Abstract
The gain of a selective advantage in cancer as well as the establishment of complex traits during evolution require multiple genetic alterations, but how these mutations accumulate over time is currently unclear. There is increasing evidence that a mutator phenotype perpetuates the development of many human cancers. While in some cases, the increased mutation rate is the result of a genetic disruption of DNA repair and replication or environmental exposures, other evidence suggests that endogenous DNA damage induced by AID/APOBEC cytidine deaminases can result in transient localized hypermutation generating simultaneous, closely-spaced (i.e. “clustered”) multiple mutations. Here, we discuss mechanisms that lead to mutation cluster formation, the biological consequences of their formation in cancer and evidence suggesting that APOBEC mutagenesis can also occur genome-wide. This raises the possibility that dysregulation of these enzymes may enable rapid malignant transformation by increasing mutation rates without the loss of fitness associated with permanent mutators.
Introduction
The processes that underlie complex genome alterations that occur during evolution and carcinogenesis are under extensive investigation. While DNA damage and errors during DNA transactions are generally accepted as key sources of mutation, the robustness of DNA repair mechanisms and high fidelity of DNA replication minimizes these errors, resulting in a stable genome. Adaptive evolution or cancer development, however, often require multiple mutations. As mutation rates are normally low and most individual mutations are neutral or deleterious, several hypotheses have been proposed to explain how beneficial genetic alterations are accumulated during germ line and cancer cell evolution. Here we discuss the potential effect of one of these mechanisms, transient hypermutation, and its potential to generate simultaneous mutations during carcinogenesis.
Evolutionary processes generally occur in the background of rare genetic changes. Recent whole-genome sequencing of parent-offspring trios estimated the rate of single nucleotide variations in the human germ line as 1×10−8 mutations per base pair per generation [1]. Such low mutation rates limit the number of productive evolutionary paths because insufficient mutations are accumulated to cross “fitness valleys.” Despite low mutation rates across a variety of organisms and viruses, reports of regionally high mutation densities suggest that locally increased mutation rates may occur transiently [2,3] and play a role in producing the multiple mutations needed for evolution. In addition, recent large scale sequencing of cancer genomes has indicated that many tumors have accumulated more mutations than would be expected from the spontaneous somatic mutation rate [4], supporting the hypothesis that many cancers acquire a mutator phenotype that facilitates their development [5]. Somatically acquired alterations of DNA repair capacity and replication fidelity appear to be relatively rare causes of increased mutation rates, occurring primarily in a subset of colorectal and endometrial cancers [6]. Analysis of spatial mutation distributions and mutation spectra in a variety of cancers indicates that some “mutation clusters” (i.e. closely-spaced base substitutions and single nucleotide insertions or deletions) can occur simultaneously [7,8], suggesting that transiently elevated mutation rates may play an important role in carcinogenesis.
How does hypermutation occur?
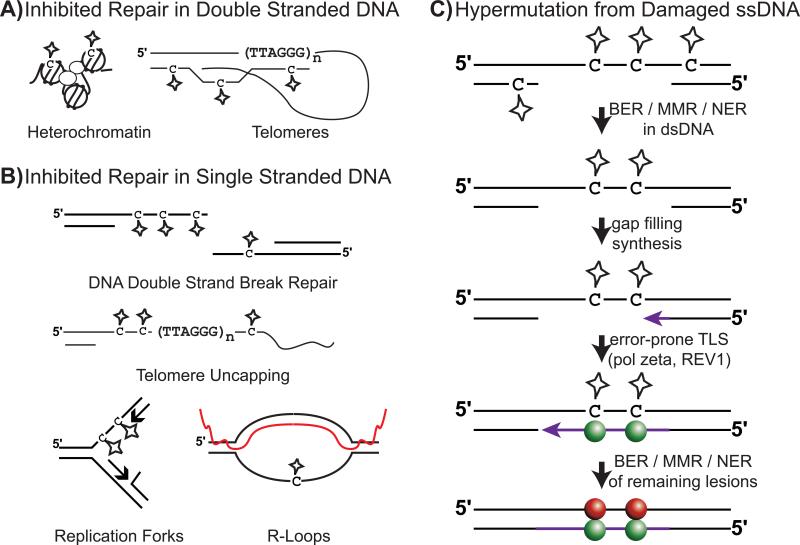
Regionally elevated mutation rates can be generated by increased susceptibility of particular areas of the genome to damage and error, or through local inhibition of DNA repair (Figure 1). Inherent chromosome features such as heterochromatic regions and telomeres can impact mutation incidence by both of these means. DNA bound within nucleosomes has been reported to be resistant to damage, but also less accessible to repair [9], whereas human telomeres accumulate more UV lesions while also being resistant to repair [10]. Studies in Escherichia coli [11,12] and yeast [13-17] have shown that transient increases in mutation rates can occur anywhere in the genome that is involved in homology-directed DNA double strand break (DSB) repair. The requirement of the trans-lesion synthesis polymerase, pol zeta to mediate hypermutation in DNA flanking DSBs [13,14] suggested that DNA lesions persisting in single stranded (ss) DNA regions formed by 5′ to 3′ resection during the break repair may contribute to the elevated mutation frequencies in these regions (Figure 1c).
Figure 1. Sources and mechanisms of DNA damage-induced transient hyper-mutation.
Repair of DNA lesions (stars) is inhibited in A: double stranded DNA by heterochromatin and telomeres and in B: single stranded DNA by the lack of a template strand to complete excision repair. DSB repair intermediates, uncapped telomeres, uncoupled replication forks and R-loop forming sequences (Red line indicates RNA) contain single strand DNA regions known to be targets of transient hypermutation. C: Mutation clusters are produced by genome-wide DNA damage. Inhibited DNA repair in ssDNA regions results in persistent DNA lesions in these areas. Error-prone gap-filling synthesis, mediated in part by specific trans-lesion synthesis polymerases in a lesion specific manner (e.g. pol zeta and REV1function in bypass of abasic sites), mis-inserts nucleotides across from the modified bases. Spheres indicate base changes which are colored based on the identity of the nucleotide prior to mutation (C = red and G = green). In cases of high lesion density, multiple mis-insertions occur in a single ssDNA gap, resulting in the formation of a mutation cluster.
Damage to ssDNA is a source of hypermutation
Yang et al. directly tested this possibility by exposing ssDNA regions generated during DSB repair to exogenous DNA base damage and measuring the resulting mutation frequencies [18,19]. Exposure to UV light or the base alkylating agent methyl methanesulfonate (MMS) strongly increased CAN1 mutation frequencies near DSBs. Sequencing of UV-induced CAN1 mutations provided further support to the notion that lesions in ssDNA initiate hyper-mutability. In this system, the UV light induced strand biased mutations occurring exclusively at pyrimidines 5′ of the DSB and at purines 3′ of the DSB [19]. The lack of the complementary mutations in these spectra suggested an absence of the complementary DNA strand, whereas the specific bases mutated were consistent with mutagenesis stemming from pyrimidine dimers in the ssDNA overhangs remaining after 5′ to 3′ resection of the DSB. MMS produced a similar switching of strand bias spectra (mutated cytosines 5′ of the DSB and guanines 3′ of the DSB), consistent with the mutations being induced by a single-strand-specific lesion, N3-methyl cytosine. Damage-induced hypermutation also extends to other sources of ssDNA and forms of DNA damage. Sub-telomeric locations prone to telomere uncapping and consequently single strandedness have been shown to be mutagenized by UV light, MMS, and even APOBEC3G cytidine deaminase [19,20] in a manner consistent with lesion accumulation in the persisting DNA strand.
Enzymatic deamination of ssDNA causes hypermutation
In mammalian B-cells, hypermutation of chromosomal DNA is strongly linked to ssDNA formed during transcription. However, in this case, the DNA damage is induced by activation-induced cytosine deaminase (AID). During the establishment of high-affinity antibodies, activated B-cells express AID to initiate somatic hypermutation (SHM) of immunoglobulin genes, resulting in localized mutation clusters with densities approaching 1 mutation/kbp [21]. Initiation of this process is dependent on transcription of the targeted regions [22] which can include non-immunoglobulin genes [23]. Biochemical and genetic analysis of AID activity suggests this dependence is largely a result of AID's specificity for ssDNA over dsDNA [24,25]. This enzyme efficiently deaminates transient ssDNA associated with transcription bubbles [24,26,27] as well as in R-loop forming sequences [28,29]. AID can also deaminate ssDNA intermediates generated during other DNA transactions (e.g. DSB repair) [30,31], and in some cases by recruitment to specific sequences [32,33]. Hence, the general association of this enzyme with transcription intermediates in cells is likely due to this source of ssDNA being more abundant, as well as to specific interactions with RPA [34,35], Spt5 [36], other RNA PolII associated factors [37] and the exosome [38] that target AID to both strands of actively transcribed regions. Similar enzymatic deamination of cytidine in ssDNA may be a widespread source of damage-induced hypermutation. AID belongs to a family of eleven cytidine deaminases, including APOBEC1 and seven APOBEC3 enzymes, which normally function in editing mRNA [39] and retroelement (i.e. retroviruses and retrotransposons) restriction [40], respectively. Other AID/APOBEC family members share AID's specificity for ssDNA [41,42], the ability to deaminate ssDNA in transcription bubbles in vitro [26,43,44], and the ability to damage nuclear DNA, hence leading to elevated mutation rates [45,46].
