Abstract
Background
Sphingolipids (Sls) are not only key components of cellular membranes, but also play an important role as signaling molecules in orchestrating both cell growth and apoptosis. In Saccharomyces cerevisiae, three complex SLs are present and hydrolysis of either of these species is catalyzed by the inositol phosphosphingolipid phospholipase C (Isc1p). Strikingly, mutants deficient in Isc1p display several hallmarks of mitochondrial dysfunction such as the inability to grow on a non-fermentative carbon course, increased oxidative stress and aberrant mitochondrial morphology.
Scope of Review
In this review, we focus on the pivotal role of Isc1p in regulating mitochondrial function via SL metabolism, and on Sch9p as central signal transducer. Sch9p is one of the main effectors of the target of rapamycin complex 1 (TORC1), which is regarded as a crucial signaling axis for regulation of Isc1p-mediated events. Finally, we describe the retrograde response, a signaling event originating from mitochondria to the nucleus which results in the induction of nuclear target genes. Intriguingly, the retrograde response also interacts with SL homeostasis.
Major Conclusions
All the above suggests a pivotal signaling role for SLs in maintaining correct mitochondrial function in budding yeast.
General Significance
Studies with budding yeast provide insight on SL signaling events that affect mitochondrial function.
Keywords: S. cerevisiae, sphingolipids, mitochondrial function, Isc1p, Sch9p, retrograde response
1. Introduction1
Sphingolipids (SLs) are lipid species characterized by the presence of a sphingoid base as structural backbone. These sphingoid bases are either sphingosine, dihydrosphingosine (DHS) or phytosphingosine (PHS) [1]. In yeast research, DHS and PHS are termed long chain bases (LCBs). In general, SLs serve as important parts of membranes, but also take part as signaling molecules in the regulation of cell division [2], cell death [3], lifespan [4] and autophagy [5].
SL biosynthetic pathways are highly conserved between mammalian and yeast cells [6-8]. In Saccharomyces cerevisiae de novo SL biosynthesis (Fig. 1) typically starts by the condensation of serine and palmitoyl Coenzyme A (palmitoyl CoA) to generate 3-ketodihydrosphingosine (3-keto DHS) by the target of Myriocin, namely serine palmitoyltransferase (SPT) [9-12]. As for yeast, 3-keto DHS is reduced to DHS, which then is processed into either dihydroceramide (dhCer) or PHS. Subsequently, both dhCer and PHS are converted into the central yeast SL phytoceramide (phytoCer). PhytoCer serves as precursor in the formation of the three complex SLs by addition of polar headgroups: (i) addition of phospho-inositol to phytoCer by inositolphosphoceramide (IPC) synthase yields IPC; (ii) addition of mannose to IPC by mannose inositolphosphoceramide (MIPC) synthase yields MIPC; (iii) addition of a second phospho-inositol residue to MIPC by inositolphosphotransferase (Ipt1p) leads to the generation of the end product mannose diinositolphosphoceramide (M(IP)2C) [13-15]. These three complex SLs can be hydrolyzed by the inositol phosphosphingolipid phospholipase C (Isc1p) back to phytoCer [16]. Functionally, Isc1p is essential in the coordination of cellular morphology [17] and cell cycle [18]. Also, Isc1p is involved in tolerance or sensitivity to toxic agents such as Na+ and Li+ [19], H2O2 [20], acetic acid [21], methyl methanosulfate and hydroxyurea [22]. Furthermore, studies with Δisc1 mutants have implicated a pivotal role for SLs in the regulation of mitochondrial function and dysfunction [20, 21, 23-27].
Fig. 1.
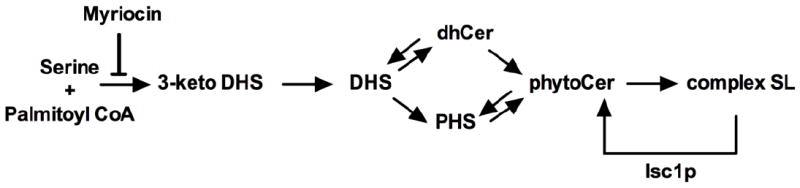
De novo SL biosynthetic pathway in S. cerevisiae. Myriocin inhibits SPT.
The documented role for Isc1p in coordinating mitochondrial function seems to be independent of the retrograde response [28]. The retrograde response is a signaling event originating from the mitochondria that results in the induction of various nuclear target genes by signal transduction proteins [29, 30]. The retrograde response in S. cerevisiae, however, also affects SL homeostasis [31, 32] and interacts with additional signaling pathways [29].
Next to the link between SLs and mitochondrial function derived from studies with Δisc1 mutants and the retrograde response, a few additional reports have linked SLs to mitochondrial function. For instance, Myriocin-induced cell death, and thus decreased de novo SL biosynthesis, is abrogated in ρ0 cells [33], which lack mitochondria DNA (mtDNA) and a functional respiratory chain. Likewise, ρ0 cells are insensitive to Suloctidil and dihydromotuporamine C [33], both compounds that are known to affect SL biosynthesis in mammalian cells [34, 35]. In addition, whereas sub-lethal LCB doses restore viability of yeast mutants defective in SL biosynthesis [36-38] and affect gene expression [39], exogenously added LCBs can kill several fungal species [39-43]. In S. cerevisiae however, loss of the mtDNA increases tolerance to LCBs [44], which is dependent on the retrograde response [44]. Such results indicate that SLs indeed are important players in mitochondrial function.
2. Inositol phosphosphingolipid phospholipase C (Isc1p) and mitochondrial function
The inositol phosphosphingolipid phospholipase C (Isc1p) is well documented as the enzyme responsible for the hydrolysis of complex SLs to generate phytoCer [16, 45]. Isc1p activity increases from early exponential to late exponential/post-diauxic growth phase and is regulated by phosphatidylglycerolphosphate synthase (Pgs1p) [23, 25], which is required for the synthesis of phosphatidylglycerol phosphate and subsequent synthesis of phosphatidylglycerol (PG) and cardiolipin (CL) [46]. The mitochondria-associated lipids PG and CL themselves, as well as phosphatidylserine are known activators of Isc1p [45, 47]. Isc1p mainly resides in the ER, but localizes to the outer mitochondrial membrane (OMM) during the late exponential and post-diauxic growth phase [23, 25, 27, 48].
2.1 Δisc1 mutants display characteristics of mitochondrial dysfunction
Several studies with Δisc1 mutants have pointed to inherent compromised mitochondrial function. One of the main initial observations with Δisc1 mutants is their decreased growth rate during late logarithmic and stationary growth phase [23]. Cells lacking Isc1p show decreased chronological lifespan (CLS) [20], a measure of survival of a non-dividing yeast population [49]. This decreased CLS or premature ageing of Δisc1 mutants is associated with increased oxidative stress and apoptosis [20, 50]. Indicative for mitochondrial dysfunction, these mutants display defective growth on a nonfermentable carbon source [24-26, 28, 50]. In addition, Δisc1 mutants exhibit an increased frequency of petite formation [27], a hallmark of yeast cells with mitochondrial defects [51]. Additional observations from Δisc1 mutants indicating aberrant mitochondrial function are the facts that they are characterized by mitochondrial hyperpolarization and mitochondrial fragmentation [26] as well as abnormal mitochondrial morphology [21]. Moreover, Δisc1 mutants exhibit increased sensitivity to toxic stimuli, such as hydrogen peroxide (H2O2) and ethidium bromide [27], that are reported as increasingly toxic to cells with defective mitochondria [52-54]. A direct link between Isc1p and the mitochondrial respiratory chain is also suggested since Δisc1 mutants display lower cytochrome c content [21] and decreased levels of the mitochondrial electron transport chain (ETC) complex IV (cytochrome c oxidase, COX) subunits Cox3p and Cox4p [25]. Hence, decreased COX activity and oxygen consumption rate have also been observed in Δisc1 mutants [26]. Interestingly, loss of the mitochondrial genome in Δisc1 mutants attenuates the decreased CLS associated with Δisc1 mutants, indicating that mitochondrial dysfunction contributes to the shortened CLS in Δisc1 mutants [24]. Summarizing, all these reports indicate that Δisc1 mutants indeed are characterized by extensive mitochondrial dysfunction.
