Al toxicity is a major constraint to crop production in acidic soil worldwide. This study elucidates how Al may regulate root growth inhibition through mediating local auxin biosynthesis and signaling in the root-apex transition zone and provides insights into how environmental cues affect root growth plasticity through influencing local auxin biosynthesis and signaling.
Abstract
The transition zone (TZ) of the root apex is the perception site of Al toxicity. Here, we show that exposure of Arabidopsis thaliana roots to Al induces a localized enhancement of auxin signaling in the root-apex TZ that is dependent on TAA1, which encodes a Trp aminotransferase and regulates auxin biosynthesis. TAA1 is specifically upregulated in the root-apex TZ in response to Al treatment, thus mediating local auxin biosynthesis and inhibition of root growth. The TAA1-regulated local auxin biosynthesis in the root-apex TZ in response to Al stress is dependent on ethylene, as revealed by manipulating ethylene homeostasis via the precursor of ethylene biosynthesis 1-aminocyclopropane-1-carboxylic acid, the inhibitor of ethylene biosynthesis aminoethoxyvinylglycine, or mutant analysis. In response to Al stress, ethylene signaling locally upregulates TAA1 expression and thus auxin responses in the TZ and results in auxin-regulated root growth inhibition through a number of auxin response factors (ARFs). In particular, ARF10 and ARF16 are important in the regulation of cell wall modification–related genes. Our study suggests a mechanism underlying how environmental cues affect root growth plasticity through influencing local auxin biosynthesis and signaling.
INTRODUCTION
Al is the most abundant metal in the earth’s crust. In acidic soils (pH < 5), the phytotoxic Al3+ ion becomes increasingly soluble and becomes a significant constraint on crop productivity. Acidic soils are widespread, especially in the tropics and subtropics (Von Uexküll and Mutert, 1995; Kochian et al., 2004). The inhibition of root elongation has been widely used as a bioassay for Al toxicity (Delhaize and Ryan, 1995). The root apex is the major target site of Al toxicity (Ryan and Kochian, 1993); in maize (Zea mays), the distal part of the root-apex transition zone (TZ), located between the apical meristem and the basal elongation region, is the most Al-sensitive part of the root (Sivaguru and Horst, 1998), and a similar zone is involved in both common bean (Phaseolus vulgaris) (Rangel et al., 2007) and Arabidopsis thaliana (Illés et al., 2006). The importance of the distal part of the root TZ in the response to Al toxicity has been confirmed in sorghum (Sorghum bicolor) by the demonstration that it is the site of reactive oxygen species production (Sivaguru et al., 2013).
Tissue- or cell-specific hormone signaling is an important component of the regulation of root growth (Gifford et al., 2008; Ding and De Smet, 2013; Duan et al., 2013; Rosquete et al., 2013). The root-apex TZ is a critical site for the perception and response to both endogenous phytohormones and environmental cues (Baluška et al., 2010), particularly to auxin, the key regulator of root development (Benková and Hejátko, 2009; Overvoorde et al., 2010; Bielach et al., 2012; Jansen et al., 2012; Lavenus et al., 2013). In conjunction with auxin, ethylene is also involved in the regulation of root growth (Růzicka et al., 2007; Swarup et al., 2007). Ethylene can regulate auxin biosynthesis and basipetal auxin transport toward the elongation zone, which activates auxin signaling in the root apex, and thus causes root growth inhibition (Růzicka et al., 2007; Stepanova et al., 2007, 2011; Swarup et al., 2007). It was recently reported that ethylene signaling regulates the expression of Trp aminotransferase/Trp aminotransferase–related proteins (TAA1/TARs), which catalyze the key step of auxin biosynthesis through the conversion of Trp to indole-3-pyruvic acid, thus positively directing auxin biosynthesis (Stepanova et al., 2008; He et al., 2011).
Al-regulated inhibition of root growth is clearly regulated by auxin (Kollmeier et al., 2000; Sun et al., 2010). High-throughput sequencing analysis of the Al-regulated microRNAs in wild soybean (Glycine soja) (Zeng et al., 2012) and Medicago truncatula (Chen et al., 2012) revealed the potential regulation of microRNA160 (miR160) in the Al-induced inhibition of root growth through mediating the expression of auxin response factors (ARFs) such as ARF10 and ARF16 (Wang et al., 2005; Liu et al., 2007; Liu et al., 2010). The targeted application of Al to the distal part of the root-apex TZ has been shown to be as inhibitory as when it is applied over the entire maize root apex (Sivaguru et al., 1999; Kollmeier et al., 2000), while the localized application of Al to the root-apex elongation zone (EZ) has little effect on root elongation. The implication is that signaling between the root-apex TZ and EZ determines the extent of Al-induced inhibition of root growth. The exogenous supply of the auxin indole-3-acetic acid (IAA) to the EZ significantly reduces the extent of Al-induced root elongation inhibition, but the same treatment given to the meristematic zone has no effect (Kollmeier et al., 2000). The presence of Al promotes the accumulation of auxin in the root cap, the root apical meristem, and the distal part of the TZ (Kollmeier et al., 2000; Sun et al., 2010) as a result of the disruption caused to auxin transport from the distal part of the root-apex TZ to the EZ (Sivaguru et al., 1999; Kollmeier et al., 2000; Doncheva et al., 2005). Since an elevated level of auxin in the EZ is required for cell elongation (Teale et al., 2005, 2006), it is probable that the regulation of Al-induced root growth involves the inhibition of basipetal auxin transport, mediated by auxin signaling between the TZ and EZ (Kollmeier et al., 2000).
As yet, how auxin levels in the TZ and EZ mediate the Al-regulated inhibition of root growth remains largely unknown. Here, we exploited a green fluorescent protein (GFP)–tagged auxin-responsive DR5rev:GFP transgene, the expression of which provides a robust spatial representation of auxin distribution (Ding and Friml, 2010; Tian et al., 2013, 2014a), to monitor auxin activity in the Arabidopsis root-apex TZ as induced by externally applied Al. This tool has allowed us to show that the activity of TAA1 was essential for auxin accumulation in the root-apex TZ in response to Al stress. In addition, our investigations indicate that the local upregulation of TAA1 and the resulting local auxin accumulation in the root-apex TZ constitute an ethylene signaling–dependent process. Moreover, our data also show that auxin plays a negative role in Al stress–regulated root growth via regulating the expression of cell wall modification–related genes in Arabidopsis.
RESULTS
The Auxin Signaling Maximum in the Root-Apex TZ Is Crucial for Root Growth Inhibition in Response to Al Stress
The root-apex TZ, as expected from the findings of Sivaguru and Horst (1998) in maize, proved to be the perception site of Al toxicity in Arabidopsis as well, as demonstrated by the staining pattern of propidium iodide (PI), which accumulates only in dying and dead cells, and fluorescein diacetate (FDA) staining, which is taken up and converted into the green fluorescent compound “fluorescin” only by living cells (Supplemental Figure 1; Moore et al., 1998; Steward et al., 1999).
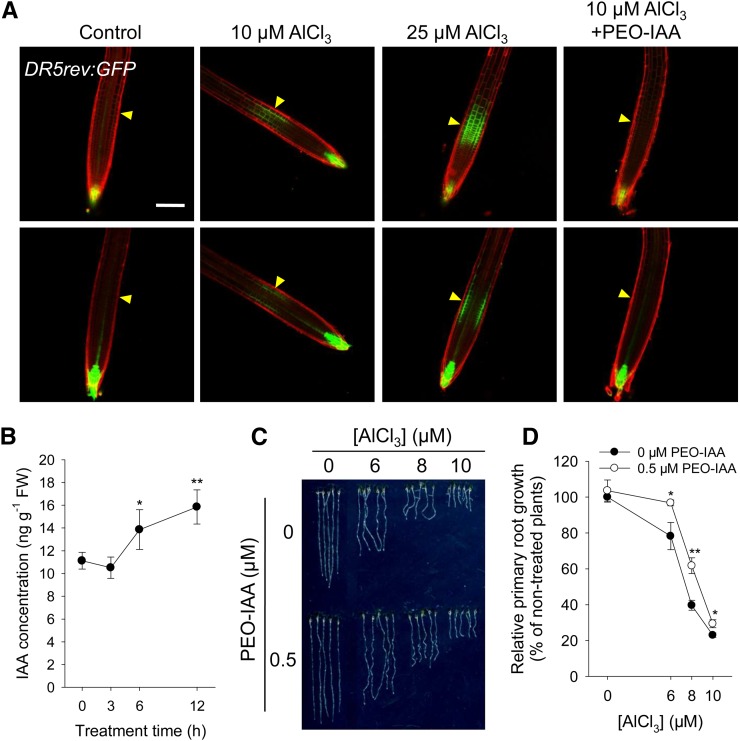
The distribution of auxin in the root tip of plants exposed to Al was revealed by employing the auxin-responsive reporter construct DR5rev:GFP. After a 2-h exposure to 10 µM Al, an increased DR5 activity was detected specifically in the epidermis and cortex region of the root TZ, and increasing the concentration of Al to 25 µM produced even stronger DR5rev:GFP signals (Figure 1A). Further time-course analysis of DR5rev:GFP signals in the presence of 6 μM Al revealed that the DR5rev:GFP signals in the TZ increased until 12 h and, thereafter, remarkably decreased and even disappeared after 7 d of exposure to toxic Al (Supplemental Figure 2), suggesting that auxin may act as an early Al-responsive signal to regulate the Al-induced root growth inhibition. To test whether the Al-induced auxin reporter DR5rev:GFP signaling maximum in the TZ reflects the endogenous increased auxin level, we measured the concentration of IAA in 1-mm-long root tips (the region including the meristematic, transition, and fast elongation zones according to Verbelen et al., 2006). In agreement with the increased DR5rev:GFP expression in the TZ, 6 μM Al significantly increased the endogenous auxin levels in root tips (1 mm from the apex) after 6 and 12 h of Al exposure (Figure 1B). The undetected increment of free IAA levels after 3 h of Al treatment might be due to the small induction of local auxin production only in the TZ, which is masked when analyzing the whole root-tip tissues.
Figure 1.
The Al-Induced Auxin Response in the TZ Is Crucial for Root Growth Inhibition in Arabidopsis.
(A) Expression of the DR5rev:GFP transgene in roots exposed for 2 h to 0, 10, or 25 μM AlCl3 or to 10 μM AlCl3 plus 15 μM PEO-IAA. Control indicates nontreated roots. The top row shows DR5rev:GFP signals in the epidermis, and the bottom row shows DR5rev:GFP signals in the cortex. Cell boundaries appear red following PI staining. The TZ is marked by yellow arrowheads. Bar = 100 μm.
(B) IAA concentration in root tips (1 mm long) exposed to 6, 8, and 10 μM AlCl3 for 0, 3, 6, and 12 h. Asterisks indicate significant differences at *P < 0.05 and **P < 0.01 (t test). Values represent means ± sd (n = 3). FW, fresh weight.
(C) and (D) Primary root growth after exposure to 0, 6, 8, and 10 μM AlCl3 and either 0 or 0.5 μM PEO-IAA for 7 d. Asterisks indicate that differences between the PEO-IAA treatments are significant at *P < 0.05 and **P < 0.01 (t test). Values in (D) represent means ± sd (n = 30).