Damage to ssDNA is the primary source of clustered mutations in yeast
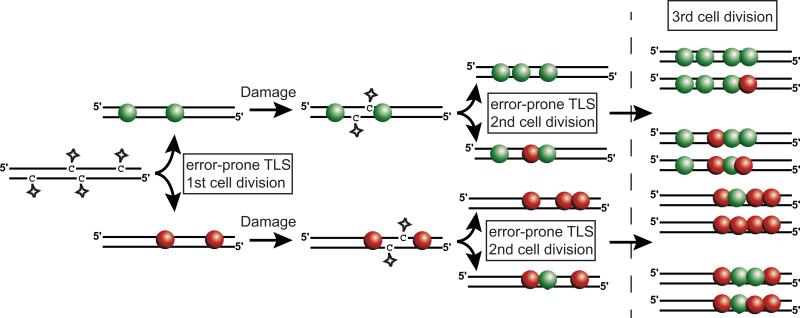
Importantly, where rates of spontaneous mutation in ssDNA are not sufficient to produce alleles with multiple mutations [3], lesion densities associated with exogenous and elevated endogenous damage to ssDNA regions can induce multiple mutations within a single or few generations. In fact, damage to ssDNA appears to be the primary mechanism by which simultaneously induced clustered mutations occur in Saccharomyces cerevisiae. Selection of MMS-induced mutations in closely-spaced URA3 and CAN1 genes [8] revealed large regions of hypermutation. These mutation clusters exhibited “strand-coordination” (i.e. runs of mutations occurring in the same type of base), a hallmark of transient hypermutation that indicates the mutations were induced simultaneously (Figure 2). Moreover, an excess of mutated C:G pairs in clusters compared to mutations in the rest of the genome also implied the contribution of the ssDNA-specific N3-methyl cytosine lesion in cluster formation. One subset of clusters again identified homology-directed DSB repair as a likely source of ssDNA targeted for hyper-mutation and simultaneous mutations. These clusters were composed of strand-coordinated mutated cytosines at the 5′ side of the cluster, switching to mutated guanines towards the 3′ side. This correlates with the MMS-induced mutational spectra 5′ and 3′ of I-SceI induced DSBs [18]. Bi-directional resection is the only known process that explains the incidence of switching clusters without invoking an extended chain of events. However, the lengths of these mutation clusters imply that these repair processes occasionally generate ssDNA regions up to 100 kbps long, even without a genetic deficiency in homologous recombination, which are known to produce ssDNA regions tens of kbps long [47,48]. Along with DSB repair, MMS can also induce clustered mutations associated with destabilized replication forks. Ablation of either TOF1 or CSM3, members of the replication fork protection complex, increased the frequency of strand-coordinated mutation clusters which additionally displayed mutational strand biases dependent on the direction of replication through the area where clusters were selected [8]. Thus, ssDNA regions exposed during normal DNA metabolism, such as DSB repair or replication, can accumulate base lesions and ultimately result in the production of simultaneously induced clustered mutations. A third class of large completely strand-coordinated clusters selected with the URA3-CAN1 mutation cluster system indicates that additional sources of ssDNA are suitable targets for damage-induced mutation clusters. While a variety of processes could potentially generate such clusters, their similarity - in length and mutation density - to switching clusters suggests that a DSB repair process involving only one DNA end may provide the ssDNA.
Figure 2. Strand-coordinated clustered mutations are induced simultaneously.
Multiple simultaneously acquired DNA lesions (stars) that occur on opposite DNA strands will be converted to mutations during replication and segregated in a strand-specific manner into daughter cells during division. When all the mutation inducing lesions are of a similar type (e.g. N3 methyl-cytosine, deamination of TCW) the resulting simultaneous mutations will be strand-coordinated. Each subsequent round of mutation accumulation reduces the probability of maintaining strand-coordination within a cluster. Spheres indicate base changes which are colored based on the identity of the nucleotide prior to mutation (C = red and G = green).
Mutation clusters accumulate in cancer genomes
Early efforts to understand the mutagenic processes that could drive carcinogenesis found evidence of closely-spaced multiple mutations resembling the damage-induced clusters described above. When sequencing the LacZ mutation reporter from spontaneous tumors derived from the Big Blue mouse [49], Wang et al. found that several tumors contained multiple mutations in the LacZ gene. Because of the short inter-mutation distances and no evidence of a permanent increase in the mutability of the reporter, the authors proposed that the mutations were unlikely to be independent in time, but rather occurred simultaneously or within a few cell divisions. Sequencing of the genomic regions flanking the reporter determined that the LacZ mutations were often part of larger “mutation showers” composed primarily of base substitutions spaced 100 to 1000 bp apart that extended up to 30 kbp in length.
More recently, analyses of mutation distributions identified by high-throughput sequencing of human cancers has uncovered frequent incidence of clustered mutations. Groups of 2 or more mutations whose spacing was unlikely based on random mutagenesis [8,50] (Figure 3) have been found among multiple myeloma, prostate, head-and-neck cancers, and colorectal cancers. Further evaluation of mutation spectra, revealed these clusters existed in three classes based on stand-coordination (A- or T-coordinated, C- or G-coordinated, and non-coordinated), the non-coordinated class predominating.
Figure 3. Identification and classification of mutation clusters in human cancers.
Mutation clusters were identified in human tumors as groups of closely-spaced changes whose distribution is unlikely to be randomly generated. Following identification, clusters can be classified by strand-coordination, genomic location, and mutated motifs as a means to suggest mutagenic factors that caused the event. Spheres indicate base changes which are colored based on the identity of the nucleotide prior to mutation (C = red, G = green, A = blue and T = yellow). Localization of some non-coordinated clusters to known regions of somatic hyper-mutation (SHM) in multiple myeloma samples and enrichment of mutations in WRC and WA sequences indicate these clusters are the result of AID and pol eta activity during antibody maturation. Likewise the localization of C- or G-coordinated clusters near chromosome rearrangements and the enrichment of mutations in the sequence TCW suggest a mechanism involving APOBEC activity on ssDNA containing DSB repair intermediates. The mechanisms producing randomly located non-coordinated and A- or T-coordinated clusters are less clear.
The majority of A- or T-coordinated clusters as well as a subclass of non-coordinated clusters occurring in known regions of SHM were found within multiple myeloma samples. The specificity for this cancer type suggests that these clusters are likely related to B-cell SHM. Supporting this, these clusters were composed primarily of mutated cytosines in the sequence context of WRC (all mutation contexts include the complementary sequence; mutated base is underlined; W = adenine or thymine; R = adenine or guanine) and mutated adenines in the context of WA [8]. The motifs correspond to the respective sequence preferences of AID [51-53] and pol eta [54,55] mediated mutations during SHM.
APOBEC cytidine deaminases induce clustered mutations in cancer
We identified clustered mutations displaying C- or G-coordination in random locations of whole-genome sequenced tumors from four cancer types: multiple myeloma, prostate, head-and-neck, and colorectal cancers [8,50]. This class of clustered mutations was composed almost exclusively of mutations occurring at the tri-nucleotide DNA sequence, TCW, suggesting that an enzymatic activity may be involved in this mutagenesis. The nearly exclusive substitution of the mutated cytosine by either T or G, suggested that this mechanism may also involve error-prone trans-lesion synthesis past abasic sites [56-58], which can be generated by glycolytic removal of a damaged base. Moreover, approximately 40% of C- or G-coordinated clusters occurred within 20 kbp of a genomic rearrangement breakpoint. As these rearrangements are often generated by aberrant DSB repair processing, the proximity of C- or G- coordinated clusters to rearrangement sites suggested that, as seen in the generation of MMS-induced clustered mutations in yeast, an ssDNA intermediate of the repair process may have been targeted. Based on the specific mutated motif (TCW) and the co-localization of clusters with rearrangement breakpoints, we proposed that at least one member of the AID/APOBEC family of cytidine deaminases was inflicting the base damage that generated the clusters. Seven APOBECs (APOBEC1, 3A, 3B, 3C, 3DE, 3F, and 3H) deaminate cytosine to uracil specifically in the DNA sequence TC (reviewed in [40]), of which several specifically target TCW. The related cytidine deaminases, AID and APOBEC3G respectively modify WRC [53] and CC [59], two sequences excluded in the identified mutation clusters [8] (see Box 1). Additionally, AID/APOBEC deaminase activities are specific for ssDNA ([24,41] and reviewed in [40]) and can induce DSBs [46,60,61], which could lead to chromosome rearrangements. These features all correlate with a potential role in mutagenizing ssDNA DSB repair intermediates.
