This work presents a genome-scale data set that precisely identifies transcription start sites for a majority of Arabidopsis genes, revealing that plant promoters are not primarily TATA based and have an unexpected structure composed of many position-specific sequence elements. This analysis identifies combinations of factors that are likely to lead to transcription initiation.
Abstract
Understanding plant gene promoter architecture has long been a challenge due to the lack of relevant large-scale data sets and analysis methods. Here, we present a publicly available, large-scale transcription start site (TSS) data set in plants using a high-resolution method for analysis of 5′ ends of mRNA transcripts. Our data set is produced using the paired-end analysis of transcription start sites (PEAT) protocol, providing millions of TSS locations from wild-type Columbia-0 Arabidopsis thaliana whole root samples. Using this data set, we grouped TSS reads into “TSS tag clusters” and categorized clusters into three spatial initiation patterns: narrow peak, broad with peak, and weak peak. We then designed a machine learning model that predicts the presence of TSS tag clusters with outstanding sensitivity and specificity for all three initiation patterns. We used this model to analyze the transcription factor binding site content of promoters exhibiting these initiation patterns. In contrast to the canonical notions of TATA-containing and more broad “TATA-less” promoters, the model shows that, in plants, the vast majority of transcription start sites are TATA free and are defined by a large compendium of known DNA sequence binding elements. We present results on the usage of these elements and provide our Plant PEAT Peaks (3PEAT) model that predicts the presence of TSSs directly from sequence.
INTRODUCTION
Transcriptional regulation is an integral process for the control of cell and organ identity, growth, development, differentiation, and response in many organisms. Knowledge of transcriptional start sites (TSSs) and promoter architecture are thus crucial for understanding the transcriptional regulation underlying these fundamental processes. For genes transcribed by RNA polymerase II (pol-II), the architecture of promoter regions has been extensively studied in many prokaryotes and eukaryotes, such as bacteria, yeast, and humans (David et al., 2006; Yamashita et al., 2011; Jorjani and Zavolan, 2014; Park et al., 2014). A major component of promoter architecture identified in animal species are DNA sequence elements, which are bound by different components of the pol-II transcription initiation machinery (Kadonaga, 2004, 2012; Thomas and Chiang, 2006). With this information, detailed models of promoter architecture have been developed in these animal species (Smale and Kadonaga, 2003; Juven-Gershon and Kadonaga, 2010; Grünberg and Hahn, 2013).
In plants, DNA sequence elements that are known to play an important role in gene expression include the pol-II binding elements TATA and Initiator, as well as additional elements that are thought to play an enhancer role in some settings. These additional elements include specific transcription factor binding sites (TFBSs), such as DOF, MYB, and MADS box, as well as general sequence enrichments including Y-Patch and GA content. Classical reviews on the subject (Grasser, 2006) follow past animal-based models for a “core promoter” region consisting of position-specific DNA sequence binding elements TATA and Initiator, with common inclusion of a CCAAT box proximal to the core promoter. However, these models of plant core promoter structure were postulated without high-resolution genome-scale TSS information, at a time when less than 100 well-characterized examples were available in plants. Since this time, there have been no major changes to views of plant core promoters. More recently, motif discovery–based analyses utilizing thousands of available promoter examples (Yamamoto et al., 2009, 2011) have included a focus on general sequence enrichments such as Y-Patch, GA, and CA elements, hypothesizing that these enrichments may play a similar role to CpG islands in mammalian promoters. CpG islands are found in the promoters of many mammalian genes (Saxonov et al., 2006), and their presence has been used as a key feature in TSS prediction models (Deaton and Bird, 2011). Yet, to date, little is known about the specific combinations of elements in pol-II promoters that likely lead to transcription in plants.
Conventional annotation of plant TSSs rely on low- to mid-throughput technologies such as EST/cDNA alignment, 5′ rapid amplification of cDNA ends (RACE), and modified versions of MPSS. Most TAIR10 annotated TSSs are at best based on the alignment of ESTs. EST/cDNA-based annotations of 5′ transcript locations are known to be inaccurate, given that they rely on a reverse transcriptase based assay. The precise identification of gene promoter regions allows for the characterization of pol-II binding elements that have positional constraints: For example, the TATA-box element is known to be found between 25 and 45 nucleotides upstream of the TSS. Accurate location of the promoter also assists in the identification of functional TFBSs, which are short, degenerate sequence motifs found in both intergenic and promoter sequences.
For pol-II transcribed noncoding RNAs, this inference problem of functional core promoter identification is more extreme, as little data on the location of the RNA primary transcript may be present. MicroRNAs (miRNAs)—small RNAs that regulate gene expression relevant to the development of many eukaryotes—are a prominent example of this problem because the TSS of each miRNA primary transcript is located in an unknown region that is at a variable distance from the mature miRNA sequence. Individual TSSs identified using 5′ RACE have been determined for ∼50 Arabidopsis thaliana miRNAs (Xie et al., 2005), but the landscape of primary transcript TSSs remains entirely unknown for the majority of the 337 known Arabidopsis miRNAs (Griffiths-Jones et al., 2006). Further complicating promoter analysis with publicly available data, such as annotated TAIR10 transcripts, is the fact that transcription does not always initiate from a single location, but for some genes, can initiate at any site within a short genomic window (Carninci et al., 2006; Ni et al., 2010). Therefore, for all pol-II genes, a more detailed data set is needed that accurately describes the TSS locations that are most highly expressed relative to neighboring locations.
In this study, we employ the paired end analysis of transcription start sites (PEAT) protocol to generate a high-throughput TSS data set because of its accuracy in other organisms, nucleotide resolution, and paired-end mapping capability (Ni et al., 2010) to analyze a pooled root sample of the first sequenced plant, the Columbia-0 ecotype of Arabidopsis (Arabidopsis Genome Initiative, 2000). The high resolution of the PEAT data set allows us to categorize the TSS tags into distinct transcription initiation patterns, then identify position-specific enrichments for known TFBS signals in proximity to each initiation pattern. We use these signals to construct an accurate machine learning model for each initiation pattern called the Plant PEAT Peaks or “3PEAT” model. 3PEAT predicts the probability of a TSS at any given nucleotide in the Arabidopsis genome solely from the DNA sequence surrounding that nucleotide. It is trained and tested on separate partitions of each initiation pattern category. Because the 3PEAT model is able to achieve a remarkably high combination of sensitivity and specificity, analysis of its features yields direct insight into the combinations of plant promoter elements that give rise to pol-II transcription.
RESULTS
Our data set is composed of short regions representing the 5′ ends of pol-II RNA transcripts, obtained using the PEAT protocol. Cap-trapping protocols such as CAGE (Carninci et al., 2005) and PEAT (Ni et al., 2010) aim to isolate pol-II mRNA transcripts and sequence the capped ends (TSS tags) with nucleotide resolution. Recent analyses using genome-scale TSS data sets produced with PEAT and CAGE in animal tissues (Ni et al., 2010; Batut et al., 2013; Nepal et al., 2013) have observed that TSSs are not simply located at one or even a few “alternative start site” locations upstream of each pol-II gene. When these TSSs are mapped back to the genome, they distribute spatially in a variety of different “TSS tag clusters” or “initiation patterns.” In animals, these initiation patterns have been associated with different core promoter elements, gene functions, and chromatin structure (Carninci et al., 2006; Rach et al., 2009, 2011). Narrow peak animal promoters are typically significantly enriched for TFBS motifs and associated with development and differentiation, while genes with weak peak initiation patterns contain fewer significant TFBS enrichments and are often housekeeping genes.
