This work uses multiple-environment expression quantitative trait locus analysis of the Brassica rapa genome, combined with comparative genomics of Arabidopsis, to identify putative vacuolar calcium transporters with potential uses in biofortification to increase the accumulation of calcium in food crops.
Abstract
Although Ca transport in plants is highly complex, the overexpression of vacuolar Ca2+ transporters in crops is a promising new technology to improve dietary Ca supplies through biofortification. Here, we sought to identify novel targets for increasing plant Ca accumulation using genetical and comparative genomics. Expression quantitative trait locus (eQTL) mapping to 1895 cis- and 8015 trans-loci were identified in shoots of an inbred mapping population of Brassica rapa (IMB211 × R500); 23 cis- and 948 trans-eQTLs responded specifically to altered Ca supply. eQTLs were screened for functional significance using a large database of shoot Ca concentration phenotypes of Arabidopsis thaliana. From 31 Arabidopsis gene identifiers tagged to robust shoot Ca concentration phenotypes, 21 mapped to 27 B. rapa eQTLs, including orthologs of the Ca2+ transporters At-CAX1 and At-ACA8. Two of three independent missense mutants of BraA.cax1a, isolated previously by targeting induced local lesions in genomes, have allele-specific shoot Ca concentration phenotypes compared with their segregating wild types. BraA.CAX1a is a promising target for altering the Ca composition of Brassica, consistent with prior knowledge from Arabidopsis. We conclude that multiple-environment eQTL analysis of complex crop genomes combined with comparative genomics is a powerful technique for novel gene identification/prioritization.
INTRODUCTION
Ca is an essential element required in large amounts by plants for structural roles and also, in lesser amounts, for cell signaling (White and Broadley, 2003; Dodd et al., 2010; White, 2014). Leaf/shoot Ca concentration (hereafter referred to as shoot-Ca) is typically in the low percentage (w/w) dry weight range, although it varies widely among terrestrial species (0.02 to 13.1%; Broadley et al., 2003, 2004; Watanabe et al., 2007). At the cellular scale, Ca is distributed to cell wall, vacuole/organelle, and cytosolic fractions by an array of transport and binding processes (White and Broadley, 2003; Dodd et al., 2010; Martinoia et al., 2012; White, 2014). For example, when a cell is perturbed by developmental cues or abiotic/biotic stresses, rapid Ca2+ influx to the cytosol occurs through Ca2+-permeable ion channels (White and Broadley, 2003; Dodd et al., 2010; White, 2014). To avoid cytotoxicity, cytosolic Ca2+ is then returned to submicromolar concentrations through the activities of Ca2+-ATPases of the P2A-ATPase (ECA) and P2B-ATPase (ACA) protein families and Ca2+/H+ antiporters of the calcium exchanger (CAX) protein family, which are located on plasma and organelle membranes (Shigaki and Hirschi, 2006; Dodd et al., 2010; Bonza and De Michelis 2011; Conn et al., 2011; Pittman, 2011; Martinoia et al., 2012; White, 2014). At the tissue scale, large differences in Ca concentrations are observed between cell types. For example, Ca is preferentially accumulated in mesophyll vacuoles and epidermal trichomes, which may protect guard cell function (Conn and Gilliham, 2010; Conn et al., 2011). At the organ scale, Ca accumulates preferentially within actively transpiring organs because it is immobile in the phloem (White and Broadley, 2003; Karley and White, 2009). The initial entry of Ca to plants through roots and the transfer to xylem tissues involve a combination of symplastic, transcellular, and apoplastic routes (White, 2001, 2014; White and Broadley, 2003), which are affected by apoplastic barriers currently being characterized (Baxter et al., 2009; Hosmani et al., 2013).
Shoot-Ca is under strong environmental and genetic control. Unsurprisingly, plants typically have greater shoot-Ca at high external Ca supply ([Ca]ext) than at low [Ca]ext (White, 2014). In Brassicaceae species, Ca sequestration in mesophyll vacuoles occurs at high [Ca]ext (Conn and Gilliham, 2010; Conn et al., 2011, 2012; Rios et al., 2012). However, relationships between shoot-Ca and [Ca]ext differ between “physiotypes” (Kinzel, 1982) associated with certain plant families. Thus, shoot-Ca responds to increased [Ca]ext to a greater extent in calcicole-like calcitroph plants (e.g., Brassicaceae and Crassulaceae) than among calcifuge-like potassium plants (e.g., Apiaceae and Asteraceae) and oxalate plants (e.g., Amaranthaceae) (Kinzel, 1982; Kinzel and Lechner, 1992; White and Broadley, 2003; White, 2014). Genetic controls of shoot-Ca have been quantified more formally at contrasting scales. For example, at higher levels of phylogenetic organization, shoot-Ca is low among commelinoid monocot species (e.g., grasses and sedges), as is shoot Mg concentration (shoot-Mg). Species of Amaranthaceae (e.g., spinach [Spinacia oleracea]) and related clades of the order Caryophyllales have low shoot-Ca but high shoot-Mg (Broadley et al., 2003, 2004; Watanabe et al., 2007; Broadley and White 2012). Phylogenetic variation in shoot-Ca may be due in part to evolutionary differences in cell wall chemistry, as up to 50% of Ca in plants is bound to pectins in cell wall lamellae (White and Broadley, 2003). At intraspecific scales, shoot-Ca is strongly heritable among leafy Brassica oleracea subspecies, with several significant quantitative trait loci (QTLs) being identified under controlled environment and field conditions (Broadley et al., 2008). In addition to natural genetic variation, functional studies have revealed key roles for specific gene products in regulating the vacuolar sequestration of Ca in the leaf mesophyll (Cheng et al., 2005; Conn et al., 2011) and the root uptake of Ca (Baxter et al., 2009) and their effects on shoot-Ca. For example, Arabidopsis thaliana cax1 cax3 double mutants have lower shoot-Ca (Cheng et al., 2005; Conn et al., 2011) due to reduced uptake into mesophyll vacuoles. Conversely, ectopic expression of a truncated At-CAX1 (sCAX1) increased shoot-Ca in carrot (Daucus carota), potato (Solanum tuberosum), tomato (Solanum lycopersicum), and tobacco (Nicotiana tabacum; Shigaki and Hirschi, 2006; Morris et al., 2008).
Despite advances in our knowledge of environmental and genetic factors controlling shoot-Ca at multiple scales, there are no reported comprehensive studies of these factors and their interactions (genetic × environment). Recent developments in genetical genomics, also known as expression quantitative trait locus (eQTL) analysis, can now measure variation in global gene expression among individuals within mapping populations (Kliebenstein, 2009; Druka et al., 2010; Hammond et al., 2011; Holloway et al., 2011; Moscou et al., 2011; Cubillos et al., 2012; Ballini et al., 2013; Huang et al., 2013). Variation in gene expression can be linked to sequence polymorphisms in target genes or, perhaps more interestingly, inherited differences in cis-regulatory (proximal) or trans-regulatory (distal) regions. eQTLs, therefore, can be used to integrate physical and genetic maps, to select genes underlying phenotypic differences for detailed characterization, and/or to identify novel regulatory networks. However, despite the increasing affordability of genetical genomic approaches, reported multiple-environment eQTLs are still limited to a few studies, such as two-condition biotic (Druka et al., 2008; Chen et al., 2010; Moscou et al., 2011) and abiotic (Hammond et al., 2011) experiments.
The primary aim of this study was to characterize the genetic, environmental, and genetic × environment effects on plant gene regulation by identifying eQTLs under altered [Ca]ext supply in Brassica rapa (2n = 2x = 20) and characterizing candidate genes. This species can accumulate relatively high Ca concentrations in its leaf mesophyll tissues (Rios et al., 2012). Furthermore, B. rapa has a sequenced genome that enables comparative genomic analyses with Arabidopsis (Wang et al., 2011) and postgenomic resources, including high-density exon-based expression arrays (Love et al., 2010), populations of mutants identified by targeting induced local lesions in genomes (TILLING) (Stephenson et al., 2010; Wang et al., 2012), and mapping lines (Iniguez-Luy et al., 2009). All of these resources were exploited in this study to test the potential of a Brassica-to-Arabidopsis-to-Brassica workflow. Multiple-environment conditions were used so that cis- and trans-eQTLs, which were constitutive or responded solely to Ca or Mg or to both elements, could be identified. Novel eQTLs were characterized in silico from phenotypes of loss-of-function Arabidopsis mutants in a large public ionomics database (Baxter et al., 2007). Mutations in the Ca2+ transporter BraA.CAX1a, identified previously by TILLING (Ó Lochlainn et al., 2011) and characterized here, show a link between eQTLs and altered shoot-Ca in B. rapa and Arabidopsis. Given that Brassica species are consumed widely as vegetable crops and are possible target crops for increasing intakes of important nutrients such as Ca (Broadley and White, 2010), this study reveals further novel candidate genes that might also be used for biofortification.
RESULTS AND DISCUSSION
Responses of B. rapa to Altered [Ca]ext and External Mg Supply
We investigated the response of shoot biomass and shoot-Ca to altered [Ca]ext and external Mg supply ([Mg]ext) (Figures 1 and 2). These responses were determined using an experimental design that included Ca and Mg supply as treatment factors, each represented by six external concentrations (i.e., plants were grown under 36 [Ca]ext × [Mg]ext conditions). Mg was used as a treatment factor so that responses specific to [Ca]ext could be separated from those due to elevated concentrations of a nutrient cation or chloride. There was a significant positive relationship between added and measured (water-extractable) compost [Ca]ext and [Mg]ext, indicating that appropriate experimental conditions were used (Supplemental Figure 1). All primary biomass and tissue concentration data are presented in Supplemental Data Set 1.
