at | STEAP family member 3 | [81] |
| KRT8 | U76549 | 209008_x_at | Keratin 8 | [82] |
| SCARA3 | NM_016240 | 219416_at | Scavenger receptor class A, member 3 | [83] |
| SYKQ | NM_003177 | 207540_s_at | Spleen tyrosine kinase | [84] |
| CCNG1 | BC000196 | 208796_s_at | Cyclin G1 | [85] |
| DUSP5 | U16996 | 209457_at | Dual specificity phosphatase 5 | [86] |
| NINJ1 | NM_004148 | 203045_at | Ninjurin 1 | [87] |
| DKK1 | NM_012242 | 204602_at | Dikkopf homolog 1 (Xenopus laevis) | [88] |
| EGR1 | AI459194 | 227404_s_at | Early growth response 1 | [89] |
| TP73L | AB010153 | 211194_s_at | Tumor protein p73 like | [90] |
| ARID3A | NM_005224 | 205865_at | AT rich interactive domain 3A (BRIGHT-like) | [81] |
| CHEK1 | NM_001274 | 205394_at | CHK1 checkpoint homolog (S. pombe) | [91] |
We note that the identified BRG1 transcriptional targets in this study may include genes regulated by another BAF complex ATPase, BRM, since BRG1 and BRM have partial functional redundancy [20], [45], [63], [64]. Additional studies will be needed to dissect the redundant and non-redundant roles of the individual ATPase subunits in gene regulation.
In conclusion, this study establishes that transcriptional response to UV damage is regulated by BRG1 in human cells. Several BRG1 up-regulated genes, including ATF3, EGFR, p21, and Gadd45a, are all involved in cellular defense against UV radiation [36]–[38]. Thus, it appears that the mechanisms by which BRG1 suppresses UV induced cell death in SW13 cells are multifaceted, playing roles in several different cell stress response pathways. Finally, our understanding of the mechanistic basis of diverse cellular processes, including cell cycle and actin cytoskeleton regulation, has been augmented by the transcriptional profiling of the BRG1 gene. There is abundant evidence that the mechanistic basis of BRG1 function is determined by the transactivation and transrepression of target gene expression [33], [41]. Our study identifies several important new transcriptional targets regulated by BRG1 and a large amount of possible new gene targets. The experiments presented here will serve as a platform for investigating the direct role of BRG1 in gene regulation, and will contribute toward the development of a wide range of intelligent hypotheses and drive a new generation of functional experiments.
Supporting Information
PCR primers used in this study.
(DOCX)
Table S2A: List of genes induced by UV in SW13+vector cells. Table S2B: List of genes induced by UV in SW13+Brg1 cells.
(DOCX)
Table S3A: Genes up- or down-regulated by BRG1 in the absence of UV irradiation. Table S3B: Genes up- or down-regulated by BRG1 6 hours after UV irradiation.
(XLSX)
Acknowledgments
We thank members in our laboratory for critical reading of the manuscript.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All the data may be found on the NCBI Gene Expression Omnibus, using the link http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59445.
Funding Statement
This work was supported by a National Institutes of Health grant R01ES017784 to FG. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Martens JA, Winston F (2003) Recent advances in understanding chromatin remodeling by Swi/Snf complexes. Curr Opin Genet Dev 13: 136–142. [DOI] [PubMed] [Google Scholar]
- 2. Hargreaves DC, Crabtree GR (2011) ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res 21: 396–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Song S, Walter V, Karaca M, Li Y, Bartlett CS, et al. (2014) Gene silencing associated with SWI/SNF complex loss during NSCLC development. Mol Cancer Res 12: 560–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tolstorukov MY, Sansam CG, Lu P, Koellhoffer EC, Helming KC, et al. (2013) Swi/Snf chromatin remodeling/tumor suppressor complex establishes nucleosome occupancy at target promoters. Proc Natl Acad Sci U S A 110: 10165–10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shain AH, Pollack JR (2013) The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 8: e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, et al. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chai B, Huang J, Cairns BR, Laurent BC (2005) Distinct roles for the RSC and Swi/Snf ATP-dependent chromatin remodelers in DNA double-strand break repair. Genes Dev 19: 1656–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bennett G, Papamichos-Chronakis M, Peterson CL (2013) DNA repair choice defines a common pathway for recruitment of chromatin regulators. Nat Commun 4: 2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Palomera-Sanchez Z, Zurita M (2011) Open, repair and close again: Chromatin dynamics and the response to UV-induced DNA damage. DNA Repair 10: 119–125. [DOI] [PubMed] [Google Scholar]
