The plant circadian clock consists of multiple interlocked transcriptional feedback loops. This work shows that LNK1 and LNK2, two NIGHT LIGHT–INDUCIBLE AND CLOCK-REGULATED genes, encode transcriptional coactivators that physically interact in the nucleus with multiple Myb transcription factors (CCA1, LHY, RVE4, and RVE8) and are necessary for full transcriptional induction of PRR5 and TOC1 by RVE8.
Abstract
Transcriptional feedback loops are central to the architecture of eukaryotic circadian clocks. Models of the Arabidopsis thaliana circadian clock have emphasized transcriptional repressors, but recently, Myb-like REVEILLE (RVE) transcription factors have been established as transcriptional activators of central clock components, including PSEUDO-RESPONSE REGULATOR5 (PRR5) and TIMING OF CAB EXPRESSION1 (TOC1). We show here that NIGHT LIGHT–INDUCIBLE AND CLOCK-REGULATED1 (LNK1) and LNK2, members of a small family of four LNK proteins, dynamically interact with morning-expressed oscillator components, including RVE4 and RVE8. Mutational disruption of LNK1 and LNK2 function prevents transcriptional activation of PRR5 by RVE8. The LNKs lack known DNA binding domains, yet LNK1 acts as a transcriptional activator in yeast and in planta. Chromatin immunoprecipitation shows that LNK1 is recruited to the PRR5 and TOC1 promoters in planta. We conclude that LNK1 is a transcriptional coactivator necessary for expression of the clock genes PRR5 and TOC1 through recruitment to their promoters via interaction with bona fide DNA binding proteins such as RVE4 and RVE8.
INTRODUCTION
Circadian clocks enable organisms to coordinate with diurnal changes in the environment; thus, clock function enhances and clock dysfunction diminishes fitness (Dodd et al., 2005; Graf and Smith, 2011). The plant circadian clock is composed of interlocked transcriptional feedback loops that include both activating and repressive components (Hsu and Harmer, 2014; McClung, 2014). The first loop to be described entails the reciprocal repression of the evening-expressed gene TIMING OF CAB EXPRESSION1 (TOC1) by two morning-expressed genes, CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and LATE ELONGATED HYPOCOTYL (LHY). The transcriptional repression of TOC1 by CCA1 and LHY requires the recruitment of the transcriptional corepressor DEETIOLATED1 (DET1), likely as part of a larger COP10-DET1-DDB1 complex (Lau et al., 2011). TOC1 (also called PRR1) is the founding member of a small PSEUDO-RESPONSE REGULATOR (PRR) family. TOC1 is a transcriptional repressor of multiple genes, including not only CCA1 and LHY but also PRR9, PRR7, PRR5, and the evening-expressed genes LUX ARRHYTHMO (LUX), EARLY FLOWERING4 (ELF4), and GIGANTEA (GI) (Gendron et al., 2012; Huang et al., 2012; Pokhilko et al., 2012). TOC1 represses CCA1 and LHY from its induction at dusk until slightly before dawn; the sequential expression of PRR9, PRR7, and PRR5 broadens the temporal domain of CCA1 and LHY repression to begin shortly after dawn and to continue through the induction of TOC1 at dusk and extend until shortly before dawn (Nakamichi et al., 2010). Thus, CCA1 and LHY transcription is limited to a narrow window around dawn. PRR9, PRR7, and PRR5 bind to the CCA1 and LHY promoters and recruit transcriptional corepressors of the Groucho/Tup1 corepressor family, TOPLESS/TOPLESS-RELATED (TPL/TPR), to repress CCA1 and LHY transcription (Wang et al., 2013). The mechanism by which TOC1 represses CCA1 and LHY transcription is less completely understood; TOC1 may possess intrinsic repressor activity, but CCA1 HIKING EXPEDITION (CHE) interacts with TOC1 and contributes to this repression (Pruneda-Paz et al., 2009).
CCA1 and LHY are members of a small gene family, including REVEILLE (RVE) genes, that encodes Myb domain transcription factors (Carré and Kim, 2002). Although CCA1 and LHY are best defined as transcriptional repressors, genetic experiments show them to positively regulate the expression of PRR9 and PRR7 (Farré et al., 2005), and chromatin immunoprecipitation shows that CCA1 and LHY bind to the PRR9 and PRR7 promoters (Portolés and Más, 2010). Some RVE genes, including RVE4, RVE6, and RVE8 (also called LHY-CCA1-LIKE5), have been placed in positive regulatory roles in clock feedback loops (Hsu and Harmer, 2014). For example, RVE8 promotes the expression of PRR5 and TOC1 as well as other evening genes, including LUX, ELF4, and GI, by directly binding to the evening element (EE) of their promoters (Farinas and Mas, 2011; Rawat et al., 2011; Hsu et al., 2013).
LNK1 and LNK2, two members of a family of NIGHT LIGHT–INDUCIBLE AND CLOCK-REGULATED genes, were recently demonstrated to be important for wild-type period determination (Rugnone et al., 2013). Here, we report that LNK1 and LNK2 physically interact in the nucleus with multiple dawn-phased transcription factors, including CCA1, LHY, RVE4, and RVE8. LNK1 and LNK2 have not been shown to bind directly to DNA, yet they possess transcriptional activator activity. Expression of PRR5 and TOC1 is perturbed in mutants lacking LNK1 and LNK2 function. We show that LNK1 is recruited to fragments of the PRR5 and TOC1 promoters that contain EEs, the binding targets of CCA1, LHY, RVE4, and RVE8. We propose that LNK1 and LNK2 are transcriptional coactivators required for the activation of PRR5 and TOC1 transcription by RVE8 and possibly by RVE4 and other transcription factors.
RESULTS
Mutation of LNK1 and LNK2 Disturbs Multiple Circadian Outputs
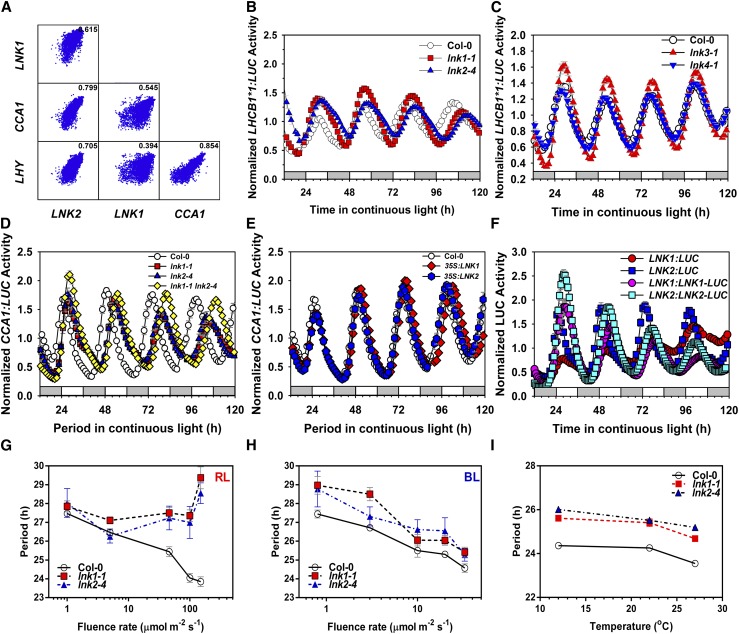
To identify components contributing to the Arabidopsis thaliana circadian transcriptional network, we examined the genome-wide expression profile of NASCArrays (Nottingham Arabidopsis Stock Centre Transcriptomics Service) for genes whose expression was highly correlated with the well-known morning-expressed clock genes CCA1 and LHY (Figure 1A). We identified a small gene family, LNK1 to LNK4, recently shown to be important for circadian clock function (Rugnone et al., 2013). LNK1 to LNK4 expression oscillates in both shoots and roots, and the LNK genes are among a small set of genes whose mRNA abundance continues to oscillate in roots under continuous darkness (James et al., 2008). To confirm that these LNK genes are important for circadian clock function, we identified lines homozygous for T-DNA insertions into each LNK gene (lnk1-1, lnk2-4, lnk3-1, and lnk4-1), introduced a circadian-expressed reporter construct in which firefly LUCIFERASE (LUC) expression is driven by the promoter of the LIGHT-HARVESTING CHLOROPHYLL a/b BINDING PROTEIN gene (LHCB1*1; also called CAB2), and measured circadian function in seedlings entrained in 12-h-light/12-h-dark cycles before release into free-running conditions of continuous white light and constant temperature. Loss of either LNK1 or LNK2 function lengthened the period ∼2 h relative to wild-type Columbia-0 (Col-0) (Figure 1B; Supplemental Table 1), while loss of either LNK3 or LNK4 function conferred no obvious clock defect (Figure 1C; Supplemental Table 1).
Figure 1.
LNK1 and LNK2 Are Required to Maintain Circadian Rhythms and Contribute to Red Light Signaling to the Clock.
(A) Correlation of expression of LNK1, LNK2, CCA1, and LHY from 5211 individual microarray experiments deposited in the NASCArrays database (http://affymetrix.arabidopsis.info). The values in each panel are Pearson correlation coefficients.
(B) to (F) Genetic analysis of the effects of LNK loss of function or overexpression on gene expression as determined with PROMOTER:LUCIFERASE (PRO:LUC) gene fusions: LHCB1*1:LUC ([B] and [C]), CCA1:LUC ([D] and [E]), and LNK:LUC transcriptional and LNK:LNK-LUC translational fusions (F). The data in (B) to (E) are summarized in Supplemental Table 1.
(G) to (I) Effects of continuous red (G) or blue (H) light intensity and of temperature (I) on circadian period measured with CCA1:LUC in lnk1-1, lnk2-4, and the wild type (Col-0). The values are means ± se from two independent biological replicates. These data are summarized in Supplemental Tables 2 and 3.