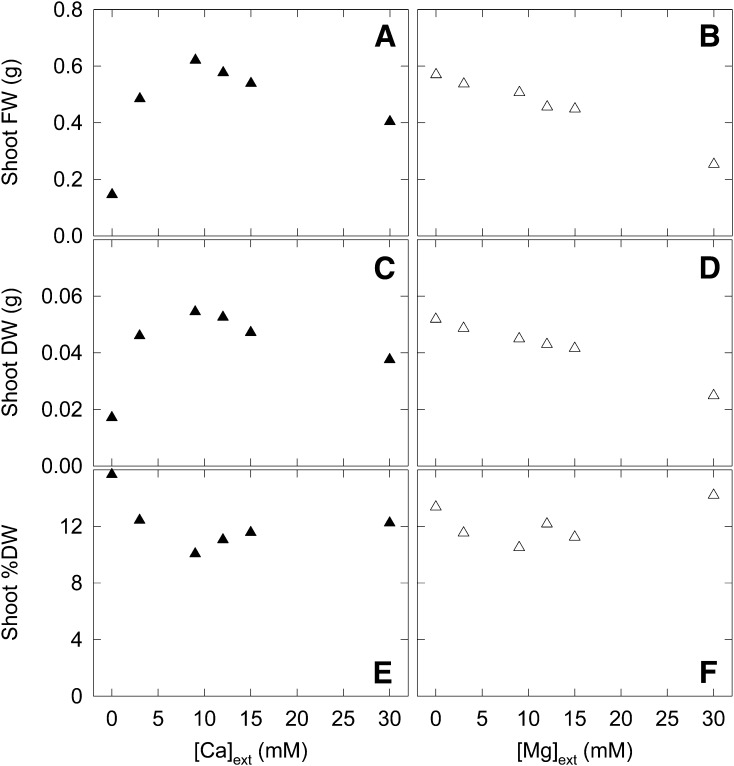
Figure 1.
Pooled Yield Responses of B. rapa as Functions of [Ca]ext and [Mg]ext.
Plants were grown in compost for 18 d. Data are means of 54 samples: 3 genotypes (IMB211, R500, and R-o-18), 6 [Mg]ext or [Ca]ext treatment levels per [Ca]ext or [Mg]ext treatment level, respectively, and 3 replicates. LSD values (P = 0.05) are as follows: 0.1368 (shoot fresh weight [FW]; [A] and [B]), 0.0055 (shoot dry weight [DW]; [C] and [D]), and 2.634 (shoot percentage dry weight [%DW]; [E] and [F]).
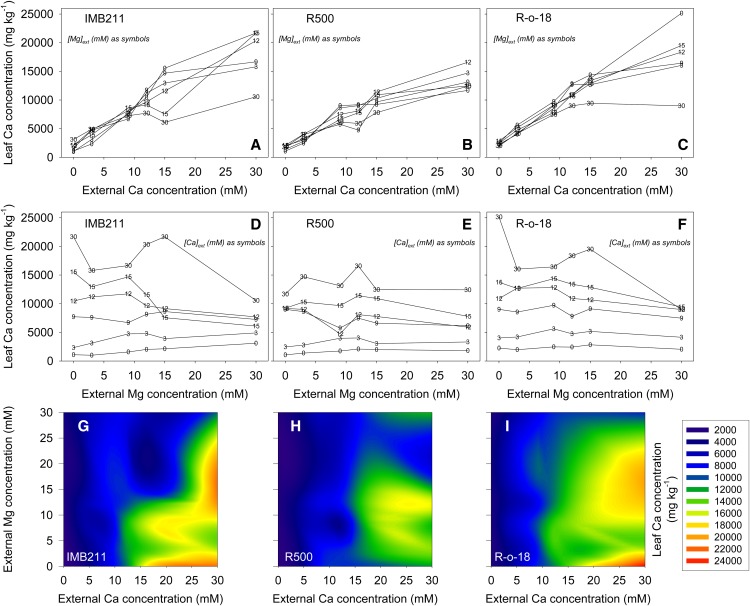
Figure 2.
Shoot Ca Concentrations of Three Genotypes of B. rapa (IMB211, R500, and R-o-18) as Functions of [Ca]ext and [Mg]ext.
Plants were grown in compost for 18 d. Data are means of three replicates analyzed using a genotype/([Ca]ext × [Mg]ext) treatment structure within an ANOVA. In (A) to (C), there are six [Mg]ext treatment levels per [Ca]ext treatment level; these are indicated as numerical symbols for each data point. Similarly, in (D) to (F), there are six [Ca]ext treatment levels per [Mg]ext treatment level. (G) to (I) are ternary plots showing interpolated data for all 36 Ca × Mg treatment combinations. Error terms (LSD values; P = 0.05) are as follows: 689, 1689, and 4136 mg/kg for genotype, [Ca]ext or [Mg]ext, and genotype/([Ca]ext × [Mg]ext) terms, respectively.
There were no significant interactions of [Ca]ext × [Mg]ext on shoot biomass production (P > 0.05); hence, the data in Figure 1 are presented as global means for each main factor of [Ca]ext and [Mg]ext. Shoot fresh and dry weights both increased when [Ca]ext increased from 0 to 9 mM (Figures 1A and 1C). The percentage dry weight (w/w) of shoots decreased when [Ca]ext increased from 0 to 9 mM and did not change significantly with further increases in [Ca]ext (Figure 1E). Shoot fresh and dry weights of B. rapa decreased with increasing Mg supply, although there was no significant effect on biomass or percentage dry weight in the range 0 to 9 mM [Mg]ext (Figures 1B, 1D, and 1F).
There was a general increase in shoot-Ca with increasing [Ca]ext across all [Mg]ext treatments (Figure 2). In IMB211 (Figures 2A, 2D, and 2G) and R-o-18 (Figures 2C, 2F, and 2I), shoot-Ca increased by >10-fold in response to increasing [Ca]ext and shoot-Ca exceeded 2% (w/w) at 30 mM [Ca]ext at low [Mg]ext. In R500 (Figure 2B), shoot-Ca increased to ∼1.5% (w/w) at 30 mM [Ca]ext. At high [Ca]ext (15 and 30 mM), a high [Mg]ext led to a substantial decrease in shoot-Ca in IMB211 and R-o-18 but not in R500 (Figure 2). There was a general increase in shoot-Mg with increasing [Mg]ext across all [Ca]ext treatments (Supplemental Figure 2). Shoot-Mg increased by ∼5-fold in response to increasing [Mg]ext, with shoot-Mg reaching 1% (w/w) at 30 mM [Mg]ext at low [Ca]ext (Supplemental Figures 2D to 2I). At higher [Mg]ext (30 mM), increasing [Ca]ext led to a substantial decrease in shoot-Mg in IMB211 and R-o-18 but not in R500 (Supplemental Figures 2D to 2F). A 2 × 2 combination of [Ca]ext × [Mg]ext treatments was selected for subsequent eQTL experiments, defined as low/low (LL), high/low (HL), low/high (LH), and high/high (HH). The high concentrations were 24 mM [Ca]ext and 15 mM [Mg]ext, and the low concentrations were 3 mM [Ca]ext and 1 mM [Mg]ext.
Genetical Genomics Analysis of Plant Responses to Ca and Mg
Novel Gene Expression Markers and Genetic Map for B. rapa
Sequence polymorphisms between the parents of the mapping population result in differences in the hybridization of transcripts to the probes on the array and subsequent differences in signal values for those probes. These differences can be used to develop markers based on gene expression. Initially, the expression data from the two parents, across all treatments, were analyzed to identify exon-level probe sets that had significantly different signal values. The linkage map reported here, and used for eQTL analyses, is based on 134 gene expression markers (GEMs), selected from 3272 potential GEMs, and mapped for 85 individuals from the Brassica rapa IMB211 × R500 Recombinant Inbred (BraIRRI) mapping population (Iniguez-Luy et al., 2009). This new map has a length of 811.6 centimorgan (cM), similar to that reported previously by Hammond et al. (2011), which spanned 881.6 cM. Of the 134 GEMs in the current map, 121 have reported genomic locations consistent with their mapping positions. An additional two have locations near the genetic position reported, but not in the same order as expected from the physical location. The remaining nine GEMs could not be located uniquely in the reported physical map at present. The median heritability of GEMs was 0.86, consistent with the high heritability of GEMs identified within this mapping population previously (Hammond et al., 2011; Supplemental Figure 3).
Global Analyses of B. rapa Gene Expression Data Using Ca and Mg Supply as Cofactors
eQTLs are genetic regions associated with variations in gene expression among individuals (Kliebenstein, 2009). This variation can arise due to sequence polymorphisms in target genes, their cis-regulatory (proximal) regions or trans-regulatory (distal) genes, leading to phenotypic differences. Identifying variation in gene expression within a segregating mapping population provides a powerful tool for resolving contributions and interactions within functional gene regulatory networks (Druka et al., 2010). We quantified gene expression levels for the 135k probe sets on the Brassica Exon 1.0 ST Affymetrix array in 85 lines of the BraIRRI mapping population under the LL, LH, and HL treatments. Since the array was designed to all Brassica sequence information available at the time, including B. rapa, B. napus, and B. oleracea, not all probe sequences correspond to unique B. rapa genomic locations (Love et al., 2010), and individual B. rapa gene models can be represented by more than one probe set. In total, 19,999 unique BRAD gene models were represented by probe sets on the array Thus, of the 35,256 significant (log of the odds [LOD] score > 3.17) differential signals, 9910 had physical locations in the B. rapa genome (1895 cis and 8015 trans) and therefore were designated as eQTLs (Figure 3). The average LOD score for cis-eQTLs was 19.89 ± 0.97 (se), and for trans-eQTLs the average LOD score was 5.85 ± 0.13 (se), consistent with previous eQTL studies (Hammond et al., 2011). The heritability of gene expression and treatment variance component effects of all of the 135k probe sets are visualized in Supplemental Figure 3.