The correlation between auxin response in the root-apex TZ and inhibition of root growth was investigated by exposing seedlings to both Al and the auxin antagonist α-(phenylethyl-2-one)-indole-3-acetic acid (PEO-IAA), a molecule that blocks the auxin binding sites of TIR1/AFB auxin receptors (Hayashi et al., 2008; Nishimura et al., 2009; Hayashi, 2012). The Al treatment–induced DR5rev:GFP signals in the root-apex TZ were markedly reduced by PEO-IAA (Figure 1A), and the effect of increasing the Al supply on root growth was reduced (Figures 1C and 1D; Supplemental Figure 3). These results clearly suggest that the Al-induced auxin response in the root-apex TZ is involved in the Al-induced inhibition of root growth.
Auxin Enhances the Al-Induced Inhibition of Root Growth
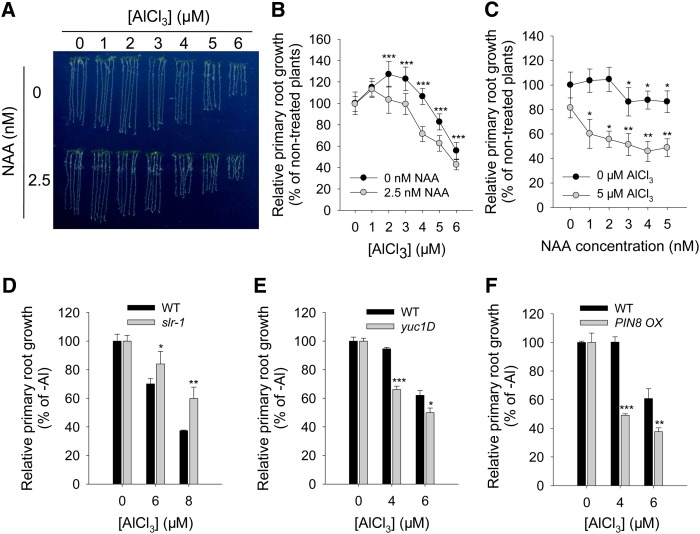
To confirm the role of auxin in Al-induced root growth inhibition, further experiments were performed via manipulating auxin levels or auxin signaling through exogenous 1-naphthaleneacetic acid (NAA) application or mutant analysis. A low concentration of Al (≤3 µM) in the growing medium tended to promote rather than to inhibit root elongation. The presence of 2.5 nM NAA had no effect on root growth without Al supply, although the expression of DR5rev:GFP increased in the whole root apex particularly in the apical meristem zone after 6 h of NAA exposure (Supplemental Figure 4). However, when the roots were exposed to levels of Al >1 μM, NAA substantially enhanced the extent of Al-induced growth inhibition (Figures 2A and 2B). At 5 μM Al, the effect of NAA on the level of inhibition was even more pronounced (Figure 2C). However, the NAA-enhanced Al-induced root growth inhibition did not result from the specific increase in auxin signaling in the TZ. Instead, the whole root apex showed higher DR5rev:GFP signals, particularly clearly after 12 h of treatment (Supplemental Figure 4). Several auxin-responsive genes, including IAA7, IAA10, IAA14, and IAA18 (in the AUXIN/IAA gene family), GH3.2, GH3.3, GH3.5, GH3.12, and AT5G13370 (in the GRETCHEN HAGEN3 gene family), and SAUR37 and SAUR59 (in the SMALL AUXIN-UP RNA gene family), were selected from the results generated by RNA sequencing (RNAseq) (Supplemental Data Set 1) to check their response to the NAA and/or Al treatments by quantitative real-time PCR (qRT-PCR). The results showed that, independently, NAA or Al treatment significantly induced the expression of most of the selected genes at 6 and 12 h, while combined NAA and Al treatments led to higher gene expression than either NAA or Al treatment alone mainly for the GH3 gene family, including GH3.2, GH3.3, GH3.5, GH3.12, and AT5G13370 (Supplemental Figure 5). This suggests an additive effect of NAA and Al on auxin signaling and the potential regulation of the GH3 gene family in Al-induced root growth inhibition at an early stage.
Figure 2.
Auxin Mediates the Al-Induced Inhibition of Root Growth.
(A) to (C) Effect of exogenous NAA on root growth following a 7-d exposure to 0 or 2.5 nM NAA in the presence of 0 to 6 μM AlCl3 ([A] and [B]) or 0 to 5 nM NAA in the presence of 0 or 5 μM AlCl3 (C). Asterisks in (B) and (C) indicate that NAA treatments differ significantly at *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
(D) to (F) Root growth after a 7-d exposure to AlCl3. Wild-type plants and gain-of-function mutants slr-1, PIN8 OX, and yuc1D were exposed to 0, 4, 6, or 8 μM AlCl3. Asterisks indicate that means within the wild type and slr-1, PIN8 OX, or yuc1D in each Al concentration differ significantly at *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
Values in (B) to (F) represent means ± sd (n = 30).
[See online article for color version of this figure.]
The gain-of-function solitary root1 (slr-1) mutant, in which a stabilized form of the AUX/IAA14 protein is present, leading to a reduced level of auxin signaling (Fukaki et al., 2002), showed a more strongly reduced root growth inhibition in the presence of Al than the wild type (Figure 2D). Both the dominant Arabidopsis yucca (yuc1D) mutant, which is characterized by a high endogenous IAA level as a result of the overexpression of a rate-limiting auxin synthesis gene (Zhao et al., 2001), and the pin-formed8 overexpressor (PIN8 OX) mutant, which also accumulates higher free IAA in its roots (Ding et al., 2012), exhibited stronger inhibition of root growth in the presence of Al than the wild-type plants (Figures 2E and 2F). These results imply that the elevated levels of free auxin enhance the Al-induced inhibition of root growth.
Both Auxin Signaling Maximum in the TZ and Al-Induced Inhibition of Root Growth Are Regulated by TAA1
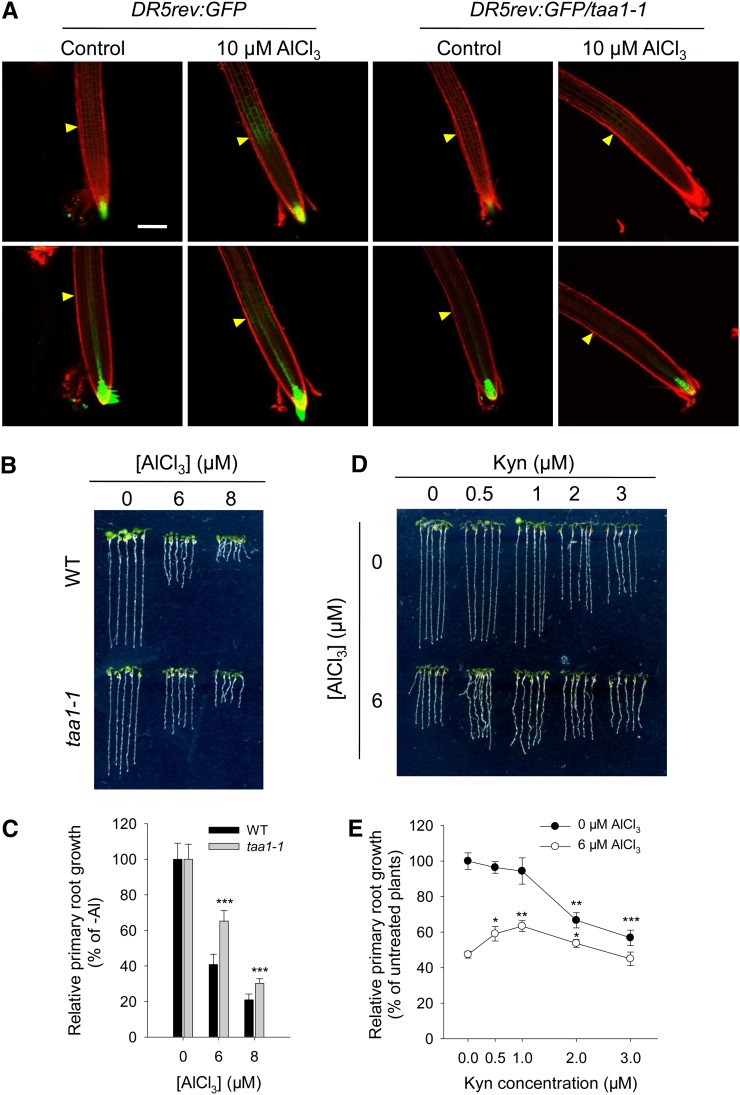
To test if auxin biosynthesis is involved in the Al-induced local auxin signaling maximum and root growth inhibition, we first examined the phenotypes of the TAA1 mutant taa1-1, which is defective in the indole-3-pyruvic acid branch of the auxin synthetic pathway and exhibits a reduced auxin level in its roots (Stepanova et al., 2008; Tao et al., 2008). In taa1-1 root tips, the Al treatment–induced boosting of DR5rev:GFP signals in the TZ shown by the wild type did not occur (Figure 3A). Consistently, the root growth inhibition in the presence of Al stress was also significantly alleviated in taa1-1 compared with the wild-type controls (Figures 3B and 3C). The small molecule l-kynurenine (Kyn) can effectively and selectively bind to the substrate pocket of TAA1/TAR proteins, and thus competitively inhibit TAA1/TAR activity, mimicking the loss of TAA1/TAR functions (He et al., 2011). Treatment of roots with Kyn only reduced root growth at >1 μM Kyn in the medium in the absence of Al (Figures 3D and 3E). However, when Al was included in the medium, 0.5 or 1 μM Kyn largely alleviated the Al-induced inhibition of root growth (Figures 3D and 3E), suggesting that the TAA1-regulated local auxin biosynthesis mediates the auxin signaling maximum in the root-apex TZ and thus root growth inhibition in response to Al stress.
Figure 3.
Both Auxin Signaling Maximum in the TZ and Al-Induced Inhibition of Root Growth Are Regulated by TAA1.
(A) Expression of the DR5rev:GFP transgene in the root tip of the wild type and the loss-of-function mutant taa1-1. Six-day-old DR5rev:GFP and DR5rev:GFP/taa1-1 seedlings were exposed for 2 h to either 0 (control) or 10 μM AlCl3. The top row shows DR5rev:GFP signals in the epidermis, and the bottom row shows DR5rev:GFP signals in the cortex. Cell boundaries appear red following PI staining. The TZ is marked by yellow arrowheads. Bar = 100 μm.
(B) and (C) Root growth of wild-type and taa1-1 plants after a 7-d exposure to 0, 6, or 8 μM AlCl3. Asterisks in (C) indicate that means within the wild type and the taa1-1 mutant in each Al concentration differ significantly at ***P < 0.001 (t test).
(D) and (E) Effect on the Al-induced inhibition of root growth of adding the TAA1 inhibitor Kyn. Root growth was measured after a 7-d exposure to 0 or 6 μM AlCl3 in the presence of 0 to 3 μM Kyn. Asterisks in (E) indicate that Kyn treatments differ significantly at *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
Values in (C) and (E) represent means ± sd (n = 30).