2.2 Δisc1 mutants show aberrant mitochondrial SL composition
The aforementioned phenotypes in Δisc1 mutants can be correlated to aberrancies in mitochondrial SL composition. Intriguingly, wild type yeast cell mitochondria are enriched in α-hydroxylated phytoCer and depleted in sphingoid bases as compared to whole cells [27]. Except for α-OH-C14-phytoCer and C26-phytoCer levels, Δisc1 mutants display decreased levels of all SLs, with the most prominent decreases in the levels of α-OH-C24-phytoCer and α-OH-C26-phytoCer species. Also, during CLS Δisc1 mutants display an abnormal SL composition: decreased levels of DHS and α-OH-phytoCer and increased levels C26-dhCer and C26-phytoCer [50]. In addition, exogenously supplied C12-phytoCer restores the ability of Δisc1 mutants to grow on a non-fermentable carbon source [25]. This suggests that Isc1p-mediated generation of phytoCer in mitochondria is important for mitochondrial function [27].
2.3 Mitochondrial dysfunction in Δisc1 mutants is caused by a misregulation of gene expression
The mitochondrial dysfunction-related phenotypes prevalent in Δisc1 mutants are, however, caused by a misregulation of gene expression, and not by an intrinsic mitochondrial defect [28]. For instance, deficient growth on a non-fermentable carbon source of Δisc1 mutants is not characterized by a loss of mtDNA, nor do isolated mitochondria from Δisc1 mutants exhibit differences in the rate of oxygen consumption [28], indicating that Δisc1 mutants have a functional respiratory chain. However, Δisc1 mutants display little oxygen consumption rate in the post-diauxic shift phase [26], suggesting that additional mechanisms repress mitochondrial function in intact cells during the post-diauxic shift phase. Nonetheless, during the diauxic shift Δisc1 mutants are unable to up-regulate genes that are predominantly involved in the utilization of non-fermentable carbon sources, and to down-regulate genes involved in nutrient uptake and amino acid metabolism, independently of the retrograde response [28]. These results suggest an indispensable role of Isc1p in metabolic adaptation and a crucial signaling role for SLs in orchestrating mitochondrial function in intact cells.
2.4 Mechanistic insights into the mitochondria-related function of Isc1p
Mechanistic insights in the underlying signaling pathways that mediate Isc1p-related changes in mitochondrial function were provided by studies showing that loss of either Sit4p, Hog1p, Tor1p or Sch9p attenuates mitochondrial dysfunction-related phenotypes in Δisc1 mutants [24, 26, 48, 50]. In the next paragraphs we will provide more details on the interplay between Isc1p and Sit4p/CAPP, the high osmolarity glycerol pathway (HOG pathway) and the TORC1/Sch9p pathway (as summarized in Fig. 2).
Fig. 2.
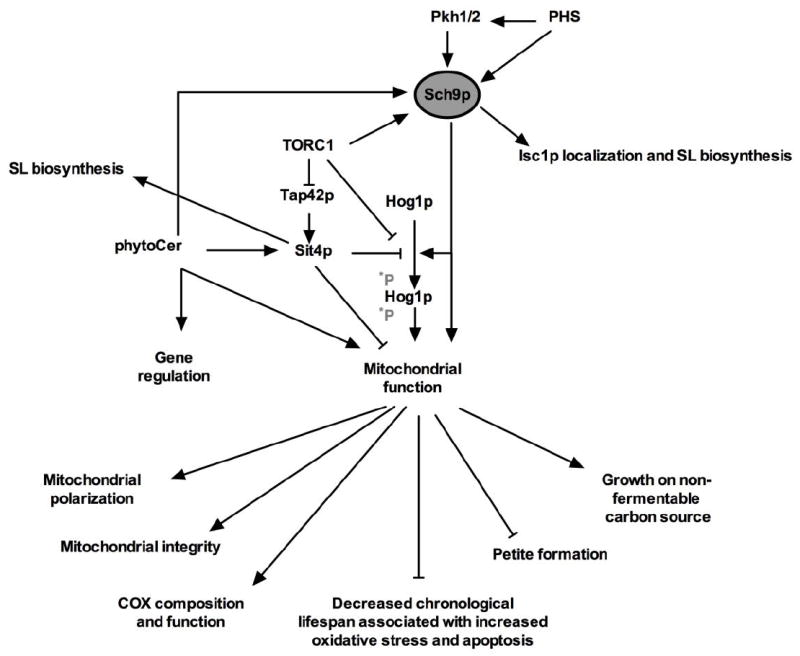
Interplay between SLs and mitochondrial function. Sch9p governs correct localization of Isc1p to the OMM during the post diauxic shift and affects SL biosynthesis. Sit4p affects SL biosynthesis, represses Hog1p phosphorylation and mitochondrial function. Sch9p and Sit4p combine Isc1p-mediated phytoCer signaling. Adapted from [26].
Sit4p/CAPP
Sit4p is the catalytic subunit of the ceramide-activated protein phosphatase (CAPP), which is activated by Cer [55]. Sit4p is inactivated by the target of rapamycin complex 1 (TORC1) [56], which is part of the highly conserved target of rapamycin (TOR) nutrient signaling pathway [57]. A regulatory role for Sit4p in SL biosynthesis was recently suggested as Δsit4 mutants display an aberrant SL composition [43]. More specifically, Sit4p was suggested to regulate Sur2p or Cer synthase activity [43]. Sur2p converts DHS to PHS [58]. Cer synthases such as Lag1p and Lac1p generate dhCer from DHS and phytoCer from PHS [6].
Barbosa and coworkers initially linked Sit4p to SLs and mitochondrial function based on the observation that cells lacking Isc1p display increased Sit4p activity [50]. Thus, Sit4p dependent Sur2p or Cer synthase activity [43] could point to a compensatory mechanism for the lack of phytoCer production in Δisc1 mutants. This compensatory mechanism could, for instance, be responsible for the increased generation of αOH-C14-phytoCer and C26-phytoCer levels in Δisc1 mutant mitochondria [27]. Also, loss of Sit4p attenuates the shortened CLS and H2O2 sensitivity in Δisc1 mutants. In addition, Δisc1Δsit4 mutants have a restored mitochondrial function during the diauxic shift and are able to grow on a non-fermentable carbon source [50]. Hence, Sit4p is a downstream target of SLs in regulating mitochondrial function.
The HOG pathway
The HOG pathway is involved in the response to hyperosmotic stress, in which the Hog1p mitogen activated protein kinase (MAPK) cascade plays a central role [59-62]. MAPK cascades are conserved signaling pathways that are crucial in e.g. the regulation of gene expression [63]. The HOG pathway in yeast is initiated in response to unfavorable osmotic conditions by activation of MAPK kinase Pbs2p through the Sln1p or Sho1p branch that activate the redundant MAPKKK Ssk2p and Ssk22p or Ste11p, respectively. Activation of Pbs2p results in phosphorylation of Hog1p, which in turn translocates to the nucleus and initiates Hog1p-mediated phosphorylation of transcriptional regulators that drive transcription and cell cycle. Finally, the action of Hog1p restores the osmotic balance, its activity decreases and is exported back to the cytoplasm. For a detailed review the reader is referred to [62].
