Abstract
The light-based control of ion channels has been transformative for the neurosciences, but the optogenetic toolkit does not stop there. An expanding number of proteins and cellular functions have been shown to be controlled by light, and the practical considerations in deciding between reversible optogenetic systems (such as systems that use light-oxygen-voltage domains, phytochrome proteins, cryptochrome proteins and the fluorescent protein Dronpa) are well defined. The field is moving beyond proof of concept to answering real biological questions, such as how cell signalling is regulated in space and time, that were difficult or impossible to address with previous tools.
With the proper reagents, light can be used to observe and perturb the spatiotemporal dynamics of signals in living cells and organisms. The first attempts to acutely control cell signalling with light chemically ‘caged’ small molecule messengers by covalently attaching photolabile chemical groups at positions that are necessary for signalling. Upon exposure to light, these groups would cleave and dissociate, thereby ‘uncaging’ the molecule to signal in the cell. However, the engineering challenges in making these tools suitable for diverse signalling pathways and the difficulty in delivering them to cells and organisms limited their use1,2. Then optogenetics came along — the genetic encoding of light-sensitive proteins that activate signalling pathways in response to light. Its first application was the use of light-gated ion channels to manipulate the excitability of neuronal cells3–5. With optogenetics, it no longer takes a chemist to produce the light-sensitive reagents, uncaging is no longer irreversible and the light-controlled proteins are much easier to deliver (and thus a greater level of spatial control is possible), because they can be expressed rather than injected. Investigators have taken advantage of the spatial precision of proteins that either hyperpolarize or depolarize neurons3–6 to non-invasively identify the pacemaker cells in the zebrafish heart7, and used the temporal precision and reversibility of these proteins to elucidate the importance of timing in neuronal activity for behavioural conditioning8.
A limitation of these neuronal optogenetic tools is that they can only control membrane potential, and there are a wide range of other cellular and developmental biology questions that require the manipulation of other processes that affect cell signalling, such as protein localization, post-translational modification, GTP loading, and so on. With the adoption of other genetically encoded light-responsive proteins, the optogenetic toolkit has markedly expanded to include a wide array of regulatory proteins, and consequently cellular functions, which can now be controlled with light. Here, we first review the various optogenetic systems and practical considerations in using them. Then, we address the types of cell signalling questions that are being investigated with these approaches. Finally, we discuss future opportunities for the development of optogenetic tools.
Overview of optogenetic systems
Proteins that change conformation in response to light have been adapted to regulate a wide array of signalling activities in living cells. Here, we discuss the optogenetic systems that are reversible and can be adopted to control a variety of signalling pathways. Three are based on photosensitive plant proteins (cryptochromes9–11, light-oxygen-voltage (LOV) domains12–15 and phytochromes16–18), and one is based on the fluorescent protein Dronpa19, which was isolated from the coral Pectiniidae20. Other recent publications discuss the use of optogenetic proteins that manipulate specific signalling events, such as those that regulate neuronal excitability4,21, cyclic nucleotides22,23 and heterotrimeric G protein signalling24,25, or proteins that are irreversibly activated26–28 or inactivated29 by light.
The PHYTOCHROME B protein
PHYTOCHROME B (PHYB) is a protein that is activated by red light (650 nm) and inactivated by infrared light (750 nm), and normally controls seedling stem elongation in Arabidopsis thaliana. When expressed in cells, the apo-PHYB protein (which is chromophore free) only becomes light sensitive when it autocatalytically ligates to PCB, a chromophore that is present in photosynthetic organisms; however, in non-photosynthetic organisms, PCB must be delivered to cells directly or through the expression of the bio-synthetic enzymes that produce it30,31. Upon exposure to red light, PHYB that is bound to PCB changes conformation and binds to a PHYTOCHROME INTERACTING FACTOR (PIF) protein16 within seconds. This association is reversed within seconds upon exposure to infrared light or is stable for hours in the dark18.
The CRYPTOCHROME 2 protein
CRYPTOCHROME 2 (CRY2) is a protein from A. thaliana that is sensitive to blue light (405–488 nm). Two changes occur upon exposure to blue light: the light-sensitive CRY2 protein homo-oligomerizes11 and binds to its binding partner, CIB1 (CRYPTOCHROME-INTERACTING BASIC HELIX–LOOP–HELIX 1)32, both within seconds10. In the dark, CRY2 previously activated with blue light resets to its initial state within ~5 minutes. CRY2 uses the ubiquitously expressed endogenous flavin as its chromophore.
The LOV domains
The LOV sensory domains from several different organisms have been successfully used as optogenetic tools. They are all sensitive to blue light (440–473 nm) and use ubiquitously expressed endogenous flavin as a chromophore. The LOV systems differ in how each one uses the light-induced conformational change to regulate cell signalling. One approach directly fuses the LOV domain to an effector protein and relies on the light-induced conformational change in the LOV domain to relieve the autoinhibition14. In some optogenetic LOV systems, the LOV domains heterodimerize with natural or engineered binding partners to recruit signalling domains15, whereas in other systems the domains homodimerize and bind to DNA, thereby regulating gene expression33,34.
The Dronpa protein
Dronpa is a well-known photoactivatable fluorescent protein. Photoactivation not only changes the fluorescence of Dronpa but also changes its quaternary structure. In the ‘dark state’ (that is, not photoactivated) Dronpa exists as a monomer, and in the fluorescent state it exists as a dimer. As Dronpa has a low affinity for itself in the dimjeric state, the system is most robustly used by fusing a copy of Dronpa to the amino and carboxyl termini of a protein of interest. When Dronpa is activated with light at a wavelength of 390 nm, the Dronpa domains at either end bind to each other, which inhibits the function of the protein of interest by obscuring or altering the active conformation. The extent of protein inhibition can be tracked by the accompanying increase in Dronpa fluorescence. This change can be reversed with light at a wavelength of 490 nm, which converts Dronpa back to a monomer19. The system requires no small-molecule chromophore.