TAA1 Is Locally Induced in the TZ by the Presence of Al
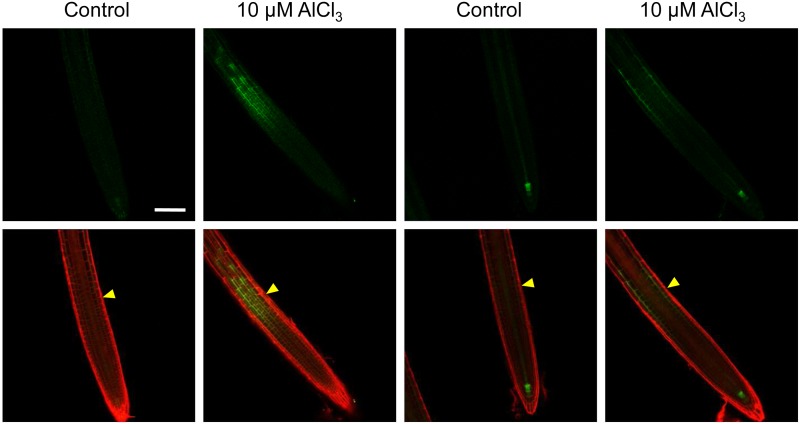
To explore how TAA1 regulates local auxin signaling in the TZ of root tips in the presence of Al, the effect of exogenous Al on the spatial expression of TAA1 was examined using the TAA1:GFP-TAA1 translational fusion transgene. The transgene is expressed mainly in the root tip, specifically in the zone surrounding the root quiescent center (Stepanova et al., 2008). A clear GFP signal was observed in the TZ after a 2-h exposure to Al (Figure 4). The dynamic expression of Al-induced TAA1:GFP at different time points of Al exposure (0 to 48 h and 7 d) was consistent with the changes in DR5rev:GFP signals, showing the highest induction of TAA1 after 12 h of treatment (compare Supplemental Figures 2 and 6). Similar to DR5rev:GFP, the local induction of TAA1 in the TZ was also absent on day 7 of Al exposure. This result confirmed the early regulatory role of TAA1 for the local auxin signaling maximum in the root-apex TZ in response to Al stress. However, no expressional change of TAA1 and its homolog TARs at the transcript level was found in Al-exposed whole roots through RNAseq analysis (Supplemental Data Set 1); the potential increase in gene expression in the TZ might be masked when analyzing the whole roots.
Figure 4.
Al Induces TAA1:GFP Expression in the TZ.
Six-day-old transgenic TAA1:GFP seedlings were exposed to either 0 (control) or 10 μM AlCl3 for 2 h. The two left columns show TAA1:GFP signals in the epidermis, and the two right columns show TAA1:GFP signals in the cortex. The top row shows the expression of TAA1:GFP, while the bottom row shows an overlay of the TAA1:GFP expression and PI staining (the latter used to highlight cell boundaries). The TZ is marked by yellow arrowheads. Bar = 100 μm.
Decreasing the pH of the medium from 5.5 to 4.2 reduced the growth of the root severely (Supplemental Figure 7) but did not affect the expression of either the DR5rev:GFP or the TAA1:GFP transgene (Supplemental Figure 8), implying that exogenous Al, not protons, upregulates TAA1, driving up the accumulation of auxin in the TZ and, finally, inhibiting root growth.
Al-Induced Local TAA1 Expression in the TZ Is Regulated by Ethylene
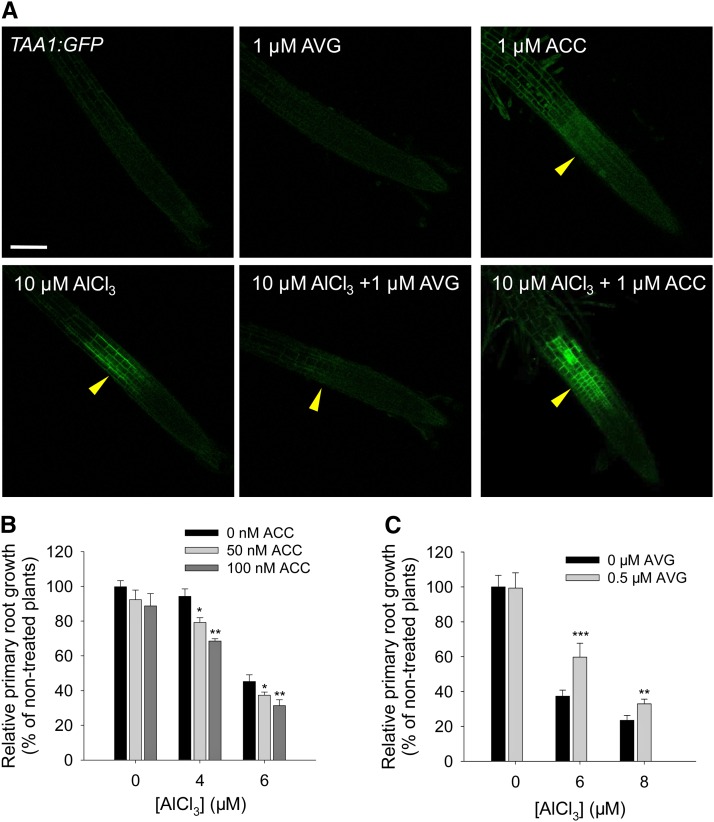
Recently, Stepanova et al. (2008) and He et al. (2011) found that TAA1-mediated auxin biosynthesis in the root apex was regulated by ethylene, and mutations in TAA1 resulted in root-specific ethylene insensitivity (Stepanova et al., 2008). To study if the Al-induced local TAA1 expression in the TZ is dependent on ethylene signaling, we examined local TAA1:GFP induction in response to Al when cotreated with the precursor of ethylene biosynthesis 1-aminocyclopropane-1-carboxylic acid (ACC) or the inhibitor of ethylene biosynthesis aminoethoxyvinylglycine (AVG). The Al-induced local TAA1:GFP signals in the TZ were strongly intensified by ACC cotreatment, while they were highly repressed by AVG (Figure 5A). Consistent with these results, application of exogenous ACC or AVG significantly enhanced or alleviated the Al-induced inhibition of primary root growth, respectively (Figures 5B and 5C).
Figure 5.
Al-Induced TAA1 Expression in the TZ Is Regulated by Ethylene.
(A) Expression of the TAA1:GFP transgene in the epidermis of the root apex in the presence of Al plus ACC or AVG treatment. Four-day-old transgenic TAA1:GFP seedlings were pretreated without or with 1 μM AVG or 1 μM ACC for 2 d, and then the seedlings were continuously treated without or with 1 μM AVG or 1 μM ACC in the absence or presence of 10 μM AlCl3 for 2 h. The TZ is marked by yellow arrowheads. Bar = 100 μm.
(B) and (C) Relative primary root growth of the wild type in the presence of 0, 4, and 6 μM AlCl3 plus 0, 50, or 100 nM ACC (B) or 0, 6, and 8 μM AlCl3 plus 0 or 0.5 μM AVG (C) for 7 d. Asterisks in (B) indicate that means within the ACC treatment differ significantly at *P < 0.05 and **P < 0.01 (t test). Asterisks in (C) indicate that means within the AVG treatment differ significantly at **P < 0.01 and ***P < 0.001 (t test). Values represent means ± sd (n = 30).
TAA1-Regulated Local Auxin Response in the TZ and Inhibition of Root Growth in Response to Al Depends on Ethylene Signaling
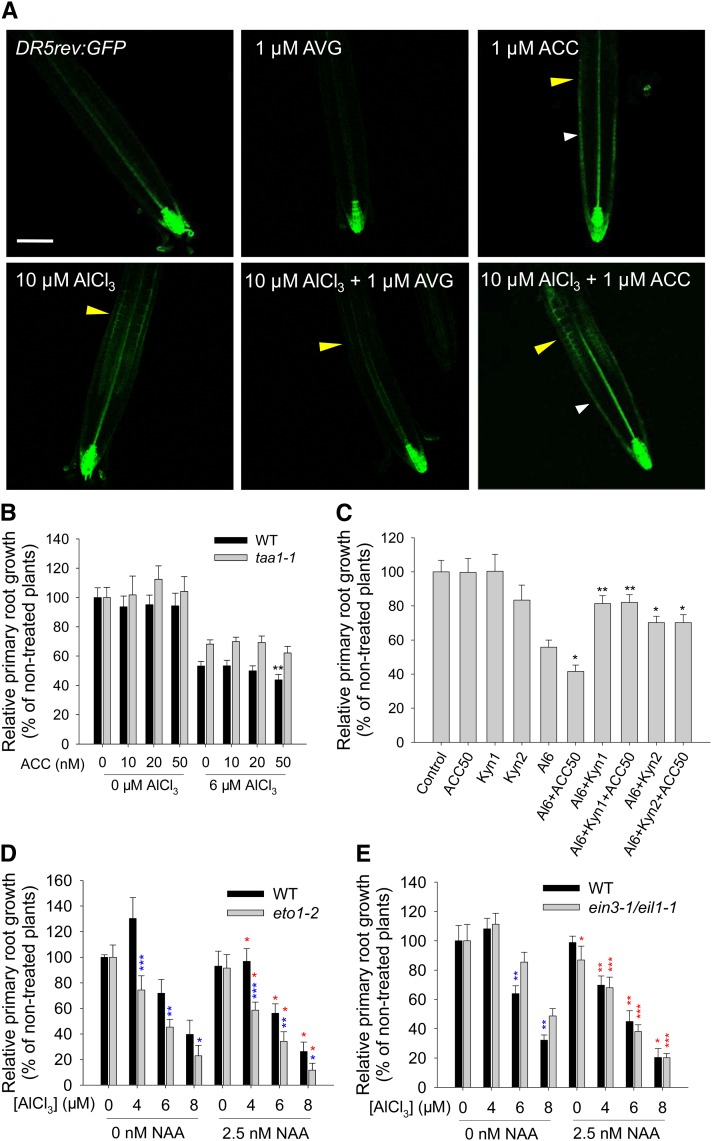
To identify if ethylene-regulated local TAA1 upregulation contributes to the local auxin signaling maximum in the TZ and thus to the inhibition of root growth in response to Al exposure, the local DR5rev:GFP signals in the TZ under cotreatment with ACC or AVG with Al were examined. The application of exogenous ACC strongly enhanced DR5rev:GFP signals, while AVG cotreatment highly repressed the local DR5rev:GFP signals in the TZ in response to Al exposure (Figure 6A). However, ACC and ACC plus Al treatment intensified the DR5rev:GFP signals along the whole root apex, indicating that the ACC-enhanced Al-induced root growth inhibition was not due to the specific increase in the auxin response in the TZ (Figure 6A; Supplemental Figure 9).
Figure 6.
Ethylene Mediates TAA1-Regulated Local Auxin Response in the TZ in Response to Al Stress.
(A) Expression of the DR5rev:GFP transgene in the root apex in the presence of Al plus ACC or AVG treatment. Four-day-old transgenic DR5rev:GFP seedlings were pretreated without or with 1 μM AVG or 1 μM ACC for 2 d, and then the seedlings were continuously treated without or with 1 μM AVG or 1 μM ACC in the absence or presence of 10 μM AlCl3 for 2 h. The TZ is marked by yellow arrowheads, and white arrowheads show DR5rev:GFP signals in the meristem zone. Bar = 100 μm.
(B) Relative primary root growth of the wild type and taa1-1 in the presence of ACC (0, 10, 20, and 50 nM) and Al (0 and 6 μM AlCl3) for 7 d. Asterisks indicate that means within the ACC treatment in the 6 μM AlCl3–exposed taa1-1 mutant differ at **P < 0.01 (t test). Values represent means ± sd (n = 30).