Additional mechanistic studies further indicate that APOBEC cytidine deaminases indeed have the biological capacity to generate mutation clusters by targeting induced as well as naturally occurring ssDNA regions. Along with specificity for ssDNA regions, which may have limited excision repair capacity, some APOBEC family members display unique biochemical characteristics that actively promote localized hypermutation. AID and APOBEC3G bind ssDNA strongly (displaying binding constants (Kd) of ~100 nM) and can translocate to catalyze multiple deaminations in a single binding event [53,62-64]. The role of this processivity in generating the observed clustered TCW mutations in human cancer, however, is currently unclear as this feature appears to not be universal among APOBEC enzymes that target TCW sequences. For example, APOBEC3A binds ssDNA much more weakly (Kd~100 μM) and displays only weak processivity in vitro [43,65]. APOBEC activity on ssDNA in yeast also produces base substitution patterns similar to those seen in C- or G-coordinated clusters in cancer. Expression of human APOBEC3G has been shown to induce multiple C to T and C to G mutations while excluding C to A transversions within regions of ssDNA through Ung1 mediated conversion of deoxyuridine to abasic sites and subsequent pol zeta and REV1 dependent error-prone trans-lesion synthesis [20]. Moreover, expressing hyper-active mutants of human AID and APOBEC3G [66] and lamprey APOBEC [67] in yeast directly induces clustered strand-coordinated mutations.
Expanding the number of cancer types displaying evidence of APOBEC-induced clustered mutations, Nik-Zainal et al. reported similar strand-coordinated clustering of mutated cytosines among 21 whole-genome sequenced breast cancers [7]. Spatially clustered mutations, referred to as “kataegis” (Greek for thunderstorm), became visually apparent by graphing the inter-mutation distance for each pair of mutations in a “rainfall” plot. (Note: in the current literature both terms, kataegis and mutation clusters, are interchangeably used, often in the same paper [66-69]). This analysis identified tracts of mutations as long as 14 Mb, where many of the mutations were separated by only 10 to 5000 bps. As with the clustered mutations observed in multiple myeloma, prostate, head-and-neck, and colorectal cancers, kataegis events were composed of strand-coordinated substitutions of cytosine to either thymine or guanine, primarily within the trinucleotide sequence of TpCpX (X = any nucleotide). Additionally, some kataegis events co-localized with various chromosome re-arrangements including complex events characteristic of chromothripsis. Recent evaluation of 507 sequenced tumor genomes identified kataegis in lung adenocarcinoma, liver cancer, B-cell lymphoma, acute lymphoid leukemia, pancreatic cancer, chronic lymphoid leukemia, and medulloblastoma [6], indicating that while such events are rare in each sample, they are spread across a large variety of cancer types.
An APOBEC mutagenesis pattern is widespread in human cancers
Analysis of exome sequenced cancers has likewise identified strand-coordinated clustered TCW mutations within a wide variety of cancer types [50]. As seen in whole genome sequenced cancers, TCW clusters were identified in head-and-neck, prostate, and breast cancers among mutations from 2680 exome sequenced cancers, primarily from The Cancer Genome Atlas (TCGA). This analysis also expanded the cancer types where these events are seen to include bladder, cervical, lung adenocarcinoma, lung squamous cell carcinoma, uterine endometrial, ovarian, colorectal, rectal, stomach, and kidney cancers [50]. Despite the widespread observation of C- or G-coordinated mutation clusters in 13 distinctly different tumor types, the frequency at which such events occur is heavily dependent upon cancer type: bladder, cervical, head-and-neck, breast, lung adenocarcinoma, and lung squamous cell carcinoma are clearly enriched in such clusters compared with other cancer types where these clusters appear to occur at a background level (Table 1). The biological impact of clustered mutations, especially in terms of cancer development, is currently unknown. While multiple mutations are necessary to deregulate cell growth and proliferation, genes whose alterations are causative in cancer are distributed across the human genome. Consequently, the inactivation of multiple cancer drivers by a single mutation cluster is unlikely, and limits the potential carcinogenic roles of this type of transient hyper-mutation to 1) increasing the likelihood an individual tumor suppressor is inactivated by a single mutagenic event, 2) inducing multiple mutations jointly required for activating an oncogene, or 3) altering the functionally of regulatory sequences. The same processes that lead to cluster formation (i.e., damage to transiently formed ssDNA) can also result in individual, scattered mutations. Thus, damage to multiple, simultaneously occurring ssDNA regions may establish a global transient hypermutation, a subset of which may be synergistic mutational events that provide a significant growth advantage for cancer cells.
Table 1.
Presence of clustered and genome-wide APOBEC mutagenesis in multiple cancer types.
| Mutation Clusters | In G- or C-coordinated clusters | Among All Mutations | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cancer Type | Samples | # | # Non- coordinated |
# A- or T- coordinated |
# G- or C- coordinated |
Mutations | APOBEC signature mutations |
Fold APOBEC enrichment |
Fisher's p-value | Fraction APOBEC signature mutations |
Mutations | APOBEC signature mutations |
Fold APOBEC enrichment |
Fisher's p-value | Fraction APOBEC signature mutations |
|
| Exomes | BLCA | 28 | 32 | 11 | 1 | 20 | 75 | 45 | 4.64 | < 2.2E-16 | 0.60 | 9167 | 3280 | 3.58 | < 2.2E-16 | 0.36 |
| BRCA | 507 | 39 | 15 | 0 | 24 | 81 | 58 | 4.21 | < 2.2E-16 | 0.72 | 29411 | 5906 | 2.29 | < 2.2E-16 | 0.20 | |
| CESC | 36 | 36 | 12 | 0 | 24 | 74 | 54 | 4.33 | < 2.2E-16 | 0.73 | 8732 | 4027 | 3.90 | < 2.2E-16 | 0.46 | |
| COAD | 155 | 9 | 6 | 1 | 2 | 7 | 1 | 1.32 | 5.66E-01 | 0.14 | 61725 | 1785 | 0.35 | 1.00E+00 | 0.03 | |
| HNSC | 74 | 16 | 7 | 0 | 9 | 24 | 14 | 4.00 | 1.02E-06 | 0.58 | 9333 | 2024 | 2.48 | < 2.2E-16 | 0.22 | |
| KIRC | 293 | 28 | 19 | 1 | 8 | 20 | 6 | 1.94 | 8.00E-02 | 0.30 | 70880 | 1641 | 1.01 | 3.92E-01 | 0.02 | |
| LAML | 199 | 5 | 4 | 1 | 0 | 0 | 0 | N/A | N/A | N/A | 5149 | 155 | 0.43 | 1.00E+00 | 0.03 | |
| LUAD | 230 | 162 | 89 | 2 | 71 | 260 | 153 | 3.71 | < 2.2E-16 | 0.59 | 209993 | 18666 | 1.96 | < 2.2E-16 | 0.09 | |
| LUSC | 178 | 63 | 34 | 0 | 29 | 101 | 39 | 3.39 | 1.36E-12 | 0.39 | 89899 | 10246 | 2.25 | < 2.2E-16 | 0.11 | |
| OV | 317 | 15 | 5 | 1 | 9 | 25 | 9 | 3.33 | 7.22E-04 | 0.36 | 15938 | 1316 | 1.12 | 6.70E-06 | 0.08 | |
| PRAD | 195 | 24 | 18 | 0 | 6 | 20 | 9 | 3.53 | 4.94E-04 | 0.45 | 13524 | 513 | 0.68 | 1.00E+00 | 0.04 | |
| READ | 69 | 8 | 6 | 0 | 2 | 8 | 5 | 2.88 | 1.62E-02 | 0.63 | 16291 | 611 | 0.45 | 1.00E+00 | 0.04 | |
| STAD | 151 | 23 | 13 | 1 | 9 | 31 | 18 | 4.70 | 7.86E-10 | 0.58 | 233733 | 2416 | 0.41 | 1.00E+00 | 0.01 | |
| UCEC | 248 | 37 | 32 | 0 | 5 | 21 | 5 | 1.62 | 1.91E-01 | 0.24 | 180472 | 6491 | 0.45 | 1.00E+00 | 0.04 | |
| Genomes | PRAD | 7 | 47 | 29 | 6 | 12 | 42 | 23 | 2.85 | 3.03E-07 | 0.55 | 24215 | 3106 | 2.14 | < 2.2E-16 | 0.13 |
| MM | 23 | 322 | 228 | 31 | 63 | 276 | 159 | 3.90 | < 2.2E-16 | 0.58 | 157247 | 21010 | 1.91 | < 2.2E-16 | 0.13 | |
| HNSC | 2 | 25 | 17 | 0 | 8 | 36 | 16 | 3.44 | 1.17E-06 | 0.44 | 25418 | 3897 | 2.04 | < 2.2E-16 | 0.15 | |
| BRCA | 21 | 1600 | 853 | 43 | 704 | 3168 | 2228 | 3.95 | < 2.2E-16 | 0.70 | 178080 | 68342 | 3.16 | < 2.2E-16 | 0.38 | |
| COAD | 9 | 167 | 134 | 2 | 31 | 158 | 104 | 3.91 | < 2.2E-16 | 0.66 | 121160 | 7466 | 0.97 | 9.99E-01 | 0.06 | |
Do APOBEC cytidine deaminases contribute to cancer by genome-wide mutagenesis?