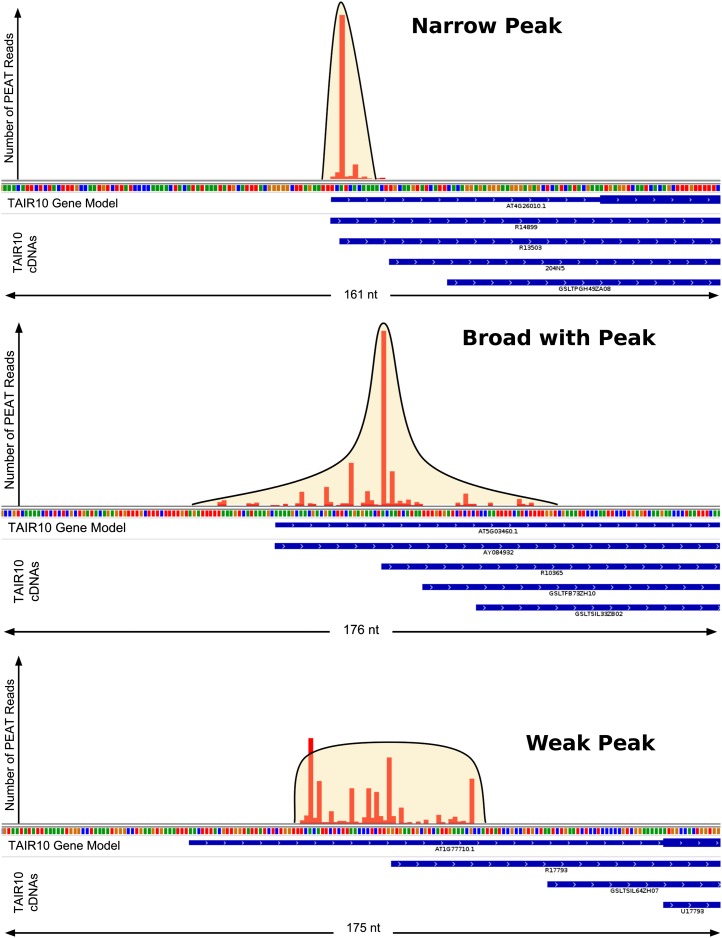
In this study, we use the nomenclature of other PEAT studies (Ni et al., 2010): (1) a narrow peak (NP) pattern represents a narrow genomic segment containing a very large number of TSS tags, (2) a broad with peak (BP) pattern is a region where many tags are spread over a relatively wide genomic segment with a narrow region containing a majority of the TSS tags, and (3) a weak peak [WP] pattern is a case where relatively fewer tags are observed within a segment and shape may be less definitive, nonetheless a clear tag cluster is present. To be clear on naming, the WP category is most similar to the “broad” category in the original CAGE-based analyses in mammalian genomes (Carninci et al., 2005, 2006). Our PEAT data set yields ∼4 million tag pairs mapping with the highest certainty, identifying 24,207 TSS tag clusters associated with 17,619 protein-coding genes and is comparable in gene expression coverage with RNA-Seq data in similar root samples (Supplemental Figure 1) (Brady et al., 2007; Li et al., 2013).
The PEAT procedure starts with TAP digestion, which removes the 5′ CAP or 5′ PPP from transcripts resulting in ones with a 5′ P. Since there are transcripts in vivo (degradation intermediates) that have 5′ P, these “background” transcripts could contribute to the observed TSS patterns. To assess this possibility, we compared the locations of TSS tag cluster peaks to the TAIR10 annotation of the associated gene. These results show that all PEAT TSS tag clusters located within the promoter region of genes closely agree with TAIR10 annotation. For example, for all peaks taken together (regardless of shape or number of reads), the median distance between peak modes and TAIR10 annotations was only 35 nucleotides. Individually, NP clusters were located a median of 50 nucleotides, BP clusters 22 nucleotides, and WP clusters 32 nucleotides from the TAIR10 annotation. With 100 reads per tag cluster specified (the minimum required for inclusion in the model), the median distance across all initiation patterns dropped to 18 nucleotides (distances within the classes were as follows: NP, 4 nucleotides; BP, 10 nucleotides; WP, 24 nucleotides) (Supplemental Figure 2). In summary, this analysis shows that all PEAT TSS tag clusters agree closely with the TAIR10 annotations, even at the minimum read level. Therefore, we consider it unlikely that any background transcripts present contribute substantially or in a biased way toward the observed tag clusters of any type.
Distinct Patterns of Transcription Initiation Exist in Plants
Similar to what has been observed in animal systems, our data show that distinct patterns of transcription initiation exist in Arabidopsis (Figure 1, Table 1; Supplemental Table 1). Previous computational studies of promoter regions upstream of similar TSS initiation patterns in animals have shown that it is possible to predict the locations of NP TSSs from DNA sequence (Frith et al., 2008; Megraw et al., 2009; Rach et al., 2011). A machine learning model called S-Peaker (Megraw et al., 2009) was constructed to use sequence content along with the position specificity and binding affinity of TFBSs to reveal which factors are important for achieving a sharp spatial peak of transcription. The S-Peaker study confirmed the hypothesis that, at least in the NP TSS case for animals, many elements can play a guiding role in pol-II transcription initiation, and these elements are most likely to do so in transcription factor (TF)-specific locations with respect to a highly transcribed TSS location.
Figure 1.
PEAT Data Set Examples of NP (Top), BP (Middle), and WP (Bottom) Tag Clusters.
The horizontal axis of each plot displays a region of genomic sequence, with TAIR10 cDNAs in the region displayed below the axis. The vertical axis displays the number of PEAT reads observed at each nucleotide location in the region. Tag cluster shape estimates are overlaid, providing an example of each peak shape as determined by the PEAT peak shape caller.
Table 1. Quality-Filtered TSS Initiation Pattern Data Set.
| Data Set/Initiation Pattern | Total Examples | Total PEAT Reads |
|---|---|---|
| NP | 1,276 (14%) | 1,382,237 (19%) |
| BP | 2,050 (22%) | 2,665,313 (36%) |
| WP | 6,000 (64%) | 3,330,821 (45%) |
| All protein coding | 9,326 (100%) | 7,378,371 (100%) |
Given that plant promoters have proven to be very difficult cases for TSS prediction models (Shahmuradov et al., 2003, 2005), we posited that TFBS model-based analysis (Megraw et al., 2009) would be an appropriate method to solve this challenging problem. Due to the lack of CpG islands in plants as compared with mammalian promoters (Kapranov, 2009), many alternative sequence enrichments must collectively be examined for efficacy. Specific positional relationships for TFBSs with respect to TSSs are poorly documented in plants beyond TATA-box. These positional relationships between TFBSs and TSSs are currently unknown for a majority of TFs and may well be serving as the key to transcription initiation. In addition, these relationships are likely not limited to single-TSS locations. A large data set containing millions of tags that map stringently to a well-sequenced and well-annotated genome is likely to be sufficient for an accurate model, at least in the case of NP promoters. We expected that a tissue sample such as the root, with its complex regulatory network (Bruex et al., 2012; Lan et al., 2013), would be enriched for rapidly dividing cells where tissue-specific patterning is taking place, a setting expected to give rise to many sharp NP TSSs, in addition to more broadly distributed gene promoters typically associated with ubiquitously expressed genes.
The Location of Transcription Initiation Can Be Accurately Modeled by DNA Binding Affinity in Plants
We next investigated whether a plant-specific TSS identification model based on DNA sequence features and machine learning can address plant-specific complexity such as the lack of CpG islands. We examined this question by using the large PEAT TSS initiation pattern data set to query whether patterns of sequence enrichments and spatial positioning of elements that lead to pol-II transcription could be identified. In this model, each PEAT tag cluster in the data set was collapsed to a single genomic location representing the most highly expressed nucleotide relative to neighboring locations (the TSS tag cluster mode), with an upstream promoter region that contains computationally identified TFBS elements at specific locations relative to this TSS mode (Figure 2). A control group of randomly chosen sequences was selected from the genome, representing a wide variety of sequences that are not pol-II transcribed. To identify sequence elements that show TFBS enrichment, we next searched the promoters for specific upstream locations where these TFBS elements could be collectively enriched (see Methods: 3PEAT TSS Peak Prediction Model). We used a standard log-likelihood TFBS scanning technique to approximate DNA binding affinity along with a set of 200 known plant elements characterized in the literature (Grasser, 2006; Megraw et al., 2006; Bryne et al., 2008; Wingender, 2008; Civán and Svec, 2009; Yamamoto et al., 2009).
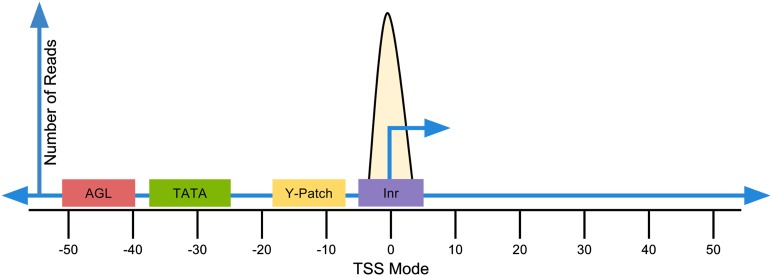
Figure 2.
Diagram Displays an Example of a PEAT Peak Promoter, as Considered by the 3PEAT Model.
3PEAT considers the mode of a PEAT peak as the genomic location for the TSS location of the peak. 3PEAT identifies known transcription factor binding site motifs upstream of TSSs and records the high-scoring locations for these elements. This information from each PEAT peak is then used as a data example on which to train or test the 3PEAT TSS prediction model.
[See online article for color version of this figure.]
We identified a subset of at least 150 elements exhibiting TFBS enrichment on at least one strand within each initiation pattern category (Supplemental Data Set 1). In accordance with what has been previously reported, we observed sharp well-defined enrichment signals for the canonical pol-II elements TATA and Initiator in the NP class. We also identified novel TFBS elements that are associated with the core promoter region or with position-specific locations with respect to a TSS. In total, we observed at least 20 such novel elements within each initiation pattern category (Supplemental Figures 3 to 5). We also observed that each initiation pattern had high levels of enrichment for a different set of elements, in some cases also with minor differences in enrichment location or enrichment region width, suggesting that different initiation patterns are regulated by different factors, as has been reported in animals (Carninci et al., 2006; Rach et al., 2009). We therefore used these different regions of enrichment separately, training a different version of our model for each initiation pattern.