(C) Relative primary root growth of the wild type in the presence of Al (0 and 6 μM AlCl3) plus ACC (0 and 50 nM) and/or plus Kyn (0, 1, and 2 μM) for 7 d. Asterisks indicate that means within the ACC, Kyn, or Kyn plus ACC treatment in the 6 μM AlCl3–exposed wild-type roots differ at *P < 0.05 and **P < 0.01 (t test). Values represent means ± sd (n = 30).
(D) and (E) Relative primary root growth of the wild type, eto1-2, and ein3-1 eil1-1 in the presence of 0, 4, 6, and 8 μM AlCl3 plus 0 and 2.5 nM NAA for 7 d. Blue asterisks indicate that wild-type and mutant means differ significantly at *P < 0.05, **P < 0.01, and ***P < 0.001; red asterisks indicate that wild-type and mutant means within the NAA treatment in each Al concentration differ significantly at *P < 0.05, **P < 0.01, and ***P < 0.001 (t test). Values represent means ± sd (n = 30).
To address if TAA1-mediated local auxin biosynthesis acts downstream of ethylene signaling in response to Al exposure, the response of taa1-1 to ACC under Al stress was tested. ACC treatment at up to 50 nM did not affect the root growth of wild-type and taa1-1 plants (Figure 6B). However, the Al-induced root growth inhibition was significantly enhanced by 50 nM ACC only in the wild type but not in taa1-1 (Figure 6B). Furthermore, the inhibitor of TAA1/TAR-dependent auxin biosynthesis, Kyn, which slightly reduced the root growth of wild-type plants at 2 µM but not at 1 µM, clearly alleviated at both concentrations the root growth–inhibiting additive effects of 50 nM ACC and 6 µM Al cotreatment (Figure 6C). In order to clarify the possible role of auxin as a downstream signal of ethylene to regulate Al-induced inhibition of root growth, the additive effects of auxin on Al-induced root growth inhibition were studied in eto1-2 (for ethylene overproducer; He et al., 2011) and the double mutant ein3-1 eil1-1 (for ethylene insensitive; Alonso et al., 2003). Confirming the role of ethylene, Al-induced root growth inhibition was enhanced in eto1-2 (Figure 6D) but alleviated in ein3-1 eil1-1 (Figure 6E) compared with the wild type. However, the application of exogenous NAA (2.5 nM) consistently and significantly enhanced the Al-induced root growth inhibition in both eto1-2 and ein3-1 eil1-1 mutants, similar to the wild type (Figures 6D and 6E), implying that auxin acts downstream of ethylene to regulate Al-induced root growth inhibition.
ARFs Are Involved in Al-Induced Inhibition of Root Growth
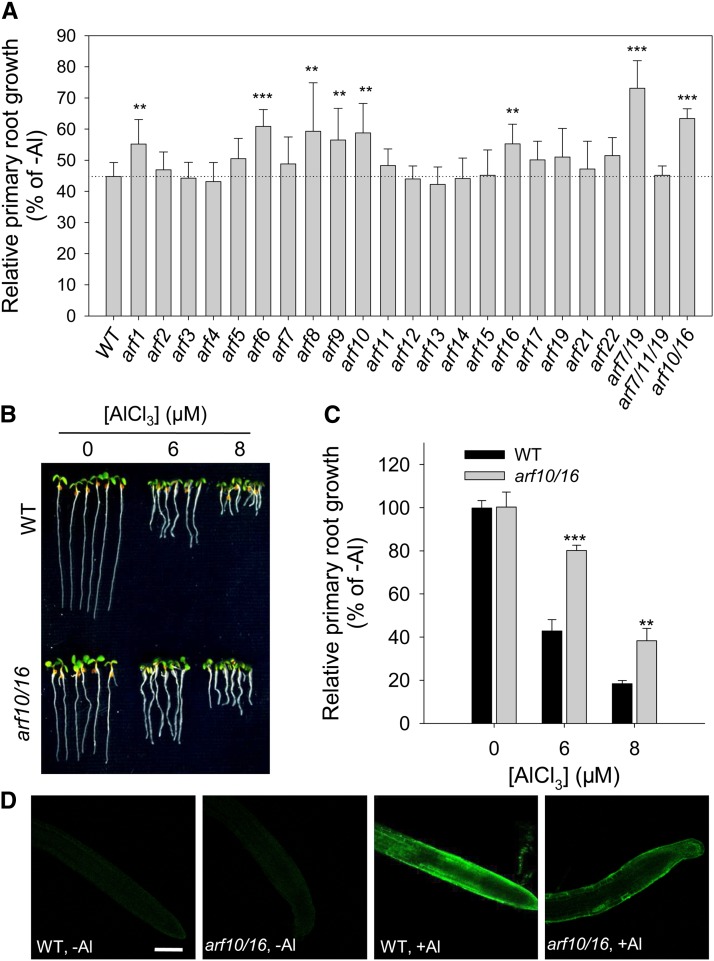
ARFs are a class of transcription factors involved in the auxin signaling pathway, which is an important component of plant development and growth responses to various environmental stimuli (Guilfoyle and Hagen, 2007; Ding and Friml, 2010; Tian et al., 2014a). To explore the role of auxin signaling in Al-induced root growth inhibition, the response to Al of a set of 24 loss-of-function ARF mutants was compared with that of the wild type. The Al-induced inhibition of root growth in the single mutants arf1, arf6, arf8, arf9, arf10, and arf16, and particularly in the double mutants arf7/19 and arf10/16, was less severe than in the wild type (Figure 7A). However, the ARF mutants did not differ in transcript levels in response to Al treatment, according to the results of RNAseq (Supplemental Data Set 1). To understand how ARFs regulate Al-induced root growth inhibition, the arf10/16 double mutant was selected for further experiments, since particularly high levels of expression of ARF10 and ARF16 were observed in the epidermis and cortex of the root apex (Supplemental Figure 10), the major perception site of Al toxicity (Ryan et al., 1993). A dose–response experiment confirmed the marked reduction of Al-induced inhibition of root growth in the arf10/16 double mutant compared with the wild type (Figures 7B and 7C). A lower intensity of morin staining (which stains Al) in the double mutant root after a 24-h exposure to 10 μM Al (Figure 7D) suggests that its reduced Al sensitivity is due to a lower Al accumulation in the TZ.
Figure 7.
ARFs Mediate the Al-Induced Inhibition of Root Growth.
(A) Relative primary root growth of arf mutants exposed for 7 d to either 0 or 6 μM AlCl3. Asterisks indicate that wild-type and mutant means differ significantly at **P < 0.01 and ***P < 0.001 (t test). Values represent means ± sd (n = 30).
(B) and (C) Root growth of wild-type and arf10/16 double mutant plants following a 7-d exposure to 0, 6, or 8 μM AlCl3. Asterisks in (C) indicate that wild-type and double mutant means differ significantly at **P < 0.01 and ***P < 0.001 (t test). Values in (C) represent means ± sd (n = 30).
(D) Morin staining illustrates the accumulation of Al in the root tip of 6-d-old wild-type and arf10/16 double mutant seedlings exposed to 10 μM AlCl3 for 24 h. Bar = 100 μm.
[See online article for color version of this figure.]
The arf10/16 Double Mutant Has a Differential Transcriptional Program in Response to Al Treatment
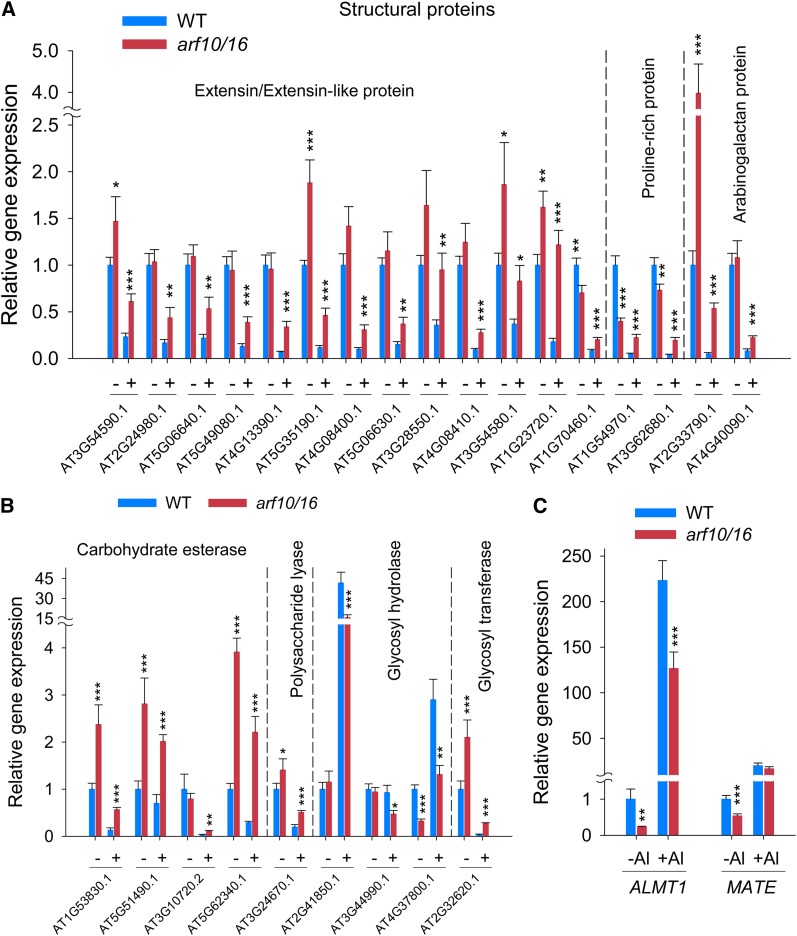
To reveal how ARF10 and ARF16 regulate Al-induced root growth inhibition, a transcriptome analysis through RNAseq was performed by comparing the arf10/16 double mutant line and the wild type in the presence and absence of Al (Supplemental Data Set 1), and a more detailed analysis of the transcriptome focused on the comparison of the Al-exposed arf10/16 double mutant line and the wild type. In total, 207 genes were upregulated by at least 2-fold and 93 genes were downregulated by at least 2-fold in the root tissue of the double mutant compared with wild-type plants, both exposed to Al (Supplemental Figures 11A and 11B and Supplemental Data Sets 2 and 3). About 21% of the upregulated genes and 27% of the downregulated genes could not be functionally assigned. Among the upregulated genes, ∼20% were involved in cell wall synthesis and organization, 12% in primary metabolic processes, 10% in each of transport and cellular protein metabolic/modification processes, 9% in each of transcription regulation and stress/defense, etc.; the downregulated genes were dominated by primary metabolic processes (15%), transport (13%), stress/defense (12%), transcription regulation (10%), cell wall synthesis and organization (7%), etc. The strong representation of cell wall synthesis and organization-related genes implied that the improved Al tolerance of the double mutant relied heavily on cell wall modifications. Among the 47 regulated cell wall–related genes, 22 encode structural cell wall proteins, specifically Hyp-rich glycoproteins (HRGPs), a group that includes the extensins, the arabinogalactan proteins, and the Pro-rich proteins (Supplemental Figure 11C and Supplemental Data Set 4). Thus, cell wall assembly appears to be important in the auxin-mediated response of the root to Al exposure. Differential transcription of genes in the categories glycosyl hydrolases (three upregulated and five downregulated), carbohydrate esterases (four upregulated), glycosyl transferases (two upregulated), and polysaccharide lyases (two upregulated) (Supplemental Figure 11C and Supplemental Data Set 4) also suggests the importance of cell wall modification.
