The extracted tetraploid wheat (ETW) containing the BBAA subgenomes of hexaploid bread wheat has a stabilized karyotype but anomalous phenotypes. Genome-wide comparisons between ETW and natural tetraploid wheat revealed a large number of differentially expressed genes in ETW; these changes showed early occurrence and evolutionary persistence during bread wheat evolution.
Abstract
Subgenome integrity in bread wheat (Triticum aestivum; BBAADD) makes possible the extraction of its BBAA component to restitute a novel plant type. The availability of such a ploidy-reversed wheat (extracted tetraploid wheat [ETW]) provides a unique opportunity to address whether and to what extent the BBAA component of bread wheat has been modified in phenotype, karyotype, and gene expression during its evolutionary history at the allohexaploid level. We report here that ETW was anomalous in multiple phenotypic traits but maintained a stable karyotype. Microarray-based transcriptome profiling identified a large number of differentially expressed genes between ETW and natural tetraploid wheat (Triticum turgidum), and the ETW-downregulated genes were enriched for distinct Gene Ontology categories. Quantitative RT-PCR analysis showed that gene expression differences between ETW and a set of diverse durum wheat (T. turgidum subsp durum) cultivars were distinct from those characterizing tetraploid cultivars per se. Pyrosequencing revealed that the expression alterations may occur to either only one or both of the B and A homoeolog transcripts in ETW. A majority of the genes showed additive expression in a resynthesized allohexaploid wheat. Analysis of a synthetic allohexaploid wheat and diverse bread wheat cultivars revealed the rapid occurrence of expression changes to the BBAA subgenomes subsequent to allohexaploidization and their evolutionary persistence.
INTRODUCTION
Polyploidy (whole-genome duplication [WGD]) is ubiquitous in the evolution of angiosperms, as all extant flowering plants harbor WGD signatures in their genomes (Soltis and Soltis, 2009; Amborella Genome Project, 2013; Madlung, 2013). Allopolyploidy (coupling WGD with interspecific hybridization) in particular has played a significant role in the speciation of vascular plants, including major agricultural crops (Feldman et al., 1995; Wendel, 2000; Soltis and Soltis, 2009; Renny-Byfield and Wendel, 2014). Given sufficient time, structural genetic diploidization or physical fractionation (selective loss and retention of genes and noncoding DNAs) at the genome-wide scale leading to paleopolyploidy is a common outcome of all WGDs (Birchler, 2012; Freeling et al., 2012; Pont et al., 2013; Roulin et al., 2013). In the initial generations following allopolyploidy, however, diverse genetic, epigenetic, and structural subgenome modifications arise, presumably to facilitate functional coordination between the duplicated divergent subgenomes (Madlung and Wendel, 2013). Indeed, an array of investigations in diverse plant taxa have documented that allopolyploidization induces a cascade of rapidly occurring genomic modifications as well as alterations in gene expression (reviewed in Wendel, 2000; Comai, 2005; Chen, 2007; Otto, 2007; Doyle et al., 2008; Jackson and Chen, 2010; Feldman and Levy, 2012; Soltis and Soltis, 2012). It has been suggested that the degree of robustness to these rapid genomic and expression changes may determine the fates of the newly formed allopolyploid individuals, as new species or on a path to extinction (Chelaifa et al., 2010; Buggs et al., 2011; Feldman and Levy, 2012; Hegarty et al., 2013; Madlung, 2013). Notwithstanding these many studies, to date the biological effects associated with specific modifications of a subgenome(s) in isolation, for a given allopolyploid species, remain unclear.
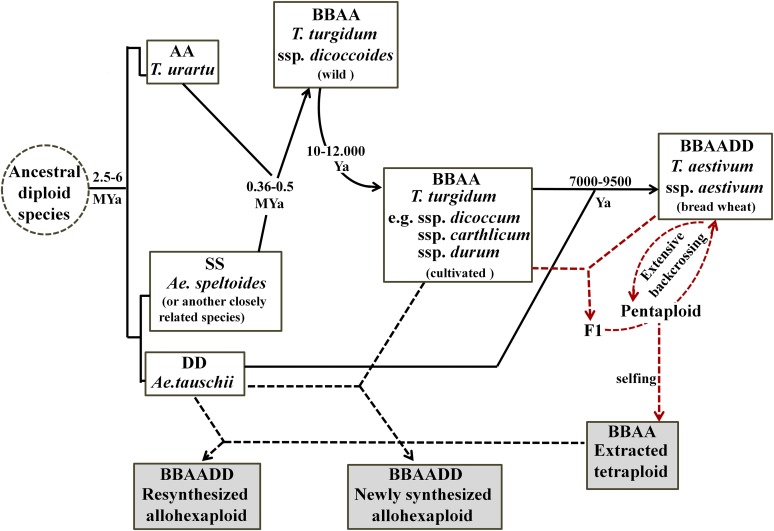
The origin of allohexaploid bread wheat (Triticum aestivum) involved two sequential allopolyploidization events. First, allotetraploidization between two diploid species, represented by modern Triticum urartu (genome AA) and a yet undiscovered or extinct goatgrass species closely related to the Sitopsis section of Aegilops (genome SS≈BB), led to the origin of allotetraploid wheat 0.36 to 0.5 million years ago (Dvořák, 1976; Huang et al., 2002; Dvorak and Akhunov, 2005) (Figure 1). Second, allohexaploidization ∼10,000 years ago between a primitive form of domesticated tetraploid wheat (closely related to Triticum turgidum subsp durum) and goatgrass (Aegilops tauschii; genome DD) led to the formation of bread wheat (Feldman et al., 1995; Salamini et al., 2002) (Figure 1). The allopolyploidization event at the hexaploid level can be reproduced in the laboratory by crossing the corresponding progenitor species and inducing WGD in the F1 hybrids. Thus, to an extent, the initial stage of the evolutionary trajectory associated with the speciation of T. aestivum can be recapitulated in newly synthesized allohexaploid wheat (Figure 1).
Figure 1.
Schematic Representation of the Evolutionary History of Wheat (Triticum) and Related Aegilops Species and the Three Synthetic Polyploid Wheat Lines Used in This Study.
Diagrammatic illustration of the divergent speciation of diploid progenitor species (containing A, S [≈B], or D genomes) of polyploid wheat, the evolutionary trajectories and/or domestication processes of natural tetraploid and hexaploid wheats, and the construction (dashed arrows) of the three synthetic polyploid wheat lines used in this study. The synthetic wheat lines (shaded) included an ETW containing the BBAA genomes of a bread wheat cultivar, a resynthesized allohexaploid (XX329) produced by crossing ETW and Ae. tauschii followed by WGD, and a newly synthesized allohexaploid (Allo960) produced by crossing T. turgidum and Ae. tauschii followed by WGD. The estimated evolution or domestication timing is according to Feldman et al. (1995), Huang et al. (2002), and Dvorak and Akhunov (2005).
[See online article for color version of this figure.]
A series of studies in newly synthesized wheats have shown that extensive changes in gene expression arise during the early generations following allopolyploidization (reviewed in Feldman and Levy, 2012; Li et al., 2014). In addition, studies of natural bread wheat cultivars indicate that homoeolog-specific gene silencing, subfunctionization, and subgenome-specific alternative splicing are prevalent (Bottley et al., 2006; Akhunova et al., 2010; Akhunov et al., 2013). Structurally, however, the three constituent subgenomes of bread wheat remain largely intact, with only a few intersubgenome translocations (Jiang and Gill, 1994), presumably owning to the presence of the Ph1 locus, which ensures exclusive homologous chromosome pairing in meiosis (Griffiths et al., 2006). Because of these attributes, it is feasible to extract the BBAA component from bread wheat by hybridization to a tetraploid wheat, followed by repeated backcrossing to the hexaploid wheat (TAA10) as the recurrent parent, to construct a ploidy-reversed (from hexaploid to tetraploid) “extracted” tetraploid wheat (ETW) with a genomic composition of BBAA that is virtually identical to the BBAA subgenomes of its bread wheat donor (Kerber, 1964).
An intriguing but barely explored issue in allopolyploid genome evolution is whether and to what extent allopolyploidy induces karyotypic, genomic, and transcriptomic changes to its constituent subgenomes, as well as ancillary questions about the timing of these changes and their biological consequences. These issues can be addressed if the constituent subgenome(s) of a given allopolyploid organism remain largely intact and, hence, can be extracted to restitute an independent organism, as shown for hexaploid bread wheat. If alterations in karyotype, genomes, and gene expression did not occur to the BBAA component of bread wheat during its history in an allohexaploid nucleus, then extracted BBAA individuals should be little modified from the original hybridizing parental species containing the BBAA genomes. If, on the other hand, alterations in karyotype, genome, and/or transcriptome have arisen during this period, then the extracted BBAA subgenomes should be phenotypically and genomically distinct from the original founder progenitor tetraploid.
Some evidence suggests that the latter scenario is most likely. First, genomic and gene expression analyses indicate that the BBAA subgenomes have been modified by the added DD subgenome following allohexaploidization (Pont et al., 2011, 2013). Second, independent structural subgenome evolution in an allopolyploid genomic environment may also generate heritable modifications to subgenome expression timing in the allohexaploid environment (Adams et al., 2003). In fact, the abnormal phenotypes of ETW (Kerber, 1964) provide de facto evidence that alterations in gene expression and/or function have occurred to the BBAA subgenomes of bread wheat during its history at the allohexaploid level. Third, resynthesized allohexaploid wheat (parented by an ETW) was found to exhibit mostly additive gene expression (Chelaifa et al., 2013; but see Akhunova et al., 2010), which contrasts with the higher proportions of nonadditive expression generally found in different newly synthesized allohexaploid wheats parented by normal tetraploid wheats (Pumphrey et al., 2009; Chagué et al., 2010; Qi et al., 2012; Li et al., 2014); these observations suggest fundamental differences between ETW and its natural counterpart.
The availability of a fully ETW, a resynthesized allohexaploid wheat (parented by ETW), a newly synthesized allohexaploid wheat (parented by natural tetraploid wheat), and representative natural tetraploid and hexaploid wheat genotypes provides a tractable system with which to systemically address the issues of whether and to what extent changes to karyotypic stability and gene expression of the BBAA component of bread wheat have occurred due to a history at the allohexaploid level. Here, we detail these changes and their phenotypic consequences.