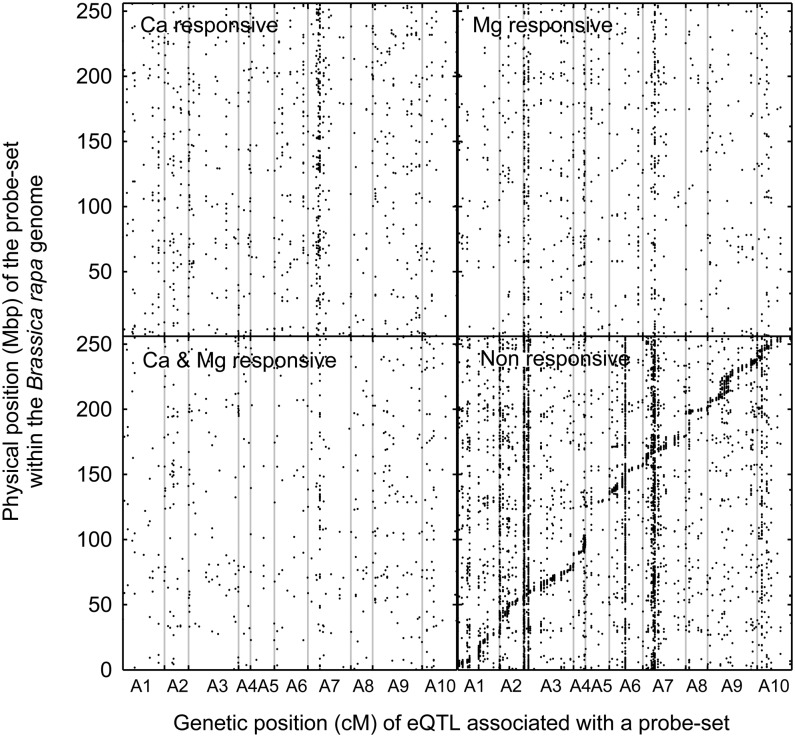
Figure 3.
Physical Positions of eQTLs as a Function of Genetic Position.
Data are shown from 85 lines of the B. rapa BraIRRI mapping population grown in compost under three treatments (LL, HL, and LH) as follows: the high concentrations were 24 mM CaCl2 and 15 mM MgCl2, and the low concentrations were 3 mM CaCl2 and 1 mM MgCl2. Each data point represents a unique eQTL (LOD score > 3.17) designated as responsive to Ca only, to Mg only, to Ca and Mg, or nonresponsive based on additive effects.
The direction (positive or negative) of the additive effect (of the IMB211 allele) for an eQTL was the same under all treatment conditions for most of the significant eQTLs associated with B. rapa gene models (7419); therefore, these were designated as nonresponsive. The remaining eQTLs associated with B. rapa gene models were designated as Ca-responsive (high Ca/low Mg; 971), Mg-responsive (low Ca/high Mg; 972), or Ca- and Mg-responsive (548), based on whether the direction of the additive effect of an eQTL changed under high Ca supply only, under high Mg supply only, or when both Ca and Mg supply were altered, respectively. Among the 1895 cis-eQTLs, 23 were Ca-responsive, 6 were Mg-responsive, and 8 were both Ca- and Mg-responsive. Among the 8015 trans-eQTLs, 948 were Ca-responsive, 966 were Mg-responsive, and 540 were both Ca- and Mg-responsive. When the physical positions of eQTLs are plotted as a function of their genetic positions, a cis-diagonal can be seen clearly among the nonresponsive group. Among the 25,346 eQTLs lacking a physical position in the genome, 3615 were Ca-responsive, 3139 were Mg-responsive, and 2490 were both Ca- and Mg-responsive.
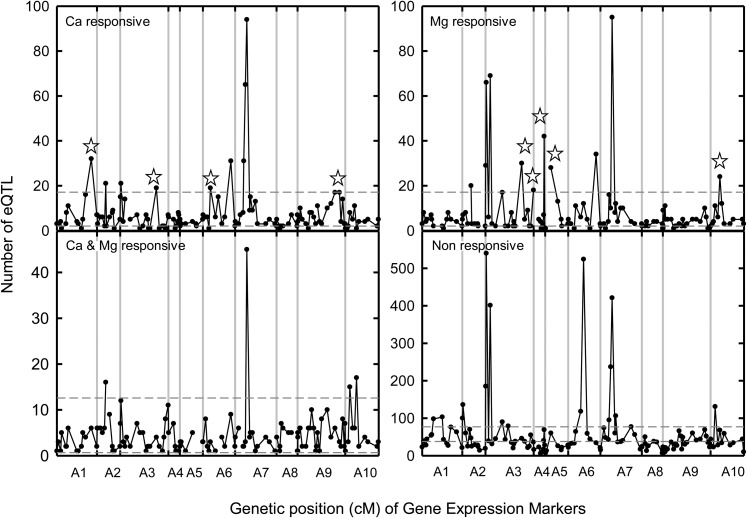
Hot spots of eQTLs can be seen when a mapping interval GEM has more eQTLs associated with it than would be expected by chance (Figure 4). Nonresponsive eQTL hot spots share a number of genomic locations with phosphorus-responsive hot spots identified previously for B. rapa in the same mapping population (Hammond et al., 2011; Supplemental Table 1). These genes might represent regulatory controls or general stress responses, given their identification across wide environmental conditions.
Figure 4.
Distribution of eQTLs Associated with Individual Markers (Black Circles).
Dashed horizontal lines represent the 99.0% upper and lower confidence intervals of the Poisson distribution, which assumes that GEMs and eQTLs are distributed equally across the genome. Data are shown from 85 lines of the B. rapa BraIRRI mapping population grown in compost under three treatments (LL, HL, and LH) as follows: the high concentrations were 24 mM CaCl2 and 15 mM MgCl2, and the low concentrations were 3 mM CaCl2 and 1 mM MgCl2. eQTLs were designated as responsive to Ca only, to Mg only, to Ca and Mg, or nonresponsive based on additive effects. Stars indicate eQTL hot spots discussed in the article.
For eQTLs defined as Ca-responsive (i.e., when the direction of the additive effect of an eQTL changed under altered [high] Ca supply only), hot spots were identified at 86 cM on A01, 92 cM on A03, 19 cM on A06, and 88 to 99 cM on A09b (Figure 4). The Ca-responsive hot spot at 19 cM on A06 contains the cis-eQTLs associated with an auxin-induced basic/helix-loop-helix transcription factor (AtTCP14; At3g47620) and a small nuclear ribonucleoprotein family protein (At1g20580). A trans-eQTL associated with the autoinhibited Ca2+-ATPase, isoform 8 (ACA8; At5g57110), was also identified under the eQTL hot spot at 92 cM on A03.
Mg-responsive eQTL hot spots were identified at 92 and 121 cM on A03, 26 cM on A04, 15 cM on A05, and 24 cM on A10 (Figure 4). No cis-eQTLs were identified under any Mg-responsive eQTL hot spots. However, within the Mg-responsive hot spot between 25 and 27 cM on A04, a significant eQTL associated with the B. rapa ortholog of the Arabidopsis CAX1 gene (At2g38170) was identified. The B. rapa ortholog (BraA.CAX1a; BRAD identifier Bra017134) is represented by two probe sets on the array, which are also associated with a nonresponsive eQTL at 2.5 cM on A03 and another Mg-responsive eQTL at 107 cM on A03. A trans-eQTL associated with the autoinhibited Ca2+-ATPase, isoform 4 (ACA4; At2g41560), was also identified under the eQTL hot spot at 92 cM on A03.
In Silico Phenotyping of eQTL Orthologs Using a Large Arabidopsis Ionomics Database
In silico phenotyping using the Purdue Ionomics Information Management System (PiiMS; Baxter et al., 2007) database of Arabidopsis leaf ionomic profiles was performed to identify gene targets associated with eQTLs in B. rapa that also had shoot-Ca phenotypes associated with mutations in the orthologous gene in Arabidopsis. Identifying Arabidopsis genotypes with mutations in known genes that have a shoot-Ca phenotype and are associated with a significant eQTL in B. rapa provides an efficient pipeline to select eQTLs for further study. We identified 39 mutant genotypes in the PiiMS database with consistent and robust altered shoot-Ca phenotypes (i.e., which had increased [z score ≥ 3] or decreased [z score ≤ 3] leaf-Ca relative to a control plant in ≥50% of the samples for an individual mutant genotype). Of these, 24 genotypes had decreased leaf-Ca and 15 had increased leaf-Ca (Supplemental Data Set 4). These 39 genotypes have 31 unique Arabidopsis Genome Initiative (AGI) code tags. These AGI codes include four potential Ca2+ transporters: ACA8, an autoinhibited Ca2+-ATPase (Geisler et al., 2000); ECA4, a P2A-type Ca2+-ATPase located in the endoplasmic reticulum (Mills et al., 2008); and two Ca2+/H+ antiporters, CAX1 and CAX3 (Shigaki and Hirschi, 2006; Conn et al., 2011). They also include a cation efflux protein of the cation diffusion facilitator family, MTP5 (Gustin et al., 2011).
The 31 AGI codes mapped to 62 probe sets on the Brassica Exon 1.0 ST Affymetrix array (Supplemental Data Set 5), of which 21 probe sets were associated with one or more significant eQTLs, for a total of 27 eQTLs. Of these, six eQTLs were designated as responsive: two Ca-responsive, three Mg-responsive, and one Ca- and Mg-responsive. Among the responsive eQTLs, two were associated with ACA8, two with CAX1, and two with an Arabidopsis FLOWERING LOCUS F MADS box protein (AT5G10140). Among the nonresponsive eQTLs, two were associated with CAX3 and one each with IAR1 (At1g68100), a member of the ZIP metal ion transporter family, and ECA4. CRYPTOCHROME2, a blue light photoreceptor (AT1G04400), was associated with four nonresponsive eQTLs and has been implicated in the regulation of circadian oscillations in the concentration of cytosolic Ca2+ in response to blue light (Xu et al., 2007). The functional significance of this association is not known.