- 10. Yu YC, Teng YM, Liu HR, Reed SH, Waters R (2005) UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proceedings of the National Academy of Sciences of the United States of America 102: 8650–8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gong F, Fahy D, Smerdon MJ (2006) Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13: 902–907. [DOI] [PubMed] [Google Scholar]
- 12. Gong F, Fahy D, Liu H, Wang W, Smerdon MJ (2008) Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle 7: 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, et al. (2006) Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J 25: 3986–3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park JH, Park EJ, Hur SK, Kim S, Kwon J (2009) Mammalian SWI/SNF chromatin remodeling complexes are required to prevent apoptosis after DNA damage. DNA Repair (Amst) 8: 29–39. [DOI] [PubMed] [Google Scholar]
- 15.Keenen B, Qi H, Saladi SV, Yeung M, de la Serna IL (2009) Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. [DOI] [PMC free article] [PubMed]
- 16. Kothandapani A, Gopalakrishnan K, Kahali B, Reisman D, Patrick SM (2012) Downregulation of SWI/SNF chromatin remodeling factor subunits modulates cisplatin cytotoxicity. Exp Cell Res 318: 1973–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao Q, Wang QE, Ray A, Wani G, Han C, et al. (2009) Modulation of nucleotide excision repair by mammalian SWI/SNF chromatin-remodeling complex. J Biol Chem 284: 30424–30432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang L, Zhang Q, Jones K, Patel M, Gong F (2009) The chromatin remodeling factor BRG1 stimulates nucleotide excision repair by facilitating recruitment of XPC to sites of DNA damage. Cell Cycle 8: 3953–3959. [DOI] [PubMed] [Google Scholar]
- 19. Roberts CW, Orkin SH (2004) The SWI/SNF complex–chromatin and cancer. Nat Rev Cancer 4: 133–142. [DOI] [PubMed] [Google Scholar]
- 20. Reisman D, Glaros S, Thompson EA (2009) The SWI/SNF complex and cancer. Oncogene 28: 1653–1668. [DOI] [PubMed] [Google Scholar]
- 21. Hoffman GR, Rahal R, Buxton F, Xiang K, McAllister G, et al. (2014) Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proceedings of the National Academy of Sciences of the United States of America 111: 3128–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wong AK, Shanahan F, Chen Y, Lian L, Ha P, et al. (2000) BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res 60: 6171–6177. [PubMed] [Google Scholar]
- 23. von Figura G, Fukuda A, Roy N, Liku ME, Morris Iv JP, et al. (2014) The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat Cell Biol 16: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, et al. (2000) A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell 6: 1287–1295. [DOI] [PubMed] [Google Scholar]
- 25. Bultman SJ, Herschkowitz JI, Godfrey V, Gebuhr TC, Yaniv M, et al. (2008) Characterization of mammary tumors from Brg1 heterozygous mice. Oncogene 27: 460–468. [DOI] [PubMed] [Google Scholar]
- 26. Koch-Paiz CA, Amundson SA, Bittner ML, Meltzer PS, Fornace AJ Jr (2004) Functional genomics of UV radiation responses in human cells. Mutat Res 549: 65–78. [DOI] [PubMed] [Google Scholar]
- 27. Adimoolam S, Ford JM (2003) p53 and regulation of DNA damage recognition during nucleotide excision repair. DNA Repair (Amst) 2: 947–954. [DOI] [PubMed] [Google Scholar]
- 28. Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, et al. (2008) Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res 68: 415–424. [DOI] [PubMed] [Google Scholar]
- 29. Medina PP, Carretero J, Ballestar E, Angulo B, Lopez-Rios F, et al. (2005) Transcriptional targets of the chromatin-remodelling factor SMARCA4/BRG1 in lung cancer cells. Human Molecular Genetics 14: 973–982. [DOI] [PubMed] [Google Scholar]
- 30. Hendricks KB, Shanahan F, Lees E (2004) Role for BRG1 in cell cycle control and tumor suppression. Mol Cell Biol 24: 362–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 98: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sancar A, Reardon JT (2004) Nucleotide excision repair in E. coli and man. Adv Protein Chem 69: 43–71. [DOI] [PubMed] [Google Scholar]