For validation, the transcription of a set of 40 of the differentially transcribed genes putatively involved in the determination of cell wall properties was characterized using qRT-PCR. The correlation between the RNAseq and qRT-PCR data was highly significant (r2 = 0.85, P < 0.001) (Supplemental Figure 11D). The rapid binding of Al to the cell wall matrix is expected to alter the properties of the cell wall and thereby affect root growth (Tabuchi and Matsumoto, 2001; Ma et al., 2004; Horst et al., 2010; Yang et al., 2013). Several genes related to cell wall structure and composition, including those encoding xyloglucanases, endotransglycosylases, polygalacturonases, and glycosyl transferases, have been identified as being involved in the regulation of Al tolerance or the Al-induced inhibition of root growth (Kumari et al., 2008; Maron et al., 2008; Tsutsui et al., 2012; Zhu et al., 2012). Among the 40 genes whose transcription was monitored using qRT-PCR in both wild-type and double mutant plants, either exposed or not exposed to Al for 24 h, 26 genes (17 encoding HRGPs, 4 encoding carbohydrate esterases, 3 encoding glycosyl hydrolases, and 1 each encoding polysaccharide lyase and glycosyl transferase) with relatively high expressional changes were identified. Nearly all of these genes were strongly downregulated by Al, especially in the wild-type plants but less so in the double mutant. The exceptions were genes encoding glycosyl hydrolases, which were either not affected by the Al treatment or were upregulated (but less so in the double mutant than in the wild type) (Figures 8A and 8B).
Figure 8.
Transcriptional Analysis Based on qRT-PCR.
(A) and (B) Gene expression of cell wall–related genes predicted to be involved in the ARF10/16-mediated Al-induced inhibition of root growth.
(C) Expression of the Al resistance–associated genes ALMT1 and MATE.
Six-day-old seedlings were exposed to 10 μM AlCl3 for 24 h. UBQ1 was used as the reference, and a nontreated wild type was used as the sample control. Values represent means ± se (n = 3). Asterisks indicate that wild-type and arf10/16 mutant means differ significantly at *P < 0.05, **P < 0.01, and ***P < 0.001 (t test).
[See online article for color version of this figure.]
It has been established that ALUMINUM-ACTIVATED MALATE TRANSPORTER1 (ALMT1), a gene that encodes a malate transporter, and MULTIDRUG AND TOXIN EXTRUSION FAMILY PROTEIN (MATE), which encodes a citrate transporter, are required for the exudation from the root of malate and citrate, respectively, a process associated with improved Al resistance (Liu et al., 2009). The RNAseq analysis revealed that the loss of both ARF10 and ARF16 function had no effect on the transcript abundance of MATE and even suppressed that of ALMT1 (Supplemental Data Sets 2 and 3). The qRT-PCR analysis showed that the Al treatment strongly upregulated ALMT1 in both the double mutant and the wild type and that the abundance of ALMT1 transcript was substantially lower in the double mutant than in the wild type, regardless of whether the plants were exposed to Al (Figure 8C). The transcription of MATE was also enhanced by exposure to Al, but it remained at a lower level in the double mutant than in the wild type in the absence of Al. Therefore, it is considered unlikely that the reduced Al-induced root growth inhibition of the double mutant compared with the wild type can be explained by a greater malate or citrate exudation.
DISCUSSION
Auxin is a prime regulator of root cell division, elongation, and differentiation and, hence, overall root growth. High concentrations of auxin, however, inhibit the elongation of certain cell types (Teale et al., 2005). Auxin signaling within the root-apex TZ is sensitive not only to developmental signals but also to environmental cues (Baluška et al., 2010), including Al (Sivaguru and Horst, 1998). Until now, how external Al interacts with the root-apex TZ to inhibit root growth has been unclear. The importance of the basipetal transport of auxin from the meristematic zone through the TZ and into the EZ in driving the Al-induced inhibition of root elongation in maize has been highlighted by Kollmeier et al. (2000). Since the external supply of auxin to the EZ is able to partially overcome the inhibition of root growth imposed by the application of Al to the TZ, the suggestion was made that the EZ suffers from auxin deficiency.
In Arabidopsis, TAA1-mediated localized auxin biosynthesis has been reported to be involved in shade avoidance and root growth and development (Stepanova et al., 2008; Tao et al., 2008). Here, a combination of appropriate mutants and the use of a specific signaling transgene have demonstrated that both manipulating the internal auxin level and interfering with the auxin signaling pathway affect the Al-induced inhibition of root growth via auxin signaling in the root-apex TZ, and this process largely depends on TAA1-regulated localized auxin biosynthesis. Auxin alleviates the Al-induced inhibition of the elongation of the maize EZ (Kollmeier et al., 2000), while, by contrast, it enhances the Al-induced inhibition of root growth in Arabidopsis. Such a fundamental difference may well reflect a diversity of target sites for Al, since in monocotyledonous species such as maize and sorghum, Al sensitivity is restricted to the TZ (Sivaguru and Horst, 1998; Sivaguru et al., 1999, 2013), while in the dicotyledonous species common bean, both the TZ and the EZ are sensitive (Rangel et al., 2007). The observation that toxic levels of Al drive up the endogenous level of auxin in the common bean root tip (including the meristematic zone, TZ, and EZ) (Yang et al., 2012), in agreement with our findings here in Arabidopsis root tips (Figure 1B), provides some corroboration for Al-induced enhanced auxin signaling in the Arabidopsis root-apex TZ.
For the moment, it cannot be excluded that polar transport of auxin could also contribute to the Al-induced accumulation of auxin in the root TZ, since root growth in pin1-5 is significantly more inhibited by Al than in the wild type (Supplemental Figure 12). How polar transport of auxin mediates Al-induced root growth inhibition needs to be substantiated by further, more detailed studies. However, this investigation shows that TAA1-mediated local auxin synthesis in the TZ is crucial for the Al-induced inhibition of root growth. The auxin accumulation in the TZ mimics the root growth inhibition induced by an external supply of auxin, much of which is taken up by the TZ (Schlicht et al., 2006). A recent report, which showed that auxin enhances Al toxicity via an alteration in ALUMINUM-SENSITIVE1–mediated Al distribution (Zhu et al., 2013), provides further support for our findings. Unfortunately, the study of Zhu et al. (2013) made no attempt to define the spatial correlations between Al and auxin effects in the root apex during the Al-induced inhibition of root growth.
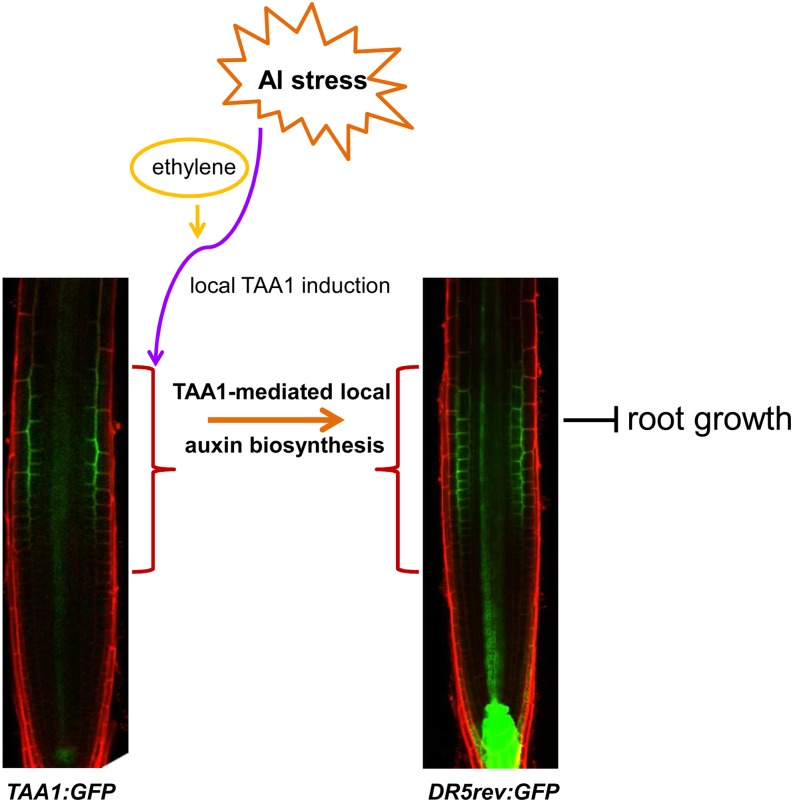
The role of the auxin–ethylene interaction in regulating the growth and development of seedlings has been proposed earlier (Pickett et al., 1990), well investigated (Růzicka et al., 2007; Stepanova et al., 2007, 2011; Swarup et al., 2007), and reviewed recently by Muday et al. (2012). Similar to auxin, which has long been known to play a critical role in Al-induced root growth inhibition (Kollmeier et al., 2000; Sun et al., 2010), ethylene has also been reported to be important in Al-induced root growth inhibition in common bean (Massot et al., 2002; Eticha et al., 2010), Lotus japonicus (Sun et al., 2007), Arabidopsis (Sun et al., 2010), and wheat (Triticum aestivum; Tian et al., 2014b). Also, the potential participation of the auxin–ethylene interaction in the regulation of Al-induced root growth inhibition has been suggested (Sun et al., 2010; Tian et al., 2014b), while the exact interaction mechanism during Al-induced inhibition of root growth is still undetermined. Here, our results indicate that, in response to Al stress, ethylene induces local TAA1 upregulation and thus the auxin signaling maximum in the root-apex TZ, inducing root growth inhibition (Figure 9).
Figure 9.
Schematic Representation of TAA1-Regulated Local Auxin Biosynthesis in the TZ Mediating Root Growth Inhibition in Response to Al Stress.
Al stress induces TAA1 local upregulation (shown by TAA1:GFP) in the TZ through an ethylene-dependent pathway, thus inducing local auxin accumulation and an auxin signaling maximum in the TZ (shown by DR5rev:GFP) and inhibition of root growth.
ARFs bind to auxin response elements in promoters of early auxin response genes and are crucial for mediating auxin signaling (Guilfoyle and Hagen, 2007). Loss-of-function ARF mutants revealed that arf1, arf6, arf8, arf9, arf10, and arf16, and particularly the double mutants arf7/19 and arf10/16, showed greater Al resistance than the wild type (Figure 7A). The analysis of tissue expression pattern demonstrated that ARF10 and ARF16, which were regulated by the Al-modulated miR160 (Wang et al., 2005; Liu et al., 2007; Liu et al., 2010; Chen et al., 2012; Zeng et al., 2012), were highly expressed in the epidermis and cortex of the root apex (Supplemental Figure 10; Rademacher et al., 2011; Baster et al., 2013), the major Al perception site (Ryan et al., 1993), suggesting the potential involvement of ARF10 and ARF16 in auxin-mediated Al-induced inhibition of root growth. However, no transcriptional change was found for ARF10 and ARF16 under Al stress on the entire root basis (Supplemental Data Set 1).