Ideally, data from a complete set of homozygous knockout mutants and overexpression lines of Arabidopsis are required to test the specificity of in silico phenotyping, but these data are not yet available. This study, which is thus biased toward transporter mutants, shows that 16 out of the 37 Brassica genes (43%) with a mapped physical location linked to an AGI code tag for shoot-Ca are linked to an eQTL. Similarly, 17 out of 32 Brassica genes (53%) linked to an AGI code tag for shoot-Mg are linked to an eQTL. A similar analysis for Fe (34%) and Zn (32%) shows these proportions to be lower, despite many mutants having multiple phenotypes. For example, 32 and 23% of Arabidopsis mutants with a phenotype of z > 3 or z < −3 for shoot-Ca concentration also have significantly altered leaf Fe and Zn concentration phenotypes, respectively.
Characterizing BraA.CAX1a Mutants
We had previously identified three missense mutations in the Ca2+ transporter BraA.CAX1a by TILLING (Ó Lochlainn et al., 2011). Therefore, we conducted further selfing, genotyping, and characterization of these lines, based on the identification of a significant eQTL associated with BraA.CAX1a under high-Mg, low-Ca conditions and the subsequent in silico identification of a Ca phenotype associated with the Arabidopsis ortholog of this gene (Supplemental Data Sets 4 to 7). The characterization of this gene in B. rapa demonstrates the potential of the Brassica-Arabidopsis-Brassica workflow but was also guided by prior studies in Arabidopsis (Hirschi et al., 1996; Pittman and Hirschi, 2001; Shigaki et al., 2001, 2002; Catalá et al., 2003; Cheng et al., 2003, 2005).
A shoot-Ca phenotype in B. rapa was observed for BraA.CAX1a (Bra017134) mutants, which corresponded to an eQTL and an Arabidopsis phenotype in the ionomics database. To isolate lines with mutations in the BraA.CAX1a gene in B. rapa, a TILLING approach was adopted previously. A 1.5-kb fragment including the transcriptional start site and the first exon was used as the target. In total, 20 mutations were identified comprising 10 missense, 1 nonsense, 2 silent, and 7 noncoding changes (Ó Lochlainn et al., 2011; Supplemental Figure 4). Lines with missense and nonsense (stop) mutations were backcrossed to the R-o-18 parent line of the TILLING population, and homozygous and segregating wild-type lines for each mutation were isolated in the BC1S1 population. Of these lines, three missense alleles designated BraA.cax1a-4 (A-to-T change at amino acid 77), BraA.cax1a-7 (R-to-K change at amino acid 44), and BraA.cax1a-12 (P-to-S change at amino acid 56) were characterized in BC1S2.
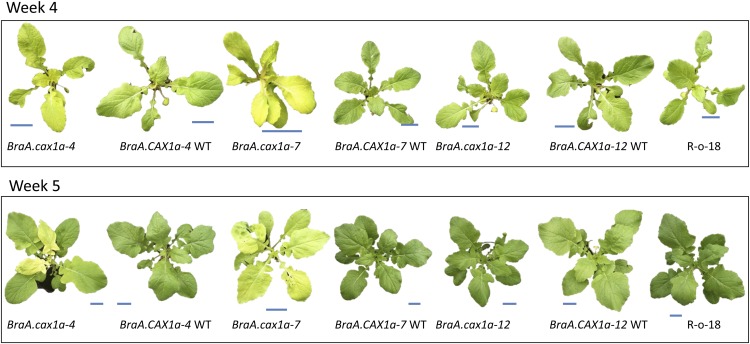
Visibly different phenotypes were observed in the homozygous mutants BraA.cax1a-4 and BraA.cax1a-7 compared with their segregating wild types (Figure 5). Homozygous plants of the BraA.cax1a-4 and BraA.cax1a-7 lines had paler/yellow leaves at both 4 and 5 weeks old (Figure 5). This phenotype was seen in expanding and fully expanded leaves in BraA.cax1a-7 at both time points. However, in BraA.cax1a-4, expanding leaves were pale at weeks 4 and 5, but fully expanded leaves were only pale at week 5. The leaves of BraA.cax1a-12 appeared similar to those of the segregating wild-type and R-o-18 plants at both time points. To quantify this phenotype, leaf chlorophyll concentration was measured indirectly at two time points using a single-photon avalanche diode (SPAD) meter. Leaf chlorophyll concentration was lower in both the expanding and fully expanded leaves of the BraA.cax1a-4 and BraA.cax1a-7 lines compared with their segregating wild types at both time points (P < 0.01; Supplemental Figure 5). The difference in chlorophyll concentration between fully expanded leaves of BraA.cax1a-4 and its segregating wild type was smaller than that between expanding leaves, especially at week 5. Further investigations are now required to elucidate the significance of the chlorotic phenotype in relation to leaf Ca homeostasis.
Figure 5.
Characterization of Independent Homozygous BraA.cax1a Lines.
Three independent homozygous BraA.cax1a lines (BC1S2s), their segregating wild types (BC1S2s), and R-o-18 were grown in compost for 4 or 5 weeks. Bars = 5 cm.
Ca phenotypes were observed in fully expanded leaves of BraA.cax1a-7 and BraA.cax1a-12 compared with their segregating wild types (Table 1). In BraA.cax1a-7, shoot-Ca was less than the wild-type value (26,036 versus 34,356 mg/kg; P = 0.05, n = 3). In BraA.cax1a-12 plants, shoot-Ca was greater than the wild-type value (44,534 versus 27,161 mg/kg; P = 0.02, n = 3). There was no significant difference in shoot-Ca in BraA.cax1a-4 and its segregating wild type. The only significant shoot-Mg phenotype was seen in BraA.cax1a-12, which had a higher shoot-Mg in the mutant compared with the wild type (18,733 versus 12,544 mg/kg; P = 0.04, n = 3). There were no mutant phenotypes observed for shoot Zn, Fe, or K concentration (Table 1).
Table 1. Mean Shoot Mineral Compositions of BraA.cax1a Mutants and Their Segregating Wild Types.
| Shoot-Ca | Shoot-Mg | Shoot-Zn | Shoot-Fe | Shoot-K | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Line | mg/kg | se | mg/kg | se | mg/kg | se | mg/kg | se | mg/kg | se |
| BraA.cax1a-12 | 44,534 | 4,215 | 18,733 | 1,892 | 74.3 | 7.9 | 55.2 | 8.8 | 54,875 | 12,435 |
| BraA.CAX1a-12 WT | 27,161 | 1,392 | 12,544 | 633 | 62.3 | 3.5 | 55.2 | 2.0 | 40,301 | 7,021 |
| P | 0.017 | 0.036 | 0.238 | 0.996 | 0.365 | |||||
| BraA.cax1a-7 | 26,036 | 2,255 | 11,835 | 1,459 | 75.2 | 8.7 | 60.2 | 2.9 | 60,011 | 9,022 |
| BraA.CAX1a-7 WT | 34,356 | 2,030 | 15,450 | 1,248 | 55.9 | 3.5 | 52.7 | 6.5 | 40,607 | 5,077 |
| P | 0.052 | 0.133 | 0.109 | 0.352 | 0.134 | |||||
| BraA.cax1a-4 | 30,335 | 8,027 | 12,970 | 3,471 | 61.3 | 13.0 | 56.8 | 14.7 | 39,834 | 8,801 |
| BraA.CAX1a-4 WT | 44,379 | 9,704 | 21,908 | 6,247 | 68.8 | 10.8 | 63.5 | 13.4 | 54,225 | 14,634 |
| P | 0.327 | 0.279 | 0.680 | 0.752 | 0.447 | |||||
Shoot samples (n = 3) were analyzed by inductively coupled plasma mass spectrometry. P values were calculated using a two-sided Student’s t test.
Further studies are needed to resolve the variation in shoot-Ca phenotypes observed among B. rapa missense mutants of BraA.CAX1a, especially given the relative complexity of the B. rapa genome compared with Arabidopsis. However, even in the simpler genome of Arabidopsis, shoot-Ca phenotypes are not always consistent in lines containing loss-of-function mutations in CAX1 (White, 2014). For example, Catalá et al. (2003) reported Arabidopsis cax1 mutants having lower shoot-Ca than wild-type plants, and Conn et al. (2011) reported a positive correlation between CAX1 expression and shoot-Ca in 15 ecotypes of Arabidopsis. By contrast, other groups have not reported reduced shoot-Ca in At-cax1 mutants (Cheng et al., 2003, 2005; Conn et al., 2011), possibly due to functional compensation by CAX3 (Cheng et al., 2005; Conn et al., 2011), although CAX3 expression does not correlate with shoot-Ca in the same way as that of CAX1 (Conn et al., 2011). Consistent with this interpretation, the shoot-Ca of cax1 cax3 double mutants is often significantly lower than that of wild-type plants, and impaired gas exchange has been observed (Cheng et al., 2005; Conn et al., 2011, 2012). No previous reports show an Arabidopsis cax1 loss-of-function mutant having increased shoot-Ca. However, there are numerous reports of altered tissue Ca concentrations arising from the overexpression of genes encoding truncated versions of At-CAX proteins lacking autoinhibitory domains (White, 2014). For example, unregulated expression of sAt-CAX1, a modified At-CAX2 gene (sAt-CAX2), or At-CAX4 has been shown to increase Ca concentrations in edible portions of several crops, including carrot, lettuce (Lactuca sativa), tomato, and potato (Park et al., 2004, 2005a, 2005b, 2009; Morris et al., 2008).
Comparative genomic studies are now feasible between plant species, including those with complex genomes such as B. rapa (King, 2013). Next-generation sequencing has facilitated the identification of paralogous and orthologous genes, resulting from genome segmentation and polyploidy events in plant genomes, even within large gene families (Cheng et al., 2012). In B. rapa, there are two paralogous copies of BraA.CAX1 but only a single version of BraA.CAX3. It will be necessary to isolate mutants of these individually and in combination to test for redundancy between gene family members and between paralogous copies to establish definitive roles for these proteins in Ca transport and storage. The three independent missense allelic variants in BraA.CAX1 characterized here target different amino acids upstream of the N-terminal autoinhibitory domain (Supplemental Figure 4; Pittman and Hirschi, 2001), offering insights into other residues critical for the regulation and function of this protein.