With the purpose of designing a highly interpretable model that effectively selects among many sequence features within a minimal set for optimal classification performance, we performed model training and 10-fold cross-validation (see Methods: 3PEAT TSS Peak Prediction Model) using L1-regularized logistic regression (Koh et al., 2007). As in Megraw et al. (2009), input features to the model reflect whether each TFBS element has a binding sequence signal present within that TFBS’s region of enrichment (ROE) with respect to the specific genomic location being examined (e.g., Chr 1:+:34,567). An ROE (see Methods: 3PEAT TSS Peak Prediction Model) is a segment of genomic sequence where a specific TF is preferentially located with respect to many observed TSSs; it represents a location of putative biological relevance for TF binding. The model output is a probability that the genomic location under examination is the mode of a TSS distribution within the given initiation pattern. Each model was trained using positive examples from its initiation pattern group, and negative examples selected from the immediate upstream regions of these TSSs as well as from annotated coding sequence. Model performance was measured by the area under the ROC curve (auROC) and area under the precision recall curve (auPRC) of the model on an independent subset of PEAT data that was not used to train the model (Supplemental Figures 6 and 7). After observing strong testing performance within each shape class (Supplemental Figures 6 and 7), we constructed an ALL model trained on the union of all initiation patterns.
We observed that the NP model has the strongest performance (auROC of 0.98 and auPRC of 0.89). Remarkably, we were able to train models that performed nearly as well for BP and WP classes. This outcome suggests that all TSS tag clusters in Arabidopsis are likely associated with position-specific element combinations and enrichments. Perhaps even more surprising was the excellent performance of the ALL model (Supplemental Figure 8), where auROC matched the peak shape models at 0.98 and auPRC of 0.88 fell within 2% of these specific models. This finding supports the hypothesis that although position-specific TFBS enrichments differ for each initiation pattern, when taken together, TF affinities for specific regions within the promoter are collectively predictive of a TSS mode regardless of pattern. Thus, the ALL model appears to successfully account for different usages of the elements in different transcriptional landscapes to create a “promoter signature” based only on TF binding affinity for promoter sequences in all three initiation patterns. This finding suggests that the locations of highly expressed TSSs are accurately distinguished using DNA sequence signals alone. We name our model design for the NP, BP, WP, and ALL TSS initiation pattern classes the 3PEAT (Plant PEAT Peaks) model.
3PEAT Enables Genomic Scans for pol-II Protein Coding and miRNA Gene TSSs
Because auROC and auPRC values on a relatively small test set of locations can be deceiving (Lobo et al., 2008), the ability to produce a high-resolution signal when scanning a model over large regions of the genome may still fail even if the model achieves high auROC and auPRC. This failure in signal detection occurs because genomic sequence usually contains an overwhelming number of negative examples, and only a tiny percentage of these examples can reasonably be included in the test sets from which auROC and auPRC values are computed. Relative to the entirety of the Arabidopsis genome, there are few genomic locations are TSS peak modes. If the 3PEAT negative training sets poorly characterize the large variety of locations that are not TSS peak modes, then when the model is scanned over genomic regions, an overwhelming number of false positive calls will be produced. Therefore, we further tested 3PEAT in genomic scanning to determine if the model design and training set together translate into a useful performance outcome.
Each 3PEAT initiation pattern model (NP, BP, WP, and ALL) was scanned over the entire set of genomic sequence locations within 4 kb of each test set example (Supplemental Figure 9). Scanning performance was evaluated by an auROC curve estimate (Supplemental Figure 9A). Sensitivity was approximated by the percentage of PEAT peak mode TSS locations “hit” by the probability output signal at a given threshold, and specificity was approximated by the number of additional TSS locations hit per kilobase (see Methods: 3PEAT TSS Peak Prediction Model). This evaluation of scanning performance is a highly conservative approximation because some of the additionally predicted TSS locations may in fact be PEAT TSS mode locations, but we consider that it provides a fair estimate. Supplemental Figure 9A mirrors our previous observations (Supplemental Figures 6 to 8). In conservative auROC estimates, individual initiation pattern models and the model combining ALL patterns performed extremely well; individual pattern models and the model for ALL pattern types also performed similarly to each other. These data validate our approach by demonstrating that even in genomic scans, the 3PEAT models do not produce a large number of false positives. In Supplemental Figure 9B, we plotted a different estimate of scanning resolution that reflects the ability of each trained model to center high probability output signals on the peak modes. Here, we observed clear differences in probability signal resolution, with the NP shape model producing the highest scanning resolution and the WP model producing the lowest. The ALL model was trained from both NP peaks (relatively fewer cases) and WP peaks (relatively many cases compared with NP); therefore, it stands to reason that the ALL model’s scanning performance at second-to-lowest is proportionally influenced. For example, at a probability threshold of 0.5, the centers of the NP model probability peaks are within about seven nucleotides of 80% of actual NP modes. For the ALL model at the same probability threshold, probability peaks are within 20 nucleotides of ∼75% of ALL initiation pattern modes. This resolution is quite remarkable. One would expect that a model trained on a specific initiation pattern type would more precisely estimate the location of the mode (most highly transcribed location) of that pattern, with the most precise estimate for NPs. Figure 3 shows an example of a scan using the WP model (all scans are provided in Supplemental Data Sets 2 to 4). For all models, probability signals agree closely with PEAT tag distributions of each pattern type as well as with the TAIR10 annotation. This agreement supports the conclusion that, when taken together on appropriate training sets, numerical performance measures such as auROC and auPRC accurately reflect predictive power in genomic scanning.
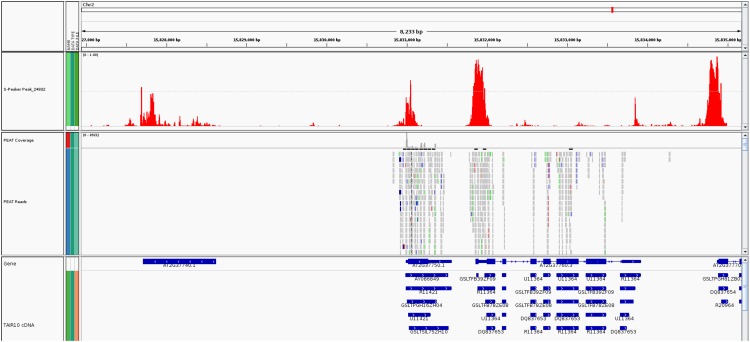
Figure 3.
Example Genomic Scan Output from 3PEAT Model, Showing Agreement of Predicted TSSs, PEAT Tag Reads, and TAIR10 Gene Annotations.
The track in red at the top of the figure displays probability output from a representative scan over the region of a WP test set TSS using the 3PEAT WP model (this particular example shows the PEAT WP tag cluster, center, at Chr2:+: 15830983-15831107). The middle track in gray displays actual PEAT reads mapped to the genome. The bottom track in blue displays TAIR10 genes and cDNAs. The model successfully calls out highly probable WPs, but also other types of PEAT tag clusters present in the sequence being scanned. Additionally, the model frequently indicates TSS tag clusters in locations that are not covered by PEAT reads in our Arabidopsis root sample but agree quite precisely with pol-II gene transcript start sites annotated in TAIR10.
Although miRNA primary transcripts are degraded after processing in the cell nucleus (Rogers and Chen, 2013), we still observed 40 PEAT peaks in our data set that were located in the near upstream regions of ∼30 TAIR10-annotated miRNA precursors (see Methods: PEAT TSS Peaks Data Set Production). To determine if the TSS tag clusters identified by the PEAT protocol were consistent with past observations derived using traditional laboratory techniques, we compared the miRNA-proximal TSS locations identified in this study with a set of 66 miRNA primary transcript TSS locations from a 5′ RACE study in Arabidopsis (Xie et al., 2005). Among this set of miRNAs with 5′ RACE–supported locations, 15 miRNA upstream regions were in common with our putative miRNA TSS peaks. We compared the precise locations of our PEAT TSS tag cluster modes with the reported locations of the 5′ RACE observations. Strikingly, 12 of 15 had close agreement: eight had a cluster mode within 1 nucleotide of the 5′ RACE location, and an additional four had a cluster mode within 7 nucleotides of the 5′ RACE location. Of the three locations that did not agree closely, all locations were within 50 nucleotides of a secondary 5′ RACE TSS observation. Most surprising was the strong agreement regardless of initiation pattern or expression level. Some tag clusters contained as little as 10 tags at the mode, the minimum number considered for PEAT tag clusters in our study; yet, mode location was identical to a reported 5′ RACE TSS location.