Optogenetic control of cell signalling
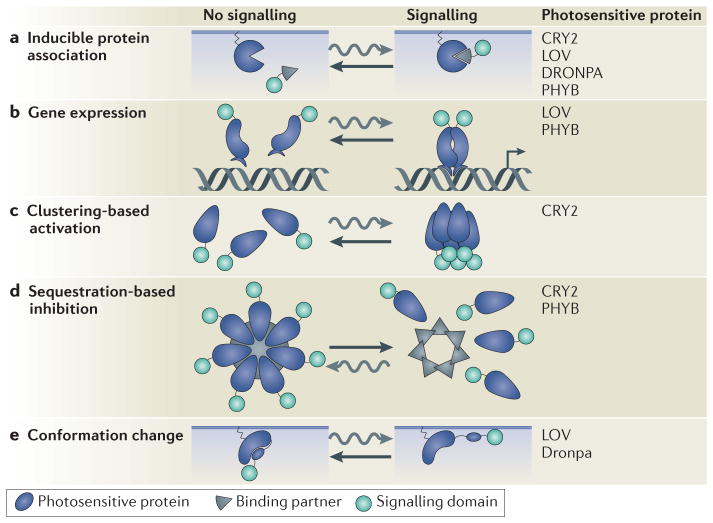
There are five general strategies to manipulate intracellular signals with optogenetic proteins (FIG. 1).
Figure 1. Different strategies for optogenetic inputs.
Optogenetically stimulated signals can be induced in various ways (the photosensitive proteins that have been used for each approach are listed). System reversion occurs either in the dark or can be stimulated with light depending on the system used (BOX 1). a | Heterodimerization is used to recruit a signalling domain to its substrate, which is commonly located on the plasma membrane. b | Homodimerization and heterodimerization techniques recruit transcriptional activators or other DNA-modifying proteins to the DNA to initiate the expression of a gene of interest. c | CRYPTOCHROME 2 (CRY2) naturally clusters when it is activated. By fusing CRY2 with signalling domains, the activities of which depend on domain density, signalling can be activated with light. d | Alternatively, signalling can be inhibited by sequestering a signalling protein away from its site of action. Proteins can be sequestered in cytosolic clusters or recruited to compartments away from their downstream effectors or upstream activators. e | Conformational changes in the photosensitive protein can expose a concealed signalling domain or relieve a protein from an allosterically autoinhibited state. LOV, light-oxygen-voltage; PHYB, PHYTOCHROME B. Curvy arrows indicate the response of the system to light, whereas straight arrows indicate dark- or light-stimulated reversion. The small arrow on the DNA represents active transcription.
Inducible protein associations
The most widely used strategy of manipulating intra-cellular signals using optogenetics takes advantage of light-induced conformational changes to promote the association of two polypeptides. These changes typically cause heterodimerization of a light-responsive protein and its effector. Signalling proteins can be either recruited to or away from their normal site of action, thereby activating or inhibiting intracellular signals (FIG. 1a). The advantage of this technique is that it quickly initiates or terminates signalling and that it is adaptable to many applications. Protein associations and dissociations, and changes in protein localization are ubiquitous modes of regulating cell signalling, and many cellular events have been brought under optogenetic control by exploiting these features. Examples include: CRY2–CIB-based regulation of inositol polyphosphate 5-phosphatase OCRL35, PI3K35 and RAF36; the PHYB–PIF-based regulation of actin polymerization37, cyclins38, CDC42 (REF. 18), lac signalling38, PI3K39, RAC18, RAS40 and RHO18; and the LOV domain heterodimerization-based regulation of CDC42 and MAPK15. A limitation of the heterodimerization-based approach is that the involvement of additional proteins increases the complexity of system optimization.
Gene expression
Cell signalling can also be induced by controlling the expression of a gene of interest. Heterodimerization strategies using CRY2–CIB10,41, PHYB–PIF17 and LOV42 domains, as well as a LOV-domain-homodimerization approach33,34, have successfully regulated transcription by localizing a transcriptional activator to a promoter (FIG. 1b). Other means of protein expression that can be modulated by optogenetics include Cre–loxP-based recombination10, chromatin modifications41, splicing43 and translation44. An advantage is that new pathways can be brought under optogenetic control by simply changing the coding sequence of the expressed protein in the DNA. However, regulating the synthesis of signalling proteins may not be fast enough to study many post-translational signalling pathways.
Clustering-based activation
Some proteins oligomerize in response to light, and this can be used to drive the clustering of signalling proteins that are required in high local concentrations to activate signalling cascades (FIG. 1c). This technique has been used for cryptochrome-based regulation of β-catenin and RHO in the context of transcription and cytoskeletal rearrangements, respectively11. A downside of this strategy is that it can be difficult to quantify the number of molecules in the induced clusters, so measuring the strength of the optogenetic input can be challenging, particularly in real time.
Sequestration-based inhibition
Alternatively, similar to the use of organelle-based recruitment, clustering can be used to sequester proteins away from their site of action38 (FIG. 1d). This technique has recently been used to inhibit several actin cytoskeleton regulators and as a general technique to regulate signalling proteins fused to GFP45. Sequestration is potentially a very general approach, because one only needs to recruit a protein away from its site of action rather than rely on the natural propensity of a protein to signal when clustered.
