Abstract
There is a need for improved biomaterials for use in treating non-healing bone defects. A number of natural and synthetic biomaterials have been used for the regeneration of bone tissue with mixed results. One approach is to modify native tissue via decellularization or other treatment for use as natural scaffolding for tissue repair. In this study, our goal was to improve on our previously published alternating solution immersion (ASI) method to fabricate a robust, biocompatible, and mechanically competent biomaterial from natural demineralized bone matrix (DBM). The improved method includes an antigen removal (AR) treatment step which improves mineralization and stiffness while removing unwanted proteins. The chemistry of the mineral in the remineralized bone matrix (RBM) was consistent with dicalcium phosphate dihydrate (brushite), a material used clinically in bone healing applications. Mass spectrometry identified proteins removed from the matrix with AR treatment to include α-2 HS-glycoprotein and osteopontin, non-collagenous proteins (NCPs) and known inhibitors of biomineralization. Additionally, the RBM supported the survival, proliferation, and differentiation of human mesenchymal stromal cells (MSCs) in vitro as well or better than other widely used biomaterials including DBM and PLG scaffolds. DNA content increased more than 10-fold on RBM compared to DBM and PLG; likewise, osteogenic gene expression was significantly increased after 1 and 2 weeks. We demonstrated that ASI remineralization has the capacity to fabricate mechanically stiff and biocompatible RBM, a suitable biomaterial for cell culture applications.
Keywords: Mineralization, Bone graft, Brushite, Collagen, Matrix, MSCs
1. Introduction
Musculoskeletal injuries remain the 2nd largest category for hospital expenditures by body system, with hospitalizations rising 15% since 1997 1. With the number of fractures projected to increase in the coming years, it is vital to have effective treatments available. The field of bone tissue engineering (BTE) presents promising alternatives to traditional clinical treatments. A central challenge to improve bone grafts is to fabricate a mechanically competent substrate that evokes a limited immune response (biocompatibility) while promoting osteogenic differentiation of progenitor cells (osteoinductivity) and encouraging integration with surrounding bone tissue (osteoconductivity) to promote healing.
Numerous studies describe the use of a variety of materials for scaffolds including polymers, metals, and ceramics 2-4. In order to improve existing approaches, design cues can be taken from the bone tissue itself in a biomimetic process where the engineer attempts to recapitulate the building of the natural tissue in vitro. Bone is essentially a two phase nanocomposite tissue in which the organic matrix is the source of bone strength and toughness and the inorganic mineral the source of stiffness 5. The use of composites containing multiple material components is a growing trend in BTE and is likely to provide the most desirable properties for a mechanically and biologically useful bone replacement scaffold 6-8. In our work, we follow this direction and build a composite of the natural organic bone matrix with an artificially created mineral phase. Our goal is to produce a biocompatible allograft material that has strength, stiffness, and toughness comparable to native bone.
In previous work, we have presented an automated method to incorporate a mineral phase into demineralized bone matrix (DBM) 9. By stiffening the natural DBM with an osteoconductive mineral phase, we have integrated the functional properties of multiple materials into one substrate. DBM also has the advantage of having the complex native extracellular matrix (ECM) structure and composition that is difficult to recreate with synthetic materials. The ECM helps regulate bone tissue at the cellular level, particularly affecting adhesion, migration, proliferation, and differentiation of cells within the tissue 10,11. For example, bone marrow derived mesenchymal stromal cells (MSCs) adhere and proliferate well on collagen matrices, demineralized bone, and calcium-phosphate mineral 12-14 suggesting our previously reported material is compatible with this important bone progenitor cell source.
In the current work we demonstrate that the remineralization of the bone matrix is significantly improved after treatment of the (soft) matrix to remove antigenic molecules. Antigen removal (AR), traditionally referred to as decellularization, is a key step towards clinical application in order to prevent or limit the immune response 15,16. This is something of a misnomer since several publications have shown that not all antigenic components are cellular in origin, and that removal of visible cells does not necessarily correlate with removal of known xenoantigens or reduction of overall antigen burden of the tissue. AR processes therefore may aim not only to remove cellular components during the cleansing process, but also extracellular molecules adherent to the ECM. Decellularization (AR) processes incorporating protein solubilization steps represent progress towards reducing the antigenicity of transplant tissues 17, but this often overlooked effect is poorly characterized in terms of changes in ECM proteins and the influence such changes may have on downstream applications utilizing decellularized soft tissues. In this study we improved our remineralized bone matrices both mechanically and chemically, studied the effects of using a published AR protocol on the preparation of RBM 18, and assessed the ability of the RBM to act as a scaffold in supporting MSC survival and osteogenic differentiation in vitro.
2. Methods
2.1 Tissue Collection
Equine third metacarpal (MC3) bones were collected post mortem at the J.D. Wheat Veterinary Orthopedic Research Lab (VORL) at UC Davis. Rectangular cortical bone beams (~2 × 2 × 22 mm) were prepared from the diaphyseal portion of MC3 using an Exakt cutting system (Exakt Technologies) and specialized cutting jigs. Cylindrical cores of trabecular bone 7 mm in diameter and 25 mm in length were cut from the distal end of the MC3 using a drill press (Model J-2530, JET) with coring tool attachment (9/32” diameter, Starlite Industries, Rosemont, PA). Trabecular disks (height = 2 mm) were cut from the cylindrical cores for use in cell culture experiments (IsoMet 1000 Precision Saw, Buehler). Tissue was frozen at −20°C when not being machined. The tissue was kept hydrated during machining with deionized (DI) water.
2.2 Specimen Preparation: Demineralization, Antigen Removal, Remineralization
Mineral was removed from MC3 bones via submersion in a demineralizing solution (formic acid (22.5%) – sodium citrate (100 g/L)) that has been reported to effectively remove mineral from bone tissue while preserving the collagen matrix 19. Following demineralization, DBM specimens were rinsed thoroughly with DI water and split into two groups, with one group undergoing a protein/antigen removal treatment and the other group receiving no further treatment prior to remineralization. AR was performed as described by Wong et al. 18. Briefly, intact demineralized bone matrix (DBM) specimens were sequentially exposed to hydrophile (containing OptSARB with no additional additives) and lipophile (OptSARB containing 1% w/v ASB-14) antigen removal solutions, for 48 hours each, designed to differentially solubilize proteins within the tissue. Nucleic acid digestion (24 hours) and a washout step (48 hours) were then performed before the specimens were hydrated with DI water and frozen for future use. As a control, a sham antigen removal process was performed on a subset of DBM specimens. These specimens were treated with the same protocol and maintained under identical conditions as the treatment group, except ultrapure water (Milli-Q filtered; EMD Millipore Corporation, Billerica, MA, USA) was substituted for the treatment solution at each step. Supernatants from each antigen removal step were collected for protein analysis via liquid chromatography–mass spectrometry (LC-MS) as described below. Demineralized specimens were then remineralized using the previously described ASI method 9. Briefly, the demineralized specimens were sequentially exposed to separate solutions containing either calcium or phosphate ions, allowing for nucleation and growth of a calcium phosphate mineral phase within the DBM.