RESULTS
Extracted Allotetraploid Wheat Has a Stable Karyotype but Exhibits Aberrant Phenotypes
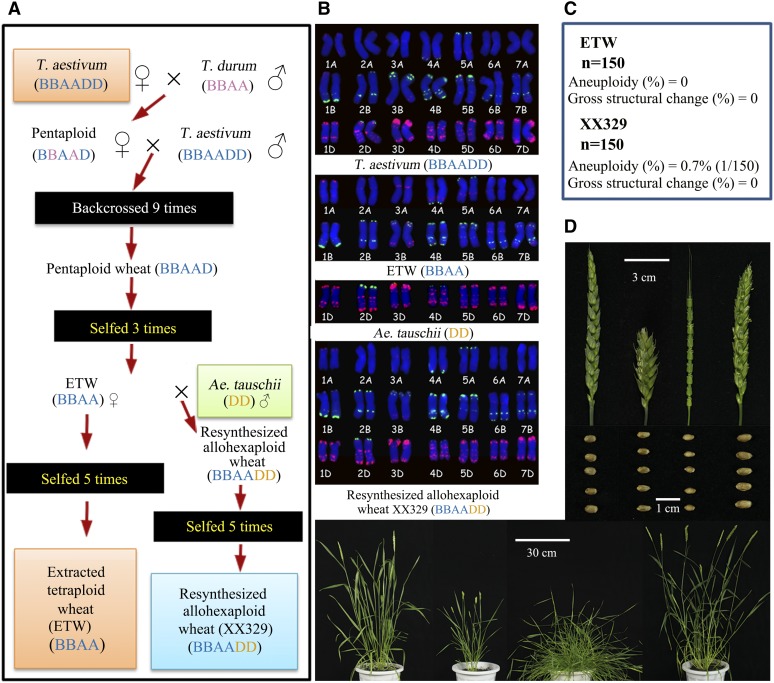
ETW contains a BBAA genome that is virtually identical to the BBAA subgenomes of its allohexaploid bread wheat donor (T. aestivum cv Canthach; designated as TAA10), from which it was extracted via hybridization and nine cycles of backcrossing (Figure 2A; Kerber, 1964). Thus, in theory, the genome (BBAA) of ETW should be >99.8% identical to the BBAA subgenomes of its bread wheat donor after the ninth backcross [1 − (1/29)]. For the same reason, other types of de novo genomic changes, if they occurred in the BBAA component during the initial hybridization between tetraploid wheat and hexaploid wheat (line TAA10) and/or in the process of backcrossing of the resultant pentaploid with TAA10 as the recurrent parent, should have been eliminated during nine cycles of backcrossing. ETW showed high stability in karyotype: no aneuploidy or gross chromosomal rearrangements were detected among 150 plants randomly taken from three consecutive selfed generations by sequential fluorescent in situ hybridization (FISH) and genomic in situ hybridization (GISH) analyses (Figures 2B and 2C; Supplemental Figure 1). This combined FISH/GISH karyotyping enables unequivocal identification of each of the 21 homologous chromosome pairs in hexaploid wheat (Zhang et al., 2013b) as well as efficient diagnosis of structural rearrangements (Xiong et al., 2011; Chester et al., 2012). For example, various kinds of small intergenomic rearrangements (including reciprocal exchanges and unidirectional homoeologous transfer) between subgenomes A and D or S (≈B) and D in two newly synthesized allotetraploid wheat lines with genome combinations AADD (T. urartu × Ae. tauschii) and SSDD (Aegilops bicornis × Ae. tauschii), respectively, can be readily detected by FISH/GISH karyotyping (Zhang et al., 2013a; Supplemental Figure 2). Therefore, the FISH/GISH results of ETW indicated that aneuploidy and intergenomic exchanges involving large chromosomal segments translocated from D to B or A, or between B and A subgenomes, did not occur after eliminating the DD subgenome (Figure 2B; Supplemental Figure 1). This is consistent with the earlier meiotic data of Kerber (1964), who found no evidence for intergenomic homoeologous paring in the intermediate pentaploid (BBAAD). Together, it is clear that elimination of the DD subgenome in bread wheat did not compromise karyotype stability. Although pollen contamination to ETW during backcrossing or selfing by a normal tetraploid wheat can explain this result, this possibility is ruled out because this would immediately be noted by the distinct phenotype of the resulting F1 hybrid (Supplemental Figure 3).
Figure 2.
Description and Characterization of the ETW and Resynthesized Allohexaploid Wheat.
(A) Diagrammatic illustration of the pedigree for the construction of ETW from allohexaploid bread wheat (cv Canthach; our designation is TAA10) by hybridization with a tetraploid line of subsp durum (cv Stewart; our designation is TTR13) and repeated backcrossing to the hexaploid parent and resynthesizing allohexaploid wheat (XX329). This is based on Kerber (1964) with additional backcrossing for two times and selfing for propagations. The same genomes in different colors denote intraspecific differentiation.
(B) Karyotypes of bread wheat (cv Canthach; our designation is TAA10), ETW, Ae. tauschii (line TQ18), and XX329 based on sequential FISH using four hybridization probes to identify each chromosome pair.
(C) Nearly complete euploidy and lack of gross structural rearrangements in both ETW and XX329 were evidenced by karyotyping 150 randomly chosen plant individuals from each line.
(D) Typical phenotypes of the bread wheat donor (line TAA10) to ETW, Ae. tauschii (line TQ18), and the resynthesized allohexaploid wheat (XX329). Severely deteriorated phenotypes in spikes and whole plants are evident in ETW, which, however, are fully restored in the resynthesized allohexaploid wheat (XX329) by crossing ETW (maternal parent) and Ae. tauschii (paternal parent). Notably, although kernels of ETW do not show apparent deterioration, its seed-setting rate is conspicuously reduced to only approximately 20%, which is also fully restored in XX329.
The karyotype stability of ETW was also manifested in the resynthesized allohexaploid wheat (XX329) with ETW as the maternal parent and Ae. tauschii as the paternal parent (Kerber, 1964; Figure 2A), as XX329 also showed high karyotype stability with a low frequency (<1%) of aneuploidy and no gross structural rearrangements in 150 karyotyped individuals randomly taken from three consecutive generations (Figure 2C; Supplemental Figure 1). This high karyotype stability in XX329 provides a contrast with newly synthesized allohexaploid wheats parented by natural tetraploid wheat (T. turgidum), in which whole-chromosome aneuploidy was found to occur frequently (Zhang et al., 2013b). Together, the karyotyping data suggested that the property of karyotype stability that evolved in allohexaploid bread wheat is maintained in its extracted BBAA component. This is in line with recent findings in the genetic control of meiotic regularity in an adapted natural allopolyploid of Arabidopsis thaliana, in the sense that the controlling genetic factor(s) exerts its effect irrespective of altered genetic backgrounds (Henry et al., 2014). Therefore, it appears that different genes or pathways have evolved to serve the purpose of karyotype stabilization by similar mechanisms in different allopolyploid species. It should be cautioned that the conclusion of “evolved karyotype stability” based on comparisons of natural and synthetic polyploidy cannot rule out the possibility that the actual diploid progenitors leading to the polyploid species formation may have different properties from their extant counterparts used to generate the synthetic polyploids (Gottlieb, 2004).
Despite karyotype stability, ETW manifested severely anomalous phenotypes at multiple growth/developmental stages relative to natural allotetraploid wheat (T. turgidum subsp durum), as originally reported by Kerber (1964). These include dwarfed stature, decreased number of tillers, compacted spikes, and reduced fertility (<20% seed set under our conditions) (Figure 2D). Given that its bread wheat donor (line TAA10) is normal (Kerber, 1964; Figure 2D), the abnormal phenotypes of ETW indicate that the DD subgenome is essential as well as sufficient to compensate for the compromised functionality of the BBAA subgenomes of bread wheat, as confirmed by the fully restored normal phenotypes in the resynthesized allohexaploid wheat (XX329) parented by ETW (Kerber, 1964; Figure 2D). Given the wide-ranging phenotypic abnormalities in ETW, we suspect that many (rather than a few) genes may have been affected in its genome. An additional phenotypic observation supporting this possibility was the extreme variability in phenotypes of a single morphological trait, spike shape, in the F2 progeny of a cross between ETW and a durum wheat cultivar (Supplemental Figure 4).
Transcriptome Changes to the BBAA Component of Bread Wheat Reflect Effects of Allohexaploidization and Selection under Domestication
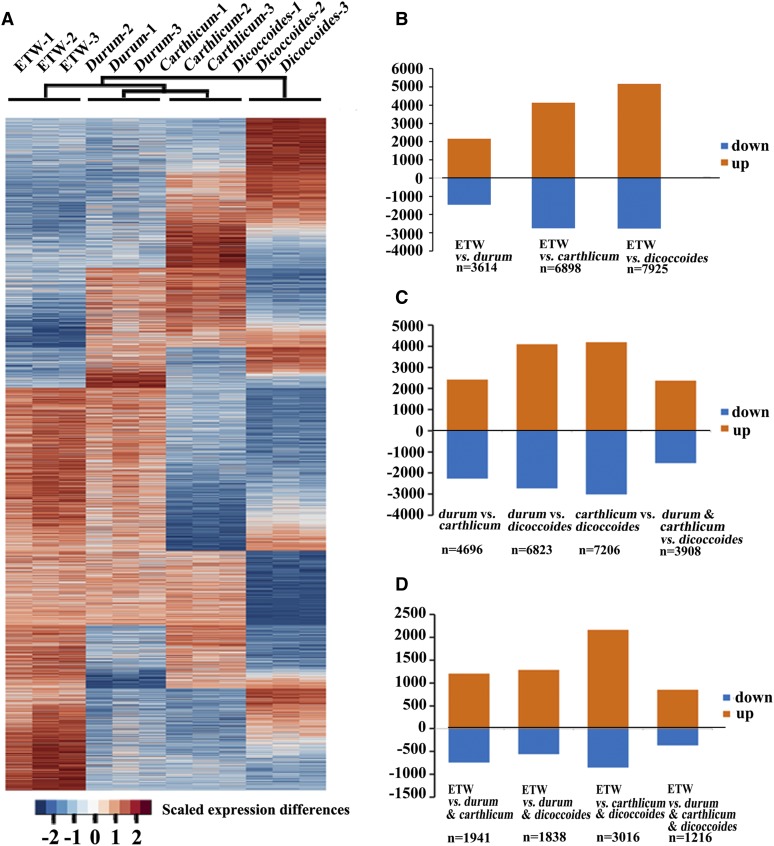
The dramatically altered phenotypes in ETW point to genomic and/or gene expression changes to the BBAA component of bread wheat since its origin. Thus, we compared the transcriptome of ETW with those of T. turgidum, represented by three subspecies, durum (cv TTR13), carthlicum (cv Blackbird), and dicoccoides (line TD265), using the Affymetrix GeneChip Wheat Genome Array with three biological replicates for each genotype. We found 3614 (13.6%), 6898 (26.0%), and 7925 (29.9%) genes being differentially expressed in ETW versus durum, carthlicum, and dicoccoides, respectively (Figures 3A and 3B; Supplemental Table 1). We then compared pairwise transcriptome differences between any two of the three natural subspecies of T. turgidum and detected similar numbers of differentially expressed genes to those of ETW versus each of the natural subspecies (Figures 3A and 3C; Supplemental Table 1). Together, transcriptome divergence, with respect to expression similarity, among all the analyzed tetraploid wheats led to the following relationships: durum was most similar to carthlicum, which as a subgroup associated with ETW, with these three then associated with dicoccoides (Figure 3A). This transcriptome-based relationship in gene expression similarity was consistent with the known evolution and/or domestication histories of these lines: the three lines, durum, carthlicum, and ETW (representing hexaploid bread wheat), are all domesticated forms originating from dicoccoides, perhaps via a common free-threshing primitive accession (Feldman et al., 1995; Figure 1); hence, their more distant relationship with their shared wild progenitor (Figure 3A) most likely reflects selection on gene expression under domestication at the tetraploid level. Interestingly, however, ETW, which in theory should be more closely related to durum (which was the most probable tetraploid progenitor of common wheat; Salamini et al., 2002), was only sister to both the domesticated subspecies of durum and carthlicum as a subgroup (Figure 3A). This unexpected relationship at the transcriptome level suggests that the gene expression changes to the BBAA component of bread wheat have, in fact, exceeded the divergence between the two domesticated subspecies, which points to the effects of allohexaploidization and selection under domestication at the hexaploid level.