Intramolecular control of protein function
Finally, light can be used to induce an intra-molecular conformational change to generate an active signalling protein (FIG. 1e). This technique has been used in conjunction with LOV domains to regulate CDC42 and RAC14, and formins46 in the context of cell migration, in association with degrons to regulate protein degradation47 and in conjunction with Dronpa to regulate CDC42 and proteinase K19. The advantage of this approach is that it only requires the expression of one protein. It is also one of the few optogenetic approaches that does not involve transcription or light-sensitive ion channels and that has been used in multicellular organisms48,49. One of the limitations of this strategy is that intra-molecular inhibition can be challenging to engineer. As a consequence, fewer tools have been developed for this approach than for the more general dimerization-based techniques.
These are the five common ways in which reversible optogenetic protein systems have been used to drive cellular signalling. However, other important differences between optogenetic approaches need to be considered to ensure that the best one is used for a particular experiment.
Choosing an optogenetic system
There are several considerations when choosing an appropriate optogenetic system. These are summarized in BOX 1 and explained below.
Box 1. Comparison of the reversible photosensitive proteins used in optogenetic systems.
The four photosensitive proteins that are at the core of current reversible optogenetic systems are compared (see the table). Note that some photosensitive proteins actually represent a collection of proteins from different organisms and have been used by different groups to control different signalling systems. This is particularly true of the light-oxygen-voltage (LOV) domains. This table summarizes the features for the entire classes of photosensitive proteins; the features will vary based on the particular protein used.
Column heading definitions
Turn-on speed
The speed with which the system activates when illuminated with stimulatory light (λon).
Turn-off speed
The speed with which the system resets in the dark or when illuminated with inhibitory light (λoff).
Chromophore requirement
Lists the small molecule, if any, that is needed to make the protein photosensitive and whether it is naturally found in the cell or has to be provided.
Compatible imaging wavelengths
These wavelengths of light are not markedly stimulatory and can be used to image other fluorophores without notably activating the optogenetic system.
λon
The wavelength (or wavelengths) of light that is most effective at activating the system. Wavelengths outside these ranges could still activate the system but may require higher intensities and/or longer exposures.
λoff
The wavelength, if any, that actively resets the system. Wavelengths outside this range could still inhibit the system but may require higher intensities and/or longer exposures.
Effector affinity
The order of magnitude approximation of the dissociation constant for a system. Heterodimerization affinities are listed for the systems that are based on PHYB–PIF18 and the LOV-domain-based TULIP (tunable, light-controlled interacting protein) system15. The homodimerization affinity for Dronpa is listed19.
| Photosensitive protein |
Turn-on speed |
Turn-off speed (t1/2) |
Chromophore requirement |
Compatible imaging wavelengths (nm) |
λon (nm) | λoff (nm) | Effector affinity |
Refs |
|---|---|---|---|---|---|---|---|---|
| PHYB | Seconds |
|
PCB; exogenous or synthesized in situ | ≤514 | 650 | 750 |
|
16–18 |
| CRY2 | Seconds | 5 minutes | Flavin; endogenous | ≥561 | 405–488 | NA | Not determined | 9–11 |
| LOV | Seconds | Tens of seconds to minutes | Flavin; endogenous | ≥514 | 440–473 | NA |
|
12–15, 67 |
| Dronpa | Seconds |
|
None | ≥600 | 390 | 490 |
|
19 |
CRY2, CRYPTOCHROME 2; NA, not applicable; PHYB, PHYTOCHROME B; PIF, PHYTOCHROME INTERACTING FACTOR.
Speed of system reversal
The faster an optogenetic system can be reversed, the more precisely intracellular signals can be manipulated in space and time. Importantly, the rate at which optogenetic systems can be turned off varies by several orders of magnitude, ranging from seconds (for phytochromes inactivated by light), tens of seconds (for LOV domains and Dronpa) and minutes (for CRY2) to hours (for phytochromes inactivated in the dark). Furthermore, some systems such as systems based on LOV domains and CRY2 spontaneously revert to the dark state, whereas others such as those involving PHYB and Dronpa can be driven to the dark state by separate illumination wavelengths. Light-driven reversal tends to reduce basal activity, expand the dynamic range and offer faster turn-off times of optogenetic systems. Faster turn-off rates increase spatial control, as the activated molecule cannot diffuse far from the excitatory light before it is turned off.
Quantifying the input
Using optogenetic tools for more than qualitative experiments requires not only a means to quantify the dose of activating light but also a way to quantify the strength of the optogenetic input — that is, the amount of optogenetic proteins that are actively signalling. Owing to non-linearities in these systems, reducing the intensity or duration of activating light by 50% does not necessarily decrease the amount of the signalling complex formed to the same degree. Quantification is easiest with two-component optogenetic systems that regulate protein recruitment to the plasma membrane, in which changes in the localization of the signalling domain of interest can be analysed by fusing it to a fluorescent protein. For example, membrane recruitment of a fluorescently tagged PI3K subunit was used to quantify the strength of optogenetic activation of this enzyme50. Single-component systems typically require a live-cell reporter immediately downstream of the input to quantify the strength of optogenetic activation. An interesting exception is the Dronpa system. When used to intra-molecularly inhibit a protein, Dronpa ‘dims’ when converted to the signalling competent, monomeric state. Thus, it is important to note that evaluating intermediate levels of Dronpa activation requires the measurement of dim fluorescence on a light background.