We then used this small set of 40 miRNA-proximal peaks as a test set for the 3PEAT ALL and NP models (Supplemental Figure 10). This test set includes many primary transcript clusters with as few as 10 reads, whereas the protein coding test and training sets contain at least 100 reads per cluster. As expected, performance on this data set was reduced compared with that of our large protein-coding test set. Performance was still better than expected, given the small number of reads associated with many of these TSSs. We observed that at reasonable mid-level probability thresholds (0.4 to 0.6 range), the models are quite specific but not very sensitive (Supplemental Figure 10). Some miRNA NPs as well as other types of peaks are missed. While this reduced performance is likely in part due to the liberal collection of miRNA primary transcript clusters, it also suggests that TSSs just upstream of mature sequences may not be the only peaks for miRNA primary transcripts and that additional miRNA proximal peaks may have a promoter composition that differs somewhat from protein-coding genes. These observations are consistent with past computational studies on plant miRNAs (Megraw et al., 2006) and may reflect the fact that miRNAs have multiple distinct heavily used TSS locations, which seems consistent with findings in mammalian systems that indicate miRNAs have multiple, complex transcripts that are related to the occurrence of numerous downstream miRNA processing steps (Saini et al., 2007; Bhattacharyya et al., 2012; Marco et al., 2013). In summary, our study indicates that 3PEAT is a useful model for the identification of high confidence, highly expressed TSS locations for miRNA primary transcripts in the absence of laboratory data in the region of interest.
PEAT Provides Significant Gains in TSS Resolution over the TAIR10 Annotation
We constructed a 3PEAT model based exclusively on TAIR10 annotation and compared its performance with the PEAT data set–based model. We observed that the TAIR10 based model achieved an auROC of 0.95. This was not substantially lower than that of the 3PEAT ALL model (0.98), but auPRC performance suffered considerably—0.69 for TAIR10 as compared with 0.88 for ALL (Supplemental Figure 11). The regions of enrichment defined by the TAIR10-based model were considerably less well defined than the enrichments of the PEAT-based models (Figure 4). Genomic scanning performance suffered as well, showing a 50% decrease in positional resolution compared with the PEAT ALL model (Supplemental Figure 12). To understand this loss of performance, we evaluated how closely the PEAT data agrees with TAIR10 annotation overall. PEAT tag clusters located within 500 nucleotides of an annotated gene are centered on average 40 nucleotides from the annotated gene start site (Supplemental Figure 2). Given this difference, enrichment locations determined using the TAIR10 annotations do not precisely reveal where pol-II must interact with an element to initiate transcription. The impact of this shift is shown in Figure 4. These differences in ROE width and location cause the TAIR10-based model to miss important combinations of elements present within their biologically relevant location. Overall, these results strongly indicate that a high-resolution data set is critical for accurately characterizing Arabidopsis promoter architecture.
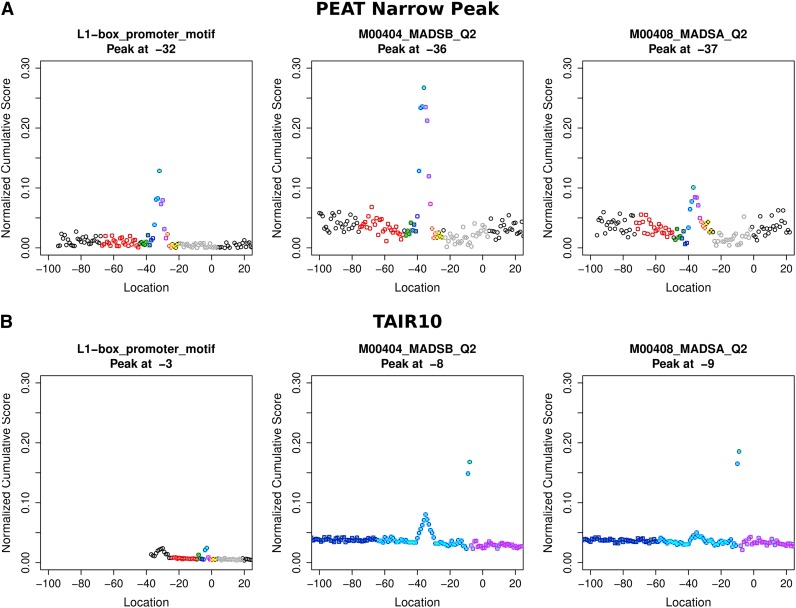
Figure 4.
Comparison of Three ROEs Defined by PEAT NP Data (Top) versus ROEs Defined by TAIR10 Data (Bottom).
Each colored region represents a portion of the region of enrichment and flanking area detected for the transcription factor. The ROEs center on different locations with respect to observed TSSs: MADSB (−36 versus −8), MADSA (−37 versus −9), and L1-box (−32 versus −3). ROEs are more pronounced and precise with PEAT data. These differences in data precision are likely to explain the success of the 3PEAT NP, WP, and BP models as opposed to the poor performance of the TAIR10 model in sequence scanning.
3PEAT Model Feature Analysis Yields Shape-Dependent Promoter Signatures
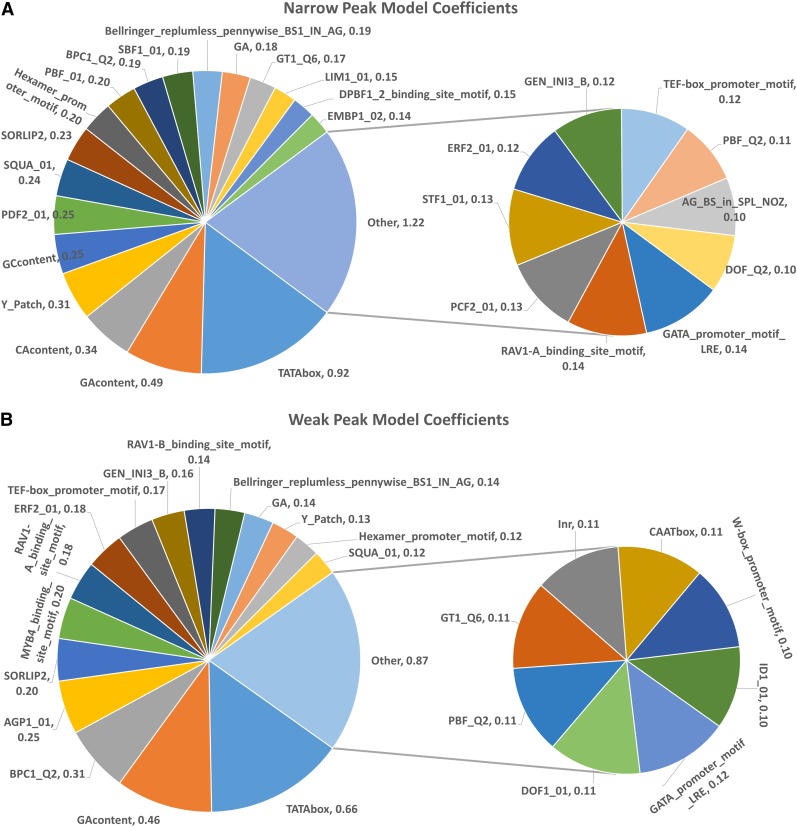
An important question in the study of gene regulation is the composition of a functional promoter. The L1-regularized logistic regression approach used in 3PEAT has a major benefit in that model features are highly interpretable. The features of the 3PEAT model measure the presence of individual binding elements (TATA, Ini, CAAT-box, MADS box, etc.) within their ROEs on each DNA strand with respect to the location under examination (see Supplemental Methods: Model Features for Details). Feature weights used by the model are directly proportional to their importance in the model’s predictive success. The weights of a successful NP model determine a promoter signature for NPs. A specific weighting of feature elements is thus produced that accurately determines whether an input sequence location is likely to be an NP. Figure 5 and Supplemental Figure 13 display detailed promoter signatures for the NP, WP, and BP models.
Figure 5.
3PEAT Model Coefficients for NP and WP Models, Forming Promoter Signatures.
Model coefficients can range from −1.0 to 1.0, only those exceeding 0.1 are displayed as part of the signature. Based on the model, heavily weighted positive coefficients indicate TFs whose presence is strongly associated with the presence of a TSS tag cluster.