The transcriptomic analysis presented here revealed that many of the differentially transcribed genes associated with cell wall modification were regulated by the transcription factors ARF10 and ARF16. The implication is that the auxin-regulated Al-induced inhibition of root growth arises from auxin signaling–regulated modification of cell wall structure or components. The importance of the properties of the cell wall in the context of Al-induced inhibition of root elongation and in Al tolerance has been emphasized (Horst et al., 2010). The binding of Al to the cell wall, and particularly to the pectic and hemicellulosic matrix in the most Al-sensitive zone of the root apex, has a major effect on cell wall properties and root growth in Arabidopsis (Yang et al., 2011a; Yang et al., 2011b; Zhu et al., 2012, 2013). The binding of Al to pectins is known to be highly dependent on their degree of methylation: binding occurs preferentially to unmethylated pectin, catalyzed by the activity of pectin methylesterase (PME) (Horst et al., 2010; Yang et al., 2011b). These enzymes are regulated by specific inhibitors (PMEIs). Group 2 PMEs contain, in addition to the catalytic domain, an N-terminal extension domain, PRO, which has some similarity to known PMEI domains such as PME2 (Gribaa et al., 2013). The clearly higher transcript abundance of two PMEI family genes and two genes coding for PMEI domain–containing proteins in the roots of the arf10/16 double mutant exposed to Al (Figure 8; Supplemental Data Set 4) may reflect a higher degree of pectin methylation and, thus, a reduced rate of Al accumulation (Figure 7D) compared with wild-type roots.
The upregulation of cell wall–related genes in the arf10/16 double mutant primarily encoding cell wall structure proteins, including HRGPs, arabinogalactan proteins, and various Pro- and Gly-rich proteins, suggests that its reduced sensitivity to Al-induced inhibition of root growth is probably the outcome of a complex network including structure assembly and remodeling. A potential role for HRGPs in Al tolerance in rice (Oryza sativa) has been suggested by Pan et al. (2011), although no mechanistic basis has yet been proposed. One possibility is that the Al-induced accumulation of HRGPs may alter cell wall porosity and, as a result, reduce the mobility of Al in the root apoplast (Yang et al., 2011b). The plant cell wall is a developmentally dynamic structure that also responds to various external stimuli (Farrokhi et al., 2006). Thus, once the cell wall structure proteins have been deposited, the expression of genes involved in wall polysaccharide remodeling (such as those encoding glycosyl hydrolases and polysaccharide lyases) can be expected to be promoted, as was indeed the case (Figure 8B; Supplemental Data Set 4). An outcome of this burst of gene expression is to facilitate cell wall enlargement (Gribaa et al., 2013). The upregulation of CELLULOSE SYNTHASE-LIKE PROTEIN B2, coding for a protein involved in cellulose synthesis, may have led to the reduced inhibition of growth in the double mutant roots, while the repression of XTH31 and XTH7, genes that encode xyloglucan endotransglucosylase/hydrolases, may have alleviated Al toxicity by reducing the xyloglucan content of the cell wall, thereby lowering its Al binding capacity (Zhu et al., 2012).
The malate transporter ALMT1 and the citrate transporter MATE are known to improve Al resistance by promoting the exudation of malate/citrate from the Arabidopsis root (Liu et al., 2009). The change in transcript abundance of ALMT1 and MATE in the roots of the arf10/16 double mutant in response to Al exposure (Figure 8C) does not support the assumption that the reduced Al-induced inhibition of root growth resulted from an enhanced malate and/or citrate exudation. Kobayashi et al. (2013) described how externally supplied IAA enhances both ALMT1 and MATE transcription, which, however, did not affect malate exudation, while in the IAA signaling double mutant line nonphototropic hypocotyls4-1 auxin-responsive factor19-1 (synonymous with arf7/19), the Al-induced expression of ALMT1 remained unaltered. Thus, it appears that the mechanism for auxin signaling–regulated Al toxicity might be independent of organic acid anion exudation.
In conclusion, this study provides evidence that the TAA1-mediated local auxin biosynthesis is responsible for the Al-induced inhibition of root growth. The mechanistic basis for the process operates via the mediation of the auxin response depending on ethylene-regulated TAA1 expression in the root-apex TZ and, thus, downstream signaling regulation via ARF transcription factors. Cell wall modification is probably a further downstream response to Al exposure and contributes to the auxin-mediated root growth inhibition in response to Al stress. The wider picture illustrates how hormone signaling can regulate root growth plasticity in response to environmental cues, providing a general mechanism for plants to adapt to a changing environment.
METHODS
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia, mutant lines yuc1D (Zhao et al., 2001), slr-1 (Fukaki et al., 2005; Vanneste et al., 2005), PIN8 OX (Ding et al., 2012), taa1-1 (CS66987), eto1-2 (He et al., 2011), ein3-1 eil1-1 (Alonso et al., 2003; He et al., 2011), arf1 to arf22 (except arf18 and arf20) (Mai et al., 2011), arf7/19 (Okushima et al., 2007), arf10/16 (Ding and Friml, 2010), arf7/11/19, DR5rev:GFP (Ding and Friml, 2010), TAA1:GFP-TAA1 (Stepanova et al., 2008), and DR5rev:GFP/taa1-1 were used in this study. Seedlings were grown hydroponically as described by Kobayashi et al. (2007) in 2% strength modified MGRL (Fujiwara et al., 1992) solution (but with Pi eliminated and the Ca concentration adjusted to 200 μM) at pH 5.0 in a growth chamber with a 16/8-h light/dark cycle at 22°C.
Treatments and Root Growth Analysis
In this study, pH 5.0 medium was selected to prevent the precipitation of Al in the medium as described by Iuchi et al. (2007) and also because of the hypersensitivity of Arabidopsis root growth to low pH (Supplemental Figure 7). Although we attempted to maintain the pH constant by frequent adjustment, we cannot exclude the possibility that the pH occasionally decreased slightly below the target pH, which may explain why Al-induced root growth inhibition varied between experiments. For root growth experiments, the seeds were sown onto polypropylene mesh floating on 2% modified MGRL solution containing Al (total [AlCl3] 0 to 8 μM, pH 5.0) or Al plus PEO-IAA (gifts from Ken-ichiro Hayashi), NAA, Kyn, ACC, or AVG (pH 5.0) or various pH treatments (pH 4.2 to 5.5) for 7 d. The solution was renewed every 2 d. At day 7, the roots were scanned and primary root length was calculated with ImageJ software.
For short-term treatments, the seedlings were pregrown in 2% modified MGRL solution for 6 d and then transferred to different Al treatment solutions ([AlCl3] 10 or 25 μM, pH 5.0) or various pH stress solutions (pH 4.2 to 5.5) for 2 h. The free Al3+ activity in 2% MGRL solution with [AlCl3] 1, 2, 3, 4, 5, 6, 8, 10, and 25 μM at pH 5.0 is 0.0006, 0.27, 0.66, 1.1, 1.4, 1.8, 2.6, 3.4, and 9.4 μM, respectively, calculated using GEOCHEM-EZ software (http://www.plantmineralnutrition.net/Geochem/geochem%20home.htm).
Confocal Microscopy
Imaging was performed on an LSM-700 laser-scanning confocal microscope (Zeiss). For visualization of the expression of DR5rev:GFP and TAA1:GFP in roots, 6-d-old plants were treated with AlCl3 ([AlCl3] 0 and 10 μM) or various pH values for 2 h, and then roots were mounted with PI. For visualization of Al in the root tips, after AlCl3 treatment ([AlCl3] 0 and 10 μM) for 24 h, the excised roots were washed in MES buffer (pH 5.5) for 10 min, stained with 100 µM morin in the same buffer for 1 h with gentle shaking, and washed several times with MES buffer, and the green Al-morin fluorescence signal was then observed. Cell viability was determined by PI/FDA staining; living cells generate green fluorescence when stained in FDA, while nuclei of membrane-compromised cells will show red PI fluorescence as a sign of cell death.
IAA Concentration Determination
Six-day-old wild-type seedlings were treated with 6 μM AlCl3 at pH 5.0 for 0, 3, 6, and 12 h. Then, 1-mm-long root tips were excised and immediately frozen in liquid nitrogen. Root tips were ground to powder in liquid nitrogen. Afterward, 1 mL of 80% (v/v) methanol was added to each sample and vortexed. The IAA in root tips was extracted, and the concentration was determined according to Zhou et al. (2010) by gas chromatography–mass spectrometry.
RNA Isolation and qRT-PCR
Approximately 500 seedlings were grown in 2% modified MGRL nutrient solution for 6 d, and then the plants were treated with AlCl3 ([AlCl3] 0 and 10 μM) for 24 h. The roots were rinsed with distilled water, harvested, and shock-frozen in liquid nitrogen. Total RNA was isolated using TriPure isolation reagent (Roche), and first-strand cDNA was synthesized from 1 µg of total RNA using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s protocol. qRT-PCR was performed using the CFX Connect Real-Time System (Bio-Rad) with FastStart Universal SYBR Green Master (Rox) (Roche). Samples for qRT-PCR were run in three biological replicates and two technical replicates. For the normalization of gene expression, the ubiquitin gene UBQ1 (AT3G52590) was used as an internal standard, and the nontreated wild type was used as a sample control. Primers were designed using Primer 5 software, and the specifications of the primers of the genes studied are given in Supplemental Data Set 5.
RNAseq Analysis
The RNAseq analysis was performed by BGI Tech. Approximately 500 seedlings (6 d old) of both the wild-type Columbia and the arf10/16 double mutant line were exposed to a 2% MGRL solution containing 0 or 10 μM AlCl3 (pH 5.0). After 24 h, the roots were sampled for RNA isolation. Total RNA was isolated using the RNeasy Plant Mini Kit (Qiagen) following the manufacturer’s protocol. The isolated total RNA samples were treated with DNase I to degrade any possible DNA contamination and enriched using oligo(dT) magnetic beads (for eukaryotes) or by removing rRNAs from the total RNA (for prokaryotes). Mixed with the fragmentation buffer, the mRNA was fragmented into short fragments (∼200 bp). Then, the first strand of cDNA was synthesized by using random hexamer primer. Buffer, deoxynucleotide triphosphates, RNase H, and DNA polymerase I were added to synthesize the second strand. The double-stranded cDNA was purified with magnetic beads. End reparation and 3′ end single nucleotide A (adenine) addition were then performed. Finally, sequencing adaptors were ligated to the fragments. The fragments were enriched by PCR amplification. The constructed sample libraries were qualified and quantified with an Agilent 2100 Bioanaylzer and the ABI StepOnePlus Real-Time PCR System and then sequenced via Illumina HiSeq 2000 or other sequencer when necessary.
The raw reads were obtained by transferring the original image data produced by the sequencer into sequences by base calling. The raw reads were then cleaned by removing the low-quality reads and/or adaptor sequences. The clean reads with high-quality sequences were mapped to reference sequences and/or the reference gene set using SOAPaligner/SOAP2 (Li et al., 2009). No more than two mismatches were allowed in the alignment.
The gene expression levels were calculated using the reads per kilobase per million reads method (Mortazavi et al., 2008). Differentially expressed genes (DEGs) within samples were screened with a modified method according to Audic and Claverie (1997). Statistical analysis of DEGs was conducted using the P value and the false discovery rate (FDR). The P value corresponds to the differential gene expression test. FDR is a method used to determine the threshold of P values in multiple tests (Benjamini and Yekutieli, 2001). FDR ≤ 0.001 and absolute value of log2 ratio ≥ 1 were used as the threshold to judge the significance of gene expression differences. The functional categories of the DEGs were BLASTed against the nonredundant GenBank, KEGG Pathway, and UniProt protein databases by the Gene Ontology annotation.