Considering dynamic range
Optogenetic systems markedly vary in their dynamic range, which is typically a function of two parameters: the basal activation of the system and the affinity of the photoproteins for their binding partners. Higher dynamic ranges are preferable for manipulating cell signalling cascades over a wide range of expression levels of the optogenetic components. Cryptochrome- and LOV-domain-based systems have higher basal activation than systems involving PHYB and Dronpa, both of which can be forced into the off state through illumination with inactivating wavelengths. The affinity of photoproteins for their binding partners varies by several orders of magnitude from 100 nM (estimated PHYB–PIF6 affinity in cells)18, to 600 nM (measured PHYB–PIF3 affinity in vitro)37 to 10 μM (for Dronpa oligomerization)19. Taken together, the largest dynamic range is expected for the phytochrome system.
Ease of titration
The ease with which an input can be titrated to and maintained at a desired level (something that is necessary for analysing the equilibrium of cell signalling) depends on how quickly the system turns off. Producing stable, intermediate levels of a signal in systems that take minutes (rather than seconds) to turn off can be challenging and requires the level of activating light to be carefully controlled to counteract spontaneous inactivation. It is easiest to achieve a stable, intermediate amount of signalling complexes with the PHYB- and Dronpa-based systems, because they can turn off in seconds and can be actively inhibited with light, whereas the CRY2- and LOV-domain-based systems spontaneously revert. Thus, constant illumination with a fixed ratio of stimulatory light to inhibitory light can keep an input at a desired level. Furthermore, the fact that stimulatory and inhibitory light buffer each other can overcome the effects of incidental excitation that can occur while imaging other chromophores.
Chromophore requirement
Optogenetic proteins that use an endogenous chromophore, such as CRY2, LOV domains and Dronpa, are well suited for multicellular and developmental models, in which the delivery of an exogenous chromophore may be impractical. Although some progress has been made in synthesizing chromophores in situ for bacterial and mammalian cells in culture by co-expressing the appropriate enzymes from plants and algae30,31, this has not yet been demonstrated for multicellular organisms. In some contexts, such as when controlling irreversible pathways (for example, differentiation or apoptosis)51, having the system only respond to light upon the addition of an exogenous chromophore may be an advantage to limit the activation of a ‘leaky’ pathway.
Compatibility with fluorescent reporters
Finally, optogenetic inputs should ideally be paired with some way of measuring both the strength of the optogenetically induced signal and the activity of a live-cell reporter of the resulting downstream response. Such reporters frequently involve the use of fluorescent proteins, and it is important to know which ones can be safely imaged without activating the optogenetic input. Luckily, most optogenetic systems include at least two ‘safe’ spectra of wavelengths that can be imaged without markedly altering the activation of the system. For example, when investigating how son of sevenless (Sos; a RAS guanine nucleotide-exchange factor (GEF)) activates Erk, light at a wavelength of 514 nm was used to measure PIF–YFP–Sos recruitment to the plasma membrane (the input); and light at a wavelength of 405 nm was used to measure how the cell responded to PIF–YFP–Sos recruitment by tracking Erk–blue fluorescent protein translocation into the nucleus40. Cells exposed to light at wavelengths of 514 nm and 405 nm are safe, because these wavelengths do not markedly alter the PHYB–PIF interaction39.
Optogenetic hardware
As the discussed optogenetic systems are fairly sensitive to visible light, various common light sources on a microscope can activate them. Epifluorescent light, FRAP (fluorescence recovery after photobleaching) lasers with the power turned to a minimum, light-emitting diodes and even glass filters in the bright-field path have activated these optogenetic systems. The intensity of light from light-emitting diodes and some laser sources can be continuously varied, which makes them ideal for precisely titrating an optogenetic input or maintaining it at a specific level with feedback control39.
These light sources can be spatially restricted by closing down the corresponding field diaphragm. Single cells can be stimulated with a low magnification objective, whereas subcellular regions can be activated with a high magnification objective. Finer spatial control requires a digital micromirror device, an array of tiled mirrors that can be individually toggled to project an arbitrary pattern of light. Using a digital micromirror device, exact regions of a cell can be targeted (for example, just the dendrites of a neuron) or the pattern of illumination can be updated as the cell changes shape. Some FRAP systems already use a digital micromirror device and can be co-opted for optogenetic experiments once the power is greatly reduced.
Quantifying signal integration
Optogenetic tools strongly synergize with existing biological approaches. Biochemistry and genetics are often used to first identify signalling molecules and to discern how they activate and inhibit one another. However, assessing how these individual components are linked together to generate complex cellular behaviours can be much more challenging. In some instances, loss-of-function perturbations of single nodes in a pathway simply break a signalling circuit and are insufficient for understanding how each node functions in the overall network. Similarly, not all pathways are sufficiently understood to enable their full biochemical reconstitution, and even those that are well understood may exhibit different behaviours in vitro than in the cell.
For pathways that are sufficiently understood to enable signalling nodes to be manipulated by precise optogenetic inputs, it is now possible to carry out in vivo biochemical experiments52 to understand the function of individual subcircuits within complex signalling networks. Optogenetics can be used to both isolate distinct subcircuits and determine their logic of signal integration40.
Isolating distinct subcircuits
The easiest way to modify signalling cascades is by the addition of extracellular ligands, but activating cellular receptors induces many arms of a signalling pathway. These arms often interact with one another in a complex manner that may be difficult to disentangle when analysed as a whole. Optogenetic inputs can be used to manipulate downstream nodes in signalling cascades, thereby breaking down a large, complex pathway into a series of smaller, easier-to-understand units (FIG. 2a).