TILLING is a powerful approach for generating an allelic series of mutations within genes to study more subtle alterations in protein expression and structure. The development of these resources in crop species will assist in the functional characterization of genes and their products beyond model systems and shed new light on gene function. Given that Brassica species are consumed widely as vegetable crops and are possible target crops for increasing intakes of important nutrients such as Ca (Broadley and White, 2010), the knowledge and resources generated here will be of use in future strategies for the biofortification of edible crops. It is important to stress that the Brassica-Arabidopsis-Brassica workflow is currently dependent on (1) extensive prior annotation and characterization (in Arabidopsis) and (2) the availability of leaf Ca concentration data from the Arabidopsis ionomics database, which is currently weighted toward well-characterized transporter mutants. A full set of phenotypic data from homozygous knockout mutants and overexpression lines of Arabidopsis would be the preferred in silico resource in the coming years to improve the workflow and identify novel genes where a priori information is not available.
METHODS
Responses of Brassica rapa to Altered [Ca]ext and [Mg]ext
Two parental lines of the B. rapa (2n = 2x = 20; A genome) BraIRRI population, IMB211 and R500 (Iniguez-Luy et al., 2009; Hammond et al., 2011), and the R-o-18 reference parent of the B. rapa TILLING population (Stephenson et al., 2010) were grown in a compost substrate in 9 × 9 × 8-cm pots (Desch Plantpak) in a controlled environment. The controlled environment was a walk-in high-specification growth room (Weiss-Gallenkamp) at Wellesbourne, UK, set to a 16-h photoperiod using metal halide lamps, giving a photon flux density between 400 and 700 nm (photosynthetically active radiation) of 250 µmol m−2 s−1 at plant height. The day/night settings for temperature were 18/15°C and those for relative humidity were 76/71%. The compost substrate comprised Shamrock Professional Range medium peat (pHwater 3.8 to 4.4) mixed with horticultural-grade silver sand (particle size < 1 mm; J. Arthur Bowers) at a ratio of 3:1 (v/v). Nutrients were added as follows per volume of substrate: 0.629 g/L NH4NO3 (Nitram; GrowHow), 0.184 g/L NH4H2PO4 (Krista MAP; Yara UK), 0.03 g/L Na2SO4, 2 mL/L 44 mM FeNaEDTA, and 4 mL/L micronutrient solution comprising 30 mM H3BO3, 10 mM MnSO4, 3 mM CuSO4, 1 mM ZnSO4, and 0.5 mM Na2MoO4. All chemicals were supplied by VWR unless stated otherwise. A 6 × 6 combination of Ca and Mg treatment levels (36 treatments in total) was imposed via the addition of 0.375 M aqueous solutions of CaCl2 and MgCl2 to achieve 0, 3, 9, 12, 15, and 30 mM each element based on compost volume (Supplemental Figure 1). The substrate solution pH was adjusted to ∼5 by the addition of 4 mL of 10 M KOH (Fisher Scientific). Compost mixes were made in bulk using a paddle mixer (model 156; St. Moritz). The experiment was replicated three times using a randomized block design, in which each block represented one of three benches in the controlled environment room and each plot represented a treatment. For each treatment, six seeds of each line were sown into individual pots (18 pots), and the pots for each treatment were placed on capillary matting within a self-contained tray to avoid cross-contamination of nutrients between treatments. The capillary matting was irrigated with deionized rainwater twice a day for 30 s, using three drippers per tray and delivering 132 mL/d. Cotyledon, leaf, and stem samples were taken 18 d after sowing. Tissue samples from each line within a treatment were bulked, and their combined fresh weight was determined. Bulked tissues were dried at 60°C for 48 h before reweighing. Dry leaf samples were milled (0.5 mm; type 529AA mill; Apex Engineering Industries), and a 0.3-g subsample was digested in a closed vessel in 2 mL of HNO3 in a microwave (MARSXpress; CEM) (Hammond et al., 2011). After dilution with 23 mL of deionized water, samples were analyzed for mineral concentration by inductively coupled plasma atomic emission spectroscopy (JY Ultima 2; Jobin Yvon). Data were subjected to ANOVA using GenStat (12th edition; VSN International).
Genetical Genomics Analysis of Plant Responses to Ca and Mg
Growth of the BraIRRI Population for eQTL Analysis
Recombinant inbred lines (RILs) (n = 85; BraIRRI_05) (Supplemental Data Sets 2 and 3) of the BraIRRI mapping population that had been selfed for a minimum of eight generations, plus two parental lines, were grown in compost as described in the previous section in a glasshouse at the Sutton Bonington campus of the University of Nottingham. Plants were grown in March and April 2011 with supplementary lighting, giving a mean photon flux density of 170 μmol m−2 s−1, provided by 600-W sodium lamps (Philips Sun-T Pia Greenpower; Philips Electricals) for 18 h/d. A combination of Ca and Mg applications was made to give four treatments (LL, HL, LH, and HH): the high concentrations were 3.5 g/L compost (24 mM) CaCl2 and 3.04 g/L (15 mM) MgCl2, and the low concentrations were 0.44 g/L (3 mM) CaCl2 and 0.2 g/L (1 mM) MgCl2. Twelve pots of each RIL and 36 pots of each parent line were sown per treatment using a randomized block design. Fully expanded leaves from six plants per RIL and 18 plants per parent were sampled 18 d after sowing, bulked (in pools of six for parent lines), and snap-frozen in liquid N2. In total, 268 biological samples were obtained, comprising three replicates for each parent line at each treatment (3 replicates × 4 treatments × 2 parents = 24) and samples from 85, 81, 65, and 13 unique samples of RIL from the LL, LH, HL, and HH treatments, respectively.
RNA Extraction and Array Analysis
For each sample, total RNA was extracted using a modified TRIzol extraction protocol (Hammond et al., 2006) and purified using RNA Cleanup for RNeasy columns with on-column DNase digestion (Qiagen). Total RNA was labeled using the Affymetrix WT Labeling Kit and hybridized to the Affymetrix Brassica Exon 1.0 ST array (Love et al., 2010) according to the manufacturer’s instructions at two service providers: Nottingham Arabidopsis Stock Centre (University of Nottingham) and Source Bioscience. The total number of RNA samples processed on the Affymetrix Brassica Exon 1.0 ST array (and therefore *.cel output files) was 292. These samples were made up of the 268 biological samples described above plus 24 technical replicates, comprising 12 samples from parent lines grown under LL and LH treatments, 8 RNA samples from selected BraIRRI lines from the LL treatment, and 4 RNA samples from selected BraIRRI lines from the LH treatment. For the technical replicates, the RNA sample was split and hybridized to arrays independently by the two service providers. All data were imported into GeneSpring GX (version 11.0.2; Agilent Technologies) and normalized using the robust multiarray average (RMA) algorithm (Bolstad et al., 2003; Irizarry et al., 2003a, 2003b). All data have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE44185. The normalized signal value for an individual probe set from an array was standardized to the median signal value for that probe set across all arrays. Comparisons between technical replicates from different service providers identified 229 differentially (2-fold; P < 0.01, Benjamini-Hochberg false discovery–corrected P value based on paired t tests between technical replicates) expressed probe sets across all technical replicates, equivalent to 0.17% of probe sets. The average correlation coefficient of raw expression values between technical pairs was 0.958 ± 0.002 (se; n = 24).
Development of a Linkage Map Using GEMs
A set of 134 GEMs was selected for the BraIRRI_05 subpopulation using an approach modified from a previous study (Hammond et al., 2011). The approach of Hammond et al. (2011) was based on the Agilent Brassica 95k 60-mer arrays with a single probe per gene model. By contrast, probe sets on the Affymetrix Exon 1.0 ST array are derived from multiple 25-mer probes (Love et al., 2010). Therefore, because signal values are averaged across multiple probes, polymorphisms affecting the signal values of individual probes are likely to be masked. To overcome this constraint, exon probe sets, consisting of fewer probes, were used to identify polymorphic GEMs between the two parent lines. First, exon-level *.cel files for the two parents under all four treatments (i.e., IMB211 [n = 18 {3 replicates × 4 treatments} + 6 technical replicates] and R500 [n = 18]) were normalized using the RMA normalization algorithm (Bolstad et al., 2003; Irizarry et al., 2003a, 2003b), and the signal value for each exon probe set on an array was standardized to the median signal value for that exon probe set across all arrays. Second, exon probe set signal values were ranked according to segregation differences between parents based on P values from a two-tailed Student’s t test. A subset of 3272 exon probe sets, representing 2358 unique unigenes, were selected for exploration as suitable GEMs based on a segregation threshold of P < 2.1 × 10−8 between parents (Supplemental Data Set 3).
For mapping using GEMs, a reiterative procedure was followed using JoinMap4.1 (Kyazma; van Ooijen, 2006) with a combination of maximum likelihood mapping (MLM) and regression mapping (RM). The midpoint between mean parental exon probe set signal values was used to provisionally classify offspring as IMB211 (a allele) or R500 (b allele). A distinctness score was calculated for each exon probe set based on the sd of each exon probe set signal value for the entire population, divided by the average of the means of the sd of the a-class offspring and the sd of the b-class offspring. High values of distinctness should give more accurate allele calling and might reflect sequence-based polymorphisms. The 350 GEMs with the highest distinctness scores were used for initial grouping using the “independence LOD” option, ranging from LOD 2 to 15 in 2-LOD steps. Generally, the final LOD stringency selected was between LOD 3 and LOD 5 based on visual inspection of linkage groups. For initial linkage group maps, default settings were used in MLM and RM (Haldane mapping function; “jump” value of 5). The robustness of GEMs was tested by comparing results from MLM and RM. Thus, an initial set of GEMs was generated through iterative removal of GEMs by examining “fit and stress” parameters for MLM and by visual inspection of graphical genotypes for MLM and RM. Miscalled GEMs were removed (e.g., double recombination events in individual lines within small genetic distances [1 to 5 cM]). Further GEM removal was conducted to give a spaced genetic map of 134 GEMs for 85 RILs with intervals of between 5 and 10 cM for the subsequent eQTL analysis.