Statistical Analysis
Statistical analysis was performed using SAS 9.2 (SAS Institute). Means were compared using Student’s t test. Asterisks in the figures denote significant differences as follows: *P < 0.05, **P < 0.01, and ***P < 0.001.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database and the GenBank/EMBL databases under the following accession numbers: SLR (AT4G14550, AF334718), YUC1 (AT4G32540, NM_119406), PIN8 (AT5G15100, NM_121514), TAA1 (AT1G70560, NM_105724), ETO1 (AT3G51770, NM_001035762), EIN3 (AT3G20770, NM_112968), EIL1 (AT2G27050, BT003344), ARF10 (AT2G28350, AF325073), ARF16 (AT4G30080, AY059792), ALMT1 (AT1G08430, NM_100716), and MATE (At1g51340, AY059793). RNA sequencing data analyzed in this study are available in the Gene Expression Omnibus database under accession number GSE57487.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. The Root-Apex TZ Is the Major Site of Al Sensitivity, as Indicated by PI and FDA Staining.
Supplemental Figure 2. Time-Course Response of the Expression of DR5rev:GFP in Root Apex Exposed to Al Stress.
Supplemental Figure 3. Short-Term Effect of PEO-IAA on Al-Induced Inhibition of Root Growth.
Supplemental Figure 4. Effect of Exogenous NAA on Al-Induced DR5rev:GFP Expression in Root Apex.
Supplemental Figure 5. Effect of Exogenous NAA on the Expression of Auxin-Responsive Genes in Al-Exposed Roots.
Supplemental Figure 6. Temporal Analysis of Al-Induced Expression of TAA1:GFP in Root Apex.
Supplemental Figure 7. The Sensitivity of the Arabidopsis Root to Low pH.
Supplemental Figure 8. The pH of the Medium Has No Effect on the Expression of Either the DR5rev:GFP or the TAA1:GFP Transgene.
Supplemental Figure 9. Effect of ACC and AVG on the Expression of DR5rev:GFP Transgene in the Root Apex under Al Stress.
Supplemental Figure 10. The Expression Pattern of ARFs:GFP Transgene in the Root Apex.
Supplemental Figure 11. Transcriptomic Analysis of the Response to Al Exposure.
Supplemental Figure 12. Phenotype Analysis of Root Growth of Polar Auxin Transport–Related Mutants in Response to Al Stress.
Supplemental Data Set 1. Comparison of the Differentially Expressed Genes in Roots of the Wild Type and arf10/16 Exposed to Al Stress though RNAseq Analysis.
Supplemental Data Set 2. Upregulated Genes in Roots of arf10/16 Double Mutants.
Supplemental Data Set 3. Downregulated Genes in Roots of arf10/16 Double Mutants.
Supplemental Data Set 4. List of Differential Expressed Genes (DEGs) Involved in Cell Wall Synthesis and Organization in Roots of arf10/16 Double Mutants.
Supplemental Data Set 5. List of Genes and Specific Primer Pairs Used for Quantitative Real-Time PCR (qRT-PCR).
Supplementary Material
Acknowledgments
We thank Ken-ichiro Hayashi for PEO-IAA chemicals. We thank Jiri Friml, Jose Alonso, Tom Beeckman, Hongwei Guo, Dolf Weijers, Guang-qin Guo, and Hong-quan Yang for published materials. We also thank the editor and anonymous reviewers for their constructive comments and insightful suggestions, which greatly improved this article. Z.D. is supported by the National Natural Science Foundation of China (Grants 31270327 and 31222005), the 1000-Talents Plan from China for young researchers (Grant 104.206.99.99), and Qilu Scholarship from Shandong University of China (Grant 11200081963024). Z.-B.Y. is supported by the Independent Innovation Fund project of Shandong University (Grant 2012HW005) and the China Postdoctoral Science Foundation (Grants 2012M521322 and 2013T60664).
AUTHOR CONTRIBUTIONS
Z.D. and Z.-B.Y. conceived the study and designed the experiments. Z.-B.Y., Z.D., X.G., C.H., and R.W. performed the experiments and analyzed the data. Z.D., Z.-B.Y., and W.J.H. wrote the article. All authors discussed the results and commented on the article.
Glossary
- TZ
transition zone
- EZ
elongation zone
- IAA
indole-3-acetic acid
- PI
propidium iodide
- FDA
fluorescein diacetate
- PEO-IAA
α-(phenylethyl-2-one)-indole-3-acetic acid
- NAA
1-naphthaleneacetic acid
- RNAseq
RNA sequencing
- qRT-PCR
quantitative real-time PCR
- Kyn
l-kynurenine
- AVG
aminoethoxyvinylglycine
- ACC
1-aminocyclopropane-1-carboxylic acid
- HRGP
Hyp-rich glycoprotein
- DEG
differentially expressed gene
- FDR
false discovery rate
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Alonso J.M., Stepanova A.N., Solano R., Wisman E., Ferrari S., Ausubel F.M., Ecker J.R. (2003). Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 2992–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S., Claverie J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7: 986–995. [DOI] [PubMed] [Google Scholar]
- Baluška F., Mancuso S., Volkmann D., Barlow P.W. (2010). Root apex transition zone: A signalling-response nexus in the root. Trends Plant Sci. 15: 402–408. [DOI] [PubMed] [Google Scholar]
- Baster P., Robert S., Kleine-Vehn J., Vanneste S., Kania U., Grunewald W., De Rybel B., Beeckman T., Friml J. (2013). SCF(TIR1/AFB)-auxin signalling regulates PIN vacuolar trafficking and auxin fluxes during root gravitropism. EMBO J. 32: 260–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Benková E., Hejátko J. (2009). Hormone interactions at the root apical meristem. Plant Mol. Biol. 69: 383–396. [DOI] [PubMed] [Google Scholar]
- Bielach A., Duclercq J., Marhavý P., Benková E. (2012). Genetic approach towards the identification of auxin-cytokinin crosstalk components involved in root development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Wang T., Zhao M., Tian Q., Zhang W.H. (2012). Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235: 375–386. [DOI] [PubMed] [Google Scholar]
- Delhaize E., Ryan P.R. (1995). Aluminum toxicity and tolerance in plants. Plant Physiol. 107: 315–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., De Smet I. (2013). Localised ABA signalling mediates root growth plasticity. Trends Plant Sci. 18: 533–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., Friml J. (2010). Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 107: 12046–12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z., et al. (2012). ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 3: 941–949. [DOI] [PubMed] [Google Scholar]
- Doncheva S., Amenós M., Poschenrieder C., Barceló J. (2005). Root cell patterning: A primary target for aluminium toxicity in maize. J. Exp. Bot. 56: 1213–1220. [DOI] [PubMed] [Google Scholar]
- Duan L., Dietrich D., Ng C.H., Chan P.M., Bhalerao R., Bennett M.J., Dinneny J.R. (2013). Endodermal ABA signaling promotes lateral root quiescence during salt stress in Arabidopsis seedlings. Plant Cell 25: 324–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eticha D., Zahn M., Bremer M., Yang Z., Rangel A.F., Rao I.M., Horst W.J. (2010). Transcriptomic analysis reveals differential gene expression in response to aluminium in common bean (Phaseolus vulgaris) genotypes. Ann. Bot. (Lond.) 105: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrokhi N., Burton R.A., Brownfield L., Hrmova M., Wilson S.M., Bacic A., Fincher G.B. (2006). Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes. Plant Biotechnol. J. 4: 145–167. [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Hirai M.Y., Chino M., Komeda Y., Naito S. (1992). Effects of sulfur nutrition on expression of the soybean seed storage protein genes in transgenic petunia. Plant Physiol. 99: 263–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H., Nakao Y., Okushima Y., Theologis A., Tasaka M. (2005). Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J. 44: 382–395. [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tameda S., Masuda H., Tasaka M. (2002). Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J. 29: 153–168. [DOI] [PubMed] [Google Scholar]
- Gifford M.L., Dean A., Gutierrez R.A., Coruzzi G.M., Birnbaum K.D. (2008). Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 105: 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribaa A., Dardelle F., Lehner A., Rihouey C., Burel C., Ferchichi A., Driouich A., Mollet J.C. (2013). Effect of water deficit on the cell wall of the date palm (Phoenix dactylifera ‘Deglet nour’, Arecales) fruit during development. Plant Cell Environ. 36: 1056–1070. [DOI] [PubMed] [Google Scholar]
- Guilfoyle T.J., Hagen G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10: 453–460. [DOI] [PubMed] [Google Scholar]
- Hayashi K. (2012). The interaction and integration of auxin signaling components. Plant Cell Physiol. 53: 965–975. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Tan X., Zheng N., Hatate T., Kimura Y., Kepinski S., Nozaki H. (2008). Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc. Natl. Acad. Sci. USA 105: 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W., et al. (2011). A small-molecule screen identifies l-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell 23: 3944–3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst W.J., Wang Y., Eticha D. (2010). The role of the root apoplast in aluminium-induced inhibition of root elongation and in aluminium resistance of plants: A review. Ann. Bot. (Lond.) 106: 185–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illés P., Schlicht M., Pavlovkin J., Lichtscheidl I., Baluška F., Ovečka M. (2006). Aluminium toxicity in plants: Internalization of aluminium into cells of the transition zone in Arabidopsis root apices related to changes in plasma membrane potential, endosomal behaviour, and nitric oxide production. J. Exp. Bot. 57: 4201–4213. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Koyama H., Iuchi A., Kobayashi Y., Kitabayashi S., Kobayashi Y., Ikka T., Hirayama T., Shinozaki K., Kobayashi M. (2007). Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 104: 9900–9905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen L., Roberts I., De Rycke R., Beeckman T. (2012). Phloem-associated auxin response maxima determine radial positioning of lateral roots in maize. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367: 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Hoekenga O.A., Itoh H., Nakashima M., Saito S., Shaff J.E., Maron L.G., Piñeros M.A., Kochian L.V., Koyama H. (2007). Characterization of AtALMT1 expression in aluminum-inducible malate release and its role for rhizotoxic stress tolerance in Arabidopsis. Plant Physiol. 145: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Kobayashi Y., Sugimoto M., Lakshmanan V., Iuchi S., Kobayashi M., Bais H.P., Koyama H. (2013). Characterization of the complex regulation of AtALMT1 expression in response to phytohormones and other inducers. Plant Physiol. 162: 732–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochian L.V., Hoekenga O.A., Pineros M.A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55: 459–493. [DOI] [PubMed] [Google Scholar]
- Kollmeier M., Felle H.H., Horst W.J. (2000). Genotypical differences in aluminum resistance of maize are expressed in the distal part of the transition zone. Is reduced basipetal auxin flow involved in inhibition of root elongation by aluminum? Plant Physiol. 122: 945–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari M., Taylor G.J., Deyholos M.K. (2008). Transcriptomic responses to aluminum stress in roots of Arabidopsis thaliana. Mol. Genet. Genomics 279: 339–357. [DOI] [PubMed] [Google Scholar]
- Lavenus J., Goh T., Roberts I., Guyomarc’h S., Lucas M., De Smet I., Fukaki H., Beeckman T., Bennett M., Laplaze L. (2013). Lateral root development in Arabidopsis: Fifty shades of auxin. Trends Plant Sci. 18: 450–458. [DOI] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. (2009). SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967. [DOI] [PubMed] [Google Scholar]