2.3 Assessing Remineralization (Imaging + Mechanical Testing)
Two groups of remineralized cortical bone beams were tested for mineral content and mechanical properties following remineralization. The first group (RBM(+); n=12) was treated with the antigen removal protocol as described above, while the second group (RBM(−); n=12) received no AR. All specimens were stored and tested using the same methods, as follows.
MicroCT analysis of remineralized cortical beams (55 kVp, 145 μA, 300 ms integration time, average of 3 images, 6 μm resolution) was performed to determine the amount (mineral volume fraction (MVF)) and apparent mineral density of the construct using standard SCANCO software (μCT 35, Scanco Medical). Regions of interest (ROIs) were selected manually for each specimen cross-section as close to the outside edge as possible. We selected a global threshold of 375 mm HA/cc, with all pixels above the threshold considered mineralized and those below classified as non-mineralized. The MVF was defined as the number of voxels above the threshold divided by the total number of voxels in the ROI.
Mechanical characterization of RBM(+) and RBM(−) constructs was performed using a Bose Enduratec ELF3200 mechanical testing system. Beams were loaded up to 0.375N via 3 point bending and the stiffness was calculated as the slope of the linear region of the force-displacement curve, making sure to avoid the toe-in portion of the curve (n=12, per group).
2.4 Characterizing the Mineral Phase and Matrix
2.4.1 Electron Microprobe analysis
The electron microprobe collects x-ray counts which are used to determine the relative abundance of elements of interest within the sample. Specimens were quantitatively analyzed using a Cameca SX-100 5-spectrometer wavelength dispersive electron microprobe (accelerating voltage: 15 kV; beam current: 10 nAmp; rastering beam diameter: 10 microns). The standards are well-characterized, homogenous, naturally occurring minerals. Apatite was used for O, Ca, and P, diopside (Ca-pyroxene) for Mg, and Albite (Na-feldspar) for Na. Data points were sampled randomly across the specimens, with 5 points collected and averaged per sample. RBM(+) and RBM(−) specimens were analyzed (n = 4 per group).
2.4.2 Fourier Transform Infrared Spectroscopy (FTIR) analysis
The KBr disc method was applied for IR analysis. RBM(+) and RBM(−) samples were lyophilized, ground manually in an agate mortar with a pestle, and mixed with KBr powder (1:150 weight ratio) before being pressed into discs. The IR spectra were then collected using a Nicolet™ iS10 FT-IR spectrometer at room temperature immediately after preparation of the discs. Each spectrum was acquired by the accumulation of 36 scans at a resolution of 4 cm−1.
2.4.3 Histological analysis
In preparation for undecalcified histology, specimens were stored in ascending concentrations of ethanol (70%, 75%, 85% EtOH, 2 days each) under vacuum. Following dehydration, scaffolds were embedded in Technovit (Kulzer, Wehrheim, Germany) under vacuum and constant agitation in a series of solutions ranging from 100% EtOH to 100% Technovit for at least 2 days at each step. Finally, the scaffolds were placed in molds with the Technovit and polymerized using an Exakt polymerizing light machine. Once embedded, each sample was attached to a slide and processed to a thickness of 40-50 μm. Briefly, the surface of each scaffold was exposed by grinding with 320 grit sandpaper under DI water rinsing (Beuhler grinding wheel), and then polished with an automated Exakt grinder using progressively finer grinding paper, down to 2500 grit. Once prepared, slides were contact x-rayed (Model 805, Faxitron Bioptics; 35 kVp for 45 minutes) and stained with VonKossa to view mineral distribution or hematoxylin and eosin (H&E) to view tissue morphology.
2.4.4 Liquid chromatography–mass spectrometry (LC-MS)
2.4.4.1 Sample Preparation and Protein Digestion
Supernatants of the various AR solutions and water were collected for protein analysis (n=4). Protein samples from the hydrophile and lipophile steps were pooled to increase concentration and then were precipitated using the Calbiochem ProteoExtract Precipitation Kit (Millipore) based on the manufacturer's instructions. Following digestion, the protein sample was resuspended in 100uL of 50mM ammonium bicarbonate at pH 8 and disulfide bonds were reduced with 10mM TCEP at 90°C for 10 minutes. The protein solution was then alkylated with 15mM iodoacetamide (IAA) in the dark for 1 hour, followed by addition of 5mM dithiothreitol to quench the IAA. Trypsin was added in a 1:30 ratio (enzyme: protein) and digested overnight at 37°C.
2.4.4.2 Liquid chromatography–mass spectrometry (LC-MS)
LC-MS/MS analysis was performed using a standard top 15 method on Thermo Scientific Q-Exactive orbitrap mass spectrometer in conjunction with a Proxeon Easy-nLC II HPLC (Thermo Scientific) and Proxeon nanospray source. The digested peptides were reconstituted in 2% acetonitrile/0.1% trifluoroacetic acid and loaded onto a Magic C18 reversed phase trap where they were desalted before being separated using a Magic C18 reverse phase column. Data were collected using higher energy collision dissociation (HCD). Peptides were eluted using a gradient of 0.1% formic acid (A) and 100% acetonitrile (B). A 90 minute gradient was run with 5% to 35% B over 70 minutes, 35% to 80% B over 9 minutes, 80% B for 1 minute, 80% to 5% B over 1 minute, and finally held at 5% B for 10 minutes.
2.4.4.3 Data Analysis
Database searching - Tandem mass spectra were extracted and charge states were deconvoluted and deisotoped. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version CYCLONE (2013.02.01.1)). X! Tandem was set up to search the NCBI Equus caballus database (June, 2013; 31659 entries) plus an equal number of reverse sequences and 64 common laboratory contaminant proteins, assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 PPM and a parent ion tolerance of 20 PPM. Carbamidomethyl of cysteine was specified in X! Tandem as a fixed modification. Glu->pyro-Glu of the n-terminus, ammonia-loss of the n-terminus, gln->pyro-Glu of the n-terminus, oxidation of proline, dioxidation of methionine and tryptophan and acetyl of the n-terminus were specified in X! Tandem as variable modifications.
Criterion for protein identification - Scaffold (version Scaffold_4.0.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80.0% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 90.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii, Al et al Anal. Chem. 2003;75(17):4646-58). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters. Using these parameters, a false discovery rate was calculated as 0.33% on the peptide level and 2.1% on the protein level for samples searched against the Equus caballus database.
2.5 hMSC Culture on Matrices
2.5.1 Matrix/Scaffold preparation
Trabecular equine bone disks were prepared as described above (section 2.1). For cell culture experiments, the matrices were separated into two groups. One group (DBM) was demineralized, rinsed thoroughly with DI water, and frozen prior to use. A second group (RBM) was demineralized, rinsed with DI water, and then remineralized via the ASI method 9. All samples from these two groups were treated with the previously described antigen removal protocol developed by Wong et al. 18. All specimens were sonicated briefly in DI water to remove particulates resulting from the cutting and decalcification processes.