Mutation of LNK1 or LNK2 also lengthened the period of clock gene expression (CCA1:LUC, LHY:LUC, and TOC1:LUC) in continuous light (Figure 1D; Supplemental Figure 1 and Supplemental Table 1). The long period phenotypes of lnk1 and lnk2 mutants were completely rescued by transgenic complementation with LNK1 and LNK2, respectively, driven by their endogenous promoters (Supplemental Figure 1A and Supplemental Table 1). LNK1 and LNK2 are at least partially redundant, because the lnk1 lnk2 double mutant had a longer period of CCA1:LUC expression than either single mutant (Figure 1D) (Rugnone et al., 2013). These observations were confirmed by quantitative RT-PCR (qRT-PCR) assay for the level of steady state mRNA abundance of CCA1, LHY, and TOC1 and extended to show period lengthening of the expression of PRR5, PRR7, and PRR9 in lnk single and double mutants (Figure 2). However, overexpression of either LNK1 or LNK2 (35S:LNK1 or 35S:LNK2) had no effect on circadian period (Figure 1E; Supplemental Figure 2A and Supplemental Table 1). Thus, although LNK1 and LNK2 are necessary for multiple clock-controlled output rhythms, overexpression of either alone is insufficient to disrupt clock function.
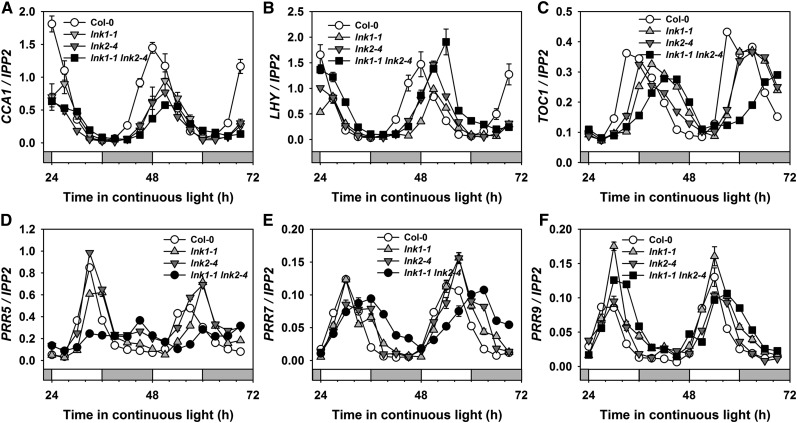
Figure 2.
qRT-PCR Analysis of Circadian Clock–Regulated Gene Expression in lnk1 and lnk2 Single and lnk1 lnk2 Double Mutants.
Expression of circadian clock genes in the wild type (Col-0) and in lnk1-1, lnk2-4, and lnk1-1 lnk2-4 mutants was compared by qRT-PCR. Seedlings were entrained at 22°C in 12-h-light/12-h-dark photocycles for 7 d before release to constant light (70 µmol m−2 s−1) at ZT0. IPP2 was used as a noncycling normalization control to quantify the relative expression of CCA1 (A), LHY (B), TOC1 (C), PRR5 (D), PRR7 (E), and PRR9 (F). The data are presented as means ± se of three technical replicates from one of two independent biological experiments; both experiments yielded congruent results.
LNK1 and LNK2 Are Clock Regulated and Contribute to Red Light Signaling to the Clock
LNK1 and LNK2, like many genes associated with the circadian clock, show robust oscillations in transcription and protein accumulation in both diurnal and free-running conditions. qRT-PCR analysis demonstrated cycling abundance of LNK1 and LNK2 steady state mRNA (Rugnone et al., 2013). LNK transcriptional (LNK1:LUC and LNK2:LUC) and translational (LNK1:LNK-LUC and LNK2:LNK-LUC) fusions show ∼24-h rhythms, with peaks occurring in early morning (1.5 to 2 h after subjective dawn) and with the peaks of fusion protein accumulation lagging slightly behind peaks in transcriptional activity (Figure 1F).
Plant circadian clock function is typically sensitive to light, and period typically shortens with increasing light intensity (Salomé and McClung, 2005a). We found that lnk1 and lnk2 mutants were insensitive to red light in that period failed to shorten with increasing intensity (Figure 1G; Supplemental Table 2). However, lnk1 and lnk2 mutants retained sensitivity to blue light, with period inversely proportional to the light intensity (Figure 1H; Supplemental Table 2). Thus, we conclude that LNK1 and LNK2 are directly or indirectly the targets of red but not blue light signaling. In contrast to its sensitivity to light, circadian period typically is relatively insensitive to temperature and remains relatively constant across a range of physiologically relevant temperatures (temperature compensation) (Salomé and McClung, 2005a). lnk1 and lnk2 mutants retained wild-type temperature compensation between 12 and 27°C (Figure 1I; Supplemental Table 3).
LNK1 and LNK2 Are Nuclear Localized and Physically Interact with CCA1, LHY, RVE4, and RVE8
LNK1 and LNK2 expression is widespread in most tissues and organs at all developmental stages tested, as measured in Arabidopsis transgenic plants expressing β‑glucuronidase (GUS) protein fusions driven by the LNK promoters and by qRT-PCR (Supplemental Figure 3). GUS staining was more intense in the vasculature of roots, hypocotyl, and cotyledons and was particularly intense at root tips and in young true leaves. By qRT-PCR analysis, LNK1 and LNK2 mRNAs were found to be most abundant in leaves and least abundant in roots. LNK is localized to the nucleus, as determined by transient expression in Nicotiana benthamiana leaves or in stable transgenic Arabidopsis lines expressing fusions of LNK1 or LNK2 to green fluorescent protein (GFP; fused in frame to either the C or N terminus of LNK) that were driven by the constitutive 35S promoter (Supplemental Figure 4).
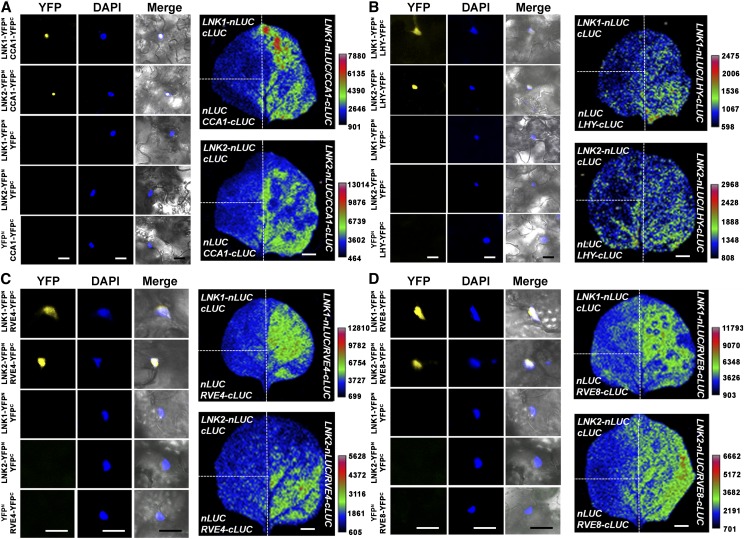
LNK1 and LNK2 show similar circadian, spatial, and developmental expression patterns to CCA1 and LHY, which also localize to the nucleus (Carré and Kim, 2002). Therefore, we asked whether these proteins might be physically associated in plants using bimolecular fluorescence complementation (BiFC) and firefly luciferase complementation imaging (LCI) in Agrobacterium tumefaciens–infiltrated N. benthamiana leaves. For BiFC, 35S:LNK1-YFPn or 35S:LNK2-YFPn was coinfiltrated with 35S:CCA1-YFPc or 35S:LHY-YFPc. For LCI, 35S:LNK1-nLUC or 35S:LNK2-nLUC was coinfiltrated with 35S:CCA1-cLUC or 35S:LHY-cLUC. Both LNK1 and LNK2 interacted with CCA1 and LHY in both assays with complemented yellow fluorescent protein (YFP) in the nucleus or complemented luciferase activity (Figures 3A and 3B; Supplemental Figure 5). We extended this study to show that the CCA1/LHY-related RVE8 and RVE4, which act as positive regulators in the Arabidopsis clock (Farinas and Mas, 2011; Rawat et al., 2011; Hsu et al., 2013), both interact with LNK1 and LNK2, as shown with BiFC and firefly LCI in transiently infected N. benthamiana leaves (Figures 3C and 3D; Supplemental Figure 5) or in stable Arabidopsis transgenic lines (Figures 4A and 4B). We note that these interactions are specific; although LNK1 and LNK2 interact with multiple morning-phased CCA1/LHY/RVE proteins, two other morning-phased clock proteins, PRR9 and PRR7, do not interact with LNK1 and LNK2 in yeast two-hybrid assays (Supplemental Figures 6A to 6C). In addition, we note that in yeast two-hybrid assays, both LNK1 and LNK2 fused to an activation domain (AD) interacted with RVE4 and RVE8 recruited to DNA via the yeast Gal4 DNA binding domain (BD) (Supplemental Figures 6D and 6E). These interactions could not be confirmed in the reciprocal experiment, because both LNK1-BD and LNK2-BD activated the target in the absence of an interaction partner, indicating that LNK1 and LNK2 each possesses intrinsic transcriptional activation activity (Figure 5A; Supplemental Figures 6D and 6E). Thus, we conclude that LNK1 and LNK2 physically interact in the nucleus with at least four known morning clock transcription factors: CCA1, LHY, RVE4, and RVE8.
Figure 3.
LNK1 and LNK2 Interact with Myb Transcription Factors CCA1, LHY, RVE4, and RVE8 in the Nucleus.
The left three panels show BiFC analysis of LNK1 and LNK2 with CCA1 (A), LHY (B), RVE4 (C), and RVE8 (D) in the nucleus (bars = 20 µm). Fusion constructs in which the N-terminal half of YFP (YFPn) was fused to LNK1 or LNK2 and the C-terminal half of YFP (YFPc) was fused to the Myb transcription factor were coinfiltrated into N. benthamiana leaves. Confocal microscopy images were captured from epidermal cell layers of transfected leaves 2 to 3 d after infiltration. Panels on the right show LCI in which both LNK1 and LNK2 interact with CCA1 (A), LHY (B), RVE4 (C), and RVE8 (D) in planta (bars = 1 cm). Overexpression fusion constructs in which the C-terminal half of firefly LUC (cLUC) was fused to LNK1 or LNK2 and the N-terminal half (nLUC) was fused to the Myb transcription factor were coinfiltrated into N. benthamiana leaves. Images were captured via a low-light charge-coupled device camera.