- Liu J., Magalhaes J.V., Shaff J., Kochian L.V. (2009). Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 57: 389–399. [DOI] [PubMed] [Google Scholar]
- Liu P.P., Montgomery T.A., Fahlgren N., Kasschau K.D., Nonogaki H., Carrington J.C. (2007). Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 52: 133–146. [DOI] [PubMed] [Google Scholar]
- Liu X., Huang J., Wang Y., Khanna K., Xie Z., Owen H.A., Zhao D. (2010). The role of floral organs in carpels, an Arabidopsis loss-of-function mutation in microRNA160a, in organogenesis and the mechanism regulating its expression. Plant J. 62: 416–428. [DOI] [PubMed] [Google Scholar]
- Ma J.F., Shen R., Nagao S., Tanimoto E. (2004). Aluminum targets elongating cells by reducing cell wall extensibility in wheat roots. Plant Cell Physiol. 45: 583–589. [DOI] [PubMed] [Google Scholar]
- Mai Y.X., Wang L., Yang H.Q. (2011). A gain-of-function mutation in IAA7/AXR2 confers late flowering under short-day light in Arabidopsis. J. Integr. Plant Biol. 53: 480–492. [DOI] [PubMed] [Google Scholar]
- Maron L.G., Kirst M., Mao C., Milner M.J., Menossi M., Kochian L.V. (2008). Transcriptional profiling of aluminum toxicity and tolerance responses in maize roots. New Phytol. 179: 116–128. [DOI] [PubMed] [Google Scholar]
- Massot N., Nicander B., Barceló J., Poschenrieder C., Tillberg E. (2002). A rapid increase in cytokinin levels and enhanced ethylene evolution precede Al3+-induced inhibition of root growth in bean seedlings (Phaseolus vulgaris L.). Plant Growth Regul. 37: 105–112. [Google Scholar]
- Moore A., Donahue C.J., Bauer K.D., Mather J.P. (1998). Simultaneous measurement of cell cycle and apoptotic cell death. Methods Cell Biol. 57: 265–278. [DOI] [PubMed] [Google Scholar]
- Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. (2008). Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5: 621–628. [DOI] [PubMed] [Google Scholar]
- Muday G.K., Rahman A., Binder B.M. (2012). Auxin and ethylene: Collaborators or competitors? Trends Plant Sci. 17: 181–195. [DOI] [PubMed] [Google Scholar]
- Nishimura T., Nakano H., Hayashi K., Niwa C., Koshiba T. (2009). Differential downward stream of auxin synthesized at the tip has a key role in gravitropic curvature via TIR1/AFBs-mediated auxin signaling pathways. Plant Cell Physiol. 50: 1874–1885. [DOI] [PubMed] [Google Scholar]
- Okushima Y., Fukaki H., Onoda M., Theologis A., Tasaka M. (2007). ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19: 118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde P., Fukaki H., Beeckman T. (2010). Auxin control of root development. Cold Spring Harb. Perspect. Biol. 2: a001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan W., Shou S., Zhou X., Guo T., Zhu M., Pan J. (2011). Al-induced cell wall hydroxyproline-rich glycoprotein accumulation is involved in alleviating Al toxicity in rice. Acta Physiol. Plant. 33: 601–608. [Google Scholar]
- Pickett F.B., Wilson A.K., Estelle M. (1990). The aux1 mutation of Arabidopsis confers both auxin and ethylene resistance. Plant Physiol. 94: 1462–1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher E.H., Möller B., Lokerse A.S., Llavata-Peris C.I., van den Berg W., Weijers D. (2011). A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 68: 597–606. [DOI] [PubMed] [Google Scholar]
- Rangel A.F., Rao I.M., Horst W.J. (2007). Spatial aluminium sensitivity of root apices of two common bean (Phaseolus vulgaris L.) genotypes with contrasting aluminium resistance. J. Exp. Bot. 58: 3895–3904. [DOI] [PubMed] [Google Scholar]
- Rosquete M.R., von Wangenheim D., Marhavý P., Barbez E., Stelzer E.H., Benková E., Maizel A., Kleine-Vehn J. (2013). An auxin transport mechanism restricts positive orthogravitropism in lateral roots. Curr. Biol. 23: 817–822. [DOI] [PubMed] [Google Scholar]
- Růzicka K., Ljung K., Vanneste S., Podhorská R., Beeckman T., Friml J., Benková E. (2007). Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19: 2197–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P.R., Kochian L.V. (1993). Interaction between aluminum toxicity and calcium uptake at the root apex in near-isogenic lines of wheat (Triticum aestivum L.) differing in aluminum tolerance. Plant Physiol. 102: 975–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P.R., Ditomaso J.M., Kochian L.V. (1993). Aluminum toxicity in roots: An investigation of spatial sensitivity and the role of the root cap. J. Exp. Bot. 44: 437–446. [Google Scholar]
- Schlicht M., Strnad M., Scanlon M.J., Mancuso S., Hochholdinger F., Palme K., Volkmann D., Menzel D., Baluška F. (2006). Auxin immunolocalization implicates vesicular neurotransmitter-like mode of polar auxin transport in root apices. Plant Signal. Behav. 1: 122–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Horst W.J. (1998). The distal part of the transition zone is the most aluminum-sensitive apical root zone of maize. Plant Physiol. 116: 155–163. [Google Scholar]
- Sivaguru M., Baluška F., Volkmann D., Felle H.H., Horst W.J. (1999). Impacts of aluminum on the cytoskeleton of the maize root apex. Short-term effects on the distal part of the transition zone. Plant Physiol. 119: 1073–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaguru M., Liu J., Kochian L.V. (2013). Targeted expression of SbMATE in the root distal transition zone is responsible for sorghum aluminum resistance. Plant J. 76: 297–307. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Doležal K., Schlereth A., Jürgens G., Alonso J.M. (2008). TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133: 177–191. [DOI] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Likhacheva A.V., Alonso J.M. (2007). Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19: 2169–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova A.N., Yun J., Robles L.M., Novak O., He W., Guo H., Ljung K., Alonso J.M. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23: 3961–3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward N., Martin R., Engasser J.M., Goergen J.L. (1999). A new methodology for plant cell viability assessment using intracellular esterase activity. Plant Cell Rep. 19: 171–176. [DOI] [PubMed] [Google Scholar]
- Sun P., Tian Q.Y., Chen J., Zhang W.H. (2010). Aluminium-induced inhibition of root elongation in Arabidopsis is mediated by ethylene and auxin. J. Exp. Bot. 61: 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun P., Tian Q.Y., Zhao M.G., Dai X.Y., Huang J.H., Li L.H., Zhang W.H. (2007). Aluminum-induced ethylene production is associated with inhibition of root elongation in Lotus japonicus L. Plant Cell Physiol. 48: 1229–1235. [DOI] [PubMed] [Google Scholar]
- Swarup R., Perry P., Hagenbeek D., Van Der Straeten D., Beemster G.T., Sandberg G., Bhalerao R., Ljung K., Bennett M.J. (2007). Ethylene upregulates auxin biosynthesis in Arabidopsis seedlings to enhance inhibition of root cell elongation. Plant Cell 19: 2186–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A., Matsumoto H. (2001). Changes in cell-wall properties of wheat (Triticum aestivum) roots during aluminum-induced growth inhibition. Physiol. Plant. 112: 353–358. [DOI] [PubMed] [Google Scholar]
- Tao Y., et al. (2008). Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Ditengou F., Palme K. (2005). Auxin and the developing root of Arabidopsis thaliana. Physiol. Plant. 123: 130–138. [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. (2006). Auxin in action: Signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7: 847–859. [DOI] [PubMed] [Google Scholar]
- Tian H., Niu T., Yu Q., Quan T., Ding Z. (2013). Auxin gradient is crucial for the maintenance of root distal stem cell identity in Arabidopsis. Plant Signal. Behav. 8: e26429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Wabnik K., Niu T., Li H., Yu Q., Pollmann S., Vanneste S., Govaerts W., Rolcík J., Geisler M., Friml J., Ding Z. (2014a). WOX5-IAA17 feedback circuit-mediated cellular auxin response is crucial for the patterning of root stem cell niches in Arabidopsis. Mol. Plant 7: 277–289. [DOI] [PubMed] [Google Scholar]
- Tian Q., Zhang X., Ramesh S., Gilliham M., Tyerman S.D., Zhang W.H. (2014b). Ethylene negatively regulates aluminium-induced malate efflux from wheat roots and tobacco cells transformed with TaALMT1. J. Exp. Bot. 65: 2415–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui T., Yamaji N., Huang C.F., Motoyama R., Nagamura Y., Ma J.F. (2012). Comparative genome-wide transcriptional analysis of Al-responsive genes reveals novel Al tolerance mechanisms in rice. PLoS ONE 7: e48197–e48205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S., et al. (2005). Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17: 3035–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbelen J.P., De Cnodder T., Le J., Vissenberg K., Baluška F. (2006). The root apex of Arabidopsis thaliana consists of four distinct zones of growth activities: Meristematic zone, transition zone, fast elongation zone and growth terminating zone. Plant Signal. Behav. 1: 296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Uexküll H.R., Mutert E. (1995). Global extent, development and economic impact of acid soils. Plant Soil 171: 1–15. [Google Scholar]
- Wang J.W., Wang L.J., Mao Y.B., Cai W.J., Xue H.W., Chen X.Y. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.L., Zhu X.F., Peng Y.X., Zheng C., Li G.X., Liu Y., Shi Y.Z., Zheng S.J. (2011a). Cell wall hemicellulose contributes significantly to aluminum adsorption and root growth in Arabidopsis. Plant Physiol. 155: 1885–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.B., Eticha D., Albacete A., Rao I.M., Roitsch T., Horst W.J. (2012). Physiological and molecular analysis of the interaction between aluminium toxicity and drought stress in common bean (Phaseolus vulgaris). J. Exp. Bot. 63: 3109–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.B., Eticha D., Rotter B., Rao I.M., Horst W.J. (2011b). Physiological and molecular analysis of polyethylene glycol-induced reduction of aluminium accumulation in the root tips of common bean (Phaseolus vulgaris). New Phytol. 192: 99–113. [DOI] [PubMed] [Google Scholar]
- Yang Z.B., Rao I.M., Horst W.J. (2013). Interaction of aluminium and drought stress on root growth and crop yield on acid soils. Plant Soil 372: 3–25. [Google Scholar]
- Zeng Q.Y., Yang C.Y., Ma Q.B., Li X.P., Dong W.W., Nian H. (2012). Identification of wild soybean miRNAs and their target genes responsive to aluminum stress. BMC Plant Biol. 12: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. (2001). A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291: 306–309. [DOI] [PubMed] [Google Scholar]
- Zhou W., Wei L., Xu J., Zhai Q., Jiang H., Chen R., Chen Q., Sun J., Chu J., Zhu L., Liu C.M., Li C. (2010). Arabidopsis tyrosylprotein sulfotransferase acts in the auxin/PLETHORA pathway in regulating postembryonic maintenance of the root stem cell niche. Plant Cell 22: 3692–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.F., Lei G.J., Wang Z.W., Shi Y.Z., Braam J., Li G.X., Zheng S.J. (2013). Coordination between apoplastic and symplastic detoxification confers plant aluminum resistance. Plant Physiol. 162: 1947–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X.F., et al. (2012). XTH31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 24: 4731–4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.