The AID/APOBEC-induced mutations are prime candidates for generating genome-wide “mutation storms” that could initiate or promote cancer. Beyond commonly inducing clustered TCW mutations in a variety of human tumors, several AID/APOBEC family members have been shown to directly induce cancer. Collateral mutagenesis by AID is known to contribute to multiple human B-cell derived cancers (reviewed in [70]). Altered expression of AID in mice can result in not only immune system malignancies but also lung [71] and liver [72] cancers, indicating that AID expression in some non-B-cell human cancers (e.g. colorectal [73], lung [74], and liver [75]) may be one underlying cause. Similarly, liver specific over-expression of APOBEC1 in mice induces hepatocarcinoma [76], suggesting that relaxed control of the AID/APOBEC family, including the TCW targeting deaminases, may constitute a powerful carcinogenic force resulting from genome-wide mutagenesis. A general presence of a TpCpX mutation motif has been noted in several pan-cancer analysis publications [68,69]: Drier et al., for example, note enrichment with this signature in areas adjacent to rearrangement breakpoints.
TCW mutations are statistically over-represented in cancer
Applying a statistical method (Box 2) to the 2680 cancer exomes described above revealed that the same cancer types that display increased incidence of C- or G-coordinated clusters (i.e. bladder, cervical, head-and-neck, breast, lung adenocarcinoma, and lung squamous cell carcinoma) also frequently display dramatic over-representation of scattered mutations occurring in the TCW motif [50]. Moreover, this method highlighted the increased presence of TCW mutagenesis in one molecular subtype of breast cancer, HER2-enriched, suggesting that mutagenesis can be associated with specific features of tumor development. Complementary de novo pattern recognition analysis of mutations in 7042 tumors similarly identified APOBEC mutagenesis as a prominent mutation signature in 16 of 30 cancer types analyzed [6], including the 6 cancer types previously highlighted in the hypothesis-driven TCGA analyses [50,77]; all together these works establish APOBEC enzymes as one of the most widespread mutational forces during cancer progression.
Genome-wide APOBEC mutagenesis correlates with APOBEC expression
Why specific cancer types display such strong mutagenesis at the TCW motif is currently unclear. Several studies indicate that increased expression of APOBEC3B likely contributes in part. Burns et al. demonstrated that breast cancer cell lines exhibiting increased APOBEC3B mRNA levels, as measured by qPCR, microarray, or RNA-seq, correlated with increased levels of DNA damage and cytidine deaminase activity in extracts [46]. Importantly, this deaminase specifically targets cytidines in the TCW context, consistent with the over-representation of mutations in TCW within both mutation clusters and whole-exomes of some breast cancer samples. Direct comparison of APOBEC3B RNA-seq to either total C to T transitions or TCW mutations within various exome sequenced cancers identified positive correlations within breast cancers and lung adenocarcinomas and generally high APOBEC3B expression in head-and-neck, bladder and lung squamous cell carcinoma [46,50,77]. This relation implied that APOBEC3B is responsible for the overwhelming mutation of TCW in multiple cancer types and that its expression is one factor dictating the extent to which this mutation pattern occurs. However, roles for other APOBECs in TCW mutagenesis cannot be excluded. Expression of several APOBECs can correlate with the number of TCW mutations across multiple cancer samples (although lesser so than 3B) [50] and polymorphic deletion of APOBEC3B results in an increase in breast [78-80] and liver cancers [81], suggesting a complicated relationship between the APOBEC enzymes, TCW mutagenesis and cancer incidence.
How is APOBEC mutagenesis limited?
The access that AID/APOBEC family cytosine deaminases have to chromosomal DNA is normally limited by mechanisms that impact expression level, cellular localization, substrate specificity, and enzymatic activity. AID exemplifies the multifaceted regulation of these enzymes. AID protein levels are tightly controlled transcriptionally (reviewed in [82]), through the activity of miR-155 and miR-181 micro-RNAs [83-85], and through active degradation through interactions with REG-γ [86]. Nuclear import and export mechanisms also control AID's distribution between the cytoplasm (AIDs primary location) and the nucleus [87-89] where its activity on chromosomal DNA is further controlled by protein co-factors like Spt5 and RPA, multiple phosphorylation events [90,91] and ubiquitination [92] that modulate the enzyme's association with DNA. Specific aspects of this regulation are clearly shared among other AID/APOBEC family members. As the APOBEC3 enzymes function in innate immunity, their transcription is inducible by traditional modulators of the immune response (i.e. viral infection [81,93,94], CpG DNA [95], Toll-like receptor signaling [96], cytokines [97] and interferons [95,98,99]) as well as by DNA damage signaling [100], and hormones like estrogen [101]. The APOBEC3 enzymes also display defined cellular localization [102], being primarily expressed in the cytoplasm with the exceptions of the pan-cellular expression of APOBECs 3A and 3C and the nuclear localization of APOBEC3B whose import into the nucleus is controlled through the same mechanism utilized by AID [103]. Once translated, protein levels of at least APOBEC3A are controlled by proteosomal targeting through an interaction with Tribbles3 [104] and the deaminase activity of APOBEC1 is modulated by protein-protein interactions. APOBEC1 specificity for the C6666 of the apoB mRNA is enhanced by an auxiliary protein, ACF1 [105,106]; however the extent to which similar co-factors or post-translational modifications limit other APOBECs’ access to chromosomal DNA is unknown.
Can transient APOBEC activation induce genome-wide hypermutation?
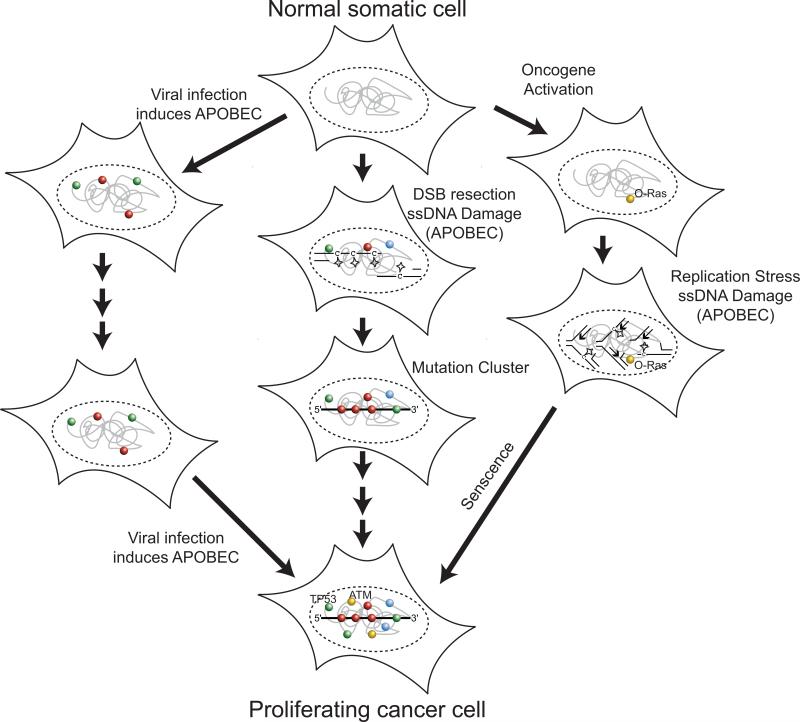
Dysregulation of any of these control mechanisms may play an important role in determining the extent of APOBEC mutagenesis that occurs within an evolving tumor. Elevated expression of APOBEC3B and potentially other APOBEC enzymes during the course of a tumor's evolution appears to be one mechanism that likely contributes in part to APOBEC mutagenesis. However, APOBEC3B expression varies greatly, spanning 3 orders of magnitude within any given cancer type [50,77]. While such variation could result from large differences in the constitutive levels of APOBEC3B mRNA between tumors, it is just as likely to result from transient induction of expression over time. Moreover, the sensitivity of APOBEC expression to such a wide range of exogenous factors suggests that environmental exposures could transiently elevate APOBEC levels resulting in transient hypermutation of TCW sequences (Figure 4). A role for the immune response in inducing AID/APOBEC expression and in turn hypermutation is particularly appealing. Viral induction of AID has been shown to contribute to carcinogenesis [107] and three of the cancer types where TCW mutagenesis is most prominent (i.e. bladder, cervical, and head-and-neck) are linked to viral infection.