Synthetic polymer scaffolds were fabricated using a gas foaming/particulate leaching method as described by He and others 7,20. Briefly, poly(lactide-co-glycolide) (PLG) microspheres were prepared using a double emulsion process. Lyophilized microspheres were mixed with NaCl (250-425 micron diameter) and compressed into solid disks (8.5 mm diameter, 1.5 mm thickness) using a stainless steel die and a Carver press (3 tons pressure, 1 min). Compressed disks were then exposed to high pressure CO2 gas for 16 hours followed by rapid pressure release. NaCl particles were leached from scaffolds by immersion in DI H2O for 24 hours.
Prior to cell culture, bone matrices and PLG scaffolds were placed in sealed 50 mL Steriflip (Millipore) conical tubes with 70% EtOH under vacuum for 45 minutes for sterilization. Matrices were then rinsed twice under vacuum with sterile phosphate-buffered saline (PBS), left overnight in culture medium, and patted dry with sterile gauze prior to cell seeding.
2.5.2 Cell seeding
Human bone marrow-derived MSCs (Lonza, Basel, Switzerland) were expanded in minimum essential alpha medium (α-MEM, Invitrogen) supplemented with 10% fetal bovine serum (FBS; JR Scientific) and 1% penicillin and streptomycin (P/S; Mediatech). Cells were utilized at passages 5-6.
Following expansion, MSCs (3 × 105 cells per matrix) were suspended in 30 μL of media and applied drop-wise to the matrix/scaffold surface (n = 4 per group, per time point, unless otherwise noted). In addition, cells (3 × 105/30uL) were cultured on standard tissue culture plastic (TCP) for 90 minutes and used to quantify initial DNA content (n=4). Matrices were placed in 24-well plates and transferred to standard cell culture incubators (37°C, 5% CO2) for 90 minutes to allow cell attachment. Following this attachment period, 2mL of osteogenic media (OM; α-MEM supplemented with 10mM β-glycerophosphate, 10 nM dexamethasone, 1mM ascorbate-2-phosphate) was added to each well and the plates were placed on an XYZ shaker (Stovall Life Sciences, Inc., Greensboro, NC) within an incubator to enhance nutrient transport. Media was changed the following day and every 3 days thereafter for the duration of culture. Matrices/scaffolds were collected at 4 or 5 time points (24 hours, 72 hours, 7 days, and 14 days for both DNA and RNA; 21 days for DNA) for each assay described below.
2.5.3 Cellular Assays
At each time point, scaffolds were collected for DNA quantification and gene expression analysis. Scaffolds were minced with a razor blade, flash-frozen in liquid nitrogen, and processed using TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Following isolation and purification, total DNA and RNA were quantified using a spectrophotometer (Nanodrop 1000, Thermo Scientific, Waltham, MA). DNA content was used to estimate cell number based on the ratio of DNA measured per number of cells seeded. RNA was passed through the Qiaquick RNeasy mini-kit (Qiagen) according to manufacturer's protocol to clean-up the RNA and to perform DNAse digestion. The clean RNA was then reverse transcribed into cDNA with the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative polymerase chain reaction (qPCR) was performed using TaqMan1 Universal PCR Master Mix (Applied Biosystems, Foster City, CA) on a Mastercycler1 realplex2 (Eppendorf, Westbury, NY); primers and probes for ALPL (Hs01029144_m1), IBSP (Hs00173720_m1), RUNX2 (Hs00231692_m1), CCND1 (Hs00765553_m1), and MRPL13 (Hs00204173_m1), were purchased from Life Technologies. Amplification conditions were 50°C for 2 min and 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Quantitative PCR results were normalized to RPL13 transcript level to yield ΔCt values. Relative expression was subsequently calculated using the formula 2−ΔCt.
2.6 Statistical Methods
GraphPad Prism 6.0 software was used for all analyses. Student's t-test was used to compare stiffness, MVF, morphological parameters between RBM(+) and RBM(−) groups, as well as LCMS spectral counts. One way analysis of variance (ANOVA) was used to compare Ca/P values between native, RBM(+) and RBM(−) specimens. Two-way ANOVA, including both time point and treatment, was used for all cellular assays performed. Tukey's multiple comparisons test was used for post hoc pairwise analyses. Values are reported as mean +/−standard error of the mean (SEM). All tests were considered significant at p<0.05.
3. Results
3.1 Changes in mineral content and stiffness following antigen removal technique
Antigen removal treatment improved mineral content and stiffness of remineralized constructs (RBM(+)) compared to matrices not treated with AR (RBM(−)) (Table 1). RBM(+) specimens had an average mineral volume fraction (MVF) of 0.481 +/- 0.006, significantly higher than the value for RBM(−) (MVF = 0.382 +/− 0.01; p<0.05). Morphological measurements calculated using Scanco software also indicate differences in the connectivity density (or degree to which mineralized regions are connected) as well as the trabecular number and spacing and degree of anisotropy (Table 1). Overall, the RBM(+) had an average apparent density of 693.55 +/− 16.22 mg HA/cc for the mineralized regions above the threshold, similar to the apparent density of RBM(−) (665.10 +/− 14.46 mg HA/cc). For comparison, native cortical specimens before treatment had a MVF of 0.928 +/− 0.008 and an apparent density of 1041.7 +/− 13.26 mg HA/cc. Demineralized specimens had a MVF of 0.005 +/− 0.003.
Table 1.
Comparison of mechanical and morphological properties between RBM(+) and RBM(−)
| Measured Property: | RBM(+) | RBM(−) |
|---|---|---|
| MVF | 0.481 +/− 0.006 *** | 0.382 +/− 0.01 |
| Apparent Density (mg HA/cc) | 693.55 +/− 16.22 | 665.11 +/− 14.46 |
| Connectivity | 1498.01 +/− 82.71 *** | 1006.27 +/− 63.83 |
| Trabecular Number | 11.30 +/− 0.25 *** | 7.82 +/− 0.36 |
| Trabecular Spacing | 0.107 +/− 0.002 *** | 0.169 +/− 0.007 |
| Degree of Anisotropy | 1.47 +/− 0.03 ** | 1.374 +/− 0.01 |
| Stiffness (N/mm) | 5.27 +/− 0.22 ** | 2.76 +/− 0.57 |
p<0.01
p<0.001.
Three-point bending tests determined the mean stiffness of RBM(+) specimens to be: 5.27 +/− 0.22 N/mm (Fig. 1) (n=12), nearly double that of RBM(−) specimens (p<0.01). Following antigen removal treatment, the standard deviation of the stiffness measured via 3 point bending decreased significantly from 1.99 (RBM(−)) to 0.75 (RBM(+)) (p<0.05).
Figure 1.
Improvement in mechanical properties and MVF of RBM following antigen removal (AR) treatment (RBM(+)) compared to those without AR (RBM(−)). Both stiffness and MVF are significantly increased with ASI remineralization following AR. n=12; *p<0.05. Error bars represent mean stiffness +/− SEM.