Hog1p is known to affect mitochondrial function as well as dysfunction [24, 64-68]. Transient activation of Hog1p is essential for cellular responses during osmotic stress [59-62] and results in the induction of transcription of genes involved in mitochondrial function [65-67]. In addition, Hog1p controls correct functioning of the respiratory metabolism in Candida albicans [68]. However, prolonged Hog1p activation results in cell death [69], inhibition of mitochondrial respiration and a concomitant increase in levels of reactive oxygen species (ROS) [64]. In addition, growth on a nonfermentable carbon source is impaired during osmotic stress [64]. These observed phenotypes resulting from prolonged Hog1p activation are also characteristic of Δisc1 mutants [20, 21, 23-27]. This implies that activation of Hog1p controls correct mitochondrial functioning, but can also result in its dysfunction upon prolonged activation. Hence, Hog1p activation should be highly controlled in order to regulate mitochondrial function and cell survival.
The HOG pathway has also been shown to involve interaction with SLs [24, 26, 70]. For instance, inhibition of de novo SL biosynthesis increases Hog1p phosphorylation [70] while Δisc1 mutants display Hog1p hyperphosphorylation [24, 26]. In addition, exogenous addition of C2-Cer increases phosphorylated Hog1p levels [24, 26]. Similarly as reported for Δisc1Δsit4 mutants [50], loss of Hog1p in Δisc1 mutants increases H2O2 tolerance, attenuates the decreased CLS, allows growth on a non-fermentable carbon source and restores mitochondrial function [24]. Thus, SL signaling affects Hog1p phosphorylation and is a negative upstream regulator of mitochondrial function.
The TORC1/Sch9p pathway
The TORC1 pathway is part of the highly conserved TOR nutrient signaling pathway and is important for metabolic adaptation during the diauxic shift [57, 71]. TORC1 activity directly reflects both nutrient availability and absence of stress factors in the growth medium. Two branches execute the majority of TORC1-mediated effects (Fig. 2). The Tap42p branch is inhibited upon phosphorylation by TORC1 and regulates Sit4p activity. The Sch9p branch is activated upon TORC1-mediated phosphorylation [57]. Alternatively, Sch9p activity is modulated by SLs via the action of Pkh1 and SLs themselves [72]. Pkh1p and its paralog Pkh2p (Pkh1/2p) are functionally redundant protein kinases homologous to mammalian 3-phosphoinositide-dependent protein kinase PDK1 [73, 74] and are important kinases in the regulation of cell processes such as aging, endocytosis and stress resistance [15]. The TORC1/Sch9p signaling axis also regulates CLS in yeast [4, 75]. For a recent review on TORC1/Sch9p the reader is referred to [57].
The TORC1/Sch9p signaling axis plays a pivotal role in Isc1p localization, SL biosynthesis and the regulation of mitochondrial function [26, 48, 76]. For instance, Sch9p represses the expression of genes involved in the ETC [76], but also affects the expression of SL biosynthetic genes in a TORC1 dependent manner [48]. Concomitantly Δsch9 mutants display altered abundance of several LCBs, Cer and complex SL [48]. Strikingly, an important role for Sch9p in Isc1p localization was demonstrated as loss of Sch9p attenuates the localization of Isc1p to the mitochondria during the post-diauxic growth phase [48]. Additionally, Teixeira and coworkers showed that the TORC1/Sch9p pathway is activated in Δisc1 mutants [26]. Moreover, loss of either Tor1p or Sch9p abolishes the aberrant mitochondrial dysfunction-related phenotypes associated with Δisc1 mutants such as decreased oxidative stress tolerance, decreased CLS, inability to grow on a non-fermentable carbon source, mitochondrial hyperpolarization, mitochondrial fragmentation and increased apoptotic cell death and ROS production during chronological aging [26, 48]. Hence, while Sch9p affects SL biosynthesis, the kinase also regulates the correct translocation of Isc1p to the OMM during the post-diauxic shift, and vice versa SL species generated by Isc1p target the TORC1/Sch9p axis as crucial signal transducer in regulating mitochondrial function.
Teixeira and coworkers also further integrated a putative connection between the TORC1/Sch9p pathway and previous mechanistic studies performed on Δisc1 mutants such as the connection with the HOG pathway and Sit4p/CAPP and reported extensive interactions (Fig. 2) [26]. For instance, Δisc1Δsit4 and Δisc1Δtor1 mutants show increased Hog1p phosphorylation as compared to Δisc1 mutant, indicating that Sit4p and TORC1 block Hog1p phosphorylation in a Δisc1 mutant [26]. Whether the Sit4p-mediated block on Hog1p phosphorylation is TORC1-dependent has not yet been addressed. Their results also indicate that Sch9p causes Hog1p phosphorylation in Δisc1 mutants, and that Sch9p phosphorylates Hog1p in response to exogenously added C2-Cer [26]. Therefore, the TORC1/Sch9p signaling axis appears to act as a crucial signal transducer between on one hand SLs and on the other hand Hog1p in regulating mitochondrial function in Δisc1 mutants. An overview of the thus far identified interactions is given in Fig. 2
Additional indications concerning a central signal transducing role for Sch9p in regulating mitochondrial function originates from Δisc1-like phenotypical observations in a yeast model for Niemann-Pick type C1 disease based on Δncr1 mutants [77, 78]. Niemann Pick type C1 is a lipid storage disorder characterized by neurodegeneration [79] and is caused by mutations in NPC1 in 95 % of all cases [80-82]. Yeast Δncr1 mutants lack the orthologue of NPC1 [83]. These Δncr1 mutants exhibit increased sensitivity to oxidative stress and decreased CLS, characterized by increased levels of oxidative stress markers. Additionally, these mutant cells are unable to grow on a non-fermentative carbon source, show decreased mitochondrial membrane potential (Δψm) and mitochondrial fragmentation [77]. In line with mammalian Niemann Pick type C1 cells [84], accumulation of LCBs is observed in the yeast Δncr1 mutants caused by an increased turnover of complex SLs. In addition, Δncr1 mutants show increased Sch9p phosphorylation due to the action of the Pkh1/2p pathway. Similarly as observed for Δisc1 mutants loss of either Sch9p or Pkh1p restores the aberrant phenotypes of Δncr1 mutants. In contrast, in Δisc1 mutants Sch9p phosphorylation is independent of Pkh1p [26]. Furthermore, Δncr1Δsch9 cells exhibit restored SL levels [77]. Such results indicate that the role of Sch9 as central signal transducer in the regulation of mitochondrial function is not restricted to the Δisc1 mutant genetic background.
3. The retrograde response interacts with sphingolipid homeostasis
The retrograde response is typically referred to as a signaling event, originating from the mitochondria, to the nucleus, resulting in the induction of various nuclear target genes by signal transduction pathways [29, 30]. In S. cerevisiae, the retrograde response is used as a sensor for mitochondrial dysfunction and results in metabolic adaptations, which are largely dependent on three signal transduction genes namely RTG1, RTG2 and RTG3 [29, 30]. Interestingly, the retrograde response is linked to SL homeostasis and multidrug resistance [31, 32, 85, 86]. In addition, the retrograde response interacts with additional pathways such as the TORC1/Sch9p signaling axis as exemplified by decreased TORC1-dependent Sch9p phosphorylation after dropping Δψm [87].
Crucial in orchestrating the link between the retrograde response and SL biosynthesis is Pdr3p. Like Pdr1p, Pdr3p is a Zn2Cys6-cluster-containing transcription factor [88, 89], which typically recognizes Pdr1p/Pdr3p response elements (PDREs). Therefore, PDREs are found in the promoters of all known genes regulated by these transcription factors [85, 90]. In addition, both Pdr1p and Pdr3p appear important players in multidrug resistance as gain of function mutations in PDR1 and PDR3 increase expression of their target genes encoding multidrug efflux pumps [85]. Furthermore, PDR3 expression increases upon loss of the mitochondrial genome during the retrograde response and is regulated by both Pdr3p itself [86] and Sit4p [91].