Figure 4. APOBEC-induced transient hyper-mutation during cancer evolution.
(middle) Generation of isolated persistent single stranded DNA regions (e.g. a resected DSB) are targets for APOBEC cytidine deamination (stars) resulting in clustered mutations (mutations are depicted as spheres: C = red, G = green, A = blue and T = yellow). Individual clusters can inactivate a single tumor suppressor gene, activate an oncogene (e.g. oncogenic Ras: “O-Ras”), or alter transcriptional profiles by modifying microRNAs or transcription factor expression. Additional genetic alterations accumulated during subsequent cell divisions lead to cancer progression. (left) Multiple viral infections may each transiently induce APOBEC activity resulting in waves of APOBEC signature mutations that ultimately lead to dis-regulated cell proliferation. (right) Formation of multiple ssDNA regions in a cell, as occurs during oncogene-induced replication stress, allows APOBEC-induced mutagenesis to inactivate multiple cancer genes simultaneously including those responsible of proliferative barriers that can hold pre-cancerous cells in a senescent state (e.g. TP53, ATM).
Is substrate availability a limiting factor for APOBEC mutagenesis?
An increased availability of ssDNA substrate is an additional parameter that may also contribute strongly to the number of TCW mutations in a sample. The enrichment of both clustered and scattered TCW mutations near chromosome rearrangement breakpoints suggests that the formation of ssDNA, potentially during aberrant DNA DSB repair is likely a limiting factor for APOBEC-induced mutagenesis. DSBs could contribute to the formation ssDNA targeted by APOBECs by at least two mechanisms dependent on the repair pathway utilized. All homology-directed repair mechanisms produce ssDNA intermediates during 5′ to 3′ resection from the broken DNA ends. In addition, break-induced replication (BIR), a specialized form of homology-directed repair, produces large amounts of ssDNA during long-tract conservative DNA synthesis following strand-invasion [108]. Such DSB-associated events are likely involved in the formation of APOBEC-induced mutation clusters. However, the number of segmental copy number alterations (a surrogate measure of the number of DSBs) failed to correlate with the total number of TCW mutations in 449 breast cancer samples [50], indicating that DSBs may be a minor source of the ssDNA target dictating the extent of genome-wide TCW mutagenesis. Transcription or DNA replication may provide alternatives. Altered levels of transcription during tumor progression could either increase the amount of ssDNA or formation of stable R-loops and G-quadruplexes that can serve as AID/APOBEC substrates [26,29]. An association between transcription and AID-induced mutagenesis is well documented. Establishing whether the level at which a sequence is transcribed influences the ability of other APOBEC enzymes to induce mutation in vivo, however requires further investigation. Similarly, the discontinuous synthesis of the lagging strand provides a consistent and sizable target for ssDNA-specific mutagenesis. During normal replication, RPA may limit the ability of some APOBECs to induce mutations [109]. However in yeast, genetic perturbation of the replication fork increases both general mutagenesis, as well as the incidence of clustered mutations originating from damaged ssDNA [8].
Does replication stress provide a substrate for APOBEC mutagenesis?
APOBEC deamination of ssDNA during replication may have a profound impact on carcinogenesis. The enhanced proliferation of cancer cells would itself increase the total APOBEC mutational load accumulated in cells by simply increasing the number of genome replications that have occurred. Moreover, oncogene-induced replication stress may also increase the amount and persistence of ssDNA. Activation of oncogenes like Ras, Cyclin E, Cdc25A, and E2F1 induces hyper-replication, increased ssDNA formation, and ultimately activation of the DNA damage checkpoint following conversion of stalled replication forks into DSBs [110-114]. ATM and p53 dependent signaling pathways activated by the formation of ssDNA appear to limit further progression towards a cancer state by inducing cell senescence [111,112]. The large amount of persistent ssDNA formed during this oncogene-induced senescent state would serve as an ideal target for APOBEC enzymes and could thereby lead to inactivation of multiple genes that contribute to the DNA damage barrier to tumorigenesis. Such a mechanism would seemingly invoke transient hypermutation, as induction of multiple mutations would likely be required during the cell generation that transitions from a non-dividing state to one of active proliferation. Several lines of evidence suggest APOBEC-mediated genome-wide transient hypermutation may be one means to overcome a senescent barrier during an oncogene-induced replication stress. Specifically, databases of p53 mutations are statistically enriched in alterations at the sequence TC[59], and most TCW mutations occur late during breast cancer progression [115] when a senescent barrier would likely be inactivated. Moreover, tumor samples that have experienced significant TCW mutagenesis are frequent among cancer subtypes associated with amplification of the HER2 oncogene [8], which is known to induce an ATM dependent DNA damage response and senescence in mouse tumors [116].
Can mutation bursts accelerate cancer development?
APOBEC hypermutation is likely often a transient event, because its substrate, ssDNA, would be present in abundance only in a small fraction of cells undergoing abnormal DNA transactions (dysfunctional replication forks, multiple DSBs, etc.). We speculate that the impact of such mutational bursts in promoting tumorigenesis may differ from endogenous mutators caused by genetic defects. APOBEC mutagenesis combines the features of an endogenous mutagen with those associated with momentary exposure to exogenous environmental agents or drugs. Importantly, transient hypermutation may be more carcinogenic than the sustained mutagenesis associated with moderate permanent mutators. While both types of mutagenesis would produce similar number of mutations over large periods of time, transient hypermutation can rapidly generate multiple mutations which can overcome multilayer growth control mechanisms and promote carcinogenic growth. As opposed to permanent mutators, transient hypermutation also decreases the likelihood of subsequent deleterious mutations reducing any proliferative advantage a cancer cell may have acquired. Moreover, along with increased mutations, permanent mutators, especially those associated with genetic disruption of DNA repair activities, elevate DNA damage loads that in turn can trigger apoptosis. Transient induction of DNA damage, however, increases mutagenesis, but also may provide time for most apoptosis triggering DNA lesions to be repaired. Consequently, cells may be able to accumulate several cancer driving mutations during a transient exposure to DNA damage while initially escaping the p53 dependent genome surveillance machinery. The ability to avoid apoptosis may thus be one reason potentially transient forms of hypermutation (i.e. APOBECs, environmental exposures) appear to be common among human tumors as compared permanent mutators involving defects in DNA repair enzymes [6].
Conclusion
Transient hypermutation enables the induction of multiple mutations within a single cell generation without the establishment of a persistent mutator phenotype that may reduce a cell's fitness through accumulation of deleterious mutations. Simultaneous timing of mutations provides the opportunity to induce synergistic changes to rapidly produce new functions or a growth advantage. Such hyper-mutability over localized regions of a genome can form mutation clusters that may be important steps during the evolution of new protein functions [117] or alteration of DNA regulatory sequences [118]. Recent efforts to sequence human tumors genomes have revealed that mutation clusters are surprisingly common during cancer development. The biological importance of mutation clusters during tumorigenesis is currently unknown. However, the incidence of clustered mutations at TCW correlates with frequent genome-wide hyper-mutation at the same DNA sequence, suggesting that the same mechanisms that induce mutation clusters may also create multiple mutations genome-wide. Moreover, the highly regulated expression of the APOBEC proteins [93,95,96,98,119] and the transient nature of their ssDNA target likely limit the opportunity of these enzymes to deaminate chromosomal DNA. This suggests that -- like clustered mutations -- scattered TCW mutations may accumulate over a short time- span. Scattered simultaneous mutations have the potential to accelerate cancer progression through the inactivation of multiple tumor suppressor genes. Knowing whether scattered TCW mutations occur simultaneously or are accumulated through time will be critical not only to our understanding of the mutator phenotype that occurs in cancers, but also to how these cancers should be monitored and treated. While analysis of allelic fractions may provide evidence regarding the timing of TCW mutations [115,120], sequencing of paired neoplasms, primary tumors, and re-growths or metastases may provide the best evidence to understand how genetic alterations are accumulated, the role of transient mutageneisis in cancer, and how tumors evolve during disease progression.