Figure 4.
LNK1 and LNK2 Show Dynamic Interactions with Myb Transcription Factors CCA1, LHY, RVE4, and RVE8 in Seedlings, and the Interactions Depend on Two Conserved Domains.
(A) and (B) Endogenous full-length LNK1/LNK2 and CCA1/LHY/RVE8/RVE4 were fused to the N-terminal half of firefly LUC (nLUC) or the C-terminal half of firefly LUC (cLUC). Constructs were independently transformed into Arabidopsis to yield stable transgenic lines that were crossed, and LUC activity was measured in F1 heterozygotes. Data collected with a TopCount luminometer are presented as absolute LUC activities (means ± se, n = 12 to 24).
(C) Amino acid alignment of LNK1, LNK2, LNK3, and LNK4 reveals two plant-specific conserved domains. Site-specific mutations of the two conserved domains of LNK1 in mutants M1 (RD/GG) and M2 (DR/GG) are indicated in red.
(D) and (E) Luciferase activity (means ± se, n = 24 to 36) of Arabidopsis expressing LNK1:LNK1-cLUC (either the wild type or mutated with M1 or M2) together with RVE4:RVE4-nLUC. Data are presented as absolute expression levels (D) or as normalized data (E), in which the absolute value was divided by the mean value for that trace to allow the visualization of low-amplitude oscillations of m1-LNK1:LNK1-cLUC and m2-LNK1:LNK1-cLUC with RVE4:RVE4-nLUC.
Figure 5.
LNK1 and LNK2 Are Transcriptional Activators and Are Recruited to the TOC1 and PRR5 Promoters.
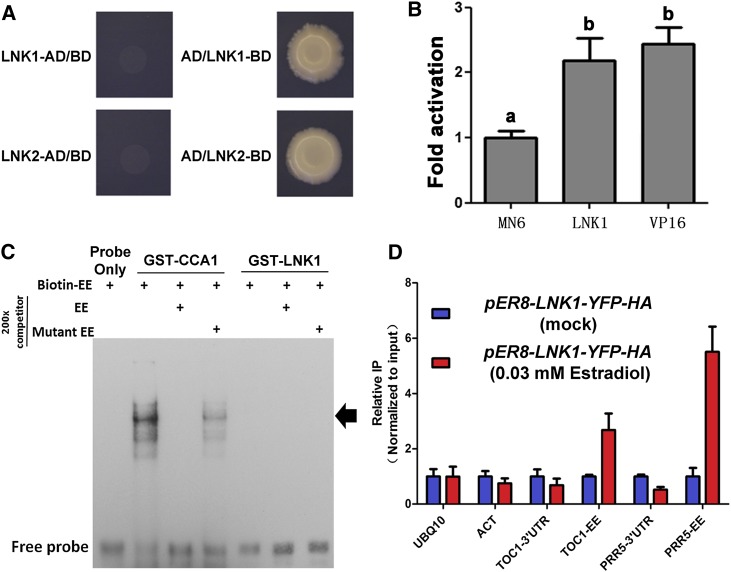
(A) LNK1 and LNK2 have transcriptional activity in yeast, as indicated by prototrophy restoration and cell growth of the yeast AH109 strain on selective medium without Leu, Trp, His, and Ade when fused to the Gal4 DNA binding domain (BD) and recruited to the Gal4 DNA binding site.
(B) LNK1 has transcription activator activity in planta when transiently expressed in plant mesophyll protoplasts. Transcriptional activity was measured with the use of a dual-luciferase assay system (Promega) in which firefly LUC is driven by the minimal cauliflower mosaic virus 35S promoter Gal4 UAS in the pGLL reporter. Transfection efficiency was normalized by coinfection with 35S-driven Renilla luciferase. Expression driven by LNK1-Gal4-BD (LNK1), presented as means ± sd (n ≥ 4), is expressed relative to that of the effector plasmid (pMN6) alone. VP16 has known transcriptional activation activity and serves as a positive control. Different letters indicate significant differences (P < 0.001) as determined by ANOVA.
(C) EMSA of a probe including the EE with CCA1-GST and LNK1-GST purified from E. coli BL21 (DE3). The arrow indicates the CCA1/EE complex.
(D) Chromatin immunoprecipitation of an estradiol-inducible YFP/HA-tagged LNK1 (pER8-LNK1-YFP-HA) with an anti-GFP antibody. Tissue from 2-week-old seedlings treated at ZT0 with DMSO (blue bars) or with 0.03 mM estradiol (red bars) was harvested at ZT28. Immunoprecipitated DNA was quantified by qRT-PCR using TOC1- and PRR5-specific primers flanking the EE. Primers specific to ACTIN7 (ACT), UBIQUITIN10 (UBQ10), PRR5 3′ UTR, and the TOC1 3′ UTR were used as negative controls. The data are presented as means ± sd of three technical replicates.
[See online article for color version of this figure.]
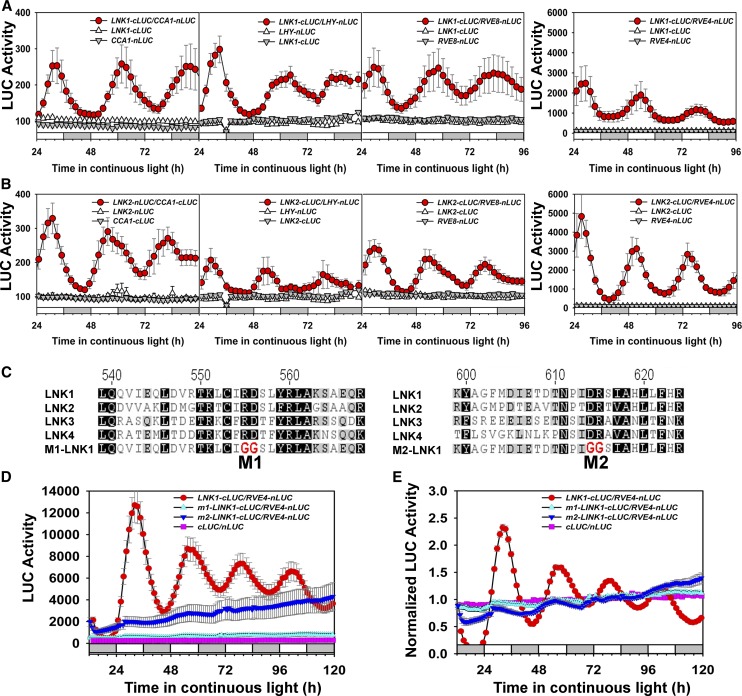
LNK1 and LNK2 Proteins Interact Dynamically with CCA1, LHY, RVE4, and RVE8 Proteins in Planta
We confirmed these interactions in stable Arabidopsis transgenic lines. We transformed endogenous full-length genomic DNA fusion constructs (LNK1:LNK1-cLUC, LNK2:LNK2-cLUC, LNK2:LNK2-nLUC, CCA1:CCA1-nLUC, CCA1:CCA1-cLUC, LHY:LHY-nLUC, RVE8:RVE8-nLUC, or RVE4:RVE4-nLUC) individually into Arabidopsis. Stable transformed lines were crossed to yield F1 seedlings coexpressing pairs of fusions for LUC activity measurement in free-running conditions. All four LNK-cLUC/RVE-nLUC combinations showed robust LUC activity oscillations with a period close to 24 h (Figures 4A and 4B). Both LNK1-cLUC/CCA1-nLUC and LNK2-nLUC/CCA1-cLUC combinations and both LNK-cLUC/LHY-nLUC combinations also showed robust oscillations, albeit with a long period (Figures 4A and 4B). For the LNK–CCA1 interaction, the increased CCA1 expression associated with the introduction of a second copy of CCA1 into the wild-type Col-0 background lengthens the period (Supplemental Figure 7), and we conclude that the expression of a second LHY copy similarly lengthens the period.
We observed much greater luciferase activity in plants expressing LNK1 or LNK2 with RVE4 compared with LNK1 or LNK2 with CCA1, LHY, or RVE8 (Figures 4A and 4B). This could result from a stronger interaction between LNK1 or LNK2 and RVE4. Alternatively, RVE4 might accumulate to higher levels than CCA1, LHY, or RVE8, which would suggest that Myb abundance is limiting to the interaction. We analyzed microarray data obtained under similar growth conditions (http://diurnal.mocklerlab.org/) and found that RVE4 mRNA accumulated to higher levels than CCA1, LHY, and RVE8 mRNAs (Supplemental Figure 8), consistent with this second hypothesis that RVE4 accumulates to higher levels than RVE8, CCA1, and LHY.
Two Plant-Specific Conserved Regions Are Important for Dynamic Protein Interactions
Although LNK family members lack known functional domains, LNK1 to LNK4 have two highly conserved regions that are plant specific (Figure 4C). Mutation of two residues in either conserved region (Arg-555Asp-556 to GlyGly or Asp-614Arg-615 to GlyGly) of LNK1 was used to test if there are specific regions of LNK1 required for interaction with RVE4. The two mutations significantly reduced LUC complementation of LNK1:LNK1-cLUC with RVE4:RVE4-nLUC in transgenic Arabidopsis seedlings (Figures 4D and 4E). Rhythmic LUC activity was detectable with both mutated LNK1s and RVE4, although the m1-LNK1/RVE4 activity was greater than that observed with m2-LNK1/RVE4. m1-LNK1-cLUC protein abundance was not diminished and may actually be increased relative to wild-type LNK1-cLUC (Supplemental Figure 9). Thus, the reduction in LUC complementation activity did not result from a loss of m1-LNK1-cLUC protein stability. We conclude that the mutations greatly reduce the ability of LNK1 to interact with RVE4, either by directly disrupting the RVE binding domain or else by generally perturbing the overall protein structure and thereby indirectly compromising the RVE binding domain. Therefore, we conclude that LNK1 and LNK2 physically interact with multiple clock transcription factors (CCA1, LHY, RVE4, and RVE8) at the core of the Arabidopsis oscillator through a novel conserved domain found in all four LNK proteins.