Figure 3.
Transcriptome Differences between or among the Four Allotetraploid Wheat Lines, ETW and T. turgidum subsp durum (cv TTR13), subsp carthlicum (cv Blackbird), and subsp dicoccoides (Line TD265), Based on Affymetrix Wheat Genome Array Analysis.
(A) Hierarchical clustering of differentially expressed genes, based on the microarray data, between ETW and each of the three natural allotetraploid subspecies, durum, carthlicum, and dicoccoides, of T. turgidum. All three biological replicates of each line are shown. The color key is indicated at the bottom.
(B) Histograms of the total numbers of differentially expressed genes in each of the three pairwise comparisons between ETW and three subspecies, durum, carthlicum, and dicoccoides, of T. turgidum and numbers of upregulated and downregulated genes (vermilion and blue bars, respectively) in each comparison.
(C) Histograms of the total numbers of differentially expressed genes between any two and between common genes of durum and carthlicum versus dicoccoides and numbers and proportions of upregulated and downregulated genes (vermilion and blue bars, respectively) in each comparison.
(D) Histograms of the total numbers of differentially expressed genes between ETW and any two or all three subspecies, durum, carthlicum, and dicoccoides, of T. turgidum and numbers and proportions of upregulated and downregulated genes (vermilion and blue bars, respectively) in each comparison.
Next, we analyzed the proportions of upregulated versus downregulated genes for a given comparison. Of the ETW versus each of the three natural subspecies of T. turgidum, durum, carthlicum, and dicoccoides, 2153 (59.6%), 4141 (60.0%), and 5161 (65.1%) genes were upregulated while 1461 (40.4%), 2757 (40.0%), and 2764 (34.9%) genes were downregulated, respectively (Figure 3B; Supplemental Table 1). These pairwise comparisons revealed an interesting observation: the BBAA transcriptome of ETW has been changed toward a generally higher expression level than those of the three natural subspecies of T. turgidum (differences of upregulation versus downregulation for all three comparisons were statistically significant according to a binomial test [P < 2.2e-16]; Supplemental Table 1). Notably, this trend was most pronounced between ETW and dicoccoides (upregulation versus downregulation = 65.1:34.9; P = 4.82e-162), implicating the combined effects of two sequential episodes of allopolyploidization and selection under domestication. To test this further, we analyzed transcriptome differences between ETW and the shared genes between any two and among all three natural subspecies of T. turgidum. We found similar proportions of more upregulated than downregulated genes as those between the pairwise comparisons (Figure 3D; Supplemental Table 1). Furthermore, we found significantly more upregulation than downregulation between durum and dicoccoides and between carthlicum and dicoccoides, although these were to a lesser extent, relative to that between ETW and dicoccoides (Figure 3C; Supplemental Table 1). Similar results of higher gene expression levels in domesticated than wild tetraploid wheats were also observed previously for a smaller number of genes (Ayal et al., 2005). By contrast, the proportion of upregulation versus downregulation in the differentially expressed genes between durum and carthlicum, both being domesticated tetraploid forms, showed the expected 1:1 ratio (Figure 3C; Supplemental Table 1). Together, these transcriptome comparisons suggest that (1) domestication at the tetraploid level might have significantly increased the expression level of a substantial proportion of genes in durum and carthlicum relative to their common wild progenitor dicoccoides, and (2) this effect may have been further reinforced at the hexaploid level in bread wheat, which has been fixed in the BBAA component of bread wheat, as reflected in ETW. Although we purposely chose three subspecies to broadly represent T. turgidum diversity, we need to acknowledge the alternative possibility that the exact founder genotype of T. turgidum that donated the BBAA genome to hexaploid bread wheat might be different from those we analyzed. This issue is inherent in all studies using model extant accessions to represent actual progenitors of naturally formed polyploids (Gottlieb, 2004).
The consistent microarray data among the three biological replicates for a given line points to the reliability of these data (Figure 3A). This was further tested by real-time quantitative RT-PCR (qRT-PCR) analysis of 36 comparisons (Supplemental Data Set 1) representing upregulation or downregulation between ETW and T. turgidum. It was found that 33 of the 36 analyzed genes (92%) showed consistency between the qRT-PCR data and the microarray data in terms of both expression level and direction (Supplemental Figure 5), thus validating the reliability of the microarray data and analysis.
To further test if the transcriptome changes to the BBAA subgenomes of bread wheat were under trans-regulation by the DD subgenome, we analyzed gene expression in a resynthesized allohexaploid wheat (XX329) relative to its mid parent values (MPVs). Our assumption was that if the transcriptome changes were not trans-regulated by the DD subgenome, then the affected genes should exhibit additive expression in the resynthesized allohexaploid; conversely, immediate subgenome interactions would occur upon reintroducing the DD subgenome, and hence, nonadditive expression of these genes would be seen in XX329. We analyzed microarray-based expression levels and patterns for the differentially expressed genes from each of the comparisons between ETW and T. turgidum in XX329 relative to its MPVs as two groups, upregulated and downregulated. We found that the vast majority (from 96.1% to 98.2%) of the genes from each pairwise comparison showed additive expression in XX329 relative to its MPV (Supplemental Figure 6 and Supplemental Table 2), confirming the lack of immediate trans-subgenome interaction on the expression of these genes after reintroducing the DD genome to the BBAA complement. This striking preponderance of gene expression additivity in XX329 is in line with the observations of Chelaifa et al. (2013). The small proportion of genes (ranging from 1.8% to 3.9%) that showed nonadditive expression in XX329 relative to its MPVs (Supplemental Figure 6 and Supplemental Table 2) suggest that the expression of this set of genes is trans-regulated by the reintroduced DD genome to the BBAA genome complement in XX329 (Supplemental Figure 6). We also compared the microarray-based overall gene expression similarity between TAA10 (the bread wheat donor to ETW) and XX329; in theory, the two lines differ only in the D subgenome of different genotypes. We found that these two hexaploid wheats are highly similar in overall gene expression, with only 2.4% (646 of 26,539 expressed genes in seedling leaf tissue) being differentially expressed genes (Supplemental Figure 7). This analysis confirmed that de novo changes in gene expression arising from the extraction process for producing ETW should be minimal or nonexistent.
To explore whether and to what extent the predominance of additive gene expression was due to the inherent limitation of the microarray platform to differentiate transcripts of the three subgenome homoeologs (e.g., nonadditive expression by individual subgenomes could be masked by reciprocal compensating effects), we selected 14 genes that harbor individual subgenome-specific single-nucleotide polymorphisms (SNPs) for cDNA pyrosequencing analysis. We found that all tested genes showed full expression additivity of the B, A, and D subgenomic gene copies (Supplemental Figure 8 and Supplemental Data Set 2). This suggests that nonadditive but compensating expression by subgenomes is not a prominent feature of XX329.
ETW versus T. turgidum Downregulated Genes Are Enriched for Distinct Gene Ontology Categories
To explore whether the differentially expressed genes between ETW and natural allotetraploid wheat are related to specific functional features, we performed a Gene Ontology (GO) analysis for each differentially expressed gene group. We found that all groups of the ETW versus T. turgidum (durum, carthlicum, and dicoccoides) downregulated genes showed overrepresentation for several categories of genes involved in chloroplast/plastid functions (Supplemental Figures 9A to 9E). This suggests that the extracted wheat may have been specifically destabilized in processes related to photosynthesis. To test this, we measured the photosynthetic capacity of ETW relative to the two domesticated subspecies (durum and carthlicum) of T. turgidum and the bread wheat (line TAA10) donor of ETW. We found that the photosynthetic capacity of ETW was indeed significantly lower than those of the two domesticated tetraploid wheats and bread wheat (Supplemental Figure 10), suggesting that the presence of the DD subgenome is important for the high photosynthetic capacity of bread wheat. We also found that, in contrast with the downregulated genes in the ETW versus T. turgidum comparison, upregulated genes in the three natural subspecies showed enrichments for GO categories involved in calcium ion binding and carboxypeptidase activity (Supplemental Figures 10F and 10G). The other groups of upregulated, differentially expressed genes between ETW and T. turgidum did not show significant category enrichment.
We next performed GO analysis for genes that were differentially expressed between any two of the three natural subspecies of T. turgidum. We found that the durum versus dicoccoides and carthlicum versus dicoccoides upregulated genes showed several enriched GO categories involved in metabolic processes (Supplemental Figures 11A and 11B), while the downregulated genes showed enriched GO categories involved in the maintenance and regulation of basic cellular activities (Supplemental Figure 11C). The differentially expressed genes between the two domesticated tetraploid wheat subspecies, durum and carthlicum, did not show any GO enrichment. Together, the GO categories shown by the differentially expressed genes between ETW and all three natural subspecies of T. turgidum were distinct from those between the natural tetraploid subspecies. This further testifies to the unique nature of the transcriptome changes that occurred to the BBAA component of bread wheat in comparison with those due to natural transcriptome divergence at the tetraploid level.