Box 1: Signatures of AID/APOBEC-induced hypermutation in human cancer genomes.
Increased mutation frequencies for di- and tri-nucleotide sequences that are the preferential targets of AID/APOBEC family cytidine deaminases has now been reported in a variety of human cancers. Different AID/APOBECs display different sequence preferences with AID favoring the sequence WRC, APOBEC3G favoring CC, and seven APOBECs (i.e. APOBEC1, 3A, 3B, 3C, 3DE, 3F,and 3H) all capable to mutating TC and TCW motifs. Current exome and whole-genome sequencing of tumors suggests that of AID/APOBEC family members, TC-specific APOBECs provide the most extensive mutagenesis of human cancers, with the enrichment of WRC mutations associated with AID being limited mainly to lymphoid cancers and little to no over-represented mutation of the CC dinucleotide associated with APOBEC3G (e.g. CC di-nucleotides are excluded from reported mutation clusters). The large contribution of TC-specific APOBECs towards the total mutation load in many tumor types underscores the importance of assessing the biochemical and cellular characteristics of these enzymes that underlie their ability to hypermutate TCW sequences genome-wide.
Box 2: A Hypothesis-driven analysis of APOBEC mutagenesis.
A hypothesis-based statistical assessment APOBEC mutagenesis employed existing mechanistic knowledge about APOBEC cytidine deaminases and mutagenesis in persistent ssDNA to generate a simplified mutation spectrum indicative of APOBEC activity (i.e. the number of TCW→TTW or TCW→TGW; termed as “TCW mutations”). This analysis provided sufficient power to assign a p-value for to most whole-genome or exome sequenced cancer samples. TCW →TTW or →TGW mutations in cancers displaying APOBEC mutagenesis are often greater than 3-fold more abundant than expected by random mutagenesis, constituting up to 68% of total mutations identified in individual exome-sequenced samples [50] and even 95% of mutations within one outstanding case of whole-genome sequenced breast cancer [7].
Acknowledgements
The authors thank Drs. Mike Resnick, Kin Chan, and Scott Lujan for helpful discussion of this manuscript. This research was supported by the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences (project ES065073: PI - Mike Resnick). S.A.R. is supported by NIH Pathway to Independence Award K99ES022633-01.
List of Abbreviations
- UV
ultraviolet
- DSB
DNA double strand break
- ss
single stranded
- ds
double stranded
- MMS
methyl methanesulfonate
- SHM
somatic hypermutation
- AID
activation-induced cytidine deaminase
- TCGA
The Cancer Genome Atlas
References
- 1.Campbell CD, Chong JX, Malig M, Ko A, et al. Estimating the human mutation rate using autozygosity in a founder population. Nature genetics. 2012;44:1277–81. doi: 10.1038/ng.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drake JW. Too many mutants with multiple mutations. Crit Rev Biochem Mol Biol. 2007;42:247–58. doi: 10.1080/10409230701495631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burch LH, Yang Y, Sterling JF, Roberts SA, et al. Damage-induced localized hypermutability. Cell Cycle. 2011;10:1073–85. doi: 10.4161/cc.10.7.15319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox EJ, Prindle MJ, Loeb LA. Do mutator mutations fuel tumorigenesis? Cancer Metastasis Rev. 2013 doi: 10.1007/s10555-013-9426-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeb LA. Mutator phenotype may be required for multistage carcinogenesis. Cancer Res. 1991;51:3075–9. [PubMed] [Google Scholar]
- 6.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, et al. Mutational Processes Molding the Genomes of 21 Breast Cancers. Cell. 2012;149:979–93. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts SA, Sterling J, Thompson C, Harris S, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Molecular cell. 2012;46:424–35. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thoma F. Repair of UV lesions in nucleosomes--intrinsic properties and remodeling. DNA Repair (Amst) 2005;4:855–69. doi: 10.1016/j.dnarep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 10.Rochette PJ, Brash DE. Human telomeres are hypersensitive to UV-induced DNA Damage and refractory to repair. PLoS genetics. 2010;6:e1000926. doi: 10.1371/journal.pgen.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez C, Hadany L, Ponder RG, Price M, et al. Mutability and importance of a hypermutable cell subpopulation that produces stress-induced mutants in Escherichia coli. PLoS genetics. 2008;4:e1000208. doi: 10.1371/journal.pgen.1000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Molecular cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 13.Holbeck SL, Strathern JN. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–24. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rattray AJ, Shafer BK, McGill CB, Strathern JN. The roles of REV3 and RAD57 in double-strand-break-repair-induced mutagenesis of Saccharomyces cerevisiae. Genetics. 2002;162:1063–77. doi: 10.1093/genetics/162.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strathern JN, Shafer BK, McGill CB. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–72. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malkova A, Haber JE. Mutations arising during repair of chromosome breaks. Annu Rev Genet. 2012;46:455–73. doi: 10.1146/annurev-genet-110711-155547. [DOI] [PubMed] [Google Scholar]
- 17.Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–5. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Gordenin DA, Resnick MA. A single-strand specific lesion drives MMS-induced hyper-mutability at a double-strand break in yeast. DNA Repair. 2010;9:914–21. doi: 10.1016/j.dnarep.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y, Sterling J, Storici F, Resnick MA, et al. Hypermutability of damaged single-strand DNA formed at double-strand breaks and uncapped telomeres in yeast Saccharomyces cerevisiae. PLoS genetics. 2008;4:e1000264. doi: 10.1371/journal.pgen.1000264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan K, Sterling JF, Roberts SA, Bhagwat AS, et al. Base damage within single-strand DNA underlies in vivo hypermutability induced by a ubiquitous environmental agent. PLoS Genet. 2012;8:e1003149. doi: 10.1371/journal.pgen.1003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu M, Schatz DG. Balancing AID and DNA repair during somatic hypermutation. Trends Immunol. 2009;30:173–81. doi: 10.1016/j.it.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 22.Peters A, Storb U. Somatic hypermutation of immunoglobulin genes is linked to transcription initiation. Immunity. 1996;4:57–65. doi: 10.1016/s1074-7613(00)80298-8. [DOI] [PubMed] [Google Scholar]
- 23.Liu M, Duke JL, Richter DJ, Vinuesa CG, et al. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 2008;451:841–5. doi: 10.1038/nature06547. [DOI] [PubMed] [Google Scholar]
- 24.Bransteitter R, Pham P, Scharff MD, Goodman MF. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci U S A. 2003;100:4102–7. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dickerson SK, Market E, Besmer E, Papavasiliou FN. AID mediates hypermutation by deaminating single stranded DNA. The Journal of experimental medicine. 2003;197:1291–6. doi: 10.1084/jem.20030481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramiro AR, Stavropoulos P, Jankovic M, Nussenzweig MC. Transcription enhances AID-mediated cytidine deamination by exposing single-stranded DNA on the nontemplate strand. Nature immunology. 2003;4:452–6. doi: 10.1038/ni920. [DOI] [PubMed] [Google Scholar]
- 27.Chaudhuri J, Tian M, Khuong C, Chua K, et al. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–30. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 28.Yu K, Roy D, Bayramyan M, Haworth IS, et al. Fine-structure analysis of activation-induced deaminase accessibility to class switch region R-loops. Mol Cell Biol. 2005;25:1730–6. doi: 10.1128/MCB.25.5.1730-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wongsurawat T, Jenjaroenpun P, Kwoh CK, Kuznetsov V. Quantitative model of R-loop forming structures reveals a novel level of RNA-DNA interactome complexity. Nucleic Acids Res. 2012;40:e16. doi: 10.1093/nar/gkr1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen H. Activation-induced cytidine deaminase acts on double-strand breaks in vitro. Molecular Immunology. 2007;44:974–83. doi: 10.1016/j.molimm.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 31.Poltoratsky V, Heacock M, Kissling GE, Prasad R, et al. Mutagenesis dependent upon the combination of activation-induced deaminase expression and a double-strand break. Mol Immunol. 2010;48:164–70. doi: 10.1016/j.molimm.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michael N, Shen HM, Longerich S, Kim N, et al. The E box motif CAGGTG enhances somatic hypermutation without enhancing transcription. Immunity. 2003;19:235–42. doi: 10.1016/s1074-7613(03)00204-8. [DOI] [PubMed] [Google Scholar]
- 33.Xu Z, Fulop Z, Wu G, Pone EJ, et al. 14-3-3 adaptor proteins recruit AID to 5′-AGCT-3′-rich switch regions for class switch recombination. Nat Struct Mol Biol. 2010;17:1124–35. doi: 10.1038/nsmb.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chaudhuri J, Khuong C, Alt FW. Replication protein A interacts with AID to promote deamination of somatic hypermutation targets. Nature. 2004;430:992–8. doi: 10.1038/nature02821. [DOI] [PubMed] [Google Scholar]