Activation of PRR5 and TOC1 Transcription by RVE8 Requires LNK1 and LNK2 as Transcriptional Coactivators
If LNK1 or LNK2 physically interacts with multiple clock transcription factors, does either LNK1 or LNK2 possess DNA binding ability? Each of the LNK interaction partners binds to the EE promoter motif. However, we failed to detect LNK1 binding to the EE via electrophoretic mobility shift assay (EMSA) employing Escherichia coli–produced GST-tagged LNK1 (Figure 5C). This, however, would not preclude either LNK1 or LNK2 binding to the EE when complexed to a binding partner or to some other DNA sequence.
As noted above, both LNK1-BD and LNK2-BD activate transcription when recruited to the Gal4 upstream activating sequence (UAS) (Figure 5A; Supplemental Figures 6D and 6E), suggesting that both LNKs possess intrinsic transcriptional activation activity in yeast. Therefore, we investigated whether LNK1 can activate transcription in Arabidopsis protoplasts. LNK1 fused to the Gal4-BD and driven by the 35S promoter activated transcription of a reporter construct consisting of a minimal 35S promoter with the Gal4 UAS driving LUC as effectively as did the strong VP16 activator domain (Sadowski et al., 1988) (Figure 5B). Thus, LNK1 functions as a transcriptional activator in planta.
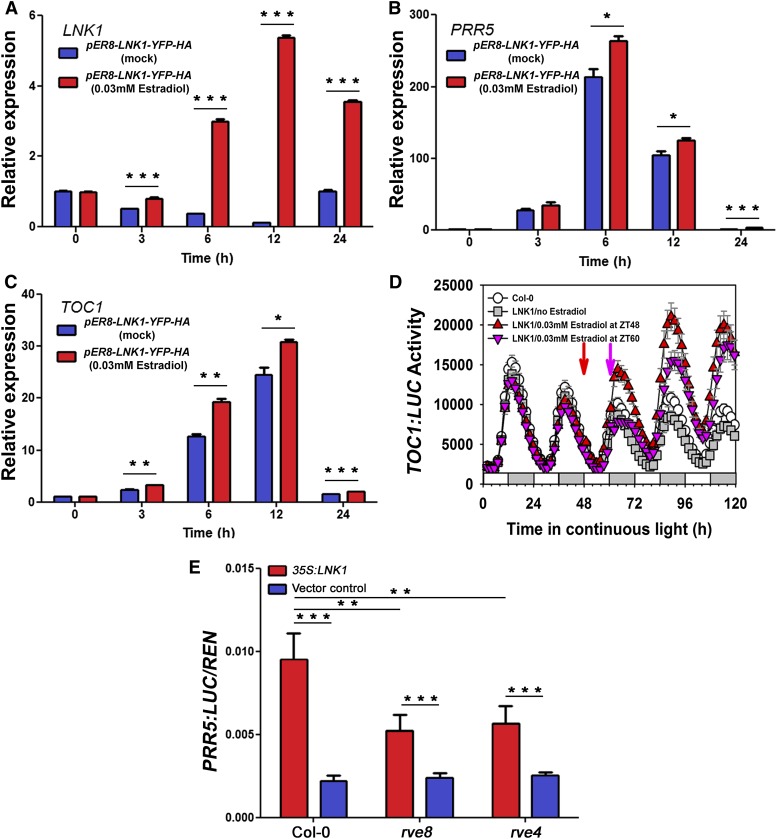
To assess potential regulatory targets of LNK1 and LNK2, we considered the effects of LNK loss of function on the expression of clock genes. mRNA accumulation for CCA1, LHY, PRR9, and PRR7 showed long period in lnk single and double mutants, but the mRNA abundance of LHY, PRR9, and PRR7 was largely unperturbed (Figures 2B, 2E, and 2F). By contrast, PRR5 and, to a lesser extent, CCA1 and TOC1 mRNA accumulation was compromised in lnk single and double mutants (Figures 2A, 2C, and 2D) (Rugnone et al., 2013), suggesting that LNK1 and LNK2 might be coactivators for PRR5, TOC1, and CCA1 transcription. We developed an estradiol-inducible version of LNK1 and showed that induction of LNK1 resulted in increased expression of both PRR5 and TOC1 mRNAs (Figure 6). When estradiol was added at Zeitgeber time 0 (ZT0), LNK1 mRNA was increased relative to mock-treated controls within 3 h, with maximal induction after 12 h (Figure 6A). Both PRR5 and TOC1 mRNAs were increased in induced relative to uninduced seedlings 6 and 12 h after LNK1 induction (Figures 6B and 6C). In both uninduced and induced seedlings, PRR5 and TOC1 mRNA abundances were maximal at ZT6 and ZT12, respectively, consistent with their normal circadian phasing. To further confirm the induction of TOC1 expression by LNK1, we added estradiol at ZT48 (subjective dawn) and observed that induction of LNK1 increased TOC1 expression (measured as luciferase activity from a TOC1:LUC transcriptional fusion) at dusk for each of the next three cycles. When LNK1 was induced by estradiol addition at ZT60 (subjective dusk), TOC1 expression was not increased until 24 and 48 h later (Figure 6D). We attribute this lag to the induction kinetics of LNK1; LNK1 was detectably increased 12 and 24 h but not 6 h after estradiol addition (Supplemental Figure 2B).
Figure 6.
LNK1 Is a Transcriptional Coactivator of PRR5 and TOC1.
(A) to (C) qRT-PCR detection of LNK1 (A), PRR5 (B), and TOC1 (C) expression in lines carrying an estradiol-inducible version of LNK1 (pER8-LNK1-YFP-HA). Seedlings were treated with DMSO (mock; blue bars) or estradiol (red bars) at ZT0, and RNA was isolated from tissue harvested at the indicated times. Horizontal lines indicate values that are significantly different (ANOVA; *P < 0.05, **P < 0.01, ***P < 0.001).
(D) Effect of LNK1 induction on TOC1:LUC expression. Seedlings carrying an estradiol-inducible YFP/HA-tagged LNK1 (pER8-LNK1-YFP-HA) were entrained under a 12-h-light/12-h-dark cycle and subsequently released to constant light conditions. LNK1 was induced by 30 µM estradiol treatment at ZT48 (red arrow) or ZT60 (pink arrow), and luciferase activity was determined via a TopCount luminometer.
(E) Relative expression of PRR5:LUC, normalized to Renilla luciferase (35S:RenLUC), in transiently transfected Arabidopsis protoplasts constitutively overexpressing LNK1-FLAG in Col-0 and in rve8 and rve4 mutant backgrounds. Horizontal lines indicate values that are significantly different (ANOVA; **P < 0.01, ***P < 0.001).
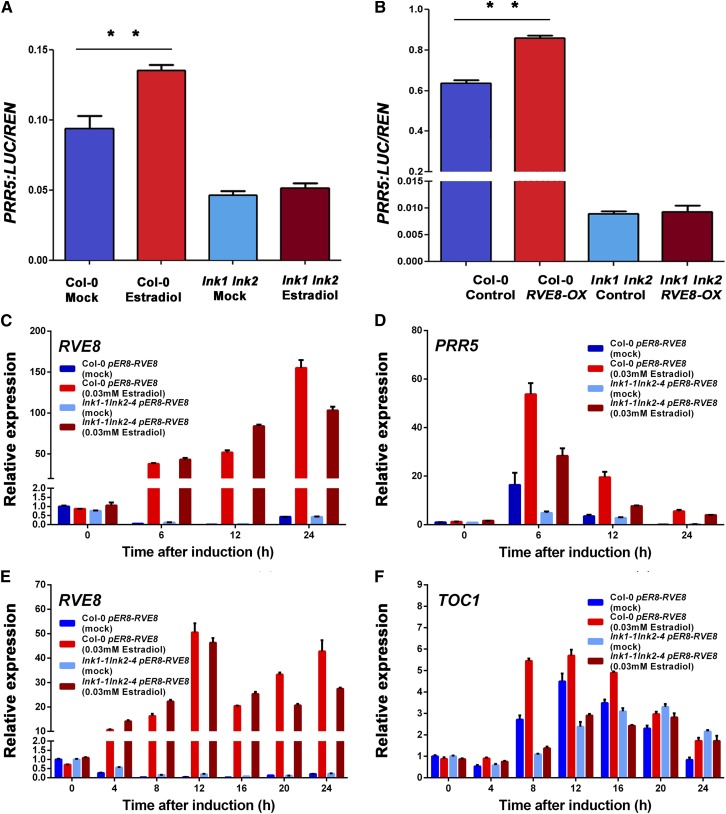
RVE8 has recently been established as an activator of PRR5 and TOC1 transcription (Rawat et al., 2011; Hsu et al., 2013). However, we noticed that RVE4 and RVE8, unlike LNK1 and LNK2, failed to activate transcription in yeast when recruited to the reporter gene promoter through fusion to the Gal4 DNA binding domain (Supplemental Figure 6). Therefore, we asked whether LNK1 and LNK2 were coactivators necessary for the induction of PRR5 and TOC1 transcription by RVE8. First, we developed an estradiol-inducible version of RVE8 and established that induction increased PRR5:LUC expression (Figure 7A). However, this induction was blocked in the lnk1 lnk2 double mutant background (Figure 7A). Second, PRR5:LUC expression is elevated in protoplasts overexpressing RVE8 from the constitutive 35S promoter, but the stimulation of PRR5:LUC by RVE8 overexpression was blocked in the lnk1 lnk2 double mutant (Figure 7B). In Arabidopsis transgenic lines carrying an estradiol-inducible RVE8, we observed estradiol-mediated induction of RVE8, PRR5, and TOC1 mRNAs in the wild-type Col-0 background. However, in the lnk1 lnk2 double mutant, estradiol addition induced RVE8 mRNA, but induction of PRR5 and TOC1 was reduced relative to that observed in Col-0 (Figures 7C to 7F).
Figure 7.
Full Activation of PRR5 and TOC1 Transcription by RVE8 Requires LNK1 and LNK2.
(A) and (B) Relative expression of PRR5:LUC, normalized to Renilla luciferase (35S:RenLUC), in transiently transfected Arabidopsis protoplasts expressing an estradiol-inducible RVE8 (A) or constitutively overexpressing RVE8 (B). Horizontal lines indicate values that are significantly different (ANOVA; **P < 0.01).