Given the unique promoter signatures for NP, WP, and BP patterns, we were interested in determining whether these different initiation patterns associate with different biological processes. For this determination, we selected the set of TAIR10 protein coding genes associated with one or more TSS tag clusters of each initiation pattern and queried these sets for statistically overrepresented (P < 0.01) Gene Ontology (GO) terms (Supplemental Data Set 5). We examined the overrepresented terms unique to each initiation pattern for common themes. We observed that the unique NP category contains terms that pertain to oxidative stress (similarly reported in Yamamoto et al., 2009), biogenesis, and membrane systems. BP themes included chromatin, biosynthesis, and metabolism, as well as ATP-related and transport terms. WP themes dealt with metabolism and catabolism, as well as general protein-related processes. In general, statistically overrepresented terms unique to the NP category tended to relate to processes that must occur at a particular time and place in a developing tissue or cell. This category contained the smallest number of unique GO terms. In contrast, the WP category contained ∼6 times as many terms (representing about six times as many peaks) with a clear tendency toward general processes associated with housekeeping cellular functions. The BP category contained an intermediate number of terms that are not typically associated with housekeeping genes but rather processes necessitating stronger gene expression routinely at certain times in the circadian cycle. These general observations suggest that an organism’s unique use of the different promoter peak shapes is largely consistent with animal studies (Hoskins et al., 2011). NPs in our study tend to associate with spatiotemporal processes in an organism, which are controlled by promoter element combinations that come together at a precise, narrow region on the genome to initiate a strong burst of transcription. WPs tend to associate with a variety of processes requiring at least a low, maintenance level of transcription. The BP promoter signature may reflect a need for both of these capabilities, including processes that must be induced rapidly at a particular routine time.
pol-II Gene Expression Depends Largely on the Presence of Location-Specific TFBS Combinations That Are TATA Free
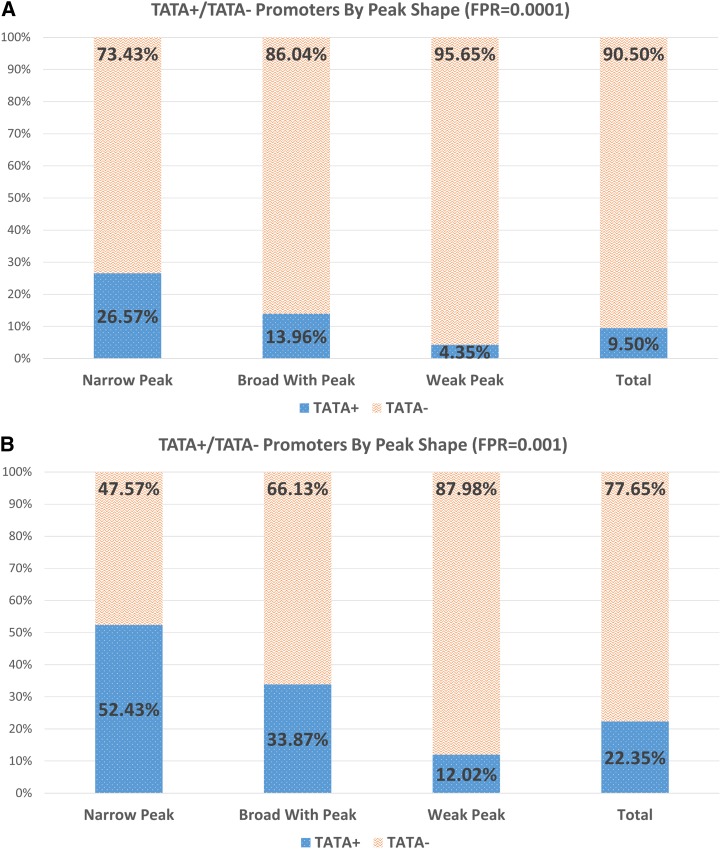
Previous studies (Yamamoto et al., 2009, 2011) reported that TATA-box and sequence enrichments, such as GA content, were associated heavily with TSS peaks of different patterns. These reports suggest that given a high-resolution data set and a model that uses location-specific enrichments, these features alone could be explanatory. We tested whether this data set would support equally strong performance using TATA-box along with GA, CA, and GC content. We trained and tested the 3PEAT NP model using only these features. This model achieved an auROC of 0.95, an auPRC of 0.68, and very poor scanning performance (Supplemental Figure 14). TATA and sequence enrichments, even position-specific versions of these features, are neither necessary nor sufficient for transcription. To understand the nature of this observation, we examined the occurrence of TATA in our data set, partitioned by initiation pattern (Figure 6; Supplemental Table 2).
Figure 6.
Comparison of the Percentage of TATA+ versus TATA- Promoters According to PEAT TSS Initiation Pattern.
Here, a TATA+ promoter is a TSS peak upstream region containing a TATA-box signal within TATA’s ROE (approximately −45:−25 with respect to the TSS peak mode). A TATA- promoter does not contain a TATA-box signal in this region. Top: The presence of a TATA-box signal is decided according to a thresholded log-likelihood score that reflects a strict false positive rate (FPR) of 0.0001. Bottom: The same view is provided using a less stringent FPR for TATA-box presence.
[See online article for color version of this figure.]
While TATA presence or absence is clearly a strong predictive factor in all of our 3PEAT promoter signatures, Figure 6 shows that promoters containing a statistically significant TATA signal within the approximate region necessary for pol-II transcription (−45:−25) (TATA+ promoters) are not in the majority for any peak shape category. The percentage of TATA+ promoters associated with an NP shape is higher than other shapes, but clearly no overwhelming association between TATA and NP promoters was observed in our data when TATA location with respect to the TSS is accounted for. Plant promoters in general have been thought to be largely TATA-containing; however, this observation is confounded by the fact that the Arabidopsis genome is AT-rich (Megraw et al., 2006). When a log-likelihood scanning threshold that controls for false positives is used (see Methods: 3PEAT TSS Peak Prediction Model), and the location of the TATA-box signal with respect to TSS is accounted for, we observe that at most ∼22% of the 9325 genes expressed in our data set have a TATA+ promoter. This result is consistent with our observation that, in fact, many other signals not limited to TATA, in the right position-specific combinations, are necessary and sufficient to induce gene expression (Figure 5).
DISCUSSION
High-Throughput TSS Sequencing Is Necessary for Detailed Promoter Analysis
As pol-II is responsible for the transcription of protein coding genes and microRNAs, one of the most expedient methods for identifying transcriptional and posttranscriptional control networks is to first identify known TF binding sequences in the promoter region around a TSS. However, the overall frequency of functional TF binding events in vivo, the functional combinations with other elements, and the positional preferences within plant gene promoters remain largely unknown. In a popular model, the TATA-box and one of several possible plant-specific forms of the Initiator element may alone form a recognizable core promoter of pol-II transcribed genes in plants (Grasser, 2006; Yamamoto et al., 2011). Yet, some well-studied and highly expressed plant genes do not contain an upstream TATA site (Nakamura et al., 2002), and this combination of short sequence elements is abundant in intergenic regions.
Several published works provide TSS analysis for data sets on the order of thousands of sites in plants (Gowda et al., 2006; Yamamoto et al., 2007), the largest of these sets uses CT-MPSS technology in a pooled sample of Arabidopsis tissues (Yamamoto et al., 2009). The CT-MPSS study suggests that plant promoters are divided into two groups, associated with TATA and GA, respectively. In this framework, TATA is the predominant element necessary for sharp, narrow peaks of TSS tags along the genome, whereas GA enrichment gives rise to other TSS landscapes. By contrast, our analysis shows that promoter shape observations and the conclusions drawn from these observations can change dramatically as sequencing depth changes.
The library sample depth and sequencing quality that can be achieved through PEAT is substantially greater than that of past technologies used to generate published plant TSS data sets, such as CT-MPSS. PEAT has benefits and limitations compared with other high-throughput TSS sequencing methods such as CAGE (Shiraki et al., 2003). Benefits include the repeatability of PEAT measurements verified over many technical replicates in published studies (Ni et al., 2010). Both PEAT and CAGE have been shown in animals to yield reproducible data at the level of promoter distribution shapes (Carninci et al., 2006; Ni et al., 2010). PEAT in particular has a reliable cap-trap system, is capable of identifying 5′ location at nucleotide resolution (Ni et al., 2010), and yields strong alignment accuracy due to its paired-end strategy. Using a technique that provides accurate mapping of the 5′ cap locations of pol-II transcripts is the key to our study, and our analysis of ROEs for TATA, Initiator, and a wide variety of additional TFBS elements bears out PEAT data quality at this level. The primary limitation of PEAT is the amount of input material necessary to create a library, as at least 30 μg of total RNA is required. When enough material is available, PEAT provides high-quality data for pol-II TSS regions that are in excellent agreement with past data sets. An advantage of PEAT is that it extends resolution to the point where a carefully designed model can recover TSS peak presence from sequence alone with high probability.