Figure 2. Optogenetics for in vivo biochemistry.
a | Assessing the function of an arm of a signalling pathway with precise, defined inputs is difficult with traditional tools, as endogenous ligands frequently activate upstream and parallel regulatory connections in addition to the signalling module under study (grey arrows). Optogenetic inputs circumvent this problem by initiating precise, defined signals at intermediate points in a pathway. By measuring the downstream response of a cell with a live-cell activity reporter, the function of the intervening signalling module can be inferred. b | Dose–response curves generated for a population of cells using western blots obscure single-cell behaviour. Single-cell techniques such as immunolabelling-based fluorescence-activated cell sorting (FACS) use fixed cells and, as one time point is not sufficient to generate a dose–response curve, the response of thousands of cells must be combined to do so. Cell-to-cell variation confounds the measurement of cell responses because of extrinsic differences in the responses, and the aggregation of data from multiple cells distorts the intrinsic behaviour of the pathway. The titratability, speed and reversibility of optogenetic inputs enable multiple measurements and thus the generation of complete dose–response curves for individual living cells40. This approach removes previous noise arising from cell heterogeneity and provides the most detailed view of the intrinsic precision of signalling pathways in living cells. Here, data are shown for light-gated activation of RAS (through PHYTOCHROME B (PHYB)–PHYTOCHROME INTERACTING FACTOR (PIF)-based recruitment of the Ras activator son of sevenless (SOS)) and its induction of ERK activation, as analysed by nuclear recruitment of a fluorescently tagged ERK40. The precision of optogenetic dose–response curves makes them ideal for understanding how a particular enzyme regulates a signalling pathway. c | Beyond determining whether drug treatment blocks a pathway, optogenetics can uncover how co-occurring cellular events regulate core pathways by carrying out several experiments on one cell, which enables it to be used as its own control. For example, kinase phosphorylation of a scaffold may increase protein binding and thus pathway sensitivity. Generating a dose–response curve using optogenetic techniques before and after kinase inhibition by the addition of a drug is a powerful approach for detecting these regulatory effects in a manner that is not confounded by cell-to-cell variation in response. d | Signalling pathways can respond to more than the steady-state amount of a signal, such as its rate of change or integrated amount (sum of input over time). As the optogenetic inputs can be varied in space, time and concentration, many types of input–output analyses can be conducted. All of these signalling attributes have been found to be important in different biological contexts40,51,54–57,68. Part b reprinted from Cell, 155/1430, Toettcher, J. E., Weiner, O. D. & Lim, W. A., Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module, 1422–1434, Copyright (2013), with permission from Elsevier.
Single-cell dose–response curves
Just as one can tell whether an enzyme functions in a cooperative manner in vitro by varying the amount of enzyme and measuring the resulting product, it is also possible to determine whether a part of a pathway functions in a graded or switch-like manner in vivo by varying the amount of upstream input and measuring the downstream response. Importantly, the speed and reversibility of optogenetic systems make it possible to generate dose–response curves for individual cells. It is well appreciated that population-level analysis tools such as western blotting obscure single-cell behaviours and give overall averages that can lead to incorrect conclusions regarding individual cell behaviour53. Single-cell analyses are preferable, but these are commonly implemented through techniques such as flow cytometry. Using this method, one datum is acquired per cell, and the overall population of individual cells is used to build a response curve. This has the disadvantage that cell-to-cell variability can obscure the true response of signalling cascades. Indeed, single-cell dose–response curves generated using optogenetics — for instance, by measuring the concentration of light-gated SOS (a RAS activator) versus the concentration of ERK (a RAS effector) — show marked smaller variability than generating a response curve using the entire cell population40 (FIG. 2b). Optogenetics can also be used in conjunction with pharmacological approaches to analyse the input–output behaviour of a signalling node before and after drug treatment, which overcomes the confounding effects of cell variability by using each cell as its own control (for example, fibroblast cells before and after treatment with PI3K inhibitors)39 (FIG. 2c).
The x axis for these single-cell dose–response curves is not restricted to the concentration of the signalling input. As optogenetic signals can be varied and measured in space and time, as well as in concentration, we can also analyse the cell response to the rate of change of the input54, the fold change of the input55,56, the spatial distribution of the input57, and so on (FIG. 2d). All of these signalling attributes have been found to be important in different biological contexts.
Optogenetic control of signals in space
The spatial control of cell signalling has a fundamental role in cell and organismal biology. To understand these processes, we need tools to actively manipulate signals in space, both at the level of individual cells in organisms and on a subcellular level.
Spatial regulation of multicellular signalling
Communication between cells can enable a group of cells to more sensitively interpret signals than cells acting alone. A classic example is visual gradient interpretation in the horseshoe crab. By spatially restricting light to only two light-sensitive cells at a time, two rules for contrast enhancement were derived58: a stronger external stimulus causes a stronger upstream signal; and the stronger the upstream signal, the stronger the inhibitory effect of the cell on its neighbours (FIG. 3a).
Figure 3. Optogenetic control of signals in space.
Both light-sensitive cells in the eye58 (part a) and groups of chemotactic cells in the Drosophila melanogaster egg chamber48 (part b) use lateral inhibition to generate strong cellular responses from shallow external gradients. Acting alone, each cell can sense the external gradient but by comparing with their neighbours a higher contrast interpretation is possible. First, the cells generate an upstream signal that is proportional to the perceived input (rule 1). Next, cells inhibit the activity of their neighbours: the stronger the perceived signal, the stronger the lateral inhibition (rule 2). The end result is that weak signals are inhibited more than strong ones, which produces an amplified representation of subtle external gradients and enhances visual contrast in the eye (part a). A similar lateral inhibition mechanism is thought to enable coordinated collective cell migration in the fly egg chamber (part b) for the interpretation of external chemoattractant gradients (triangles). Here, the light-controlled activation of Rac inhibits protrusion in adjacent cells, and the inhibition of Rac in the leader cell activates protrusions in adjacent cells. These dissections of lateral inhibition were only possible, because cells were individually activated with spatially restricted inputs.