Both Pdr1p and Pdr3p are reported to affect SL homeostasis, e.g. by their observed regulation of IPT1 expression [31]. In line with the reported activation of Pdr3p during the retrograde response [86], loss of the mitochondrial genome causes a Pdr3p-dependent increase of IPT1 expression [31]. Concomitantly, ρ0 cells display an aberrant SL composition as compared to WT cells, which is restored to normal-state levels of most complex SLs upon loss of Pdr3p [31]. Next to IPT1, expression of additional genes encoding enzymes involved in SL biosynthesis respond to increased activity of Pdr1p/Pdr3p such as (i) LAC1, (ii) SUR2 and (iii) LCB2 [32], a component of SPT [37]. These studies indicate that compromised mitochondrial function initiates signaling events that result in the Pdr3p-dependent induction of SL biosynthesis.
In addition, despite the fact that the retrograde response is not activated in Δisc1 mutants [28], several observations during the retrograde response are also observed in Δisc1 mutants and are linked to SL signaling (Fig. 3). For instance, while Rtg2p or Rtg3p upregulate expression of SWE1 [92], encoding the Swe1p protein kinase involved in G2/M transition [93], phytoCer generated by Isc1p regulates Swe1p levels [18, 94]. Additionally, the protein product of RSB1, whose expression of the encoding gene increases in a Pdr3p-dependent manner upon loss of the mitochondrial genome [44, 95, 96], removes LCBs from the cell [97] and is also increased in Δisc1 mutants [20]. Furthermore, while expression of Pdr5p is dependent on Pdr3p during the retrograde response [95], Δisc1 mutants exhibit increased expression of the gene encoding Pdr5p and its paralog Pdr15p [20, 98]. These findings suggest that SL signaling activates pathways in Δisc1 mutants that resemble responses to retrograde signaling as summarized in Fig. 3.
Fig. 3.
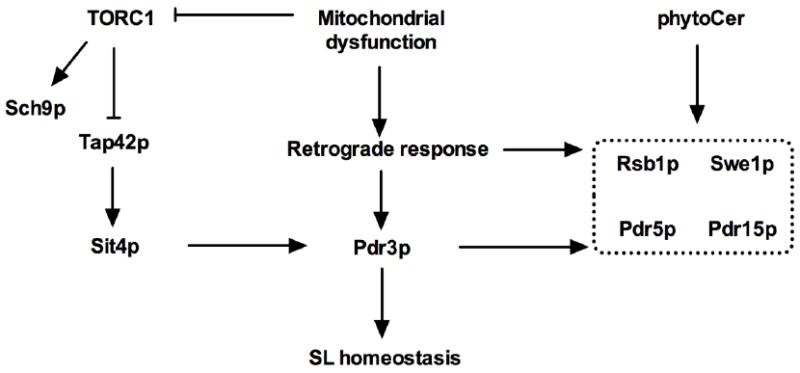
Interconnections between SLs and the retrograde response in S. cerevisiae. Mitochondrial dysfunction affects TORC1-dependent Sch9p phosphorylation and Pdr3p regulates SL homeostasis. Sit4p affects expression of the gene encoding Pdr3p. SL signaling activates pathways in Δisc1 mutants that resemble responses to retrograde signaling, and thus affect several common proteins as delineated by the dotted black line.
4. Conclusion
Thus far the most intriguing insights into a direct connection between SLs and mitochondrial function in S. cerevisiae mainly originate from studies that focused on Isc1p and the retrograde response. An overview of the thus far identified interactions is given in Fig. 2 and 3.
Several reports indicate that the translocation of Isc1p to the OMM is crucial in the regulation of mitochondrial function during the post-diauxic growth phase [20, 21, 23-27, 48]. Loss of Isc1p is detrimental for cellular mitochondrial function [20, 21, 23-26], resulting from a misregulation of gene expression [28]. Sch9p is responsible for the correct translocation of Isc1p from the ER to the OMM [48], however, Sch9p itself is a downstream signaling target of Isc1p, and affects SL biosynthesis [26, 48].
Nevertheless, the TORC1/Sch9p signaling axis acts as the central switch to pass upstream signaling events to downstream effectors, where Sch9p and Sit4p combine Isc1p-mediated phytoCer signaling. Thus far, Hog1p has been identified as their downstream target more specifically with Sch9p phosphorylating Hog1p, and Sit4p blocking Hog1p phosphorylation [26]. Intriguingly, while loss of Sit4p suppresses mitochondrial dysfunction-related phenotypes in Δisc1 mutants [50], Δisc1Δsit4 mutants exhibit increased phosphorylated Hog1p levels as compared to Δisc1 mutants [26]. Given the reports that indicate that transient Hog1p activation promotes mitochondrial function [65-68], and prolonged Hog1p activation is detrimental [64, 69], these results also imply that Hog1p phosphorylation is a delicate event in order to preserve mitochondrial function in yeast. The role of the TORC1/Sch9p axis is not restricted to the Δisc1 mutant genetic background, as Sch9p was also shown to be crucial in regulating mitochondrial function in a yeast model for Niemann Pick type C1 [77, 78]. Hence, Sch9p acts as a critical switch in the regulation of mitochondrial function in yeast; whether Hog1p is the sole downstream target in the regulation of mitochondrial function in response to SL remains to be elucidated.
The role of the retrograde response is limited in Isc1p-mediated effects on mitochondrial function [28]. Still, the retrograde response is linked to the induction of SL biosynthetic genes [31, 32]. As such, during the retrograde response, the central transcription factor Pdr3p induces expression of SL biosynthetic genes by binding to the PDRE in the promotors of its target genes [29]. Among these targets are several genes involved in SL biosynthesis such as IPT1, LCB2, SUR2 and LAC1 [31, 32]. In addition, Sit4p affects Pdr3p expression and SL biosynthesis [91]. Whether this effect is mediated via a Pdr3p mechanism is yet to be shown. In addition, loss of Δψm decreases TORC1-mediated Sch9p phosphorylation [87]. Despite the fact that the retrograde response is not activated in Δisc1 mutants [28], several cellular responses during the retrograde response are also observed in Δisc1 mutants [18, 20, 44, 92, 94-96, 98]. This indicates that loss of mitochondrial quality, which triggers the retrograde response, interacts with SL homeostasis, and SL signaling activates pathways in Δisc1 mutants that resemble responses to retrograde signaling.
Potential implications for the described connection between SLs and mitochondrial function in yeast are diverse and can serve to understand SL-related human pathologies or specific mammalian/human systems. For instance, Isc1p is homologous to mammalian neutral sphingomyelinases (nSMases) involved in the hydrolysis of the mammalian SL sphingomyelin to Cer [16]. In mammalian cells, four nSMases have been cloned and purified, with nSMase2 being the best studied isoform [99]. Only recently, the fourth nSMase was identified in mice, termed MA-nSMase (mitochondrial-associated nSMase) and was shown to localize to both ER and mitochondria, [100]. In addition the presence of a putative human MA-nSMase encoding gene has been reported [99]. As Isc1p in yeast is implicated in generating SLs that initiate signaling events that regulate mitochondrial function, the question arises whether human MA-nSMase serves the same purpose. With regard to the study of human SL-related pathologies using yeast as a model, yeast studies have revealed new insights into the neurodegenerative condition Niemann Pick type C1. Cells that lack NPC1 accumulate lipids such as cholesterol and Sph [84, 101] and exhibit markers of oxidative stress and mitochondrial dysfunction [102-106]. However, the specific mechanisms that lead to neurodegeneration during Niemann Pick type C1 are not fully elucidated. Interestingly, Δncr1 mutants not only exhibit hallmarks of oxidative stress and mitochondrial dysfunction, they also accumulate LCBs [77]. In addition, these LCBs are proposed to be crucial in regulating mitochondrial function in Δncr1 mutants [77]. Hence this suggests that yeast may reveal important insights into cellular events regarding SL signaling and mitochondrial function in higher eukaryotes.