Arabidopsis TSS Initiation Patterns Have Distinct Promoter Signatures
A striking aspect of the model promoter signatures is that they are comprised of many substantially weighted elements, only a few of which have been reported in the literature to have a strong influence on promoter type (e.g., TATA-box, Initiator, and GA content). In fact, for each initiation pattern, most of the elements in the signatures have not been previously discussed in the literature as core promoter elements having location-specific signals. The SQUAMOSA (SQUA) element is just one among many such examples. SQUA is contained in the promoter signatures for all patterns, is most heavily weighted in NP, and has a distinct ROE peak at exactly −36 in all three peak shapes. We provide the ROE tables and enrichment plots for all factors in Supplemental Data Set 1. Beyond the use of many position-specific TF elements, we further observed that although TATA and GA are important for determining the presence of all transcription initiation patterns, the relative importance of these elements differs across pattern type. In the NP case, the signature is dominated by the importance of TATA together with a large block of sequence enrichments including GA content, CA content, Y-Patch, and GC content, followed by many specific TF elements. The WP case is proportionally dominated by TATA and GA. Beyond the TATA and GA elements, a large array of nearly equally weighted TF-specific elements comprise this signature. Finally, the BP signature is in some sense a combination of these qualities; presence of TATA is proportionally important, followed by a particular GA-rich element BPC1 (also highly weighted in WP) and a mixture of lesser-weighted sequence enrichments and TF-specific elements. Our models thus show that multiple sequence enrichments are highly important in plants such as Arabidopsis and perhaps interchangeable in some cases. GA content can play an important role in plant WP cases, unlike in mammals where lowly transcribed tag clusters do not have a clear association with CpG enrichment (Megraw et al., 2009). However, the presence of a variety of TF-specific elements can also collectively contribute a high weight to the likelihood of a WP tag cluster. In this way, GA content in plants is not a perfect analogy for CpG enrichment in mammal genomes. Finally, BPs are not distinguished solely by lack of TATA signal and presence of sequence enrichments; rather, BPs appear to arise as a result of many possible complex combinations of TF elements and sequence enrichments together in a genomic region.
Functional Plant Promoters Do Not Show a Clear Division into Core and Proximal Regions
Our study demonstrates that similar to animal systems (Megraw et al., 2009), many TFs potentially involved in the regulation of genes expressed in the Arabidopsis root have positional constraints relative to the TSS. However, our results strongly suggest that Arabidopsis promoters are not divided into a core promoter region and a proximal promoter region as they are classically defined in animal organisms. The regions immediately upstream of Arabidopsis TSS tag clusters may contain many possible elements, but none of these elements appear to be an absolute requirement for expression. Common pol-II binding elements, such as TATA, do have location-specific signals, but these signals are neither necessary nor sufficient for expression to occur. In our analysis, we find that many enrichments are not only location specific, but their locations are further upstream than the canonical core region. Furthermore, most of these elements were not previously viewed as core elements. We also find that while many enrichments are location specific, others are in fact quite general. Whether these enrichments collectively take the place of CpG Islands is unclear. Our study shows that no single enrichment, including GA enrichment, has the same signal strength as CpG islands in mammals (Megraw et al., 2009). Collectively, all of the position-specific and non-position-specific enrichments appear to form a unified Arabidopsis promoter region.
Conclusion
We used a high-resolution PEAT data set to accurately model the probability of a TSS tag cluster mode, using only DNA sequences as input and our PEAT TSS tags as training locations. It remains a challenge to predict the quantity of transcript expression at any given genomic location, as opposed to the likelihood of a peak shape mode. Additional data, such as information about chromatin status, may be important for determining the absolute quantity of expression. We hope that the data and analysis provided by our study will spur follow-up investigations into open questions, such as the quantity of transcript expression, which our data pose. Our work successfully identified a large number of location-specific TF binding elements that were previously thought to be enhancers. By contrast, we show the existence of clear enrichments in the proximal promoter region within ∼40 nucleotides from the TSS mode. Furthermore, we describe different initiation patterns, in particular NP modes, which allow de novo sequence element searches to be used much more productively than in the past, due to accurate locations for highly expressed TSSs. Finally, our PEAT TSS promoter locations and position-specific TF binding locations will facilitate more accurate computational network analyses in plants, such as those aimed at predicting levels of transcript expression and TF binding to individual promoters. The nature of our high-resolution data set and knowledge of promoter architecture from our 3PEAT model will inform future studies of the control of gene expression that are vital to understanding cellular and organ identity, growth, development, differentiation, and response of plants.
METHODS
PEAT TSS Peaks Data Set Production
Sample and Library Preparation
Arabidopsis thaliana Columbia-0 wild-type seeds were sterilized using 50% (v/v) bleach and 0.1% Tween for 5 min and then rinsed five times with sterile water. Nylon mesh was placed on top of the solidified media (1.0% agar [10 g], 0.5 g of MES [M-2933; Sigma-Aldrich], 1% sucrose [10 g], and 4.33 g of Murashige and Skoog salts [catalog No. 11117-066; Invitrogen], pH to 5.7 to 5.8 with KOH). Following vernalization 2 d at 4°C, sterilized seeds were evenly plated in two dense rows with ∼500 seeds per row. Seedlings were grown vertically on plates for 7 d at 22°C in a Percival with 16 h of constant illumination. Seven days after being placed in the Percival, roots were cut and harvested from the seedlings just below the root-hypocotyl junctions. Samples were ground in liquid nitrogen and total RNA was isolated using the RNeasy kit (Qiagen) according to the manufacturer's instructions. Samples were pooled for 144 μg of total RNA, from which a PEAT library was prepared according to a previously published protocol (Ni et al., 2010). The library was sequenced on an Illumina Hi-Sequation 2000 sequencer.
PEAT TSS Data Processing
Paired-end reads were mapped to the TAIR10 transcriptome (Lamesch et al., 2012) using the same procedure as Ni et al. (2010). TSS tag clusters were then partitioned into NP, BP, and WP initiation patterns (for details, see Supplemental Methods: PEAT TSS Data Processing).
PEAT Data Quality Analysis
PEAT Sequencing Depth Analysis
To determine whether the sequencing depth achieved was sufficient to represent the gene expression state in our pooled Arabidopsis root samples, we performed a saturation analysis on our tag cluster data set. Starting with our full set of stringently mapped reads, we randomly sampled 10%, 20%, and so on up to 100% of the reads; for each sample, we reperformed the same process described above to produce our annotated tag cluster data set, starting with a requirement of 10 reads per tag cluster. For each subsampled, reclustered set of reads, we recorded the number of annotated genes associated with the resulting tag clusters (Supplemental Figure 16A). We observed that the amount of annotated gene increase between subsamples in Supplemental Figure 15A leveled off nearly to zero as the subsample size grew to 90% and then 100% of the reads, indicating that the sampling depth we achieved was within a few percent of the maximum depth that could be achieved. Additional details and analysis are described in Supplemental Methods: Sequencing Depth Analysis.
PEAT Comparison to Existing Data for Coding and Noncoding Transcripts
The list of genes expressed in the PEAT data set was compared with those genes found to be expressed in publically available microarray and RNA-Seq data sets for wild-type Arabidopsis roots (Brady et al., 2007; Li et al., 2013). The mutually expressed genes in these three data sets show excellent overlap (Supplemental Figure 1).
To compare the PEAT tag clusters in our data set associated with annotated miRNAs, we identified 40 peaks whose mode was within 4 kb upstream of an Arabidopsis miRNA precursor recorded in miRBase (Griffiths-Jones et al., 2006) and then compared the location of the mode of each peak with previously reported TSS locations determined using 5′ RACE assays (Xie et al., 2005). Fifteen miRNA precursor upstream regions contained TSSs reported in common to both sets, and we report the distances between the PEAT peak modes and 5′ RACE TSS locations in each case in Results. The outstanding agreement between 5′ RACE locations and PEAT peak modes suggests that the RACE assays effectively sampled the most highly expressed TSS locations for these miRNA-associated regions.
PEAT Peak Distance Comparison with TAIR Annotations
To evaluate how closely our PEAT data agreed with the TAIR10 gene annotation, we calculated the distance between the tag cluster modes located in gene promoter regions (within 500 nucleotides of an annotated TAIR10 gene) and their associated TAIR10 gene start locations. These distances were computed for each initiation pattern (NP, BP, and WP) and at varying cutoff requirements for the number of reads per tag cluster (Supplemental Figure 2).
3PEAT TSS Peak Prediction Model
3PEAT Model Construction
The 3PEAT model was constructed in a similar manner to that described by Megraw et al. (2009).
Data Sets
PEAT tag clusters (or “peaks”) are groups of contiguously mapping PEAT TSS tags, where each TSS tag corresponds to the 5′ end of a capped pol-II RNA transcript. We annotated tag clusters by initiation pattern and TAIR10 association and selected all highly expressed tag clusters for our analysis (see Supplemental Methods: Tag Cluster Annotation). Tag clusters were separated into training and testing partitions prior to model construction (see Supplemental Methods: Model Data Sets).