3.2 Mineral Characterization
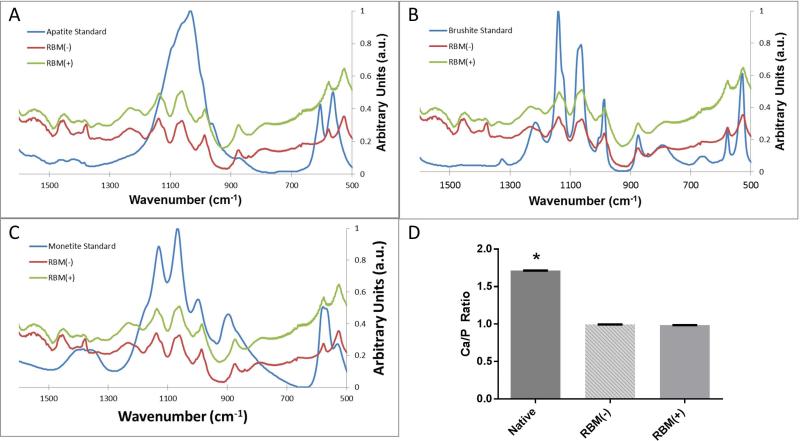
Native bone was analyzed and found to have calcium to phosphate ratio (Ca/P) of 1.71 +/− 0.006, in agreement with the expected ratio based on the chemical structure of hydroxyapatite (Ca10(PO4)6(OH)2). RBM(+) specimens had an average Ca/P = 0.986 +/− 0.005 and RBM(−) specimens had a Ca/P = 0.993 +/− 0.006. These ratios were found to be significantly different from that of native bone (p<0.001), but not significantly different from each other (Fig. 2).
Figure 2.
FTIR spectra of RBM(+) and RBM(−) mineral, compared to known mineral standards (A) apatite, (B) brushite, and (C) monetite. Mineral from both RBM(+) and RBM(−) specimens have nearly identical spectra, and share characteristic ν1,ν3 spectral peaks between 900 and 1,200 cm−1 (P-O stretching).
Average Ca/P ratios (D) show no difference between RBM(+) and RBM(−) samples, both of which are significantly different than native bone (*p<0.0001). Error bars represent mean Ca/P ratio +/− SEM; n=4.
FTIR spectra for remineralized specimens were compared against the hydroxyapatite spectrum, as well as those for dicalcium phosphate dihydrate (brushite) and dicalcium phosphate anhydrous (monetite), known calcium phosphate minerals with Ca/P ratios similar to those measured with electron microprobe. Due to the alignment of characteristic ν1,ν3 spectral peaks between 900 and 1,200 cm−1 (P-O stretching), it was determined that the ASI method leads to the formation of brushite within demineralized matrices (Fig. 2). Both RBM(+) and RBM(−) specimens exhibited the same spectrum.
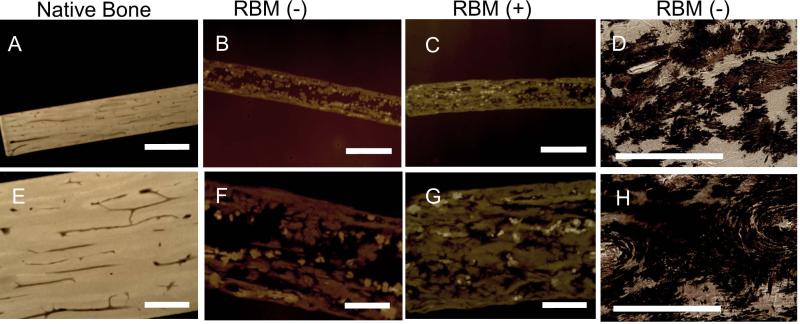
Contact x-ray images and histological slides stained with VonKossa indicate diffuse, heterogeneous mineralization for all remineralized matrices, although RBM(+) has more robust, complete mineralization compared to RBM(−). Mineral is present in various regions throughout the RBM including osteonal matrix, interstitial matrix, Haversian canals, and osteocyte lacunae (Fig 3).
Figure 3.
Contact X-ray of thin (40-60um) sections allows visualization of mineral in native equine bone (A,E), as well as RBM(−) (B,F) and RBM(+) (C,G) specimens. Note the difference in grayscale intensity between remineralized specimens and native bone, indicating a difference in mineralization. Additionally, RBM(+) (C,G) has more robust, complete mineralization compared to RBM(−) (B,F). Panels A,B,C: 1×, scale bar = 2mm; Panels E,F,G: 4×, scale bar = 0.5mm.
VonKossa staining (D,H) allows visualization of mineral present throughout RBM(−). Dark (brown) staining shows mineral nucleation is heterogeneous throughout the matrix, crystalizing within osteoid regions or interstitial regions. Panels D,H: 20×, scale bar = 250μm.
3.3 Matrix characterization
The relative abundance of individual proteins within a sample can be determined from the total spectrum count, essentially the number of times the mass spectrometer identifies a specific protein during analysis 21,22. A number of unique proteins were identified by the Scaffold Software as present in the AR supernatant, the nanopure water (H2O) control, or both. ColIα1 and ColIα2 were identified at lower spectrum counts in the treatment solution compared to the H2O control solution (p<0.0001). Proteins identified at greater spectrum counts in the treatment solution than the H2O solution include: α-2 HS-glycoprotein (fetuin) (p<0.0001), osteopontin-1 (p<0.05), and osteonectin (p<0.001). A more comprehensive list of proteins identified is presented in Table 2.
Table 2.
Molecules identified by mass spectrometry in supernatant of AR solution or sham H2O treatment solution. Known mineralization inhibitors (OPN, fetuin, ON) have higher threshold count in AR treatment solution compared to sham (H2O) solution. Note that AR treatment more effectively removes smaller molecular weight proteins. Proteins identified from Equus caballus database.
| Protein | Molecular Weight (kDa) | Average spectrum count (control) | Average spectrum count (treatment) | P-value |
|---|---|---|---|---|
| Col I-α2 | 129 | 1738.75 +/− 339.55 * | 390 +/− 93.29 | <0.0001 |
| Col I-α1 | 125 | 883 +/− 188.55 * | 139.75 +/− 42.87 | <0.0001 |
| Prothrombin-like | 70 | 0 | 237 +/− 35.83 * | <0.0001 |
| Alkaline phosphatase | 57 | 0 | 17.75 +/− 4.50 * | <0.001 |
| Biglycan precursor | 42 | 0 | 259.5 +/− 65.55 * | <0.0001 |
| Alpha-2-HS glycoprotein (fetuin) | 39 | 61.25 +/− 42.59 | 2670.5 +/− 596.43 * | <0.0001 |
| OPN-like Isoform 1 | 35 | 283.75 +/− 88.20 | 978.5 +/− 140.94 * | <0.05 |
| SPARC precursor (ON) | 35 | 0.25 +/− 0.25 | 283 +/− 30.75 * | <0.001 |
| Bone sialoprotein 2-like | 34 | 0 | 45.25 +/− 13.21 * | <0.05 |
| Insulin-like growth factor binding protein-5 | 30 | 2.25 +/− 1.93 | 32.5 +/− 3.80 * | <0.001 |
indicates significantly higher spectrum count
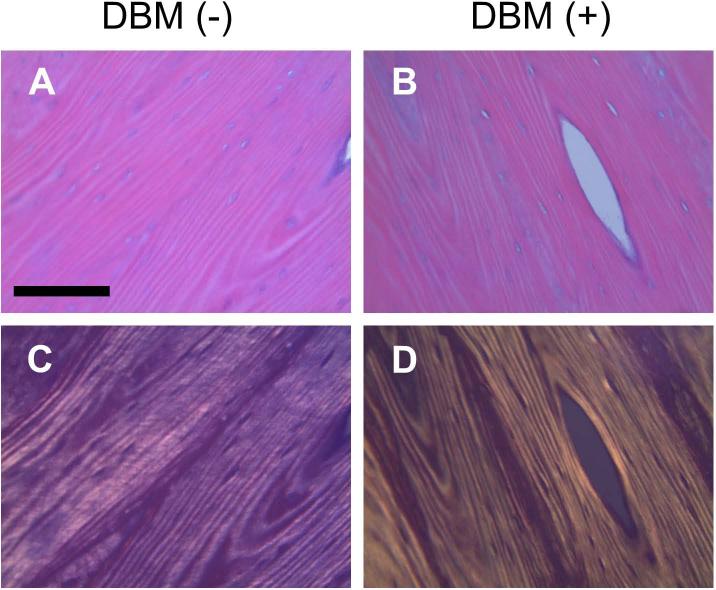
Analysis of histological slides stained with H&E demonstrates no apparent change in collagen fibril morphology resulting from the antigen removal treatment (Fig. 4). These data agree with previous assessments of the effect of antigen removal process on soft tissue (bovine pericardium) collagen 18.