Global Analyses of B. rapa Expression Data Using Ca and Mg Supply Treatments as Cofactors
Gene expression data from three treatments (n = 273 *.cel files) were again normalized using RMA. These data represented (1) 105 LL *.cel files from 85 RILs plus 8 technical replicates and 12 parent files comprising biological (n = 3) and technical (n = 3) replicates from two parents; (2) 97 LH *.cel files from 81 RILs plus 4 technical replicates and 12 parent files as above; and (3) 71 HL *.cel files from 65 RILs plus biological (n = 3) replicates from two parents. The HH treatment was excluded due to low sample numbers. Log2-normalized signal values were exported as a data matrix with 273 columns (RNA samples) and 135,264 (135k) rows (array probe sets). Custom GenStat scripts (available on request from the authors) were used to batch-process the log2-normalized signal values in groups of up to 1500 genes. Genotypic and environmental variance components were estimated for each probe set using a residual maximum likelihood procedure (Patterson and Thompson, 1971; Robinson, 1987; Hammond et al., 2011), where the genotypic variance component is the effect of line within the population and the environment variance component is the treatment effect of altered Ca and Mg supply. Thus, the variance components model included gene expression as the variate, treatment (Vtreat) as a fixed model term, and line + (line × treatment) [Vline + (Vline × Vtreat)] and a residual (Vres) variance random model term. For each probe set, the treatment effect was defined as follows:
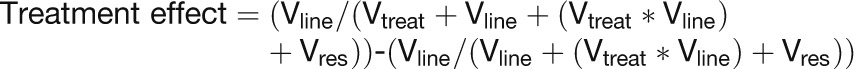 |
The heritability of the expression of each probe set was interpreted as a broad-sense mean line heritability based on Cullis et al. (2006). A variance–covariance matrix (_vp) was derived from the residual maximum likelihood for each probe set in turn, which is the line × line matrix yielding an expression variance term for each line (Vline). This matrix thereby yields a diagonal (_dpev) of variance terms across all lines. Subsequently, the mean of this diagonal is used [mean(_dpev)], for each gene in turn, to estimate the heritability of gene expression across the population (H2) as follows:
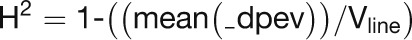 |
Identifying and Positioning Significant eQTLs
Data for parental lines were removed from the above (273 *.cel files) data set to generate a data matrix of 243 columns and 135k rows (probe sets). These data were used to identify eQTLs using a two-stage process. First, the GenStat procedure QIBDPROBABILITIES (Boer and Thissen, 2009) was used to calculate a set of genetic predictors at the marker positions on the GEMs genetic map (BraIRRI_05_2012a) for 85 RILs. A parallel regression model was used to identify possible QTL effects across 1500 probe sets simultaneously, by testing for a combined QTL + (QTL × environment) effect, where QTL indicates a genetic predictor used as a covariate. This model was fitted for each genetic predictor in turn. Second, for probe sets with a significant (LOD score > 3.3, P < 0.0005) result in this first step, simple interval mapping was performed using QTL procedures in GenStat using genetic predictors within the mixed model framework and a LOD threshold > 3.17 (Boer et al., 2007). Briefly, a mixed model was fitted with fixed environment effects and QTL × environment interaction for each genetic predictor in turn and random terms comprising Vline, Vline × Vtreat, and Vres. A compound symmetry structure was used to account for genetic covariance across environments. The additive allelic effect was defined as half of the difference in probe set expression between parental alleles at each marker and reported with respect to the IMB211 (a allele).
Individual probes sets on the Affymetrix Brassica Exon 1.0 ST array were aligned to the B. rapa Chiifu-401 genome sequence (version 1.0; 255.9 Mb, representing 90% of the assembled sequences) (Wang et al., 2011). Probe sets were assigned to specific chromosome sequence coordinates when >70% of probes within the probe set aligned with a match of ≥98% within the genome sequence. A cis-eQTL was defined when the physical position of the probe set and its associated GEM were in the same region (i.e., at the midpoint between two GEMs or between a GEM and the end of the chromosome). eQTLs outside of these regions were defined as trans-eQTLs. eQTL hot spots were defined when the number of eQTLs associated with a GEM exceeded an empirical threshold (the upper 99% confidence interval for the Poisson distribution, assuming an equal distribution of GEMs and eQTLs). Genesect (Virtual Plant 1.3; Katari et al., 2010) was used to test for overlaps in the genes associated with eQTL hot spots identified both here and by Hammond et al. (2011), based on z scores.
Four nonredundant gene lists were defined according to whether the direction (positive, the IMB211 allele resulted in a greater expression value of the gene; or negative, the IMB211 allele resulted in a lower expression value of the gene) of the additive effect of an eQTL was the same under all treatment conditions (nonresponsive) or whether it changed under altered Ca supply only (Ca-responsive), under altered Mg supply only (Mg-responsive), or when both Ca and Mg supply were altered (Ca- and Mg-responsive).
In Silico Phenotyping of eQTLs Using Arabidopsis thaliana Databases
Probe Set Annotation and Comparative Genomics
Each probe set on the Affymetrix Brassica Exon 1.0 ST array was mapped to the B. rapa gene lists from the BRAD database (version 1.2; plantgdb.org/BrGDB/) (Cheng et al., 2011). A match between a probe set and a gene was created if at least one probe in that probe set had an exact, full-length, ungapped alignment to that gene. The probes were aligned using TimeLogic DeCypher Tera-BLASTN. Brassica genes most similar in sequence to a particular Arabidopsis gene were found by aligning Brassica protein sequences (BRAD version 1.2) to Arabidopsis protein sequences from TAIR (release 10; http://www.arabidopsis.org) using TimeLogic DeCypher Tera-BLASTP (e value cutoff = 10e-4). For each Brassica protein sequence, a single match was defined as the Arabidopsis sequence aligning with the highest bit score. Gene Ontology enrichment was performed using the Gene Ontology category analysis tool in Gene Spring GX, using a hypergeometric test, P < 0.05 with a Benjamini-Yekutieli correction for multiple testing (Benjamini and Yekutieli, 2001).
Comparative Analysis of an Arabidopsis Ionomics Data Set
An ionomics database of 152,601 unique samples (“tubes”) representing shoot tissue concentrations of Ca, Mg, and 21 other elements in Arabidopsis (hosted by PiiMS [Baxter et al., 2007]; downloaded October 26, 2011) was used for in silico phenotyping. Of these samples, 7992 had an altered shoot-Ca phenotype as defined by a z score of less than −3.0 (decreased shoot-Ca) or greater than +3.0 (increased shoot-Ca) compared with a control; 3048 and 4944 samples had increased and decreased shoot-Ca, respectively. Among these 7992 samples, 3272 were of mutant genotypes/lines tagged with one or more Arabidopsis gene identifiers; 1154 and 2118 had increased and decreased shoot-Ca, respectively (Supplemental Data Set 6). These 3272 samples represented 682 Arabidopsis genotypes/lines where one or more samples had a leaf-Ca phenotype, of which 562 were single gene mutants and the remainder were multiple mutants (Supplemental Data Set 7). Among these 682 Arabidopsis genotypes/lines, 39 had at least 50% of their samples in the PiiMS database showing altered shoot-Ca, demonstrating a consistent shoot-Ca phenotype; 24 and 15 genotypes/lines had increased and decreased shoot-Ca, respectively (Supplemental Data Set 4). There were 31 unique Arabidopsis gene identifiers associated with these 39 genotypes/lines (Supplemental Data Set 5).
Characterizing BraA.CAX1a Mutants
Mutations in the BraA.CAX1a (Bra017134) gene were identified in a B. rapa TILLING population as described by Ó Lochlainn et al. (2011) using the RevGenUK service (http://revgenuk.jic.ac.uk). A 1.5-kb fragment, including the transcriptional start site and the first exon, was used as the target for TILLING. In total, 20 mutations were identified, including 10 missense, 1 nonsense, 2 silent, and 7 noncoding changes. M3 lines with missense and nonsense mutations were back-crossed to the R-o-18 parent line of the TILLING population. The BC1 plants were grown and individual plants genotyped using high-resolution melt analysis (Ó Lochlainn et al., 2011), modified to include MeltDoctor HRM Master Mix (Applied Biosystems) according to the manufacturer’s instructions. Plants heterozygous for their mutation were selfed, and 20 individual BC1S1 plants were grown and genotyped. Homozygous mutants of three missense alleles designated BraA.cax1a-4 (A-to-T change at amino acid 77), BraA.cax1a-7 (R-to-K change at amino acid 44), and BraA.cax1a-12 (P-to-S change at amino acid 56) and their segregating wild-type lines were identified and selfed to BC1S2 for phenotyping.
Homozygous mutants and the corresponding wild type from segregation for each of BraA.cax1a-4, BraA.cax1a-7, and BraA.cax1a-12, and the original R-o-18 parent line, were phenotyped (i.e., seven lines in total). Plants were grown in a glasshouse at Sutton Bonington, UK, in July and August 2012. Plants were grown in 13-cm-diameter pots containing 1 liter of Levington M2 compost (J. Arthur Bowers). Supplementary lighting was provided as described above, and plants were watered with tap water as required. Total leaf Ca, Mg, and chlorophyll concentrations were determined on a single fully expanded leaf (true leaf number 4 or 5) after 5 weeks from triplicate plants of each line. Total leaf Ca and Mg concentrations were determined by a commercial foliar analysis laboratory (Yara Analytical Services, Lancrop Laboratories, Yara UK) using inductively coupled plasma atomic emission spectrometry. Leaf chlorophyll concentration was measured indirectly on an expanding leaf and a fully expanded leaf using a SPAD meter (SPAD-502; Konica Minolta); three measurements were taken per leaf at 4 and 5 weeks. All data were analyzed using GenStat.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: BraA.CAX1a, Bra017134; and At-CAX1, At2g38170.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Target and Measured [Ca]ext and [Mg]ext in Compost.