Regions of Enrichment and Transcription Factor Selection
To identify regions where transcription factor binding sequences are enriched with respect to the TSS, we started with a set of 203 TF positional weight matrices (PWMs) collected from a variety of sources (Grasser, 2006; Megraw et al., 2006; Bryne et al., 2008; Wingender, 2008; Civán and Svec, 2009; Yamamoto et al., 2009). While the PWMs used in this study were derived using experimentally supported data (rather than strictly computational approaches), PWMs are rarely a perfect characterization of TF binding domains or their sequence specificity. Due to their short length, the sequences described by PWMs appear commonly throughout the genome, but many of these identified sequences are not involved in gene regulation. Secondly, PWMs cannot directly capture higher-order relationships between nucleotides. These issues were mitigated through the use of ROEs and background correction.
PWMs were first processed with pseudocounts as described (Megraw et al., 2009). Using the Scanner Toolset for TFBS Discovery (Megraw et al., 2009, 2013; Morton and Megraw, 2014) to compute log-likelihood scores, each PWM was scanned over an 8-kb region centered around the tag cluster mode, identifying the regions where each TF binding site was most likely to occur across all examples in the training data set. ROEs were defined on both strands by identifying the highest scoring region for each PWM across all training examples. PWMs with log-likelihood score peaks (maximum score) up to 4 kb from the TSS were considered. If the scores computed at nucleotides surrounding the peak dropped below the background score for more than 5 nucleotides, the PWM was discarded. Procedural details are previously described (Megraw et al., 2009). We identified ROEs associated with downstream as well as upstream elements (for details, see Supplemental Data Set 1 and Supplemental Methods: Analysis of Downstream Promoter Elements).
Feature Set
Sequence content information and PWMs describing TF binding sequences and their associated ROEs were used together to build a set of features describing each positive and negative training example. The included sequence content features were produced from the GC, CA, and GA content in the 200-nucleotide windows surrounding the peak mode (for details, see Supplemental Methods: Model Features).
PEAT Peaks 3PEAT Model Training and Testing
Our model uses L1-regularized logistic regression, a method which performs automatic feature selection by removing the least significant features in the model. We use the l1_logreg package, an efficient C implementation of logistic regression (Koh et al., 2007). All performance statistics reported here were computed from an independent test set. Cross-validation performance statistics are provided in Supplemental Table 3. See Supplemental Methods (3PEAT Model Training and Testing) for details on the parameter selection and cross-validation procedure.
Gene Scanning
The final 3PEAT models (NP, BP, WP, and ALL) were used to classify each nucleotide in the 8-kb region surrounding each PEAT tag cluster in the model’s test data set, calculating the probability that this location is the mode of a TSS tag cluster (for details, see Supplemental Methods: 3PEAT Model Gene Scanning Procedure).
Analyses of PEAT Peak Sets
PEAT GO Analysis by Peak Type
We determined the overrepresented GO terms (P < 0.01) for the genes associated with each initiation pattern using GOstat (Beissbarth and Speed, 2004). The TAIR10 gene set was selected as the background set (Berardini et al., 2004). The genes expressed in the complete data sets used in model construction and evaluation (combined training and testing) of each initiation pattern (NP, WP, and BP) were considered. Overrepresented GO terms that were unique to each initiation pattern were extracted and are shown in Supplemental Data Set 5, along with the genes associated to each term.
3PEAT Model Feature Analysis
The model feature coefficients were extracted from the L1-Logistic Regression models using a modified version of the l1_logreg tool. The coefficients produced by l1_logreg are on a −1.0 to 1.0 scale. Within a single TF feature group, the coefficients of all ROE windows with non-zero coefficients from both strands were averaged. Supplemental Data Set 6 contains these average coefficient values for all TFBSs that had at least one non-zero coefficient in the model. Coefficients with a value of 0.1 or greater are displayed in Figure 5 and Supplemental Figure 13.
3PEAT Model Analysis of TATA-Less Transcription Start Sites
TATA-Only Model
NP promoters classically have been strongly associated with TATA-box binding sequences. To test the predictive value of this element for NP promoters, we constructed a model containing only the TATA-box and dinucleotide sequence enrichment (GC, GA, and CA) features from our NP tag cluster data set. This model was used to scan for TSS locations in the 8-kb regions surrounding NP tag clusters. Supplemental Figure 14 compares the performance of the full model (labeled NP_100_PS) with this model (NP_100_PS_TATA), revealing its poor predictive performance.
TATA-Less Transcription Start Sites
We queried the proportion of PEAT tag clusters in our TSS data set used in 3PEAT model construction/evaluation (training and test sets were combined) that could be considered to contain a TATA binding site within the TATA-box ROE. A TSS was considered TATA+ if the log-likelihood score at any nucleotide within the TATA-box ROE exceeded a threshold reflective of a particular false positive rate with respect to sequences drawn from the background nucleotide distribution. Each false positive rate for the TATA PWM was computed as described (Megraw et al., 2013).
Data and Software Availability
The full data set of mapped, annotated PEAT tag clusters is available in Supplemental Data Set 7 and at http://megraw.cgrb.oregonstate.edu/suppmats/3PEAT. All 3PEAT model training and test data sets, along with the 3PEAT model classifiers and 3PEAT TFBS-Scanner tool set used in this project for identifying transcription factor binding sites within promoter sequences are publicly available as open source command line tools at http://megraw.cgrb.oregonstate.edu/software/3PEAT.
Accession Numbers
PEAT read alignments (.bam file) have been deposited in the National Center for Biotechnology Information Sequence Read Archive repository under accession number SRR1425301.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. RNA-Seq Gene Expression Comparison.
Supplemental Figure 2. Distance of PEAT Peak Modes to TAIR10 Annotated TSSs.
Supplemental Figure 3. Sharp, Well-Defined Regions of Enrichment for NP Promoters.
Supplemental Figure 4. Sharp, Well-Defined Regions of Enrichment for WP Promoters.
Supplemental Figure 5. Sharp, Well-Defined Regions of Enrichment for BP Promoters.
Supplemental Figure 6. Model Performance on Narrow Peak Modes.
Supplemental Figure 7. Model Performance on Broad with Peak and Weak Peak Modes.
Supplemental Figure 8. Model Performance on ALL Modes.
Supplemental Figure 9. Model Performance on Genomic Sequence Scans.
Supplemental Figure 10. Model Performance on miRNA-Proximal Modes.
Supplemental Figure 11. Model Performance on TAIR10 annotated TSSs.
Supplemental Figure 12. Model Performance on TAIR10 Genomic Sequence Scans.
Supplemental Figure 13. 3PEAT Promoter Signatures, Detailed for All Shape Models.
Supplemental Figure 14. Model Performance with TATA-Box and Enrichment Features Only.
Supplemental Figure 15. Tag Cluster Shape Definitions.
Supplemental Figure 16. Sampled Read Depth Saturation Analysis.
Supplemental Figure 17. Proportions of TATA+/TATA- Promoters.
Supplemental Table 1. Counts of Tag Clusters in Each Peak Shape Data Set Used in 3PEAT Model.
Supplemental Table 2. Comparison of the Number of TATA+ versus TATA- Promoters by PEAT TSS Initiation Pattern.
Supplemental Table 3. Cross-Validation Performance: auROC and auPRC Statistics for Each Cross-Validation Fold of Each 3PEAT Model Trained.
Supplemental Methods. Data Set Processing and Partitioning, Model Construction, and Model Evaluation.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.r2342.
Supplemental Data Set 1. Regions of Enrichment for all 3PEAT Models.
Supplemental Data Set 2. Genome Scanning Results: Narrow Peak.
Supplemental Data Set 3. Genome Scanning Results: Broad with Peak.
Supplemental Data Set 4. Genome Scanning Results: Weak Peak.
Supplemental Data Set 5. Overrepresented GO Terms Uniquely Associated with NP, WP, and BP Shape Categories.
Supplemental Data Set 6. 3PEAT Logistic Regression Coefficients for Each Model.
Supplemental Data Set 7. PEAT Peak Complete Data Set and Data Sets for 3PEAT Model Training/Testing.
Supplementary Material
Acknowledgments
M.M. was supported by an NIH K99-R00 Pathway to Independence Award GM097188. T.M. was supported by startup funds from Oregon State University. J.P. is supported by a National Institutes of Health Ruth L. Kirschstein NRSA award (F32GM086976). P.N.B. and U.O. are funded by National Science Foundation Award IOS- 1021619. U.O. additionally acknowledges support from National Science Foundation Award MCB-0822033. We thank Jun Zhu and his laboratory for PEAT protocol insights and sequencing support. We thank Andrea Gosset for her thoughtful conversation and assistance in vetting PEAT raw data processing methods.
AUTHOR CONTRIBUTIONS
M.M. and U.O. designed the study. D.L.C. contributed ideas for the design of the study. M.M. carried out the experiments, data analysis, and algorithm design. T.M. performed algorithm implementation and contributed to analysis of results. J.P., C.M.W., A.C., and P.N.B. contributed to laboratory experiments and troubleshooting. D.L.C. and S.L. contributed to computational methods for data analysis. U.O. and P.N.B. contributed to the evaluation of results. M.M. and T.M. wrote the article. All authors contributed to editing of the article and approved the final version of the article.