(C) and (D) qRT-PCR detection of RVE8 (C) and PRR5 (D) expression in mock-treated (blue and cyan bars) and estradiol-treated (red and purplish red bars) Col-0 and lnk1 lnk2, respectively, transgenic lines carrying an estradiol-inducible version of RVE8.
(E) and (F) qRT-PCR detection of RVE8 (E) and TOC1 (F) expression in mock-treated (blue and cyan bars) and estradiol-treated (red and purplish red bars) Col-0 and lnk1 lnk2, respectively, transgenic lines carrying an estradiol-inducible version of RVE8.
The data are presented as means ± sd of three technical replicates.
The data presented above indicate that LNK1 and LNK2 serve as transcriptional coactivators for PRR5 and TOC1 transcription, which suggests that LNK1 and LNK2 should be recruited to the PRR5 and TOC1 promoters. We used chromatin immunoprecipitation to show that an estradiol-inducible version of LNK1 is recruited to EE-containing fragments of both the PRR5 and TOC1 promoters (Figure 5D). Because LNK1 does not bind to the EE in vitro (Figure 5C), we conclude that it is recruited to the PRR5 and TOC1 promoters via protein–protein interaction with DNA binding proteins such as RVE8 and RVE4. Are RVE4 and RVE8 necessary for the induction of PRR5 transcription by LNK1? When we transiently expressed 35S:LNK1-FLAG in Col-0 or in rve8 or rve4 mutant protoplasts carrying the PRR5:LUC reporter, we found that the activation of PRR5 transcription by LNK1 was significantly reduced in the mutants compared with Col-0 (Figure 6E; Supplemental Figure 2C).
We conclude that LNK1 and LNK2 are coactivators for the induction of PRR5 transcription that are recruited to the PRR5 promoter through their interaction with the DNA binding proteins, RVE8 and RVE4. We note, however, that induction of RVE8 elevated PRR5 and mRNA abundance even in the lnk1 lnk2 background, suggesting either that RVE8 has intrinsic transcriptional activation activity or that there are additional coactivators capable of recruitment to the PRR5 and TOC1 promoters either via interaction with RVE8 or via an indirect means. For example, RVE8 could induce a second transcriptional activator that is recruited to the PRR5 and TOC1 promoters.
DISCUSSION
LNK1 and LNK2 Play Roles in Red Light Input to the Clock
The circadian clock is entrainable by environmental light/dark cycles through signaling pathways associated with multiple photoreceptors. Fluence response curves show that lnk1 and lnk2 are impaired in red light signaling to the circadian clock compared with the wild type under constant red light, as period fails to shorten with increasing fluence rate (Figure 1G; Supplemental Table 2). No such defect was detected in blue light signaling, as the period of lnk1 or lnk2 mutants shortened in response to increased fluence rate similarly to the wild type; although the period of either lnk mutant was longer than that of Col-0, the sensitivity to blue light, as indicated by the slope of the curve, was similar to that of Col-0 (Figure 1H; Supplemental Table 2). In a separate analysis of light sensitivity, lnk1 and lnk2 single mutants had longer hypocotyls than wild-type seedlings under constant white and red light conditions and were late flowering under long-day conditions (16 h of light and 8 h of dark) (Rugnone et al., 2013). LNK1 and LNK2 function in red light signaling is at least partially redundant, as the lnk1 lnk2 double mutant had significantly longer hypocotyls than single lnk1 and lnk2 mutants (Rugnone et al., 2013). Collectively, these data establish that LNK1 and LNK2 play a role in red light input to the clock, in photomorphogenesis, and in developmental (flowering) timing.
LNK1 and LNK2 Serve as Transcriptional Coactivators
The plant circadian clock, like those of fungi and animals, includes multiple interlocked transcriptional feedback loops with both activating and repressive components (Dunlap, 1999; Zhang and Kay, 2010). The Arabidopsis clock is the best studied among plant clocks, and many clock components have been identified (Hsu and Harmer, 2014; McClung, 2014). Although many clock components encode DNA binding transcription factors, the mechanistic details of how these transcription factors modulate transcription are not fully understood. Among examples of transcriptional repression, multiple diverse mechanisms have been described. For example, CCA1 and LHY repress evening-phased TOC1 and GI transcription via recruitment of the COP10-DET1-DDB1 complex, in which DET1 serves as a transcriptional corepressor (Lau et al., 2011). CCA1 and LHY repress ELF4 transcription in the morning by binding to and antagonizing three transcriptional activators, FAR-RED ELONGATED HYPOCOTYL3 (FHY3), FAR-RED IMPAIRED RESPONSE1 (FAR1), and LONG HYPOCOTYL5 (HY5) (Li et al., 2011). Transcription of morning-phased genes, including PRR9 and the PHYTOCHROME INTERACTING FACTORS, are repressed by the evening complex consisting of ELF3, ELF4, and the DNA binding protein LUX (also called PHYTOCLOCK1) (Helfer et al., 2011; Nusinow et al., 2011; Herrero et al., 2012). Transcription of the morning-phased CCA1 and LHY is repressed later in the day by the sequential binding of PRR9, PRR7, and PRR5 to the CCA1 and LHY promoters (Nakamichi et al., 2010); this repression requires a corepressor, encoded by members of the TPL/TPR gene family (Wang et al., 2013).
Examples of transcriptional activation in the plant circadian clock are also accumulating. As indicated above, FHY3, FAR1, and HY5 activate the expression of ELF4. In mutants lacking function of both LIGHT-REGULATED WD1 (LWD1) and LWD2, the period is shortened and the expression of multiple clock genes is greatly reduced (Wu et al., 2008; Wang et al., 2011). LWD1 binds directly to the PRR9, PRR5, and TOC1 promoters, suggesting that it is a transcriptional activator of these genes (Wang et al., 2011). The CCA1/LHY relatives RVE4, RVE6, and RVE8 have been defined as activating transcriptional factors in clock feedback loops (Hsu and Harmer, 2014). Morning-phased RVE8 promotes the expression of evening-phased PRR5 and TOC1 as well as other evening components, including LUX, ELF4, and GI, by directly binding to the EE of their promoters (Farinas and Mas, 2011; Rawat et al., 2011; Hsu et al., 2013). Here, we report that LNK1 and LNK2 physically interact with RVE8 and RVE4 coincident with their morning-phased peak in expression. Transcriptional activation of PRR5 or TOC1 by RVE8 is attenuated in the lnk1 lnk2 double mutant. Although the LNK proteins lack known DNA binding motifs and LNK1 fails to bind to the EE in vitro, LNK1 is recruited to EE-containing elements of the PRR5 and TOC1 promoters, as shown by chromatin immunoprecipitation. Therefore, we propose that LNK1, and by extension LNK2, serves as a transcriptional coactivator in a “morning complex” (or set of complexes) with the DNA binding proteins RVE8 and RVE4.
Our luciferase complementation data (Figures 4A and 4B; Supplemental Figure 5) suggest that the LNKs interact with the RVEs whenever the proteins are coexpressed, which is consistent with their common circadian phasing. However, there is considerable precedent for posttranslational modification of Arabidopsis clock proteins, and modifications such as phosphorylation are known to change the binding affinities of proteins for one another. For example, phosphorylation of PRR5 and TOC1 increases their affinity for the F-box protein ZTL (Fujiwara et al., 2008). It also is possible that other protein interactors may participate in these LNK/RVE complexes and might alter (either enhancing or attenuating) the affinities of LNK/RVE binding or perhaps sequester the LNKs from their RVE binding partners. ELF3 interaction with COP1 enables the interaction of COP1 with GI (Yu et al., 2008). When both TOC1 and PRR3 are highly phosphorylated, they interact and inhibit ZTL-TOC1 binding (Fujiwara et al., 2008). Other modes of regulation by protein interaction could further modulate transcriptional activation by LNK/RVE complexes. For example, ELF4 binds to GI, preventing the binding of GI to its DNA targets by subnuclear sequestration (Kim et al., 2013). PRR5 regulates the phosphorylation and nuclear import of TOC1, which markedly modulates DNA binding and the repressor function of TOC1 (Wang et al., 2010).
We also note that LNK1 and LNK2 each interacts with CCA1 and LHY as well as with RVE4 and RVE8. CCA1 and LHY, although best characterized as transcriptional repressors, have been implicated as activators of the three C-REPEAT BINDING FACTOR (CBF) genes (Seo et al., 2012) as well as of PRR7 and PRR9 (Farré et al., 2005). We speculate that the interaction of the LNKs with CCA1 and LHY may be sufficient to convert them from transcriptional repressors to transcriptional activators, either simply by the recruitment of the LNK activation domain or possibly by concomitant masking of an intrinsic CCA1/LHY repressor domain or displacement of a bound corepressor.
Potential Mechanism of LNK in Circadian Transcriptional Control
The circadian clock of mammals, like that of plants, is an autoregulatory transcriptional network that consists of interlocked feedback loops. The core loop employs a heterodimer of two basic helix-loop-helix-PER-ARNT-SIM (PAS) transcription factors, CLOCK (CLK) or its brain-expressed paralog, NEURONAL PAS DOMAIN PROTEIN2, and BRAIN AND MUSCLE ARNT-LIKE1 (BMAL1), that activate transcription of the Period (Per1 and Per2) and Cryptochrome (Cry1 and Cry2) genes during the day (Mohawk et al., 2012). Associated with the CLK-BMAL1 induction of target genes is rhythmic chromatin modification. CLK has intrinsic histone acetyltransferase (HAT) activity (Doi et al., 2006), and the CLK-BMAL heterodimer also recruits coactivators, including the CREB binding protein and P300, which also possess HAT activity (Koike et al., 2012; Aguilar-Arnal and Sassone-Corsi, 2013). Thus, CLK-BMAL1 binding mediates rhythmic chromatin reconfiguration, permitting the recruitment of additional clock-regulated transcription factors and allowing broad transcriptional programming (Menet et al., 2014).