- 33. Trotter KW, Archer TK (2008) The BRG1 transcriptional coregulator. Nucl Recept Signal 6: e004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, et al. (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728. [DOI] [PubMed] [Google Scholar]
- 35. Gong F, Fahy D, Liu H, Wang WD, Smerdon MJ (2008) Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle 7: 1067–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taketani K, Kawauchi J, Tanaka-Okamoto M, Ishizaki H, Tanaka Y, et al. (2012) Key role of ATF3 in p53-dependent DR5 induction upon DNA damage of human colon cancer cells. Oncogene 31: 2210–2221. [DOI] [PubMed] [Google Scholar]
- 37. Insinga A, Cicalese A, Pelicci PG (2014) DNA damage response in adult stem cells. Blood Cells Mol Dis 52: 147–151. [DOI] [PubMed] [Google Scholar]
- 38. Salvador JM, Brown-Clay JD, Fornace AJ Jr (2013) Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol 793: 1–19. [DOI] [PubMed] [Google Scholar]
- 39. Kang H, Cui K, Zhao K (2004) BRG1 controls the activity of the retinoblastoma protein via regulation of p21CIP1/WAF1/SDI. Mol Cell Biol 24: 1188–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsu CC, Lee YC, Yeh SH, Chen CH, Wu CC, et al. (2012) 58-kDa Microspherule Protein (MSP58) Is Novel Brahma-related Gene 1 (BRG1)-associated Protein That Modulates p53/p21 Senescence Pathway. Journal of Biological Chemistry 287: 22533–22548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu YZ, Thuraisingam T, Marino R, Radzioch D (2011) Recruitment of SWI/SNF complex is required for transcriptional activation of the SLC11A1 gene during macrophage differentiation of HL-60 cells. J Biol Chem 286: 12839–12849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U (1999) ATF3 and stress responses. Gene Expr 7: 321–335. [PMC free article] [PubMed] [Google Scholar]
- 43. Turchi L, Aberdam E, Mazure N, Pouyssegur J, Deckert M, et al. (2008) Hif-2alpha mediates UV-induced apoptosis through a novel ATF3-dependent death pathway. Cell Death and Differentiation 15: 1472–1480. [DOI] [PubMed] [Google Scholar]
- 44. Fan FY, Jin SQ, Amundson SA, Tong T, Fan WH, et al. (2002) ATF3 induction following DNA damage is regulated by distinct signaling pathways and over-expression of ATF3 protein suppresses cells growth. Oncogene 21: 7488–7496. [DOI] [PubMed] [Google Scholar]
- 45. Strobeck MW, DeCristofaro MF, Banine F, Weissman BE, Sherman LS, et al. (2001) The BRG-1 subunit of the SWI/SNF complex regulates CD44 expression. J Biol Chem 276: 9273–9278. [DOI] [PubMed] [Google Scholar]
- 46.Gong F, Fahy D, Liu H, Wang W, Smerdon MJ (2008) Role of the mammalian SWI/SNF chromatin remodeling complex in the cellular response to UV damage. Cell Cycle 7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Banine F, Bartlett C, Gunawardena R, Muchardt C, Yaniv M, et al. (2005) SWI/SNF chromatin-remodeling factors induce changes in DNA methylation to promote transcriptional activation. Cancer Research 65: 3542–3547. [DOI] [PubMed] [Google Scholar]
- 48. Dhillon AS, Hagan S, Rath O, Kolch W (2007) MAP kinase signalling pathways in cancer. Oncogene 26: 3279–3290. [DOI] [PubMed] [Google Scholar]
- 49. Wu GS (2004) The functional interactions between the p53 and MAPK signaling pathways. Cancer Biology & Therapy 3: 156–161. [DOI] [PubMed] [Google Scholar]
- 50. Buschmann T, Potapova O, Bar-Shira A, Ivanov VN, Fuchs SY, et al. (2001) Jun NH2-terminal kinase phosphorylation of p53 on Thr-81 is important for p53 stabilization and transcriptional activities in response to stress. Molecular and Cellular Biology 21: 2743–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, et al. (1994) The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell 79: 119–130. [DOI] [PubMed] [Google Scholar]