Glossary
- TSS
transcriptional start site
- TFBS
transcription factor binding site
- RACE
rapid amplification of cDNA ends
- TF
transcription factor
- miRNA
microRNA
- PEAT
paired end analysis of transcription start sites
- NP
narrow peak
- BP
broad with peak
- WP
weak peak
- TF
transcription factor
- ROE
region of enrichment
- auROC
area under the ROC curve
- auPRC
area under the precision recall curve
- GO
Gene Ontology
- PWM
positional weight matrix
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
References
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Batut P., Dobin A., Plessy C., Carninci P., Gingeras T.R. (2013). High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23: 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T., Speed T.P. (2004). GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20: 1464–1465. [DOI] [PubMed] [Google Scholar]
- Berardini T.Z., et al. (2004). Functional annotation of the Arabidopsis genome using controlled vocabularies. Plant Physiol. 135: 745–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya M., Das M., Bandyopadhyay S. (2012). miRT: a database of validated transcription start sites of human microRNAs. Genomics Proteomics Bioinformatics 10: 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady S.M., Orlando D.A., Lee J.Y., Wang J.Y., Koch J., Dinneny J.R., Mace D., Ohler U., Benfey P.N. (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806. [DOI] [PubMed] [Google Scholar]
- Bruex A., Kainkaryam R.M., Wieckowski Y., Kang Y.H., Bernhardt C., Xia Y., Zheng X., Wang J.Y., Lee M.M., Benfey P., Woolf P.J., Schiefelbein J. (2012). A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet. 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. (2008). JASPAR, the open access database of transcription factor-binding profiles: new content and tools in the 2008 update. Nucleic Acids Res. 36: D102–D106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P., et al. (2006). Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 38: 626–635. [DOI] [PubMed] [Google Scholar]
- Carninci P., et al. FANTOM Consortium. RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) (2005). The transcriptional landscape of the mammalian genome. Science 309: 1559–1563. [DOI] [PubMed] [Google Scholar]
- Civán P., Svec M. (2009). Genome-wide analysis of rice (Oryza sativa L. subsp. japonica) TATA box and Y Patch promoter elements. Genome 52: 294–297. [DOI] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M. (2006). A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 103: 5320–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deaton A.M., Bird A. (2011). CpG islands and the regulation of transcription. Genes Dev. 25: 1010–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. (2008). A code for transcription initiation in mammalian genomes. Genome Res. 18: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowda M., Li H., Alessi J., Chen F., Pratt R., Wang G.L. (2006). Robust analysis of 5′-transcript ends (5′-RATE): a novel technique for transcriptome analysis and genome annotation. Nucleic Acids Res. 34: e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser, K.E., ed (2006). Regulation of Transcription in Plants. (Oxford, UK: Wiley-Blackwell). [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. (2006). miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 34: D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünberg S., Hahn S. (2013). Structural insights into transcription initiation by RNA polymerase II. Trends Biochem. Sci. 38: 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins R.A., et al. (2011). Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 21: 182–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjani H., Zavolan M. (2014). TSSer: an automated method to identify transcription start sites in prokaryotic genomes from differential RNA sequencing data. Bioinformatics 30: 971–974. [DOI] [PubMed] [Google Scholar]
- Juven-Gershon T., Kadonaga J.T. (2010). Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 339: 225–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J.T. (2004). Regulation of RNA polymerase II transcription by sequence-specific DNA binding factors. Cell 116: 247–257. [DOI] [PubMed] [Google Scholar]
- Kadonaga J.T. (2012). Perspectives on the RNA polymerase II core promoter. Wiley interdisciplinary reviews. Dev. Biol. 1: 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P. (2009). From transcription start site to cell biology. Genome Biol. 10: 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh K., Kim S.-J., Boyd S. (2007). An interior-point method for large-scale l1-regularized logistic regression. Mach. Learn. Res. 8: 1519–1555. [Google Scholar]
- Lamesch P., et al. (2012). The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 40: D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan P., Li W., Lin W.-D., Santi S., Schmidt W. (2013). Mapping gene activity of Arabidopsis root hairs. Genome Biol. 14: R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Liberman L.M., Mukherjee N., Benfey P.N., Ohler U. (2013). Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res. 23: 1730–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo J.M., Jiménez-Valverde A., Real R. (2008). AUC: a misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 17: 145–151. [Google Scholar]
- Marco A., Ninova M., Ronshaugen M., Griffiths-Jones S. (2013). Clusters of microRNAs emerge by new hairpins in existing transcripts. Nucleic Acids Res. 41: 7745–7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Baev V., Rusinov V., Jensen S.T., Kalantidis K., Hatzigeorgiou A.G. (2006). MicroRNA promoter element discovery in Arabidopsis. RNA 12: 1612–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Mukherjee S., Ohler U. (2013). Sustained-input switches for transcription factors and microRNAs are central building blocks of eukaryotic gene circuits. Genome Biol. 14: R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megraw M., Pereira F., Jensen S.T., Ohler U., Hatzigeorgiou A.G. (2009). A transcription factor affinity-based code for mammalian transcription initiation. Genome Res. 19: 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton, T., and Megraw, M. (2014). 3PEAT TFBS-Scanner Toolset, http://megraw.cgrb.oregonstate.edu/software/3PEAT/.
- Nakamura M., Tsunoda T., Obokata J. (2002). Photosynthesis nuclear genes generally lack TATA-boxes: a tobacco photosystem I gene responds to light through an initiator. Plant J. 29: 1–10. [DOI] [PubMed] [Google Scholar]
- Nepal C., et al. (2013). Dynamic regulation of the transcription initiation landscape at single nucleotide resolution during vertebrate embryogenesis. Genome Res. 23: 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni T., Corcoran D.L., Rach E.A., Song S., Spana E.P., Gao Y., Ohler U., Zhu J. (2010). A paired-end sequencing strategy to map the complex landscape of transcription initiation. Nat. Methods 7: 521–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D., Morris A.R., Battenhouse A., Iyer V.R. (2014). Simultaneous mapping of transcript ends at single-nucleotide resolution and identification of widespread promoter-associated non-coding RNA governed by TATA elements. Nucleic Acids Res. 42: 3736–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach E.A., Winter D.R., Benjamin A.M., Corcoran D.L., Ni T., Zhu J., Ohler U. (2011). Transcription initiation patterns indicate divergent strategies for gene regulation at the chromatin level. PLoS Genet. 7: e1001274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rach E.A., Yuan H.-Y., Majoros W.H., Tomancak P., Ohler U. (2009). Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 10: R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K., Chen X. (2013). Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25: 2383–2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini H.K., Griffiths-Jones S., Enright A.J. (2007). Genomic analysis of human microRNA transcripts. Proc. Natl. Acad. Sci. USA 104: 17719–17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxonov S., Berg P., Brutlag D.L. (2006). A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 103: 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmuradov I.A., Solovyev V.V., Gammerman A.J. (2005). Plant promoter prediction with confidence estimation. Nucleic Acids Res. 33: 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahmuradov I.A., Gammerman A.J., Hancock J.M., Bramley P.M., Solovyev V.V. (2003). PlantProm: a database of plant promoter sequences. Nucleic Acids Res. 31: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraki T., et al. (2003). Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 100: 15776–15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T. (2003). The RNA polymerase II core promoter. Annu. Rev. Biochem. 72: 449–479. [DOI] [PubMed] [Google Scholar]
- Thomas M.C., Chiang C.M. (2006). The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41: 105–178. [DOI] [PubMed] [Google Scholar]
- Wingender E. (2008). The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief. Bioinform. 9: 326–332. [DOI] [PubMed] [Google Scholar]
- Xie Z., Allen E., Fahlgren N., Calamar A., Givan S.A., Carrington J.C. (2005). Expression of Arabidopsis MIRNA genes. Plant Physiol. 138: 2145–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Ichida H., Abe T., Suzuki Y., Sugano S., Obokata J. (2007). Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 35: 6219–6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Yoshioka Y., Hyakumachi M., Obokata J. (2011). Characteristics of core promoter types with respect to gene structure and expression in Arabidopsis thaliana. DNA Res. 18: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y.Y., Yoshitsugu T., Sakurai T., Seki M., Shinozaki K., Obokata J. (2009). Heterogeneity of Arabidopsis core promoters revealed by high-density TSS analysis. Plant J. 60: 350–362. [DOI] [PubMed] [Google Scholar]
- Yamashita R., Sathira N.P., Kanai A., Tanimoto K., Arauchi T., Tanaka Y., Hashimoto S., Sugano S., Nakai K., Suzuki Y. (2011). Genome-wide characterization of transcriptional start sites in humans by integrative transcriptome analysis. Genome Res. 21: 775–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.