Supplemental Figure 2. Leaf Mg Concentrations of Three Genotypes of B. rapa (IMB211, R500, and R-o-18) as Functions of External CaCl2 and MgCl2 Supply.
Supplemental Figure 3. Summary of Heritability (H2) and Treatment Effect Variance Components for 135k Brassica Array Probe Sets.
Supplemental Figure 4. Putative Structure of BraA.CAX1a.
Supplemental Figure 5. Single-Photon Avalanche Diode (SPAD) Measurements of Expanding and Fully Expanded Leaves of Three Independent Homozygous BraA.cax1a Lines and Their Segregating Wild Types.
Supplemental Table 1. Analysis of Nonresponsive eQTL Hot Spots.
Supplemental Data Set 1. All Raw Primary Biomass and Tissue Concentration Data for Plants Grown under 36 [Ca]ext × [Mg]ext Conditions.
Supplemental Data Set 2. Selection of 3272 GEMs Based on Exon-Level Probe Sets.
Supplemental Data Set 3. BraIRRI_05_2012a Map from 134 GEMs.
Supplemental Data Set 4. Arabidopsis Genotypes with Altered Leaf-Ca in ≥50% of Their Ionomics Samples.
Supplemental Data Set 5. B. rapa Probe Sets and eQTLs Linked to 31 Arabidopsis Genes Whose Mutants Have Altered Shoot-Ca Phenotypes.
Supplemental Data Set 6. Ionomics Data for 3272 Samples with Altered Shoot-Ca.
Supplemental Data Set 7. Arabidopsis Genotypes/Lines with Altered Shoot-Ca in the Ionomics Database (n = 682).
Supplementary Material
Acknowledgments
We thank David Salt and colleagues for assisting with access to data in the PiiMS. Funding was provided by the Biotechnology and Biological Sciences Research Council Agri-Food Committee through an Industry Partnering Award (Grant BB-G013969-1). P.J.W. and L.X.D. were supported by the Rural and Environment Science and Analytical Services Division of the Scottish Government through Workpackages 3.3, 3.4, and 7.2 (2011 to 2016). Rothamsted Research receives strategic funding from the Biotechnology and Biological Sciences Research Council.
AUTHOR CONTRIBUTIONS
N.S.G., J.P.H., G.J.K., P.J.W., and M.R.B. conceived and designed the project. J.P.H. and H.C.B. performed growth-response experiments. N.S.G., J.P.H., S.Ó.L., and M.R.B. performed eQTL experiments. S.M. conducted GEM and mapping analyses. N.S.G., S.Ó.L., B.B., and J.J.R. performed TILLING experiments. N.S.G., J.P.H., A.L., C.J.R., S.W., P.W.C.C., L.X.D., and M.R.B. conducted data analyses. All authors contributed to the writing and editing of the article. All authors have read and approved the final article.
Glossary
- [Ca]ext
external Ca supply
- QTL
quantitative trait locus
- eQTL
expression quantitative trait locus
- TILLING
targeting induced local lesions in genomes
- [Mg]ext
external Mg supply
- LL
low/low
- HL
high/low
- LH
low/high
- HH
high/high
- GEM
gene expression marker
- cM
centimorgan
- LOD
log of the odds
- PiiMS
Purdue Ionomics Information Management System
- AGI
Arabidopsis Genome Initiative
- SPAD
single-photon avalanche diode
- RIL
recombinant inbred line
- RMA
robust multiarray average
- MLM
maximum likelihood mapping
- RM
regression mapping
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Ballini E., Lauter N., Wise R. (2013). Prospects for advancing defense to cereal rusts through genetical genomics. Front. Plant Sci. 4: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I., Hosmani P.S., Rus A., Lahner B., Borevitz J.O., Muthukumar B., Mickelbart M.V., Schreiber L., Franke R.B., Salt D.E. (2009). Root suberin forms an extracellular barrier that affects water relations and mineral nutrition in Arabidopsis. PLoS Genet. 5: e1000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter I., Ouzzani M., Orcun S., Kennedy B., Jandhyala S.S., Salt D.E. (2007). Purdue Ionomics Information Management System. An integrated functional genomics platform. Plant Physiol. 143: 600–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Boer M.P., Thissen J.T.M. (2009). Procedure QIBDPROBABILITIES. GenStat Release 12 Reference Manual, Part 3: Procedure Library PL20. (Hemel Hemstead, UK: VSN International; ). [Google Scholar]
- Boer M.P., Wright D., Feng L., Podlich D.W., Luo L., Cooper M., van Eeuwijk F.A. (2007). A mixed-model quantitative trait loci (QTL) analysis for multiple-environment trial data using environmental covariables for QTL-by-environment interactions, with an example in maize. Genetics 177: 1801–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Åstrand M., Speed T.P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19: 185–193. [DOI] [PubMed] [Google Scholar]
- Bonza M.C., De Michelis M.I. (2011). The plant Ca2+-ATPase repertoire: Biochemical features and physiological functions. Plant Biol. (Stuttg.) 13: 421–430. [DOI] [PubMed] [Google Scholar]
- Broadley M.R., White P.J. (2010). Eats roots and leaves. Can edible horticultural crops address dietary calcium, magnesium and potassium deficiencies? Proc. Nutr. Soc. 69: 601–612. [DOI] [PubMed] [Google Scholar]
- Broadley M.R., White P.J. (2012). Some elements are more equal than others: Soil-to-plant transfer of radiocaesium and radiostrontium, revisited. Plant Soil 355: 23–27. [Google Scholar]
- Broadley M.R., Bowen H.C., Cotterill H.L., Hammond J.P., Meacham M.C., Mead A., White P.J. (2003). Variation in the shoot calcium content of angiosperms. J. Exp. Bot. 54: 1431–1446. [DOI] [PubMed] [Google Scholar]
- Broadley M.R., Bowen H.C., Cotterill H.L., Hammond J.P., Meacham M.C., Mead A., White P.J. (2004). Phylogenetic variation in the shoot mineral concentration of angiosperms. J. Exp. Bot. 55: 321–336. [DOI] [PubMed] [Google Scholar]
- Broadley M.R., Hammond J.P., King G.J., Astley D., Bowen H.C., Meacham M.C., Mead A., Pink D.A.C., Teakle G.R., Hayden R.M., Spracklen W.P., White P.J. (2008). Shoot calcium and magnesium concentrations differ between subtaxa, are highly heritable, and associate with potentially pleiotropic loci in Brassica oleracea. Plant Physiol. 146: 1707–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalá R., Santos E., Alonso J.M., Ecker J.R., Martínez-Zapater J.M., Salinas J. (2003). Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15: 2940–2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., et al. (2010). An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE 5: e8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Liu S., Wu J., Fang L., Sun S., Liu B., Li P., Hua W., Wang X. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F., Wu J., Fang L., Wang X. (2012). Syntenic gene analysis between Brassica rapa and other Brassicaceae species. Front. Plant Sci. 3: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.-H., Pittman J.K., Barkla B.J., Shigaki T., Hirschi K.D. (2003). The Arabidopsis cax1 mutant exhibits impaired ion homeostasis, development, and hormonal responses and reveals interplay among vacuolar transporters. Plant Cell 15: 347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng N.-H., Pittman J.K., Shigaki T., Lachmansingh J., LeClere S., Lahner B., Salt D.E., Hirschi K.D. (2005). Functional association of Arabidopsis CAX1 and CAX3 is required for normal growth and ion homeostasis. Plant Physiol. 138: 2048–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S., Gilliham M. (2010). Comparative physiology of elemental distributions in plants. Ann. Bot. (Lond.) 105: 1081–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn S.J., Berninger P., Broadley M.R., Gilliham M. (2012). Exploiting natural variation to uncover candidate genes that control element accumulation in Arabidopsis thaliana. New Phytol. 193: 859–866. [DOI] [PubMed] [Google Scholar]
- Conn S.J., et al. (2011). Cell-specific vacuolar calcium storage mediated by CAX1 regulates apoplastic calcium concentration, gas exchange, and plant productivity in Arabidopsis. Plant Cell 23: 240–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos F.A., Coustham V., Loudet O. (2012). Lessons from eQTL mapping studies: Non-coding regions and their role behind natural phenotypic variation in plants. Curr. Opin. Plant Biol. 15: 192–198. [DOI] [PubMed] [Google Scholar]
- Cullis B.R., Smith A.B., Coombes N.E. (2006). On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 11: 381–393. [Google Scholar]
- Dodd A.N., Kudla J., Sanders D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61: 593–620. [DOI] [PubMed] [Google Scholar]
- Druka A., et al. (2008). Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia graminis f. sp. tritici in barley. Theor. Appl. Genet. 117: 261–272. [DOI] [PubMed] [Google Scholar]
- Druka A., Potokina E., Luo Z., Jiang N., Chen X., Kearsey M., Waugh R. (2010). Expression quantitative trait loci analysis in plants. Plant Biotechnol. J. 8: 10–27. [DOI] [PubMed] [Google Scholar]
- Edgar R., Domrachev M., Lash A.E. (2002). Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Food Standards Agency. (2002). McCance and Widdowson’s The Composition of Foods, 6th ed. (Cambridge, UK: Royal Society of Chemistry). [Google Scholar]
- Geisler M., Axelsen K.B., Harper J.F., Palmgren M.G. (2000). Molecular aspects of higher plant P-type Ca2+-ATPases. Biochim. Biophys. Acta 1465: 52–78. [DOI] [PubMed] [Google Scholar]