- 52. Strober BE, Dunaief JL, Guha, Goff SP (1996) Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol 16: 1576–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mikryukov A, Moss T (2012) Agonistic and Antagonistic Roles for TNIK and MINK in Non-Canonical and Canonical Wnt Signalling. Plos One 7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mahmoudi T, Li VSW, Ng SS, Taouatas N, Vries RGJ, et al. (2009) The kinase TNIK is an essential activator of Wnt target genes. Embo Journal 28: 3329–3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Javaid S, Zhang J, Anderssen E, Black JC, Wittner BS, et al. (2013) Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep 5: 1679–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Prokocimer M, Davidovich M, Nissim-Rafinia M, Wiesel-Motiuk N, Bar DZ, et al. (2009) Nuclear lamins: key regulators of nuclear structure and activities. J Cell Mol Med 13: 1059–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Harada T, Swift J, Irianto J, Shin JW, Spinler KR, et al. (2014) Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. Journal of Cell Biology 204: 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Oh J, Sohn DH, Ko M, Chung H, Jeon SH, et al. (2008) BAF60a interacts with p53 to recruit the SWI/SNF complex. J Biol Chem 283: 11924–11934. [DOI] [PubMed] [Google Scholar]
- 59. Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, et al. (2002) SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. Journal of Biological Chemistry 277: 22330–22337. [DOI] [PubMed] [Google Scholar]
- 60. Yan C, Boyd DD (2006) ATF3 regulates the stability of p53: a link to cancer. Cell Cycle 5: 926–929. [DOI] [PubMed] [Google Scholar]
- 61. Kurki S, Latonen L, Laiho M (2003) Cellular stress and DNA damage invoke temporally distinct Mdm2, p53 and PML complexes and damage-specific nuclear relocalization. J Cell Sci 116: 3917–3925. [DOI] [PubMed] [Google Scholar]
- 62. Xu Y, Zhang J, Chen X (2007) The activity of p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF chromatin remodeling complexes. J Biol Chem 282: 37429–37435. [DOI] [PubMed] [Google Scholar]
- 63. Strobeck MW, Reisman DN, Gunawardena RW, Betz BL, Angus SP, et al. (2002) Compensation of BRG-1 function by Brm: insight into the role of the core SWI-SNF subunits in retinoblastoma tumor suppressor signaling. J Biol Chem 277: 4782–4789. [DOI] [PubMed] [Google Scholar]
- 64. Reisman DN, Strobeck MW, Betz BL, Sciariotta J, Funkhouser W, et al. (2002) Concomitant down-regulation of BRM and BRG1 in human tumor cell lines: differential effects on RB-mediated growth arrest vs CD44 expression. Oncogene 21: 1196–1207. [DOI] [PubMed] [Google Scholar]
- 65. Yin X, Fontoura BM, Morimoto T, Carroll RB (2003) Cytoplasmic complex of p53 and eEF2. J Cell Physiol 196: 474–482. [DOI] [PubMed] [Google Scholar]
- 66. McKinney K, Prives C (2002) Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol Cell Biol 22: 6797–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Brazda V, Coufal J, Liao JCC, Arrowsmith CH (2012) Preferential binding of IFI16 protein to cruciform structure and superhelical DNA. Biochemical and Biophysical Research Communications 422: 716–720. [DOI] [PubMed] [Google Scholar]
- 68. Hoffman WH, Biade S, Zilfou JT, Chen J, Murphy M (2002) Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 277: 3247–3257. [DOI] [PubMed] [Google Scholar]
- 69. Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, et al. (2006) A global map of p53 transcription-factor binding sites in the human genome. Cell 124: 207–219. [DOI] [PubMed] [Google Scholar]
- 70. Hamard PJ, Barthelery N, Hogstad B, Mungamuri SK, Tonnessen CA, et al. (2013) The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes & Development 27: 1868–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shin TH, Paterson AJ, Kudlow JE (1995) P53 Stimulates Transcription from the Human Transforming Growth-Factor-Alpha Promoter - a Potential Growth-Stimulatory Role for P53. Molecular and Cellular Biology 15: 4694–4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. An HJ, Ryu SJ, Kim SY, Choi HR, Chung JH, et al. (2009) Age associated high level of major vault protein is p53 dependent. Cell Biochemistry and Function 27: 289–295. [DOI] [PubMed] [Google Scholar]
- 73. Ikeda R, Nishizawa Y, Tajitsu Y, Minami K, Mataki H, et al. (2014) Regulation of major vault protein expression by upstream stimulating factor 1 in SW620 human colon cancer cells. Oncology Reports 31: 197–201. [DOI] [PubMed] [Google Scholar]
- 74. Lohr K, Moritz C, Contente A, Dobbelstein M (2003) p21/CDKN1A mediates negative regulation of transcription by p53. J Biol Chem 278: 32507–32516. [DOI] [PubMed] [Google Scholar]
- 75. Smith ML, Chen IT, Zhan Q, Bae I, Chen CY, et al. (1994) Interaction of the p53-regulated protein Gadd45 with proliferating cell nuclear antigen. Science 266: 1376–1380. [DOI] [PubMed] [Google Scholar]