- 35.Yamane A, Resch W, Kuo N, Kuchen S, et al. Deep-sequencing identification of the genomic targets of the cytidine deaminase AID and its cofactor RPA in B lymphocytes. Nat Immunol. 2011;12:62–9. doi: 10.1038/ni.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pavri R, Gazumyan A, Jankovic M, Di Virgilio M, et al. Activation-induced cytidine deaminase targets DNA at sites of RNA polymerase II stalling by interaction with Spt5. Cell. 2010;143:122–33. doi: 10.1016/j.cell.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Willmann KL, Milosevic S, Pauklin S, Schmitz KM, et al. A role for the RNA pol II-associated PAF complex in AID-induced immune diversification. J Exp Med. 2012;209:2099–111. doi: 10.1084/jem.20112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Basu U, Meng FL, Keim C, Grinstein V, et al. The RNA exosome targets the AID cytidine deaminase to both strands of transcribed duplex DNA substrates. Cell. 2011;144:353–63. doi: 10.1016/j.cell.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–9. doi: 10.1126/science.8511591. [DOI] [PubMed] [Google Scholar]
- 40.Refsland EW, Harris RS. The APOBEC3 family of retroelement restriction factors. Curr Top Microbiol Immunol. 2013;371:1–27. doi: 10.1007/978-3-642-37765-5_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suspene R, Sommer P, Henry M, Ferris S, et al. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic acids research. 2004;32:2421–9. doi: 10.1093/nar/gkh554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu Q, Konig R, Pillai S, Chiles K, et al. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–42. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 43.Love RP, Xu H, Chelico L. Biochemical Analysis of Hypermutation by the Deoxycytidine Deaminase APOBEC3A. Journal of Biological Chemistry. 2012;287:30812–22. doi: 10.1074/jbc.M112.393181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell. 2002;10:1247–53. doi: 10.1016/s1097-2765(02)00742-6. [DOI] [PubMed] [Google Scholar]
- 45.Suspene R, Aynaud MM, Guetard D, Henry M, et al. Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism. Proc Natl Acad Sci U S A. 2011;108:4858–63. doi: 10.1073/pnas.1009687108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burns MB, Lackey L, Carpenter MA, Rathore A, et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–70. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Westmoreland J, Ma W, Yan Y, Van Hulle K, et al. RAD50 is required for efficient initiation of resection and recombinational repair at random, gamma-induced double-strand break ends. PLoS Genet. 2009;5:e1000656. doi: 10.1371/journal.pgen.1000656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung WH, Zhu Z, Papusha A, Malkova A, et al. Defective resection at DNA double-strand breaks leads to de novo telomere formation and enhances gene targeting. PLoS Genet. 2010;6:e1000948. doi: 10.1371/journal.pgen.1000948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Gonzalez KD, Scaringe WA, Tsai K, et al. Evidence for mutation showers. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8403–8. doi: 10.1073/pnas.0610902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mayorov VI, Rogozin IB, Adkison LR, Frahm C, et al. Expression of human AID in yeast induces mutations in context similar to the context of somatic hypermutation at G-C pairs in immunoglobulin genes. BMC Immunol. 2005;6:10. doi: 10.1186/1471-2172-6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogozin IB, Pavlov YI. The cytidine deaminase AID exhibits similar functional properties in yeast and mammals. Mol Immunol. 2006;43:1481–4. doi: 10.1016/j.molimm.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 53.Pham P, Bransteitter R, Petruska J, Goodman MF. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 2003;424:103–7. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 54.Pavlov YI, Rogozin IB, Galkin AP, Aksenova AY, et al. Correlation of somatic hypermutation specificity and A-T base pair substitution errors by DNA polymerase eta during copying of a mouse immunoglobulin kappa light chain transgene. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:9954–9. doi: 10.1073/pnas.152126799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, et al. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nature immunology. 2001;2:530–6. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 56.Gibbs PE, McDonald J, Woodgate R, Lawrence CW. The relative roles in vivo of Saccharomyces cerevisiae Pol eta, Pol zeta, Rev1 protein and Pol32 in the bypass and mutation induction of an abasic site, T-T (6-4) photoadduct and T-T cis-syn cyclobutane dimer. Genetics. 2005;169:575–82. doi: 10.1534/genetics.104.034611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gibbs PE, Lawrence CW. Novel mutagenic properties of abasic sites in Saccharomyces cerevisiae. Journal of molecular biology. 1995;251:229–36. doi: 10.1006/jmbi.1995.0430. [DOI] [PubMed] [Google Scholar]
- 58.Simonelli V, Narciso L, Dogliotti E, Fortini P. Base excision repair intermediates are mutagenic in mammalian cells. Nucleic acids research. 2005;33:4404–11. doi: 10.1093/nar/gki749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, et al. Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. Journal of molecular biology. 2004;337:585–96. doi: 10.1016/j.jmb.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 60.Landry S, Narvaiza I, Linfesty DC, Weitzman MD. APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 2011;12:444–50. doi: 10.1038/embor.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Petersen S, Casellas R, Reina-San-Martin B, Chen HT, et al. AID is required to initiate Nbs1/gamma-H2AX focus formation and mutations at sites of class switching. Nature. 2001;414:660–5. doi: 10.1038/414660a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chelico L, Pham P, Goodman MF. Mechanisms of APOBEC3G-catalyzed processive deamination of deoxycytidine on single-stranded DNA. Nature structural & molecular biology. 2009;16:454–5. doi: 10.1038/nsmb0509-454. author reply 5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chelico L, Pham P, Goodman MF. Stochastic properties of processive cytidine DNA deaminases AID and APOBEC3G. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:583–93. doi: 10.1098/rstb.2008.0195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pham P, Chelico L, Goodman MF. DNA deaminases AID and APOBEC3G act processively on single-stranded DNA. DNA Repair (Amst) 2007;6:689–92. doi: 10.1016/j.dnarep.2007.01.001. author reply 93-4. [DOI] [PubMed] [Google Scholar]
- 65.Byeon IJ, Ahn J, Mitra M, Byeon CH, et al. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Taylor BJ, Nik-Zainal S, Wu YL, Stebbings LA, et al. DNA deaminases induce break-associated mutation showers with implication of APOBEC3B and 3A in breast cancer kataegis. Elife. 2013;2:e00534. doi: 10.7554/eLife.00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lada AG, Dhar A, Boissy RJ, Hirano M, et al. AID/APOBEC cytosine deaminase induces genome-wide kataegis. Biol Direct. 2012;7:47. doi: 10.1186/1745-6150-7-47. discussion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Drier Y, Lawrence MS, Carter SL, Stewart C, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Res. 2013;23:228–35. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lawrence MS, Stojanov P, Polak P, Kryukov GV, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park S-R. Activation-induced Cytidine Deaminase in B Cell Immunity and Cancers. Immune Network. 2012;12:230. doi: 10.4110/in.2012.12.6.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Okazaki IM, Hiai H, Kakazu N, Yamada S, et al. Constitutive expression of AID leads to tumorigenesis. J Exp Med. 2003;197:1173–81. doi: 10.1084/jem.20030275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takai A, Toyoshima T, Uemura M, Kitawaki Y, et al. A novel mouse model of hepatocarcinogenesis triggered by AID causing deleterious p53 mutations. Oncogene. 2009;28:469–78. doi: 10.1038/onc.2008.415. [DOI] [PubMed] [Google Scholar]
- 73.Endo Y, Marusawa H, Chiba T. Involvement of activation-induced cytidine deaminase in the development of colitis-associated colorectal cancers. J Gastroenterol 46 Suppl. 2011;1:6–10. doi: 10.1007/s00535-010-0326-1. [DOI] [PubMed] [Google Scholar]
- 74.Shinmura K, Igarashi H, Goto M, Tao H, et al. Aberrant expression and mutation-inducing activity of AID in human lung cancer. Ann Surg Oncol. 2011;18:2084–92. doi: 10.1245/s10434-011-1568-8. [DOI] [PubMed] [Google Scholar]
- 75.Kou T, Marusawa H, Kinoshita K, Endo Y, et al. Expression of activation-induced cytidine deaminase in human hepatocytes during hepatocarcinogenesis. Int J Cancer. 2007;120:469–76. doi: 10.1002/ijc.22292. [DOI] [PubMed] [Google Scholar]