- Gustin J.L., Zanis M.J., Salt D.E. (2011). Structure and evolution of the plant cation diffusion facilitator family of ion transporters. BMC Evol. Biol. 11: 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J.P., Bowen H.C., White P.J., Mills V., Pyke K.A., Baker A.J.M., Whiting S.N., May S.T., Broadley M.R. (2006). A comparison of the Thlaspi caerulescens and Thlaspi arvense shoot transcriptomes. New Phytol. 170: 239–260. [DOI] [PubMed] [Google Scholar]
- Hammond J.P., Mayes S., Bowen H.C., Graham N.S., Hayden R.M., Love C.G., Spracklen W.P., Wang J., Welham S.J., White P.J., King G.J., Broadley M.R. (2011). Regulatory hotspots are associated with plant gene expression under varying soil phosphorus supply in Brassica rapa. Plant Physiol. 156: 1230–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschi K.D., Zhen R.-G., Cunningham K.W., Rea P.A., Fink G.R. (1996). CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc. Natl. Acad. Sci. USA 93: 8782–8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway B., Luck S., Beatty M., Rafalski J.A., Li B. (2011). Genome-wide expression quantitative trait loci (eQTL) analysis in maize. BMC Genomics 12: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosmani P.S., Kamiya T., Danku J., Naseer S., Geldner N., Guerinot M.L., Salt D.E. (2013). Dirigent domain-containing protein is part of the machinery required for formation of the lignin-based Casparian strip in the root. Proc. Natl. Acad. Sci. USA 110: 14498–14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.-F., Bertrand Y., Guiraud J.-L., Vialet S., Launay A., Cheynier V., Terrier N., This P. (2013). Expression QTL mapping in grapevine—Revisiting the genetic determinism of grape skin colour. Plant Sci. 207: 18–24. [DOI] [PubMed] [Google Scholar]
- Iniguez-Luy F.L., Lukens L., Farnham M.W., Amasino R.M., Osborn T.C. (2009). Development of public immortal mapping populations, molecular markers and linkage maps for rapid cycling Brassica rapa and B. oleracea. Theor. Appl. Genet. 120: 31–43. [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. (2003a). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Hobbs B., Collin F., Beazer-Barclay Y.D., Antonellis K.J., Scherf U., Speed T.P. (2003b). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264. [DOI] [PubMed] [Google Scholar]
- Karley A.J., White P.J. (2009). Moving cationic minerals to edible tissues: Potassium, magnesium, calcium. Curr. Opin. Plant Biol. 12: 291–298. [DOI] [PubMed] [Google Scholar]
- Katari M.S., Nowicki S.D., Aceituno F.F., Nero D., Kelfer J., Thompson L.P., Cabello J.M., Davidson R.S., Goldberg A.P., Shasha D.E., Coruzzi G.M., Gutiérrez R.A. (2010). VirtualPlant: A software platform to support systems biology research. Plant Physiol. 152: 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, G.J. (2013). Genome analysis. In Biotechnology of Crucifers, S.K. Gupta, ed (New York: Springer), pp. 91–109. [Google Scholar]
- Kinzel, H. (1982). Pflanzenökologie und Mineralstoffwechsel. (Stuttgart, Germany: Ulmer). [Google Scholar]
- Kinzel H., Lechner I. (1992). The specific mineral metabolism of selected plant species and its ecological implications. Bot. Acta 105: 355–361. [Google Scholar]
- Kliebenstein D. (2009). Quantitative genomics: Analyzing intraspecific variation using global gene expression polymorphisms or eQTLs. Annu. Rev. Plant Biol. 60: 93–114. [DOI] [PubMed] [Google Scholar]
- Love C.G., Graham N.S., O Lochlainn S., Bowen H.C., May S.T., White P.J., Broadley M.R., Hammond J.P., King G.J. (2010). A Brassica exon array for whole-transcript gene expression profiling. PLoS ONE 5: e12812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinoia E., Meyer S., De Angeli A., Nagy R. (2012). Vacuolar transporters in their physiological context. Annu. Rev. Plant Biol. 63: 183–213. [DOI] [PubMed] [Google Scholar]
- Mills R.F., Doherty M.L., López-Marqués R.L., Weimar T., Dupree P., Palmgren M.G., Pittman J.K., Williams L.E. (2008). ECA3, a Golgi-localized P2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol. 146: 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J., Hawthorne K.M., Hotze T., Abrams S.A., Hirschi K.D. (2008). Nutritional impact of elevated calcium transport activity in carrots. Proc. Natl. Acad. Sci. USA 105: 1431–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou M.J., Lauter N., Steffenson B., Wise R.P. (2011). Quantitative and qualitative stem rust resistance factors in barley are associated with transcriptional suppression of defense regulons. PLoS Genet. 7: e1002208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ó Lochlainn S.O., Amoah S., Graham N.S., Alamer K., Rios J.J., Kurup S., Stoute A., Hammond J.P., Ostergaard L., King G.J., White P.J., Broadley M.R. (2011). High resolution melt (HRM) analysis is an efficient tool to genotype EMS mutants in complex crop genomes. Plant Methods 7: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Cheng N.H., Pittman J.K., Yoo K.S., Park J., Smith R.H., Hirschi K.D. (2005a). Increased calcium levels and prolonged shelf life in tomatoes expressing Arabidopsis H+/Ca2+ transporters. Plant Physiol. 139: 1194–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Elless M.P., Park J., Jenkins A., Lim W., Chambers E., IV, Hirschi K.D. (2009). Sensory analysis of calcium-biofortified lettuce. Plant Biotechnol. J. 7: 106–117. [DOI] [PubMed] [Google Scholar]
- Park S., Kang T.-S., Kim C.-K., Han J.-S., Kim S., Smith R.H., Pike L.M., Hirschi K.D. (2005b). Genetic manipulation for enhancing calcium content in potato tuber. J. Agric. Food Chem. 53: 5598–5603. [DOI] [PubMed] [Google Scholar]
- Park S., Kim C.-K., Pike L.M., Smith R.H., Hirschi K.D. (2004). Increased calcium in carrots by expression of an Arabidopsis H+/Ca2+ transporter. Mol. Breed. 14: 275–282. [Google Scholar]
- Patterson H.D., Thompson R. (1971). Recovery of inter-block information when block sizes are unequal. Biometrika 58: 545–554. [Google Scholar]
- Pittman J.K. (2011). Vacuolar Ca2+ uptake. Cell Calcium 50: 139–146. [DOI] [PubMed] [Google Scholar]
- Pittman J.K., Hirschi K.D. (2001). Regulation of CAX1, an Arabidopsis Ca2+/H+ antiporter. Identification of an N-terminal autoinhibitory domain. Plant Physiol. 127: 1020–1029. [PMC free article] [PubMed] [Google Scholar]
- Rios J.J., Lochlainn S.O., Devonshire J., Graham N.S., Hammond J.P., King G.J., White P.J., Kurup S., Broadley M.R. (2012). Distribution of calcium (Ca) and magnesium (Mg) in the leaves of Brassica rapa under varying exogenous Ca and Mg supply. Ann. Bot. (Lond.) 109: 1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson D.L. (1987). Estimation and use of variance components. Statistician 36: 3–14. [Google Scholar]
- Shigaki T., Hirschi K.D. (2006). Diverse functions and molecular properties emerging for CAX cation/H+ exchangers in plants. Plant Biol. (Stuttg.) 8: 419–429. [DOI] [PubMed] [Google Scholar]
- Shigaki T., Cheng N.-H., Pittman J.K., Hirschi K. (2001). Structural determinants of Ca2+ transport in the Arabidopsis H+/Ca2+ antiporter CAX1. J. Biol. Chem. 276: 43152–43159. [DOI] [PubMed] [Google Scholar]
- Shigaki T., Sreevidya C., Hirschi K.D. (2002). Analysis of the Ca2+ domain in the Arabidopsis H+/Ca2+ antiporters CAX1 and CAX3. Plant Mol. Biol. 50: 475–483. [DOI] [PubMed] [Google Scholar]
- Stephenson P., Baker D., Girin T., Perez A., Amoah S., King G.J., Østergaard L. (2010). A rich TILLING resource for studying gene function in Brassica rapa. BMC Plant Biol. 10: 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ooijen, J.W. (2006). JoinMap 4, Software for the Calculation of Genetic Linkage Maps in Experimental Populations. (Wageningen, The Netherlands: Kyazma). [Google Scholar]
- Wang T.L., Uauy C., Robson F., Till B. (2012). TILLING in extremis. Plant Biotechnol. J. 10: 761–772. [DOI] [PubMed] [Google Scholar]
- Wang X., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Broadley M.R., Jansen S., White P.J., Takada J., Satake K., Takamatsu T., Tuah S.J., Osaki M. (2007). Evolutionary control of leaf element composition in plants. New Phytol. 174: 516–523. [DOI] [PubMed] [Google Scholar]
- White P.J. (2001). The pathways of calcium movement to the xylem. J. Exp. Bot. 52: 891–899. [DOI] [PubMed] [Google Scholar]
- White, P.J. (2014). Calcium. In A Handbook of Plant Nutrition, A.V. Barker and D.J. Pilbeam, eds (Boca Raton, FL: CRC Press), in press. [Google Scholar]
- White P.J., Broadley M.R. (2003). Calcium in plants. Ann. Bot. (Lond.) 92: 487–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Hotta C.T., Dodd A.N., Love J., Sharrock R., Lee Y.W., Xie Q., Johnson C.H., Webb A.A. (2007). Distinct light and clock modulation of cytosolic free Ca2+ oscillations and rhythmic CHLOROPHYLL A/B BINDING PROTEIN2 promoter activity in Arabidopsis. Plant Cell 19: 3474–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.