During visual contrast enhancement, although all cells inhibit the activity of each other to some extent, cells in the brightest areas reduce the activity of their neighbour the most, which emphasizes slight differences in an external gradient. This so-called lateral inhibition functions in the eyes, from horseshoe crabs to humans, and increases the contrast in our vision before action potentials enter the optic nerve.
More recently, optogenetic approaches have shown how cellular comparison can sharpen signal interpretation in other multicellular contexts. In Drosophila melanogaster, small groups of border cells collectively migrate about 175 μm up a chemical gradient to a maturing oocyte in the egg chamber48. This collection of cells uses lateral inhibition to ensure that only the one or two cells at the highest point in the gradient actively protrude. Using photoactivatable Rac to override normal chemotactic cues and initiate movement in cells at the side of the collective caused the normally leading cells to stop protruding. Thus, the activity of one cell inhibits the activity of its neighbour. Although all of the border cells are capable of protrusion, lateral inhibition ensures that only one or two leading cells are active at a time and that a collective decision is possible (FIG. 3b).
The rules derived from these two examples show how regulation at a distance increases the ability of a system to properly respond to external cues. They were only discovered because spatially restricted inputs enabled the investigation of how the activity of one cell affects the activity of neighbouring cells. It is probably no coincidence that biology converged on lateral inhibition to increase gradient sensing of both light and chemoattractants.
Spatial regulation of subcellular signalling
On a multicellular level, non-optogenetic tools have been used to study the spatial control of signalling — for instance, the transplantation of cells or the generation of single-cell clones. However, optogenetic tools enable much more precise and dynamic control of cell signalling than non-optogenetic tools. There have been fewer tools to manipulate spatial signals on a subcellular level, and optogenetic systems are enabling some of the first demonstrations of the spatial sufficiency of signals to coordinate cellular processes, including the molecules that direct cell polarization and movement14,15,18,35,39,49. For example, LOV-domain-based control of RAC activity showed that asymmetries in the activation of this GTPase are sufficient to specify the migratory direction of individual neutrophils in zebrafish49 and the collective migration of border cells in D. melanogaster48.
Optogenetic control of signals in time
Although some signalling cascades only respond to the current level of stimulus, other cascades respond to the timing of the input59. For example, adaptive cascades, like most sensory inputs, reset themselves to the current level of stimulus and primarily sense changes in, as opposed to absolute levels of, signalling inputs54. Other cascades seem to monitor the dynamics of the input. For example, sustained or transient pathway activity can drive different cellular decisions. Differences in the duration of MAPK signalling can regulate whether cells decide to proliferate or differentiate60, whereas differences in the signal duration of the tumour suppressor protein p53 can regulate whether cells arrest their cell cycle or apoptose61. In this section, we address the challenges in analysing these processes together with the opportunities that optogenetic systems present for probing the role of timing in cell decisions.
The dynamics of intracellular signalling has been implicated in the regulation of cellular decisions. For example, different ligands trigger different dynamics of ERK activation epidermal growth factor (EGF) drives transient ERK activation, whereas nerve growth factor (NGF) drives sustained ERK activation), and these different dynamics correlate with different downstream responses (EGF drives the proliferation of PC12 cells, whereas NGF drives the differentiation of PC12 cells). To investigate whether differences in the timing of MAPK activation are causative for downstream behaviours, one group used pharmacological activators and inhibitors of protein kinase C to produce sustained activation of MAPK in response to EGF and transient activation of MAPK in response to NGF60. Exchanging the MAPK dynamics downstream of these ligands also exchanged their downstream responses, which is consistent with the idea that the timing of MAPK activation determines the resulting cellular outcome (FIG. 4a). As many signalling cascades are not sufficiently understood to use pharmacological or genetic means to influence intracellular signalling dynamics, and because these pharmacological perturbations also affect the topology of the signalling network, it would be preferable to directly manipulate intracellular signalling dynamics in a user-defined manner. This is now possible with optogenetic inputs, and multiple groups have used this approach to demonstrate that the MAPK signal duration is sufficient to regulate the differentiation decision in PC12 cells40,62 and to identify the signalling pathways that are differentially activated by sustained versus transient MAPK activation40 (FIG. 4b).
Figure 4. Optogenetic control of signals in time.
a | Signalling dynamics can affect cellular decisions. Normally, treatment of PC12 cells with epidermal growth factor (EGF) causes transient ERK activity and cellular proliferation, whereas treatment with nerve growth factor (NGF) causes sustained ERK activity and differentiation into neuron-like cells. Adding drugs that decrease or increase ERK signalling can convert the effect of NGF on ERK activity into an EGF-like effect, and vice versa. This perturbation causes an exchange in the outcome (EGF now drives differentiation and NGF drives proliferation)60. Subsequent studies in which ERK-signalling dynamics were directly controlled with optogenetic inputs confirmed that differences in MAPK dynamics suffice to specify cellular differentiation in PC12 cells40,62. b | Using optogenetics, researchers showed that upstream signalling dynamics affect downstream pathways. In particular, signal transducer and activator of transcription 3 (STAT3) phosphorylation functioned as a persistent detector of ERK activation. In 3T3 fibroblast cells expressing an optically controllable son of sevenless homologue (SOS) construct (opto-SOS; an activator of the RAS GTPase), 2 hours of light stimulation caused a robust STAT3 phosphorylation. However, the same stimulus broken into two, 1-hour pulses did not cause STAT3 phosphorylation despite the activation of other ERK-responsive pathways40. Thus, optogenetic approaches have directly shown that cells respond to specific upstream signalling dynamics and have identified some of the pathways that respond to a particular signalling pattern. Part b reprinted from Cell, 155/1430, Toettcher, J. E., Weiner, O. D. & Lim, W. A., Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module, 1422–1434, Copyright (2013), with permission from Elsevier.