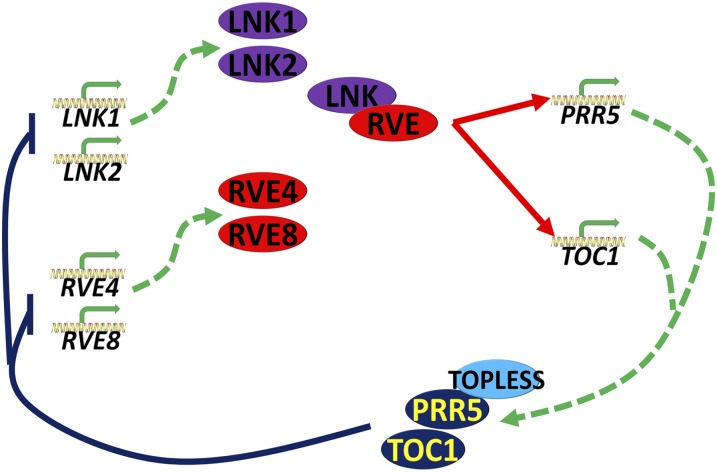
Circadian transcriptional control in plants is known to include chromatin modification; CCA1, LHY, and TOC1 transcription is positively correlated with levels of histone H3 acetylation and H3 Lys-4 trimethylation and inversely correlated with levels of H3 Lys-36 dimethylation (Hemmes et al., 2012; Malapeira et al., 2012; Song and Noh, 2012). RVE8 binding to the TOC1 promoter is associated with increased histone H3 acetylation levels (Farinas and Mas, 2011). H3 Lys-4 trimethylation levels at the promoters of clock genes are increased by the histone methyltransferase SET DOMAIN GROUP2/ARABIDOPSIS TRITHORAX RELATED (SDG2/ATXR3) (Malapeira et al., 2012), but the identity of the responsible HAT(s) is not known (Henriques and Mas, 2013). LNK1 and LNK2 are plant-specific proteins that lack any recognized functional domains, so it is unlikely that they themselves have chromatin-modifying capacity. We postulate that LNK1 and LNK2 serve as coactivators connecting the clock–associated transcription factors CCA1, LHY, RVE4, and RVE8 to other components of the transcription complex, likely including chromatin-modifying complexes. Taken together, our results show that LNK1 serves as a transcriptional coactivator necessary for proper expression of the clock genes PRR5 and TOC1 through recruitment to their promoters via interaction with bona fide DNA binding proteins such as CCA1, LHY, RVE4, and RVE8 (Figure 8).
Figure 8.
A Model Proposing a Role for the LNKs as Transcriptional Coactivators in the Arabidopsis Clock Feedback Loops of RVE4/RVE8 with PRR5 and TOC1.
LNK1 and LNK2 serve as transcriptional coactivators recruited to the promoters of target genes (here, PRR5 and TOC1) via protein–protein interaction with the DNA binding transcription factors RVE8 and RVE4. Expression of LNK1 and LNK2 cycles in phase with RVE4 and RVE8 and peaks in mid morning. The LNK/RVE morning complex activates PRR5 and TOC1 transcription (red arrows). TOC1 and a PRR5/TOP/TPL corepressor complex feed back on the LNK1, LNK2, RVE4, and RVE8 promoters to repress transcription (blue lines ending in perpendicular bars). Dashed green arrows represent transcription and translation.
[See online article for color version of this figure.]
METHODS
Plant Materials and Growth Conditions
Plant materials used in this study were in the Arabidopsis thaliana Col-0 background, except cca1-11 (Wassilewskija) and lhy-21 (Wassilewskija). T-DNA insertion lines lnk1-1 (SALK_024353), lnk2-4 (CS807006), lnk3-1 (SALK_085551C), and lnk4-1 (CS120858) were obtained from the ABRC at Ohio State University. Seeds were sterilized in 20% bleach, placed on half-strength Murashige and Skoog medium (Murashige and Skoog, 1962) with 0.8% agar and 1% Suc, and then stratified for 3 d at 4°C in the dark. Plates were transferred to white light (70 µmol m−2 s−1) in a Percival CU36L5 growth chamber (Percival Scientific). Agrobacterium tumefaciens–mediated transformation of Arabidopsis was by floral dip (Clough and Bent, 1998).
Constructs
LNK1 and LNK2 promoter-driven firefly LUC reporter plasmids were made through insertion of PCR-amplified (using Phusion High-Fidelity DNA Polymerase; New England Biolabs) promoter regions into a modified pENTR vector with the LUC+ gene at multiple cloning sites. The LNK1 and LNK2 promoter regions (2157- and 1809-bp fragments before the start codon) were amplified from Col-0 genomic DNA with primer pairs LNK1-SA5_locus_F1/LNK1-SA5_Promoter_R and LNK2-CS1_locus_F1/LNK2-CS1_Promoter_R (Supplemental Table 4). The promoter:LUC+ fragments were recombined into a modified pH2GW7∆ (Karimi et al., 2002) from which the 35S promoter had been deleted via Gateway LR Clonase II enzyme (Life Technologies) (Supplemental Table 4).
To make LNK1 and LNK2 promoter-driven firefly LUC translational fusions to the LNK coding sequences (LNK1:LNK1-LUC and LNK2:LNK2-LUC), PCR products of full-length LNK1 and LNK2 genomic DNA including promoter and 3′ untranslated region (UTR) were amplified from Col-0 genomic DNA with primer pairs LNK1-SA5_locus_F1/LNK1-SA5_locus_R2 and LNK2-CS1_locus_F1/LNK2-CS1_locus_R1 (Supplemental Table 4) and then cloned into pENTR. The LNK stop codons were then replaced with paired SfiI restriction sites through PCR amplification (primer pairs were LNK1-SA5-SF/LNK1-SA5-SR and LNK2-CS1-SF/LNK2-CS1-SR; Supplemental Table 4), and then a modified LUC+ gene flanked by SfiI sites was inserted to create in-frame LNK-LUC+ translational fusions (primers used to amplify LUC+ were Luc+SF and Luc+SR; Supplemental Table 4). The resultant LNK:LNK-LUC fusions were then recombined into pH2GW7∆. The same strategy was used to make the PRR5pro:LUC+ plasmids and for the LUC complementation experiments. Full-length LNK1/LNK2/CCA1/LHY/RVE4/RVE8 genomic DNAs were cloned into pENTR or pCR8 vectors and then modified to replace the stop codons with two SfiI sites between which either nLUC or cLUC was cloned to create in-frame translational fusions. The sense primers used for nLUC or cLUC amplification were Luc+SF and cLuc+SF, and the antisense primers were nLuc+SR and Luc+SR (Supplemental Table 4). The target genes were recombined into pH2GW7∆ (Deleted 35S) or pMDC123 (Curtis and Grossniklaus, 2003). A related strategy was used to make the PRR5:LUC+ transcriptional fusion, except that the entire PRR5 coding sequence was replaced with paired SfiI sites (the PRR5 promoter region used in this study was 3871 bp, and primers used to amplify the promoter were PRR5_locus_F and PRR5_NSF-NEW; Supplemental Table 4).
For gene expression pattern (promoter:GUS) assays, PCR-amplified LNK promoters were cloned into pMDC163 (GUS) (Curtis and Grossniklaus, 2003). The sense primers used in the amplification were LNK1-SA5_G_F and LNK2-CS1_G_F, and the antisense primers were LNK1-SA5_Prom_R and LNK2-CS1_Prom_R (Supplemental Table 4). For subcellular localization (C-terminal and N-terminal GFP fusions), PCR-amplified LNK promoter/coding sequences were cloned into pMDC43 (N-terminal GFP) (Curtis and Grossniklaus, 2003) or into p35S:CDS-GFP (C-terminal GFP with a pCAMBIA1300 [GenBank AF234296.1] vector backbone). The sense primers used in the amplification were LNK1-SA5_G_F and LNK2-CS1_G_F, and the antisense primers were LNK1-SA5_G_R_ns and LNK2-CS1_G_R_ns (Supplemental Table 4). Full-length coding sequences of LNK1, LNK2, CCA1, LHY, RVE4, and RVE8 were cloned into pSPYNE-35S and pSPYCE-35S (Walter et al., 2004) for BiFC and into pCAMBIA-NLuc and pCAMBIA-CLuc (Chen et al., 2008) for LUC complementation.
To make RVE8 overexpression and inducible constructs, RVE8 cDNAs were amplified by PCR and inserted into pENTR/SD/D-TOPO (Invitrogen) and then transferred into pER8-GW (Papdi et al., 2008) or pMDC32 (Curtis and Grossniklaus, 2003).
LNK-GST and CCA1-GST fusion constructs were made by inserting PCR-amplified products into the EcoRI and XhoI sites of pGEX 4T-1 (GE Healthcare). PCR-amplified LNK1 and VP16 were inserted into pMN6 (Huq et al., 2004) at the SmaI and KpnI sites to test activator activity. pGLL was used as the reporter LUC and pRNL as the internal reference Renilla LUC reporter (Promega).
Gene Expression Assays
LNK1:GUS and LNK2:GUS transgenic (T3) lines were used to determine the expression pattern of LNK1/LNK2 via histochemical GUS reporter activity (Jefferson et al., 1987). For RT-PCR analysis, total RNA was isolated from Arabidopsis samples using RNAiso Plus (TaKaRa) and treated with DNase I. After quantification of RNA, 3 µg of total RNA was used for cDNA synthesis using an oligo(dT) and RevertAid First Strand cDNA Synthesis Kit (Fermantas Thermo Fisher). TaKaRa SYBR Premix Ex Taq and a 7500 Fast Real-Time PCR instrument (Applied Biosystems) were used for qRT-PCR as described (Wang et al., 2012). LNK1-FLAG and LNK1-YFP-HA protein accumulation was assessed by immunoblot analysis. Protein extracts prepared from whole seedlings were separated on 8% SDS-polyacrylamide gels and transferred to a polyvinylidene fluoride membrane (Immobilon-P; Millipore) by electroblotting. Immunoblot analysis was performed with monoclonal GFP or FLAG antibody (Sigma-Aldrich). Horseradish peroxidase–conjugated anti-mouse secondary antibody (Bio-Rad) was used to detect the primary antibody.