Figure 4.
H&E stained histology slides of collagen matrix (A: DBM(−), B: DBM(+)). Polarized images (C: DBM(−), D: DBM(+)) highlight characteristic collagen fibril structure and indicate no apparent obvious change to structure with antigen removal treatment. Magnification = 20×; scale bar = 125μm.
3.4 Cellular assays
3.4.1 DNA content estimates cell number
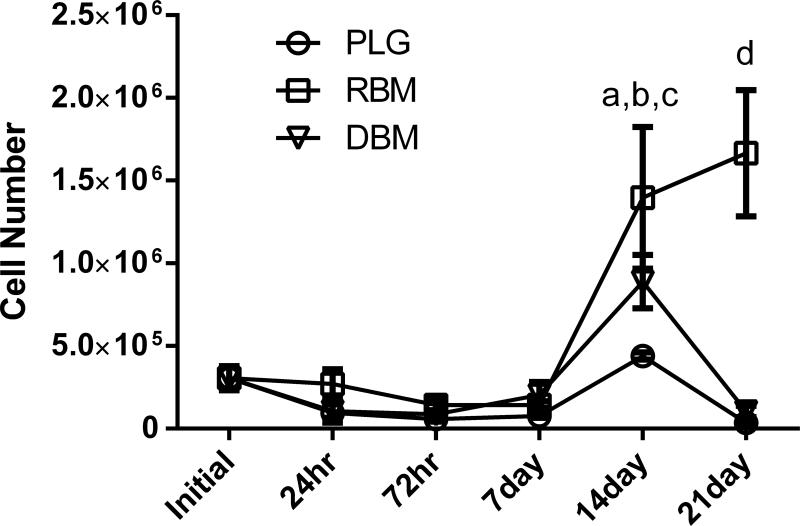
DNA content was used as an indicator of the number of cells present on each scaffold at each time point. There was no significant difference between the number of cells initially seeded on each matrix and the number of cells present after 24 hours. Additionally, there was no difference in cell number between DBM, RBM, and PLG through the first week in culture. After 14 days in culture, both DBM (p<0.01) and RBM (p<0.001) demonstrated increased cell number compared to the 24 hour time point. This significant increase was no longer apparent by day 21 for DBM, but remained in RBM samples. When comparing DNA content between groups, there was a significantly greater number of cells present on the RBM compared to PLG at the 14 day time point (p<0.01), and this difference remained intact at 21 days, with the RBM showing significantly higher cell number than both DBM and PLG matrices (p<0.001) (Fig. 5).
Figure 5.
Average cell number per matrix, estimated by DNA content. No significant differences in cell number within or between groups are present until the 14 day time point, when DBM was increased compared to 24 hours (ap<0.01) and RBM was increased compared to both initial cell count and 24 hours (bp<0.001). RBM had significantly greater cell number than PLG at 14 days (cp<0.01) and was significantly greater than both PLG and DBM at 21 days (dp<0.001). Both DBM and RBM groups had AR treatment prior to in vitro study.
3.4.2 Gene expression measured via RT-PCR
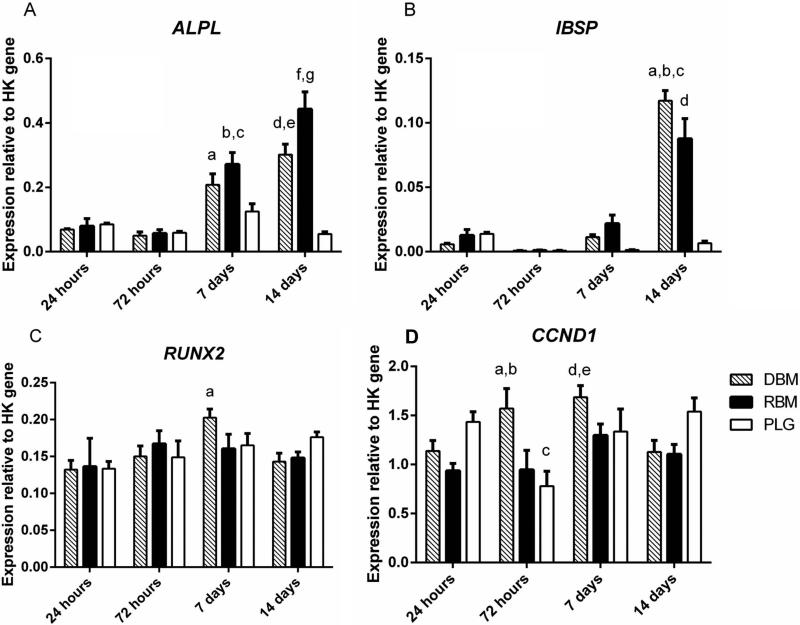
qPCR was used to measure the expression of several osteogenic genes in MSCs, as well as CCND1 (Cyclin D1), a gene involved in the progression of cells through the G1 and S phases of the cell cycle (Fig 6).
Figure 6.
Quantitative polymerase chain reaction analysis of hMSC gene expression. DBM, RBM, and PLG scaffolds seeded with MSCs and collected after 24 hours, 72 hours, 14 days, or 21 days.