- 76. Ludes-Meyers JH, Subler MA, Shivakumar CV, Munoz RM, Jiang P, et al. (1996) Transcriptional activation of the human epidermal growth factor receptor promoter by human p53. Mol Cell Biol 16: 6009–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hu WW, Feng ZH, Teresky AK, Levine AJ (2007) p53 regulates maternal reproduction through LIF. Nature 450: 721–U728. [DOI] [PubMed] [Google Scholar]
- 78. de Stanchina E, Querido E, Narita M, Davuluri RV, Pandolfi PP, et al. (2004) PML is a direct p53 target that modulates p53 effector functions. Mol Cell 13: 523–535. [DOI] [PubMed] [Google Scholar]
- 79. Fei PW, Wang WG, Kim SH, Wang SL, Burns TF, et al. (2004) Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell 6: 597–609. [DOI] [PubMed] [Google Scholar]
- 80. De Giovanni C, Nanni P, Sacchi A, Soddu S, Manni I, et al. (1998) Wild-type p53-mediated down-modulation of interleukin 15 and interleukin 15 receptors in human rhabdomyosarcoma cells. British Journal of Cancer 78: 1541–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Passer BJ, Nancy-Portebois V, Amzallag N, Prieur S, Cans C, et al. (2003) The p53-inducible TSAP6 gene product regulates apoptosis and the cell cycle and interacts with Nix and the Myt1 kinase. Proceedings of the National Academy of Sciences of the United States of America 100: 2284–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mukhopadhyay T, Roth JA (1996) p53 involvement in activation of the cytokeratin 8 gene in tumor cell lines. Anticancer Research 16: 105–112. [PubMed] [Google Scholar]
- 83. Han HJ, Tokino T, Nakamura Y (1998) CSR, a scavenger receptor-like protein with a protective role against cellular damage caused by UV irradiation and oxidative stress. Human Molecular Genetics 7: 1039–1046. [DOI] [PubMed] [Google Scholar]
- 84. Okamura S, Ng CC, Koyama K, Takei Y, Arakawa H, et al. (1999) Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncology Research 11: 281–285. [PubMed] [Google Scholar]
- 85. Endo Y, Fujita T, Tamura K, Tsuruga H, Nojima H (1996) Structure and chromosomal assignment of the human cyclin G gene. Genomics 38: 92–95. [DOI] [PubMed] [Google Scholar]
- 86. Ueda K, Arakawa H, Nakamura Y (2003) Dual-specificity phosphatase 5 (DUSP5) as a direct transcriptional target of tumor suppressor p53. Oncogene 22: 5586–5591. [DOI] [PubMed] [Google Scholar]
- 87. Cho SJ, Rossi A, Jung YS, Yan WS, Liu G, et al. (2013) Ninjurin1, a target of p53, regulates p53 expression and p53-dependent cell survival, senescence, and radiation-induced mortality. Proceedings of the National Academy of Sciences of the United States of America 110: 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Shou J, Ali-Osman F, Multani AS, Pathak S, Fedi P, et al. (2002) Human Dkk-1, a gene encoding a Wnt antagonist, responds to DNA damage and its overexpression sensitizes brain tumor cells to apoptosis following alkylation damage of DNA. Oncogene 21: 878–889. [DOI] [PubMed] [Google Scholar]
- 89. Eichhorn T, Hiller C, Hirschfelder K, Frank M, Krauth-Siegel RL, et al. (2013) Identification by high-throughput in silico screening of radio-protecting compounds targeting the DNA-binding domain of the tumor suppressor p53. Cancer Genomics Proteomics 10: 35–45. [PubMed] [Google Scholar]
- 90. Harmes DC, Bresnick E, Lubin EA, Watson JK, Heim KE, et al. (2003) Positive and negative regulation of Delta N-p63 promoter activity by p53 and Delta N-p63-alpha contributes to differential regulation of p53 target genes. Oncogene 22: 7607–7616. [DOI] [PubMed] [Google Scholar]
- 91. Gali-Muhtasib H, Kuester D, Mawrin C, Bajbouj K, Diestel A, et al. (2008) Thymoquinone triggers inactivation of the stress response pathway sensor CHEK1 and contributes to apoptosis in colorectal cancer cells. Cancer Research 68: 5609–5618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers used in this study.
(DOCX)
Table S2A: List of genes induced by UV in SW13+vector cells. Table S2B: List of genes induced by UV in SW13+Brg1 cells.
(DOCX)
Table S3A: Genes up- or down-regulated by BRG1 in the absence of UV irradiation. Table S3B: Genes up- or down-regulated by BRG1 6 hours after UV irradiation.
(XLSX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All the data may be found on the NCBI Gene Expression Omnibus, using the link http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE59445.