Expression Changes to the BBAA Component of Bread Wheat Are Distinct from Those Due to Divergent Evolution at the Tetraploid Level
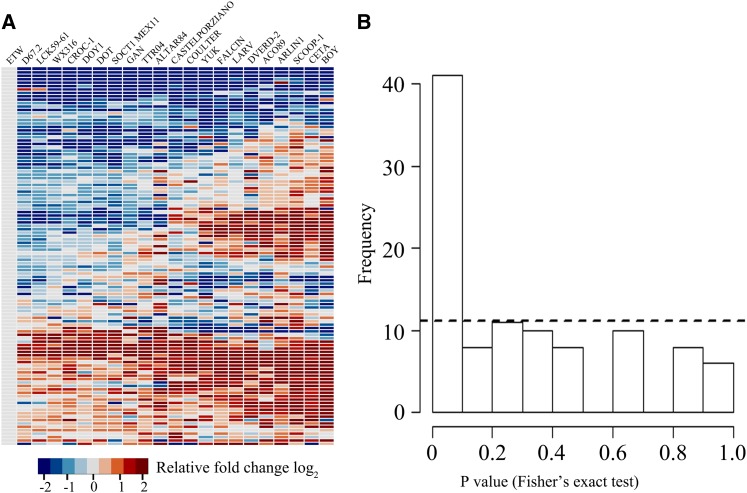
The foregoing results documented that substantial, D subgenome–independent, and to an extent functionally distinct transcriptome changes have occurred to the BBAA component of bread wheat (being reflected by ETW) compared with natural tetraploid wheat. The question arises of how the distinct gene expression differences between ETW and T. turgidum compare with that of “normal” divergent evolution of gene expression at the tetraploid level. To address this, we quantified the expression of 111 genes by qRT-PCR in a set of 21 durum wheat cultivars of diverse origins (Figure 4A; Supplemental Data Set 3). Notably, these genes were randomly chosen among the ETW versus T. turgidum subsp durum differentially expressed genes identified by the microarray-based transcriptome profiling (Supplemental Data Set 3). Therefore, it is reasonable to assume that results based on this set of genes can be extrapolated to the whole transcriptome. For each of the 111 genes, a P value was calculated to evaluate the statistical significance of the expression level difference between ETW and each of the 21 durum wheat cultivars and each pairwise comparison among the 21 cultivars. For each gene, 21 ETW versus durum and 210 durum versus durum P values were computed by using two-tailed t tests. Our null hypothesis was that the gene expression difference is equal between the ETW versus durum comparisons and the durum versus durum comparisons. Therefore, the 21 P values from ETW versus durum comparisons should contain the same proportion of low P values (<0.05) as those of the durum versus durum comparisons. A Fisher’s exact test using these distance-measuring P values was then applied to test whether the above two proportions are equal on a per gene basis, and thus another set of 111 P values were generated. Finally, a P value was obtained by testing the null hypothesis that this proportion is equal to 0.05.
Figure 4.
Expression Level Differences between ETW and 21 durum Wheat Cultivars of Diverse Origins Based on qRT-PCR Assay in the Seedling Leaf Tissue.
(A) Heat maps of expression of 111 genes (Supplemental Data Set 3) in ETW relative to each of 21 durum wheat cultivars of diverse origins based on qRT-PCR assay with three biological replicates. The color key is indicated.
(B) A Fisher’s exact test was conducted to test whether the gene expression level difference between ETW and each durum cultivar versus that of normal gene expression divergence among the diverse durum cultivars was equal.
If the null hypothesis is correct, then the 111 P values obtained from the Fisher’s exact test should follow a uniform distribution, meaning that there should be 111 × 0.05 (<6) P values lower than 0.05. Instead, however, we found 35 P values lower than 0.05, rendering the probability in support of the null hypothesis that the two proportions are equal to 0.05 very unlikely (P < 2.2e-16). This statistical analysis indicated that the ETW versus durum comparisons were significantly different from the durum versus durum comparisons for expression level differences of the 111 genes analyzed (Figure 4B), suggesting that the changes in gene expression in the BBAA component of bread wheat are distinct from those due to the divergent evolution of gene expression at the tetraploid level among diverse durum wheat cultivars.
Expression Changes to the BBAA Component of Bread Wheat Can Be Either Preferentially to One Homoeolog or Equally to Both Homoeologs
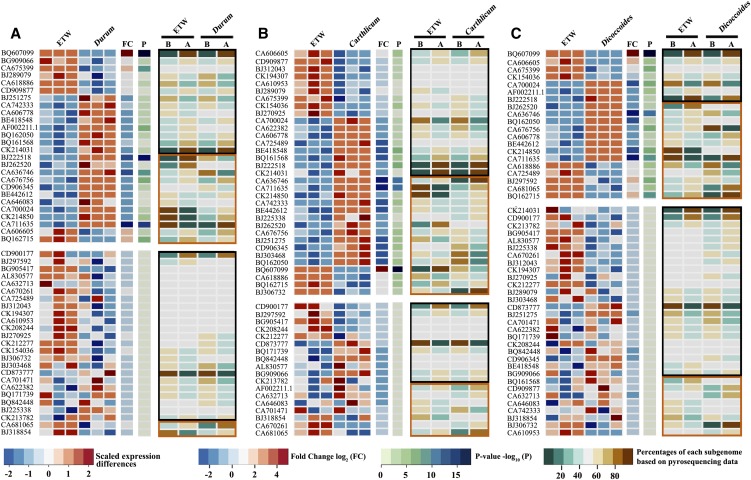
Due to the intrinsic technical limitations of the microarray- or qRT-PCR–based assays, it was unclear whether the transcriptome changes to the BBAA component of bread wheat have preferentially affected transcripts from the B or A subgenome. To gain insight into this issue, we performed cDNA pyrosequencing for a set of low-copy genes in which diagnostic SNPs exist between the B and A homoeologs to enable unequivocal quantification of their relative contributions to the collective transcripts of a given gene. In total, we analyzed 51 genes for which pyrosequencing primers were successfully designed (Supplemental Data Set 2). According to the microarray data, 36 of these 51 genes were differentially expressed between ETW and at least one of the three natural subspecies of T. turgidum, while 15 genes were equally expressed across all lines (Figures 5A to 5C, left panels). We conducted cDNA pyrosequencing for each of these genes in the three pairwise comparisons between ETW and durum, carthlicum, and dicoccoides. Thus, a total of 153 gene expression data points (51 genes × 3 pairwise comparisons) were generated, which included both differential and equal overall expression depending on a given comparison.
Figure 5.
Difference in Homoeolog-Specific Expression between ETW and Natural Tetraploid Wheat, T. turgidum.
Heat maps of equal or differential expression for each of 51 genes (Supplemental Data Set 2) in ETW relative to each of the three natural subspecies of T. turgidum, durum (cv TTR13) (A), carthlicum (cv Blackbird) (B), and dicoccoides (line TD265) (C), as collective transcripts for each gene based on the microarray data of three biological replicates (left panels) and the relative transcript contribution by the B and A subgenomes for each gene based on cDNA pyrosequencing data (right panels). Differential (top part of each panel) or equal (bottom part of each panel) expression between ETW and each of the three natural subspecies for each of the 51 analyzed genes were determined by statistically significant (FDR, P < 0.05) fold changes (FC) of the microarray data (three biological replications). The relative transcript contribution by the B and A subgenomes for each gene is calculated based on mean ratios of pyrosequencing data of three biological replicates using the same cDNAs as for the microarray analysis. Equal or preferential changes to the B and A subgenome transcripts were determined according to statistically insignificant (t test, P > 0.05) or significant (t test, P < 0.05) changes of the B versus A subgenome transcript ratios between the two partners of each pairwise comparison, which are marked by black and vermilion rectangles for the 51 analyzed genes, respectively. The color keys are indicated.
In the pairwise comparison of ETW versus durum (cv TTR13), 26 of the 51 genes were differentially expressed according to the microarray analysis (Figure 5A, left panel). In 14 of these 26 genes, the ratios of the relative expression of the B and A homoeologs were statistically the same in ETW and durum (t test P > 0.05; Figure 5A, right panel, boxed by black rectangles), indicating equal alteration of the B and A transcripts in ETW; in the remaining 12 genes, the ratios of relative expression of B and A were statistically different (t test, P < 0.05; Figure 5A, right panel, boxed by vermilion rectangles), indicating preferential alteration of either the B and A homoeologs for these genes in ETW. The remaining 25 genes were equally expressed in ETW and durum (Figure 5A, left panel), with 23 of these 25 genes showing unaltered B/A expression in ETW relative to durum (t test, P > 0.05; Figure 5A, right panel, boxed by black rectangles), while for the other two genes, ratios of B and A expression levels were significantly altered but in opposite directions in ETW (t test, P < 0.05; Figure 5A, right panel, boxed by vermilion rectangles), rendering the total transcript amount unaltered (Figure 5A, left panel). Broadly similar results were obtained in the pairwise comparisons of ETW versus carthlicum (cv Blackbird) and ETW versus dicoccoides (line TD265), although the exact proportions of genes showing equal alteration to both B and A homoeologs or preferential alteration to one of the homoeologs varied across the comparisons (Figures 5B and 5C). Taking all three pairwise comparisons together, of the 51 genes or 153 expression data points assessed by cDNA pyrosequencing for the differentially expressed genes between ETW and T. turgidum, the proportions showing equal alteration to both homoeologs and preferential alteration to one of the homoeologs is ∼1:1. Of this set of analyzed genes or expression data points, we did not observe a differential propensity to expression alteration between the B and A subgenomes. For those genes that were equally expressed between ETW and T. turgidum, in most cases transcripts of both homoeologs remained unchanged (Figure 5).