In conclusion, all the above findings clearly support the importance of a feedback/feed-forward loop between SLs and mitochondrial function.
Highlights.
Sphingolipids (SLs) are components of cellular membranes and signaling molecules
Isc1p-mediated phytoCer generation is crucial for mitochondrial function
In response to SLs Sch9p transduces signals to regulate mitochondrial function
The retrograde response affects SL homeostasis
SLs are important signaling molecules in the regulation of mitochondrial function
Acknowledgments
P.S. is supported by IWT-Vlaanderen and K.T. by ‘Industrial Research Fund’ of KU Leuven (IOF-M). This work is also partly supported by grants GM63265 and GM43825 from the NIH.
Footnotes
Abbreviations in order of appearance: SLs; sphingolipids; DHS, dihydrosphingosine; PHS, phytosphingosine; LCBs, long chain bases; palmitoyl CoA, palmitoyl coenzyme A; SPT, serine palmitoyltransferase; dhCer, dihydroceramide; IPC, inositolphosphoceramide; MIPC, mannose inositolphosphoceramide; Ipt1, inositolphosphotransferase; M(IP)2C, mannose diinositolphosphoceramide; Isc1p, inositol phosphosphingolipid phospholipase C; PG, phosphatidylglycerol; CL, cardiolipin; OMM, outer mitochondrial membrane; CLS, chronological lifespan; ETC, electron transport chain; COX, cytochrome c oxidase; HOG pathway, high osmolarity glycerol pathway; CAPP, ceramide-activated protein phosphatase; TORC1, target of rapamycin complex 1; TOR, target of rapamycin; MAPK, mitogen activated protein kinase; ROS, reactive oxygen species; Δψm, mitochondrial membrane potential; PDRE, Pdr1p/Pdr3p response elements; nSMase, neutral sphingomyelinase; MA-nSMase, mitochondrial-associated nSMase
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karlsson KA. Sphingolipid long chain bases. Lipids. 1970;5(11):878–91. doi: 10.1007/BF02531119. [DOI] [PubMed] [Google Scholar]
- 2.Epstein S, Castillon GA, Qin Y, Riezman H. An essential function of sphingolipids in yeast cell division. Mol Microbiol. 2012;84(6):1018–32. doi: 10.1111/j.1365-2958.2012.08087.x. [DOI] [PubMed] [Google Scholar]
- 3.Eisenberg T, Buttner S. Lipids and cell death in yeast. FEMS Yeast Res. 2013;14(1):179–197. doi: 10.1111/1567-1364.12105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang X, Withers BR, Dickson RC. Sphingolipids and lifespan regulation. Biochim Biophys Acta. 2013;1841(5):657–664. doi: 10.1016/j.bbalip.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yamagata M, Obara K, Kihara A. Sphingolipid synthesis is involved in autophagy in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2011;410(4):786–91. doi: 10.1016/j.bbrc.2011.06.061. [DOI] [PubMed] [Google Scholar]
- 6.Rego A, Trindade D, Chaves SR, Manon S, Costa V, Sousa MJ, Corte-Real M. The yeast model system as a tool towards the understanding of apoptosis regulation by sphingolipids. FEMS Yeast Res. 2013;14(1):160–178. doi: 10.1111/1567-1364.12096. [DOI] [PubMed] [Google Scholar]
- 7.Hannun YA, Luberto C, Argraves KM. Enzymes of sphingolipid metabolism: from modular to integrative signaling. Biochemistry. 2001;40(16):4893–903. doi: 10.1021/bi002836k. [DOI] [PubMed] [Google Scholar]
- 8.Sims KJ, Spassieva SD, Voit EO, Obeid LM. Yeast sphingolipid metabolism: clues and connections. Biochem Cell Biol. 2004;82(1):45–61. doi: 10.1139/o03-086. [DOI] [PubMed] [Google Scholar]
- 9.Wadsworth JM, Clarke DJ, McMahon SA, Lowther JP, Beattie AE, Langridge-Smith PR, Broughton HB, Dunn TM, Naismith JH, Campopiano DJ. The chemical basis of serine palmitoyltransferase inhibition by myriocin. J Am Chem Soc. 2013;135(38):14276–85. doi: 10.1021/ja4059876. [DOI] [PubMed] [Google Scholar]
- 10.Chen JK, Lane WS, Schreiber SL. The identification of myriocin-binding proteins. Chem Biol. 1999;6(4):221–35. doi: 10.1016/S1074-5521(99)80038-6. [DOI] [PubMed] [Google Scholar]
- 11.Lowther J, Naismith JH, Dunn TM, Campopiano DJ. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem Soc Trans. 2012;40(3):547–54. doi: 10.1042/BST20110769. [DOI] [PubMed] [Google Scholar]
- 12.Hanada K. Serine palmitoyltransferase, a key enzyme of sphingolipid metabolism. Biochim Biophys Acta. 2003;1632(1-3):16–30. doi: 10.1016/s1388-1981(03)00059-3. [DOI] [PubMed] [Google Scholar]
- 13.Montefusco DJ, Matmati N, Hannun YA. The yeast sphingolipid signaling landscape. Chem Phys Lipids. 2014;177:26–40. doi: 10.1016/j.chemphyslip.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dickson RC. Roles for sphingolipids in Saccharomyces cerevisiae. Adv Exp Med Biol. 2010;688:217–31. doi: 10.1007/978-1-4419-6741-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickson RC. Thematic review series: sphingolipids. New insights into sphingolipid metabolism and function in budding yeast. J Lipid Res. 2008;49(5):909–21. doi: 10.1194/jlr.R800003-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matmati N, Hannun YA. Thematic review series: sphingolipids. ISC1 (inositol phosphosphingolipid-phospholipase C), the yeast homologue of neutral sphingomyelinases. J Lipid Res. 2008;49(5):922–8. doi: 10.1194/jlr.R800004-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi K, Matmati N, Zheng WJ, Hannun YA, Mohanty BK. Cellular morphogenesis under stress is influenced by the sphingolipid pathway gene ISC1 and DNA integrity checkpoint genes in Saccharomyces cerevisiae. Genetics. 2011;189(2):533–47. doi: 10.1534/genetics.111.132092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matmati N, Kitagaki H, Montefusco D, Mohanty BK, Hannun YA. Hydroxyurea sensitivity reveals a role for ISC1 in the regulation of G2/M. J Biol Chem. 2009;284(13):8241–6. doi: 10.1074/jbc.M900004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Betz C, Zajonc D, Moll M, Schweizer E. ISC1-encoded inositol phosphosphingolipid phospholipase C is involved in Na+/Li+ halotolerance of Saccharomyces cerevisiae. Eur J Biochem. 2002;269(16):4033–9. doi: 10.1046/j.1432-1033.2002.03096.x. [DOI] [PubMed] [Google Scholar]
- 20.Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, Almeida B, Rodrigues P, Ludovico P, Hohmann S, Moradas-Ferreira P, Corte-Real M, Costa V. Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol Biol Cell. 2008;19(3):865–76. doi: 10.1091/mbc.E07-06-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rego A, Costa M, Chaves SR, Matmati N, Pereira H, Sousa MJ, Moradas-Ferreira P, Hannun YA, Costa V, Corte-Real M. Modulation of mitochondrial outer membrane permeabilization and apoptosis by ceramide metabolism. PLoS One. 2012;7(11):e48571. doi: 10.1371/journal.pone.0048571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A. 2002;99(26):16934–9. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaena de Avalos S, Okamoto Y, Hannun YA. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J Biol Chem. 2004;279(12):11537–45. doi: 10.1074/jbc.M309586200. [DOI] [PubMed] [Google Scholar]