Conclusions and perspectives
Optogenetic systems have moved beyond the control of light-gated ion channels to include a wide range of other light-responsive proteins and cellular pathways that can now be manipulated by light. All of these systems started with proof-of-principle demonstrations, activating signalling pathways that others had previously activated by protein overexpression, microinjection, and so on. However, investigators have recently taken advantage of the capacity of optogenetics to dynamically manipulate and measure signals in space and time to address questions that have been difficult or impossible to address with other tools.
We end with a wish list for the field. The optogenetics field is based on light-responsive proteins from plants and other systems. We are fortunate that these systems work as well as they do, but we should consider them like the first generation of GFPs — systems with a huge potential to evolve and to be optimized. Some engineering work has been done for the LOV domains, but far more work could be done to optimize the affinities, the photoactivation and photoreversion rates, the wavelengths for photoswitching and the light sensitivity of other systems. This optimization will be aided by advances in the structure and mechanism of these photosensors63,69. Furthermore, we would like these systems to be completely orthogonal to cell signalling cascades, but we do not yet know whether post-translational modifications affect their function in optogenetic applications. To understand these constraints, it will be useful to follow advances in the physiology and regulation of these proteins in their normal context64–66. On a similar note, many of these proteins were chosen for their light-responsive interactions in plants, and a greater understanding of their regulators could expand our strategies for photoregulation. For instance, cryptochromes not only form heterodimers with downstream effectors in response to light (such as CIB1)9 but also oligomerize in response to light32. Both of these properties have been used to regulate cell signalling10,11, and a greater knowledge of phototransduction mechanisms is likely to reveal more surprises and opportunities for optogenetics.
Acknowledgments
The authors thank members of the Weiner laboratory for helpful discussions. This work was supported by a Genentech Fellowship (D.T.) and US National Institutes of Health (NIH) grants GM084040, GM096164 and GM109899 (O.D.W.).
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Kaplan JH, Forbush B, Hoffman JF. Rapid photolytic release of adenosine 5-triphosphate from a protected analogue: utilization by the Na:K pump of human red blood cell ghosts. Biochemistry. 1978;17:1929–1935. doi: 10.1021/bi00603a020. [DOI] [PubMed] [Google Scholar]
- 2.Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci USA. 1993;90:7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zemelman BV, Lee GA, Ng M, Miesenbock G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 4.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 5.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 7.Arrenberg AB, Stainier DY, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 8.Tsai HC, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy MJ, et al. Rapid blue-light-mediated induction of protein interactions in living cells. Nature Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, Schaffer DV. Optogenetic protein clustering and signaling activation in mammalian cells. Nature Methods. 2013;10:249–252. doi: 10.1038/nmeth.2360. [DOI] [PubMed] [Google Scholar]
- 12.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 14.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strickland D, et al. TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nature Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–784. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- 17.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nature Biotech. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 18.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou XX, Chung HK, Lam AJ, Lin MZ. Optical control of protein activity by fluorescent protein domains. Science. 2012;338:810–814. doi: 10.1126/science.1226854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 21.Fenno L, Yizhar O, Deisseroth K. The development and application of optogenetics. Annu Rev Neurosci. 2011;34:389–412. doi: 10.1146/annurev-neuro-061010-113817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285:41501–41508. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stierl M, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2011;286:1181–1188. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 25.Karunarathne WK, Giri L, Patel AK, Venkatesh KV, Gautam N. Optical control demonstrates switch-like PIP3 dynamics underlying the initiation of immune cell migration. Proc Natl Acad Sci USA. 2013;110:E1575–E1583. doi: 10.1073/pnas.1220755110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen D, Gibson ES, Kennedy MJ. A light-triggered protein secretion system. J Cell Biol. 2013;201:631–640. doi: 10.1083/jcb.201210119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crefcoeur RP, Yin R, Ulm R, Halazonetis TD. Ultraviolet-B-mediated induction of protein–protein interactions in mammalian cells. Nature Commun. 2013;4:1779. doi: 10.1038/ncomms2800. [DOI] [PubMed] [Google Scholar]
- 28.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nature Biotech. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 29.Bonger KM, Rakhit R, Payumo AY, Chen JK, Wandless TJ. General method for regulating protein stability with light. ACS Chem Biol. 2014;9:111–115. doi: 10.1021/cb400755b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gambetta GA, Lagarias JC. Genetic engineering of phytochrome biosynthesis in bacteria. Proc Natl Acad Sci USA. 2001;98:10566–10571. doi: 10.1073/pnas.191375198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller K, et al. Synthesis of phycocyanobilin in mammalian cells. Chem Commun (Camb ) 2013;49:8970–8972. doi: 10.1039/c3cc45065a. [DOI] [PubMed] [Google Scholar]