Transcriptome Changes to the BBAA Component of Bread Wheat May Have Occurred Rapidly Subsequent to Allohexaploidization but with Evolutionary Persistence across Genotypes
A pertinent question to ask is when did the transcriptome changes to the BBAA subgenomes in bread wheat arise? To address this question, we analyzed the expression patterns (based on microarray data) for the upregulated and downregulated genes as two groups for each of the three pairwise comparisons involving ETW (ETW versus durum, carthlicum, or dicoccoides) in a newly synthesized, eight-generation-old allohexaploid wheat (Allo960; genome, BBAADD) relative to its MPVs (Zhang et al., 2013b). The rationale is that if immediate modification occurred to the expression of these genes upon allohexaploidization, then the affected genes should show nonadditive expression patterns in the synthetic allohexaploid wheat; otherwise, additive expression is expected. We found several things. (1) The majority of both (upregulated and downregulated) groups of genes of each pairwise comparison showed an additive expression pattern in Allo960 relative to its MPV, which ranged from 84.5 to 89.6% (Supplemental Figure 12 and Supplemental Table 3). This suggests that the majority of these ETW versus T. turgidum expression-altered genes did not show an immediate change in expression upon allohexaploidization; therefore, their altered expression in the BBAA subgenomes, as reflected in ETW (Figure 3; Supplemental Table 1), entailed a longer time to accomplish. (2) A small but substantial portion of genes from each upregulated and downregulated group for these genes did show nonadditive expression in Allo960 relative to its MPVs (from 10.4 to 15.5%; Supplemental Figure 12 and Supplemental Table 3). These proportions of nonadditive expression were significantly higher (χ2 = 92.11026, P < 0.001) than the proportions expected from random sampling of a similar number of genes from the total of 26,539 expressed genes with a nonadditive expression frequency of 9.4% in Allo960. This suggests that the ETW versus T. turgidum differentially expressed genes were more prone to immediate modification in expression following allohexaploidization than those that did not differentiate in expression between the two types of tetraploid wheats. (3) The downregulated group of the nonadditively expressed genes showed the altered expression pattern that mirrors their relative expression levels in the ETW versus each of the three natural tetraploid wheat subspecies of T. turgidum (Supplemental Figures 12E to 12H and Supplemental Table 3). That is, the ETW versus T. turgidum downregulated genes also showed more downregulation than upregulation (73.7 to 86.0% versus 14.0 to 26.3%; t test, P < 0.05) in Allo960 relative to its MPVs.
To address the question of whether the modified expression in the BBAA component of bread wheat might have been selected for during allohexaploid wheat evolution, we tested the extent of expression level conservation for a set of the ETW versus T. turgidum subsp durum downregulated genes among different bread wheat genotypes. The rationale to choose the ETW versus durum downregulated genes for this purpose, apart from their distinct expression patterns in Allo960, described above, was because these genes showed unique GO categories (Supplemental Figure 9) and, therefore, were more likely to reflect the distinct expression changes to the BBAA subgenomes of bread wheat. We conducted qRT-PCR analysis coupled with cDNA pyrosequencing for nine ETW versus durum (cv TTR13) downregulated genes in which transcripts of each subgenome could be unequivocally distinguished in all 21 bread wheat cultivars of diverse origins (Supplemental Figure 13 and Supplemental Data Set 2), generating 189 expression data comparisons (9 genes × 21 genotypes). The qRT-PCR results indicated that in the majority of the comparisons (126 of 189), expression in the bread wheat cultivars showed significantly lower levels (t test, P < 0.05) than in durum (cv TTR13) (Supplemental Figure 13A). For the remaining comparisons (63 of 189), total expression levels in bread wheat were similar to or higher than those in durum (cv TTR13) (Supplemental Figure 13A), which might be due to compensation by the DD subgenome transcripts. This was confirmed after subtracting the transcripts contributed by the DD subgenome (determined by cDNA pyrosequencing; Supplemental Figure 13B), as substantially more comparisons (159 of 189) showed significantly lower expression levels in the BBAA subgenomes (t test, P < 0.05) than in durum (cv TTR13) (Supplemental Figure 13A). These results suggest that expression levels of the BBAA subgenomes for a majority of the 189 gene × genotype comparisons were conserved in the course of natural and human selection in these bread wheat cultivars, implying that at least some of the gene expression modifications to the BBAA component of bread wheat occurred early and are under selective constraint. The cDNA pyrosequencing results also revealed that expression changes for these genes can be either in one or both of the B and A homoeologs, depending on a given gene × genotype combination across the 21 diverse bread wheat cultivars (Supplemental Figure 13B), similar to the changes observed in ETW versus T. turgidum (Figure 5). We reiterate here the same caution regarding the unknown exact parentage of the polyploids used (Gottlieb, 2004).
DISCUSSION
Karyotype stability is usually a hallmark of established allopolyploid species (Comai, 2005; Hollister et al., 2012; Yant et al., 2013; Bomblies and Madlung, 2014; Henry et al., 2014). Although exceptions to this general rule have been documented recently in Brassica (Xiong et al., 2011) and Tragopogon (Chester et al., 2012), in which both structural chromosomal rearrangements and aneuploidy are abundant and even transgenerationally persistent, stabilized euploidy has selective advantages over aneuploid individuals in the long run. For polyploid wheat at two ploidy levels, extensive whole-chromosome aneuploidy was found to be generally and persistently associated with nascent allohexaploid wheats mimicking bread wheat in genome composition (Zhang et al., 2013b). Given that bread wheat has a stable karyotype, and assuming that the actual founder cross combination giving rise to bread wheat behaved the same as the synthetic lines studied (Zhang et al., 2013b), a mutation-based karyotype stabilization mechanism likely has evolved to confer this phenotype, as in other plants (Hollister et al., 2012; Yant et al., 2013; Bomblies and Madlung, 2014; Henry et al., 2014). If this is the case, we show here that the evolved karyotype stabilization mechanism in bread wheat is not only fully preserved in the extracted BBAA component of bread wheat but also exerts control in resynthesized allohexaploid wheat (XX329). Although it is known that the Ph1 locus located on the long arm of chromosome 5B controls chromosome stability in bread wheat (Griffiths et al., 2006, and references cited therein), we argue that Ph1 is unlikely to have played a major role in the initial karyotype stabilization in nascent allohexaploid wheats, because T. turgidum harbors a functional Ph1 locus and yet the allohexaploid wheats it parented have unstable karyotypes (Zhang et al., 2013b). Therefore, additional mechanism(s) might be required for the initial karyotype stabilization following allohexaploidization in bread wheat evolution, which may function collaboratively with, or be eventually replaced by, the Ph1 locus inherited from T. turgidum. In parallel, Ph1 also cannot be responsible for the intrinsically stabilized karyotypes of newly formed tetraploid wheats with a genome composition SSAA (analogous to BBAA of T. turgidum), because the diploid parental species involved did not contain this gene (Zhang et al. 2013a). Moreover, immediate numerical karyotype stability following allopolyploidization appears also to be the case in Nicotiana (Kar et al., 2004), although in this genus, no Ph1-like gene has been found. Regardless, our documentation of karyotype stability in the extracted BBAA component of bread wheat has implications for further exploring the initial karyotype stabilization mechanism(s) responsible for the establishment of bread wheat as a stable euploid species.
The evolution of gene expression is a driving force for phenotype diversification in all organisms. Polyploidy, being ubiquitous in the evolutionary histories of higher plants (Soltis and Soltis, 2009), has provided novel avenues for the innovation of gene expression. Recent studies have shown that allopolyploidization may induce a cascade of rapid as well as evolutionarily accruing alterations in gene expression (Wendel, 2000; Adams et al., 2003; Hegarty et al., 2006; Wang et al., 2006; Chaudhary et al., 2009; Pumphrey et al., 2009; Rapp et al., 2009; Akhunova et al., 2010; Chagué et al., 2010; Buggs et al., 2011; Grover et al., 2012; Li et al., 2014). This phenomenon has been collectively termed “transcriptome shock” (Hegarty et al., 2006; Buggs et al., 2011), sensu the “genome shock” proposed by McClintock (1984), referring to situations of dramatic restructuring of a hybrid genome. Although the molecular mechanisms remain elusive, allopolyploidy-induced transcriptome shock is suggested to involve (1) interactions between diverged parental regulatory networks, (2) stoichiometric disruptions due to the incongruence between WGD and differential dosage sensitivity among genes and pathways, and; (3) de novo genetic and heritable epigenetic alterations in allopolyploids (Wendel, 2000; Osborn et al., 2003; Riddle and Birchler, 2003; Adams and Wendel, 2005; Comai, 2005; Chen, 2007; Doyle et al., 2008; Soltis and Soltis, 2009; Jackson and Chen, 2010; Birchler, 2012; Madlung and Wendel, 2013). With respect to subgenome(s) of a given allopolyploid, transcriptome alterations can be either “reversible,” if they were due to causes 1 and 2, or “irreversible,” if causally linked to cause 3. Of course, if the causes are interwoven, the situation will be more complex.
These observations of novel gene expression patterns in allopolyploids have advanced our understanding of allopolyploid evolution and suggested key features that in at least some circumstances may be advantageous (reviewed in Comai, 2005; Chen, 2007; Otto, 2007; Soltis and Soltis, 2009; Madlung and Wendel, 2013). However, all the results obtained thus far regarding allopolyploidization-associated alterations in gene expression have been based on the analysis of existing allopolyploid species in their genomic entirety. That is, altered expression of a given subgenome(s) within an allopolyploid species has been studied only in the presence of its cohabiting subgenome(s), thus obstructing any assessment of transcriptome changes to a specific subgenome(s), because it is not possible to disengage subgenome-specific changes from confounding effects (e.g., trans-acting factors) by the other subgenome(s) at the organismal level. This issue, however, is important not only with respect to dissecting possible subgenome-specific transcriptome changes and their attendant biological effects but also in terms of designing novel biotechnological manipulations targeting specific homoeologs.
That ETW with the BBAA component of bread wheat is phenotypically compromised points to its fundamentally altered functionality. Given the short evolutionary time span of bread wheat since allohexaploidization and the stable karyotype of ETW, it is conceivable that transcriptome alterations likely contribute to phenotypic abnormality. Here, we show that extensive and, to an extent, functionally distinct changes in gene expression to the BBAA component of bread wheat have indeed occurred during its evolutionary residence at the allohexaploid level. This conclusion is reached based on multiple lines of evidence: (1) the greater than expected global gene expression difference between ETW and its closest natural counterpart, durum wheat (cv TTR13), based on direct transcriptome comparisons; (2) a collectively distinct expression pattern in ETW compared with a set of diverse durum wheat cultivars for >100 genes by qRT-PCR assay, which were randomly selected among the ETW versus T. turgidum subsp durum differentially expressed genes; (3) enriched distinct GO categories by the ETW versus T. turgidum downregulated genes; and (4) the near absence of trans-regulation on the ETW versus T. turgidum differentially expressed genes by the newly added DD genome in a resynthesized allohexaploid wheat (XX329). Together, our results indicate that the transcriptome changes in the BBAA subgenomes of bread wheat bear signatures distinct from normal divergent transcriptome evolution at the tetraploid level, thus pointing to a unique impact of the allohexaploid trajectory on the transcriptome evolution of its constituent subgenomes.