- 24.Barbosa AD, Graca J, Mendes V, Chaves SR, Amorim MA, Mendes MV, Moradas-Ferreira P, Corte-Real M, Costa V. Activation of the Hog1p kinase in Isc1p deficient yeast cells is associated with mitochondrial dysfunction, oxidative stress sensitivity and premature aging. Mech Ageing Dev. 2012;133(5):317–30. doi: 10.1016/j.mad.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J Biol Chem. 2005;280(8):7170–7. doi: 10.1074/jbc.M411058200. [DOI] [PubMed] [Google Scholar]
- 26.Teixeira V, Medeiros TC, Vilaça R, Moradas-Ferreira P, Costa V. Reduced TORC1 signaling abolishes mitochondrial dysfunctions and shortened chronological lifespan of Isc1p-deficient cells. Microbial Cell. 2014;1(1):21–36. doi: 10.15698/mic2014.01.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitagaki H, Cowart LA, Matmati N, Vaena de Avalos S, Novgorodov SA, Zeidan YH, Bielawski J, Obeid LM, Hannun YA. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim Biophys Acta. 2007;1768(11):2849–61. doi: 10.1016/j.bbamem.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kitagaki H, Cowart LA, Matmati N, Montefusco D, Gandy J, de Avalos SV, Novgorodov SA, Zheng J, Obeid LM, Hannun YA. ISC1-dependent metabolic adaptation reveals an indispensable role for mitochondria in induction of nuclear genes during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem. 2009;284(16):10818–30. doi: 10.1074/jbc.M805029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jazwinski SM. The retrograde response: when mitochondrial quality control is not enough. Biochim Biophys Acta. 2013;1833(2):400–9. doi: 10.1016/j.bbamcr.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14(1):1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- 31.Hallstrom TC, Lambert L, Schorling S, Balzi E, Goffeau A, Moye-Rowley WS. Coordinate control of sphingolipid biosynthesis and multidrug resistance in Saccharomyces cerevisiae. J Biol Chem. 2001;276(26):23674–80. doi: 10.1074/jbc.M101568200. [DOI] [PubMed] [Google Scholar]
- 32.Kolaczkowski M, Kolaczkowska A, Gaigg B, Schneiter R, Moye-Rowley WS. Differential regulation of ceramide synthase components LAC1 and LAG1 in Saccharomyces cerevisiae. Eukaryot Cell. 2004;3(4):880–92. doi: 10.1128/EC.3.4.880-892.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemmer D, McHardy LM, Hoon S, Reberioux D, Giaever G, Nislow C, Roskelley CD, Roberge M. Combining chemical genomics screens in yeast to reveal spectrum of effects of chemical inhibition of sphingolipid biosynthesis. BMC Microbiol. 2009;9:9. doi: 10.1186/1471-2180-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornhuber J, Tripal P, Reichel M, Terfloth L, Bleich S, Wiltfang J, Gulbins E. Identification of new functional inhibitors of acid sphingomyelinase using a structureproperty- activity relation model. J Med Chem. 2008;51(2):219–37. doi: 10.1021/jm070524a. [DOI] [PubMed] [Google Scholar]
- 35.Roskelley CD, Williams DE, McHardy LM, Leong KG, Troussard A, Karsan A, Andersen RJ, Dedhar S, Roberge M. Inhibition of tumor cell invasion and angiogenesis by motuporamines. Cancer Res. 2001;61(18):6788–94. [PubMed] [Google Scholar]
- 36.Buede R, Rinker-Schaffer C, Pinto WJ, Lester RL, Dickson RC. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol. 1991;173(14):4325–32. doi: 10.1128/jb.173.14.4325-4332.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nagiec MM, Baltisberger JA, Wells GB, Lester RL, Dickson RC. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc Natl Acad Sci U S A. 1994;91(17):7899–902. doi: 10.1073/pnas.91.17.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wells GB, Lester RL. The isolation and characterization of a mutant strain of Saccharomyces cerevisiae that requires a long chain base for growth and for synthesis of phosphosphingolipids. J Biol Chem. 1983;258(17):10200–3. [PubMed] [Google Scholar]
- 39.Montefusco DJ, Newcomb B, Gandy JL, Brice SE, Matmati N, Cowart LA, Hannun YA. Sphingoid bases and the serine catabolic enzyme CHA1 define a novel feedforward/feedback mechanism in the response to serine availability. J Biol Chem. 2012;287(12):9280–9. doi: 10.1074/jbc.M111.313445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cheng J, Park TS, Chio LC, Fischl AS, Ye XS. Induction of apoptosis by sphingoid long-chain bases in Aspergillus nidulans. Mol Cell Biol. 2003;23(1):163–77. doi: 10.1128/MCB.23.1.163-177.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thevissen K, Hillaert U, Meert EM, Chow KK, Cammue BP, Van Calenbergh S, Francois IE. Fungicidal activity of truncated analogues of dihydrosphingosine. Bioorg Med Chem Lett. 2008;18(13):3728–30. doi: 10.1016/j.bmcl.2008.05.067. [DOI] [PubMed] [Google Scholar]
- 42.Chung N, Mao C, Heitman J, Hannun YA, Obeid LM. Phytosphingosine as a specific inhibitor of growth and nutrient import in Saccharomyces cerevisiae. J Biol Chem. 2001;276(38):35614–21. doi: 10.1074/jbc.M105653200. [DOI] [PubMed] [Google Scholar]
- 43.Woodacre A, Lone MA, Jablonowski D, Schneiter R, Giorgini F, Schaffrath R. A novel Sit4 phosphatase complex is involved in the response to ceramide stress in yeast. Oxid Med Cell Longev. 2013;2013:129645. doi: 10.1155/2013/129645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Panwar SL, Moye-Rowley WS. Long chain base tolerance in Saccharomyces cerevisiae is induced by retrograde signals from the mitochondria. J Biol Chem. 2006;281(10):6376–84. doi: 10.1074/jbc.M512115200. [DOI] [PubMed] [Google Scholar]
- 45.Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem. 2000;275(50):39793–8. doi: 10.1074/jbc.M007721200. [DOI] [PubMed] [Google Scholar]
- 46.Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J Biol Chem. 1998;273(16):9829–36. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- 47.Okamoto Y, Vaena De Avalos S, Hannun YA. Structural requirements for selective binding of ISC1 to anionic phospholipids. J Biol Chem. 2002;277(48):46470–7. doi: 10.1074/jbc.M207779200. [DOI] [PubMed] [Google Scholar]
- 48.Swinnen E, Wilms T, Idkowiak-Baldys J, Smets B, De Snijder P, Accardo S, Ghillebert R, Thevissen K, Cammue B, De Vos D, Bielawski J, Hannun YA, Winderickx J. The protein kinase Sch9 is a key regulator of sphingolipid metabolism in Saccharomyces cerevisiae. Mol Biol Cell. 2014;25(1):196–211. doi: 10.1091/mbc.E13-06-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Longo VD, Shadel GS, Kaeberlein M, Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16(1):18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barbosa AD, Osorio H, Sims KJ, Almeida T, Alves M, Bielawski J, Amorim MA, Moradas-Ferreira P, Hannun YA, Costa V. Role for Sit4p-dependent mitochondrial dysfunction in mediating the shortened chronological lifespan and oxidative stress sensitivity of Isc1p-deficient cells. Mol Microbiol. 2011;81(2):515–27. doi: 10.1111/j.1365-2958.2011.07714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Day M. Yeast petites and small colony variants: for everything there is a season. Adv Appl Microbiol. 2013;85:1–41. doi: 10.1016/B978-0-12-407672-3.00001-0. [DOI] [PubMed] [Google Scholar]