ALP: ap<0.01 compared to DBM at 24/72 hours; bp<0.001 compared to RBM at 24/72 hours; cp<0.01 compared to PLG at 7 days; dp<0.001 compared to DBM at 24/72 hours; ep<0.001 compared to PLG at 14 days; fp<0.001 compared to RBM at 24/72 hours, 14 days; gp<0.001 compared to DBM/PLG at 14 days
IBSP: ap<0.001 compared to DBM at 24/72 hours, 7days; bp<0.001 compared to PLG at 14 days; cp<0.05 compared to RBM at 14 days; dp<0.001 compared to PLG at 14 days
Runx2: ap<0.05 compared to DBM at 24
CyclinD1: ap<0.05 compared to RBM at 72 hours; bp<0.01 compared to PLG at 72 hours; cp<0.05 compared to PLG at 24 hours; dp<0.05 compared to RBM at 24/72 hours; ep<0.01 compared to PLG at 72 hours
ALP expression was initially steady, showing no difference between or within groups at 24 or 72 hours. After 7 days in culture, ALP expression significantly increased in MSCs cultured on both DBM and RBM compared to their expression at 72 hours (p<0.001). ALP expression on both RBM (p<0.001) and DBM (p<0.01) was significantly greater than ALP expression on PLG at 72 hours. ALP gene expression on RBM continued to increase and by day 14 was significantly greater than both DBM (p<0.05) and PLG (p<0.001). ALP expression in cells cultured on PLG matrices was unchanged at all measured time points.
IBSP gene expression showed a similar trend to ALP, with no differences present between or within groups up to the first 72 hours. Expression began to trend upward by day 7, although no significant increases in IBSP occurred until day 14 when both DBM and RBM were significantly increased compared to all other time points within the group (p<0.001), as well as all PLG time points (p<0.001). At day 14, IBSP expression was increased in the DBM group compared to RBM (p<0.05). PLG showed no significant change in IBSP expression across all measured time points.
Expression of Runx2 remained stable for the duration of culture, with the only significant change being an increase in expression on DBM from 72 hours to 7 days (p<0.05).
Within the groups, there was no significant change in CCND1 (Cyclin D1) in RBM from 24 hours up to 14 days, while DBM had a slight increasing trend up to day 7 before decreasing at day 14 (p<0.05). PLG demonstrated a more cyclic expression pattern, decreasing between 24 and 72 hours (p<0.05) before increasing at 7 and 14 days (p<0.05). There was no difference in Cyclin D1 expression between groups at 24 hours, although by 72 hours DBM had increased compared to both RBM (p<0.05) and PLG (p<0.01). The difference in expression between groups disappeared by 7 and 14 days.
4. Discussion
We previously published a method to fabricate RBMs with increased mechanical properties compared to DBM, a commonly used material in orthopaedic medical procedures 23,24. The goals of this study were to improve and further characterize the RBM generated with our protocol. The remineralization process was modified to include an AR step and the capacity of RBM to be used as a scaffold for BTE applications was examined in vitro.
In this study we determined that ASI-remineralization introduces a brushite mineral phase throughout DBM. The inorganic phase of native bone is hydroxyapatite (Ca10(PO4)6OH2), a naturally occurring mineral that forms within the collagen fibrils. This material imparts the stiffness and compressive strength characteristic of the skeleton. It has also been shown to be biocompatible, with the ability to support attachment, proliferation, and viability of osteoprogenitor cells 16. Furthermore, there are a number of different calcium phosphate minerals of varying chemical formulas, crystallinity, and maturity which form under various conditions 25, some of which have demonstrated mildly osteogenic characteristics in vitro due to the ability to induce differentiation of progenitor cells down an osteogenic lineage 26. Brushite in particular has been studied as a substrate for bone repair in vitro and in vivo and has demonstrated biocompatibility and the ability to support cell proliferation and differentiation 27,28. Brushite cement is often used in surgical applications due to its biocompatibility as well as fast resorption times compared to apatite cements 29,30. Drawbacks reported for the surgical use of brushite cement include difficulty handling, quick setting time, and low mechanical properties. By nucleating and growing a brushite phase within natural bone matrix, our ASIRBM mitigates these issues and provides increased opportunity for clinical applications. The similar spectra between RBM(+) and RBM(−) specimens indicated that although the AR treatment enables the ASI remineralization process to incorporate significantly more mineral leading to a significantly stiffer substrate, the mineral type remains the same.
We observed a significant unanticipated result in that RBM(+) specimens that underwent AR accumulated significantly more mineral within the bulk of the collagen matrix, resulting in stiffer matrices with less variance between specimens. MicroCT analysis demonstrated that not only do RBM(+) specimens have more mineral (increased MVF), but this mineral is more highly connected, has less space between mineral clusters, and is more directionally dependent, all of which explain increased stiffness following treatment. While these morphometry measures are generally used to calculate parameters associated with native trabecular bone, they also represent fundamental topological properties. Just as measurements of architecture can complement measures of bone mass for assessing fracture risk, we believe the morphological data can be used to complement MVF and apparent density in order to characterize RBM and provide information about the effect of AR on ASI remineralization.
Consistent with our initial hypothesis, the AR process was able to remove proteins from equine bone matrix that act as physiological mineralization inhibitors. Five proteins which have implications in mineralization processes showed greater spectrum counts in the AR supernatant vs. control. These proteins were: α-2 HS-glycoprotein (nearly 50-fold increase in AR supernatant compared to control), osteonectin, alkaline phosphatase, and SIBLING proteins: osteopontin-1 (greater than 3-fold increase) and bone sialoprotein (present in AR supernatant, not control), α-2 HS-glycoprotein (fetuin) is a common mineralization inhibitor – a naturally occurring serum protein that works in vivo to inhibit pathological mineralization 31. In addition to known in vivo functions, fetuin has also been used in vitro and in BTE applications to eliminate unwanted solution calcification during remineralization experiments 32. This protein is embedded within bone matrix where it can transiently bind mineral species, creating a diffusion barrier which limits crystal growth 31. By removing this barrier to mineralization, the matrix is more effectively remineralized and stiffened with the ASI treatment. Likewise, osteopontin (OPN) inhibits the formation of HA in vitro 33, and there is evidence to suggest that, in vivo, OPN might prevent premature precipitation of calcium phosphate crystals lacking the well-defined structure of HA 34. This function could certainly have an effect on the nucleation and growth of the less-crystalline brushite in our system. The SIBLING proteins are known to affect bone formation in vivo and hydroxyapatite formation in solution 33. Osteonectin (ON), also known as SPARC (secreted protein, acidic and rich in cysteine), binds to collagen and contains multiple calcium-binding domains which allow it to play a role in matrix mineralization. It has been shown to inhibit apatite formation in vitro, likely by blocking mineral growth sites. Nearly all of these NCPs have either inhibitory or supportive effects on mineral formation, depending on conditions. Effects can vary based on attachment to a substrate or presence in solution, as well as the identity of other proteins and mineral ions in the local microenvironment. By removing a number of these proteins, we have demonstrated a method to improve functional mechanical outcomes resulting from our ASI remineralization, while reducing the variability inherent in the process.
We successfully cultured hMSCs on DBM and RBM, as well as a porous PLG polymer scaffold. Seeding efficiency can be determined as the percentage of cells on each matrix at 24 hours, compared to the initial number of cells seeded (3×105). While differences were not quite statistically significant, the RBM had a seeding efficiency of 90%, compared to 35% and 32% for DBM and PLG, respectively. This could indicate that the presence of mineral within the matrix improved cell adhesion. For all matrices, the cell number remained steady initially, with only non-significant decreases during the first week. After this time, the number of cells on both DBM and RBM began to increase, demonstrating the ability of these natural matrices to support cell proliferation. By 21 days, the DNA content on the RBM continued to increase, resulting in an approximate 5-fold increase from initial seeding. By demonstrating the capacity to support cell survival and proliferation over time, we can conclude that our RBMs have suitable biocompatibility for use as cell culture scaffolds in vitro.