Generally, both genetic and epigenetic mechanisms (Madlung and Wendel, 2013) might have been responsible for the heritable transcriptome changes to the BBAA component of bread wheat. First, rapid and substantial DNA loss has been shown to accompany allohexaploidization in wheat (Feldman et al., 1997; Liu et al., 1998; Ozkan et al., 2001, 2003; Eilam et al., 2008). Consistent with these earlier findings, recent genomics-based studies of bread wheat revealed that gene loss occurred extensively (Brenchley et al., 2012), particularly from the B and A subgenomes (Pont et al., 2013). Moreover, other kinds of genetic alterations, such as gene conversion, copy number variation, and transposition (Saintenac et al., 2011), may also have contributed to permanent changes of the BBAA transcriptome, although unlike other allopolyploid plants, such as Brassica (Pires et al., 2004) and Tragopogon (Buggs et al., 2012), “canonical” intergenomic rearrangements are infrequent in bread wheat due to its exclusive diploid-like meiosis. However, in this respect, two recent studies in allotetraploid cotton (Gossypium hirsutum), which also shows typical diploid-like meiosis, are illuminating in that they document incidents of nonreciprocal homoeologous exchanges, perhaps via some novel mechanism (Salmon et al., 2010; Flagel et al., 2012). Second, extensive alternative splicing (Akhunova et al., 2010; Akhunov et al., 2013) and heritable epigenetic alterations may have contributed to the transcriptome changes to the BBAA component of bread wheat (Shaked et al., 2001; Zhao et al., 2011). Indeed, it has been documented in bread wheat that permanent silencing of gene homoeologs can be caused by altered DNA methylation (Shitsukawa et al., 2007; Hu et al., 2013). Interestingly, in most of these documented individual cases, both genetic and epigenetic alterations were found to be involved in loss of function or silencing of different homoeologs of the same gene, suggesting the remarkable ability of bread wheat to partition genetic and epigenetic mechanisms for silencing of different homoeologous alleles. The generality of this conclusion merits further investigation.
The observation that for a given gene the transcripts can be changed in either or both of the B and A homoeologs, or even to opposite expression directions, points to the remarkable variation in subgenome expression modifications. This suggests that the intuitive thought of convergent evolution between or among the subgenomes conditioned by allopolyploidy (Feldman et al., 2012) does not necessarily compromise the “degree of freedom” for expression diversification of gene homoeologs. It also is conceivable that constraints imposed by the stoichiometry of gene products, which at the gene regulatory level entails balanced expression of genes with functional connectivity (Birchler and Veitia, 2007, 2012), might impose limitations on the spectrum of expression diversification that the subgenomes may explore.
Although the majority of the transcriptome changes of the BBAA subgenomes appear to have accrued since bread wheat’s origin via allohexaploidization, a substantial portion of these genes are immediately regulated following allohexaploidization, as reflected by the significantly higher than expected proportion of nonadditively expressed genes in the newly synthesized allohexaploid wheat (Allo960). A particularly interesting observation is that the downregulated genes in the nascent allohexaploid wheat (due to nonadditive expression) are comparable to those seen in ETW (showing additive expression). This suggests that immediate trans-regulation on the expression of the BBAA subgenomes due to addition of the DD subgenome is likely a transitory, compensatory mechanism, which would be replaced by more stable cis-acting regulation of additive expression in the course of bread wheat evolution. This suggests that functional “independency” of the subgenomes in bread wheat (Brenchley et al., 2012) has been “restored” by creative tinkering in the course of the allohexaploid evolutionary trajectory rather than being merely passive legacies of the parental state. This hypothesis may reconcile the conundrum between the chaotic gene expression state widely associated with nascent allopolyploidy and the stabilized expression profiles in established allopolyploids. Furthermore, we show by coupled qRT-PCR and pyrosequencing analysis of a subset of ETW versus durum (cv TTR13) downregulated genes that a great majority of the modified expression levels in the BBAA subgenomes are shared by the 21 studied bread wheat cultivars of diverse origins, which suggests the early occurrence and evolutionary persistence of altered expression. Although a similar phenomenon of rapid occurrence and evolutionary conservation of altered gene expression has been observed previously in other plant taxa, such as Arabidopsis (Wang et al., 2006), cotton (Grover et al., 2012), Senecio (Hegarty et al., 2006), and Tragopogon (Buggs et al., 2011), our results represent unequivocal documentation of rapid and functionally distinct transcriptome alterations in subgenomes of any extant allopolyploid organism without the complication of a coexisting subgenome. This can be accomplished only by reliberating the genetically “colonized” subgenomes as fully “independent” organismal genomes.
METHODS
Plant Materials
An allotetraploid wheat (designated as ETW) with an “extracted” genome (BBAA) of the allohexaploid bread wheat (Triticum aestivum cv Canthach; designated as TAA10) was produced and kindly provided by E. Kerber. Details for the production of ETW were described (Kerber, 1964) and are outlined in Figure 2. ETW was backcrossed to TAA10 as the recurrent parent for two additional times and then propagated via self-pollination in our hands for five more generations. The resynthesized allohexaploid wheat (XX329; Kerber designation RL 5405) was produced by crossing ETW (as maternal parent) with Aegilops tauschii subsp strangulata (line RL 5288; our designation is TQ18) (as paternal parent), followed by genome doubling with colchicine treatment by E. Kerber. XX329 was also propagated via self-pollination in our hands for five more generations. The newly synthesized allohexaploid wheat (Allo960) was produced by crossing Triticum turgidum subsp carthlicum (cv Blackbird) (as maternal parent) with Ae. tauschii line 30A (as paternal parent), followed by genome doubling with colchicine treatment, which were then self-pollinated for up to eight generations. All plants, both synthetic and natural, were grown in chambers under controlled conditions at 24/20°C day/night of 16-h daylength.
Sequential FISH and GISH
This protocol was essentially as described by Kato et al. (2004) with minor modifications (Zhang et al., 2013b).
Microarray Hybridization
Total RNA was extracted using Trizol reagent (Invitrogen) and purified by RNeasy Mini Spin Columns (Qiagen). The integrity of RNA was checked with the Agilent Bioanalyser 2100 Eukaryote Total RNA Nano Series II system. The microarray analysis was performed using RNA isolated from the second leaf of 3-week-old seedlings. Pooled seedlings (10) were used for each line, with three biological replicates. The RNAs of the parental lines (T. turgidum and Ae. tauschii or ETW and Ae. tauschii) were mixed at a ratio of 2:1 to generate the empirical MPVs for each of the synthetic allohexaploid lines. Microarray transcriptional profiling was performed by Affymetrix at the Gene Company, as described in the GeneChip Expression Analysis Technical Manual. The microarray data have been submitted to the National Center for Biotechnology Information’s Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) and are available under the accession number GSE47617.
Microarray Data Normalization and Analysis
The raw CEL data were normalized with the robust multichip average method using the R software limma package. Genes that were differentially expressed among genotypes were identified by performing the t-moderated test ebayes (Smyth, 2004), and the raw P values were adjusted for multiple testing effects by the Benjamini and Yekutieli method (false discovery rate [FDR] < 0.05). The present or absent calls of each probe set were determined by the MAS5 method using R software. Differently expressed genes that did not show present calls in all three biological replications of at least one genotype were excluded from further analysis (Aprile et al., 2009), and 26,539 genes were detected as expressed in the current study.
Real-Time qRT-PCR
Five micrograms of total RNA was isolated from triplicate samples of each genotype and reverse transcribed to cDNA using the SuperScript first-strand synthesis kit (Invitrogen) according to the manufacturer’s instructions. PCR amplification was performed with SYBR Green Real-Time PCR Master Mix reagent (Toyobo) on a StepOne Plus Real-Time PCR apparatus (Applied Biosystems), and the amplification conditions were as follows: 95°C for 1 min, followed by 40 cycles of 95°C for 5 s and 60°C for 1 min. Primers for all analyzed genes were designed by Primer Premier 5 using the consensus sequence of each gene (probe set) available at the Affymetrix website (http://www.affymetrix.com/estore/browse/products.jsp?navMode=34000andproductId=131517andnavAction=jumpandaId=productsNav#1_3). Two housekeeping genes, encoding GADPH and actin (Schreiber et al., 2009), were used to normalize the expression data. Primers are listed in Supplemental Data Set 1.
Subgenome-Specific cDNA Pyrosequencing
The protocol was essentially as reported by Mochida et al. (2003) with modifications detailed by Zhang et al. (2013b). Primers of single-copy genes for which diagnostic SNPs exist between the two subgenomes (B and A) or among all three subgenomes (B, A, and D) were designed (Supplemental Data Set 2) and verified by the pyrosequencing system (PyroMarkID Q96; Biotage). The diploid progenitors of bread wheat, Triticum urartu (AA) and Ae. tauschii (DD), were used to assign SNPs to the A and D subgenomes, respectively, while the B subgenome SNPs were determined using tetraploid wheat, T. turgidum. Biotin-labeled PCR products were immobilized on streptavidin-coated paramagnetic beads. Capture of biotinylated single-stranded PCR products, annealing of the sequencing primer, and solid-phase pyrosequencing were performed following the manufacturer’s recommendations.
Gene Annotation
GO annotations were performed using agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php); the Singular Enrichment Analysis tool was used to do the GO annotations and significant GO term enrichment analysis (Du et al., 2010), which computed GO term enrichment in one set of genes by comparing it with another set, named the target and reference list, respectively. Enrichment was calculated by Fisher’s exact test with Hochberg’s multitest adjustment (FDR, P < 0.05).
Measurements of Photosynthetic Capacity
Net photosynthetic rate in ETW relative to the two domesticated subspecies of natural allotetraploid wheat, durum and carthlicum, and bread wheat (line TAA10), was measured by a portable open-flow gas-exchange system (LI-6400; LICOR). Net photosynthetic rate was determined on fully expanded leaves at 9:30 to 10:30 am.
Accession Numbers
Detailed information including gene accession numbers of all genes studied in this article is listed in Supplemental Data Sets 1 to 3. The microarray data have been submitted to the National Center for Biotechnology Information’s Omnibus repository (http://www.ncbi.nlm.nih.gov/geo/) and are available under the accession number GSE47617.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequential FISH- and GISH-Based Karyotypes of Common Wheat (Line TAA10), ETW, Ae. tauschii (Line TQ18), and Resynthesized Allohexaploid Wheat XX329.
Supplemental Figure 2. Examples of Intersubgenome Rearrangements Revealed by FISH/GISH-Based Karyotyping of Two Newly Synthesized Allotetraploid Wheat Lines with Genome Combinations AADD (T. urartu × Ae. tauschii) and SSDD (Ae. bicornis × Ae. tauschii).