- 52.Demasi AP, Pereira GA, Netto LE. Cytosolic thioredoxin peroxidase I is essential for the antioxidant defense of yeast with dysfunctional mitochondria. FEBS Lett. 2001;509(3):430–4. doi: 10.1016/s0014-5793(01)03215-x. [DOI] [PubMed] [Google Scholar]
- 53.Slonimski PP, Perrodin G, Croft JH. Ethidium bromide induced mutation of yeast mitochondria: complete transformation of cells into respiratory deficient non-chromosomal “petites”. Biochem Biophys Res Commun. 1968;30(3):232–9. doi: 10.1016/0006-291x(68)90440-3. [DOI] [PubMed] [Google Scholar]
- 54.Dunn CD, Jensen RE. Suppression of a defect in mitochondrial protein import identifies cytosolic proteins required for viability of yeast cells lacking mitochondrial DNA. Genetics. 2003;165(1):35–45. doi: 10.1093/genetics/165.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev. 1996;10(4):382–94. doi: 10.1101/gad.10.4.382. [DOI] [PubMed] [Google Scholar]
- 56.Tate JJ, Feller A, Dubois E, Cooper TG. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J Biol Chem. 2006;281(49):37980–92. doi: 10.1074/jbc.M606973200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swinnen E, Ghillebert R, Wilms T, Winderickx J. Molecular mechanisms linking the evolutionary conserved TORC1-Sch9 nutrient signalling branch to lifespan regulation in Saccharomyces cerevisiae. FEMS Yeast Res. 2013;14(1):17–32. doi: 10.1111/1567-1364.12097. [DOI] [PubMed] [Google Scholar]
- 58.Grilley MM, Stock SD, Dickson RC, Lester RL, Takemoto JY. Syringomycin action gene SYR2 is essential for sphingolipid 4-hydroxylation in Saccharomyces cerevisiae. J Biol Chem. 1998;273(18):11062–8. doi: 10.1074/jbc.273.18.11062. [DOI] [PubMed] [Google Scholar]
- 59.Hohmann S, Krantz M, Nordlander B. Yeast osmoregulation. Methods Enzymol. 2007;428:29–45. doi: 10.1016/S0076-6879(07)28002-4. [DOI] [PubMed] [Google Scholar]
- 60.Bilsland E, Molin C, Swaminathan S, Ramne A, Sunnerhagen P. Rck1 and Rck2 MAPKAP kinases and the HOG pathway are required for oxidative stress resistance. Mol Microbiol. 2004;53(6):1743–56. doi: 10.1111/j.1365-2958.2004.04238.x. [DOI] [PubMed] [Google Scholar]
- 61.Rep M, Proft M, Remize F, Tamas M, Serrano R, Thevelein JM, Hohmann S. The Saccharomyces cerevisiae Sko1p transcription factor mediates HOG pathway-dependent osmotic regulation of a set of genes encoding enzymes implicated in protection from oxidative damage. Mol Microbiol. 2001;40(5):1067–83. doi: 10.1046/j.1365-2958.2001.02384.x. [DOI] [PubMed] [Google Scholar]
- 62.Saito H, Posas F. Response to hyperosmotic stress. Genetics. 2012;192(2):289–318. doi: 10.1534/genetics.112.140863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schaeffer HJ, Weber MJ. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19(4):2435–44. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vendrell A, Martinez-Pastor M, Gonzalez-Novo A, Pascual-Ahuir A, Sinclair DA, Proft M, Posas F. Sir2 histone deacetylase prevents programmed cell death caused by sustained activation of the Hog1 stress-activated protein kinase. EMBO Rep. 2011;12(10):1062–8. doi: 10.1038/embor.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pastor MM, Proft M, Pascual-Ahuir A. Mitochondrial function is an inducible determinant of osmotic stress adaptation in yeast. J Biol Chem. 2009;284(44):30307–17. doi: 10.1074/jbc.M109.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Timon-Gomez A, Proft M, Pascual-Ahuir A. Differential regulation of mitochondrial pyruvate carrier genes modulates respiratory capacity and stress tolerance in yeast. PLoS One. 2013;8(11):e79405. doi: 10.1371/journal.pone.0079405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275(12):8290–300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- 68.Alonso-Monge R, Carvaihlo S, Nombela C, Rial E, Pla J. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology. 2009;155(Pt 2):413–23. doi: 10.1099/mic.0.023309-0. [DOI] [PubMed] [Google Scholar]
- 69.Maeda T, Tsai AY, Saito H. Mutations in a protein tyrosine phosphatase gene (PTP2) and a protein serine/threonine phosphatase gene (PTC1) cause a synthetic growth defect in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13(9):5408–17. doi: 10.1128/mcb.13.9.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanigawa M, Kihara A, Terashima M, Takahara T, Maeda T. Sphingolipids regulate the yeast high-osmolarity glycerol response pathway. Mol Cell Biol. 2012;32(14):2861–70. doi: 10.1128/MCB.06111-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Galdieri L, Mehrotra S, Yu S, Vancura A. Transcriptional regulation in yeast during diauxic shift and stationary phase. OMICS. 2010;14(6):629–38. doi: 10.1089/omi.2010.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu K, Zhang X, Sumanasekera C, Lester RL, Dickson RC. Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem Soc Trans. 2005;33(Pt 5):1170–3. doi: 10.1042/BST20051170. [DOI] [PubMed] [Google Scholar]
- 73.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, Riezman H. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. EMBO J. 2000;19(12):2824–33. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Casamayor A, Torrance PD, Kobayashi T, Thorner J, Alessi DR. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr Biol. 1999;9(4):186–97. doi: 10.1016/s0960-9822(99)80088-8. [DOI] [PubMed] [Google Scholar]
- 75.Huang X, Liu J, Dickson RC. Down-regulating sphingolipid synthesis increases yeast lifespan. PLoS Genet. 2012;8(2):e1002493. doi: 10.1371/journal.pgen.1002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lavoie H, Whiteway M. Increased respiration in the sch9Delta mutant is required for increasing chronological life span but not replicative life span. Eukaryot Cell. 2008;7(7):1127–35. doi: 10.1128/EC.00330-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vilaca R, Silva E, Nadais A, Teixeira V, Matmati N, Gaifem J, Hannun YA, Sa Miranda MC, Costa V. Sphingolipid signaling mediates mitochondrial dysfunctions and reduced chronological lifespan in the yeast model of Niemann-Pick type C1. Mol Microbiol. 2013;91(3):438–451. doi: 10.1111/mmi.12470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Berger AC, Hanson PK, Wylie Nichols J, Corbett AH. A yeast model system for functional analysis of the Niemann-Pick type C protein 1 homolog, Ncr1p. Traffic. 2005;6(10):907–17. doi: 10.1111/j.1600-0854.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 79.Vanier MT, Millat G. Niemann-Pick disease type C. Clin Genet. 2003;64(4):269–81. doi: 10.1034/j.1399-0004.2003.00147.x. [DOI] [PubMed] [Google Scholar]