Additionally, we probed the expression of several genes relevant to MSC osteogenic differentiation, as well as cell cycle processes. We did not detect a sustained difference in the expression of Cyclin D1 within or between groups. This indicates that neither DBM nor RBM have a detrimental effect on temporal regulation of MSC mitotic events compared to PLG, a common FDA approved bone tissue replacement material. We also did not detect a significant difference in the expression of Runx2 between groups over time. As the “master transcription factor” for osteogenic differentiation, Runx2 is responsible for beginning the process of differentiating osteogenic precursors (MSCs) into pre-osteoblasts. Once activated, a cascade is initiated which induces additional osteogenic genes including ALPL and IBSP. Runx2 does not cause increased expression of bone matrix genes in mature osteoblasts. Once expressed above a certain threshold sufficient to begin differentiation processes, Runx2 remains active until later stages of differentiation; however, expression of the gene remains steady during this early time period 35. In our study, it is likely that this gene was stimulated very early in culture for all substrates studied, potentially as a result of MSC exposure to an osteogenic medium. The ALPL gene encodes for the tissue non-specific alkaline phosphatase enzyme, a molecule implicated in bone formation due to its breakdown of pyrophosphate, an inhibitor of mineralization 36. We monitored changes in ALP expression as an indicator of early osteogenic differentiation and saw significant increases in both DBM and RBM after 1 week in culture, with differences remaining at 2 weeks. Both DBM and RBM also saw increased expression of IBSP, an osteogenic gene associated with mature osteoblast function. This difference first became significant following two weeks in culture. A number of investigators have shown increased osteogenic gene expression in MSCs cultured on PLG in the presence of osteogenic supplements. It is possible that the differences seen between DBM and PLG are a result of the natural osteoinductivity of DBM. Additionally, the failure of a large portion of cells to adhere initially to PLG and DBM likely delayed the significant expression of osteogenic genes, as surviving cells focused on processes such as adhesion and proliferation at early time points.
By incorporating an AR technique in the preparation of RBM, we improved the functional outcomes of ASI remineralization and demonstrated the ability to remove NCPs from the matrix that are known to inhibit mineralization. Our next steps include identifying specific antigens removed from the tissue and assessing the safety and efficacy of RBM as a biomaterial using an in vivo animal model. The current work shows increased expression of osteogenic markers from MSCs cultured on DBM and RBM, indicating that these naturally derived matrices support MSC osteogenic differentiation in 3D culture. This, coupled with demonstrated biocompatibility and increased mechanical properties following ASI-remineralization, makes RBM an attractive substrate for further study regarding bone tissue engineering applications.
Acknowledgements
We thank Tanya Garcia-Nolen for training on x-ray and microCT; Maelene Wong for contributions to AR; Beko Binder, Alice Wong and Chrisoula Toupadakis Skouritakis for assistance with RNA isolation and PCR; Ryoji Shiraki for help with FTIR; and Darren Weber for guidance with LC-MS. This study was supported by National Institutes of Health (NIH) grant AR040776, the David Linn Endowed Chair in Orthopaedic Surgery (DPF), a Schwall Medical Research Fellowship, and Howard Hughes Medical Institute Training Fellowship: Integrating Medicine into Basic Science (HHMI: IMBS) (MAS).
References
- 1.Healthcare Cost and Utilization Project (U.S.), United States. Agency for Healthcare Research and Quality . HCUP facts and figures statistics on hospital-based care in the United States. Agency for Healthcare Research and Quality; Rockville, Md.: [Google Scholar]
- 2.Kim SW, Jung HD, Kang MH, Kim HE, Koh YH, Estrin Y. Fabrication of porous titanium scaffold with controlled porous structure and net-shape using magnesium as spacer. Mater Sci Eng C Mater Biol Appl. 2013;33(5):2808–15. doi: 10.1016/j.msec.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 3.Selvam S, Chang WV, Nakamura T, Samant DM, Thomas PB, Trousdale MD, Mircheff AK, Schechter JE, Yiu SC. Microporous poly(L-lactic acid) membranes fabricated by polyethylene glycol solvent-cast/particulate leaching technique. Tissue Eng Part C Methods. 2009;15(3):463–74. doi: 10.1089/ten.tec.2008.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viateau V, Manassero M, Sensebe L, Langonne A, Marchat D, Logeart-Avramoglou D, Petite H, Bensidhoum M. Comparative study of the osteogenic ability of four different ceramic constructs in an ectopic large animal model. J Tissue Eng Regen Med. 2013 doi: 10.1002/term.1782. [DOI] [PubMed] [Google Scholar]
- 5.Martin RB, Burr DB, Sharkey NA. Skeletal tissue mechanics. xiv. Springer; New York: 1998. p. 392. [Google Scholar]
- 6.Giannoudis PV, Dinopoulos H, Tsiridis E. Bone substitutes: an update. Injury. 2005;36(Suppl 3):S20–7. doi: 10.1016/j.injury.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 7.He J, Genetos DC, Leach JK. Osteogenesis and trophic factor secretion are influenced by the composition of hydroxyapatite/poly(lactide-co-glycolide) composite scaffolds. Tissue Eng Part A. 2010;16(1):127–37. doi: 10.1089/ten.tea.2009.0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanungo BP, Silva E, Van Vliet K, Gibson LJ. Characterization of mineralized collagen-glycosaminoglycan scaffolds for bone regeneration. Acta Biomaterialia. 2008;4(3):490–503. doi: 10.1016/j.actbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 9.Soicher MA, Christiansen BA, Stover SM, Leach JK, Fyhrie DP. Remineralization of demineralized bone matrix (DBM) via alternating solution immersion (ASI). J Mech Behav Biomed Mater. 2013 doi: 10.1016/j.jmbbm.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allori AC, Sailon AM, Warren SM. Biological basis of bone formation, remodeling, and repair-part II: extracellular matrix. Tissue Eng Part B Rev. 2008;14(3):275–83. doi: 10.1089/ten.teb.2008.0083. [DOI] [PubMed] [Google Scholar]