Supplemental Figure 3. Typical Spike Morphology of ETW, T. turgidum subsp durum (cv TTR13), and Their F1 Hybrid.
Supplemental Figure 4. Representative Segregated Spike Shapes in F2 Progeny of a Cross between ETW and T. turgidum subsp durum (cv TTR13).
Supplemental Figure 5. Validation of the Microarray Data by qRT-PCR.
Supplemental Figure 6. Graphical Distribution of Gene Expression Patterns in the Resynthesized Allohexaploid Wheat XX329 Relative to Their Corresponding MPVs for the ETW versus Natural Allotetraploid Upregulated and Downregulated Genes.
Supplemental Figure 7. Overall Gene Expression Similarity between TAA10 (the Bread Wheat Donor to ETW) and the Resynthesized Hexaploid Wheat XX329 (Parented by ETW) Based on the Affymetrix GeneChip Wheat Genome Array with Three Biological Replicates for Each Genotype.
Supplemental Figure 8. Dissecting the Subgenome Contribution to Each of 14 Selected Genes (Supplemental Data Set 2) That Showed Additive Expression in the Resynthesized Allohexaploid Wheat XX329 by Gene-Specific cDNA Pyrosequencing.
Supplemental Figure 9. GO Enrichment (>50%) for Genes That Are Differentially Expressed between ETW and the Three Natural Subspecies of T. turgidum, durum, carthlicum, and dicoccoides (Blue Bars).
Supplemental Figure 10. Net Photosynthetic Rate in ETW Relative to Its Bread Wheat Donor (Line TAA10) and the Two Domesticated Natural Subspecies of T. turgidum, durum (cv TTR13) and carthlicum (cv Blackbird).
Supplemental Figure 11. GO Enrichment (>50%) Analysis for Genes That Are Differentially Expressed between Any Two of the Three Natural Subspecies of T. turgidum, durum (cv TTR13), carthlicum (cv Blackbird), and dicoccoides (Line TD265).
Supplemental Figure 12. Graphical Distribution of Gene Expression Patterns in the Newly Synthesized Allohexaploid Wheat Allo960 Relative to Their Corresponding MPVs for the ETW versus Natural Allotetraploid Wheat Upregulated and Downregulated Genes.
Supplemental Figure 13. Clustering of Expression of Nine ETW versus durum Downregulated Genes in 21 Bread Wheat Cultivars of Diverse Origins as Collective Transcripts for Each Gene Based on qRT-PCR.
Supplemental Table 1. Differentially Expressed Genes between ETW and Each or All Three Subspecies of T. turgidum, durum (cv TTR13), carthlicum (cv Blackbird), and dicoccoides (Line TD265), Based on the Microarray Data.
Supplemental Table 2. Expression Pattern of the Upregulated and Downregulated Genes between ETW and Each of the Three T. turgidum Subspecies, durum (cv TTR13), carthlicum (cv Blackbird), and dicoccoides (Line TD265), as Well as between Any Two of the Three Subspecies, in the Resynthesized Allohexaploid Wheat XX329.
Supplemental Table 3. Expression Pattern of the Upregulated and Downregulated Genes between ETW and Each of the Three T. turgidum Subspecies, durum (cv TTR13), carthlicum (cv Blackbird), and dicoccoides (Line TD265), as Well as between Any Two of the Three Subspecies, in a Newly Synthesized Allohexaploid Line (Allo960).
Supplemental Data Set 1. Information for the 36 Comparisons (25 Genes) Used to Validate the Microarray Data through the Approach of qRT-PCR.
Supplemental Data Set 2. Pyrosequencing Primers Harboring Diagnostic SNPs between the Two Subgenomes (B and A) of Tetraploid Wheat or among All Three Subgenomes (B, A, and D) of Hexaploid Wheat Used in This Study.
Supplemental Data Set 3. Information for the 111 ETW versus durum (Line TR113) Differentially Expressed Genes Assayed by Real-Time qRT-PCR across 21 durum Wheat Genotypes.
Supplementary Material
Acknowledgments
We thank G. Fedak and X.E. Wang for providing seeds and F.P. Han for providing the FISH probes. We also thank all the anonymous reviewers and the coeditor, James Birchler, for critical comments and constructive suggestions that enabled substantial improvement to this article. This study was supported by the National Natural Science Foundation of China (Grants 31290210 and 11301064), the 863 Program of China (Grant 2011AA100101), and the Program for Introducing Talents to Universities (Grant B07017).
AUTHOR CONTRIBUTIONS
B.L. and M.F. designed the research. H.K.Z., B.Z., B.Q., X.G., Y.D., B.J.Z., C.L., H.Z., and C.Y. performed the experiments. B.Q., B.Z., H.K.Z., W.H., C.X., X.W., K.K., B.L., and J.F.W. analyzed the data. B.L., M.F., and J.F.W. wrote the article.
Glossary
- WGD
whole-genome duplication
- ETW
extracted tetraploid wheat
- FISH
fluorescent in situ hybridization
- GISH
genomic in situ hybridization
- qRT-PCR
quantitative RT-PCR
- MPV
mid parent value
- SNP
single-nucleotide polymorphism
- GO
Gene Ontology
- FDR
false discovery rate
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Adams K.L., Wendel J.F. (2005). Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 8: 135–141. [DOI] [PubMed] [Google Scholar]
- Adams K.L., Cronn R., Percifield R., Wendel J.F. (2003). Genes duplicated by polyploidy show unequal contributions to the transcriptome and organ-specific reciprocal silencing. Proc. Natl. Acad. Sci. USA 100: 4649–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunov E.D., et al. (2013). Comparative analysis of syntenic genes in grass genomes reveals accelerated rates of gene structure and coding sequence evolution in polyploid wheat. Plant Physiol. 161: 252–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhunova A.R., Matniyazov R.T., Liang H., Akhunov E.D. (2010). Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genomics 11: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amborella Genome Project (2013). The Amborella genome and the evolution of flowering plants. Science 342: 1241089. [DOI] [PubMed] [Google Scholar]
- Aprile A., Mastrangelo A.M., De Leonardis A.M., Galiba G., Roncaglia E., Ferrari F., De Bellis L., Turchi L., Giuliano G., Cattivelli L. (2009). Transcriptional profiling in response to terminal drought stress reveals differential responses along the wheat genome. BMC Genomics 10: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayal S., Ophir R., Levy A.A. (2005). Genomics of tetraploid wheat domestication. Wheat Inf. Serv. 100: 185–203. [Google Scholar]
- Birchler J.A. (2012). Insights from paleogenomic and population studies into the consequences of dosage sensitive gene expression in plants. Curr. Opin. Plant Biol. 15: 544–548. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2007). The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell 19: 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2012). Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc. Natl. Acad. Sci. USA 109: 14746–14753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K., Madlung A. (2014). Polyploidy in the Arabidopsis genus. Chromosome Res. 22: 1–18. [DOI] [PubMed] [Google Scholar]
- Bottley A., Xia G.M., Koebner R.M.D. (2006). Homoeologous gene silencing in hexaploid wheat. Plant J. 47: 897–906. [DOI] [PubMed] [Google Scholar]
- Brenchley R., et al. (2012). Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggs R.J., Zhang L., Miles N., Tate J.A., Gao L., Wei W., Schnable P.S., Barbazuk W.B., Soltis P.S., Soltis D.E. (2011). Transcriptomic shock generates evolutionary novelty in a newly formed, natural allopolyploid plant. Curr. Biol. 21: 551–556. [DOI] [PubMed] [Google Scholar]
- Buggs R.J.A., Chamala S., Wu W., Tate J.A., Schnable P.S., Soltis D.E., Soltis P.S., Barbazuk W.B. (2012). Rapid, repeated, and clustered loss of duplicate genes in allopolyploid plant populations of independent origin. Curr. Biol. 22: 248–252. [DOI] [PubMed] [Google Scholar]
- Chagué V., Just J., Mestiri I., Balzergue S., Tanguy A.M., Huneau C., Huteau V., Belcram H., Coriton O., Jahier J., Chalhoub B. (2010). Genome-wide gene expression changes in genetically stable synthetic and natural wheat allohexaploids. New Phytol. 187: 1181–1194. [DOI] [PubMed] [Google Scholar]
- Chaudhary B., Flagel L., Stupar R.M., Udall J.A., Verma N., Springer N.M., Wendel J.F. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182: 503–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H., et al. (2013). Prevalence of gene expression additivity in genetically stable wheat allohexaploids. New Phytol. 197: 730–736. [DOI] [PubMed] [Google Scholar]
- Chelaifa H., Monnier A., Ainouche M. (2010). Transcriptomic changes following recent natural hybridization and allopolyploidy in the salt marsh species Spartina × townsendii and Spartina anglica (Poaceae). New Phytol. 186: 161–174. [DOI] [PubMed] [Google Scholar]
- Chen Z.J. (2007). Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu. Rev. Plant Biol. 58: 377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M., Gallagher J.P., Symonds V.V., Cruz da Silva A.V., Mavrodiev E.V., Leitch A.R., Soltis P.S., Soltis D.E. (2012). Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Doyle J.J., Flagel L.E., Paterson A.H., Rapp R.A., Soltis D.E., Soltis P.S., Wendel J.F. (2008). Evolutionary genetics of genome merger and doubling in plants. Annu. Rev. Genet. 42: 443–461. [DOI] [PubMed] [Google Scholar]
- Du Z., Zhou X., Ling Y., Zhang Z., Su Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38: W64–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvořák J. (1976). The relationship between the genome of Triticum urartu and the A and B genomes of Triticum aestivum. Can. J. Genet. Cytol. 18: 371–377. [Google Scholar]
- Dvorak J., Akhunov E.D. (2005). Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 171: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam T., Anikster Y., Millet E., Manisterski J., Feldman M. (2008). Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum. Genome 51: 616–627. [DOI] [PubMed] [Google Scholar]
- Feldman M., Levy A.A. (2012). Genome evolution due to allopolyploidization in wheat. Genetics 192: 763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman M., Levy A.A., Fahima T., Korol A. (2012). Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 63: 5045–5059. [DOI] [PubMed] [Google Scholar]
- Feldman M., Liu B., Segal G., Abbo S., Levy A.A., Vega J.M. (1997). Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 147: 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman, M., Lupton, F.G.H., and Miller, T.E. (1995). Wheats. In Evolution of Crop Plants, 2nd ed, J. Smartt and N.W. Simmonds, eds (London: Longman Scientific), pp. 184–192. [Google Scholar]