Protein–Protein Interaction
BiFC assays (Walter et al., 2004) were performed by coinfiltration of Agrobacterium carrying the N-terminal half of YFP (YFPn) fused to LNK1/LNK2 and the C-terminal half of YFP (YFPc) fused to CCA1/LHY/RVE4/RVE8 vectors into Nicotiana benthamiana leaves. Confocal microscopy images were captured from epidermal cell layers of transfected leaves 2 to 3 d after infiltration. YFP and 4′,6-diamidino-2-phenylindole (DAPI) fluorescence was observed by confocal microscopy: YFP/DAPI, excitation at 515/405 nm, emission at 525 to 560 nm/420 to 470 nm. For LNK-GFP fusion fluorescence: GFP/DAPI, excitation at 405 nm, emission at 505 to 530 nm/420 to 470 nm.
Firefly LCI and quantification were via transient expression of cLUC and nLUC fusions in N. benthamiana leaves via Agrobacterium-mediated coinfiltration as described (Chen et al., 2008). 35S overexpression fusion constructs in which the N-terminal half of firefly LUC (nLUC) was fused to LNK1/LNK2 and the C-terminal half (cLUC) was fused to the Myb transcription factor were coinfiltrated into N. benthamiana plants. Images were captured via a low-light cooled CCD camera (Andor Technology) 3 d after infiltration. A Packard TopCount luminometer (PerkinElmer) was used to quantify the LUC signal. Full-length gene fusion constructs in which cLUC was fused to LNK1/LNK2/CCA1 and nLUC was fused to (left to right) LNK2, CCA1, LHY, RVE8, or RVE4 were independently transformed into Arabidopsis to yield stable transgenic lines that were crossed for the measurement of LUC activity in F1 hybrids entrained to 12-h-light/12-h-dark cycles and transferred to constant light and temperature at ZT0. Data collected with a TopCount luminometer are presented as absolute LUC activity or normalized LUC activity, in which case the individual values were normalized to the average value for that trace. This facilitates the comparison of cycling between traces with different signal strengths.
Yeast two-hybrid analysis used a Gal4-based yeast hybrid system (Matchmaker two-hybrid system; Clontech). Full-length cDNAs of LNK1/LNK2 and RVE4/RVE8 were cloned into bait vector and prey vector (pGADT7 and pGBKT7). β-Galactosidase assays used the manufacturer’s protocols.
Dual-Luciferase Transient Expression
Firefly LUC is driven by the minimal cauliflower mosaic virus 35S promoter Gal4 UAS in the pGLL reporter. Transfection efficiency was normalized by coinfection with 35S-driven Renilla luciferase. Expression driven by LNK1-Gal4-BD (LNK1) is relative to that of the effector plasmid (pMN6) alone. VP16 (Sadowski et al., 1988) served as a positive control. The Dual-Luciferase Reporter Assay System (Promega) was used to determine the relative expression of PRR5:LUC. Arabidopsis protoplasts were isolated from leaves of 4-week-old, soil-grown plants (entrained to 8-h-light/16-h-dark cycles at 22°C). Preparation of protoplasts (derived from the Col-0 wild type or the lnk1 lnk2 double mutant) and subsequent polyethylene glycol–mediated transformation with reporter (PRR5:LUC), effector (RVE8, estradiol-inducible RVE8 [pER8-RVE8-YFP-HA], or constitutively overexpressing RVE8 [35S:RVE8]), and internal control (35S:RLUC) plasmids were as described (Yoo et al., 2007). LUC activity was assayed using a Packard TopCount luminometer.
Firefly LUC Measurement and Data Analysis
Seedlings expressing the firefly luciferase reporter gene (LUC) under the control of promoters from CCA1, LHY, TOC1 (Salomé and McClung, 2005b), and LHCB1*1 (CAB2) (−199/+1) (Anderson et al., 1994) were entrained for 7 to 10 d in 12-h-light/12-h-dark cycles (22°C) before release into continuous light (22°C) conditions for LUC measurement with a TopCount luminometer. For temperature compensation assays, seedlings were entrained in 12-h-light/12-h-dark cycles at 22°C for 7 to 10 d before transfer to continuous light at 12, 22, and 27°C for LUC activity measurement. Circadian rhythms were assayed with BRASS 2.1.4, which employs fast Fourier transform nonlinear least squares (Plautz et al., 1997; Southern and Millar, 2005). The strength of a circadian rhythm is expressed as relative amplitude error (RAE). An ideal cosine wave is defined as RAE = 0, and RAE = 1 defines the statistically detectable limit of rhythmicity.
For fluence response curves, CCA1:LUC transgenic seedlings were entrained to 12-h-light/12-h-dark cycles for 7 d before transfer to constant red or blue light at the indicated fluence rates. On the first day in continuous light, seedlings were transferred to 96-well microplates (PerkinElmer) for LUC activity measurement; microplates were transferred manually to the Packard TopCount at 3-h intervals. The response of period to the fluence rate of constant red or blue light was analyzed by linear regression followed by analysis of covariance using GraphPad Prism software (http://www.graphpad.com/). For temperature compensation, Q10 was calculated as Q10 = [RateT2/RateT1]10/(T2-T1) = [(1/period@T2)/(1/period@T1)]10/(T2-T1).
EMSA
EMSAs were performed using biotin-labeled double-stranded probes and the Lightshift Chemiluminescent EMSA Kit (Pierce). Probes used in this study, including TOC1-EE-F, TOC1-EE-R, TOC1-mutEE-F, and TOC1-mutEE-R (Supplemental Table 4), were described previously (Harmer et al., 2000; Harmer and Kay, 2005; Pruneda-Paz et al., 2009).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation assays were performed using 3-week-old seedlings stably transformed with pER8-LNK1-YFP-HA that were grown on Murashige and Skoog agar plates under 12-h-light/12-h-dark cycles at 22°C. According to the LCI results in stable Arabidopsis transgenic lines, the strongest interaction between LNK1 and RVE4/RVE8 is at ∼ZT4, so leaf tissue samples were collected 28 h after induction with 30 µM estradiol (Mizoi et al., 2006; Li et al., 2013) at ZT0. An EZ-ChIP Chromatin Immunoprecipitation Kit (Millipore) and an anti-GFP antibody (Roche) were used for chromatin immunoprecipitation assays. Primer pairs that amplified the indicated regions of PRR5-EE, TOC1-EE, TOC1-3′UTR, ACTIN7, and UBIQUITIN10 (UBQ10) (Supplemental Table 4) were used to assess immunoprecipitation enrichment by PCR. All primer pairs were described previously (Pruneda-Paz et al., 2009; Rawat et al., 2011).
Accession Numbers
Sequence data for the genes described in this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ACT7 (At5g09810), CBF1 (At4g25490), CBF2 (At4g25470), CBF3 (At4g25480), CCA1 (At2g46830), CHE (At5g08330), COP1 (At2g32950), DET1 (At4g10180), ELF3 (At2g25930), ELF4 (At2g40080), FAR1 (At4g15090), FHY3 (At3g22170), GI (At1g22770), HY5 (At5g11260), IPP2 (At3g02780), LHCB1*1 (At1g29920), LHY (At1g01060), LNK1 (At5g64170), LNK2 (At3g54500), LNK3 (At3g12320), LNK4 (At5g06980), LUX (At3g46640), LWD1 (At1g12910), LWD2 (At3g26640), TPL/TPR (At1g15750, At1g80490, At3g16830, At5g27030, and At3g15880), PRR3 (At5g60100), PRR5 (At5g24470), PRR7 (At5g02810), PRR9 (At2g46790), RVE4 (At5g02840), RVE8 (At3g09600), SDG2/ATXR3 (At4g15180), TOC1 (At5g61380), UBQ10 (At4g05320), and ZTL (At5g57360).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. LNK1 and LNK2 Are Required for Circadian Period Determination.
Supplemental Figure 2. Accumulation of LNK1 mRNA and Protein in Lines Constitutively or Inducibly Overexpressing LNK1.
Supplemental Figure 3. LNK1 and LNK2 Expression in Arabidopsis Seedlings.
Supplemental Figure 4. Subcellular Localization of LNK-GFP Fusion Proteins.
Supplemental Figure 5. Quantification of Luciferase Complementation Imaging Assays.
Supplemental Figure 6. Yeast Two-Hybrid Analysis of LNK1 and LNK2 Interaction with RVE4, RVE8, PRR7, and PRR9.
Supplemental Figure 7. Effect of Slight CCA1 Overexpression on Circadian Period Length.
Supplemental Figure 8. Expression (mRNA Abundance) Analysis of LNK1, LNK2, CCA1, LHY, RVE4, and RVE8 in Circadian Free Run.
Supplemental Figure 9. Protein Stability of LNK1 and Mutated m1-LNK1.
Supplemental Table 1. Summary of Effects of Perturbed LNK Expression on Circadian Gene Expression.
Supplemental Table 2. Effect of Loss of LNK Function on Light Sensitivity of the Circadian Clock (Figures 1G and 1H).
Supplemental Table 3. Effect of Loss of LNK Function on Temperature Sensitivity of the Clock (Figure 1I).
Supplemental Table 4. Oligonucleotides (Shown 5′ → 3′) Used in This Study.
Supplementary Material
Acknowledgments
We thank Jun-Xian He for the pMN6 transient assay system and Bingchun Zhao and Rui Li for technical help with Arabidopsis mesophyll protoplast preparation and transient expression. This work was supported by grants from the Special Program for Key Basic Research of the Ministry of Science and Technology of China (Grant 2012CB126303), the National Science Foundation of China (Grant 31071247), the Program for New Century Excellent Talents in University of the Ministry of Education of China (Grant NCET-13-0771), and the Hebei Science Fund for Distinguished Young Scholars (Grants C2011205034, CPRC036, and 20100502) to X.X. and from the U.S. National Science Foundation (Grants IOS-0923752 and IOS-1025965) to C.R.M.