- 76.Yamanaka S, Balestra ME, Ferrell LD, Fan J, et al. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A. 1995;92:8483–7. doi: 10.1073/pnas.92.18.8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nature genetics. 2013;45:977–83. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Komatsu A, Nagasaki K, Fujimori M, Amano J, et al. Identification of novel deletion polymorphisms in breast cancer. Int J Oncol. 2008;33:261–70. [PubMed] [Google Scholar]
- 79.Xuan D, Li G, Cai Q, Deming-Halverson S, et al. APOBEC3 deletion polymorphism is associated with breast cancer risk among women of European ancestry. Carcinogenesis. 2013 doi: 10.1093/carcin/bgt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Long J, Delahanty RJ, Li G, Gao YT, et al. A common deletion in the APOBEC3 genes and breast cancer risk. J Natl Cancer Inst. 2013;105:573–9. doi: 10.1093/jnci/djt018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang T, Cai J, Chang J, Yu D, et al. Evidence of associations of APOBEC3B gene deletion with susceptibility to persistent HBV infection and hepatocellular carcinoma. Hum Mol Genet. 2013;22:1262–9. doi: 10.1093/hmg/dds513. [DOI] [PubMed] [Google Scholar]
- 82.Zan H, Casali P. Regulation of Aicda expression and AID activity. Autoimmunity. 2013;46:83–101. doi: 10.3109/08916934.2012.749244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.de Yebenes VG, Belver L, Pisano DG, Gonzalez S, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–8. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teng G, Hakimpour P, Landgraf P, Rice A, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–9. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Uchimura Y, Barton LF, Rada C, Neuberger MS. REG-gamma associates with and modulates the abundance of nuclear activation-induced deaminase. J Exp Med. 2011;208:2385–91. doi: 10.1084/jem.20110856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McBride KM. Somatic Hypermutation Is Limited by CRM1-dependent Nuclear Export of Activation-induced Deaminase. Journal of Experimental Medicine. 2004;199:1235–44. doi: 10.1084/jem.20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ito S, Nagaoka H, Shinkura R, Begum N, et al. Activation-induced cytidine deaminase shuttles between nucleus and cytoplasm like apolipoprotein B mRNA editing catalytic polypeptide 1. Proc Natl Acad Sci U S A. 2004;101:1975–80. doi: 10.1073/pnas.0307335101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Patenaude AM, Orthwein A, Hu Y, Campo VA, et al. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol. 2009;16:517–27. doi: 10.1038/nsmb.1598. [DOI] [PubMed] [Google Scholar]
- 90.Basu U, Chaudhuri J, Alpert C, Dutt S, et al. The AID antibody diversification enzyme is regulated by protein kinase A phosphorylation. Nature. 2005;438:508–11. doi: 10.1038/nature04255. [DOI] [PubMed] [Google Scholar]
- 91.McBride KM, Gazumyan A, Woo EM, Schwickert TA, et al. Regulation of class switch recombination and somatic mutation by AID phosphorylation. Journal of Experimental Medicine. 2008;205:2585–94. doi: 10.1084/jem.20081319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun J, Keim CD, Wang J, Kazadi D, et al. E3-ubiquitin ligase Nedd4 determines the fate of AID-associated RNA polymerase II in B cells. Genes Dev. 2013;27:1821–33. doi: 10.1101/gad.210211.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pauli E-K, Schmolke M, Hofmann H, Ehrhardt C, et al. High level expression of the anti-retroviral protein APOBEC3G is induced by influenza A virus but does not confer antiviral activity. Retrovirology. 2009;6:38. doi: 10.1186/1742-4690-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Komohara Y, Yano H, Shichijo S, Shimotohno K, et al. High expression of APOBEC3G in patients infected with hepatitis C virus. J Mol Histol. 2006;37:327–32. doi: 10.1007/s10735-006-9059-0. [DOI] [PubMed] [Google Scholar]
- 95.Stenglein MD, Burns MB, Li M, Lengyel J, et al. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nature structural & molecular biology. 2010;17:222–9. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhou L, Wang X, Wang YJ, Zhou Y, et al. Activation of toll-like receptor-3 induces interferon-lambda expression in human neuronal cells. Neuroscience. 2009;159:629–37. doi: 10.1016/j.neuroscience.2008.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stopak KS, Chiu YL, Kropp J, Grant RM, et al. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J Biol Chem. 2007;282:3539–46. doi: 10.1074/jbc.M610138200. [DOI] [PubMed] [Google Scholar]
- 98.Vazquez N, Schmeisser H, Dolan MA, Bekisz J, et al. Structural variants of IFNalpha preferentially promote antiviral functions. Blood. 2011;118:2567–77. doi: 10.1182/blood-2010-12-325027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tanaka Y, Marusawa H, Seno H, Matsumoto Y, et al. Anti-viral protein APOBEC3G is induced by interferon-alpha stimulation in human hepatocytes. Biochem Biophys Res Commun. 2006;341:314–9. doi: 10.1016/j.bbrc.2005.12.192. [DOI] [PubMed] [Google Scholar]
- 100.Menendez D, Nguyen TA, Freudenberg JM, Mathew VJ, et al. Diverse stresses dramatically alter genome-wide p53 binding and transactivation landscape in human cancer cells. Nucleic Acids Res. 2013;41:7286–301. doi: 10.1093/nar/gkt504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pauklin S, Sernandez IV, Bachmann G, Ramiro AR, et al. Estrogen directly activates AID transcription and function. Journal of Experimental Medicine. 2009;206:99–111. doi: 10.1084/jem.20080521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bogerd HP, Wiegand HL, Hulme AE, Garcia-Perez JL, et al. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc Natl Acad Sci U S A. 2006;103:8780–5. doi: 10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lackey L, Demorest ZL, Land AM, Hultquist JF, et al. APOBEC3B and AID have similar nuclear import mechanisms. J Mol Biol. 2012;419:301–14. doi: 10.1016/j.jmb.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aynaud MM, Suspene R, Vidalain PO, Mussil B, et al. Human Tribbles 3 protects nuclear DNA from cytidine deamination by APOBEC3A. J Biol Chem. 2012;287:39182–92. doi: 10.1074/jbc.M112.372722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol. 2000;20:1846–54. doi: 10.1128/mcb.20.5.1846-1854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chester A, Weinreb V, Carter CW, Jr., Navaratnam N. Optimization of apolipoprotein B mRNA editing by APOBEC1 apoenzyme and the role of its auxiliary factor, ACF. RNA. 2004;10:1399–411. doi: 10.1261/rna.7490704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chiba T, Marusawa H. A novel mechanism for inflammation-associated carcinogenesis; an important role of activation-induced cytidine deaminase (AID) in mutation induction. J Mol Med (Berl) 2009;87:1023–7. doi: 10.1007/s00109-009-0527-3. [DOI] [PubMed] [Google Scholar]
- 108.Saini N, Ramakrishnan S, Elango R, Ayyar S, et al. Migrating bubble during break-induced replication drives conservative DNA synthesis. Nature. 2013;502:389–92. doi: 10.1038/nature12584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lada AG, Waisertreiger IS, Grabow CE, Prakash A, et al. Replication protein A (RPA) hampers the processive action of APOBEC3G cytosine deaminase on single-stranded DNA. PLoS One. 2011;6:e24848. doi: 10.1371/journal.pone.0024848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–42. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 111.Bartkova J, Rezaei N, Liontos M, Karakaidos P, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–7. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 112.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–5. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 113.Neelsen KJ, Zanini IM, Herrador R, Lopes M. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol. 2013;200:699–708. doi: 10.1083/jcb.201212058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jones RM, Mortusewicz O, Afzal I, Lorvellec M, et al. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2012 doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- 115.Nik-Zainal S, Van Loo P, Wedge DC, Alexandrov LB, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Reddy JP, Peddibhotla S, Bu W, Zhao J, et al. Defining the ATM-mediated barrier to tumorigenesis in somatic mammary cells following ErbB2 activation. Proc Natl Acad Sci U S A. 2010;107:3728–33. doi: 10.1073/pnas.0910665107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Camps M, Herman A, Loh E, Loeb LA. Genetic constraints on protein evolution. Crit Rev Biochem Mol Biol. 2007;42:313–26. doi: 10.1080/10409230701597642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frankel N, Erezyilmaz DF, McGregor AP, Wang S, et al. Morphological evolution caused by many subtle-effect substitutions in regulatory DNA. Nature. 2011;474:598–603. doi: 10.1038/nature10200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Thielen BK, McNevin JP, McElrath MJ, Hunt BV, et al. Innate immune signaling induces high levels of TC-specific deaminase activity in primary monocyte-derived cells through expression of APOBEC3A isoforms. J Biol Chem. 2010;285:27753–66. doi: 10.1074/jbc.M110.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Carter SL, Cibulskis K, Helman E, McKenna A, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012 doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]