- Flagel L.E., Wendel J.F., Udall J.A. (2012). Duplicate gene evolution, homoeologous recombination, and transcriptome characterization in allopolyploid cotton. BMC Genomics 13: 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Woodhouse M.R., Subramaniam S., Turco G., Lisch D., Schnable J.C. (2012). Fractionation mutagenesis and similar consequences of mechanisms removing dispensable or less-expressed DNA in plants. Curr. Opin. Plant Biol. 15: 131–139. [DOI] [PubMed] [Google Scholar]
- Gottlieb L.D. (2004). Rethinking classic examples of recent speciation in plants. New Phytol. 161: 71–82. [Google Scholar]
- Griffiths S., Sharp R., Foote T.N., Bertin I., Wanous M., Reader S., Colas I., Moore G. (2006). Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Grover C.E., Gallagher J.P., Szadkowski E.P., Yoo M.J., Flagel L.E., Wendel J.F. (2012). Homoeolog expression bias and expression level dominance in allopolyploids. New Phytol. 196: 966–971. [DOI] [PubMed] [Google Scholar]
- Hegarty M., Coate J., Sherman-Broyles S., Abbott R., Hiscock S., Doyle J. (2013). Lessons from natural and artificial polyploids in higher plants. Cytogenet. Genome Res. 140: 204–225. [DOI] [PubMed] [Google Scholar]
- Hegarty M.J., Barker G.L., Wilson I.D., Abbott R.J., Edwards K.J., Hiscock S.J. (2006). Transcriptome shock after interspecific hybridization in Senecio is ameliorated by genome duplication. Curr. Biol. 16: 1652–1659. [DOI] [PubMed] [Google Scholar]
- Henry I.M., Dilkes B.P., Tyagi A., Gao J., Christensen B., Comai L. (2014). The BOY NAMED SUE quantitative trait locus confers increased meiotic stability to an adapted natural allopolyploid of Arabidopsis. Plant Cell 26: 181–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister J.D., Arnold B.J., Svedin E., Xue K.S., Dilkes B.P., Bomblies K. (2012). Genetic adaptation associated with genome-doubling in autotetraploid Arabidopsis arenosa. PLoS Genet. 8: e1003093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Han Z., Song N., Chai L., Yao Y., Peng H., Ni Z., Sun Q. (2013). Epigenetic modification contributes to the expression divergence of three TaEXPA1 homoeologs in hexaploid wheat (Triticum aestivum). New Phytol. 197: 1344–1352. [DOI] [PubMed] [Google Scholar]
- Huang S., Sirikhachornkit A., Su X., Faris J., Gill B., Haselkorn R., Gornicki P. (2002). Genes encoding plastid acetyl-CoA carboxylase and 3-phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. USA 99: 8133–8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S., Chen Z.J. (2010). Genomic and expression plasticity of polyploidy. Curr. Opin. Plant Biol. 13: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Gill B.S. (1994). Different species-specific chromosome translocations in Triticum timopheevii and T. turgidum support the diphyletic origin of polyploid wheats. Chromosome Res. 2: 59–64. [DOI] [PubMed] [Google Scholar]
- Kar Y.L., Matyasek R., Kovarik A., Leitch A.R. (2004). Genome evolution in allotetraploid Nicotiana. Biol. J. Linn. Soc. 82: 599–606. [Google Scholar]
- Kato A., Lamb J.C., Birchler J.A. (2004). Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc. Natl. Acad. Sci. USA 101: 13554–13559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerber E.R. (1964). Wheat: Reconstitution of the tetraploid component (AABB) of hexaploids. Science 143: 253–255. [DOI] [PubMed] [Google Scholar]
- Li A., et al. (2014). mRNA and small RNA transcriptomes reveal insights into dynamic homoeolog regulation of allopolyploid heterosis in nascent hexaploid wheat. Plant Cell 26: http://dx.doi.org/10.1105/tpc.114.124388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Vega J.M., Feldman M. (1998). Rapid genomic changes in newly synthesized amphiploids of Triticum and Aegilops. II. Changes in low-copy coding DNA sequences. Genome 41: 535–542. [DOI] [PubMed] [Google Scholar]
- Madlung A. (2013). Polyploidy and its effect on evolutionary success: Old questions revisited with new tools. Heredity (Edinb.) 110: 99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madlung A., Wendel J.F. (2013). Genetic and epigenetic aspects of polyploid evolution in plants. Cytogenet. Genome Res. 140: 270–285. [DOI] [PubMed] [Google Scholar]
- McClintock B. (1984). The significance of responses of the genome to challenge. Science 226: 792–801. [DOI] [PubMed] [Google Scholar]
- Mochida K., Yamazaki Y., Ogihara Y. (2003). Discrimination of homoeologous gene expression in hexaploid wheat by SNP analysis of contigs grouped from a large number of expressed sequence tags. Mol. Genet. Genomics 270: 371–377. [DOI] [PubMed] [Google Scholar]
- Osborn T.C., Pires J.C., Birchler J.A., Auger D.L., Chen Z.J., Lee H.S., Comai L., Madlung A., Doerge R.W., Colot V., Martienssen R.A. (2003). Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 19: 141–147. [DOI] [PubMed] [Google Scholar]
- Otto S.P. (2007). The evolutionary consequences of polyploidy. Cell 131: 452–462. [DOI] [PubMed] [Google Scholar]
- Ozkan H., Levy A.A., Feldman M. (2001). Allopolyploidy-induced rapid genome evolution in the wheat (Aegilops-Triticum) group. Plant Cell 13: 1735–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan H., Tuna M., Arumuganathan K. (2003). Nonadditive changes in genome size during allopolyploidization in the wheat (Aegilops-Triticum) group. J. Hered. 94: 260–264. [DOI] [PubMed] [Google Scholar]
- Pires J.C., Zhao J., Schranz M.E., Leon E.J., Quijada P.A., Lukens L.N., Osborn T.C. (2004). Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae). Biol. J. Linn. Soc. 82: 675–688. [Google Scholar]
- Pont C., Murat F., Confolent C., Balzergue S., Salse J. (2011). RNA-seq in grain unveils fate of neo- and paleopolyploidization events in bread wheat (Triticum aestivum L.). Genome Biol. 12: R119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pont C., et al. (2013). Wheat syntenome unveils new evidences of contrasted evolutionary plasticity between paleo- and neoduplicated subgenomes. Plant J. 76: 1030–1044. [DOI] [PubMed] [Google Scholar]
- Pumphrey M., Bai J., Laudencia-Chingcuanco D., Anderson O., Gill B.S. (2009). Nonadditive expression of homoeologous genes is established upon polyploidization in hexaploid wheat. Genetics 181: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B., Huang W., Zhu B., Zhong X., Guo J., Zhao N., Xu C., Zhang H., Pang J., Han F., Liu B. (2012). Global transgenerational gene expression dynamics in two newly synthesized allohexaploid wheat (Triticum aestivum) lines. BMC Biol. 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapp R.A., Udall J.A., Wendel J.F. (2009). Genomic expression dominance in allopolyploids. BMC Biol. 7: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renny-Byfield S., Wendel J.F. (2014). Doubling down on genomes: Polyploidy and crop plants. Am. J. Bot. 101: in press. [DOI] [PubMed] [Google Scholar]
- Riddle N.C., Birchler J.A. (2003). Effects of reunited diverged regulatory hierarchies in allopolyploids and species hybrids. Trends Genet. 19: 597–600. [DOI] [PubMed] [Google Scholar]
- Roulin A., Auer P.L., Libault M., Schlueter J., Farmer A., May G., Stacey G., Doerge R.W., Jackson S.A. (2013). The fate of duplicated genes in a polyploid plant genome. Plant J. 73: 143–153. [DOI] [PubMed] [Google Scholar]
- Saintenac C., Jiang D., Akhunov E.D. (2011). Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 12: R88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamini F., Özkan H., Brandolini A., Schäfer-Pregl R., Martin W. (2002). Genetics and geography of wild cereal domestication in the Near East. Nat. Rev. Genet. 3: 429–441. [DOI] [PubMed] [Google Scholar]
- Salmon A., Flagel L., Ying B., Udall J.A., Wendel J.F. (2010). Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 186: 123–134. [DOI] [PubMed] [Google Scholar]
- Schreiber A.W., Sutton T., Caldo R.A., Kalashyan E., Lovell B., Mayo G., Muehlbauer G.J., Druka A., Waugh R., Wise R.P., Langridge P., Baumann U. (2009). Comparative transcriptomics in the Triticeae. BMC Genomics 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N., Tahira C., Kassai K., Hirabayashi C., Shimizu T., Takumi S., Mochida K., Kawaura K., Ogihara Y., Murai K. (2007). Genetic and epigenetic alteration among three homoeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19: 1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G.K. (2004). Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3: e3. [DOI] [PubMed] [Google Scholar]
- Soltis P.S., Soltis D.E. (2009). The role of hybridization in plant speciation. Annu. Rev. Plant Biol. 60: 561–588. [DOI] [PubMed] [Google Scholar]
- Soltis, P.S., and Soltis, D.E., eds (2012). Polyploidy and Genome Evolution. (New York: Springer). [Google Scholar]
- Wang J., Tian L., Lee H.S., Wei N.E., Jiang H., Watson B., Madlung A., Osborn T.C., Doerge R.W., Comai L., Chen Z.J. (2006). Genomewide nonadditive gene regulation in Arabidopsis allotetraploids. Genetics 172: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel J.F. (2000). Genome evolution in polyploids. Plant Mol. Biol. 42: 225–249. [PubMed] [Google Scholar]
- Xiong Z., Gaeta R.T., Pires J.C. (2011). Homoeologous shuffling and chromosome compensation maintain genome balance in resynthesized allopolyploid Brassica napus. Proc. Natl. Acad. Sci. USA 108: 7908–7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Hollister J.D., Wright K.M., Arnold B.J., Higgins J.D., Franklin F.C.H., Bomblies K. (2013). Meiotic adaptation to genome duplication in Arabidopsis arenosa. Curr. Biol. 23: 2151–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Bian Y., Gou X., Dong Y., Rustgi S., Zhang B., Xu C., Li N., Qi B., Han F., von Wettstein D., Liu B. (2013a). Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. Proc. Natl. Acad. Sci. USA 110: 19466–19471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., et al. (2013b). Persistent whole-chromosome aneuploidy is generally associated with nascent allohexaploid wheat. Proc. Natl. Acad. Sci. USA 110: 3447–3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao N., Zhu B., Li M., Wang L., Xu L., Zhang H., Zheng S., Qi B., Han F., Liu B. (2011). Extensive and heritable epigenetic remodeling and genetic stability accompany allohexaploidization of wheat. Genetics 188: 499–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.