- 32.Mas P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nature Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 34.Motta-Mena LB, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nature Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Idevall-Hagren O, Dickson EJ, Hille B, Toomre DK, De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc Natl Acad Sci USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wend S, et al. Optogenetic control of protein kinase activity in mammalian cells. ACS Synth Biol. 2013;3:280–285. doi: 10.1021/sb400090s. [DOI] [PubMed] [Google Scholar]
- 37.Leung DW, Otomo C, Chory J, Rosen MK. Genetically encoded photoswitching of actin assembly through the Cdc42–WASP–Arp2/3 complex pathway. Proc Natl Acad Sci USA. 2008;105:12797–12802. doi: 10.1073/pnas.0801232105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X, Jost AP, Weiner OD, Tang C. A light-inducible organelle targeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell. 2013;24:2419–2430. doi: 10.1091/mbc.E13-03-0126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toettcher JE, Gong D, Lim WA, Weiner OD. Light-based feedback for controlling intracellular signaling dynamics. Nature Methods. 2011;8:837–839. doi: 10.1038/nmeth.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Konermann S, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lungu OI, et al. Designing photoswitchable peptides using the AsLOV2 domain. Chem Biol. 2012;19:507–517. doi: 10.1016/j.chembiol.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyszkiewicz AB, Muir TW. Activation of protein splicing with light in yeast. Nature Methods. 2008;5:303–305. doi: 10.1038/nmeth.1189. [DOI] [PubMed] [Google Scholar]
- 44.Cao J, et al. Light-inducible activation of target mRNA translation in mammalian cells. Chem Commun. 2013;49:8338–8340. doi: 10.1039/c3cc44866e. [DOI] [PubMed] [Google Scholar]
- 45.Lee S, et al. Reversible protein inactivation by optogenetic trapping in cells. Nature Methods. 2014;11:633–636. doi: 10.1038/nmeth.2940. [DOI] [PubMed] [Google Scholar]
- 46.Rao MV, Chu PH, Hahn KM, Zaidel-Bar R. An optogenetic tool for the activation of endogenous diaphanous-related formins induces thickening of stress fibers without an increase in contractility. Cytoskeleton. 2013;70:394–407. doi: 10.1002/cm.21115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renicke C, Schuster D, Usherenko S, Essen LO, Taxis C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem Biol. 2013;20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, He L, Wu YI, Hahn KM, Montell DJ. Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nature Cell Biol. 2010;12:591–597. doi: 10.1038/ncb2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoo SK, et al. Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish. Dev Cell. 2010;18:226–236. doi: 10.1016/j.devcel.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toettcher JE, Gong D, Lim WA, Weiner OD. Light control of plasma membrane recruitment using the Phy–PIF system. Methods Enzymol. 2011;497:409–423. doi: 10.1016/B978-0-12-385075-1.00017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Non-genetic origins of cell-to-cell variability in TRAIL-induced apoptosis. Nature. 2009;459:428–432. doi: 10.1038/nature08012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haber JE. In vivo biochemistry: physical monitoring of recombination induced by site- specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 53.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 54.Shimizu TS, Tu Y, Berg HC. A modular gradient-sensing network for chemotaxis in Escherichia coli revealed by responses to time-varying stimuli. Mol Syst Biol. 2010;6:382. doi: 10.1038/msb.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goentoro L, Shoval O, Kirschner MW, Alon U. The incoherent feedforward loop can provide fold-change detection in gene regulation. Mol Cell. 2009;36:894–899. doi: 10.1016/j.molcel.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wartlick O, et al. Dynamics of Dpp signaling and proliferation control. Science. 2011;331:1154–1159. doi: 10.1126/science.1200037. [DOI] [PubMed] [Google Scholar]
- 57.Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: a focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- 58.Hartline HK, Wagner HG, Ratliff F. Inhibition in the eye of Limulus. J Gen Physiol. 1956;39:651–673. doi: 10.1085/jgp.39.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Purvis JE, Lahav G. Encoding and decoding cellular information through signaling dynamics. Cell. 2013;152:945–956. doi: 10.1016/j.cell.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nature Cell Biol. 2007;9:324–330. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 61.Purvis JE, et al. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang K, et al. Light-mediated kinetic control reveals the temporal effect of the Raf/MEK/ERK pathway in PC12 cell neurite outgrowth. PLoS ONE. 2014;9:e92917. doi: 10.1371/journal.pone.0092917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takala H, et al. Signal amplification and transduction in phytochrome photosensors. Nature. 2014;509:245–248. doi: 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim JI, et al. Phytochrome phosphorylation modulates light signaling by influencing the protein–protein interaction. Plant Cell. 2004;16:2629–2640. doi: 10.1105/tpc.104.023879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Medzihradszky M, et al. Phosphorylation of phytochrome B inhibits light-induced signaling via accelerated dark reversion in Arabidopsis. Plant Cell. 2013;25:535–544. doi: 10.1105/tpc.112.106898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes RM, Vrana JD, Song J, Tucker CL. Light-dependent, dark-promoted interaction between Arabidopsis cryptochrome 1 and phytochrome B proteins. J Biol Chem. 2012;287:22165–22172. doi: 10.1074/jbc.M112.360545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Strickland D, et al. Rationally improving LOV domain-based photoswitches. Nature Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loewer A, Karanam K, Mock C, Lahav G. The p53 response in single cells is linearly correlated to the number of DNA breaks without a distinct threshold. BMC Biol. 2013;11:114. doi: 10.1186/1741-7007-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD. Crystal structure of the photosensing module from a red/far-red light-absorbing plant phytochrome. Proc Natl Acad Sci USA. doi: 10.1073/pnas.1403096111. (in the press) [DOI] [PMC free article] [PubMed] [Google Scholar]