AUTHOR CONTRIBUTIONS
Q.X. and X.X. conceived the project and designed the experiments. Q.X. and P.W. generated the constructs. Q.X., X.X., X.L., L.W., P.W., L.Y., Y.L., and Z.Y. screened the mutants, generated the transgenic and hybrid lines, and characterized the circadian phenotype in various conditions. P.W., Q.X., H.X., and C.Z. performed the LCI analysis. P.W. and C.Z. performed the BiFC, chromatin immunoprecipitation, EMSA, and dual-luciferase transient expression in protoplasts. X.L. analyzed the clock gene expression with qRT-PCR. P.W. and X.L. generated yeast two-hybrid plasmids, and X.L. performed the β-galactosidase analysis. X.X., L.Y., Y.L., and L.Z. did the fluence response curves assay. Q.X., C.R.M., and X.X. wrote the article.
Glossary
- EE
evening element
- Col-0
Columbia-0
- qRT-PCR
quantitative RT-PCR
- BiFC
bimolecular fluorescence complementation
- LCI
luciferase complementation imaging
- EMSA
electrophoretic mobility shift assay
- UAS
upstream activating sequence
- UTR
untranslated region
- DAPI
4′,6-diamidino-2-phenylindole
- RAE
relative amplitude error
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Aguilar-Arnal L., Sassone-Corsi P. (2013). The circadian epigenome: How metabolism talks to chromatin remodeling. Curr. Opin. Cell Biol. 25: 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S.L., Teakle G.R., Martino-Catt S.J., Kay S.A. (1994). Circadian clock- and phytochrome-regulated transcription is conferred by a 78 bp cis-acting domain of the Arabidopsis CAB2 promoter. Plant J. 6: 457–470. [DOI] [PubMed] [Google Scholar]
- Carré I.A., Kim J.-Y. (2002). MYB transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 53: 1551–1557. [DOI] [PubMed] [Google Scholar]
- Chen H., Zou Y., Shang Y., Lin H., Wang Y., Cai R., Tang X., Zhou J.M. (2008). Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 146: 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd A.N., Salathia N., Hall A., Kévei E., Tóth R., Nagy F., Hibberd J.M., Millar A.J., Webb A.A.R. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Doi M., Hirayama J., Sassone-Corsi P. (2006). Circadian regulator CLOCK is a histone acetyltransferase. Cell 125: 497–508. [DOI] [PubMed] [Google Scholar]
- Dunlap J.C. (1999). Molecular bases for circadian clocks. Cell 96: 271–290. [DOI] [PubMed] [Google Scholar]
- Farinas B., Mas P. (2011). Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 66: 318–329. [DOI] [PubMed] [Google Scholar]
- Farré E.M., Harmer S.L., Harmon F.G., Yanovsky M.J., Kay S.A. (2005). Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 15: 47–54. [DOI] [PubMed] [Google Scholar]
- Fujiwara S., Wang L., Han L., Suh S.S., Salomé P.A., McClung C.R., Somers D.E. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J. Biol. Chem. 283: 23073–23083. [DOI] [PubMed] [Google Scholar]
- Gendron J.M., Pruneda-Paz J.L., Doherty C.J., Gross A.M., Kang S.E., Kay S.A. (2012). Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl. Acad. Sci. USA 109: 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf A., Smith A.M. (2011). Starch and the clock: the dark side of plant productivity. Trends Plant Sci. 16: 169–175. [DOI] [PubMed] [Google Scholar]
- Harmer S.L., Kay S.A. (2005). Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell 17: 1926–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer S.L., Hogenesch J.B., Straume M., Chang H.S., Han B., Zhu T., Wang X., Kreps J.A., Kay S.A. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Helfer A., Nusinow D.A., Chow B.Y., Gehrke A.R., Bulyk M.L., Kay S.A. (2011). LUX ARRHYTHMO encodes a nighttime repressor of circadian gene expression in the Arabidopsis core clock. Curr. Biol. 21: 126–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmes H., Henriques R., Jang I.-C., Kim S., Chua N.-H. (2012). Circadian clock regulates dynamic chromatin modifications associated with Arabidopsis CCA1/LHY and TOC1 transcriptional rhythms. Plant Cell Physiol. 53: 2016–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R., Mas P. (2013). Chromatin remodeling and alternative splicing: Pre- and post-transcriptional regulation of the Arabidopsis circadian clock. Semin. Cell Dev. Biol. 24: 399–406. [DOI] [PubMed] [Google Scholar]
- Herrero E., et al. (2012). EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.Y., Harmer S.L. (2014). Wheels within wheels: the plant circadian system. Trends Plant Sci. 19: 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P.Y., Devisetty U.K., Harmer S.L. (2013). Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2: e00473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Pérez-García P., Pokhilko A., Millar A.J., Antoshechkin I., Riechmann J.L., Mas P. (2012). Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336: 75–79. [DOI] [PubMed] [Google Scholar]
- Huq E., Al-Sady B., Hudson M., Kim C., Apel K., Quail P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305: 1937–1941. [DOI] [PubMed] [Google Scholar]
- James A.B., Monreal J.A., Nimmo G.A., Kelly C.L., Herzyk P., Jenkins G.I., Nimmo H.G. (2008). The circadian clock in Arabidopsis roots is a simplified slave version of the clock in shoots. Science 322: 1832–1835. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kim Y., Lim J., Yeom M., Kim H., Kim J., Wang L., Kim W.Y., Somers D.E., Nam H.G. (2013). ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Reports 3: 671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N., Yoo S.H., Huang H.C., Kumar V., Lee C., Kim T.K., Takahashi J.S. (2012). Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau O.S., Huang X., Charron J.-B., Lee J.-H., Li G., Deng X.W. (2011). Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol. Cell 43: 703–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., et al. (2011). Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat. Cell Biol. 13: 616–622. [DOI] [PubMed] [Google Scholar]
- Li Z., Peng J., Wen X., Guo H. (2013). Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell 25: 3311–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malapeira J., Khaitova L.C., Mas P. (2012). Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proc. Natl. Acad. Sci. USA 109: 21540–21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung C.R. (2014). Wheels within wheels: New transcriptional feedback loops in the Arabidopsis circadian clock. F1000Prime Rep. 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J.S., Pescatore S., Rosbash M. (2014). CLOCK:BMAL1 is a pioneer-like transcription factor. Genes Dev. 28: 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J., Nakamura M., Nishida I. (2006). Defects in CTP:PHOSPHORYLETHANOLAMINE CYTIDYLYLTRANSFERASE affect embryonic and postembryonic development in Arabidopsis. Plant Cell 18: 3370–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohawk J.A., Green C.B., Takahashi J.S. (2012). Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 35: 445–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T.R., Skoog F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15: 473–497. [Google Scholar]
- Nakamichi N., Kiba T., Henriques R., Mizuno T., Chua N.-H., Sakakibara H. (2010). PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22: 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow D.A., Helfer A., Hamilton E.E., King J.J., Imaizumi T., Schultz T.F., Farré E.M., Kay S.A. (2011). The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C., Abrahám E., Joseph M.P., Popescu C., Koncz C., Szabados L. (2008). Functional identification of Arabidopsis stress regulatory genes using the controlled cDNA overexpression system. Plant Physiol. 147: 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plautz J.D., Straume M., Stanewsky R., Jamison C.F., Brandes C., Dowse H.B., Hall J.C., Kay S.A. (1997). Quantitative analysis of Drosophila period gene transcription in living animals. J. Biol. Rhythms 12: 204–217. [DOI] [PubMed] [Google Scholar]
- Pokhilko A., Fernández A.P., Edwards K.D., Southern M.M., Halliday K.J., Millar A.J. (2012). The clock gene circuit in Arabidopsis includes a repressilator with additional feedback loops. Mol. Syst. Biol. 8: 574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S., Más P. (2010). The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet. 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz J.L., Breton G., Para A., Kay S.A. (2009). A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 323: 1481–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat R., Takahashi N., Hsu P.Y., Jones M.A., Schwartz J., Salemi M.R., Phinney B.S., Harmer S.L. (2011). REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 7: e1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugnone M.L., Faigón Soverna A., Sanchez S.E., Schlaen R.G., Hernando C.E., Seymour D.K., Mancini E., Chernomoretz A., Weigel D., Más P., Yanovsky M.J. (2013). LNK genes integrate light and clock signaling networks at the core of the Arabidopsis oscillator. Proc. Natl. Acad. Sci. USA 110: 12120–12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. (1988). GAL4-VP16 is an unusually potent transcriptional activator. Nature 335: 563–564. [DOI] [PubMed] [Google Scholar]
- Salomé P.A., McClung C.R. (2005a). What makes the Arabidopsis clock tick on time? A review on entrainment. Plant Cell Environ. 28: 21–38. [Google Scholar]
- Salomé P.A., McClung C.R. (2005b). PSEUDO-RESPONSE REGULATOR 7 and 9 are partially redundant genes essential for the temperature responsiveness of the Arabidopsis circadian clock. Plant Cell 17: 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P.J., Park M.-J., Lim M.-H., Kim S.-G., Lee M., Baldwin I.T., Park C.-M. (2012). A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 24: 2427–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.-R., Noh Y.-S. (2012). Rhythmic oscillation of histone acetylation and methylation at the Arabidopsis central clock loci. Mol. Cells 34: 279–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern M.M., Millar A.J. (2005). Circadian genetics in the model higher plant, Arabidopsis thaliana. Methods Enzymol. 393: 23–35. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438. [DOI] [PubMed] [Google Scholar]
- Wang L., Fujiwara S., Somers D.E. (2010). PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J. 29: 1903–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Kim J., Somers D.E. (2013). Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc. Natl. Acad. Sci. USA 110: 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. (2012). SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24: 3278–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Wu J.-F., Nakamichi N., Sakakibara H., Nam H.-G., Wu S.-H. (2011). LIGHT-REGULATED WD1 and PSEUDO-RESPONSE REGULATOR9 form a positive feedback regulatory loop in the Arabidopsis circadian clock. Plant Cell 23: 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.-F., Wang Y., Wu S.-H. (2008). Two new clock proteins, LWD1 and LWD2, regulate Arabidopsis photoperiodic flowering. Plant Physiol. 148: 948–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu J.-W., et al. (2008). COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol. Cell 32: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang E.E., Kay S.A. (2010). Clocks not winding down: Unravelling circadian networks. Nat. Rev. Mol. Cell Biol. 11: 764–776. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.