- 80.Loftus SK, Morris JA, Carstea ED, Gu JZ, Cummings C, Brown A, Ellison J, Ohno K, Rosenfeld MA, Tagle DA, Pentchev PG, Pavan WJ. Murine model of Niemann-Pick C disease: mutation in a cholesterol homeostasis gene. Science. 1997;277(5323):232–5. doi: 10.1126/science.277.5323.232. [DOI] [PubMed] [Google Scholar]
- 81.Carstea ED, Morris JA, Coleman KG, Loftus SK, Zhang D, Cummings C, Gu J, Rosenfeld MA, Pavan WJ, Krizman DB, Nagle J, Polymeropoulos MH, Sturley SL, Ioannou YA, Higgins ME, Comly M, Cooney A, Brown A, Kaneski CR, Blanchette-Mackie EJ, Dwyer NK, Neufeld EB, Chang TY, Liscum L, Strauss JF, 3rd, Ohno K, Zeigler M, Carmi R, Sokol J, Markie D, O’Neill RR, van Diggelen OP, Elleder M, Patterson MC, Brady RO, Vanier MT, Pentchev PG, Tagle DA. Niemann-Pick C1 disease gene: homology to mediators of cholesterol homeostasis. Science. 1997;277(5323):228–31. doi: 10.1126/science.277.5323.228. [DOI] [PubMed] [Google Scholar]
- 82.Naureckiene S, Sleat DE, Lackland H, Fensom A, Vanier MT, Wattiaux R, Jadot M, Lobel P. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 2000;290(5500):2298–301. doi: 10.1126/science.290.5500.2298. [DOI] [PubMed] [Google Scholar]
- 83.Malathi K, Higaki K, Tinkelenberg AH, Balderes DA, Almanzar-Paramio D, Wilcox LJ, Erdeniz N, Redican F, Padamsee M, Liu Y, Khan S, Alcantara F, Carstea ED, Morris JA, Sturley SL. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. J Cell Biol. 2004;164(4):547–56. doi: 10.1083/jcb.200310046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lloyd-Evans E, Morgan AJ, He X, Smith DA, Elliot-Smith E, Sillence DJ, Churchill GC, Schuchman EH, Galione A, Platt FM. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat Med. 2008;14(11):1247–55. doi: 10.1038/nm.1876. [DOI] [PubMed] [Google Scholar]
- 85.Moye-Rowley WS. Retrograde regulation of multidrug resistance in Saccharomyces cerevisiae. Gene. 2005;354:15–21. doi: 10.1016/j.gene.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 86.Hallstrom TC, Moye-Rowley WS. Multiple signals from dysfunctional mitochondria activate the pleiotropic drug resistance pathway in Saccharomyces cerevisiae. J Biol Chem. 2000;275(48):37347–56. doi: 10.1074/jbc.M007338200. [DOI] [PubMed] [Google Scholar]
- 87.Kawai S, Urban J, Piccolis M, Panchaud N, De Virgilio C, Loewith R. Mitochondrial genomic dysfunction causes dephosphorylation of Sch9 in the yeast Saccharomyces cerevisiae. Eukaryot Cell. 2011;10(10):1367–9. doi: 10.1128/EC.05157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262(35):16871–9. [PubMed] [Google Scholar]
- 89.Delaveau T, Delahodde A, Carvajal E, Subik J, Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994;244(5):501–11. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- 90.Moye-Rowley WS. Transcriptional control of multidrug resistance in the yeast Saccharomyces. Prog Nucleic Acid Res Mol Biol. 2003;73:251–79. doi: 10.1016/s0079-6603(03)01008-0. [DOI] [PubMed] [Google Scholar]
- 91.Miranda MN, Masuda CA, Ferreira-Pereira A, Carvajal E, Ghislain M, Montero-Lomeli M. The serine/threonine protein phosphatase Sit4p activates multidrug resistance in Saccharomyces cerevisiae. FEMS Yeast Res. 2010;10(6):674–86. doi: 10.1111/j.1567-1364.2010.00656.x. [DOI] [PubMed] [Google Scholar]
- 92.Chen S, Liu D, Finley RL, Jr, Greenberg ML. Loss of mitochondrial DNA in the yeast cardiolipin synthase crd1 mutant leads to up-regulation of the protein kinase Swe1p that regulates the G2/M transition. J Biol Chem. 2010;285(14):10397–407. doi: 10.1074/jbc.M110.100784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12(9):3417–26. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Matmati N, Metelli A, Tripathi K, Yan S, Mohanty BK, Hannun YA. Identification of C18:1-phytoceramide as the candidate lipid mediator for hydroxyurea resistance in yeast. J Biol Chem. 2013;288(24):17272–84. doi: 10.1074/jbc.M112.444802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Devaux F, Carvajal E, Moye-Rowley S, Jacq C. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 2002;515(1-3):25–8. doi: 10.1016/s0014-5793(02)02387-6. [DOI] [PubMed] [Google Scholar]
- 96.Traven A, Wong JM, Xu D, Sopta M, Ingles CJ. Interorganellar communication. Altered nuclear gene expression profiles in a yeast mitochondrial dna mutant. J Biol Chem. 2001;276(6):4020–7. doi: 10.1074/jbc.M006807200. [DOI] [PubMed] [Google Scholar]
- 97.Kihara A, Igarashi Y. Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. J Biol Chem. 2002;277(33):30048–54. doi: 10.1074/jbc.M203385200. [DOI] [PubMed] [Google Scholar]
- 98.Wolfger H, Mamnun YM, Kuchler K. Fungal ABC proteins: pleiotropic drug resistance, stress response and cellular detoxification. Res Microbiol. 2001;152(3-4):375–89. doi: 10.1016/s0923-2508(01)01209-8. [DOI] [PubMed] [Google Scholar]
- 99.Airola MV, Hannun YA. Sphingolipid metabolism and neutral sphingomyelinases. Handb Exp Pharmacol. 2013;(215):57–76. doi: 10.1007/978-3-7091-1368-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu BX, Rajagopalan V, Roddy PL, Clarke CJ, Hannun YA. Identification and characterization of murine mitochondria-associated neutral sphingomyelinase (MA-nSMase), the mammalian sphingomyelin phosphodiesterase 5. J Biol Chem. 2010;285(23):17993–8002. doi: 10.1074/jbc.M110.102988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bi X, Liao G. Cholesterol in Niemann-Pick Type C disease. Subcell Biochem. 2010;51:319–35. doi: 10.1007/978-90-481-8622-8_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vazquez MC, Balboa E, Alvarez AR, Zanlungo S. Oxidative stress: a pathogenic mechanism for Niemann-Pick type C disease. Oxid Med Cell Longev. 2012;2012:205713. doi: 10.1155/2012/205713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kennedy BE, LeBlanc VG, Mailman TM, Fice D, Burton I, Karakach TK, Karten B. Pre-symptomatic activation of antioxidant responses and alterations in glucose and pyruvate metabolism in Niemann-Pick Type C1-deficient murine brain. PLoS One. 2013;8(12):e82685. doi: 10.1371/journal.pone.0082685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klein A, Maldonado C, Vargas LM, Gonzalez M, Robledo F, Perez de Arce K, Munoz FJ, Hetz C, Alvarez AR, Zanlungo S. Oxidative stress activates the c-Abl/p73 proapoptotic pathway in Niemann-Pick type C neurons. Neurobiol Dis. 2011;41(1):209–18. doi: 10.1016/j.nbd.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 105.Fu R, Yanjanin NM, Bianconi S, Pavan WJ, Porter FD. Oxidative stress in Niemann-Pick disease, type C. Mol Genet Metab. 2010;101(2-3):214–8. doi: 10.1016/j.ymgme.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yu W, Gong JS, Ko M, Garver WS, Yanagisawa K, Michikawa M. Altered cholesterol metabolism in Niemann-Pick type C1 mouse brains affects mitochondrial function. J Biol Chem. 2005;280(12):11731–9. doi: 10.1074/jbc.M412898200. [DOI] [PubMed] [Google Scholar]