- 11.Gentili C, Bianco P, Neri M, Malpeli M, Campanile G, Castagnola P, Cancedda R, Cancedda FD. Cell proliferation, extracellular matrix mineralization, and ovotransferrin transient expression during in vitro differentiation of chick hypertrophic chondrocytes into osteoblast-like cells. J Cell Biol. 1993;122(3):703–12. doi: 10.1083/jcb.122.3.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makhluf HA, Mueller SM, Mizuno S, Glowacki J. Age-related decline in osteoprotegerin expression by human bone marrow cells cultured in three-dimensional collagen sponges. Biochem Biophys Res Commun. 2000;268(3):669–72. doi: 10.1006/bbrc.2000.2182. [DOI] [PubMed] [Google Scholar]
- 13.Murphy WL, Hsiong S, Richardson TP, Simmons CA, Mooney DJ. Effects of a bone-like mineral film on phenotype of adult human mesenchymal stem cells in vitro. Biomaterials. 2005;26(3):303–10. doi: 10.1016/j.biomaterials.2004.02.034. [DOI] [PubMed] [Google Scholar]
- 14.Tsiridis E, Ali Z, Bhalla A, Gamie Z, Heliotis M, Gurav N, Deb S, DiSilvio L. In vitro proliferation and differentiation of human mesenchymal stem cells on hydroxyapatite versus human demineralised bone matrix with and without osteogenic protein-1. Expert Opin Biol Ther. 2009;9(1):9–19. doi: 10.1517/14712590802622473. [DOI] [PubMed] [Google Scholar]
- 15.Goncalves AC, Griffiths LG, Anthony RV, Orton EC. Decellularization of bovine pericardium for tissue-engineering by targeted removal of xenoantigens. J Heart Valve Dis. 2005;14(2):212–7. [PubMed] [Google Scholar]
- 16.Shahabipour F, Mahdavi-Shahri N, Matin MM, Tavassoli A, Zebarjad SM. Scaffolds derived from cancellous bovine bone support mesenchymal stem cells’ maintenance and growth. In Vitro Cell Dev Biol Anim. 2013;49(6):440–8. doi: 10.1007/s11626-013-9591-7. [DOI] [PubMed] [Google Scholar]
- 17.Wong ML, Leach JK, Athanasiou KA, Griffiths LG. The role of protein solubilization in antigen removal from xenogeneic tissue for heart valve tissue engineering. Biomaterials. 2011;32(32):8129–38. doi: 10.1016/j.biomaterials.2011.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Wong ML, Wong JL, Athanasiou KA, Griffiths LG. Stepwise solubilization-based antigen removal for xenogeneic scaffold generation in tissue engineering. Acta Biomater. 2013;9(5):6492–501. doi: 10.1016/j.actbio.2012.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Yeni YN, Schaffler MB, Gibson G, Fyhrie DP. Prestress due to dimensional changes caused by demineralization: A potential mechanism for microcracking in bone. Annals of Biomedical Engineering. 2002;30(2):217–225. doi: 10.1114/1.1451078. [DOI] [PubMed] [Google Scholar]
- 20.Harris LD, Kim BS, Mooney DJ. Open pore biodegradable matrices formed with gas foaming. J Biomed Mater Res. 1998;42(3):396–402. doi: 10.1002/(sici)1097-4636(19981205)42:3<396::aid-jbm7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 21.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Analytical and Bioanalytical Chemistry. 2007;389(4):1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 22.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76(14):4193–201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 23.Iwata H, Sakano S, Itoh T, Bauer TW. Demineralized bone matrix and native bone morphogenetic protein in orthopaedic surgery. Clin Orthop Relat Res. 2002;(395):99–109. doi: 10.1097/00003086-200202000-00010. [DOI] [PubMed] [Google Scholar]
- 24.Zimmermann G, Moghaddam A. Allograft bone matrix versus synthetic bone graft substitutes. Injury. 2011;42(Suppl 2):S16–21. doi: 10.1016/j.injury.2011.06.199. [DOI] [PubMed] [Google Scholar]
- 25.Nancollas GH, LoRe M, Perez L, Richardson C, Zawacki SJ. Mineral phases of calcium phosphate. Anat Rec. 1989;224(2):234–41. doi: 10.1002/ar.1092240213. [DOI] [PubMed] [Google Scholar]
- 26.Barradas AM, Monticone V, Hulsman M, Danoux C, Fernandes H, Tahmasebi Birgani Z, Barrere-de Groot F, Yuan H, Reinders M, Habibovic P. Molecular mechanisms of biomaterial-driven osteogenic differentiation in human mesenchymal stromal cells. Integr Biol (Camb) 2013;5(7):920–31. doi: 10.1039/c3ib40027a. others. [DOI] [PubMed] [Google Scholar]
- 27.Klammert U, Reuther T, Jahn C, Kraski B, Kubler AC, Gbureck U. Cytocompatibility of brushite and monetite cell culture scaffolds made by three-dimensional powder printing. Acta Biomater. 2009;5(2):727–34. doi: 10.1016/j.actbio.2008.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Tamimi F, Kumarasami B, Doillon C, Gbureck U, Le Nihouannen D, Cabarcos EL, Barralet JE. Brushite-collagen composites for bone regeneration. Acta Biomater. 2008;4(5):1315–21. doi: 10.1016/j.actbio.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Apelt D, Theiss F, El-Warrak AO, Zlinszky K, Bettschart-Wolfisberger R, Bohner M, Matter S, Auer JA, von Rechenberg B. In vivo behavior of three different injectable hydraulic calcium phosphate cements. Biomaterials. 2004;25(7-8):1439–51. doi: 10.1016/j.biomaterials.2003.08.073. [DOI] [PubMed] [Google Scholar]
- 30.Schneider G, Blechschmidt K, Linde D, Litschko P, Korbs T, Beleites E. Bone regeneration with glass ceramic implants and calcium phosphate cements in a rabbit cranial defect model. J Mater Sci Mater Med. 2010;21(10):2853–9. doi: 10.1007/s10856-010-4143-0. [DOI] [PubMed] [Google Scholar]
- 31.Heiss A, DuChesne A, Denecke B, Grotzinger J, Yamamoto K, Renne T, Jahnen-Dechent W. Structural basis of calcification inhibition by alpha 2-HS glycoprotein/fetuin-A. Formation of colloidal calciprotein particles. J Biol Chem. 2003;278(15):13333–41. doi: 10.1074/jbc.M210868200. [DOI] [PubMed] [Google Scholar]
- 32.Price PA, Toroian D, Lim JE. Mineralization by inhibitor exclusion: the calcification of collagen with fetuin. J Biol Chem. 2009;284(25):17092–101. doi: 10.1074/jbc.M109.007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen CJ. Primer on the metabolic bone diseases and disorders of mineral metabolism. American Society for Bone and Mineral Research; Washington, D.C.: 2009. American Society for Bone and Mineral Research. [Google Scholar]
- 34.Roach HI. Why does bone matrix contain non-collagenous proteins? The possible roles of osteocalcin, osteonectin, osteopontin and bone sialoprotein in bone mineralisation and resorption. Cell Biol Int. 1994;18(6):617–28. doi: 10.1006/cbir.1994.1088. [DOI] [PubMed] [Google Scholar]
- 35.Franceschi RT, Ge CX, Xiao GZ, Roca H, Jiang D. Transcriptional regulation of osteoblasts. Skeletal Biology and Medicine, Pt A. 2007;1116:196–207. doi: 10.1196/annals.1402.081. [DOI] [PubMed] [Google Scholar]
- 36.Risteli L, Risteli J. Biochemical markers of bone metabolism. Ann Med. 1993;25(4):385–93. doi: 10.3109/07853899309147301. [DOI] [PubMed] [Google Scholar]